Abstract
Many proteins are post-translationally modified by acylation targetting them to lipid membranes. While methods such as X-ray crystallography and NMR are available to determine the structure of folded proteins in solution, the precise position of folded domains relative to a membrane remains largely unknown. We used neutron and X-ray reflection methods to measure the displacement of the core domain of HIV Nef from lipid membranes upon insertion of the N-terminal myristate group. Nef is one of several HIV-1 accessory proteins and an essential factor in AIDS progression. Upon insertion of the myristate and residues from the N-terminal arm, Nef transitions from a closed to open conformation that positions the core domain 70 Å from the lipid headgroups. This work rules out speculation that the Nef core remains closely associated with the membrane to optimize interactions with the cytoplasmic domain of MHC-1.
INTRODUCTION
There is abundant evidence that enzyme activity or protein-protein interactions can be dependent upon association with lipid membranes. The positioning of proteins and protein motifs relative to either the membrane or to other membrane-bound proteins is, for some proteins, critical and may depend upon conformational changes induced upon membrane association (Kim et al., 2009; Osterhout et al., 2003; Schlessinger, 2000; Subramanian et al., 2006; Xue et al., 2004; Zha et al., 2000). Lipid modification serves to target many proteins to specific membranes or submembrane locations. Hundreds of proteins are known to be modified with covalently bound lipid groups, the most common of which are fatty acids, isoprenoids, and glycosylphosphatidylinositol anchors (Farazi et al., 2001; Jeromin et al., 2004; Perinpanayagan et al., 2013; Resh, 2006; Steinhauer and Treisman, 2009). Many of these proteins are involved in signaling, and require membrane association to signal efficiently. In addition to intra cellular membrane location, the structure adopted by proteins at membranes is also critical for certain functions. As an example, myristoyl or farnesyl switch mechanisms are known for more than a dozen proteins, with Arf GTPase being a hallmark example (Goldberg, 1998; Resh, 2006). These mechanisms cause proteins to switch between conformational states in which the myristoyl or farnesyl moiety is either sequestered or exposed, and can promote membrane binding (Ames et al., 1996; Goldberg, 1998) facilitate release from the membrane-bound state (Ames et al., 1996; Goldberg, 1998; Hantschel et al., 2003; Matsubara et al., 2004; McLaughlin and Aderem, 1995; Resh, 2006), and regulate protein-protein interactions (Hantschel et al., 2003; Matsubara et al., 2004).
Despite the obvious importance of acylated proteins in biology (e.g., kinases and phosphatases (Kim et al., 2009; Resh, 2006; Schlessinger, 2000), G proteins (Resh, 2006), GPCRs (Resh, 2006), morphogens (Steinhauer and Treisman, 2009), neuronal calcium sensors (Jeromin et al., 2004), pro- and anti-apoptotic proteins (Perinpanayagan et al., 2013)), standard approaches for studying their structure at membranes are neither adequate nor appropriate. In addition, understanding signaling mechanisms involving these proteins at the molecular level and developing pharmaceutical interventions has been limited by the absence of structural detail for these proteins in the membrane-bound state. Many structural studies of membrane-associated proteins have consisted of crystallization of soluble proteins with and without bound ligand or in complex with other proteins but in absence of a membrane (Ames et al., 1996; Flaherty et al., 1993; Goldberg, 1998; Matsubara et al., 2004). NMR, EPR, Fluorescence resonance energy transfer (FRET), Fourier transform infrared-attenuated total reflection spectroscopy (FTIR-ATR) and other methods can be applied to provide some structural details for proteins associated with membranes or membrane mimics, but do not give the full residue distribution with respect to the membrane.
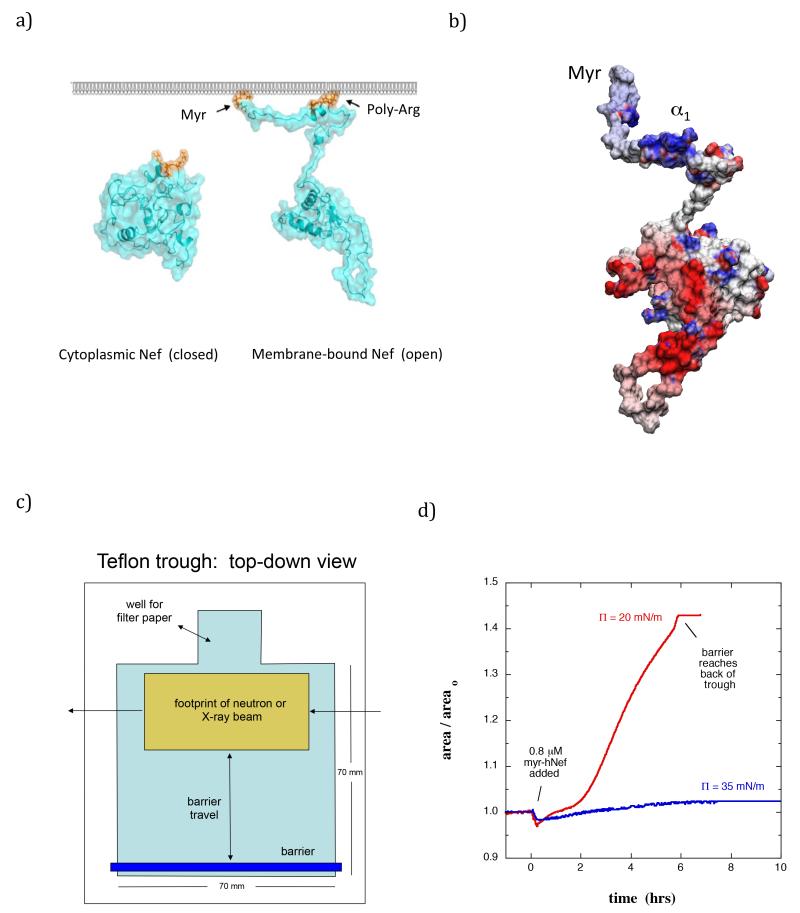
A case in which it is essential to define the structural details of a protein at the membrane is the Nef protein from HIV-1. Nef is one of several HIV-1 accessory proteins and is essential for AIDS progression (Baur, 2004; Das and Jameel, 2005). Nef is expressed in high concentrations shortly after viral infection (Klotman et al., 1991), and is required for achieving and maintaining high viral loads in vivo (Goldsmith et al., 1995). Nef lacks catalytic activity but instead realizes its functions by interacting with host proteins – more than 30 proteins that interact with Nef have been identified (Arold and Baur, 2001; Baur, 2004; Renkema and Saksela, 2000). Nef exists in both membrane-associated and cytosolic fractions (Coates et al., 1997) and shuttling may occur between the cytosolic fractions and the membrane-associated form (Fig 1a) (Bentham et al., 2006). Membrane-association is achieved by an N-terminal myristoylation essential for the virus in vivo (Harris, 1995) as well as a cluster of basic residues (17-22) within the N-terminal arm (Fig 1b) (Bentham et al., 2006; Curtain et al., 1998; Gerlach et al., 2010). The myristoylation motif (residues 2-7) is essential and highly conserved in Nef alleles from both laboratory HIV-1 strains and in primary isolates from AIDS patients (Geyer et al., 1999). Deletion of the myristate (myr) group from Nef dramatically reduces infectivity (Goldsmith et al., 1995), cripples downregulation of CD4 and MHC-1 (Goldsmith et al., 1995; Peng and Robert-Guroff, 2001), and prevents formation of an AIDS-like disease in mice transfected with Nef (Hanna et al., 2004). Both the myr group and the basic cluster are required for Nef virion incorporation (Welker et al., 1998). Nef residues 5-22 form an amphipathic helix with hydrophobic residues Trp5, Trp13, Ile16, and Met20 located on one side of the helix. Gerlach et al reported significantly decreased binding affinity to lipid membranes and impaired helix formation upon mutation of Trp5 and Trp 13 (Gerlach et al., 2010).
Figure 1.
(a) Hypothetical molecular models of Nef in solution and bound to a membrane. Left: Nef (cyan) adopts a tightly closed conformation in the cytoplasm. Right: Nef interacts with lipid bilayers via an N-terminal myristate group and poly-Arg cluster (orange). The open Nef model was adapted from Arold and Baur (Arold and Baur, 2001). (b) Distribution of surface charge on myr-Nef displayed from −8 (red) to +8 (blue) kbT (open form model of Geyer and Peterlin (Geyer and Peterlin, 2001)). (c) Diagram (top down view) of the trough used in the NR and XR measurements. The beam footprint in the NR (XR) studies was 25 mm (10 mm). (d) Change in surface area upon binding of myr-Nef (0.83 μM) at 20 mN/m and 35 mN/m. Areao is the lipid-containing surface area prior to injecting protein.
