Dear Editor,
The flow of ions through channels in cell membranes, particularly the entry of calcium ions into the cytosol, serves as a cue for environmental responses in eukaryotes. In animals, calcium-permeable transient receptor potential (TRP) channels act as sensors for temperature, osmotic potential, and other environmental conditions1. Although plants also respond to myriad environmental perturbations with cytosolic calcium fluxes2, stress-gated calcium-permeable channels like TRPs in animals are not found in land plants. Several families of proteins, including glutamate receptor-like (GLR) proteins, cyclic nucleotide-gated channels (CNGCs), and annexins, have been linked to calcium fluxes in plant cells3. However, none of them has been shown to respond directly to stress signals. Because cytosolic calcium elevation is one of the earliest responses of plant cells to stress treatments1,4, and calcium-binding proteins are required for several environmental stress responses5, stress-activated calcium channels are candidates that may link stress stimuli to calcium-dependent downstream responses. We thus seek to isolate plant genes that encode possible calcium channels that are gated by stress signals, such as osmotic stress.
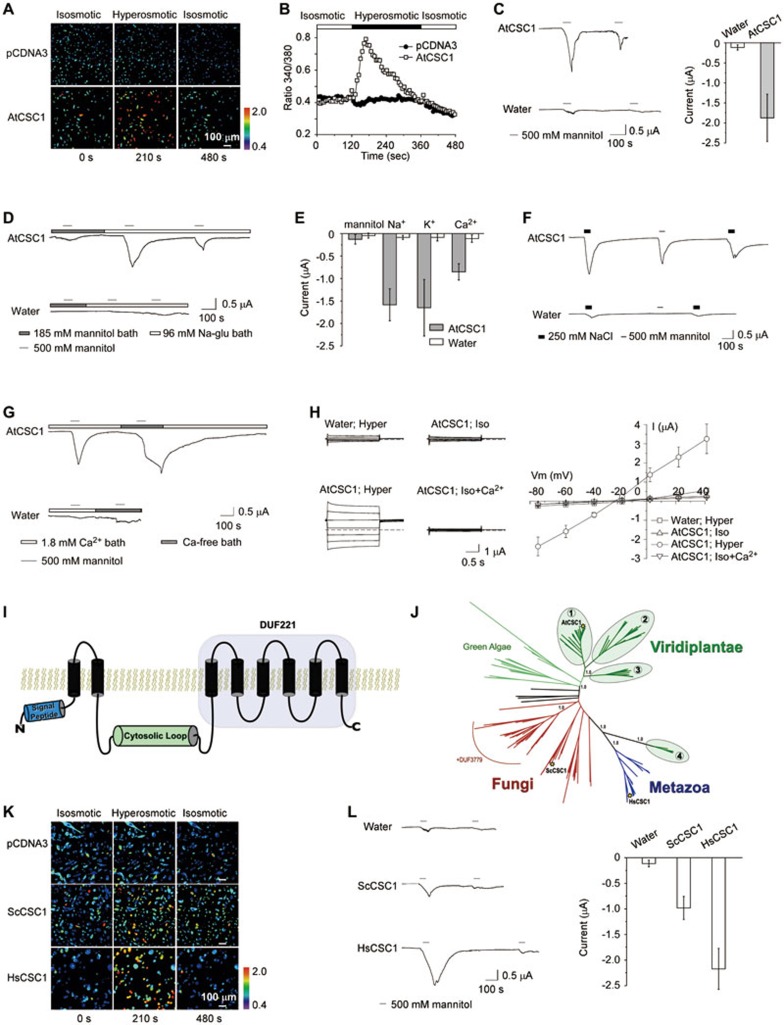
Using heterologous expression of Arabidopsis genes in Chinese Hamster Ovary (CHO) cells loaded with the calcium-responsive dye Fura-2, we screened uncharacterized integral membrane proteins for osmosensitive calcium conductance. These screens led to the identification of the Arabidopsis gene At4G22120, which encodes an integral membrane protein with eight or more predicted transmembrane helices. Ratiometric monitoring of Fura-2 fluorescence indicated that hyperosmotic shock treatment applied by 300 mM mannitol treatment induced calcium elevation in CHO cells expressing At4G22120 (UniProt: Q5XEZ5), whereas cells containing the empty vector control lacked discernible calcium elevation (Figure 1A). The observed calcium elevation occurred rapidly, peaking several seconds after hyperosmotic shock, and the calcium level sharply declined within 1 minute of osmotic shock (Figure 1B).
