FORWARD
The first issue of Biopolymers appeared in February, 1963. John F. Kennedy was president and would be killed in November of that year. The Beatles released their debut album "Please Please me". The Nobel Prize in Chemistry went to Karl Ziegler and Giullo Natta "for their discoveries in the field of the chemistry and technology of high polymers".
The goal of Biopolymers was to present the best macromolecular research in the physics, chemistry, and biology, of the life processes. Determining the molecular weights of proteins and nucleic acids was a hot field. The main methods were light scattering, a combination of sedimentation velocity and diffusion, and equilibrium sedimentation. Preparing pure samples for measurement was one of the most difficult problems; separation of one protein from the complex mixture in a cell was not easy. Egg white proteins or hemoglobin were popular choices for study. DNA was also popular; the main sources were calf thymus and salmon sperm. In fact the molecular weight of DNA seemed to be independent of the species it came from, whether it was E. coli, yeast, or salmon. It turned out that during preparation the DNA was being sheared by pipetting, pouring, etc. from its actual molecular weight of millions to less than 100,000. X-ray diffraction could give detailed structural information for crystals of small proteins such as myoglobin, but in solution a 60 MHz NMR spectrometer, the latest available, was not very useful for macromolecules.
Obviously progress has been made in the last 50 years in the tools we now use in research. It is up to the present readers and writers of Biopolymers to invent the methods, and make the discoveries, that will be used in the next 50 years. The goal of this review is to point out some of the problems waiting to be solved in understanding, predicting, and controlling how ribosomes synthesize proteins. During normal translation of a messenger RNA (mRNA) the ribosome initiates at a start codon, then reads codons three nucleotides at a time until it reaches a stop codon. We will concentrate on frameshifting: the slippage of the ribosome as it reads the mRNA. If it slips by a number of nucleotides not divisible by three, the ribosome gets out of frame. It will now synthesize a new sequence of amino acids until it reaches a stop codon present in the new frame. Thus, frameshifting leads to a very different sequence and size from the non-frameshifted protein. Random frameshifting is deleterious to the cell, but programmed frameshifts are used by the cell to make required proteins, and by viruses to efficiently make multiple proteins from one messenger RNA. Understanding frameshifting, and the regulation of translation in general, can lead to the ability to manipulate and control protein synthesis for the betterment of health.
INTRODUCTION
Frameshifting in Translation
The ribosome is the molecular machine that synthesizes proteins coded by mRNAs, which in turn are coded by DNA. This protein translation process occurs in three stages: initiation, elongation, and termination. Initiation occurs when an initiator tRNA is placed at a start codon (in bacteria typically an AUG, sometimes a GUG and less often a UUG) in the peptidyl, P, site, of the ribosome. Termination occurs at one of three stop codons: UAA, UAG, UGA. Each of the 61 remaining three-letter codons specifies one of the twenty naturally occurring amino acids in proteins (AUG also codes for methionine). During elongation Watson-Crick base pairing between the codons and the anticodons on the aminoacyl transfer RNAs translates the mRNA sequence to a polypeptide sequence. Therefore, the mRNA is read three letters at a time from start to stop signal. Obviously, errors are sometimes made; a codon can be misread to substitute one amino acid for another in the protein. Also, synthesis sometimes starts at a non-initiation AUG or continues past a stop codon. Finally, the ribosome may skip a few nucleotides in the mRNA, or back up and re-read a few. Clearly, if the number of nucleotides missed or repeated is not a multiple of three, the amino acid sequence after the shift will be completely different from the sequence coded before the shift. This is called a frameshift. There are only three reading frames possible; they are designated as in-frame (0-frame) or out-of-frame, (either −1-frame or +1-frame). The original stop will no longer be read, but usually a stop codon will be encountered earlier (because 3 of 64 codons are stops).
Translation is the most error prone of the three processes (replication, transcription, and translation) involved in preserving and propagating a genome,1 but it still occurs with high fidelity. Translation errors occur with a frequency of less than 0.1%, except when a programmed frameshift is encoded in the mRNA. A programmed frameshifting signal causes frameshifts with a frequency greater than 1% that are tightly controlled in magnitude to produce a desired ratio of in- and out-of-frame proteins. Critical retroviral proteins, such as in HIV, are made as polyproteins by successive frameshifts.
Programmed frameshifting has been studied for years as described in many reviews.2–6 The process occurs in all kingdoms, and has been studied in vivo and in vitro. The efficiency of programmed frameshifting is measured by quantitating the amounts of translated products from different reading frames.
The mRNA sequences and structures that signal and control programmed frameshifting during the translation elongation phase are well-characterized, but the mechanisms by which they determine the frameshifting are poorly understood. Here we will focus on bacterial systems, first describing the general elongation process, which is highly similar across prokaryotes and eukaryotes.7,8 Known effects on translation that are signaled by frameshift-promoting mRNA sequences will be discussed. This will lead to descriptions of specific ribosome translation dynamics relevant to mRNA reading-frame slippage. Finally, we will concentrate on the contributions of single-molecule methods to determining the causes of frameshifting.
The Elongation Cycle during Translation
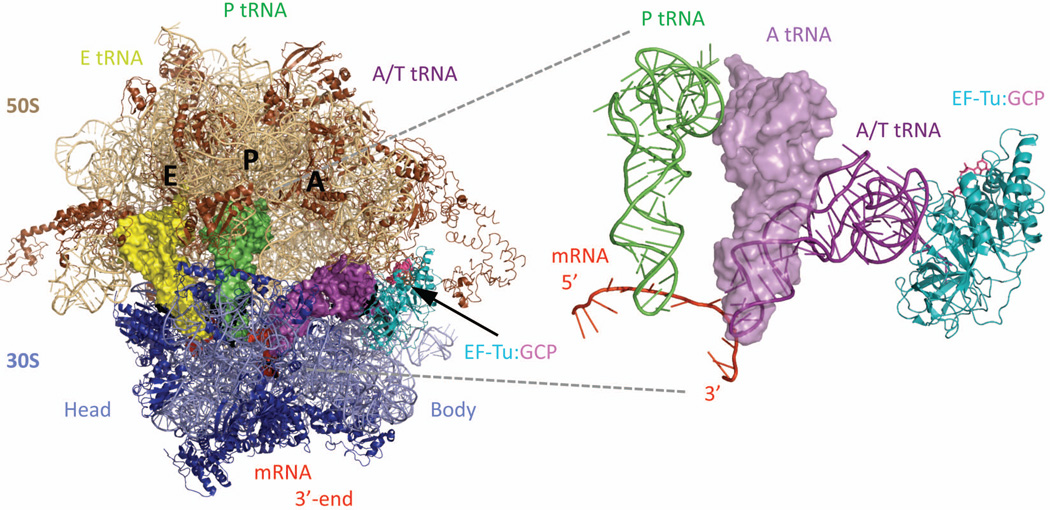
A 2.5 MDa bacterial ribosome is made of two subunits, 50S and 30S (Figure 1). The 50S subunit contains two ribosomal RNAs (rRNAs), 23S and 5S, and ~36 ribosomal proteins; the 30S subunit is composed of 16S rRNA and ~21 proteins. On the 50S ribosomal subunit, there are three adjacent binding pockets for the aminoacyl acceptor ends of transfer RNAs (tRNAs). A trench-like region on the 30S subunit, namely the 30S neck, forms the codon•anticodon base-pairing sites, where the mRNA sequence to be read binds to the anticodons of the tRNAs (Figure 1B). Thus there are three sites on both subunits that bind the tRNAs: the A- (aminoacyl), P- (peptidyl), and E- (exit) sites.9 These sites are at the inter-subunit interface, formed when the two subunits bind to the mRNA and fMet-tRNA during initiation of translation.
Figure 1.
Overall architecture of the 70S bacterial ribosome, in its classical, unrotated state (PDB: 3I8G,36 3I8F,36 2WRJ37) (A) The 70S bacterial ribosome is made of two subunits, 30S (blue, also in B) and 50S (brown, also in C). The two subunits stack together with their A- (aminoacyl), P- (peptidyl), and E- (exit) sites facing inward toward each other, thus defining an intersubunit interface where cycles of translation occur. Ribosomal proteins are shown as ribbons; ribosomal RNAs are lighter color, and the mRNA is space filling in red. The tRNAs are violet for A-, green for P-, and yellow for E-tRNA; the same color-coding for these classical state tRNAs is used in all figures. Ribosomal proteins L1 and L9 are identified on the left side of the 50S subunit (L designates proteins from the 50S subunit, S from the 30S). The decoded mRNA lies between the 30S head and body domains, wrapped in an inverted U-shape around the 30S neck (shown in panel B). (B) 30S, view from the mRNA entrance (3'-end shown) with the 50S removed. A trench-like region on the 30S is its neck, which serves as a saddle to display the decoded mRNA (red) codons. The appropriate interactions between the anticodon stem-loop of the tRNAs and the decoded codons on the mRNA are probed by highly conserved resides (detailed in Figure 4), which are adjacent to the neck and come both sides from the 30S head and body. The L-shape tRNAs have their aminoacyl-acceptor ends oriented toward the upper-right corner, revealing how their corresponding binding-sites on the 50S are configured parallel in space. (C) 50S, view of its tRNA-binding/intersubunit interface. The codon-anticodon duplex (red) is out from the paper plane toward us, whereas the aminoacyl-acceptor ends of the tRNAs are bound tightly within each of their designated cavities, clustering around the center of the 50S. Note that the A- and PtRNA acceptorends are positioned relatively close to each other and to the nearby peptidyl-transferase center (PTC). This is where elongations of the polypeptide chain take place.
A translating ribosome adopts several conformational states during the elongation cycle.10 The ribosome is defined to be in its classical state when the two ribosomal subunits, 50S and 30S, have their three tRNA binding sites aligned. Namely, a P-site tRNA (P-tRNA) bound with a classical state ribosome has its peptidylated acceptor end situated inside the 50S P-site binding pocket, adjacent to the ribosomal peptidyl-transferase center (PTC) (Figure 1C). Meanwhile, the anticodon stem loop of this P-tRNA is interacting with the mRNA codon displayed at the 30S P-site.
The translation elongation cycle begins with the delivery of the correct aminoacyl-tRNA (aa-tRNA) into the empty A-site on the classical state ribosome. Cellular aa-tRNAs are carried as ternary complexes, TC, composed of elongation factor EF-Tu, GTP, and tRNA (TC: EF-Tu•GTP•aa-tRNA). The binding on-rates for different ternary complexes into the empty A-site are quite similar, but the selection for the correct aa-tRNA lies in the dissociation off-rates.11–13 Proper base pairing of the cognate tRNA anticodon with the mRNA codon induces conformational changes in the ribosome that result in a slower off rate. The anticodon stem loop interacts with the 30S subunit while the rest of the tRNA remains bound to EF-Tu•GTP. This A/T state triggers hydrolysis of the GTP and dissociation of EF-Tu from the aa-tRNA, thus enabling an A-site tRNA (A-tRNA) to adopt the classical A/A state (Figure 2) (letter before the slash denotes the binding site position on 30S, and after the slash specifies the 50S site; T for EF-Tu). Occasionally, errors occur at the base-pairing selection step, but the ribosome can proofread and reject the mis-selected aa-tRNA. The classical state ribosome is now bound with a peptidyl-tRNA in the P/P state and an aminoacyl-tRNA in the A/A state.