It has been postulated that Nef undergoes a transition from a solution conformation to a membrane-associated conformation (Fig 1a), and this conformational rearrangement enables membrane-associated Nef to interact with host proteins (Arold and Baur, 2001; Geyer and Peterlin, 2001; Jia et al., 2012; Raney et al., 2007). In particular it has been suggested that insertion of the N-terminal arm and subsequent displacement of the core domain from the lipid membrane will expose binding sites on the core, facilitating interaction with host proteins (Arold and Baur, 2001). On the other hand, based on the crystal structure of Nef with the cytoplasmic tail of MHC-I others have suggested that association of Trp13 and Met20 on the N-terminal arm with the core domain persists upon membrane binding, and that this positions the Nef core close to the membrane for optimal interaction with the cytoplasmic domain of the MHC-I receptor (Jia et al., 2012). Others have proposed that association of the core domain of Nef with negatively-charged membranes through its basic surface (Fig 1b) orients Nef to provide optimal exposure of the dileucine sorting motif in the flexible loop (residues 152-184) that mediates interactions with adaptor protein complexes (Horenkamp et al., 2011). Nef is known to upregulate several but not all Src family kinases through interaction with their SH3 domains (Narute and Smithgall, 2012), critical to many downstream functions. These kinases are also bound to the membrane through N-terminal acylation and positioning of the Nef core domain relative to the SH3 domains may play a role in the varying binding affinities. Despite the vital importance to the pathogenicity of Nef, to date there is no information regarding the position of the core of Nef upon membrane association due to the limitations of current structural methods.
FRET has been used to detect membrane binding and insertion of Nef by Gerlach et al. (Gerlach et al., 2010). From kinetic studies they identified two processes and proposed a model for the stages of interaction, but structural information for full Nef was absent from their model. Prior structural work, while providing critical insights, has been confined to solution-based analyses even though membrane association is critical for Nef function (Arold et al., 1997; Grzesiek et al., 1996; Grzesiek et al., 1997; Jia et al., 2012; Jung et al., 2011; Lee et al., 1996). While methods such as circular dichroism (Gerlach et al., 2010) and FTIR-ATR are available to assay changes in secondary structure upon membrane binding, full global conformational characterization including distances of motifs relative to the membrane are not provided by these methods.
In the current work neutron and X-ray reflectometry (NR and XR) were used to resolve conformational changes in myristoylated Nef (myr-Nef) upon membrane insertion. Reflectivity is one of very few methods that can resolve structural details of membrane-associated proteins in physiological conditions, and may be unique in the ability to directly resolve details of the full membrane-bound protein structure, in contrast to techniques that probe only labeled residues or secondary structural elements. NR and XR involve measuring the ratio of reflected to incident intensity as a function of momentum transfer qz = 4πsinθ/λ, where θ is the angle of incidence with respect to the plane of the membrane and λ is the wavelength (Penfold and Thomas, 1990). The form of this curve is determined by the in-plane averaged scattering length density (SLD) profile normal to the surface. The neutron SLD is determined by the properties of the nuclei present, whereas the X-ray SLD is determined by the electronic properties. In both cases the SLD is directly related to the atomic composition and the density. Therefore, for a protein bound to a planar lipid membrane, NR and XR determine the in-plane averaged distribution of amino acid residues normal to the membrane in a complementary way. Typically, XR covers a qz range that extends to higher values than achievable by NR, and hence XR provides greater insight into the effect of myr-Nef binding and insertion on the structure of the lipid layer. The contrast for the protein in buffer with XR is comparable to, but slightly weaker than, that for NR with protonated myr-Nef, yet is still sufficiently high to resolve large changes in the residue profile. The NR contrast for deuterium-enriched myr-Nef in buffer is substantially greater than that with XR and NR with protonated myr-Nef. Langmuir monolayers and lipid bilayers supported on a solid substrate can both be used as model lipid membranes in NR/XR studies of membrane-bound proteins (Chen et al., 2009; Datta et al., 2010; Kent et al., 2010; McGillivray et al., 2009; Nanda et al., 2010; Shenoy et al., 2012). For biophysical studies Langmuir monolayers provide an advantage in that the membrane pressure can be controlled, and are especially suitable when proteins are known to insert into only the outer leaflet of lipid bilayers.
Myr-Nef (strain SF2) was injected underneath a Langmuir monolayer of deuterated dipalmitoylphosphatidylglycerol (dDPPG) and its conformation was resolved by NR and XR as a function of membrane conditions. The structural details of membrane-bound Nef as a function of solution concentration, membrane pressure, and Nef coverage are described below. The data demonstrate a large conformational change from a closed to an open form that displaces the Nef core domain 70 Å from the lipid headgroups upon insertion of the myristate group and residues of the N-terminal arm. This large conformational change is likely to affect its ability to interact with host proteins by exposing binding motifs on the core domain or by optimally positioning the core domain for interaction with motifs of membrane-associated host proteins.
Results
The NR and XR data in the present study indicated that soon after myr-Nef was introduced underneath the lipid membrane, a process of insertion into the membrane occurred accompanied by a large conformational transition. Because the affinity of myr-Nef for lipid membranes increases with the percentage of negatively-charged lipid, lipid monolayers composed entirely of dDPPG were used in most of this work to maximize the binding affinity, although some experiments were performed using a more biologically relevant ratio of 30% negatively-charged lipid to 70% neutral lipid. When myr-Nef was circulated underneath the monolayer, insertion of myr-Nef into the membrane was evident by the backward movement of the trough barrier maintaining the monolayer pressure (increase in surface area at fixed number of lipid molecules, see Fig 1c and Fig 1d and described in more detail in the Methods section). The insertion was dependent on the membrane pressure, with insertion readily occurring at 25 mN/m and lower but not at 35 mN/m (Figure 1d, and see also below). Due to the larger area occupied by the core domain relative to that of the myristate group, insertion of the myristate moiety alone can account for an increase in surface area of at most 5%; increases in surface area greater than 5% therefore indicate insertion of residues of the protein in addition to the myristate group. Others have reported membrane insertion and evidence for formation of an amphipathic helix within the N-terminal 27 residues of Nef upon association to lipid membranes (Gerlach et al., 2010). Upon insertion, myr-Nef remained associated with the membrane throughout extensive exchange of the subphase underneath the surface layer to remove noninserted, loosely-bound myr-Nef. On the other hand, when insertion was inhibited (35 mN/m) membrane-associated myr-Nef was readily removed upon subphase exchange. The rate and extent of insertion of residues varied both when membrane pressure was held constant and the concentration of myr-Nef was changed, and vice versa.
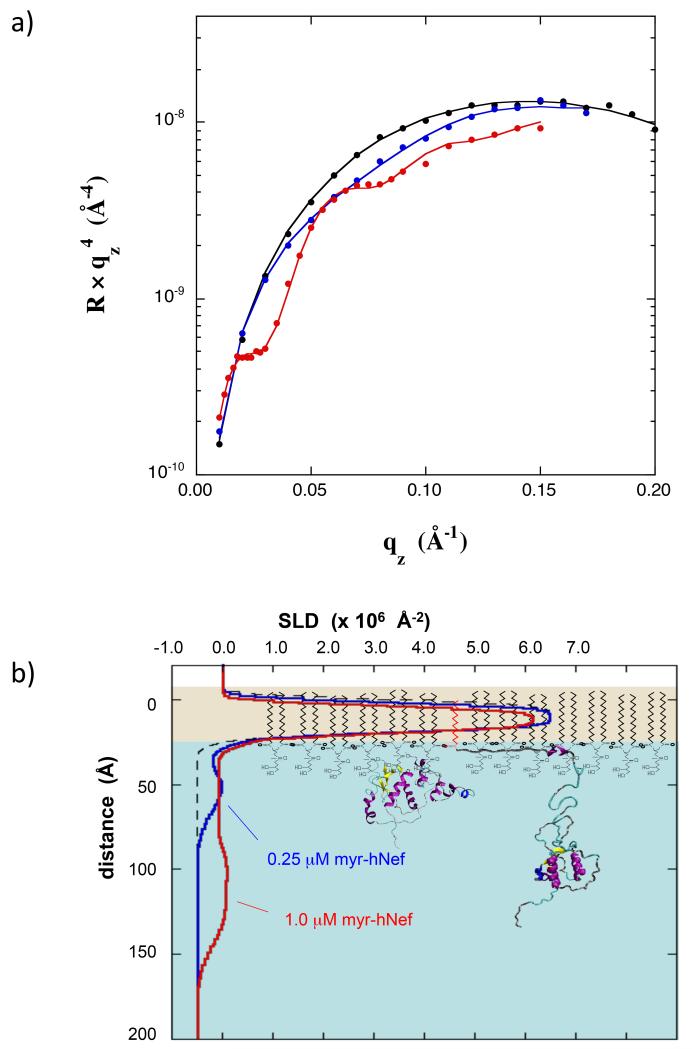
To interrogate the membrane-associated conformation of Nef, several NR studies were performed at a fixed membrane surface pressure of 30 mN/m and variable myr-Nef concentration, shown in Fig 2 (NR data in panel a and the SLD profiles resulting from the fitting analysis in panel b). At a myr-Nef concentration of 0.25 μM a fractional surface coverage (f) of 0.21 resulted (the fractional surface coverage is arbitrarily defined such that when f=1.0, the core domains of all Nef molecules would just come into contact with one another in the open conformation shown in Fig 2b), little change in surface area occurred, and a form of Nef that was compact with the core domain adjacent to the lipid headgroups (hereafter referred to as the closed form) was observed. At a myr-Nef concentration of 1.0 μM, the fractional coverage was 0.62, a large increase in area occurred (25%), and a completely different conformation was observed for myr-Nef. For this conformational state (hereafter referred to as the open form) the core domain was displaced ~70 Å below the lipid headgroups (Fig 2b).
Figure 2.
(a) NR data for pure dDPPG monolayer at 30 mN/m on Tris-buffered H2O subphase (black) and with myr-hNef adsorbed from solutions at 0.25 μM (blue) and at 1.0 μM (red). Error bars at the highest qz values are the size of the data points. (b) SLD profiles corresponding to the data in (a). The molecular models of dDPPG and of Nef are not drawn precisely to scale but were scaled to coincide approximately with the corresponding features in the SLD profiles. See also Figure S1.