Figure 1.
(A) Representative images taken from CHO cells transfected with empty pCDNA3 vector or the same vector harboring AtCSC1. These images show that AtCSC1-expressing CHO cells respond to hyperosmotic stress by cytosolic [Ca2+]i elevation not seen in control cells. [Ca2+]i changes are indicated by Fura-2 emission ratios (F340 nm/F380 nm) scaled by a pseudo-color bar in relative units. Hyperosmotic stress was applied by including 300 mM mannitol in isoosmotic saline solution. (B) Time course of intracellular calcium [Ca2+]i changes in AtCSC1-expressing CHO cells. Open and filled bars show the isoosmotic and hyperosmotic conditions, respectively. The average response of 10-20 cells from a representative experiment is presented. (C) (left) Typical whole-cell current traces recorded from AtCSC1-expressing Xenopus laevis oocytes (AtCSC1) and the water-injected control oocyte (water) with periodic perfusion of saline solution containing 500 mM D-mannitol. (right) Current amplitudes evoked by the first application of hypersosmotic stress (n = 5). (D) Typical whole-cell current traces recorded from the AtCSC1-expressing oocytes perfused with bath solution containing 185 mM D-mannitol or 96 mM Na-gluconate (Na-glu) plus 500 mM D-mannitol. (E) Current amplitudes produced by 96 mM Na+ or K+ or 80 mM Ca2+. (F) AtCSC1-produced inward current was activated by equal osmolality of NaCl or mannitol. (G) AtCSC1 failed to inactivate under calcium-free extracellular conditions, implicating calcium-dependent channel closure. (H) (left) Typical whole-cell current traces recorded from the water-injected control and AtCSC1-expressing oocytes perfused with calcium-free isoosmotic buffer (Iso), calcium-free hyperosmotic buffer (Hyper) or isoosmotic buffer with 1.8 mM Ca2+ (Iso + Ca2+). (right) Current/voltage (I/V) relationship of the same recordings. The current values were extracted at the end of 2 s voltage pulses (n = 5). The bars represent means ± SD. The membrane potential was held at 0 mV and stepped in 20 mV increments from −80 to +40 mV. (I) Predicted transmembrane topology of AtCSC1 and CSC homologs; the first predicted transmembrane helix is hypothetically a cleavable signal peptide. (J) ML phylogenetic tree of DUF221-containing proteins created using PhyML v.2.2.0. Yellow stars mark the positions of AtCSC1, ScCSC1, and HsCSC1. Land plants contain four clades of CSCs, including one clade that clusters with metazoan homologs. One fungal clade contains an additional DUF3779, which is located at the C-terminus of these homologs. Support values (aLRT) are labeled for select clades. (K) Representative images taken from Fura-2-loaded CHO cells expressing ScCSC1 or HsCSC1 after they were treated with isoosmotic saline solution or hyperosmotic solution containing 300 mM mannitol. These data indicate that yeast and human CSC proteins are also osmosensitive, calcium-permeable cation channels. (L) (left) Typical whole-cell current traces recorded from water-injected or ScCSC1- or HsCSC1-expressing oocytes with periodic application of 500 mM D-mannitol. (right) The current amplitudes evoked by the first application of 500 mM D-mannitol (n = 5). The bars represent means ± SD. The holding potential was −60 mV.