Figure 2.
A bent A/T state adopted by an aa-tRNA during its accommodation (PDB: 2XQD, 2XQE)104 The first step in each translation elongation cycle is the binding of the correct aminoacyltRNA (aa-tRNA, space-filling in purple), specified by the mRNA (space-filling in red) codon displayed in the 30S A-decoding site. The delivery of aa-tRNAs comes in the form of ternary complexes (TC: EF-Tu•GTP•aatRNA, in cyan, pink, and purple respectively), with the elongation factor protein EF-Tu (ribbons in cyan) bound at the amino-acid end of the aa-tRNA, thus preventing deacylation. The use of a non-hydrolysable GTP analog, GDPCP (spheres/sticks in bright pink) allowed the A/T state of an A-tRNA during accommodation to be determined by X-ray diffraction. A zoom-in view on the right superimposes the A/T state aa-tRNA (thin helical cartoon in purple) to its final fully-bound A/A state conformation (space-filling in violet). While the tRNA anticodon stem loop has mostly configured within the 30S A-decoding site, upon GTP hydrolysis and EF-Tu release, its amino acid acceptor end will dock into the 50S A-site pocket. Although the aa-tRNA undergoes significant rearrangement from its A/T to A/A state, the 70S ribosome remains in the classical and unrotated state, evidenced by the P/P (space-filling in green) and E/E (space-filling in yellow) t-RNAs bound within the same ribosomal complex.
The ribosome further catalyzes the transfer of the oligopeptide chain to the A-site tRNA. This is done through displacement of the peptidyl group on the P-tRNA via nucleophilic attack by the alpha-amino group of the A-tRNA. The net result is a lengthened oligopeptide chain by one amino acid at its carboxyl terminus. Now the ribosome adopts a hybrid state, defined by the staggered A/P, P/E configurations of the two tRNAs. In addition to the partial shifting and tilting of these tRNAs, the 30S subunit rotates by ~8–9° counterclockwise with respect to the 50S.14
The transition to the A/P, P/E-tRNA hybrid state is reversible; upon peptidyl-transfer, the acceptor ends of A- and P-tRNAs can rapidly fluctuate back and forth between the 50S A-, P-sites (classical) and the P-, E-sites (hybrid).15,16 The hybrid state ribosome is preferentially stabilized by the binding of another GTP-consuming elongation factor, as EF-G•GTP.17,18 The binding of EF-G•GTP to the ribosome and its catalysis of GTP hydrolysis commence the translocation process. With the cost of one GTP, the ribosome is now proposed to be in an unlocked state.19,20 The state is defined by its ability to efficiently allow the rate-limiting passage of the codon•anticodon duplexes from the 30S A-, P-sites to the 30S P-, E-sites. Once both tRNAs, and their associated mRNA codons have fully translocated forward, the ribosome again returns to the classical state. This time it is occupied with a peptidyl-tRNA in P/P and a deacylated tRNA in E/E, also displaying a new mRNA codon at its 30S A-site decoding spot. Now the ribosome is said to be in a locked state, prohibiting reversible backward translocation movements.19,20 Such “relocking” of the ribosome further states that it has reinitialized for the next round of translation elongation. However, whether a correlation exists between the binding of the next incoming aa-tRNA and the release of deacylated E-tRNA remains unsettled.21–24
Programmed Frameshifting Signals during Elongation
In bacteria there are three mRNA sequence components that together promote programmed frameshifting, as shown in Figure 3 for the frameshifting region of the E. coli dnaX gene that codes for two subunits of DNA polymerase.25 There is an internal Shine-Dalgarno sequence, a slippery sequence, and a stable stem-loop (hairpin).6 These special sequences are arranged from 5' to 3' in the mRNA and the spacing between them is important.
Figure 3.
Three sequence signals that are found in programmed frameshifting mRNAs from bacteria. The internal Shine-Dalgarno and the hairpin position the slippery sequence at the 30S codon•anticodon binding sites where the mRNA•tRNA base-pairing occurs. The example sequences and the spacing shown are from the dnaX gene in E. coli.25
The Shine-Dalgarno (SD) sequence on the mRNA is complementary to the 3'-end of the 16S rRNA of the 30S subunit, forming a ~6–8-bp long mini-helix. During initiation of protein synthesis the cellular function of the SD sequence is to guide the start codon to the P-site on the 30S where the initiator formylmethionine tRNA will be bound. Similarly, as a frameshifting promoter, the internal SD positions the slippery sequence at the codon•anticodon base-pairing sites on the 30S. The slippery sequence obviously provides alternative base-pairing that allows the tRNA anticodons to slip with respect to the mRNA while minimizing Watson-Crick mismatches. Finally, the beginning of the hairpin (the single-strand-to-double-strand junction) is at the entrance tunnel to the ribosome when the slippery sequence is at the 30S P- and A-sites. Translation forward by the ribosome will thus require it to unwind the hairpin as well as relieving the base-pair interactions with the internal SD sequence.26 Reducing the complementarity of the Shine-Dalgarno sequence, making the slippery sequence less slippery, destabilizing the hairpin, or changing the spacing from the optimum decreases the programmed frameshifting efficiency. Many viruses that infect eukaryotes use programmed frameshifting to make a fusion polyprotein of essential viral proteins; the ratio of in-frame to out-of-frame products is critical for viral viability. Example slippery sequences include UUUUUUA in HIV,27 AAAUUUA in RSV,28 and UUUAAAC in IBV.29 In eukaryotes there is no internal Shine-Dalgarno involved for promoting frameshifts; a pseudoknot, instead of a hairpin, is often the structure present downstream of the slippery sequence that impedes the mRNA entrance to the ribosome.29
In contrast to normal translation, where the codon•anticodon duplex moves in the 5'-direction by precisely three nucleotides, a frameshift requires the slip of the mRNA relative to the A-, P-, E- sites on the 30S. Furthermore the tRNAs can base-pair in a different frame on the mRNA. The number of nucleotides on the mRNA that the tRNAs can skip or repeat during translation depends on the possible alternative base-pairing combinations offered by the slippery sequence, and also the presence and precise location of the SD sequence and secondary structures on the upstream and downstream sides.
DECIPHERING THE PROGRAMMED FRAMESHIFTING SIGNALS
Many plausible frameshifting mechanisms have been suggested and new ones are continually proposed.30–34 However, the complexity of a ribosome has hampered attempts to unravel its mechanism of translation, and of frameshifting.
To understand programmed frameshifting dynamics, specifically we want to ask:
When, in time within one translation cycle, does a frameshift occur? Is it before, during, or after the peptidyl-transfer, or along the EF-G-promoted translocation step?
Where, in codon position(s) around the slippery sequence, does the ribosome actually slip? Namely, where do the last old-frame codons end and the first new-frame codons begin?
In the following, we will review structural studies of ribosomal complexes with emphasis on programmed frameshifting environments as determined by the mRNA sequence signals. Furthermore, experiments for examining various frameshifting proposals will be discussed, in particular the informative single-molecule experiments that permit direct observation of the frameshifting dependence on mRNA templates.
INSIGHTS FROM STRUCTURAL STUDIES
Detailed structural information of the ribosome and its substrates during different states of translation can help us determine when and where the ribosome shifts reading frame. Several critical motions of a translocating ribosome have been revealed recently in x-ray crystals,10,14,35–37 in cryo-EM refined structures,38,39 and in kinetics measurements on fluorophore-labeled ribosomes.40,41 Intriguingly, one essential dynamical rearrangement frequently centers around the 30S head domain.
A Kinked mRNA Sits on the 30S A-, P-, E- Sites
As seen in Figure 4A the three mRNA codons, instead of stacking consecutively next to each other, are kinked as three segments to wrap around the 30S codon•anticodon base-pairing sites (also see Fig. 1B). Highly conserved residues from 16S rRNA of 30S are proposed to act as codon•anticodon base-pairing sensors and gating partitions between each codon segment. To highlight a few: C1400, G1401, C1402, and C1403 between A-P codons; G926, A790, and G1339 between P-E codons; C1054 and G530 at the 3'-end of the A-codon; G693 at the 5'-end of the E-codon (details in Figure 4B and 4C).8,42–45 This suggests how the mRNA reading frame could be spatially defined by the ribosome. Within such tightly structured codon•anticodon binding sites on a classical state ribosome, namely during the aa-tRNA binding and the peptidyl-transfer steps, a shift in reading frame may not be likely as this requires a tRNA anticodon to base-pair across two adjacent kinked codons.
Figure 4.
The tightly structured codon•anticodon binding sites in a classical state 70S ribosome (PDB: 3I8G, 3I8F)36 (A) As the mRNA template (cartoon in gray) wraps around the 30S neck (not shown) into an inverted Ushape, the three decoded codons are distinctively kinked into segments specifically oriented toward their corresponding base-pairing anticodons (nucleotide residue 34–36 highlighted in red) from the tRNAs. A Shine-Dalgarno sequence located ~9-nt upstream from the P-site codon is also highlighted in red. (B) The kinked conformation of the mRNA and the spacing between the three base-pairing tRNAs reveals how the translation reading frame is kept in registry. Highly conserved nucleotide residues from the 16S rRNA (shown in sticks and color-coded for their corresponding interacting codon•anticodon binding sites) help to stabilize the interactions around the codon•anticodon duplex, as shown in C. (C) Close-up views at each partitioning interface along the three codon•anticodon binding sites. From left to right: 5'-end of E-site codon, between P-E sites (mRNA 5'-end facing us), between A-P sites (mRNA 3'-end facing us), and 3'-end of A-site codon. It is proposed that the 45°-kink between A- and P-site codons is stabilized by the nearby A1400 and A1401 (middle right figure). Also, shown in middle left, G1338 to U1341 and A790 collectively form a block between the P- and E-sites, with a gap of ~14Å at the narrowest. This effectively prevents movement or slippage of the P-tRNA, particularly by its anticodon stemloop portion. Another closing “latch” securing the A-tRNA is between G530 and C1054. Many of these residues have to move apart in order to permit mRNA•tRNA translocation. This is a static view of the
The Swiveling 30S Subunit Head during mRNA•tRNA Translocation
The dynamics of the 30S head domain is an important agent in translocation (Figure 5A).10,14,35–41 Its swiveling motion, including a counterclockwise rotation around the 30S neck and a backward tilt widening the 30S mRNA binding groove, allows mRNA•tRNA translocation across the 30S binding sites. Such dynamic 30S head not only accomplishes translocation, it may also permit a time window for frameshifts to occur during translocation. This is because the aforementioned highly conserved 16S nucleotide residues, which are used to keep the codon•anticodon in the in-frame registry, have moved away from the base pairs. The critical movements by the 30S head lead to displacements of the gating nucleotide residues out of the way, e.g. A790 and G1339 for P-tRNA (Figure 5C), allowing the anticodon stem loops of the A- and P-tRNAs to pass over into the 30S P- and E-sites. However, 30S head motion is the fundamental mode used by the ribosome during normal translation, thus this motion cannot be the specific cause for tRNA slippage, but rather it offers a kinetically feasible pathway to achieve frameshifting. For instance, 30S head swiveling may allow tRNA anticodons to transiently sample alternative base-pairing positions along the nearby mRNA sequence. Then what are the factors that can preferably bias a frameshifting slip?