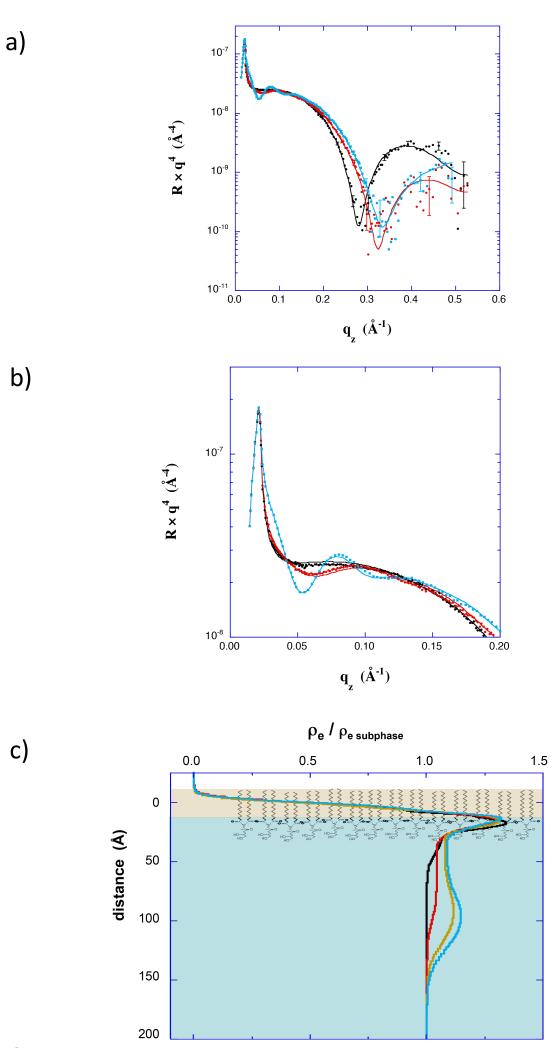
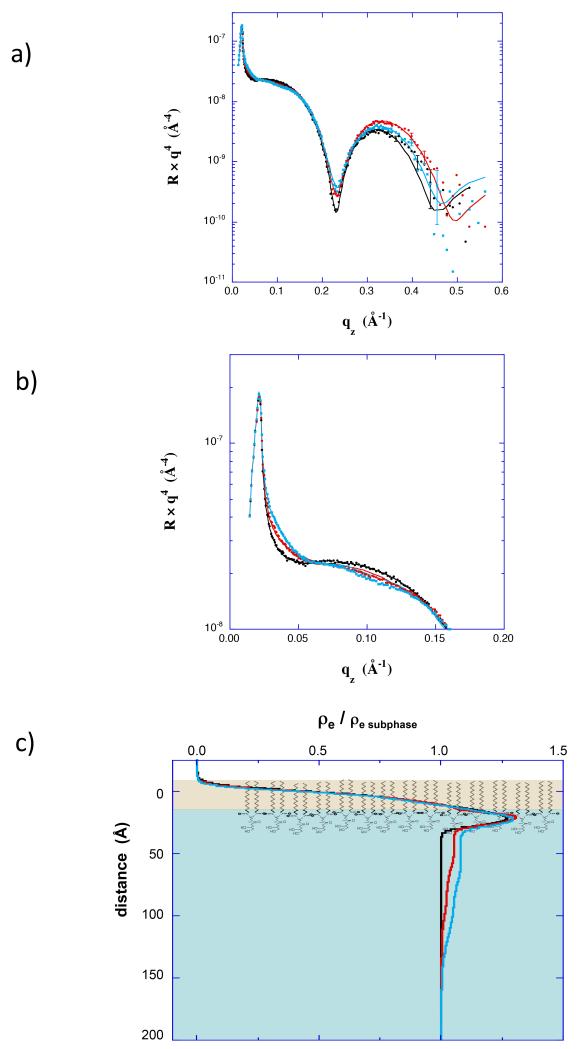
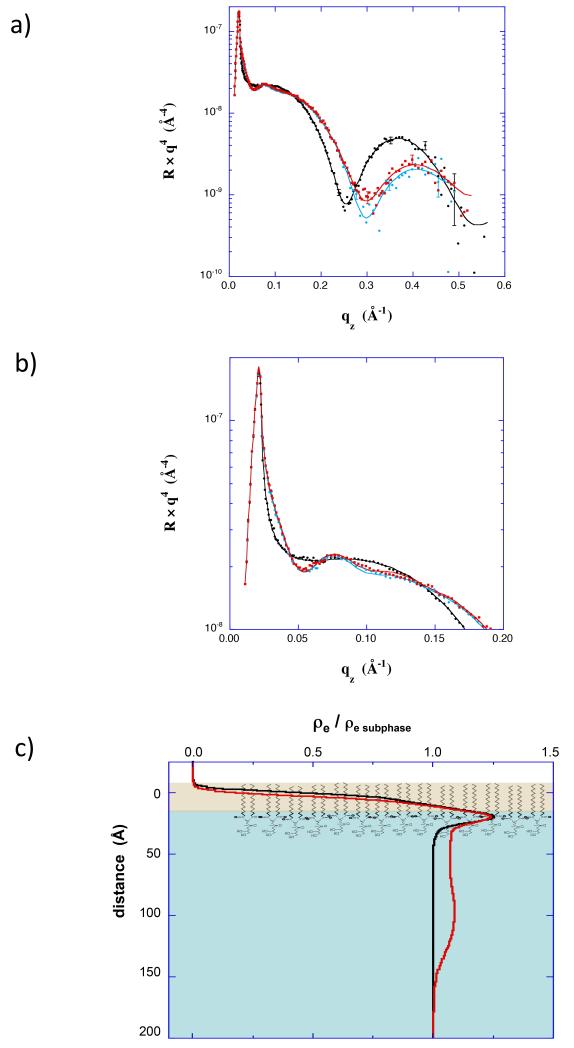
Further XR and NR studies were performed in which insertion and Nef conformation were controlled by adjusting the surface pressure of the lipid membrane. XR (Fig 3 and Fig 4) and surface area data (Fig 1d) both indicate little insertion of residues at 35 mN/m, but substantial insertion of residues at 20 mN/m. In the XR data, the conformation of adsorbed protein is evidenced by the variation at low qz (expanded in Fig 3b and Fig 4b). In the low qz region the XR curves show distinctly different patterns, and hence indicate different conformations of myr-Nef (0.83 μM), at the two membrane pressures. As shown in the electron density profiles (Fig 3c and Fig 4c) the core domain of myr-Nef was directly adjacent to the lipid headgroups at 35 mN/m, but was displaced 70 Å below the lipid headgroups at 20 mN/m, similar to the profile for 1μM myr-Nef in Figure 2. At higher qz the XR curves primarily reflect the structure of the dDPPG monolayer. In particular, the minimum in the data prior to myr-Nef addition indicates the thickness of the dDPPG layer. At 20 mN/m (Fig 3a) the minimum shifted from 0.28 Å−1 to 0.33 Å−1 after injecting myr-Nef. This shift in the minimum to higher qz indicates that the thickness of the lipid layer decreased by 4 Å, consistent with tilting of the tails upon insertion of Nef residues. This is another indication of insertion of Nef residues into the lipid layer. At 35 mN/m (Fig 4a) the minimum was unchanged after injecting myr-Nef. Insertion of residues into the membrane and the open extended form of Nef was also observed at 25 mN/m (Fig S2) and again for adsorption to 70/30 dDPPC/dDPPG membranes at 20 mN/m (Fig 5). Others have shown that myr-Nef only binds with high affinity to membranes containing at least 30% negatively-charged lipids (Gerlach et al., 2010). The XR measurements are described further in the Supporting Information.
Figure 3.
(a) XR data for a dDPPG monolayer at 20 mN/m (black) and scans initiated 2 hr (red) and 16 hrs (cyan) after addition of myr-hNef at 0.83 μM. (b) Expanded view of the XR data in a) showing the low qz region. (c) Electron density profiles corresponding to XR data in a). Also included is the profile for a scan initiated 14 hrs after addition of myr-hNef (yellow). See also Figure S2.
Figure 4.
(a) XR data for a dDPPG monolayer at 35 mN/m (black) and scans initiated 4 hr (red) and 6 hr (cyan) after addition of myr-hNef at 0.83 μM. (b) Expanded view of the XR data in a) showing the low qz region. (c) Electron density profiles corresponding to XR data in a).
Figure 5.
(a) XR data for a 70/30 dDPPC/dDPPG monolayer at 20 mN/m (black) compared with a scan collected 22 hours after injecting 0.87 μM myr-Nef (cyan) and a scan collected after exchanging the subphase with buffer (red). (b) Expanded view of the XR data in a) showing the low qz region. (c) Electron density profiles corresponding to XR data in a).
