Summary
Prdm16 is a transcription factor that regulates the thermogenic gene program in brown and beige adipocytes. However, whether Prdm16 is required for the development or physiological function of brown adipose tissue (BAT) in vivo has been unclear. By analyzing mice that selectively lacked Prdm16 in the brown adipose lineage, we found that Prdm16 was dispensable for embryonic BAT development. However, Prdm16 was required in young mice to suppress the expression of white fat-selective genes in BAT through recruitment of the histone methyltransferase Ehmt1. Additionally, Prdm16-deficiency caused a severe adult-onset decline in the thermogenic character of interscapular BAT. This resulted in BAT dysfunction and cold sensitivity but did not predispose the animals to obesity. Interestingly, the loss of brown fat identity due to ablation of Prdm16 was accelerated by concurrent deletion of the closely related Prdm3 gene. Together, these results show that Prdm16 and Prdm3 control postnatal BAT identity and function.
Keywords: Prdm16, Prdm3, brown fat, brown adipose tissue, obesity, Ucp1, Myf5
Introduction
The global rise in obesity and type-2 diabetes has precipitated the need for novel approaches to reduce adiposity. Obesity is caused by prolonged periods of positive energy balance in which energy taken in from food exceeds energy expenditure. Brown and beige adipose cells expend chemical energy in the form of heat and can thus oppose obesity in mice. Higher levels of brown and/or beige adipose activity are also correlated with reduced adiposity in people (Saito et al., 2009).
Brown adipocytes reside in discrete brown adipose tissue (BAT) depots whereas beige adipocytes are intermingled with white adipocytes in white adipose tissue (WAT). Both cell types have a multilocular morphology, large numbers of mitochondria, and express a common set of brown fat (versus white fat)-selective genes such as Uncoupling protein-1 (Ucp1) (Harms and Seale, 2013). Upon activation, Ucp1 dissipates the mitochondrial proton gradient, which results in loss of respiratory control and the production of substantial amounts of heat from the combustion of available substrates (Cannon and Nedergaard, 2004). The heat produced by brown and/or beige fat is necessary for the survival of small mammals in the cold and also reduces fat deposition in animals fed a high fat/low protein diet (Cannon and Nedergaard, 2004; Feldmann et al., 2009; Rothwell and Stock, 1979).
The development of therapies aimed at increasing the amount of brown or beige adipocytes will require a detailed mechanistic understanding of how these cell types are formed. PR (PRD1-BF1-RIZ1 homologous)-domain containing 16 (Prdm16) is a transcriptional co-regulator that has been shown to powerfully regulate the differentiation of brown and beige fat cells (Harms and Seale, 2013). Notably, increased expression of Prdm16 in mouse WAT promotes beige adipocyte development and suppresses metabolic disease (Seale et al., 2011). By contrast, deletion of Prdm16 in adipocytes causes a profound loss of beige adipocyte function in mice, leading to aggravated metabolic disease upon exposure to high fat diet (Cohen et al., 2014). Surprisingly, the deletion of Prdm16 in adipocytes, at relatively late stages of their differentiation, does not affect BAT function (Cohen et al., 2014).
BAT forms during embryonic development before other fat depots, and is an essential source of heat production in neonates. Lineage analyses indicate that brown adipocytes and skeletal myocytes originate from precursor cells in the somite that express Engrailed-1, Pax7 and Myf5 (Atit et al., 2006; Lepper and Fan, 2010; Seale et al., 2008). Previous studies showed that Prdm16 controls a bidirectional cell fate switch between brown fat and skeletal muscle in this somite-derived lineage (Kajimura et al., 2009; Ohno et al., 2013; Seale et al., 2008; Sun et al., 2011; Yin et al., 2013). However, the requirement for Prdm16 in regulating brown adipocyte development and function in vivo had not been thoroughly assessed.
In this study, we used the Myf5Cre mouse strain to delete Prdm16 in the progenitors for brown adipocytes and muscle but not beige adipose cells. BAT developed normally in the absence of Prdm16, without evidence of a cell fate switch into muscle. In BAT from young mice, Prdm16-deficiency had little effect on the expression of key BAT-selective genes but elicited a dramatic rise in expression of many WAT-selective genes. As the knockout animals aged, however, there was a striking loss of thermogenic character in the interscapular BAT (iBAT). This collapse of iBAT identity was accelerated by concurrent deletion of the closely related Prdm3 gene. Importantly, adult mice with Prdm16-depleted BAT had severely reduced BAT function but did not gain more weight than wildtype animals. Altogether, our results indicate that Prdm16 and Prdm3 play a critical role in the postnatal maintenance and function of BAT.
Results
Prdm16 is dispensable for embryonic BAT development
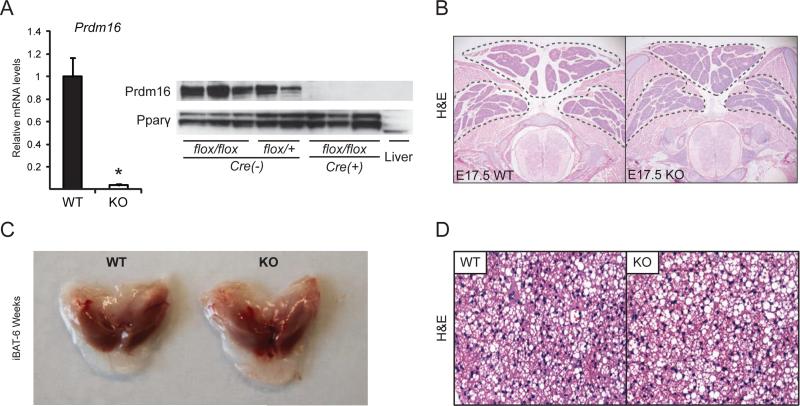
To investigate the genetic requirement for Prdm16 in BAT development and function, we deleted Prdm16 in the brown fat lineage by intercrossing Myf5Cre mice (Tallquist et al., 2000) with Prdm16flox/flox mice (Figure S1A). Myf5Cre is activated in the somitic precursors that give rise to brown adipocytes. The resulting Myf5Cre/+;Prdm16flox/flox (Myf5-ΔPrdm16) mice were born in normal Mendelian ratios and were grossly indistinguishable from their wildtype (WT) littermates. Prdm16 mRNA and protein expression were almost completely ablated in Myf5-ΔPrdm16 BAT (Figure 1A). Surprisingly, there were no differences in the morphology or size of BAT depots between WT and Myf5-ΔPrdm16 mice at E17.5 of development (Figure 1B). At 6 weeks of age, WT and Prdm16-deficient iBAT depots were also grossly and histologically similar (Figures 1C, D). The other major site of Myf5Cre-mediated DNA recombination during development is skeletal muscle where Prdm16 mRNA is not normally detected (Seale et al., 2007). Consistent with this, Prdm16 mRNA levels were equivalent in WT and Myf5-ΔPrdm16 muscles (Figure S1B). Previous studies indicated that Prdm16 can suppress the expression of certain muscle-specific genes (Seale et al., 2008); however, we did not observe elevated expression of muscle-specific genes in the iBAT of Myf5-ΔPrdm16 mice (Figure S1C). Taken together, these results indicate that Prdm16 is dispensable for BAT formation in mice.
Figure 1. Prdm16 is dispensable for embryonic BAT development.
(A) Prdm16 mRNA and protein levels from wildtype (WT) and Myf5ΔPrdm16 (KO) mice (mean ± SEM; n=5, 8 (WT, KO); *p<0.05). (B) Hematoxylin and eosin (H&E) staining of representative sections from the interscapular regions of E17.5 WT and KO embryos. (C) Interscapular brown adipose tissue (iBAT) from 6-week-old WT and KO mice. (D) H&E staining of representative sections from the iBAT of 6-week-old WT and KO mice.