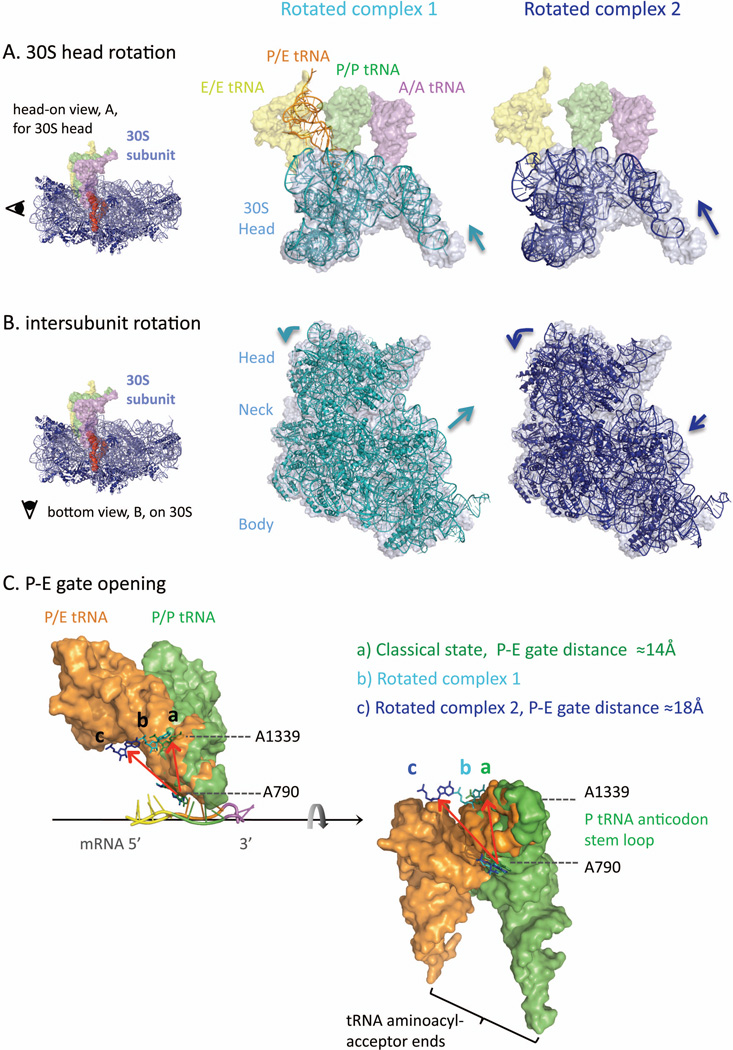
Figure 5.
Ribosome rotation dynamics (PDB: 3I8G,36 classical; 4GD1,10 rotated complex 1; 3SFS,35 rotated complex 2) Depending on the protein factors and tRNAs engaged, the ribosome has been captured in different rotation states. For instance, rotated complex 1 is crystallized with a P/E-hybrid tRNA (thin helical in orange) and recycling factor protein, RRF (not shown), while complex 2 is bound with a release factor protein 3, RF3 (not shown). (A) Overlays of 30S head domain (shown as 16S rRNA residue 930-1390): rotated state (thin helical in cyan for complex 1; dark blue for complex 2) versus classical, unrotated state (space-filling in light gray; classical state tRNAs shown in spacing-filling for comparison). The 30S head can rotate forward (counterclockwise) as large as ~15° (complex 2, right), while a smaller 4°-rotation in the same direction (complex 1, left) is enough to accompany the P-tRNA transition into its P/E-hybrid state (thin helical in orange). The fact that the P-tRNA acceptor end can swing by ~37° and dock into the 50S E-site indicates that the P-E gating residues, e.g. A790 and A1339, have moved apart upon 30S head forward rotation (detailed in panel C).
The most essential elements for frameshifting are the slippery sequences, which are designed to provide a new thermodynamically stable reading frame upon slipping by the tRNA anticodons. As demonstrated in many mRNA mutation experiments, a characteristic XXXYYYZ slippery sequence is necessary to attain efficient −1 frameshifting in producing detectable amounts of out-of-frame protein product.27,30–34,46–51 However, this sequence is not sufficient; other promoting factors are required. Indeed, the two frameshift-inducing structural barriers, i.e. an internal SD before the slippery sequence and a stable hairpin after the sequence, enhance frameshifting by interfering with the ribosome translocation step. The primary mechanism for this enhancement may be the change in dynamics of the 30S head rotation.
The Shine-Dalgarno•Anti Shine-Dalgarno Mini Helix
The frameshift-enhancing internal SD sequence52–55 forms a ~6-bp double-stranded RNA helix with a complementary anti-SD sequence on the 3'-end of 16S rRNA (Figure 6A). The resultant mini-helix remains stable as the ribosome translates from as close as two codons (counting from the first nucleotide of the P-site codon) after the SD sequence (counting from the final G of the internal SD sequence shown in Figure 3: AGGGAG) to as far as four codons after.53,54 This creates a somewhat springy mRNA downstream from the mini-helix, particularly for the E-site codon.53,54 When the spacing between the mini-helix and the P-site codon is small (6 nucleotides), the nucleotide backbone of the E-site codon is relatively stretched 5’-ward, disrupting proper orientations of the three E-site codon bases to hydrogen bond with an E-tRNA anticodon (Figure 6B).53,54 With larger spacing (~9–11-nt), the E-site codon is now relaxed and able to stack and form a stable codon•anticodon interaction. Hence, the presence of an SD•antiSD helix could tune the availability of the E-site codon to participate in base-pairing during translation. In fact, it has been reported that the E-site base-pairing condition affects the mRNA reading frame maintenance.47,48 This suggests how the spacing between the mini-helix and the P-site codon can affect the position of a programmed frameshift. Moreover, the dynamic motions during ribosomal translocation may take part in disrupting the SD•antiSD helix, thereby actively promoting frameshifting.
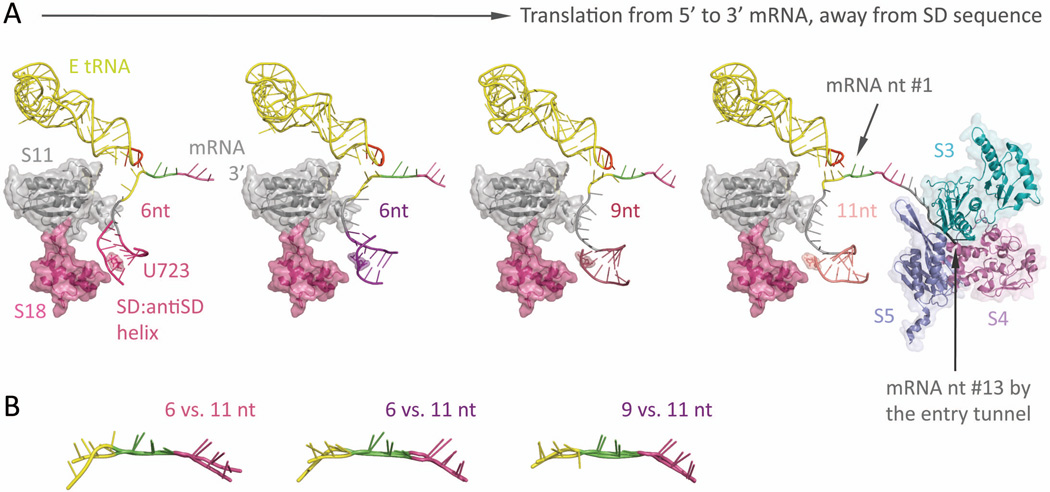
Figure 6.
The springy Shine-Dalgarno•anti Shine-Dalgarno mini-helix (PDB: 2HGR,54 2QNH,52 6-nt; 3I8G,36 9-nt; 2HGP,54 11-nt) (A) Left-most: Around the mRNA 5’-end exit pore, a ~6-bp long SD•antiSD mini-helix (thin helical cartoon in magenta) resides within a pocket, surrounded by ribosomal proteins S11 and S18 (semitransparent space filling, with ribbon; in gray and magenta, respectively) from the 30S. Left two figures: The mini-helix (in magenta and purple) readily forms while the ribosome translates only 6-nt downstream from it (P-site codon colored in green, A-site codon in violet). Under such short spacing, the backbone of E-site codon (highlighted in yellow) is stretched and distinctively distorted from proper basepairing toward the E-tRNA anticodon (residue 34–36 highlighted in red, the rest in yellow); meanwhile, the center of the mini-helix roughly sits on top of the highly conserved residue, U723 (semitransparent spacefilling, with sticks; same color as the mini-helix), though the detailed orientation of the helix may vary slightly. Right two figures: As the ribosome translates forward (seen as the mini-helix sliding away from the residue U723), the nucleotide spacing increases (9-nt, middle right; 11-nt, right-most), thus relaxing the E-site codon (yellow) to resume hydrogen bonding with the anticodon (red). This illustrates how the formation of a SD•antiSD mini-helix, as well as its spacing between the codon•anticodon binding sites, can tune the availability of E-site condon•anticodon basepairing, which directly interferes with the mRNA registry and reading frame maintenance during translation. Right-most: The mRNA entry tunnel is composed of ribosomal proteins S3, S4, and S5 (semi-transparent space-filling, with ribbon; in cyan, dark pink, and slate, respectively). Counting from the first base in the Psite codon, nucleotide #13–15 would be located at the mRNA entry pore. This suggests a minimal spacing of 7 to 9-nt between a slippery sequence placed at the P- and A-site and a downstream hairpin at the entry pore. (B) Overlays of the decoded mRNA: various stretched forms of the E-site codon (at spacing of 6-nt and 9- nt) versus the relaxed form (11-nt).
SINGLE-MOLECULE OPTICAL TWEEZERS
An obvious way to capture the frameshift timing is to watch a single ribosome translating in real-time. Such pioneering work was done by Wen, et al. with the use of optical tweezers.56 Measurements were done by monitoring the gradual opening of a hairpin mRNA that is tethered between two optically trapped beads (Figure 7A); as the ribosome translates and unwinds the hairpin, double-stranded RNA is released into single strand. This shows up as a step-wise end-to-end distance increase as a function of time, in the mRNA at a distinct size of 6-nt per codon translated (Figure 7B). Simply by counting the number of steps, the ribosome was followed in real-time through multiple translocation cycles. Measuring translation of an mRNA hairpin is ideal for studying frameshifting. Not only does the hairpin situated at the mRNA entrance to the ribosome favor frameshifting, the dwell time and translocation dynamics at each codon can be directly accessed. The motion of the ribosome relative to the mRNA, in particular special dynamics during frameshifting, can be seen directly.