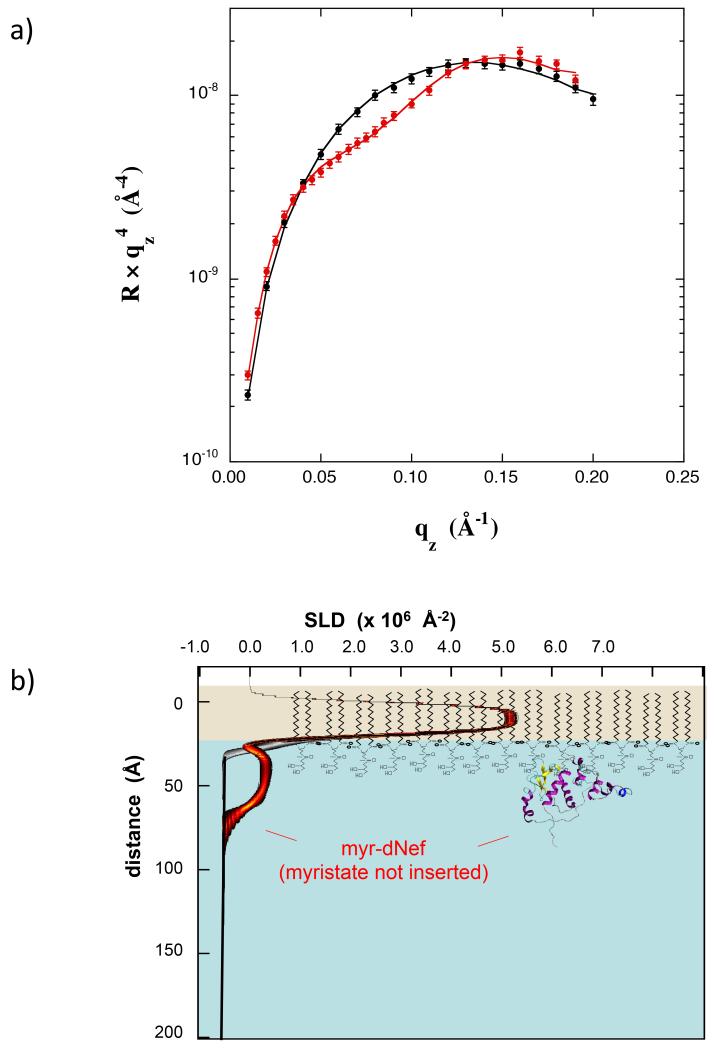
Deuterium enrichment of the protein being analyzed in NR substantially increases the SLD contrast, allowing for higher resolution data and more precise fitting with molecular models. Myr-Nef in which 80% of the non-exchangeable hydrogen atoms were replaced by deuterium (myr-dNef) was prepared. Fig 6a compares the NR data for a monolayer of dDPPG at 35 mN/m on H2O buffer compared with a scan after adsorption of myr-dNef. In this case 0.5 μM myr-dNef was incubated against the monolayer for 8 hrs and then the concentration was increased to 1.0 uM for 1 hr, at which point adsorption had slowed dramatically and a full scan was collected. The fractional coverage was 0.15. The best-fit SLD profile using a free form slab model is shown in Fig 6b. The profile band indicates that the core domain lies directly against the lipid headgroups with an uncertainty of +/− 5 Å.
Figure 6.
(a) NR data for a dDPPG monolayer at 35 mN/m on Tris–buffered H2O subphase (black) and with bound myr-dNef adsorbed from solution at 1.0 μM (red). Best fit is shown using a free-form slab model. (b) SLD profiles corresponding to the best-fits in (a). The black/grey and red/black bands correspond to the best-fit profiles for dDPPG alone and dDPPG with bound myr-dNef, respectively, with uncertainty limits using a free form slab model. The molecular models of dDPPG and of Nef are not drawn precisely to scale but were scaled to coincide approximately with the corresponding features in the SLD profiles.
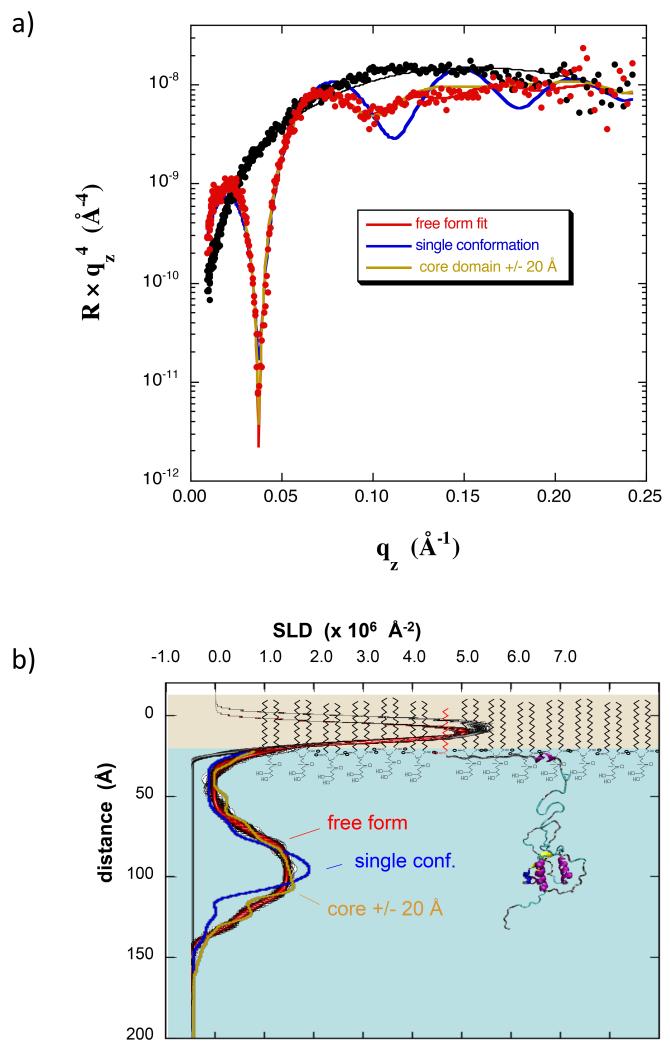
The measurement was repeated for a monolayer of dDPPG at 20 mN/m, formed initially by spreading to a pressure of ~10 mN/m and then compressing to 20 mN/m. Myr-dNef (0.28 μM) was injected under the monolayer and a scan initiated 4 hours later (Fig 7a). A large increase in area resulted, similar to that seen for myr-hNef (Fig 1d), indicating substantial insertion of residues into the lipid membrane. The fractional coverage was 0.61. Relative to the NR data for myr-Nef in Fig 2a, a larger change was observed which allowed a much more refined model to be generated. The best-fit SLD profile using a free form slab model is shown in Fig 7b. The red/black profile band contains a broad maximum, indicating the core domain, again displaced roughly 70 Å from the lipid headgroups. Clearly, in the open conformation of Nef, as now derived from multiple NR and XR experiments, the bulk of Nef does not reside next to the membrane but rather is significantly displaced.
Figure 7.
(a) NR data for a dDPPG monolayer at 20 mN/m on Tris–buffered H2O subphase (black) and with bound myr-dNef adsorbed from solution at 0.28 μM (red). Best fits are shown using a free-form slab model (red), a model of Nef with residues 2-22 located in the membrane (blue), and an ensemble of three Nef structures in which the core domain distance from the membrane was adjusted +/− 20 Å relative to that of the structure shown in b) (yellow). (b) SLD profiles corresponding to the best-fits in (a). The red/black band corresponds to the best-fit profile with uncertainty limits using a free form slab model. The other curves have the same color coding as in a). The molecular model of Nef used in the SLD calculations is shown scaled such that the core domain coincides with the peak in the SLD profile. The lipids are not drawn precisely to scale. See also Figure S3.
Further analysis was performed using molecular models of Nef in which the fractional area coverage and the position of the core domain normal to the membrane were adjusted as free parameters during fitting. As prior work by others indicated that, in addition to the myristate group, a cluster of basic residues within the N-terminal arm (17-22) interacts with negatively-charged lipid membranes to facilitate Nef adsorption (Gerlach et al., 2010), only molecular models in which residues 2-22 resided on or within the lipid headgroups were considered. Molecular structures in which the core domain was located at varying distances from residue 22 were examined. In these calculations a single orientation of the core domain was chosen arbitrarily, as it is not possible to resolve the distribution of core domain orientation from the present NR data. The structure giving the best agreement with the data is shown in Fig 7b (blue line), where the core domain is separated from residue 22 by 70 Å and the peak in SLD corresponding to residues 2-22 is located within the lipid headgroups. The fit to the data, however, is poor as the best-fit curve shown in Fig 7a (blue line) contains greater oscillations at higher qz values than are present in the data and the calculated SLD profile contains a maximum that is considerably narrower than that of the profile from the free-form fit. This result indicates that the core domains are distributed over a range of depth. Combining the myr-dNef structure shown in Fig 7b with structures in which the core domain is displaced 20 Å closer and also 20 Å further from residue 22 (weighting of 1:2:1) resulted in a good fit to the data as shown in Fig 7a and Fig 7b (yellow lines). For the same procedure but with the core displaced only +/− 10 Å, the fit was still poor (Fig S3).