Prdm16 recruits Ehmt1 to repress the expression of white fat-selective genes
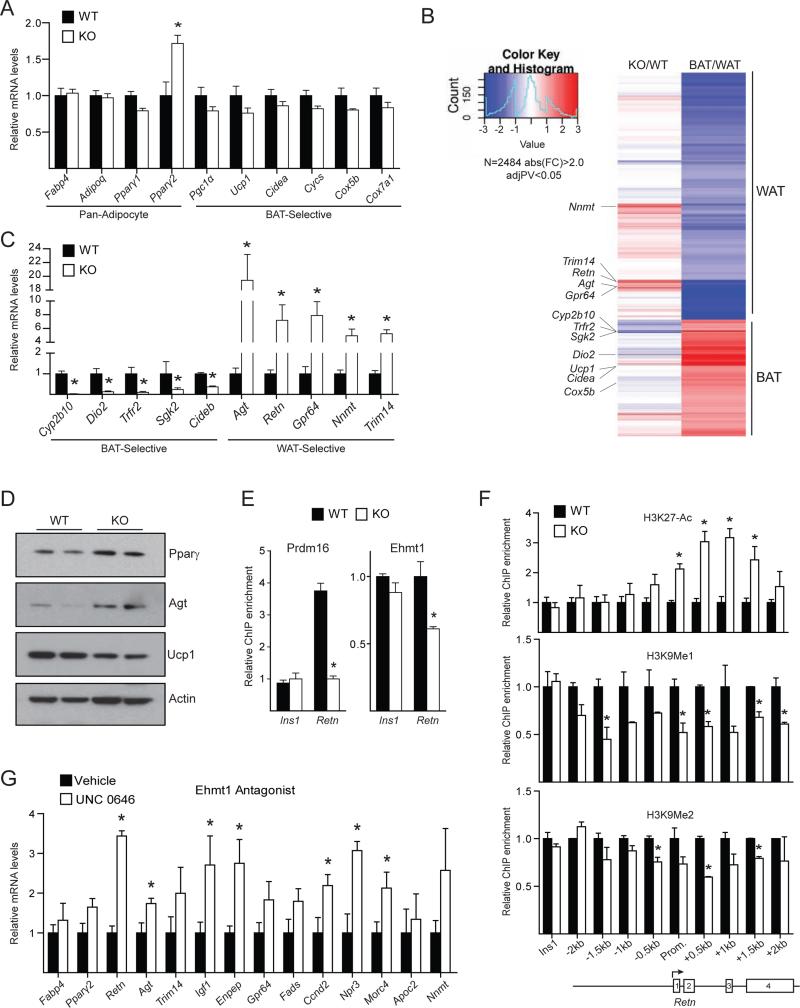
We next analyzed the molecular phenotype of iBAT from 6-week-old WT and Myf5-ΔPrdm16 mice. Pan-adipocyte genes like Fabp4 and Adipoq were equivalently expressed in WT and Prdm16 knock-out (KO) iBAT (Figure 2A), although KO tissue expressed higher levels ofPparγ2, the adipose-selective isoform of Pparγ. The levels of brown fat-selective (Pgc1α, Ucp1 and Cidea) and mitochondrial (Cycs, Cox5b, Cox7a1) genes were mildly but not significantly decreased in Prdm16 KO tissue (Figure 2A).
Figure 2. Prdm16 represses the expression of white fat-selective genes.
(A) mRNA levels of pan-adipocyte and BAT-selective genes in BAT from 6-week-old wildtype (WT) and Myf5ΔPrdm16 (KO) mice (mean ± SEM, WT n=5, KO n=4, *p<0.05). (B) Heat map depicting the mRNA levels of white and brown fat-selective genes (WAT/BAT) in BAT from 6-week-old WT and KO mice (KO/WT) (n=4). (C) qPCR validation of BAT- and WAT-selective mRNAs identified in (B) from WT and KO BAT (mean ± SEM, WT n=5, KO n=4, *p<0.05. (D) Western blot analysis of Pparγ, Agt, Ucp1 and Actin (loading control) protein levels in BAT from 6-week-old WT and KO mice. (E) ChIP-qPCR analysis of endogenous Prdm16 and Ehmt1 binding at the Retn promoter (mean ± SEM; n=3; *p<0.05). (F) ChIP-qPCR analysis of H3K27-Ac, H3K9-Me1 and H3K9-Me2 enrichment in a 4 kb region spanning the transcriptional start site of Retn. Ins1 was used a non-specific control binding site (mean ± SEM; n=3; *p<0.05). (G) mRNA levels of WAT-selective genes in brown adipocytes treated with Ehmt1 antagonist UNC 0646, or vehicle control (mean ± stdev; n=3; *p<0.05).
To search for genes/pathways that were sensitive to Prdm16 levels in BAT, we compared the global gene expression profiles of iBAT from 6-week-old WT and Myf5-ΔPrdm16 mice using cDNA microarrays. Gene ontology analysis revealed that genes involved in lipid-metabolism, including “lipid biosynthetic process” and “lipid metabolic process”, were increased in the absence of Prdm16 (Figure S2A). This suggested that loss of Prdm16 shifted adipocyte metabolism to favor a white fat-like energy storage phenotype. We thus specifically analyzed the impact of Prdm16-deficiency on the complete set of BAT- and WAT-selective genes (Figure 2B). Consistent with qPCR analysis, most typical brown-selective genes (e.g. Ucp1, Cidea, Cox5b) were only slightly reduced in Prdm16-deficient BAT. However, the expression of a few BAT-selective genes were dramatically diminished in KO BAT, including Dio2 (deiodinase, iodothyronine, type II), an important regulator of brown adipocyte function (de Jesus et al., 2001) (Figure 2B, C). In line with the mRNA data, western blots showed that Prdm16-deficient BAT expressed higher levels of Pparγ and Agt and slightly reduced levels of Ucp1 protein (Figure 2D). Additionally, the global expression analyses revealed a broad increase in the expression of white fat-selective genes in KO BAT (Figure 2B). Real-time PCR analysis validated the increased expression of many of these genes, including a 20-fold increase in Agt, and 6- to 8-fold increases in Retn, Gpr64, Nnmt and Trim14 (Figure 2C). These results reveal that Prdm16 is required in BAT to suppress the expression of many white fat-selective genes.
We used Retn as a model locus to investigate the mechanism by which Prdm16 represses white fat-selective genes. Chromatin immunoprecipitation (ChIP) for Prdm16 in WT and KO BAT showed that it was specifically enriched at the Retn promoter relative to non-specific control sites (Figure 2E). The repressive chromatin modifier, Ehmt1 (G9a-like protein), an interacting partner of Prdm16 (Kajimura et al., 2009; Ohno et al., 2013), was also bound to the Retn promoter in BAT and its binding there was reduced by ~40% in Prdm16 KO BAT relative to WT BAT (Figure 2E). Importantly, the loss of Prdm16 and Ehmt1 binding at Retn was associated with increased levels of H3K27-Ac, a histone mark correlated with active transcription; and decreased levels of H3K9-Me1 and H3K9-Me2, modifications laid down by Ehmt1 and associated with gene repression (Figure 2F). Ehmt1 binding at the Agt promoter was also significantly decreased in Myf5-ΔPrdm16 BAT (Figure S2B). These data suggest that Prdm16 recruits Ehmt1 to certain white fat-selective genes to decrease their transcription. In accordance with this, treatment of cultured brown adipocytes with UNC 0646, an Ehmt1 antagonist, increased the expression of many white fat-selective genes, including Retn and Agt (Figure 2G).
