Abstract
[ISP+] is a prion form of the global transcriptional regulator Sfp1 in Saccharomyces cerevisiae that manifests phenotypically as an anti-suppressor of specific sup35 nonsense suppressor mutations. Although SUP35 is a Sfp1 target, the mechanism of anti-suppression is unclear. Here we show that the level of SUP35 transcription in [ISP+] cells containing the sup35 mutation is increased relative to [isp−] cells and cells with a SFP1 deletion. As a result, [ISP+] cells have increased amounts of Sup35 encoded by the mutant allele. Indeed, additional experiments showed that increased amounts of mutant Sup35 may cause anti-suppression. Remarkably, [ISP+] effects are not equivalent to those produced by SFP1 deletion, so [ISP+] represents an obvious example of a functionally active prion form of a protein. This feature distinguishes [ISP+] from other yeast prions, where prion switch often has the same effect as inactivation of a prion host gene. We suggest that enhancement of SUP35 expression in [ISP+] cells is caused by specific interaction of Sfp1 in its prion form with some negative SUP35 regulator. We also demonstrate that the advantage of [ISP+] strains over [isp−] strains described in our earlier work is specific for certain genetic background and growth conditions.
Key words: yeast prion, SFP1, SUP35, nonsense suppression, regulation of gene expression
Introduction
The non-Mendelian determinant [ISP+] is a prion form of the global transcriptional regulator Sfp1.1 [ISP+] was initially detected as a non-chromosomal anti-suppressor that appears at a high rate in strains containing specific sup35 mutations that suppress the effects of the nonsense mutations his7-1 (UAA) and lys2-87 (UGA).2 As a result, strains containing [ISP+] express the dominant non-suppressor (His−Lys−) phenotype, while isogenic [isp−] strains have the His+Lys+ phenotype. The [ISP+] status of a strain can be changed to [isp−] in medium containing 5 mM guanidine chloride (GuHCl), which both cures cells of [ISP+] and also selects [isp−] cells, due to their higher resistance to GuHCl relative [ISP+] cells.2 The ability of GuHCl to cure cells of [ISP+] is contradictory to the independence of its propagation on the Hsp104 chaperone protein,1,2 since GuHCl cures yeast prions by inhibition of Hsp104 activity that is necessary for prion shearing (reviewed in refs. 3–5). This Hsp104 independence may indicate that chaperones other than Hsp104p participate in [ISP+] propagation.
We recently showed that the [ISP+] status of a strain depends on SFP1 expression.2 SFP1 deletion causes irreversible [ISP+] loss, whereas increased SFP1 expression levels induce the appearance of [ISP+]. Notably, [ISP+] cells contain the aggregated form of Sfp1, which is localized largely in the nuclei. This nuclear location may be the reason for the low efficiency of [ISP+] cytoduction and its independence of Hsp104 for its maintenance.
The conversion of proteins in yeast to their prion forms typically causes an effect that is similar to that caused by the complete or partial inactivation of the corresponding gene, since the conversion results in the loss of protein function (reviewed in refs. 6–8). However, the strains used in our earlier work illustrated another situation, with [ISP+] strains and strains containing the SFP1 deletion differing in several ways: (1) [ISP+] strains demonstrated anti-suppression, while sfp1Δ strains did not; (2) compared with isogenic sfp1Δ strains, [ISP+] strains grew faster and were more resistant to antibiotics that inhibit translation and (3) deletion of SFP1 significantly reduced cell size, whereas cells of [ISP+] strains were of normal size.1 Importantly, in the same manner, [ISP+] strains differed from [isp−] strains, albeit less obviously. The last observation would support an advantage for [ISP+] strains over [isp−] strains.
This study seeks to elucidate the nature of the anti-suppression caused by [ISP+] and to examine [ISP+] effects in the background that differs from that required to manifest the [ISP+] phenotype. We present data demonstrating that the anti-suppression observed in [ISP+] strains is caused by increased levels of Sup35 and discuss possible mechanisms that underlie this effect. Our results also indicate that advantages for [ISP+] strains appear only in certain genetic backgrounds, where increased expression of particular genes is needed for optimal adaptation to growth conditions.
