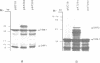Abstract
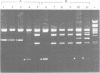
Excision is probably the initial and rate-limiting step of the movements of conjugative transposons of Gram-positive bacteria such as Tn916 and Tn1545. We have shown, by molecular cloning and DNA sequencing, that a 2058 bp Sau3A right-junction fragment of transposon Tn1545 specifies two gene products that are involved in the excision of the element. The DNA sequence of these genes, designated orf1 and orf2, has been determined and the corresponding proteins, ORF1 and ORF2, have been identified in a bacterial cell-free coupled transcription-translation system. These proteins are freely diffusible since they are able to trans-complement in vivo a deletion derivative of Tn1545 defective for excision. Using an in vivo complementation assay, we have demonstrated that ORF2 alone is able to catalyse excision and that ORF1 strongly stimulates the activity of ORF2. We also found that ORF1 and ORF2 display local homology with, respectively, proteins Xis and Int from lamboid phages, which suggests that these excision systems have a common origin. Based on the functional properties of the integrase of bacteriophage lambda, on the analysis of the nucleotide sequence of the junction fragments and of the target before insertion and after excision, a model is proposed for ORF2-catalysed excision of Tn1545 and related conjugative transposons.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrews B. J., Proteau G. A., Beatty L. G., Sadowski P. D. The FLP recombinase of the 2 micron circle DNA of yeast: interaction with its target sequences. Cell. 1985 Apr;40(4):795–803. doi: 10.1016/0092-8674(85)90339-3. [DOI] [PubMed] [Google Scholar]
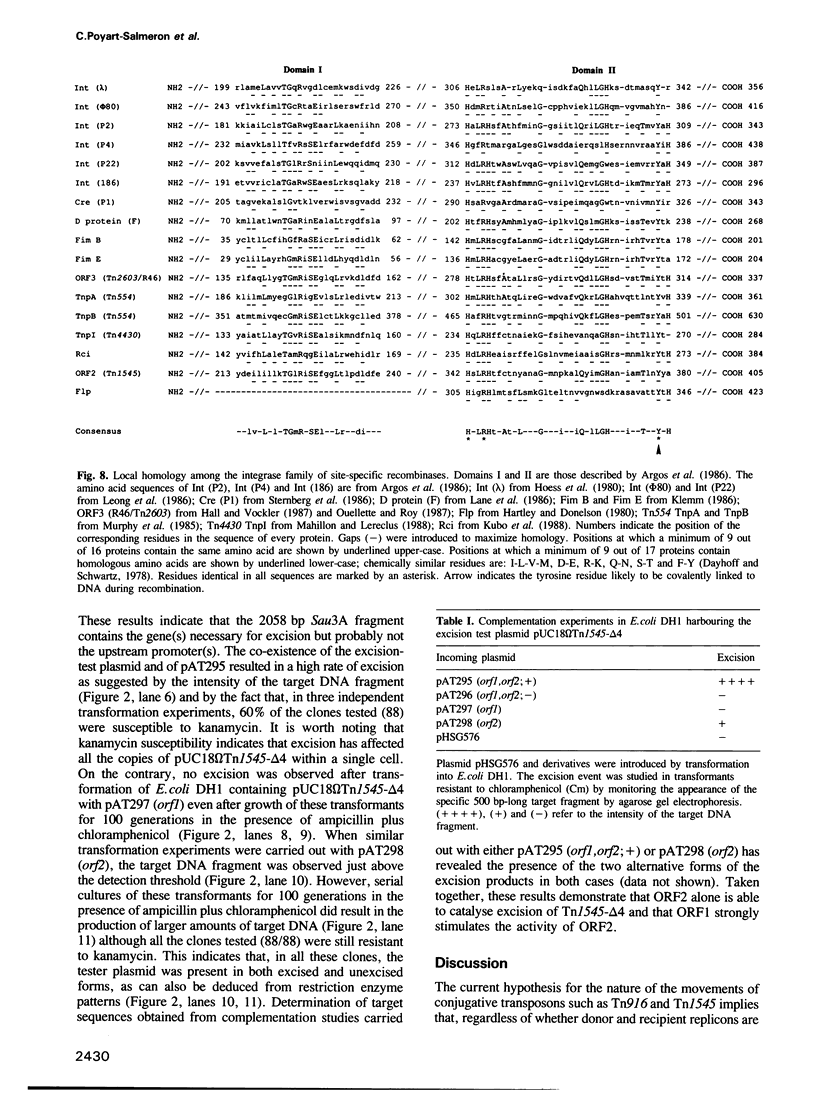
- Argos P., Landy A., Abremski K., Egan J. B., Haggard-Ljungquist E., Hoess R. H., Kahn M. L., Kalionis B., Narayana S. V., Pierson L. S., 3rd The integrase family of site-specific recombinases: regional similarities and global diversity. EMBO J. 1986 Feb;5(2):433–440. doi: 10.1002/j.1460-2075.1986.tb04229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccard F., Smokvina T., Pernodet J. L., Friedmann A., Guérineau M. Structural analysis of loci involved in pSAM2 site-specific integration in Streptomyces. Plasmid. 1989 Jan;21(1):59–70. doi: 10.1016/0147-619x(89)90087-5. [DOI] [PubMed] [Google Scholar]
- Boccard F., Smokvina T., Pernodet J. L., Friedmann A., Guérineau M. The integrated conjugative plasmid pSAM2 of Streptomyces ambofaciens is related to temperate bacteriophages. EMBO J. 1989 Mar;8(3):973–980. doi: 10.1002/j.1460-2075.1989.tb03460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillaud F., Carlier C., Courvalin P. Physical analysis of the conjugative shuttle transposon Tn1545. Plasmid. 1987 Jan;17(1):58–60. doi: 10.1016/0147-619x(87)90009-6. [DOI] [PubMed] [Google Scholar]
- Caillaud F., Courvalin P. Nucleotide sequence of the ends of the conjugative shuttle transposon Tn1545. Mol Gen Genet. 1987 Aug;209(1):110–115. doi: 10.1007/BF00329844. [DOI] [PubMed] [Google Scholar]
- Caillaud F., Trieu-Cuot P., Carlier C., Courvalin P. Nucleotide sequence of the kanamycin resistance determinant of the pneumococcal transposon Tn1545: evolutionary relationships and transcriptional analysis of aphA-3 genes. Mol Gen Genet. 1987 May;207(2-3):509–513. doi: 10.1007/BF00331623. [DOI] [PubMed] [Google Scholar]