Because At4G22120 encodes a protein with multiple predicted transmembrane helices, we tested the possibility that it may form an influx cation channel permeable to calcium. We expressed the protein by injecting cRNA of this gene into Xenopus oocytes and performed whole-cell two-electrode voltage clamp (TEVC) analysis. Oocytes expressing the cRNA of the gene were placed in ND96 bath solution, and hyperosmotic shock was applied by perfusion with ND96 solution containing 500 mM mannitol. Hyperosmotic shock induced an inward current indicative of channel activation, and the observed current disappeared upon removal of osmotic shock, implicating channel closure (Figure 1C). The second osmotic shock induced a smaller inward current, suggesting response dampening upon repeated stimulation. These results indicate that the At4G22120 protein is an ion channel that can be activated by hyperosmotic shock. We therefore named this protein AtCSC1 as Arabidopsis Calcium permeable Stress-gated cation Channel 1.
As expected of a channel, the current was barely detectable when equal molarity of mannitol (185 mM) was used to replace the major charge carrier NaCl in the ND96 solution, even after the same hyperosmotic shock (Figure 1D). Replacement of 185 mM mannitol with 96 mM Na+-gluconate restored channel conductance in response to the hyperosmotic treatment (Figure 1D), suggesting that the channel specifically conducts cations (i.e., Na+) rather than the much larger gluconate anions. Further analyses revealed that AtCSC1 is permeable to various cations, including Ca2+, K+, and Na+ (Figure 1E).
To further investigate AtCSC1 activation, we applied hyperosmotic stress to AtCSC1-expressing oocytes using various concentrations of mannitol or NaCl. Channel conductance showed dependence on D-mannitol concentration in the extracellular buffer, with 300 mM or higher concentrations sufficient for channel activation (Supplementary information, Figure S1). Hyperosmotic shock could be applied with 250 mM NaCl substituted for 500 mM mannitol, and it similarly activated AtCSC1 (Figure 1F). The currents induced by mannitol or NaCl treatment abated quickly after removal of hyperosmotic shock. This observation piqued our interest in AtCSC1 channel closure, therefore we tested the hyperosmotic shock responses of AtCSC1-expressing oocytes with 500 mM mannitol under calcium-free conditions. The channel activated in response to hyperosmotic shock but failed to inactivate after removal of osmotic shock until perfusion was resumed with ND96 solution containing 1.8 mM Ca2+(Figure 1G), indicating that inactivation or closure of the AtCSC1 channel is calcium-dependent. The currents conducted by AtCSC1 occurred instantaneously upon application of hyperosmotic shock and were time-independent (i.e., activation was not gradual). Stepwise voltage analysis indicated that the channel is non-rectifying (Figure 1H). A previous proteomics study indicates that AtCSC1 localizes to the plasma membrane6, which is consistent with its apparent localization in our CHO and oocyte experiments.
According to hydrophobicity plots and the MetaTM webserver (http://metatm.sbc.su.se), which predicts transmembrane protein topology using a consensus approach of popular transmembrane helix prediction algorithms, AtCSC1 contains two clusters of transmembrane (TM) helices (Figure 1I). The N-terminal region contains three predicted TM helices, however we postulate that the first predicted helix may act as a cleavable signal peptide. The C-terminal region comprising domain of unknown function 221 (DUF221) contains six predicted TM helices, therefore the AtCSC1 protein likely contains eight TM helices in total and may constitute the pore-forming entity of an ion channel. The two clusters of TM helices appear to be linked by a roughly 200 amino acid cytosolic loop.