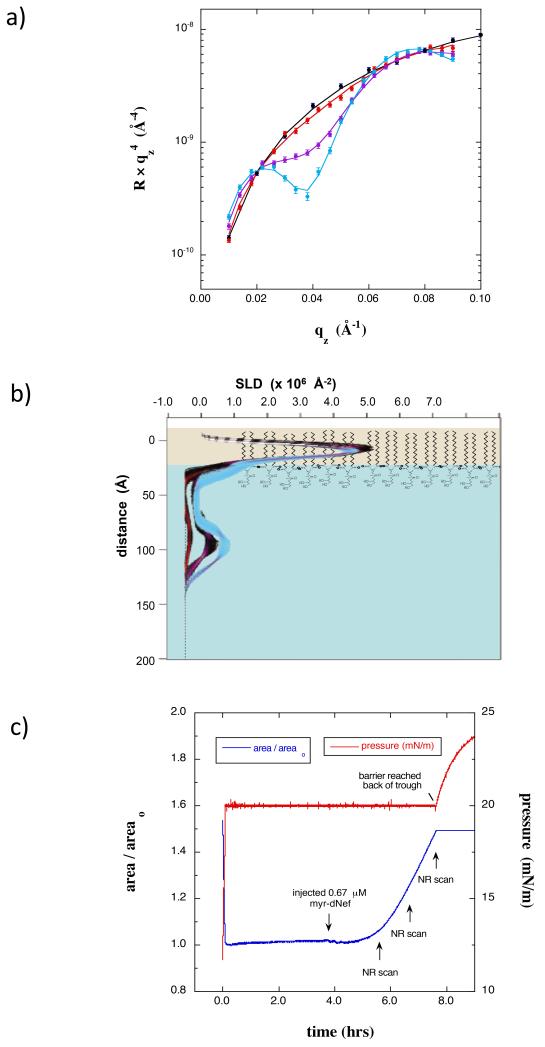
The results shown thus far were obtained after the adsorption process advanced to near completion. To gain insight into the kinetics of the process and how quickly Nef inserts and undergoes conformational change, the conformation of myr-dNef was also studied at early stages of adsorption by injecting myr-Nef at 0.67 μM underneath a monolayer of dDPPG at 20 mN/m and collecting successive scans over a limited qz range during the adsorption process. The NR results given in Fig 8a and 8b reveal that at early stages of adsorption (coverages of f = 0.07, 0.23, and 0.37) membrane bound myr-dNef was predominantly in the open form. The trough area continued to increase steadily during these scans (Fig 8c), indicating insertion of residues into the membrane.
Figure 8.
(a) NR data for a dDPPG monolayer at 20 mN/m on on Tris-buffered H2O subphase (black) along with scans collected 2 h (red), 3 h (purple), and 4 h (cyan) after injecting 0.67 μM myr-dNef. The minimum at qz = 0.04 Å−1 is indicative of the thickness of the adsorbed protein layer. b) SLD profiles corresponding to the data in a). Red, purple, and cyan profile bands correspond to 2 hr, 3hr, and 4 hr after addition of myr-hNef, respectively.. The profiles indicate that at the earliest stages of adsorption membrane-bound Nef is predominantly in the open form. C) Normalized trough area versus time showing the expansion of the area upon injection of myr-dNef and the time at which the NR scans were initiated.
Finally, the importance of the myristate group to the conformational change was examined by studying adsorption of nonmyristoylated Nef (nonmyr-Nef) to monolayers of dDPPG at 30 mN/m. In this case a deuterated Nef construct containing a N-terminal His tag was used, as described previously (Kent et al., 2010). The dDPPG monolayer was spread to 16 mN/m and then compressed to 30 mN/m before introduction of nonmyr-Nef. For this construct of Nef lacking the myristate group, the affinity for the dDPPG membrane was substantially reduced compared to myr-Nef, and adsorption to relatively high coverage (f = 0.21) required 9 hrs. at 1 μM. The NR data and corresponding profiles (Fig S1a and S1b) of nonmyr-Nef indicated no insertion and no movement of the barrier (Fig S1c). The SLD profile is that of a compact form with the core domain displaced only 10 Å from the lipid headgroups. Thus, in absence of the myristate group, insertion of N-terminal arm residues and the transition to the open form do not occur. Comparison of these results for nonmyr-Nef with the results in Figure 2 for myrNef at the same conditions (1 μM Nef and dDPPG at 30 mN/m) demonstrates that insertion of the N-terminal arm and the transition to the open form is promoted by insertion of the myristate group.
Discussion
Resolving the structure of membrane-associated proteins is extremely challenging yet critically important as positioning of residues and motifs relative to the membrane can strongly impact function. While important progress has been made recently (Chen et al., 2009; Datta et al., 2010; Kent et al., 2010; Krepkiy et al., 2012; McGillivray et al., 2009; Nanda et al., 2010; Shenoy et al., 2012), not much is known about the precise distribution of residues of membrane-associated proteins with respect to lipid membranes due to a lack of adequate tools and methods. The present NR and XR data have revealed that membrane-bound myr-Nef adopts a very different conformation depending upon the ability of residues to insert into the lipid membrane.
Our data are entirely consistent with previous hypotheses (Bentham et al., 2006; Curtain et al., 1998; Gerlach et al., 2010) that myr-Nef adsorbs through a combination of electrostatic interactions between basic residues in the N-terminal arm and the negatively-charged lipid headgroups, followed by insertion of the hydrophobic myristate group and amphipathic helix. We observed a substantially lower binding affinity with decreasing fraction of negatively-charged lipids or in absence of the myristate group, arguing that both lipid association and electrostatic attraction affect the ability of Nef to associate and insert.
Our results indicate that insertion of residues into the membrane is the key step initiating the transition to the open form. In the absence of insertion, here as a result of high membrane pressure, membrane-bound Nef adopts a closed form with the core domain directly against the lipid headgroups. At lower membrane pressure where the myristate and amino acid residues are readily able to insert into the membrane, Nef adopts an open form in which the core domain is displaced into solution ~70 Å from the lipid headgroups. From the extent of the increase in area, it is clear that a substantial number of residues, presumably residues 5-22 on the N-terminal arm known to form an amphipathic helix (Gerlach et al., 2010), insert in addition to the myristate group. Fitting the NR data with molecular models of Nef indicates that the 70 Å average distance of the core domain from the membrane in the open conformation is fully consistent with residues 5-22 residing within the lipid headgroup region. The data suggests that interactions between the N-terminal arm and the core domain that exist in solution are broken upon insertion of a portion of the N-terminal arm into the lipid monolayer, and that the latter is facilitated by the insertion of myristate group. The rate and extent of residue insertion are influenced by the density of adsorbed Nef, by the membrane pressure (lipid packing density), and by the presence of the N-terminal myristate. At a membrane pressure of 30 mN/m, insertion and the open form resulted only at higher myr-Nef concentration whereas at 20 mN/m, insertion and the open form resulted even at low myr-Nef concentration. Furthermore, at a membrane pressure of 30 mN/m and a concentration of 1 μM, insertion and the open form resulted for myrNef but not for Nef lacking the N-terminal myristate.
The open form is not triggered by high coverage of Nef on the membrane. Time dependent scans collected at early stages of adsorption and insertion at 20 mN/m show that Nef is predominantly in the open form even at low coverages. Indeed, high coverage (f=0.37) and yet very little insertion resulted in a prior study involving His-Nef adsorption to lipid monolayers containing a synthetic metal chelating lipid (Kent et al., 2010), and in that case Nef remained in the closed form. The transition to the open form does not appear to be triggered by electrostatic repulsion of the core from the membrane, as the open form was obtained for membranes containing as little as 30% negatively-charged lipids, and the displacement distance of the core domain from the lipid headgroups was nearly identical for 30% and 100% negatively-charged lipids. Furthermore, the open form did not result when insertion of residues was blocked by a high membrane pressure, despite a greater lipid packing density and therefore greater electrostatic repulsion than at lower membrane pressures.
Gerlach et al reported a kinetic study of myr-Nef binding to fluid phase membranes of DOPC and DOPG using FRET (Gerlach et al., 2010). Strong myr-Nef binding required the presence of negatively-charged lipids, as also observed in the present study. The kinetic data indicated two processes: a fast process that was attributed to electrostatic-driven association followed by myristate insertion, and a slower process that was attributed to insertion and formation of an amphipathic helix within the N terminal 27 residues. They showed that the rate of the fast process increased with membrane curvature, consistent with more rapid insertion of myristate into more loosely packed lipids. This is analogous to and entirely consistent with the present study where surface pressure and packing density were used to alter the energy barrier for insertion.
While the kinetic study of Gerlach et al revealed two processes, no information was provided on the conformation of Nef corresponding to those processes. The present study provides this new insight. While both processes in the study of Gerlach et al occurred on time scales much faster than can be resolved by NR and XR, by increasing the energetic barrier for residue insertion we isolated the membrane-bound conformation in absence of helix insertion. In that case, corresponding to the conformation at the end of the fast process of Gerlach et al, Nef is in the closed form (Fig 9). The present data reveal that upon insertion of the amphipathic helix (the slow process of Gerlach et al) Nef adopts an open conformation in which the core domain is displaced on average 70 Å from the lipid headgroups. In addition to providing insight into the conformation of Nef during the processes elucidated by Gerlach et al, the present data also reveal a much longer time scale process in which the coverage of open form Nef on the membrane increases (Fig 9). The present data thus inform the Gerlach model with respect to the conformations of Nef and also extend it to include a longer time scale process of increasing coverage.