- Christie P. J., Korman R. Z., Zahler S. A., Adsit J. C., Dunny G. M. Two conjugation systems associated with Streptococcus faecalis plasmid pCF10: identification of a conjugative transposon that transfers between S. faecalis and Bacillus subtilis. J Bacteriol. 1987 Jun;169(6):2529–2536. doi: 10.1128/jb.169.6.2529-2536.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverie J. M., Bricault L. PseqIP: a nonredundant and exhaustive protein sequence data bank generated from 4 major existing collections. Proteins. 1986 Sep;1(1):60–65. doi: 10.1002/prot.340010110. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., An F. Y., White B. A., Gawron-Burke C. Streptococcus faecalis sex pheromone (cAM373) also produced by Staphylococcus aureus and identification of a conjugative transposon (Tn918). J Bacteriol. 1985 Jun;162(3):1212–1220. doi: 10.1128/jb.162.3.1212-1220.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Flannagan S. E., Ike Y., Jones J. M., Gawron-Burke C. Sequence analysis of termini of conjugative transposon Tn916. J Bacteriol. 1988 Jul;170(7):3046–3052. doi: 10.1128/jb.170.7.3046-3052.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Gawron-Burke C. Conjugative transposons and the dissemination of antibiotic resistance in streptococci. Annu Rev Microbiol. 1986;40:635–659. doi: 10.1146/annurev.mi.40.100186.003223. [DOI] [PubMed] [Google Scholar]
- Courvalin P., Carlier C. Tn1545: a conjugative shuttle transposon. Mol Gen Genet. 1987 Feb;206(2):259–264. doi: 10.1007/BF00333582. [DOI] [PubMed] [Google Scholar]
- Courvalin P., Carlier C. Transposable multiple antibiotic resistance in Streptococcus pneumoniae. Mol Gen Genet. 1986 Nov;205(2):291–297. doi: 10.1007/BF00430441. [DOI] [PubMed] [Google Scholar]
- Craig N. L., Nash H. A. The mechanism of phage lambda site-specific recombination: site-specific breakage of DNA by Int topoisomerase. Cell. 1983 Dec;35(3 Pt 2):795–803. doi: 10.1016/0092-8674(83)90112-5. [DOI] [PubMed] [Google Scholar]
- Dodd I. B., Egan J. B. Systematic method for the detection of potential lambda Cro-like DNA-binding regions in proteins. J Mol Biol. 1987 Apr 5;194(3):557–564. doi: 10.1016/0022-2836(87)90681-4. [DOI] [PubMed] [Google Scholar]
- Fitzgerald G. F., Clewell D. B. A conjugative transposon (Tn919) in Streptococcus sanguis. Infect Immun. 1985 Feb;47(2):415–420. doi: 10.1128/iai.47.2.415-420.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flamm E. L., Weisberg R. A. Primary structure of the hip gene of Escherichia coli and of its product, the beta subunit of integration host factor. J Mol Biol. 1985 May 25;183(2):117–128. doi: 10.1016/0022-2836(85)90206-2. [DOI] [PubMed] [Google Scholar]
- Franke A. E., Clewell D. B. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of "conjugal" transfer in the absence of a conjugative plasmid. J Bacteriol. 1981 Jan;145(1):494–502. doi: 10.1128/jb.145.1.494-502.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier T., Saurin W., Cole S. T. Molecular characterization of the resolvase gene, res, carried by a multicopy plasmid from Clostridium perfringens: common evolutionary origin for prokaryotic site-specific recombinases. Mol Microbiol. 1987 Nov;1(3):371–376. doi: 10.1111/j.1365-2958.1987.tb01944.x. [DOI] [PubMed] [Google Scholar]
- Gawron-Burke C., Clewell D. B. A transposon in Streptococcus faecalis with fertility properties. Nature. 1982 Nov 18;300(5889):281–284. doi: 10.1038/300281a0. [DOI] [PubMed] [Google Scholar]
- Gawron-Burke C., Clewell D. B. Regeneration of insertionally inactivated streptococcal DNA fragments after excision of transposon Tn916 in Escherichia coli: strategy for targeting and cloning of genes from gram-positive bacteria. J Bacteriol. 1984 Jul;159(1):214–221. doi: 10.1128/jb.159.1.214-221.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronostajski R. M., Sadowski P. D. The FLP recombinase of the Saccharomyces cerevisiae 2 microns plasmid attaches covalently to DNA via a phosphotyrosyl linkage. Mol Cell Biol. 1985 Nov;5(11):3274–3279. doi: 10.1128/mcb.5.11.3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall R. M., Vockler C. The region of the IncN plasmid R46 coding for resistance to beta-lactam antibiotics, streptomycin/spectinomycin and sulphonamides is closely related to antibiotic resistance segments found in IncW plasmids and in Tn21-like transposons. Nucleic Acids Res. 1987 Sep 25;15(18):7491–7501. doi: 10.1093/nar/15.18.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley J. L., Donelson J. E. Nucleotide sequence of the yeast plasmid. Nature. 1980 Aug 28;286(5776):860–865. doi: 10.1038/286860a0. [DOI] [PubMed] [Google Scholar]