Prdm16 maintains iBAT identity during aging
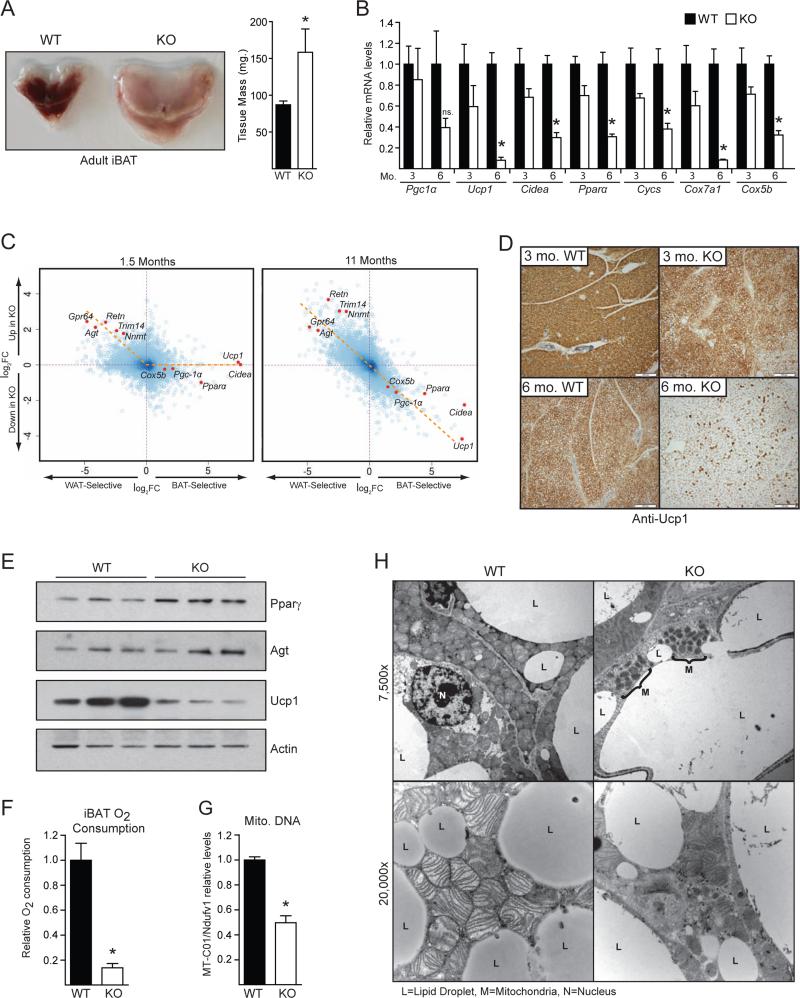
In contrast to juvenile mice, we noticed that older (>6 months of age) Myf5-ΔPrdm16 animals exhibited a profound morphological “whitening” of their iBAT. This included a ~50% increase in the size of the tissue, a switch from multilocular to unilocular morphology, and increased lipid content (Figures 3A, S3A). To determine when this phenotype emerged, we analyzed gene expression in iBAT from WT and Myf5-ΔPrdm16 mice at 3 and 6 months of age. In 3-month-old animals, Prdm16-deficiency resulted in a modest decline in the expression of brown fat-selective genes, including ~30-40% reductions in the levels of Ucp1, Pparα and many mitochondrial genes (Figure 3B). The reduction of brown fat-specific gene expression in KO BAT was much more pronounced in 6-month-old mice. At that age, Ucp1 mRNA levels were decreased by >90% and Cidea and Pparα levels were reduced by ~70% (Figure 3B). Hematoxylin and eosin staining showed that lipid droplet size increased dramatically in the KO BAT from 3 to 6 months of age (Figure S3B). Microarray analyses revealed that Myf5-ΔPrdm16 iBAT from 11-month-old mice expressed substantially reduced levels of a broad brown fat-selective program (Figure 3C). The elevation of white fat-selective gene expression caused by Prdm16-deficiency was further exaggerated in older mice (Figure 3C). Immunohistochemical analysis showed that Ucp1 protein levels decreased dramatically in the iBAT of Myf5-ΔPrdm16 animals between 3 and 6 months of age (Figure 3D). Western blot analysis confirmed that Ucp1 protein levels were much lower in KO relative to WT iBAT at 6 months of age, coincident with elevated levels of Pparγ and Agt (Figure 3E). DNA isolated from the iBAT of 11-month-old Myf5-ΔPrdm16 mice was >90% recombined at the Prdm16 locus (ie. exon 9-deleted) (Figure S3C), indicating that the “whitened” KO tissue was composed of Myf5Cre lineage-derived adipocytes.
Figure 3. Prdm16 is required for the maintenance of iBAT fate in adult animals.
(A) Gross morphology and mass of iBAT depots from one-year-old wildtype (WT) andMyf5ΔPrdm16 (KO) mice (mean ± SEM, n=7, *p<0.05). (B) mRNA levels of BAT-selective genes in BAT from 3- and 6-month-old WT and KO mice (mean ± SEM, n=4, *p<0.05). (C) Global analysis of BAT- and WAT-selective mRNA levels in BAT from 6-week-old and 11-month-old mice. Dashed orange line illustrates the change in gene expression pattern. Data is presented as a log2FC scatter plot. (n=4). (D) Immunohistochemical staining for Ucp1 protein in sections of BAT from 3- and 6-month-old WT and KO mice. (E) Western blot analysis of Pparγ, Agt, Ucp1 and Actin (loading control) protein levels in BAT from 11-month-old WT and KO mice. (F) Oxygen consumption of isolated and minced BAT from 9-month-old WT and KO mice. Data is presented as nmol of oxygen consumed per minute per mg of tissue (mean ± SEM, WT n=4, KO n=3, *p<0.05). (G) Mitochondrial DNA levels in BAT from 9-month-old WT and KO mice (mean ± SEM, WT n=4, KO n=3, *p<0.05. (H) Transmission electron micrographs of BAT from 9-month-old WT and KO mice (L=lipid droplet; M=mitochondria; N=nucleus).
We next examined the consequences of Prdm16-deficiency on mitochondrial mass and function. Using a Clark electrode, we measured oxygen consumption in isolated iBAT from 9-month-old WT and Myf5-ΔPrdm16 mice. Remarkably, basal (unstimulated) respiration in the KO iBAT was ~85% lower than in WT tissue (Figure 3F), indicative of a loss of mitochondrial mass. Indeed, PCR analysis revealed that KO BAT had 50% less mitochondrial DNA than WT tissue (Figure 3G). Transmission electron microscopy studies showed that WT adipocytes were packed with mitochondria containing well-ordered cristae, whereas KO adipocytes had a paucity of mitochondria, of which the remainder had poorly organized cristae and exhibited signs of degeneration (Figure 3H). Taken together, these data establish an important role for Prdm16 in maintaining brown adipocyte identity (including high levels of mitochondria) in adult mice.
The interscapular depot is the largest BAT depot in adult mice and was the focus of our study. However, we also investigated the impact of Prdm16-deficiency on the axillary and cervical BAT (aBAT and cBAT). The aBAT and cBAT depots in 6-month-old Myf5-ΔPrdm16 appeared paler than those in WT mice, but there was no difference in aBAT or cBAT mass between WT and Myf5-ΔPrdm16 mice (Figure S3D). Interestingly, the white fat-selective genes were markedly elevated in Prdm16-deficient aBAT and cBAT but brown fat-specific gene levels were not affected (Figure S3E). These results suggest that interscapular BAT is particularly reliant on Prdm16 for maintaining the expression of brown fat-selective genes during aging.
Prdm16 is required for induction of the brown fat gene program in isolated precursors
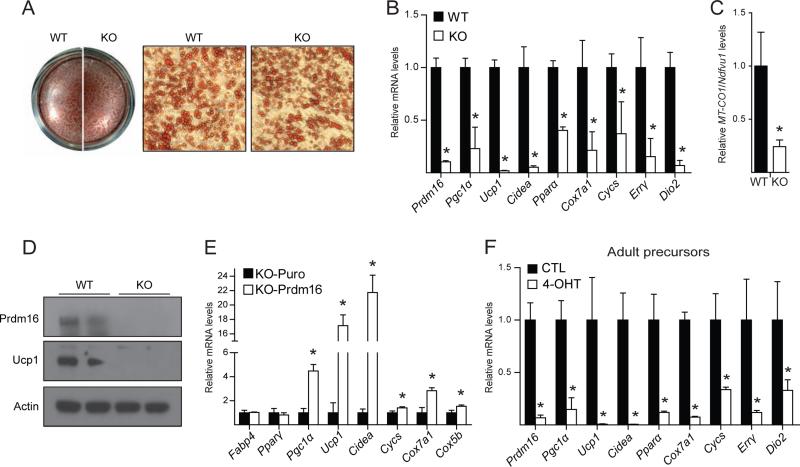
The aging-associated decline of iBAT identity in Myf5-ΔPrdm16 mice raised the question of whether Prdm16 was required cell autonomously for proper brown adipocyte differentiation in this depot. To study this, we isolated primary brown adipocyte precursors from the iBAT of WT and Myf5-ΔPrdm16 mice and examined their differentiation into adipocytes under defined culture conditions. WT and KO precursor cells from newborn iBAT differentiated with equivalent efficiently into mature lipid droplet-containing adipocytes that expressed similar levels of panadipocyte genes, including Fabp4 and Adipoq (Figure 4A, S4A). KO cultures displayed a >90% reduction in Prdm16 mRNA and protein levels (Figure 4B, D), indicating that most or all of the precursor cells in BAT descend from Myf5Cre-expressing cells. Strikingly, Prdm16-deficient cultures expressed dramatically lower levels of brown adipocyte-specific genes as compared to WT cultures, including 90-95% reductions in the mRNA levels of Ucp1, Cidea and Dio2; and 60-80% decreases in Pgc1α, Pparα, Cox7a1, Cycs, and Errγ (Figure 4B). WT adipocytes also had four times more mitochondrial DNA than KO cells (Figure 4C). Western blot analysis showed that Ucp1 protein, like its mRNA, was dramatically lower in KO adipocytes (Figure 4D). Importantly, retroviral expression of Prdm16 in KO preadipocytes efficiently activated the expression of thermogenic genes like Ucp1 and Cidea (Figure 4E), indicating that the KO cells were competent to induce the brown-selective gene program. Immortalized brown fat cell lines from newborn BAT of Myf5-ΔPrdm16 mice displayed a similarly severe defect in activating the differentiation-linked brown fat-specific gene program (Figure S4B).