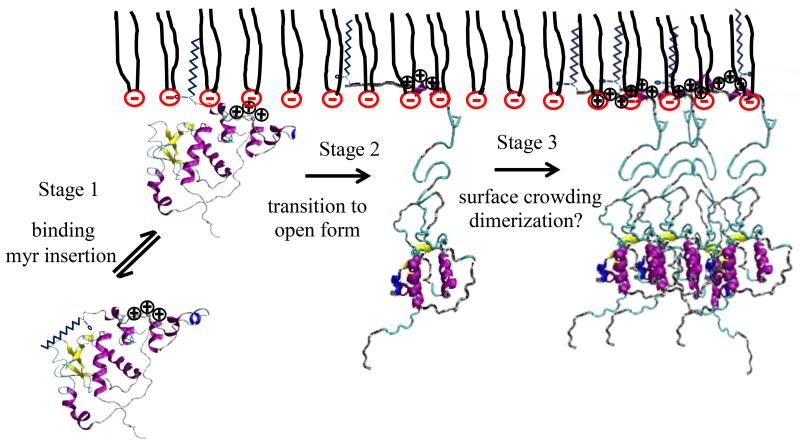
Figure 9.
Mechanism of myr-Nef binding to a dDPPG membrane. Myr-Nef is adsorbed through electrostatic attraction and myristate insertion at the first stage. This is identical to the fast process observed by Gerlach et al (Gerlach et al., 2010). At higher surface pressures (35 mN/m), there is no change in the conformation of myr-Nef after the first stage. At lower pressures (20 mN/m) with the insertion of the N-terminal arm into the membrane the (slow process of Gerlach et al) the core domain is displaced 70 A away from the membrane. Surface coverage of myr-Nef in open conformation increases very slowly as a function of time, perhaps involving membrane-driven dimerization (Poe and Smithgall, 2009). Stages I and II occur on timescales that are too fast to be detected by NR or XR.
It is interesting to speculate about how the present results may be tied to Nef biology and what role Nef insertion and conformation change may play in the ability of Nef to associate with binding partners that lead to Nef signaling/function. Recently, Jia et al determined the crystal structure of a complex of Nef with the cytoplasmic domain of MHC-I using a construct in which the MHC-I cytoplasmic domain was fused to the N-terminus of Nef (Jia et al., 2012). In the crystal structure the N-terminal helix of Nef (residues 6-22) was attached to the core domain of Nef through interactions involving Trp13 and Met20. The authors speculated that this association persists upon membrane binding and positions the Nef core close to the membrane for optimal interaction with the cytoplasmic domain of the MHC-I receptor. The present data are at odds with the assertion that the N-terminal helix of Nef remains attached to the core domain upon membrane binding. Rather, our results show that the N-terminal arm inserts into lipid membranes and the core domain is displaced 70 Å from the membrane in absence of a binding partner protein. However, this fact is not in any way inconsistent with Nef interaction with the receptor, as each core domain is free to explore the full range of distance from the membrane. The 70 Å distance is the average of the distribution of displacements. Jia et al reported that mutations W13A or M20A abolished Nef-induced downregulation of MHC-I in human T lymphocytes, and this was presented as further support for their assertion that interaction of Trp 13 and Met20 with the core domain of Nef is critical for downregulation of MHC-I. However, these residues likely play important roles in membrane binding and insertion. Therefore, it is entirely possible that the effects of these mutations observed in T lymphocytes are due to decreased membrane association or altered insertion and helix formation.
The present data also provide strong evidence against the assertion (Horenkamp et al., 2011) that association of the core domain of Nef with negatively-charged membranes through its basic surface (Fig 1b) orients Nef to provide optimal exposure of the dileucine sorting motif in the flexible loop (residues 152-184) known to mediate interactions with adaptor protein complexes. Since, as we now show, the core domain is displaced 70 Å from the lipid membrane in its final resting position, it is unlikely that the membrane affects Nef orientation.
Extensive evidence indicates that at least some functions of Nef in vivo require dimerization (Poe and Smithgall, 2009), yet we (using glutaraldehyde crosslinking) and others (Breuer et al., 2006; Horenkamp et al., 2011) have found no evidence for dimerization of free Nef in solution at 1 μM. It was shown elsewhere using analytical gel filtration that in solution, truncated Nef lacking N-terminal residues 2-44 contains significant dimeric and multimeric fractions, whereas myr-Nef and nonmyr-Nef exist primarily as monomers (Breuer et al., 2006). This suggests that dimerization of Nef may be inhibited by association of the N-terminal arm with the core domain. Residues on α-helix 4 and the adjacent loop (R109-D127) have been identified as promoting Nef dimer and trimer association (Resh, 2006). Others have proposed that membrane insertion of the myristate group causes the N-terminal arm to separate from the core domain and thereby promotes Nef dimerization (Arold and Baur, 2001; Geyer et al., 2001). Unfortunately, reflectivity methods are unable to detect structural changes that occur in the plane of the membrane; thus our results do not directly inform the dimeric status of myr-Nef at the membrane. However, our results with NR and XR show that substantially higher coverages are ultimately achieved with Nef in the open form, and thus are consistent with the hypothesis that the arm must separate from the core of the protein to promote multimerization.
In summary, we report the measurement of the precise location of the folded domain of terminally acylated Nef with respect to a lipid membrane. Hundreds of proteins are known to be lipidated, including many that are related to signaling and disease states, and many are potential targets for therapeutic intervention. The present approach will be useful to resolve the membrane-bound conformations of these proteins and will provide new insights into signaling mechanisms. It can also inform on the effects of protein-protein interaction at the membrane and disruption of said interactions with pharmacological agents.
MATERIALS AND METHODS
Materials
dDPPG and dDPPC in which the 62 protons in the aliphatic tails were replaced with deuterons were purchased from Avanti Polar Lipids. These deuterated lipids were used for both NR and XR measurements. Tris buffer salts and dithiothreitol (DTT) were purchased from Sigma-Aldrich and used as received.
Proteins
Protonated myristolated-Nef (myr-Nef) was expressed in E. coli as described elsewhere (Morgan et al., 2011) using a pET-Duet-1 vector that contained both h-NMT-1 and SF2 Nef (with a C-terminal histidine purification tag). Expression was carried out in 1L M9 minimal media until the O.D. reached 0.6, supplemented afterwards for 10 minutes with 10 ml of 5 mM myristic acid with 0.6 μM BSA. Cells were induced with 1 mM IPTG overnight at 16 °C. Purification was performed with Ni-NTA agarose, as described previously (Morgan et al., 2011) and the final purified species was >95% myristoylated, as determined by mass spectrometry. Deuterated myr-Nef (myr-dNef) was prepared by expression in a modified M9 media made with 99.8% D2O; deuterium incorporation was checked by mass spectrometry and showed that the protein was 80% deuterated (data not shown).
Methods
Adsorption studies
The Langmuir trough and monolayer system are illustrated in Fig 1c. In a typical adsorption run, dDPPG was spread from a 70/30 (by vol.) mixture of chloroform and methanol on the surface of 20 mM Tris buffered H2O subphase (pH 8.2, 100 mM NaCl) held within the Teflon trough (70 mm × 70 mm × 2 mm, Fig 1c). After allowing the chloroform and methanol to evaporate, the surface layer was compressed to the final target pressure by a movable barrier. Sufficient lipid was deposited such that after reaching the target pressure the barrier remained outside of the footprint of the neutron or X-ray beam (Fig 1c). After collecting NR or XR data for the lipid monolayer alone, myr-Nef was then injected into the subphase underneath the lipid monolayer. Successive reflectivity scans were then initiated until adsorption was completed. In some cases the subphase was then exchanged Tris buffer containing 1 mM DTT using a peristaltic pump and Teflon tubing with an inlet and outlet submerged at opposite ends of the trough. All tubing and fittings were made of Teflon and cleaned using water and Tris buffer after each experiment. The trough was maintained at 20 +/− 2 °C.
Neutron and X-ray reflection
NR measurements were performed on the NG7 (NCNR/NIST) and Liquids (SNS/ORNL) reflectometers. Details of these spectrometers and the measurement protocols are given in the Supporting information. XR measurements were performed using an X-ray reflectometer (Bruker, D8 Advance) employing Cu Kα radiation at NCNR/NIST (Gaithersburg, MD). The copper source was operated at 40 kV and 40 mA, and the wavelength was 0.154 nm. The beam width was 10 mm and the beam height was 0.1 mm.
The NR and XR data were analyzed using the Ga_refl program based on the optical matrix method. Ga_refl is available at www.ncnr.nist.gov. Simultaneous fits of the data were performed at different stages of a single adsorption run (for example lipids only, with adsorbed protein, and after subphase exchange). The SLD of the subphase was held constant for all the fits. Analyses were performed with free form models involving a small number of slabs, as well using molecular structures. The molecular structure of myr-Nef was generated from 1QA5 and 2NEF and manipulated in NAMD2 using the CHARMM22 force field. For fitting NR data the free form models consisted of one layer each for the lipid tails and the lipid headgroups, and one to four layers for the protein as required to achieve a good fit to the data. When no insertion occurred as indicated by little or no movement of the barrier, the thickness and SLD of the lipid tail layer after adsorption of myr-dNef were constrained to the same values as determined for the data taken prior to adsorption. This is based on the XR results (Fig 4) where lack of change in thickness of the lipid layers is demonstrated by absence of a shift in the fringes. For the freeform fit to the NR data in Fig 7 the thickness of the lipid tail layer after insertion of myr-dNef was constrained to be 4 Å less than that measured for dDPPG alone, based on the XR results in Fig 3. Based on the relative areas occupied by the core domain of Nef and a DPPG molecule, and the fact that the myristate group has only a single aliphatic chain, the SLD of the lipid tail layer was constrained to be greater than or equal to 0.95 × SLDtails dDPPG + 0.05 × SLDmyr. For the fits to the data in Fig 7 involving molecular models of Nef, the number of Nef molecules per area and z-position of the core domain varied in the fits, in addition to the thickness and SLD of the lipid tails and headgroups layers. The SLD of the lipid tail layer was constrained to be equal to (1.0 - 0.05 × f) × SLDtails dDPPG + 0.05 × f × SLDmyr to account for insertion of the protonated myristate group. In the Ga_refl program the roughness parameter is the full width at half maximum (FWHM = 2.35 σ, where σ is the standard deviation) of a Gaussian distribution and was constrained in the fitting to be less than the smallest thickness of the two adjacent layers.