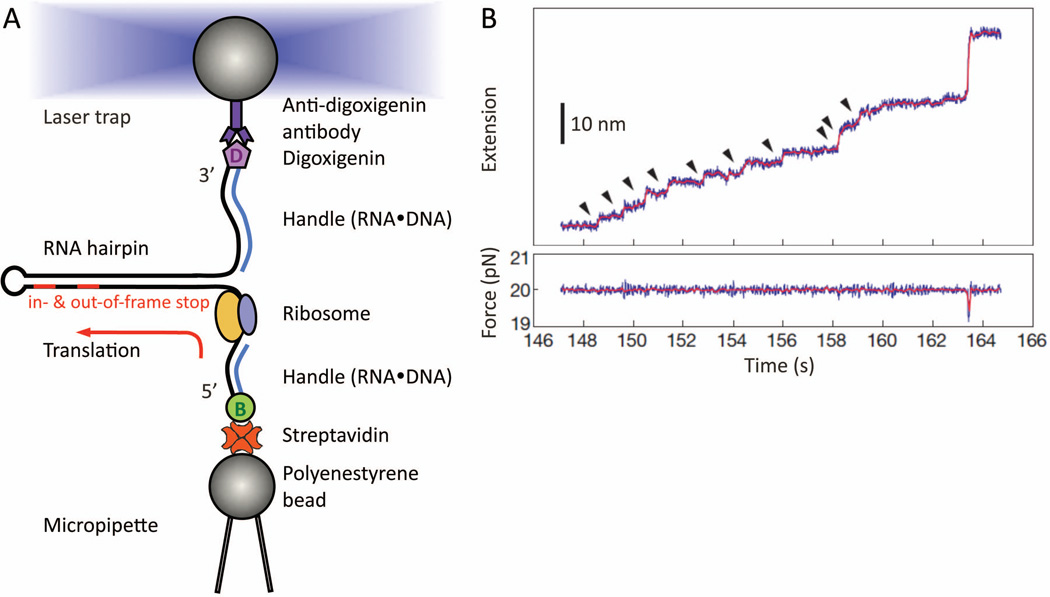
Figure 7.
Real-time step-by-step translation in an optical-tweezers experiment (A) Schematic drawing of the hairpin helicase assay used to follow single-ribosome translation. A selfcomplementary mRNA template (black line), which forms a hairpin, is held by its two ends. The tethering is done by attaching the RNA ends hybridized with either biotin- or digoxiginin- labeled DNA oligos, to either streptavidin- or anti-digoxigin antibody- coated polystyrene beads. While the bottom bead is fixed by suction onto the micropipette tip, the other is controlled by the focused laser trap. This allows us to adjust the force applied to the mRNA, thus tuning the mechanical stability of the hairpin that the ribosome will translate and unwind in 3-bp per codon-step. Note that no modifications on the ribosome, or other translation-essential components, are made. At the end of translation, the residual mRNA hairpin size provides a crosscheck confirming which codon the ribosome has reached, e.g. either the in- or out-of-frame stop codon on a frameshift-promoting mRNA. (B) Example of a step-pause-step translation trajectory from a single ribosome recorded as a function of time. This is captured as a series of fixed-size step-wise mRNA lengthening, i.e. extension, over time (top panel, ~2.7 nm per codon translated at ~20 pN). Here, a total of ten consecutive translation elongation cycles (arrow-highlighted) has been successfully followed in real-time. Each increment (vertical step) signifies the EF-G•GTP catalyzed ribosome translocation. If a frameshift occurs in parallel with the translocation step, a different extension step size will be observed. (Panel B is reproduced with permission from Figure 2A in reference 56.)
If one bead is attached to the ribosome and the other bead is attached to the 3'- or 5'-end of the mRNA, the effect of an opposing force or an assisting force on translation can be measured. With an opposing force, the stall force of the ribosome can be determined. The stall force multiplied by the step size gives the maximum work the ribosome can do.57 Applying force directly to the ribosomal subunits allows measurement of the mechanical work done during translocation by the subunits or their domains.
Resolving the Time Point in Frameshifting
In a step-pause-step translation trajectory (Figure 7B), the pause represents the codon-specific dwell time the ribosome takes to incorporate the next aa-tRNA and to catalyze a peptide bond linking two amino acids. At the end of each pause the vertical step (~20 ms) shows when EF-G catalyzes the ribosome to translocate by one codon, unzipping three base pairs and releasing six nucleotides from the mRNA hairpin. Real-time translation trajectories can reveal when a ribosome slips. If the frameshift occurs during a pause, the ribosome shifts along the mRNA during the dwell time; an increase or decrease in end-to-end distance of the mRNA occurs, but it may be difficult to detect. Such a slip may either be triggered upon aa-tRNA binding, or by the succeeding peptidyl-transfer reaction. That is, the new aa-tRNA may be bound out-of-frame, or be bound in frame, but then causes the mRNA to slip relative to the ribosome. The following translocation is normal, 3 nucleotides per step. Contrastingly, if the frameshift occurs instead during the translocation phase, we see an altered translation step size within the trajectory. This directly indicates a frameshifting event occurred during the translocation step. The amino acid incorporated at the pause right before this step is the last one read in-frame. At the next pause the first amino acid added is specified by the newly established reading frame. This is a scenario where both the timing and the codon position of a programmed frameshift can be explicitly distinguished.
MASS SPECTROMETRY ANALYSIS AND DETECTION
Mass spectrometry (MS) can provide the sequences of the translated peptides and thus reveal where a frameshift occurred along the mRNA. While the real-time tweezers experiments can track the onset of a frameshift event, the final protein products can be thoroughly characterized by MS. For instance, a mixture of protein products can be resolved through liquid chromatography/mass spectrometry (LC/MS) intact peptide detection, since each species has a unique atomic composition and molecular weight (Figure 8A).58,59 When desired, a species of interest can be selected by its mass, fragmented, and subjected to a second MS for tandem MS amino acid sequencing (Figure 8B).59–62
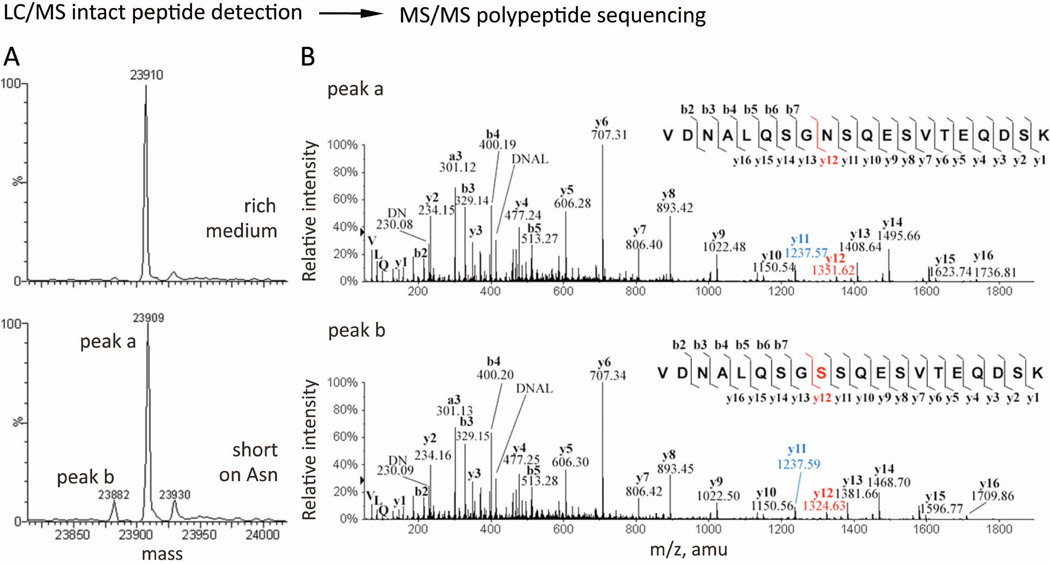
Figure 8.
Mass spectrometry analysis to identify mis-incorporation during recombinant protein overexpression in mammalian cells In addition to programmed frameshifting mRNA signals, it is known that hungry or rare codons can also induce frameshifting and miscoding with appreciable efficiencies.60,62,105 A nutritional stress was experienced when Chinese hamster ovary cells (CHO) were transfected to over-express a humanized monoclonal antibody for clinical use. This results in unexpected yet specific mis-incorporations of serine (Ser, S) for asparagine (Asn, N) encoded within the antibody, at a rate of 1–2 % per Asn residue. (A) Deconvoluted mass spectra of the synthesized antibody light chain, examined through LC/MS intact peptide detection. In low-expressing cell lines, only the predicted mass for the antibody light chain (top panel, ~23909 Da) was observed. Upon over-expression, a lighter, “-27 Da,” species (bottom panel; peak b, 23882 Da) had emerged unexpectedly and was persistently seen, suggesting one of four possible amino acid substitutions was made (Arg→Glu, Gln/Lys→Thr, or Asn→Ser) during protein overexpression. (The 23930- Da species was not assigned in this study.) (B) Tandem MS polypeptide sequencing to resolve the identity and position of the unexpected amino acid substitution. The two species detected (bottom panel A, peak a and b) were selected (based on their molecular mass and LC elution time), fragmented, and further subject to a second run of MS. As the fragment ions are generated by breaking amide bonds along the polypeptide backbone (corner insets), thus producing C-terminal bearing y-ions and N-terminal bearing b-ions of various residue length, the identity of an amino acid at each position can be determined by the difference in m/z values of two adjacent y- or bions. The peptide sequence for the expected antibody light chain is assigned (top panel inset) and confirmed to bear two Asn residues as predicted (@m/z = 114.04 for N at both b3-b2 and y12-y11). For the unexpected lighter species (bottom panel), its “-27 Da” lower mass was revealed as an Asn → Ser substitution at one of the two Asn positions (@m/z = 87.03 for S at b3-b2). Ser substitution at the other Asn position (@m/z = 87.03 for S at y12-y11) also exists and was resolved (eluted differently in LC) and assigned. (Panels are obtained with permission from Figure 1A, C and Figure 4 in reference 59.)
Resolving the Codon Position in Frameshifting
In a study of the −1-frameshifted protein product translated from the HIV-1 programmed frameshifting gene with a slippery sequence:  , MS analysis showed that the ribosome translated the 7-nt slippery sequence in more than one way. Two full length frameshifted products were produced; ~70% contained "Phe-Leu" at the slippery sequence, while ~30% contained a "Phe-Phe" pair. Both cases are followed by amino acids decoded in the −1-frame, e.g. Arg, specified by codon AGG, is the next amino acid. The amino acid sequence Phe(UUU)-Leu(UUA)-Arg(AGG), reveals that the mRNA was translated as U2U3U4-U5U6A7 in zero-frame, then −1-switched to A7GG, reading the same adenine (A7) twice. The alternative Phe(UUU)-Phe(UUU)-Arg(AGG) sequence could come either from U2U3U4 in zero-frame, then −1-switched to U4U5U6-A7GG, reading U4 twice, or from reading U1 twice to translate U1U2U3-U4U5U6-A7GG all in the −1-frame. This implies as many as three slipping paths differing in codon positions (Phe0-Leu0-Arg−1, Phe0-Phe−1-Arg−1, and Phe−1-Phe−1-Arg−1), can be utilized in the HIV-1 programmed frameshifting. This finding supports the idea of a highly adaptive ribosome in switching reading frames.