Figure 4. Prdm16 is required for activation of the brown fat-selective gene program in cultured brown fat precursors.
(A,B) Primary precursor cells from the iBAT of newborn wildtype (WT) and Myf5ΔPrdm16 (KO) mice induced to differentiate into adipocytes and stained with Oil-red-o (A) or analyzed by qPCR for their expression of brown fat-selective genes (mean ± stdev, n=3, *p<0.05) (B). (C) Mitochondrial DNA levels in WT and KO adipocytes from immortalized brown fat-derived precursor cells (mean ± SEM, n=6, *p<0.05). (D) Western blot analysis of Prdm16, Ucp1 and Actin (loading control) in WT and KO adipocytes from immortalized brown fat-derived precursor cells. (E) Primary precursor cells from iBAT of newborn KO mice infected with puromycin (KO-puro; control) or Prdm16 (KO-Prdm16) retrovirus, were induced to differentiate into adipocytes. Gene expression analysis for general adipocyte (Fabp4, Pparγ) and brown fat-selective genes (mean ± stdev, n=3, *p<0.05). (F) Primary precursor cells from the iBAT of adult Rosa26Cre/+;Prdm16flox/flox mice were treated with 4-hydroxytamoxifen (4-OHT) to delete Prdm16 or vehicle (ethanol) (CTL) before being induced to undergo adipocyte differentiation. Adipocyte cultures were analyzed for their expression of brown fat-selective genes (mean ± stdev, n=3, *p<0.05).
We also tested whether acute Prdm16 deletion affected the brown fat differentiation program of preadipocytes isolated from adult animals. To this end, brown fat precursor cells were isolated from 8-week-old Prdm16flox/flox;Rosa26CreERT mice that ubiquitously express a tamoxifen-inducible Cre recombinase. Precursor cells from the iBAT of these mice were isolated and then treated with 4-hydroxytamoxifen (4-OHT) or vehicle (ethanol). The acute loss of Prdm16 caused by 4-OHT treatment completely blocked the differentiation-linked induction of brown-selective genes (including Ucp1 and Cidea) while leaving the general adipogenic program intact (Figure 4F).
The brown-specific gene program was not significantly affected by loss of Prdm16 in BAT from embryos and young mice. This raised the possibility that the embryonic precursors may not require Prdm16 to execute a normal brown fat differentiation program. To investigate this, we purified brown adipocyte precursors from the dorsal body walls of WT and Myf5-ΔPrdm16 mouse embryos at E14.5 days post-coitum - a stage when differentiated brown adipocytes are first appearing. Precursor cells were purified by flow cytometry based on cell-surface expression of platelet-derived growth factor receptor alpha (Pdgfrα), which enriches for adipogenic precursors in BAT (Wang and Seale, in preparation) and other tissues (Berry and Rodeheffer, 2013; Lee et al., 2012; Uezumi et al., 2010). The Prdm16-deficient embryonic cells, like the cells from newborn and adult BAT, displayed an ex vivo deficit in brown fat-specific gene expression during adipogenesis (Figure S4C). Taken together, these data demonstrate that Prdm16 is required to activate the brown-specific adipogenic program in isolated BAT precursors.
Reduced BAT function in Myf5-ΔPrdm16 mice
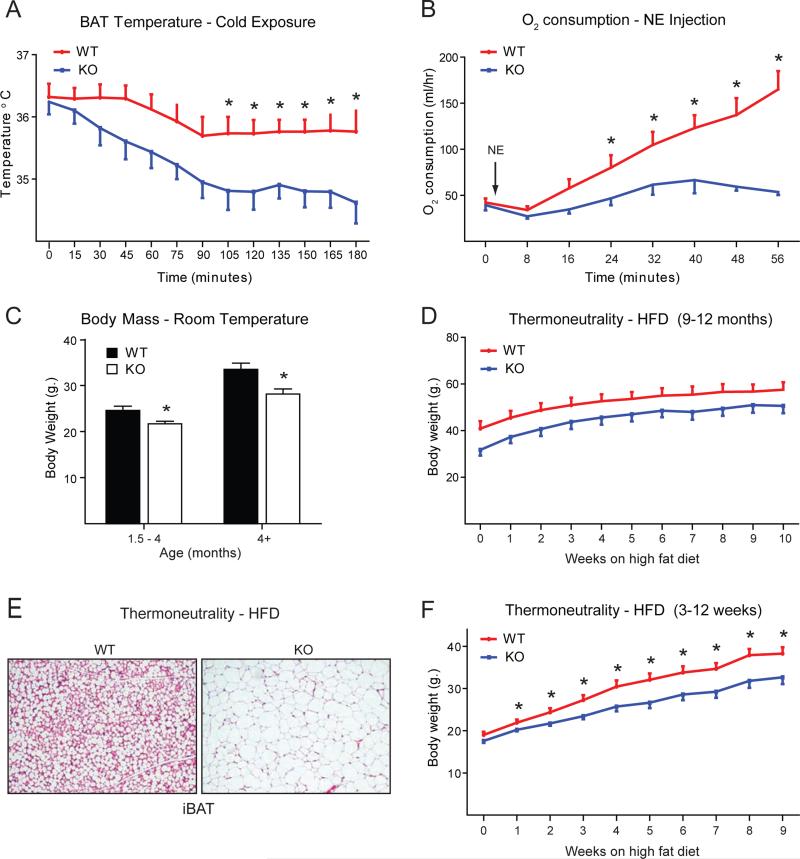
A key question is whether the loss of Prdm16 in BAT has physiological consequences for the animals. To assess BAT function in mice, we surgically implanted temperature probes subcutaneously in the interscapular region of 10-month-old WT and Myf5-ΔPrdm16 mice. After allowing the animals to recover for one week, we exposed them to cold (4°C) and monitored tissue temperature over a period of 3 hours. Under room temperature conditions (at T=0 prior to cold exposure), WT and Myf5-ΔPrdm16 mice had similar core and interscapular temperatures (Figure 5A; S5A). However, upon cold exposure, tissue temperature dropped precipitously in Myf5-ΔPrdm16 mice, declining >1°C more than in WT animals after 3 hours (Figure 5A). Since mice rely substantially on shivering thermogenesis during acute cold exposure (Cannon and Nedergaard, 2011), we further analyzed in vivo BAT function by measuring whole-animal oxygen consumption before and at several time points after injection of norepinephrine (NE), the classic activator of BAT-mediated thermogenesis. As evidenced through the study of Ucp1 KO animals, this method provides a more stringent measurement of BAT activity (Cannon and Nedergaard, 2011; Golozoubova et al., 2006). In WT mice, oxygen consumption increased by ~4-fold in response to NE (Figure 5B). By contrast, NE only marginally raised the oxygen consumption of Myf5-ΔPrdm16 mice, which diverged significantly from than that of WT mice by 24 minutes post-NE treatment (Figure 5B). Taken together, these data demonstrate that Prdm16 is required for the thermogenic function of BAT in vivo.
Figure 5. Myf5-ΔPrdm16 mice have severely deficient BAT function but are not prone to obesity or metabolic disease.