Homologs of AtCSC1 with conserved membrane topology were identified among nearly all studied eukaryotes, including baker's yeast, fruit flies, and humans. Phylogenetic analyses revealed that land plants contain 4 ancient clades of AtCSC1 homologs and that Arabidopsis contains at least 15 CSCs (Figure 1J and Supplementary information, Figure S2). We identified four CSC homologs in the model yeast Saccharomyces cerevisiae and cloned one of these homologs, ScYLR241W, for heterologous expression and electrophysiological characterization. Like AtCSC1, ScYLR241W showed osmotically gated calcium conductance when heterologously expressed in CHO cells and similar electrophysiological properties to AtCSC1 in oocytes (Figure 1K and 1L), therefore we named this protein ScCSC1. Three AtCSC1 orthologs were identified among all sequenced vertebrates. We characterized one homolog from human, HsTM63C (UniProt: Q9P1W3), in CHO cells and Xenopus oocytes, as described for AtCSC1 and ScYLR241W. HsTM63C showed similar gating properties and conductance to AtCSC1 and ScYLR241W, therefore we designated this gene HsCSC1 (Figure 1K and 1L). These data for ScCSC1 and HsCSC1 provide further support for our hypothesis that CSC homologs constitute a previously unidentified family of osmosensitive, calcium-permeable cation channels in eukaryotes.
Published expression data indicate that several Arabidopsis CSCs are transcriptionally upregulated in response to various abiotic stresses and biotic stresses, which both entail mechanical perturbation. Indeed, some CSC proteins were previously named Early Response to Dehydration (ERD) 4 proteins based on their expression profile7. According to Pfam, DUF221 belongs to the anoctamin-like clan and is homologous to domains in anoctamin/TMEM16 channels, which are calcium-activated chloride channel (CaCC) components8, and transmembrane channel-like (TMC) proteins. TMCs are cation channel components, and certain TMCs are required for salt chemosensation in C. elegans9 and hearing in mammals10. Our data establish that CSCs are a family of cation channels that are permeable to calcium and gated by physical signals such as hyperosmotic stress. These properties are reminiscent of TRPs in animals and make CSCs intriguing candidates for involvement in osmo- or mechano-sensitive calcium signaling processes in plants. Given their conservation in distantly related eukaryotes including fungi and animals, this study opens new avenues for research towards understanding the function of this novel family of ion channels not only in plants but also in other eukaryotes.
Detailed methods are described in the Supplementary information, Data S1.
Acknowledgments
We thank Dr Xiaojiang Zheng for helpful discussions. This work was supported by the National Program on Key Basic Research Projects (2012CB113900), the National Natural Science Foundation of China (31270297, 30830070) (to LL) and by a grant from the National Science Foundation (to SL).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Concentration-dependent Activation of AtCSC1 by D-mannitol.
ML phylogenetic analysis of DUF221-containing proteins reveals four clades of land plant homologs.
Experimental Procedures
References
- Venkatachalam K, Montell C. Annu Rev Biochem. 2007. pp. 387–417. [DOI] [PMC free article] [PubMed]
- Knight H, Trewavas AJ, Knight MR. Plant J. 1997. pp. 1067–1078. [DOI] [PubMed]
- Swarbreck SM, Colaço R, Davies JM. Plant Physiol. 2013. pp. 514–522. [DOI] [PMC free article] [PubMed]
- Knight MR, Campbell AK, Smith SM, et al. Nature. 1991. pp. 524–526. [DOI] [PubMed]
- Luan S, Kudla J, Rodriguez-Concepcion M, et al. Plant Cell. 2002. pp. S389–S400. [DOI] [PMC free article] [PubMed]
- Nuhse TS, Stensballe A, Jensen ON, et al. Mol Cell Proteomics. 2003. pp. 1234–1243. [DOI] [PubMed]
- Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K. Plant Mol Biol. 1994. pp. 791–798. [DOI] [PubMed]
- Schroeder BC, Cheng T, Jan YN, et al. Cell. 2008. pp. 1019–1029. [DOI] [PMC free article] [PubMed]
- Chatzigeorgiou M, Bang S, Hwang SW, et al. Nature. 2013. pp. 95–99. [DOI] [PMC free article] [PubMed]
- Pan B, Géléoc GS, Asai Y, et al. Neuron. 2013. pp. 504–515. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Concentration-dependent Activation of AtCSC1 by D-mannitol.
ML phylogenetic analysis of DUF221-containing proteins reveals four clades of land plant homologs.
Experimental Procedures