Fitting reflectivity data results in defining a family of SLD curves that are consistent with the data. The uncertainty in the fitted profiles was determined by a Monte Carlo resampling procedure in which a large number (1000) of statistically independent sets of reflectivity data were created from the original data set and the error bars from the counting statistics. The result is a range of values for each fit parameter that is consistent with the statistics of the original data. The uncertainty in a fitted profile is represented by a color-coded band (Figs 6-8). This method has been reported in detail elsewhere (Heinrich et al., 2009). The analysis focuses on the location of structural motifs (N-terminal arm and core domain) relative to the membrane. The positions of individual atoms cannot be determined due to insufficient spatial resolution.
Nef coverages were obtained by first converting the SLD or electron density profiles to amino acid (aa) volume fraction profiles using SLD = φaa (SLDaa) + (1 − φaa)(SLDwater) or ρe = φaa (ρe aa) + (1 − φaa)(ρe water), and then integrating the aa volume fraction profiles to obtain the volume (or mass) of Nef per unit area. A coverage of 1.0 was defined as the area per Nef molecule for the open form conformation shown in Fig 2 and Fig 7. The average neutron SLD values (SLDaa) for myr-Nef and myr-dNef (80% deuteration) are 2.02 × 10−6 Å−2 and 5.19 × 10−6 Å−2, respectively. The X-ray SLD is directly proportional to the electron density, where the constant of proportionality is the classical electron radius (2.82 × 10−5 Å). The calculated average electron density (ρe aa) and X-ray SLD (λ = 1.54 Å) for myr-Nef are 0.426 e−/Å3 and 1.201 × 10−5 Å−2, respectively.
Supplementary Material
Highlights.
Structural insights into membrane-associated HIV-1 Nef
Insertion of the myristate and residues on the N-terminal arm lead to an open form
In the open form, the core domain is displaced 70 Å from the lipid headgroups
When insertion is blocked, membrane-associated Nef is in a closed form
ACKNOWLEDGMENTS
Sandia is a multiprogram laboratory operated by Sandia Corporation, a Lockheed Martin Company, for the United States Department of Energy under contract DE-AC04-94AL85000. We acknowledge the support of the National Institute of Standards and Technology, U.S. Department of Commerce, in providing the neutron research facilities used in this work. Research conducted at ORNL’s Spallation Neutron Source was sponsored by the Scientific User Facilities Division, Office of Basic Energy Sciences, US Department of Energy. This work was supported by NIH grant R01-GM086507 (M.S.K. and J.R.E.) and by NIH grant R01-GM101647-02 (H.N.) Commercial materials, instruments and equipment are identified in this paper to specify the experimental procedure as completely as possible. In no case such identification imply a recommendation or endorsement by the National Institute of Standards and Technology nor does it imply that the materials, instruments, or equipment identified are necessarily the best available for the purpose.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ames JB, Tanaka T, Stryer L, Ikura M. Portrait of a myristoyl switch protein. Current Opinion in Structural Biology. 1996;6:432–438. doi: 10.1016/s0959-440x(96)80106-0. [DOI] [PubMed] [Google Scholar]
- Arold S, Franken P, Strub MP, Hoh F, Benichou S, Benarous R, Dumas C. The crystal structure of HIV-1 Nef protein bound to the Fyn kinase SH3 domain suggests a role for this complex in altered T cell receptor signaling. Structure. 1997;5:1361–1372. doi: 10.1016/s0969-2126(97)00286-4. [DOI] [PubMed] [Google Scholar]
- Arold ST, Baur AS. Dynamic Nef and Nef dynamics: how structure could explain the complex activities of this small HIV protein. Trends Biochem Sci. 2001;26:356–363. doi: 10.1016/s0968-0004(01)01846-1. [DOI] [PubMed] [Google Scholar]
- Baur A. Functions of the HIV-1 Nef protein. Curr Drug Targets Immune Endocr Metabol Disord. 2004;4:309–313. doi: 10.2174/1568008043339749. [DOI] [PubMed] [Google Scholar]
- Bentham M, Mazaleyrat S, Harris M. Role of myristoylation and N-terminal basic residues in membrane association of the human immunodeficiency virus type 1 Nef protein. The Journal of general virology. 2006;87:563–571. doi: 10.1099/vir.0.81200-0. [DOI] [PubMed] [Google Scholar]
- Breuer S, Gerlach H, Kolaric B, Urbanke C, Opitz N, Geyer M. Biochemical indication for myristoylation-dependent conformational changes in HIV-1 Nef. Biochemistry. 2006;45:2339–2349. doi: 10.1021/bi052052c. [DOI] [PubMed] [Google Scholar]
- Chen C-H, Malkova S, Pingali SV, Long F, Garde S, Cho W, Schlossman ML. Configuration of PKCa-C2 domain bound to mixed SOPC/SOPS lipid monolayers. Biophysical Journal. 2009;97:2794–2802. doi: 10.1016/j.bpj.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coates K, Cooke SJ, Mann DA, Harris MP. Protein kinase C-mediated phosphorylation of HIV-I nef in human cell lines. J Biol Chem. 1997;272:12289–12294. doi: 10.1074/jbc.272.19.12289. [DOI] [PubMed] [Google Scholar]
- Curtain CC, Lowe MG, Macreadie IG, Gentle IR, Lawrie GA, Azad AA. Structural requirements for the cytotoxicity of the N-terminal region of HIV type 1 Nef. AIDS Res Hum Retroviruses. 1998;14:1543–1551. doi: 10.1089/aid.1998.14.1543. [DOI] [PubMed] [Google Scholar]
- Das SR, Jameel S. Biology of the HIV Nef protein. Indian J Med Res. 2005;121:315–332. [PubMed] [Google Scholar]
- Datta SAK, Heinrich F, Raghunandan S, Krueger S, Curtis JE, Rein A, Nanda H. HIV-1 Gag extension: Conformational changes require simultaneous interaction with membrane and nucleic acid. J Mol Biol. 2010;406:205–214. doi: 10.1016/j.jmb.2010.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farazi TA, Waksman G, Gordon JI. The biology and enzymology of protein N-myristoylation. J Biol Chem. 2001;276:39501–39504. doi: 10.1074/jbc.R100042200. [DOI] [PubMed] [Google Scholar]
- Flaherty KM, Zozulya S, Stryer L, McKay DB. Three-dimensional structure of recoverin, a calcium sensor in vision. Cell. 1993;75:709–716. doi: 10.1016/0092-8674(93)90491-8. [DOI] [PubMed] [Google Scholar]
- Gerlach H, Laumann V, Martens S, Becker CFW, Goody RS, Geyer M. HIV-1 Nef membrane association depends on charge, curvature, composition and sequence. Nature Chemical Biology. 2010;6:46–53. doi: 10.1038/nchembio.268. [DOI] [PubMed] [Google Scholar]
- Geyer M, Fackler OT, Peterlin BM. Structure--function relationships in HIV-1 Nef. EMBO Rep. 2001;2:580–585. doi: 10.1093/embo-reports/kve141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer M, Munte CE, Schorr J, Kellner R, Kalbitzer HR. Structure of the anchor-domain of myristoylated and non-myristoylated HIV-1 Nef protein. J Mol Biol. 1999;289:123–138. doi: 10.1006/jmbi.1999.2740. [DOI] [PubMed] [Google Scholar]
- Geyer M, Peterlin BM. Domain assembly, surface accessibility and sequence conservation in full length HIV-1 Nef. FEBS Lett. 2001;496:91–95. doi: 10.1016/s0014-5793(01)02394-8. [DOI] [PubMed] [Google Scholar]
- Goldberg J. Structural basis for activation of ARF GTPase: Mechanisms of guanine nucleotide exchange and GTP-myristoyl switching. Cell. 1998;95:237–248. doi: 10.1016/s0092-8674(00)81754-7. [DOI] [PubMed] [Google Scholar]
- Goldsmith MA, Warmerdam MT, Atchison RE, Miller MD, Greene WC. Dissociation of the CD4 downregulation and viral infectivity enhancement functions of human immunodeficiency virus type 1 Nef. J Virol. 1995;69:4112–4121. doi: 10.1128/jvi.69.7.4112-4121.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzesiek S, Bax A, Clore M, Gronenborn AM, Hu J-S, Kaufman J, Palmer I, Stahl SJ, Wingfield PT. The solution structure of HIV-1 Nef reveals an unexpected fold and permits delineation of the binding surface for the SH3 domain of Hck tyrosine protein kinase. Nat Struct Biol. 1996;3:340–345. doi: 10.1038/nsb0496-340. [DOI] [PubMed] [Google Scholar]
- Grzesiek S, Bax A, Hu JS, Kaufman J, Palmer I, Stahl SJ, Tjandra N, Wingfield PT. Refined solution structure and backbone dynamics of HIV-1 Nef. Protein Sci. 1997;6:1248–1263. doi: 10.1002/pro.5560060613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna Z, Priceputu E, Kay DG, Poudrier J, Chrobak P, Jolicoeur P. In vivo mutational analysis of the N-terminal region of HIV-1 Nef reveals critical motifs for the development of an AIDS-like disease in CD4C/HIV transgenic mice. Virology. 2004;327:273–286. doi: 10.1016/j.virol.2004.06.028. [DOI] [PubMed] [Google Scholar]