(A) Temperature recordings from probes implanted into the interscapular (subcutaneous) region of 1-year-old WT and Myf5ΔPrdm16 (KO) mice. Data was collected over 3 hours after moving animals from room temperature (~22°C) to 4°C (T=0) (mean ± SEM, n=10, *p<0.05). (B) Whole-body oxygen consumption in 1-year-old WT and KO mice before and after treatment with 1 mg/kg norepinephrine (NE) (mean ± SEM, n=10, *p<0.05). (C) Body weights of WT and KO mice at different ages maintained under standard housing conditions and fed a regular chow diet (mean ± SEM, n=10-22, *p<0.05). (D) Body weights of 9-month-old WT and KO mice that were housed at 28°C and fed a high fat diet (HFD) for ten weeks (mean ± SEM, n=5, *p<0.05). (E) Hematoxylin and eosin (HE) staining of sections from the interscapular BAT of 3-week-old mice housed at 28°C on HFD. (F) Body weights of WT and KO mice that were kept at 28°C and fed a high fat diet for 9 weeks starting at weaning (3-weeks-old) (mean ± SEM, WT n=6, KO n=8, *p<0.05).
The reduced thermogenic capacity of Myf5-ΔPrdm16 mice suggested that these animals may be susceptible to weight gain and metabolic disease. However, contrary to our expectation, Myf5-ΔPrdm16 mice weighed less than their WT counterparts at all ages studied (Figure 5C). MRI examination revealed that Myf5-ΔPrdm16 mice had less lean and fat mass than WT mice (Figure S5B). Unexpectedly, Myf5-ΔPrdm16 mice were also slightly, but significantly shorter than WT animals (Figure S5C) – this likely explains their proportional reduction in lean and fat mass. Consistent with their reduced fat mass, Myf5-ΔPrdm16 mice also had improved glucose tolerance as compared to WT mice (Figure S5D).
The contribution of BAT thermogenesis to energy balance can be masked at room temperature in mice due to the cold-mediated activation of other thermogenic pathways (Cannon and Nedergaard, 2011; Feldmann et al., 2009). We therefore placed 9-month-old WT and Myf5-ΔPrdm16 mice at 28°C to exempt them from cold stress, and fed them a high fat diet for 10 weeks. Under these conditions, the weight gained by WT and Myf5-ΔPrdm16 mice was remarkably similar (Figure 5D). We also placed 3-week-old WT and Myf5-ΔPrdm16 mice on a high fat diet and housed them at 28°C for 10 weeks. Under these conditions, Myf5-ΔPrdm16 iBAT developed a severe loss of normal brown adipocyte morphology (Figure 5E) and reduced levels of brown fat-specific genes (Figure S5E). Despite this, the KO mice gained less weight than WT mice (Figure 5F), although the percent weight gained by WT and KO mice per week and overall was very similar. Consistent with this, oxygen consumption (Figure S5F) and food intake (Figure S5G) was not significantly different between WT and KO mice.
The loss of BAT function in adult Myf5-ΔPrdm16 mice was not accompanied by increased browning of WAT depots. Specifically, there was: (1) no increase in the expression levels of Ucp1 or other brown fat-selective gene markers in the inguinal or epididymal WAT of Myf5-ΔPrdm16 KO mice (Figure S5H,I); and (2) no difference between the morphology of WT and KO WAT in mice housed at 22°C (not shown) or 28°C (Figure S5J). Altogether, these data show that loss of Prdm16 activity in BAT does not impair diet-induced thermogenesis in mice.
Prdm3 compensates for the loss of Prdm16 to preserve brown fat fate in young mice
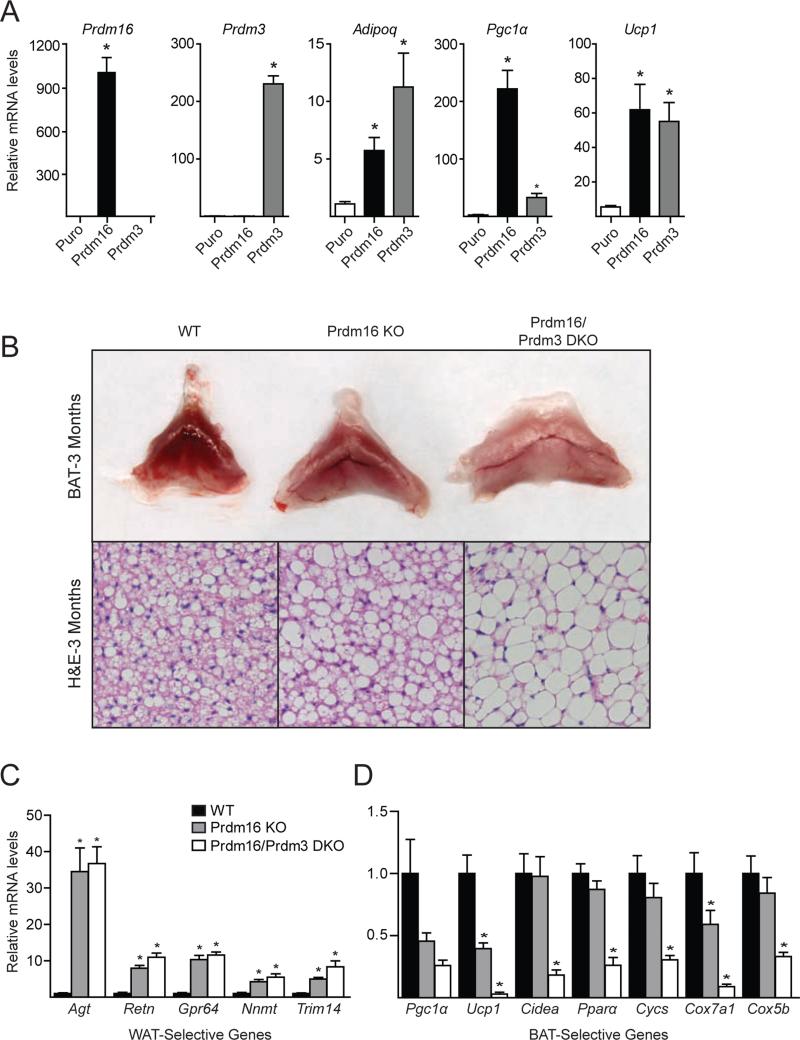
Prdm16 is most closely related in sequence and structure to Prdm3 (also called Mecom [Mds1 and Evi1 complex locus]) (Hohenauer and Moore, 2012). We previously reported that Prdm3 regulates white adipocyte differentiation through its association with c/EBPβ (Ishibashi et al., 2012). To test whether Prdm3 could induce brown fat-selective genes, we used retrovirus to ectopically express Prdm16, Prdm3 (Evi1 isoform), or empty vector (MSCV-Puro) in C2C12 cells (Figure 6A). In this context, both Prdm16 and Prdm3 robustly induced adipocyte differentiation and the expression of Adipoq, a general adipocyte marker (Figure 6A). Prdm16 and Prdm3 also potently activated Ucp1 and Pgc1α expression (Figure 6A). Interestingly, we also noted that Prdm3 expression in iBAT was higher in E18 embryos compared to 1.5-, 3- and 6-month-old mice (Figure S6A). Prdm3 mRNA levels were also somewhat reduced in 6-month-old BAT relative to 3-month-old BAT. Together, these data suggested that Prdm3 might be compensating for the loss Prdm16 during brown fat development.
Figure 6. Prdm3 compensates for the loss of Prdm16 in juvenile BAT.
(A) C2C12 muscle cells were transduced with retrovirus expressing puromycin (ctl), Prdm3 or Prdm16 and induced to undergo adipocyte differentiation. Cultures were then analyzed by qPCR for their expression levels of Adipoq (adipocyte marker) and brown fat-selective genes (Pgc1α, Ucp1). (B) Gross appearance (top) and hematoxylin/eosin (HE) staining (bottom) of interscapular BAT from 3-month-old WT, Myf5-ΔPrdm16 (KO) and Myf5-ΔPrdm16/Prdm3 (DKO) mice. (C,D) Expression analysis of WAT-selective (B) and BAT-selective (C) transcripts in WT, KO, and DKO BAT (mean ± SEM, n=5-7, *p < 0.01).