- Hoess R. H., Abremski K. Mechanism of strand cleavage and exchange in the Cre-lox site-specific recombination system. J Mol Biol. 1985 Feb 5;181(3):351–362. doi: 10.1016/0022-2836(85)90224-4. [DOI] [PubMed] [Google Scholar]
- Hoess R. H., Foeller C., Bidwell K., Landy A. Site-specific recombination functions of bacteriophage lambda: DNA sequence of regulatory regions and overlapping structural genes for Int and Xis. Proc Natl Acad Sci U S A. 1980 May;77(5):2482–2486. doi: 10.1073/pnas.77.5.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Härtlein M., Madern D., Leberman R. Cloning and characterization of the gene for Escherichia coli seryl-tRNA synthetase. Nucleic Acids Res. 1987 Feb 11;15(3):1005–1017. doi: 10.1093/nar/15.3.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm P. Two regulatory fim genes, fimB and fimE, control the phase variation of type 1 fimbriae in Escherichia coli. EMBO J. 1986 Jun;5(6):1389–1393. doi: 10.1002/j.1460-2075.1986.tb04372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A., Kusukawa A., Komano T. Nucleotide sequence of the rci gene encoding shufflon-specific DNA recombinase in the IncI1 plasmid R64: homology to the site-specific recombinases of integrase family. Mol Gen Genet. 1988 Jul;213(1):30–35. doi: 10.1007/BF00333394. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lane D., de Feyter R., Kennedy M., Phua S. H., Semon D. D protein of miniF plasmid acts as a repressor of transcription and as a site-specific resolvase. Nucleic Acids Res. 1986 Dec 22;14(24):9713–9728. [PMC free article] [PubMed] [Google Scholar]
- Leong J. M., Nunes-Düby S. E., Oser A. B., Lesser C. F., Youderian P., Susskind M. M., Landy A. Structural and regulatory divergence among site-specific recombination genes of lambdoid phage. J Mol Biol. 1986 Jun 20;189(4):603–616. doi: 10.1016/0022-2836(86)90491-2. [DOI] [PubMed] [Google Scholar]
- Levin J. M., Robson B., Garnier J. An algorithm for secondary structure determination in proteins based on sequence similarity. FEBS Lett. 1986 Sep 15;205(2):303–308. doi: 10.1016/0014-5793(86)80917-6. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Low B. Formation of merodiploids in matings with a class of Rec- recipient strains of Escherichia coli K12. Proc Natl Acad Sci U S A. 1968 May;60(1):160–167. doi: 10.1073/pnas.60.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahillon J., Lereclus D. Structural and functional analysis of Tn4430: identification of an integrase-like protein involved in the co-integrate-resolution process. EMBO J. 1988 May;7(5):1515–1526. doi: 10.1002/j.1460-2075.1988.tb02971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin P., Trieu-Cuot P., Courvalin P. Nucleotide sequence of the tetM tetracycline resistance determinant of the streptococcal conjugative shuttle transposon Tn1545. Nucleic Acids Res. 1986 Sep 11;14(17):7047–7058. doi: 10.1093/nar/14.17.7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin J. R., Murray C. L., Rabinowitz J. C. Unique features in the ribosome binding site sequence of the gram-positive Staphylococcus aureus beta-lactamase gene. J Biol Chem. 1981 Nov 10;256(21):11283–11291. [PubMed] [Google Scholar]