, MS analysis showed that the ribosome translated the 7-nt slippery sequence in more than one way. Two full length frameshifted products were produced; ~70% contained "Phe-Leu" at the slippery sequence, while ~30% contained a "Phe-Phe" pair. Both cases are followed by amino acids decoded in the −1-frame, e.g. Arg, specified by codon AGG, is the next amino acid. The amino acid sequence Phe(UUU)-Leu(UUA)-Arg(AGG), reveals that the mRNA was translated as U2U3U4-U5U6A7 in zero-frame, then −1-switched to A7GG, reading the same adenine (A7) twice. The alternative Phe(UUU)-Phe(UUU)-Arg(AGG) sequence could come either from U2U3U4 in zero-frame, then −1-switched to U4U5U6-A7GG, reading U4 twice, or from reading U1 twice to translate U1U2U3-U4U5U6-A7GG all in the −1-frame. This implies as many as three slipping paths differing in codon positions (Phe0-Leu0-Arg−1, Phe0-Phe−1-Arg−1, and Phe−1-Phe−1-Arg−1), can be utilized in the HIV-1 programmed frameshifting. This finding supports the idea of a highly adaptive ribosome in switching reading frames.
SINGLE-MOLECULE FLUORESCENCE STUDIES
Single-molecule fluorescence studies have provided much of the information about the dynamics of the translation.63–70 Real time observations by single-molecule fluorescence resonance energy transfer (smFRET) discovered that many of the conformational states that occur during translation are in highly dynamic equilibrium, readily accessible through thermal fluctuations at room temperature.64–66 The speed and accuracy of ribosomal translation may be related to the dynamics of conformational changes, which are determined by many factors including ligand binding/release, identities of tRNAs bound, and Mg2+ concentration.65 The frameshift promoting signals on the mRNA also critically modify the translation energetics; an upstream SD-like sequence and downstream secondary structures slow the translation rates.71–73 However, it is unknown in what ways the dynamics of each translation elongation step are affected by the presence of these signals. Real time tracking of individual molecules by smFRET makes it possible to follow ribosome dynamics step-by-step during translation. Measuring the timing of binding and release of elongation factors and tRNAs can reveal information about their distinctive effects on a frameshifting event.
Ribosomal Dynamics Observed during Selection and Accommodation of A-site tRNA
Frameshifting may take place during initial selection of TC (EF-Tu•GTP•aa-tRNA) and accommodation of aa-tRNA from the A/T state to the A/A state (Figure 2).31,32,74,75 During initial selection, correct codon•anticodon base-pairing in the 30S A-decoding site is proposed to allosterically initiate the GTPase activity in the TC.76,77 Upon GTP hydrolysis, further structural changes in the aa-tRNA and the ribosome are promoted for proofreading.76,77 Fidelity is achieved by the differences in interactions comparing cognate and non- or near-cognate tRNAs within the A site during the above two kinetic processes (initial selection and proofreading).76,77
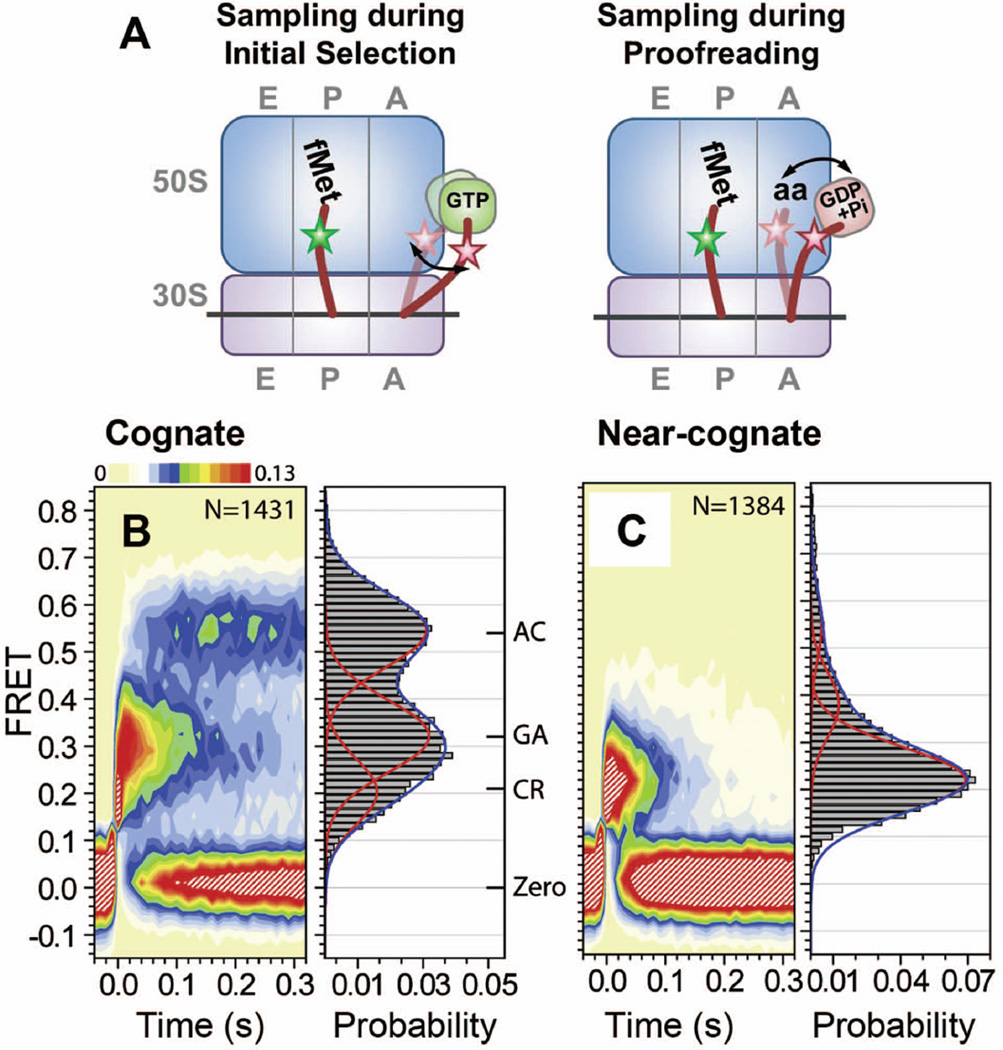
smFRET studies on ribosome dynamics that observed fluorescently labeled aa-tRNA binding into the A-site in real time support an allosteric effect of codon•anticodon interactions on the rates of TC binding and aa-tRNA accommodation.78–80 Upon delivery of cognate Phe-(Cy5)tRNAPhe in the form of a TC, ribosomal complexes bearing fMet-(Cy3)tRNAfMet in the P site sampled a series of FRET states, individually assigned as codon recognition (CR), formation of a GTPase-activated intermediate (GA), and finally, a completely accommodated state (AC) (Figure 9A and 9B).78,80 In contrast, ribosomal complexes translating a near-cognate codon showed a transient CR state with rare and short-lived excursions to GA and AC states (Figure 9C). With time resolutions of 2.5–10 ms, Geggier et al. discovered that aa-tRNA undergoes fast reversible transitions between the CR and the GA, and between the GA and the AC states.80 Their observations show that sequential steps of conformational rearrangements during the A-tRNA selection and accommodation are triggered by codon•anticodon interactions and correlate with fidelity enhancement.
Figure 9.
smFRET observations on tRNA dynamics during aa-tRNA selection and accommodation into Asite. 78,80 (A) Schematic drawings of proposed molecular basis for the observed FRET fluctuations during initial selection and proofreading. The 50S and 30S subunits are depicted in blue and purple rectangles, respectively, with mRNA in a black line, and tRNAs in brown curves. Green and purple round squares denote EF-Tu•GTP and EF-Tu•GDP+Phosphate, respectively. Green and pink stars are donor and acceptor fluorophores, respectively. Stepwise and fluctuating FRET evolved upon delivery of TC(Phe- (Cy5)tRNAPhe•EFTu• GTP) to a ribosomal complex bearing fMet-(Cy3)tRNAfMet in the P-site. Contour plots generated by superimposing smFRET trajectories, post-synchronized to the first observation of FRET above a noise threshold, and FRET histograms with (B) cognate (C) near-cognate ribosomal complexes. FRET histograms were fitted with three Gaussian functions. Cognate complexes fluctuated between three FRET states assigned as codon recognition (CR), GTPase activated (GA), accommodated (AC) states. The assignments were confirmed by control experiments using a non-hydrolyzable GTP analogue (GDPNP), and the antibiotic kirromycin, which traps ternary complex (TC) right after GTP hydrolysis. Figure A is reproduced with permission from Figure 6C in reference 80; Figure B and C are modified with permission from Figure 3A in reference 80.
The frameshift promoting signals on the mRNA may interfere with proper codon•anticodon interactions and impede conformational changes in the 30S that are critical for efficient aa-tRNA accommodation or even fidelity maintenance. FRET measurements of translation on a programmed frameshifting mRNA thus may show distinct behaviors from normal in-frame translation during the selection and accommodation steps.
Potential to Frameshift during Translocation
Translocation is often proposed as the step where frameshifts occur.31,81,82 Upon aa-tRNA accommodation into the A-site, the peptidyl-transfer reaction occurs rapidly, forming the pretranslocation state (PRE), with a deacylated P-tRNA and a peptidylated A-tRNA. For the next cycle of the elongation reaction, the tRNAs and mRNA have to translocate forward. Structural and biochemical studies have shown that translocation involves large-scale conformational rearrangements including intersubunit rotation, which entails a counterclockwise rotation of the 30S relative to the 50S (Figure 5B).10,83–85 What accompanies such an intersubunit-rotated state is a pair of hybrid tRNAs (P/E, A/P) residing in a hybrid ribosomal state, where as the classical tRNAs (P/P, A/A) are observed in an intersubunit-unrotated classical ribosomal state.10,83–85 In addition, the L1 stalk, a highly mobile domain comprised of L1 protein and 23S rRNA helices 76–78 from the 50S subunit, was found to move ~20 Å toward the subunit interface and to interact with the elbow of the P/E-hybrid tRNA upon EF-G binding (Figure 10A).44,83,86 These observations suggest strong correlations between the aforementioned motions and their vital roles in translocation. Frameshifting during translocation requires breaking the existing codon•anticodon base-pairings in the P- and A-sites, and reforming them in a new frame. Provided that the free energy difference in codon•anticodon base-pairings is minimized before and after frameshifting on the slippery sequence, a frameshift event could be effectively facilitated through any uncoupled movements between the mRNA and tRNAs during the dynamic conformational changes involved in translocation. Additionally, translocation in the programmed frameshifting environments involves the ribosome working actively as a helicase to unwind the downstream secondary structure of the mRNA, which is one of the crucial signals for efficient frameshifting. It has been observed that secondary structures effectively slow down overall translation rate.72,73 Interactions between the proposed helicase proteins (i.e. S3-S5) and the secondary structure may allosterically affect the dynamics of the conformational changes during translocation, or simply elevate the activation barrier for ribosome translocation, thus allowing frameshifting to become one of the favorable translation pathways.
Figure 10.