To explore this possibility, we created mice lacking both Prdm16 and Prdm3 in the brown fat lineage by intercrossing Myf5-ΔPrdm16 mice with Prdm3flox mice (Goyama et al., 2008). At 3 months of age, the iBAT of Myf5-ΔPrdm16/Prdm3 double knockout (DKO) mice was visibly paler than Prdm16-KO, Prdm3-KO or WT BAT (Figure 6B, S6B). H&E staining of iBAT sections revealed that DKO adipocytes had larger lipid droplets than adipocytes in WT or Prdm16 KO tissue (Figure 6B). Gene expression analysis showed that white fat-selective genes were increased to a similar extent in KO and DKO iBAT relative to their levels in WT iBAT (Figure 6C). However, the levels of brown fat-selective genes (including Pgc1α, Ucp1, Cidea, and Pparα) were dramatically reduced in DKO BAT at this age while only modestly reduced in Prdm16 KO BAT relative to WT controls. We did not detect any changes in the expression of brown or white genes in Prdm3 KO relative to WT iBAT (Figure S6C). Skeletal muscle-enriched genes were not significantly increased in DKO relative to WT BAT (Figure S6D). Notably, at two weeks of age, WT and DKO iBAT expressed nearly equivalent levels of most brown fat-selective genes, including Ucp1 (Figure S6D). These results indicate that, while both factors are dispensable for establishing a BAT-specific gene program during embryonic development, Prdm16 or Prdm3 is required for the postnatal/adult expansion and maintenance of iBAT.
Discussion
A detailed understanding of the mechanisms that control brown adipocyte development may reveal new approaches to reduce obesity and associated disorders. Prdm16, a zinc-finger containing transcription factor, is a key regulator of brown fat cell development and function. Using conditional knockout mice, we found that Prdm16 is required for suppressing a white fat-selective gene profile in all BAT depots and for maintaining the brown fat-specific attributes of iBAT in adult animals. As a result, animals lacking Prdm16 in BAT have a dramatically reduced capacity to produce heat.
The major BAT depots in mice arise from a multipotent cell population in the somites that expresses Myf5, Pax7 and En1 at early stages of development (Atit et al., 2006; Lepper and Fan, 2010; Seale et al., 2008). Surprisingly, deletion of Prdm16 in this early embryonic precursor pool caused an adult-onset loss of brown fat-specific characteristics in the interscapular depot, with little effect on the embryonic and early postnatal tissue. BAT is a critical source of heat in eutherian mammals and, as such, is required for the survival of newborn and young mice in the cold. Consequently, redundant mechanisms have likely evolved to safeguard early BAT development.
Prdm16 is most closely related in sequence and structure to Prdm3. Notably, Prdm3, like Prdm16, activates the expression of brown fat-selective genes when expressed in C2C12 muscle cells (Figure 6). Importantly, simultaneous loss of both Prdm16 and Prdm3 in the Myf5Cre-descendent brown fat lineage caused a profound loss of iBAT fate in young mice, much earlier and more completely than observed with Prdm16-deficiency alone (Figure 6). Loss of Prdm3 alone had no detectable impact on BAT development, suggesting that Prdm3 has no non-redundant functions in BAT. Altogether, our results suggest that Prdm16 or Prdm3 is required for the function of brown fat stem/precursor cells that mediate tissue expansion after birth. In other words, as BAT grows and undergoes turnover postnatally, the differentiation of new brown adipocytes requires the presence of Prdm3 or Prdm16. The distinctive requirement for Prdm16 in iBAT maintenance, which is revealed as animals get older, correlates with a decline in Prdm3 levels (Figure S6). Brown fat precursor cells also appear to lose Prdm3 expression after their isolation into culture (not shown), which could explain why Prdm16-deficient brown fat precursor cells display such a profound differentiation deficit ex vivo (Figure 4).
The precursor cells in the embryo that give rise to brown adipocytes and skeletal muscle are developmentally related (Atit et al., 2006; Lepper and Fan, 2010; Seale et al., 2008). Interestingly, Prdm16 and several other factors have been shown to regulate a muscle/brown fat cell fate switch (Kajimura et al., 2009; Seale et al., 2008; Derecka et al., 2012; Ohno et al., 2013; Park et al., 2013; Sun et al., 2011; Yin et al., 2013). In particular, ectopic expression of Prdm16 in myogenic precursor cells promotes brown fat differentiation (Kajimura et al., 2009; Seale et al., 2008; Yin et al., 2013), whereas knockdown of Prdm16 in brown preadipose cells increases muscle differentiation (Seale et al., 2008). Consistent with this, loss of the Ehmt1, a histone methyltransferase that mediates Prdm16 action in BAT, leads to the ectopic expression of a broad set of muscle-specific genes in iBAT (Ohno et al., 2013).
Unexpectedly, we found that deletion of Prdm16 and Prdm3 in the brown adipose lineage did not elevate muscle-specific gene expression. These results diverge from those of earlier studies in demonstrating that Prdm16 is, in fact, not required in brown fat cells to repress the expression of muscle genes. The increased expression of muscle genes observed in other models of Prdm16-depleted brown adipocytes may be related to differences in mouse strain used, off-target effects of the shRNA, non-brown fat cell-autonomous functions of Prdm16 or variable levels of muscle contamination during dissection. While it will be important to investigate these possible explanations, our current findings using cell-type selective genetic ablation in vivo provide compelling evidence that Prdm16 and Prdm3 are dispensable for the determination of brown fat versus muscle cell identity. Given this, we speculate that there is additional redundancy in the Prdm16/Prdm3 pathway that activates the brown-selective program whilst suppressing skeletal muscle gene expression during the first wave of brown fat differentiation in the embryo. Future studies should thus consider whether another Prdm family member or functionally related protein can participate in the same complexes as Prdm16/Prdm3 to recruit Ehmt1 during embryonic BAT development.
Prdm16 is uniquely required at all stages of BAT development to repress the transcription of many WAT (versus BAT)-selective genes. Deletion of Prdm16, but not Prdm3, caused a large increase in the expression of these genes in all the BAT depots we examined and at all ages. It makes sense that Prdm3 lacks this repressive function since it is also expressed in all WAT depots (Ishibashi et al., 2012). Our data suggest that Prdm16 represses the transcription of “white” genes (eg. Retn, Agt) by binding to their promoters and recruiting Ehmt1 to cause histone methylation (Figure 2). The consequences of having these genes over-expressed in BAT remain unknown.
A key question was whether the selective loss of Prdm16 activity in BAT had physiological consequences for the animals. Indeed, we found that Myf5-ΔPrdm16 mice had a severely crippled thermogenic response to norepinephrine, the dominant physiological activator of BAT. Despite this, Myf5-ΔPrdm16 mice did not gain more weight than their WT counterparts under a variety of conditions. By contrast, mice lacking Prdm16 in all adipose cells (AdipoQ-ΔPrdm16) display a selective loss of beige fat activity, gain more weight and become much more severely insulin resistant than WT mice (Cohen et al., 2014). Taken together, these results suggest that beige fat cells play a larger role than BAT in high fat diet-induced thermogenesis and in modulating systemic insulin sensitivity.
However, it is important to consider that alternative thermogenic mechanisms may be recruited in Myf5-ΔPrdm16 animals to compensate for their diminished iBAT function. While there was no compensatory browning of WAT in Myf5-ΔPrdm16 mice, future studies should examine whether adaptive decreases in skeletal muscle efficiency could compensate for loss of BAT function as has been observed in other models of BAT-insufficiency (Bal et al., 2012). Additionally, it is conceivable that BAT depots which express normal levels of Ucp1 in the absence of Prdm16 are recruited under the influence of high fat diet to compensate for defective iBAT function. In this regard, surgical removal of iBAT has been shown to promote expansion of the remaining BAT depots in animals exposed to a low protein diet (Connolly et al., 1982; Rothwell and Stock, 1989). On the other hand, the aBAT and cBAT in adult Myf5-ΔPrdm16 mice is pale and expresses high levels of white fat-specific genes, so it is not clear that these depots are functional even though they have normal amounts of Ucp1.
The body weight and metabolic studies led us to discover that Myf5-ΔPrdm16 mice are smaller than controls, involving proportionate decreases in fat mass, lean mass, and body length. We do not know if the reduced stature of Myf5-ΔPrdm16 mice is secondary to the BAT defects. Interestingly, BAT has been suggested to affect bone development and metabolism (Motyl and Rosen, 2011); the relevance of this will require further study. It is also incumbent upon us to note that Myf5Cre activity traces to tissues other than BAT, including muscle (where Prdm16 is not expressed) and also regions in the developing brain (Daubas et al., 2009; Tajbakhsh and Buckingham, 1995). Prdm16 may therefore regulate stature directly through its expression in non-adipose tissues.