- Hantschel O, Nagar B, Guettler S, Kretzschmar J, Dorey K, Kuriyan J, Superti-Furga G. A myristoyl/phosphotyrosine switch regulates c-Abl. Cell. 2003;112:845–857. doi: 10.1016/s0092-8674(03)00191-0. [DOI] [PubMed] [Google Scholar]
- Harris M. The role of myristoylation in the interactions between human immunodeficiency virus type I Nef and cellular proteins. Biochem Soc Trans. 1995;23:557–561. doi: 10.1042/bst0230557. [DOI] [PubMed] [Google Scholar]
- Heinrich F, Ng T, Vanderah DJ, Shekhar P, Mihailescu M, Nanda H, Losche M. A new lipid anchor for sparsely tethered bilayer lipid membranes. Langmuir. 2009;25:4219–4229. doi: 10.1021/la8033275. [DOI] [PubMed] [Google Scholar]
- Horenkamp FA, Breuer S, Schulte A, Lulf S, Weyland M, Saksela K, Geyer M. Conformation of the dileucine-based sorting motif in HIV-1 nef revelated by intermolecular domain assembly. Traffic. 2011;12:867–877. doi: 10.1111/j.1600-0854.2011.01205.x. [DOI] [PubMed] [Google Scholar]
- Jeromin A, Muralidhar D, Parameswaran MN, Roder J, Fairwell T, Scarlata S, Dowal L, Mustafi SM, Chary KV, Sharma Y. N-terminal myristoylation regulates calcium-induced conformational changes in neuronal calcium sensor-1. J Biol Chem. 2004;279:27158–27167. doi: 10.1074/jbc.M312172200. [DOI] [PubMed] [Google Scholar]
- Jia X, Singh R, Homann S, Yang H, Guatelli J, Xiong Y. Structural basis of evasion of cellular adaptive immunity by HIV-1 Nef. Nature Structural & Molecular Biology. 2012;19:701–706. doi: 10.1038/nsmb.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JJ, Byeon I-JL, Ahn J, Gronenborn AM. Structure, dynamics, and Hck Interaction of full-length HIV-1 Nef. Proteins: Structure, Function, and Bioinformatics. 2011;79:1609–1622. doi: 10.1002/prot.22986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent MS, Murton JK, Sasaki DY, Satija S, Akgun B, Nanda H, Curtis JE, Majewski J, Morgan CR, Engen JR. Neutron reflectometry study of the conformation of HIV Nef bound to lipid membranes. Biophysical Journal. 2010;99:1940–1948. doi: 10.1016/j.bpj.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim LC, Song L, Haura EB. Src kinases as therapeutic targets for cancer. Nature Reviews Clinical Oncology. 2009;6:587–595. doi: 10.1038/nrclinonc.2009.129. [DOI] [PubMed] [Google Scholar]
- Klotman ME, Kim S, Buchbinder A, DeRossi A, Baltimore D, Wong-Staal F. Kinetics of expression of multiply spliced RNA in early human immunodeficiency virus type 1 infection of lymphocytes and monocytes. Proc Natl Acad Sci U S A. 1991;88:5011–5015. doi: 10.1073/pnas.88.11.5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krepkiy D, Gawrisch K, Swartz KJ. Structural interactions between lipids, water and S1-S4 voltage-sensing domains. Journal of Molecular Biology. 2012;423:632–647. doi: 10.1016/j.jmb.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C-H, Saksela K, Mirza UA, Chait BT, Kuriyan J. Crystal structure of the conserved core of HIV-1 Nef complexed with a Src family SH3 domain. Cell. 1996;85:931–942. doi: 10.1016/s0092-8674(00)81276-3. [DOI] [PubMed] [Google Scholar]
- Matsubara M, Nakatsu T, Kato H, Taniguchi H. Crystal structure of a myristoylated CAP-23/NAP-22 N-terminal domain complexed with Ca2+/calmodulin. The EMBO Journal. 2004;23:712–718. doi: 10.1038/sj.emboj.7600093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGillivray DJ, Valincius G, Heinrich F, Robertson JWF, Vanderah DJ, Febo-Ayala W, Ignatjev I, Losche M, Kasianowicz JJ. Structure of functional Staphylococcus aureus alpha-hemolysin channels in tethered bilayer lipid membranes. Biophysical Journal. 2009;96:1547–1553. doi: 10.1016/j.bpj.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin S, Aderem A. The myristoyl-electrostatic switch: a modulator of reversible protein-membrane interactions. Trends in Biochemical Sciences. 1995;20:272–276. doi: 10.1016/s0968-0004(00)89042-8. [DOI] [PubMed] [Google Scholar]
- Morgan CR, Miglionico BV, Engen JR. The effects of HIV-1 Nef on human N-myristoyl transferase 1. Biochemistry. 2011;50:3394–3403. doi: 10.1021/bi200197e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda H, Datta SAK, Heinrich F, Losche M, Rein A, Krueger S, Curtis JE. Electrostatic interactions and binding orientation of HIV-1 matrix studied by neuron reflectivity. Biophysical Journal. 2010;99:2516–2524. doi: 10.1016/j.bpj.2010.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narute PS, Smithgall TE. Nef alleles from all major HIV-1 clades activate src-family kinases and enhance HIV-1 replication in an inhibitor-sensitive manner. PLoS ONE. 2012;7:e32561. doi: 10.1371/journal.pone.0032561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterhout JL, Waheed AA, Hiol A, Ward RJ, Davey PC, Nini L, Wang J, Milligan G, Jones TLZ, Druey KM. Palmitoylation regulates regulator of G-protein signaling (RGS) 16 function. Journal of Biological Chemistry. 2003;278:19309–19316. doi: 10.1074/jbc.M210124200. [DOI] [PubMed] [Google Scholar]
- Penfold J, Thomas R. The application of the specular reflection of neutrons to the study of surfaces and interfaces. JPhys: CondensMatter. 1990;2:1369–1412. [Google Scholar]
- Peng B, Robert-Guroff M. Deletion of N-terminal myristoylation site of HIV Nef abrogates both MHC-1 and CD4 down-regulation. Immunol Lett. 2001;78:195–200. doi: 10.1016/s0165-2478(01)00250-4. [DOI] [PubMed] [Google Scholar]
- Perinpanayagan MA, Beauchamp E, Martin DDO, Sim JYW, Yap MC, Berthiaume LG. Regulation of co-and post-translational myristoylation of proteins during apoptosis: interplay of N-myristoyltransferases and capases. The FASEB Journal. 2013;27:811–821. doi: 10.1096/fj.12-214924. [DOI] [PubMed] [Google Scholar]
- Poe JA, Smithgall TE. HIV-1 Nef dimerization is required for Nef-mediated receptor downregulation and viral replication. Journal of Molecular Biology. 2009;394:329–342. doi: 10.1016/j.jmb.2009.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raney A, Shaw AY, Foster JL, Garcia JV. Structural constraints on human immunodeficiency virus type 1 Nef function. Virology. 2007;368:7–16. doi: 10.1016/j.virol.2007.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renkema GH, Saksela K. Interactions of HIV-1 Nef with cellular signal transducing proteins. Frontiers in Bioscience. 2000;5:d268–283. doi: 10.2741/renkema. [DOI] [PubMed] [Google Scholar]
- Resh MD. Trafficking and signaling by fatty-acylated and prenylated proteins. Nature Chemical Biology. 2006;2:584–590. doi: 10.1038/nchembio834. [DOI] [PubMed] [Google Scholar]
- Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- Shenoy SS, Nanda H, Losche M. Membrane association of the PTEN tumor suppressor: Electrostatic interaction with phosphatidylserine-containing bilayers and regulatory role of the C-terminal tail. journal of Structural Biology. 2012;180:394–408. doi: 10.1016/j.jsb.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauer J, Treisman JE. Lipid-modified morphogens: functions of fats. Curr Opin Genet Dev. 2009;19:308–314. doi: 10.1016/j.gde.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian K, Dietrich LEP, Hou H, LaGrassa TJ, Meiringer CTA, Ungermann C. Palmitoylation determines the function of Vac8 at the yeast vacuole. Journal of Cell Science. 2006;119:2477–2485. doi: 10.1242/jcs.02972. [DOI] [PubMed] [Google Scholar]
- Welker R, Harris M, Cardel B, Krausslich HG. Virion incorporation of human immunodeficiency virus type 1 Nef is mediated by a bipartite membrane-targeting signal: analysis of its role in enhancement of viral infectivity. J Virol. 1998;72:8833–8840. doi: 10.1128/jvi.72.11.8833-8840.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L, Gollapalli DR, Maiti P, Jahng WJ, Rando RR. A palmitoylation switch mechanism in the regulation of the visual cycle. Cell. 2004;117:761–771. doi: 10.1016/j.cell.2004.05.016. [DOI] [PubMed] [Google Scholar]
- Zha J, Weiler S, Oh KJ, Wei MC, Korsmeyer SJ. Posttranslational N-myristoylation of BID as a molecular switch for targeting mitochondria and apoptosis. Science. 2000;290:1761–1765. doi: 10.1126/science.290.5497.1761. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.