Single molecule studies on dynamics of pretranslocation (PRE) complexes bearing deacylated PtRNA and peptidyl/analogue of petidyl A-tRNA. (A) A x-ray crystal structure of a hybrid state ribosomal complex (PDB: 4GD1, 3R8S).10 Comparing to the classical state (Figure 1), the two subunits have rotated relative to each other; the hybrid P/E-tRNA (orange) interacts closely with the L1 stalk (magenta), which has moved inward toward the 50S E-site. For illustration purposes, an A/P-tRNA mimic (cyan) is manually built in to better depict the doubly-occupied hybrid state ribosome. The structural and positioning reference for such reconstruction is based on a crystallized A-site anticodon stem-loop (17-nucleotide long) inside a rotated state ribosomal complex (PDB: 3I1Z).14 Single molecule FRET studies investigated the dynamics of the conformational changes as indicated in the arrows. (B) Intersubunit rotation dynamics were probed with (Cy3)L9 in the 50S subunit and (Cy5)S6 in the 30S subunit.91 Pretranslocation (PRE) complex contained tRNAfMet in the P site and N-Ac-Phe-tRNAPhe (Ac) in the A site. Fluorescence time traces displayed high (0.58) and low (0.4) FRET states, assigned as unrotated and rotated states, respectively. (C) L1 stalk interactions with P/E-tRNA were studied with a FRET pair on L1 protein in the 50S subunit and tRNAPhe in the P site.92 Ribosomal complex with (Cy5)L1 enzymatically formed a posttranslocation (POST) complex bearing fMet-Phe-(Cy3)tRNAPhe in the P site. FRET between L1 and tRNAPhe presented steady low (0.21) FRET state. Real time stopped-flow delivery of a cognate tRNA in the form of TC (Lys-tRNALys •EFTu• GTP) in the absence of EF-G to the POST complex triggered fluctuating FRET states between low (0.21) and high (0.84), presumably upon peptidyl-transfer reaction forming pretranslocation (PRE) complex (left fluorescence time traces). Low and high FRET states were assigned as an L1 stalk open state with classical P/P tRNA and an L1 stalk closed state interacting with hybrid P/E tRNA, respectively. Co-delivery of LystRNALys• EF-Tu•GTP and EF-G•GTP evolved a steady high (0.84) FRET state, suggesting that EF-G stabilizes the L1 stalk closed state and drives translocation forward from the state (see right fluorescence time traces). Fluorescence time traces in Figure B is modified with permission from Figure 2A in reference 91; Fluorescence time traces in Figure C is modified with permission from Figure 5 in reference 92.
A Dynamically Fluctuating PRE Complex Prior to Translocation
Dynamics of the three major conformational changes during translocation have been best studied by smFRET: tRNAs transitions between classical (A/A, P/P) and hybrid (A/P, P/E) states,15,87–90 intersubunit rotation,91 and L1 stalk opening and closing (Figure 10A).88,92–94 tRNA dynamics, probed with a FRET dye-pair placed on P- and A-tRNAs, have shown that peptide bond formation triggers spontaneous fluctuations of the PRE complex between two distinct FRET states, i.e. classical and hybrid tRNAs (τclassical = 0.20 s, τ1,hybrid = 0.39 s, τ2,hybrid = 0.08 s).15 Presence of a peptidyl-tRNA in the A site favors hybrid state formation by ~6-fold compared to the presence of an aa-tRNA. More recent studies, with improved time resolution, employing systematic mutations on tRNA binding pockets within the 50S subunit have revealed the existence of more than one hybrid state,87,89 suggesting that the movements of P and A site tRNAs may not necessarily be tightly coupled during translocation.
Cornish et al. explored the intersubunit rotation dynamics using two sets of FRET pairs on L9 in the 50S and either S6 or S11 in the 30S (Figure 10B).91 They showed that binding deacylated-tRNA in the P-site stimulated spontaneous fluctuations between rotated (τ = 5.3 s) and unrotated (τ = 3.7 s) intersubunit states. Binding elongator tRNAs (tRNAPhe, tRNATyr) instead of the initiator tRNAfMet in the P-site sped up the forward rotation rates by 2–5 fold, demonstrating that the elongation phase and the identity of the tRNA affect the ribosome dynamics. Binding EF-G also stabilizes the rotated state. However, presenting only a tRNA anticodon stem loop in the P-site is not effective in forming the rotated state even in the presence of EF-G. Their observations indicate that EF-G itself, without proper tRNA interactions with the 50S E-site, is not enough to form the stabilized rotated state. Specifically the interactions between the 50S E-site and the elbow and/or acceptor end of the P/E-hybrid tRNA plays a key role in stabilizing the rotated state.
Interactions between the L1 stalk and P/E-hybrid tRNA were interrogated with a FRET pair on L1 protein and P-tRNA (Figure 10C).92 Fluctuations between high (τ = 0.78 s) and low (τ = 0.22 s) FRET correspond to forming and disrupting L1 interactions with the P/E-hybrid tRNA upon peptidyl transfer. Occupancy of the A-site and identity of the A-tRNA altered the distributions, thus energetics, between the two conformations. Binding EF-G promotes the L1 stalk interacting with hybrid-P/E tRNA. Munro et al. reported relatively rapid P/E-hybrid tRNA formation compared to the L1 stalk closure by parallel FRET measurements on the dynamics of L1-tRNA and tRNA-tRNA,88 suggesting that L1 stalk closing may require P/E-tRNA formation, but not always be coupled with it. Independently from the P/E tRNA formation, the opening and closing motions of the L1 stalk relative to the 50S subunits were further examined.93,94 High correspondence of the L1 stalk closing rate with the intersubunit forward (counterclockwise) rotation rate suggests tight coupling of the two motions in the PRE complex.94 However, L1 stalk opening depended more on the E-site tRNA occupancy, implying a potential role of the L1 stalk guiding tRNA transits from P- to E-site and then exit from the ribosome.
Collectively, all these studies illustrate a highly dynamic PRE ribosomal complex; thermal energy is sufficient to drive the motions described above, as they form spontaneously. A hybrid PRE state, where the tRNAs adopts the hybrid state in an intersubunit-rotated ribosome and the closed L1 stalk is interacting with P/E tRNA, may be a prerequisite state for translocation. It has not been clearly resolved yet how such hybrid PRE state is attained, whether the motions are concerted or stochastic. Different smFRET studies have found different kinetics for the same motions in different conditions.88,92,94 The hybrid PRE state perhaps is achievable through multiple pathways with many transiently visited intermediate states. smFRET studies will enable us to test how individual frameshift promoting signals on mRNA alter the dynamics of the motions, especially in the context of enhancing uncoupling of the codon•anticodon base-pair interactions.
An smFRET study on the effect of downstream stable secondary structures on the ribosomal dynamics has very recently been published.95 Using three different FRET constructs with dye pairs on tRNA-tRNA, L11-tRNA, and L1-tRNA, they demonstrated that downstream secondary structures can slow both the translocation step and E-site tRNA release by as high as a factor of two.95 However, it should be pointed out that the observed decrease in E-site tRNA release rate may result from a slower codon•anticodon translocation in the 30S subunit, which cannot be distinguished using their FRET construct designs. Nonetheless, their study starts to shed light on the role of downstream secondary structure, one of the critical frameshifting promoting signals. Similar experiments but with the presence of a slippery sequence and an internal SD sequence should provide further insights on the detail mechanism of frameshifting.
Codon•Anticodon translocation catalyzed by EF-G
The ribosome requires EF-G for rapid translocation, although it can spontaneously translocate with a much slower rate in the absence of EF-G.96 smFRET studies described earlier demonstrated that EF-G binding rectifies the thermally fluctuating PRE ribosomal complexes into the hybrid PRE state, hence favoring forward translocation reactions.91,92 They also confirmed that GTP hydrolysis is not required for stabilizing the hybrid PRE state, as the stable hybrid PRE state can be formed in the presence of EF-G bound with a non-hydrolysable GTP analogue.91,92 However, it has been found, by smFRET between an A-site fMet-Phe-tRNAPhe and EF-G, that GTP hydrolysis is essential for later sub-steps during translocation, which involves appropriate EF-G engagement and its own conformational changes.97 Through these dynamical steps, the ribosome may form the unlocked state, permitting translocation of the codon•anticodon duplex in the 30S subunit.
Structural studies have indicated the rotation of the 30S head relative to the 30S body being a critical player in translocating the codon•anticodon duplex on the 30S subunit, while EF-G catalyzes GTP hydrolysis to promote translocation.14,24,39,44 Identifying crucial interactions between the codon•anticodon duplex and the 30S will enlighten the frameshifting mechanism. The first bulk FRET study of 30S head rotation was reported recently employing dye pairs on the 30S head and body domains.41 Combining with stopped-flow experiments, the authors showed that forward (counterclockwise) 30S head rotation occurs more slowly than forward intersubunit rotation: intersubunit forward (>200 s−1) > 30S head forward (80 s−1) > 30S head reverse (10 s−1) ≥ intersubunit reverse (4.5 s−1).41 Additionally, from the fluorescence quenching rate of a 3'-pyrene labeled mRNA, they observed the mRNA translocation had similar rates as that of the reverse 30S head rotation, suggesting that mRNA translocation takes place along with 30S head rotation. Studying the 30S head rotation dynamics at a single-molecule level would provide key insights into the unlocking process in the 30S subunit driven by EF-G binding and GTP hydrolysis. Correlating the unlocking process with the frameshifting signals may uncover the critical dynamics for frameshifting.
Real-Time Observation on Multiple Rounds of Translational Elongation
By annealing a FRET probe on mutated rRNA helixes in the 30S and 50S, Aitken and Puglisi have successfully monitored ribosomal intersubunit rotations over multiple rounds of elongation cycles in real time.98 During each round of elongation, the ribosomal complex transits between two distinct FRET states: a PRE state (low FRET) of an intersubunit-rotated ribosome upon peptidyl transfer and a re-locked classical ribosome state (high FRET) after EF-G driven translocation. Interestingly, their identified ribosomal transitions are not thermally accessible, unlike the other intersubunit FRET measurements showing spontaneous fluctuations as described earlier.91 It could be that the two studies were monitoring distinctly different motions particular to the dye labeling sites.70 Differences in their experimental conditions may also be one of the causes.
The number of high-low-high FRET cycles corresponds to irreversible intersubunit rotations and thus to the number of codons that are translated, enabling one to follow elongation steps codon by codon along the mRNA. Frameshifted translation could then be distinguished from in-frame translation by the number of codons translated before reaching an in-frame or out-of frame stop. By measuring the dwell time at each step, one can exam the effects of frameshifting sequences on each elongation cycle. Combining these studies with direct observation of binding and release of fluorophore-labeled EF-G99 and tRNAs100 would allow us to properly correlate the consequential dynamical events on frameshifting translation cycles monitored by FRET.