In conclusion, our analyses of Myf5-ΔPrdm16 and -ΔPrdm16/Prdm3 mice have revealed that Prdm16 and Prdm3 control the postnatal growth and maintenance of BAT. Moreover, genetic loss of iBAT activity in adult mice is not necessarily associated with obesity or metabolic disease. However, ectopically increasing BAT function remains likely to have therapeutic value, and our observation that Prdm16 and Prdm3 maintain brown fat identity will be important for designing persistent thermogenesis-based treatments.
Materials and Methods
Animals
Myf5Cre (stock no. 010529) and Rosa26Cre (stock no. 004847) mice were obtained from the Jackson Laboratory. Prdm16flox (Figure S1A) and Prdm3flox (Goyama et al., 2008) conditional-null mice were generated by standard gene-targeting techniques. Myf5Cre;Prdm16flox mice were backcrossed for 10 generations into the C57Black6 strain. The Myf5Cre;Prdm16flox;Prdm3flox mice studied were on a mixed 129Sv/C57Black6 genetic background. Mice in weight-gain studies were fed a 45% high-fat diet (Research Diets). For norepinephrine injections, mice were first placed in metabolic chambers at 22°C, then sedated with 75 mg/kg Nembutal, followed 15 min later by injection with 1 mg/kg norepinephrine. Data were collected until mice recovered from barbituate sedation. Temperature probes (IPTT 300; BioMedic Data Systems) were implanted into a subcutaneous position on top of the BAT of sedated mice. All animal experiments were approved by the University of Pennsylvania's Institutional Animal Care and Use Committee.
Histology
For immunohistochemistry, BAT was fixed in 4% PFA overnight, dehydrated, and embedded in paraffin for sectioning. Sections were stained with hematoxyin and eosin; or were probed with antibodies for Ucp1 (R&D Systems; MAB6158) . For transmission electron microscopy, tissues were fixed with 2.5% glutaraldehyde, 2.0% paraformaldehyde in 0.1M sodium cacodylate buffer, pH7.4, overnight at 4°C; then post-fixed with 2.0% osmium tetroxide for 1 hour at room temperature. Thin sections were stained with uranyl acetate and lead citrate and examined with a JEOL 1010 electron microscope.
Cell culture
Primary brown preadipocytes were isolated as described previously (Seale et al., 2007). Preadipocytes were immortalized through retroviral expression of SV40 large-T antigen. For differentiation assays, confluent preadipocytes were treated with medium containing 10% FBS, 0.5 mM isobutylmethylxanthine, 125 nM indomethacin, 1 μM dexamethosone, 20 nM insulin, and 1 nM T3. After 48 hrs, cells were switched to medium containing 10% FBS, 20 nM insulin, and 1 nM T3. Thermogenesis was stimulated by treatment with 10 μM isoproterenol. Recombination in R26Cre/Prdm16floxEx9 adipocytes was induced by treating cells with 1 μM of 4-hydroxy-tamoxifen (Sigma) for 3 days in growth phase. The GLP antagonist UNC 0646 (Tocris Bioscience) was added to medium throughout differentiation at 1 μM. Oil-red-o staining and retrovirus production was performed as decribed previously (Seale et al., 2007).
Real-Time PCR and Western blot analysis
Total RNA was extracted by TRIzol (Invitrogen) followed by purification using PureLink RNA columns (Invitrogen). Isolated mRNA was reverse transcribed using the High-Capacity cDNA Synthesis kit (Applied Biosystems) and used in real-time PCR reactions with SYBR Green master mix (Applied Biosystems) on a 7900 HT (Applied Biosystems). Tata-binding protein (Tbp) was used as an internal normalization control. Primer sequences are in Table S1. Protein extracts were prepared as previously described (Rajakumari et al., 2013). Proteins were separated in 4-12% Bis-Tris NuPAGE gels (Invitrogen), and transferred to PVDF membranes. Primary antibodies were: anti-Prdm16 (Seale et al., 2007), anti-Pparγ (E8; Santa Cruz Biotechnology; sc-7273), anti-Agt (IBL; 28101), anti-Ucp1 (R&D Systems; MAB6158), and anti-pan-actin (Chemicon; MAB1501).
Chromatin Immunoprecipitation (ChIP)
For ChIP, fat depots from WT and KO mice (6 weeks old) were dissected and washed with PBS. Chromatin was purified from the isolated fat tissue and immunoprecipitated as previously described (Rajakumari et al., 2013). Target enrichment was calculated as percent input and normalized to WT. Primer sequences are in Table S1. Anti-Prdm16 for ChIP was produced by inoculating rabbits with a Prdm16 peptide (RMDKRPEIQDLDSNPPC) to generate a polyclonal antiserum (Pierce). Commercial antibodies were anti-GLP (Abcam; ab41969), anti-H3K27-Me3 (Abcam; ab6002), anti-H3K9-Me1 (Millipore; 17 680) or anti-H3K9-Me2 (Abcam; ab1220).
Microarray Analyses
We used Agilent cDNA microarrays to profile gene expression in WT and Prdm16-deficient BAT from young (6-week-old) and old mice (11-month-old) (GSE55080). To identify depot-specific genes for BAT and WAT, we compared data sets from epididymal WAT and iBAT (GDS2813) (Seale et al., 2007). Gene expression comparisons were performed using Linear Models for Microarray Data (Smyth et al., 2005). Genes with fold-changes >2 and FDR <0.05 in each direction were selected as brown or white depot-specific genes. Hierarchical clustering was done by Euclidean distance and Ward's criterion using Fastcluster (Mullner, 2013). Gene ontology analysis was performed on differentially expressed genes in Myf5ΔPrdm16 BAT from 6-week-old mice using HOMER (Heinz et al., 2010).
Mitochondrial DNA Quantification
BAT was isolated and digested overnight in a buffer containing 100 mM Tris pH 8, 5 mM EDTA, 200 mM NaCl, 0.5% SDS, and 100 μg/ml proteinase K. DNA was ethanol precipitated and resuspended in TE. DNA was quantified by real-time PCR by comparing the ratios of Mt-Co1 and Ndufv1 (Amthor et al., 2007). Primer sequences are in Table I.
Tissue O2 Consumption
BAT was isolated, weighed, and 15-25 mg of tissue was minced in a buffer comprised of 2% BSA, 1.1 mM sodium pyruvate, and 25 mM glucose in PBS. Samples were placed in an MT200A Respirometer Cell (Strathkelvin) and oxygen consumption was measured for approximately 5 minutes. Oxygen consumption was normalized to minced tissue weight.
Fluorescence Activated Cell Sorting
Excised tissue was digested as described above. Cells were resuspended in DMEM with 5% FBS, and stained with Pdgfrα-APC (Biolegend, 135907) for 30 min at 4 °C in the dark. Stained cells were sorted with BD FACS Aria. Debris and dead cells were excluded by forward scatter, side scatter and DAPI gating. The purity was greater than 95%. Data analysis was performed using FlowJo.
Statistical Analysis
All data derived from tissues are reported as mean ± SEM. Data from cells are reported as mean ± standard deviation. Student's t-test was used to calculate significance (*p < 0.05) using Excel or Prism software packages. Data from mice in metabolic chambers was tested for significant differences using a Two-Way Anova (Prism).
Supplementary Material
Highlights.