- Mertens G., Klippel A., Fuss H., Blöcker H., Frank R., Kahmann R. Site-specific recombination in bacteriophage Mu: characterization of binding sites for the DNA invertase Gin. EMBO J. 1988 Apr;7(4):1219–1227. doi: 10.1002/j.1460-2075.1988.tb02934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K., Weisberg R., Enquist L., Mizuuchi M., Buraczynska M., Foeller C., Hsu P. L., Ross W., Landy A. Structure and function of the phage lambda att site: size, int-binding sites, and location of the crossover point. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):429–437. doi: 10.1101/sqb.1981.045.01.057. [DOI] [PubMed] [Google Scholar]
- Murphy E., Huwyler L., de Freire Bastos M. do C. Transposon Tn554: complete nucleotide sequence and isolation of transposition-defective and antibiotic-sensitive mutants. EMBO J. 1985 Dec 1;4(12):3357–3365. doi: 10.1002/j.1460-2075.1985.tb04089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pargellis C. A., Nunes-Düby S. E., de Vargas L. M., Landy A. Suicide recombination substrates yield covalent lambda integrase-DNA complexes and lead to identification of the active site tyrosine. J Biol Chem. 1988 Jun 5;263(16):7678–7685. [PubMed] [Google Scholar]
- Plasterk R. H., Brinkman A., van de Putte P. DNA inversions in the chromosome of Escherichia coli and in bacteriophage Mu: relationship to other site-specific recombination systems. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5355–5358. doi: 10.1073/pnas.80.17.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richet E., Abcarian P., Nash H. A. Synapsis of attachment sites during lambda integrative recombination involves capture of a naked DNA by a protein-DNA complex. Cell. 1988 Jan 15;52(1):9–17. doi: 10.1016/0092-8674(88)90526-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J. R., Kirchman P. A., Caparon M. G. An intermediate in transposition of the conjugative transposon Tn916. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4809–4813. doi: 10.1073/pnas.85.13.4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senghas E., Jones J. M., Yamamoto M., Gawron-Burke C., Clewell D. B. Genetic organization of the bacterial conjugative transposon Tn916. J Bacteriol. 1988 Jan;170(1):245–249. doi: 10.1128/jb.170.1.245-249.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. A new computer method for the storage and manipulation of DNA gel reading data. Nucleic Acids Res. 1980 Aug 25;8(16):3673–3694. doi: 10.1093/nar/8.16.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg N., Sauer B., Hoess R., Abremski K. Bacteriophage P1 cre gene and its regulatory region. Evidence for multiple promoters and for regulation by DNA methylation. J Mol Biol. 1986 Jan 20;187(2):197–212. doi: 10.1016/0022-2836(86)90228-7. [DOI] [PubMed] [Google Scholar]
- Takeshita S., Sato M., Toba M., Masahashi W., Hashimoto-Gotoh T. High-copy-number and low-copy-number plasmid vectors for lacZ alpha-complementation and chloramphenicol- or kanamycin-resistance selection. Gene. 1987;61(1):63–74. doi: 10.1016/0378-1119(87)90365-9. [DOI] [PubMed] [Google Scholar]
- Thompson J. F., Moitoso de Vargas L., Koch C., Kahmann R., Landy A. Cellular factors couple recombination with growth phase: characterization of a new component in the lambda site-specific recombination pathway. Cell. 1987 Sep 11;50(6):901–908. doi: 10.1016/0092-8674(87)90516-2. [DOI] [PubMed] [Google Scholar]
- Wilbur W. J., Lipman D. J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci U S A. 1983 Feb;80(3):726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yin S., Bushman W., Landy A. Interaction of the lambda site-specific recombination protein Xis with attachment site DNA. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1040–1044. doi: 10.1073/pnas.82.4.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubay G. The isolation and properties of CAP, the catabolite gene activator. Methods Enzymol. 1980;65(1):856–877. doi: 10.1016/s0076-6879(80)65079-4. [DOI] [PubMed] [Google Scholar]