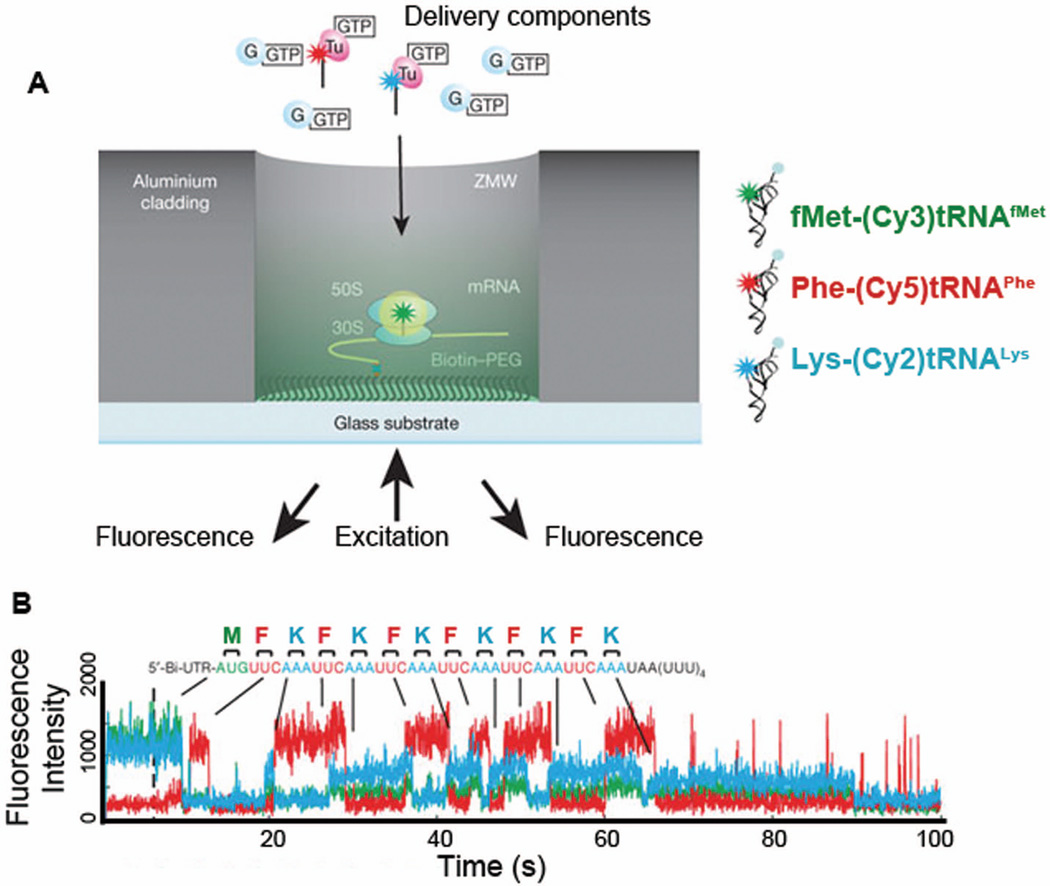
Zero-mode waveguides (ZMWs) have served as a significant breakthrough for dynamical fluorescence studying in ribosome translation. Owing to the small illumination depth in ZMWs, simultaneous detection of multiple fluorophores at higher and thus more physiological realistic concentrations has become possible (Figure 11A). Uemura et al. observed real-time tRNA transits within the translating ribosomal complex during multiple rounds of elongation cycles using ZMWs (Figure 11B).101 ZMWs enabled them to observe the micromolar range of concentrations of fluorophore-labeled tRNAs. As the molecules freely diffuse into the ZMWs (wells of 50–200 nm in diameter), binding tRNAs are not limited by tRNA availability. Such high concentrations cannot be achieved in typical total internal reflection fluorescence (TIRF) imaging simply due to high background fluorescence. By monitoring the timing and order of tRNA binding and release, they demonstrated that release of E-site tRNA may not necessarily be coupled with the arrival of A-site tRNA.
Figure 11.
Real-time observation of translation using Zero-mode waveguides (ZMW).101 (A) Ribosomal initiation complex bearing fMet-(Cy3)tRNAfMet in the P-site was immobilized in a ZMW well, cylindrical aluminum nano-well (50–200 nm in diameter), enabling imaging in the presence of high concentrations of fluorophore-labeled molecules. (B) A representative trace of fluorescence intensities from a ribosomal complex translating six repeating Phe and Lys codons after initiation codon: M(FK)6. Ternary complexes (TC) with fluorescently labeled tRNAs, 200 nM Phe-(Cy5)tRNAPhe and 200 nM Lys- (Cy2)tRNALys, in the presence of 500 nM EF-G•GTP were delivered to the ribosomal initiation complex. Simultaneous illumination and detection of multi-color fluorophores Cy2 (blue), Cy3 (green), and Cy5 (red) allowed them to image tRNAs binding and release from the ribosomal complex during active translation elongation, thereby following translation codon by codon. Figure A and B are modified with permission from Figure 1 and 3A,
ZMWs will allow direct observation of frameshifting by monitoring binding of tRNAs specific to frameshifted mRNA codons. The importance of E-site occupancy can be explored by monitoring the binding of A-tRNA and release of E-tRNA. Effects on frameshifting efficiency of binding and release of other factors, including elongation factors, can also be determined. Comparing kinetic events between the frameshifting and non-frameshifting mRNAs will provide further insight into the key steps dictating frameshifting.
CONCLUSIONS
The applications of single-molecule methods to investigate translation and ribosome dynamics are just beginning. Beads can be attached to various parts of the ribosome, to the mRNA, and to the nascent polypeptide being synthesized, to learn about the kinetics and thermodynamics of the motions.102 Fluorophores can be attached to the tRNAs, the elongation factors, and the ribosome to learn about the local dynamics near the fluorophores. Combining the two measurements (force and FRET)103 produces a very powerful tool for answering the many questions necessary to predict when and where frameshifting occurs.
Acknowledgment
We are very grateful to Prof. Harry Noller, U. California, Santa Cruz, for carefully reading the manuscript and saving us from incorrect and confusing statements. The work was supported by NIH grant GM10840 to IT.
References
- 1.Drummond DA, Wilke CO. Nat Rev Genet. 2009;10:715–724. doi: 10.1038/nrg2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weiss RB, Dunn DM, Atkins JF, Gesteland RF. Cold Spring Harb Symp Quant Biol. 1987;52:687–693. doi: 10.1101/sqb.1987.052.01.078. [DOI] [PubMed] [Google Scholar]
- 3.Atkins JF, Weiss RB, Gesteland RF. Cell. 1990;62:413–423. doi: 10.1016/0092-8674(90)90007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brierley I, Pennell S. Cold Spring Harb Symp Quant Biol. 2001;66:233–248. doi: 10.1101/sqb.2001.66.233. [DOI] [PubMed] [Google Scholar]
- 5.Dinman JD. Wiley interdisciplinary reviews RNA. 2012;3:661–673. doi: 10.1002/wrna.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farabaugh PJ. Microbiol Rev. 1996;60:103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melnikov S, Ben-Shem A, Garreau de Loubresse N, Jenner L, Yusupova G, Yusupov M. Nat Struct Mol Biol. 2012;19:560–567. doi: 10.1038/nsmb.2313. [DOI] [PubMed] [Google Scholar]
- 8.Schmeing TM, Ramakrishnan V. Nature. 2009;461:1234–1242. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- 9.Moazed D, Noller HF. Cell. 1989;57:585–597. doi: 10.1016/0092-8674(89)90128-1. [DOI] [PubMed] [Google Scholar]
- 10.Dunkle JA, Wang L, Feldman MB, Pulk A, Chen VB, Kapral GJ, Noeske J, Richardson JS, Blanchard SC, Cate JH. Science. 2011;332:981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gromadski KB, Daviter T, Rodnina MV. Mol Cell. 2006;21:369–377. doi: 10.1016/j.molcel.2005.12.018. [DOI] [PubMed] [Google Scholar]
- 12.Gromadski KB, Rodnina MV. Mol Cell. 2004;13:191–200. doi: 10.1016/s1097-2765(04)00005-x. [DOI] [PubMed] [Google Scholar]
- 13.Rodnina MV, Pape T, Fricke R, Kuhn L, Wintermeyer W. J Biol Chem. 1996;271:646–652. doi: 10.1074/jbc.271.2.646. [DOI] [PubMed] [Google Scholar]
- 14.Zhang W, Dunkle JA, Cate JH. Science. 2009;325:1014–1017. doi: 10.1126/science.1175275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blanchard SC, Kim HD, Gonzalez RL, Jr, Puglisi JD, Chu S. Proc Natl Acad Sci USA. 2004;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim HD, Puglisi JD, Chu S. Biophys J. 2007;93:3575–3582. doi: 10.1529/biophysj.107.109884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dorner S, Brunelle JL, Sharma D, Green R. Nat Struct Mol Biol. 2006;13:234–241. doi: 10.1038/nsmb1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sergiev PV, Lesnyak DV, Kiparisov SV, Burakovsky DE, Leonov AA, Bogdanov AA, Brimacombe R, Dontsova OA. Nucleic Acids Res. 2005;33:6048–6056. doi: 10.1093/nar/gki910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Savelsbergh A, Katunin VI, Mohr D, Peske F, Rodnina MV, Wintermeyer W. Mol Cell. 2003;11:1517–1523. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- 20.Rodnina MV, Wintermeyer W. Biochem Soc Trans. 2011;39:658–662. doi: 10.1042/BST0390658. [DOI] [PubMed] [Google Scholar]
- 21.Chen CL, Stevens B, Kaur J, Smilansky Z, Cooperman BS, Goldman YE. Proc Natl Acad Sci USA. 2011;108:16980–16985. doi: 10.1073/pnas.1106999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petropoulos AD, Green R. J Biol Chem. 2012;287:11642–11648. doi: 10.1074/jbc.C111.330068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nierhaus KH, Pech M. J Biol Chem. 2012;287:27049. doi: 10.1074/jbc.L112.381848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frank J, Gao H, Sengupta J, Gao N, Taylor DJ. Proc Natl Acad Sci USA. 2007;104:19671–19678. doi: 10.1073/pnas.0708517104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsuchihashi Z, Kornberg A. Proc Natl Acad Sci USA. 1990;87:2516–2520. doi: 10.1073/pnas.87.7.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qu X, Wen JD, Lancaster L, Noller HF, Bustamante C, Tinoco I., Jr Nature. 2011;475:118–121. doi: 10.1038/nature10126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacks T, Power MD, Masiarz FR, Luciw PA, Barr PJ, Varmus HE. Nature. 1988;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 28.Jacks T, Madhani HD, Masiarz FR, Varmus HE. Cell. 1988;55:447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brierley I, Digard P, Inglis SC. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Atkins JF, Bjork GR. Microbiol Mol Biol R. 2009;73:178–210. doi: 10.1128/MMBR.00010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harger JW, Meskauskas A, Dinman JD. Trends Biochem Sci. 2002;27:448–454. doi: 10.1016/s0968-0004(02)02149-7. [DOI] [PubMed] [Google Scholar]
- 32.Urbonavicius J, Qian O, Durand JMB, Hagervall TG, Bjork GR. EMBO J. 2001;20:4863–4873. doi: 10.1093/emboj/20.17.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horsfield JA, Wilson DN, Mannering SA, Adamski FM, Tate WP. Nucleic Acids Res. 1995;23:1487–1494. doi: 10.1093/nar/23.9.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yelverton E, Lindsley D, Yamauchi P, Gallant JA. Mol Microbiol. 1994;11:303–313. doi: 10.1111/j.1365-2958.1994.tb00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou J, Lancaster L, Trakhanov S, Noller HF. RNA. 2012;18:230–240. doi: 10.1261/rna.031187.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jenner LB, Demeshkina N, Yusupova G, Yusupov M. Nat Struct Mol Biol. 2010;17:555–560. doi: 10.1038/nsmb.1790. [DOI] [PubMed] [Google Scholar]
- 37.Gao YG, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. Science. 2009;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramrath DJF, Yamamoto H, Rother K, Wittek D, Pech M, Mielke T, Loerke J, Scheerer P, Ivanov P, Teraoka Y, Shpanchenko O, Nierhaus KH, Spahn CMT. Nature. 2012;485:526-U140. doi: 10.1038/nature11006. [DOI] [PubMed] [Google Scholar]
- 39.Ratje AH, Loerke J, Mikolajka A, Brunner M, Hildebrand PW, Starosta AL, Donhofer A, Connell SR, Fucini P, Mielke T, Whitford PC, Onuchic JN, Yu Y, Sanbonmatsu KY, Hartmann RK, Penczek PA, Wilson DN, Spahn CM. Nature. 2010;468:713–716. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ermolenko DN, Noller HF. Nat Struct Mol Biol. 2011;18:457-U492. doi: 10.1038/nsmb.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo ZJ, Noller HF. Proc Natl Acad Sci USA. 2012;109:20391–20394. doi: 10.1073/pnas.1218999109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stahl G, McCarty GP, Farabaugh PJ. Trends Biochem Sci. 2002;27:178–183. doi: 10.1016/S0968-0004(02)02064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Selmer M, Dunham CM, Murphy FV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 44.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 45.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 46.Tsuchihashi Z, Brown PO. Gene Dev. 1992;6:511–519. doi: 10.1101/gad.6.3.511. [DOI] [PubMed] [Google Scholar]