Prdm16 is dispensable for embryonic brown adipose tissue (BAT) development
Prdm16 recruits Ehmt1 to repress white fat-selective gene expression in BAT
Prdm16 is required to maintain brown fat identity/function during aging
Prdm3 compensates for the loss of Prdm16 to preserve BAT identity in young mice
Acknowledgements
We thank Dr. Bruce Spiegelman for generously sharing the Prdm16flox animals; Dr. Mitch Lazar and Dr. Rex Ahima for their expert guidance; Matthew Brown, Li Huang and Jim Davis for technical help; Min Min Lu, Lan Cheng and the Histology and Gene Expression Core of the Penn Cardiovascular Institute for immunohistochemistry; and Ray Meade in the electron microscopy core. We are grateful to the Functional Genomics Core and the Mouse Metabolic Phenotyping Core of the Penn Diabetes and Endocrinology Research Center (DK19525) for microarray studies and mouse metabolic analyses, respectively. This work was supported by NIGMS/NIH award DP2OD007288, and a Searle Scholars Award to P.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amthor H, Macharia R, Navarrete R, Schuelke M, Brown SC, Otto A, Voit T, Muntoni F, Vrbova G, Partridge T, et al. Lack of myostatin results in excessive muscle growth but impaired force generation. Proc Natl Acad Sci U S A. 2007;104:1835–1840. doi: 10.1073/pnas.0604893104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atit R, Sgaier SK, Mohamed OA, Taketo MM, Dufort D, Joyner AL, Niswander L, Conlon RA. Beta-catenin activation is necessary and sufficient to specify the dorsal dermal fate in the mouse. Developmental biology. 2006;296:164–176. doi: 10.1016/j.ydbio.2006.04.449. [DOI] [PubMed] [Google Scholar]
- Bal NC, Maurya SK, Sopariwala DH, Sahoo SK, Gupta SC, Shaikh SA, Pant M, Rowland LA, Bombardier E, Goonasekera SA, et al. Sarcolipin is a newly identified regulator of muscle-based thermogenesis in mammals. Nat Med. 2012;18:1575–1579. doi: 10.1038/nm.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry R, Rodeheffer MS. Characterization of the adipocyte cellular lineage in vivo. Nat Cell Biol. 2013;15:302–308. doi: 10.1038/ncb2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- Cannon B, Nedergaard J. Nonshivering thermogenesis and its adequate measurement in metabolic studies. The Journal of experimental biology. 2011;214:242–253. doi: 10.1242/jeb.050989. [DOI] [PubMed] [Google Scholar]
- Cohen P, Levy JD, Zhang Y, Frontini A, Kolodin DP, Svensson KJ, Lo JC, Zeng X, Ye L, Khandekar MJ, et al. Ablation of PRDM16 and Beige Adipose Causes Metabolic Dysfunction and a Subcutaneous to Visceral Fat Switch. Cell. 2014;156:304–316. doi: 10.1016/j.cell.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly E, Morrisey RD, Carnie JA. The effect of interscapular brown adipose tissue removal on body-weight and cold response in the mouse. Br J Nutr. 1982;47:653–658. doi: 10.1079/bjn19820077. [DOI] [PubMed] [Google Scholar]
- Daubasf P, Crist CG, Bajard L, Relaix F, Pecnard E, Rocancourt D, Buckingham M. The regulatory mechanisms that underlie inappropriate transcription of the myogenic determination gene Myf5 in the central nervous system. Developmental biology. 2009;327:71–82. doi: 10.1016/j.ydbio.2008.11.031. [DOI] [PubMed] [Google Scholar]
- de Jesus LA, Carvalho SD, Ribeiro MO, Schneider M, Kim SW, Harney JW, Larsen PR, Bianco AC. The type 2 iodothyronine deiodinase is essential for adaptive thermogenesis in brown adipose tissue. J Clin Invest. 2001;108:1379–1385. doi: 10.1172/JCI13803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann HM, Golozoubova V, Cannon B, Nedergaard J. UCP1 ablation induces obesity and abolishes diet-induced thermogenesis in mice exempt from thermal stress by living at thermoneutrality. Cell Metab. 2009;9:203–209. doi: 10.1016/j.cmet.2008.12.014. [DOI] [PubMed] [Google Scholar]
- Golozoubova V, Cannon B, Nedergaard J. UCP1 is essential for adaptive adrenergic nonshivering thermogenesis. American journal of physiology Endocrinology and metabolism. 2006;291:E350–357. doi: 10.1152/ajpendo.00387.2005. [DOI] [PubMed] [Google Scholar]
- Goyama S, Yamamoto G, Shimabe M, Sato T, Ichikawa M, Ogawa S, Chiba S, Kurokawa M. Evi-1 is a critical regulator for hematopoietic stem cells and transformed leukemic cells. Cell stem cell. 2008;3:207–220. doi: 10.1016/j.stem.2008.06.002. [DOI] [PubMed] [Google Scholar]
- Harms M, Seale P. Brown and beige fat: development, function and therapeutic potential. Nat Med. 2013;19:1252–1263. doi: 10.1038/nm.3361. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenauer T, Moore AW. The Prdm family: expanding roles in stem cells and development. Development. 2012;139:2267–2282. doi: 10.1242/dev.070110. [DOI] [PubMed] [Google Scholar]
- Ishibashi J, Firtina Z, Rajakumari S, Wood KH, Conroe HM, Steger DJ, Seale P. An Evi1-C/EBPbeta complex controls peroxisome proliferator-activated receptor gamma2 gene expression to initiate white fat cell differentiation. Mol Cell Biol. 2012;32:2289–2299. doi: 10.1128/MCB.06529-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura S, Seale P, Kubota K, Lunsford E, Frangioni JV, Gygi SP, Spiegelman BM. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature. 2009;460:1154–1158. doi: 10.1038/nature08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Petkova AP, Mottillo EP, Granneman JG. In vivo identification of bipotential adipocyte progenitors recruited by beta3-adrenoceptor activation and high-fat feeding. Cell Metab. 2012;15:480–491. doi: 10.1016/j.cmet.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepper C, Fan CM. Inducible lineage tracing of Pax7-descendant cells reveals embryonic origin of adult satellite cells. Genesis. 2010;48:424–436. doi: 10.1002/dvg.20630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motyl KJ, Rosen CJ. Temperatures rising: brown fat and bone. Discovery medicine. 2011;11:179–185. [PMC free article] [PubMed] [Google Scholar]
- Mullner D. fastcluster: Fast Hierarchical, Agglomerative Clustering Routines for R and Python. Journal of Statistical Software. 2013;53:1–18. [Google Scholar]
- Ohno H, Shinoda K, Ohyama K, Sharp LZ, Kajimura S. EHMT1 controls brown adipose cell fate and thermogenesis through the PRDM16 complex. Nature. 2013 doi: 10.1038/nature12652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumari S, Wu J, Ishibashi J, Lim HW, Giang AH, Won KJ, Reed RR, Seale P. EBF2 determines and maintains brown adipocyte identity. Cell Metab. 2013;17:562–574. doi: 10.1016/j.cmet.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwell NJ, Stock MJ. A role for brown adipose tissue in diet-induced thermogenesis. Nature. 1979;281:31–35. doi: 10.1038/281031a0. [DOI] [PubMed] [Google Scholar]
- Rothwell NJ, Stock MJ. Surgical removal of brown fat results in rapid and complete compensation by other depots. Am J Physiol. 1989;257:R253–258. doi: 10.1152/ajpregu.1989.257.2.R253. [DOI] [PubMed] [Google Scholar]
- Saito M, Okamatsu-Ogura Y, Matsushita M, Watanabe K, Yoneshiro T, Nio-Kobayashi J, Iwanaga T, Miyagawa M, Kameya T, Nakada K, et al. High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes. 2009;58:1526–1531. doi: 10.2337/db09-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Conroe HM, Estall J, Kajimura S, Frontini A, Ishibashi J, Cohen P, Cinti S, Spiegelman BM. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J Clin Invest. 2011;121:96–105. doi: 10.1172/JCI44271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Kajimura S, Yang W, Chin S, Rohas LM, Uldry M, Tavernier G, Langin D, Spiegelman BM. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007;6:38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21:2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- Sun L, Xie H, Mori MA, Alexander R, Yuan B, Hattangadi SM, Liu Q, Kahn CR, Lodish HF. Mir193b-365 is essential for brown fat differentiation. Nat Cell Biol. 2011;13:958–965. doi: 10.1038/ncb2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajbakhsh S, Buckingham ME. Lineage restriction of the myogenic conversion factor myf-5 in the brain. Development. 1995;121:4077–4083. doi: 10.1242/dev.121.12.4077. [DOI] [PubMed] [Google Scholar]
- Tallquist MD, Weismann KE, Hellstrom M, Soriano P. Early myotome specification regulates PDGFA expression and axial skeleton development. Development. 2000;127:5059–5070. doi: 10.1242/dev.127.23.5059. [DOI] [PubMed] [Google Scholar]
- Uezumi A, Fukada S, Yamamoto N, Takeda S, Tsuchida K. Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nat Cell Biol. 2010;12:143–152. doi: 10.1038/ncb2014. [DOI] [PubMed] [Google Scholar]
- Yin H, Pasut A, Soleimani VD, Bentzinger CF, Antoun G, Thorn S, Seale P, Fernando P, van Ijcken W, Grosveld F, et al. MicroRNA-133 controls brown adipose determination in skeletal muscle satellite cells by targeting Prdm16. Cell Metab. 2013;17:210–224. doi: 10.1016/j.cmet.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.