- 47.Sanders CL, Curran JF. RNA. 2007;13:1483–1491. doi: 10.1261/rna.638707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leger M, Dulude D, Steinberg SV, Brakier-Gingras L. Nucleic Acids Res. 2007;35:5581–5592. doi: 10.1093/nar/gkm578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brierley I. J Gen Virol. 1995;76:1885–1892. doi: 10.1099/0022-1317-76-8-1885. [DOI] [PubMed] [Google Scholar]
- 50.Blinkowa AL, Walker JR. Nucleic Acids Res. 1990;18:1725–1729. doi: 10.1093/nar/18.7.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brierley I, Meredith MR, Bloys AJ, Hagervall TG. J Mol Biol. 1997;270:360–373. doi: 10.1006/jmbi.1997.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Korostelev A, Trakhanov S, Asahara H, Laurberg M, Lancaster L, Noller HF. Proc Natl Acad Sci USA. 2007;104:16840–16843. doi: 10.1073/pnas.0707850104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jenner L, Rees B, Yusupov M, Yusupova G. EMBO Rep. 2007;8:846–850. doi: 10.1038/sj.embor.7401044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yusupova G, Jenner L, Rees B, Moras D, Yusupov M. Nature. 2006;444:391–394. doi: 10.1038/nature05281. [DOI] [PubMed] [Google Scholar]
- 55.Kaminishi T, Wilson DN, Takemoto C, Harms JM, Kawazoe M, Schluenzen F, Hanawa-Suetsugu K, Shirouzu M, Fucini P, Yokoyama S. Structure. 2007;15:289–297. doi: 10.1016/j.str.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 56.Wen JD, Lancaster L, Hodges C, Zeri AC, Yoshimura SH, Noller HF, Bustamante C, Tinoco I. Nature. 2008;452:598–603. doi: 10.1038/nature06716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu T, Kaplan A, Wickersham CE, Wen J, Lancaster L, Fredrick K, Noller NF, Tinoco JI, Bustamante C. Nature. Submitted. [Google Scholar]
- 58.Khetan A, Huang YM, Dolnikova J, Pederson NE, Wen D, Yusuf-Makagiansar H, Chen P, Ryll T. Biotechnol Bioeng. 2010;107:116–123. doi: 10.1002/bit.22771. [DOI] [PubMed] [Google Scholar]
- 59.Wen D, Vecchi MM, Gu S, Su L, Dolnikova J, Huang YM, Foley SF, Garber E, Pederson N, Meier W. J Biol Chem. 2009;284:32686–32694. doi: 10.1074/jbc.M109.059360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McNulty DE, Claffee BA, Huddleston MJ, Kane JF. Protein Expres Purif. 2003;27:365–374. doi: 10.1016/s1046-5928(02)00610-1. [DOI] [PubMed] [Google Scholar]
- 61.Aguirre B, Costas M, Cabrera N, Mendoza-Hernandez G, Helseth DL, Jr, Fernandez P, de Gomez-Puyou MT, Perez-Montfort R, Torres-Larios A, Gomez Puyou A. PLoS One. 2011;6:e21035. doi: 10.1371/journal.pone.0021035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kerrigan JJ, McNulty DE, Burns M, Allen KE, Tang X, Lu Q, Trulli JM, Johanson KO, Kane JF. Protein Expres Purif. 2008;60:110–116. doi: 10.1016/j.pep.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 63.Marshall RA, Aitken CE, Dorywalska M, Puglisi JD. Annu Rev Biochem. 2008;77:177–203. doi: 10.1146/annurev.biochem.77.070606.101431. [DOI] [PubMed] [Google Scholar]
- 64.Blanchard SC. Curr Opin Struct Biol. 2009;19:103–109. doi: 10.1016/j.sbi.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Frank J, Gonzalez RL., Jr Annu Rev Biochem. 2010;79:381–412. doi: 10.1146/annurev-biochem-060408-173330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aitken CE, Petrov A, Puglisi JD. Annu Rev Biophys. 2010;39:491–513. doi: 10.1146/annurev.biophys.093008.131427. [DOI] [PubMed] [Google Scholar]
- 67.Perez CE, Gonzalez RL., Jr Curr Opin Chem Biol. 2011;15:853–863. doi: 10.1016/j.cbpa.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Petrov A, Kornberg G, O'Leary S, Tsai A, Uemura S, Puglisi JD. Curr Opin Struct Biol. 2011;21:137–145. doi: 10.1016/j.sbi.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Petrov A, Chen J, O'Leary S, Tsai A, Puglisi JD. Cold Spring Harb Perspect Biol. 2012;4:a011551. doi: 10.1101/cshperspect.a011551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen J, Tsai A, O'Leary SE, Petrov A, Puglisi JD. Curr Opin Struct Biol. 2012;22:804–814. doi: 10.1016/j.sbi.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li GW, Oh E, Weissman JS. Nature. 2012;484:538–541. doi: 10.1038/nature10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kontos H, Napthine S, Brierley I. Mol Cell Biol. 2001;21:8657–8670. doi: 10.1128/MCB.21.24.8657-8670.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Somogyi P, Jenner AJ, Brierley I, Inglis SC. Mol Cell Biol. 1993;13:6931–6940. doi: 10.1128/mcb.13.11.6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Plant EP, Jacobs KL, Harger JW, Meskauskas A, Jacobs JL, Baxter JL, Petrov AN, Dinman JD. RNA. 2003;9:168–174. doi: 10.1261/rna.2132503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Leger M, Sidani S, Brakier-Gingras L. RNA. 2004;10:1225–1235. doi: 10.1261/rna.7670704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ogle JM, Ramakrishnan V. Annu Rev Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- 77.Rodnina MV, Gromadski KB, Kothe U, Wieden HJ. FEBS Lett. 2005;579:938–942. doi: 10.1016/j.febslet.2004.11.048. [DOI] [PubMed] [Google Scholar]
- 78.Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. Nat Struct Mol Biol. 2004;11:1008–1014. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- 79.Lee TH, Blanchard SC, Kim HD, Puglisi JD, Chu S. Proc Natl Acad Sci USA. 2007;104:13661–13665. doi: 10.1073/pnas.0705988104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Geggier P, Dave R, Feldman MB, Terry DS, Altman RB, Munro JB, Blanchard SC. J Mol Biol. 2010;399:576–595. doi: 10.1016/j.jmb.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Namy O, Moran SJ, Stuart DI, Gilbert RJ, Brierley I. Nature. 2006;441:244–247. doi: 10.1038/nature04735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Giedroc DP, Cornish PV. Virus Res. 2009;139:193–208. doi: 10.1016/j.virusres.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 84.Horan LH, Noller HF. Proc Natl Acad Sci USA. 2007;104:4881–4885. doi: 10.1073/pnas.0700762104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Noeske J, Cate JH. Curr Opin Struct Biol. 2012;22:743–749. doi: 10.1016/j.sbi.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Agirrezabala X, Lei J, Brunelle JL, Ortiz-Meoz RF, Green R, Frank J. Mol Cell. 2008;32:190–197. doi: 10.1016/j.molcel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Munro JB, Altman RB, O'Connor N, Blanchard SC. Mol Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Munro JB, Altman RB, Tung CS, Cate JH, Sanbonmatsu KY, Blanchard SC. Proc Natl Acad Sci USA. 2010;107:709–714. doi: 10.1073/pnas.0908597107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang L, Altman RB, Blanchard SC. RNA. 2011;17:2189–2200. doi: 10.1261/rna.029033.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen C, Stevens B, Kaur J, Cabral D, Liu H, Wang Y, Zhang H, Rosenblum G, Smilansky Z, Goldman YE, Cooperman BS. Mol Cell. 2011;42:367–377. doi: 10.1016/j.molcel.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cornish PV, Ermolenko DN, Noller HF, Ha T. Mol Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fei J, Kosuri P, MacDougall DD, Gonzalez RL., Jr Mol Cell. 2008;30:348–359. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 93.Fei J, Bronson JE, Hofman JM, Srinivas RL, Wiggins CH, Gonzalez RL., Jr Proc Natl Acad Sci USA. 2009;106:15702–15707. doi: 10.1073/pnas.0908077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cornish PV, Ermolenko DN, Staple DW, Hoang L, Hickerson RP, Noller HF, Ha T. Proc Natl Acad Sci USA. 2009;106:2571–2576. doi: 10.1073/pnas.0813180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen C, Zhang H, Broitman SL, Reiche M, Farrell I, Cooperman BS, Goldman YE. Nat Struct Mol Biol. 2013 doi: 10.1038/nsmb.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gavrilova LP, Kostiashkina OE, Koteliansky VE, Rutkevitch NM, Spirin AS. J Mol Biol. 1976;101:537–552. doi: 10.1016/0022-2836(76)90243-6. [DOI] [PubMed] [Google Scholar]
- 97.Munro JB, Wasserman MR, Altman RB, Wang LY, Blanchard SC. Nat Struct Mol Biol. 2010;17:1470-U1102. doi: 10.1038/nsmb.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Aitken CE, Puglisi JD. Nat Struct Mol Biol. 2010;17:793–800. doi: 10.1038/nsmb.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen J, Petrov A, Tsai A, O'Leary SE, Puglisi JD. Nat Struct Mol Biol. 2013 doi: 10.1038/nsmb.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen J, Tsai A, Petrov A, Puglisi JD. J Am Chem Soc. 2012;134:5734–5737. doi: 10.1021/ja2119964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Uemura S, Aitken CE, Korlach J, Flusberg BA, Turner SW, Puglisi JD. Nature. 2010;464:1012–1017. doi: 10.1038/nature08925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tinoco I, Jr, Bustamante C. Biophys Chem. 2002:101–102. 513–533. doi: 10.1016/s0301-4622(02)00177-1. [DOI] [PubMed] [Google Scholar]
- 103.Comstock MJ, Ha T, Chemla YR. Nat Methods. 2011;8:335–340. doi: 10.1038/nmeth.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. Science. 2010;330:835–838. doi: 10.1126/science.1194460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Temperley R, Richter R, Dennerlein S, Lightowlers RN, Chrzanowska-Lightowlers ZM. Science. 2010;327:301–301. doi: 10.1126/science.1180674. [DOI] [PubMed] [Google Scholar]