Results
SUP35 expression is increased in [ISP+] strain compared with sfp1Δ and [isp−] strains.
SUP35 encodes the translation termination factor eRF3,9,10 and termination defects caused by sup35 mutation can be neutralized to some extent in [ISP+] cells.2 Indeed, the SFP1 and SUP35 genes are functionally related, since data from transcriptome analysis indicate that SUP35 expression is positively regulated by Sfp1.11,12 To determine whether the efficiency of SUP35 expression in [ISP+] strains differs from that of sfp1Δ and [isp−] strains, quantitative real-time RT-PCR (see Materials and Methods) was performed. First, it was performed on the initial strain 2V-P3982 containing the wild-type SUP35 allele and its SFP1-deleted derivative. Both strains compared were [isp−], since [ISP+] arises in strains containing specific sup35 mutations.2 This experiment confirmed the data from the transcriptome analysis, as the level of SUP35 transcription in the strain lacking SFP1 was essentially lower than in the same strain containing SFP1 (upper line in Table 1).
Table 1.
The level of SUP35 transcription depends on Sfp1 production and on [ISP+] status
| Strains compared | Allele expressed | Relation of transcription levels |
| SFP1[isp−] to sfp1Δ | SUP35 | 6.4 ± 1.27 |
| SFP1[ISP+] to SFP1[isp−] | sup35-25 | 2.6 ± 0.95 |
| SFP1[isp−] to sfp1Δ | sup35-25 | 2.7 ± 0.25 |
Results of at least three estimations of transcription levels were processed in each case; data are mean ± SEM.
Then, the level of SUP35 transcription was examined in the [ISP+] variant of 25-25-2V-P3982, and in its [isp−] and sfp1Δ derivatives. These strains contained the sup35-25 mutation, which is one of two sup35 nonsense suppressor mutations for which an anti-suppressor effect of [ISP+] was registered.2 The analysis of sup35-25 expression performed for these strains revealed that in the sfp1Δ strain, as with strains containing wild-type SUP35, this allele was transcribed less efficiently than in the [isp−] strain (lower line in Table 1). At the same time, the level of sup35-25 transcription in the [ISP+] variant exceeded that shown for the [isp−] strain (middle line in Table 1).
We also used protein gel blotting to show that in [ISP+], [isp−] and sfp1Δ strains containing sup35-25, the increased level of SUP35 transcription in the [ISP+] strain is accompanied by an increase in Sup35p production. Again, the initial strain 2V-P3982 bearing wild-type SUP35 and its sfp1Δ derivative were used as additional controls. The results show that: (1) deletion of SFP1 in strain 2V-P3982 decreases the amount of Sup35 (Fig. 1A and Table 2) and (2) the amount of Sup35 in the [ISP+] strain is higher compared with the [isp−] and sfp1Δ derivative (Fig. 1B and Table 2). Thus, the manifestation of the anti-suppressor effect in [ISP+] strains correlates with increased transcription of the sup35-25 allele that is accompanied by increased amounts of Sup35. This increase in Sup35 levels may account for the anti-suppression observed in [ISP+] strains.
Figure 1.
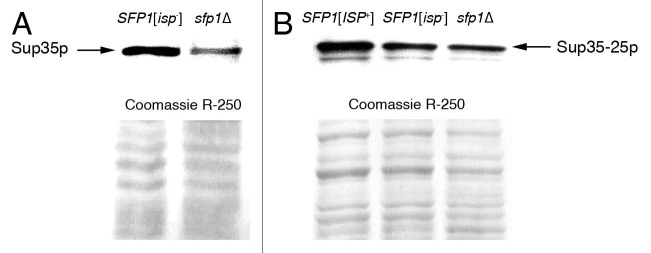
The amount of Sup35 is decreased in strains with deletion of SFP1 and is increased in [ISP+] strains comparatively to isogenic [isp−] strains. Detection of Sup35 by protein gel blot in strains that express the wild-type SUP35 allele and do not contain [ISP+] (A) and in [ISP+], [isp−] and sfp1Δ strains expressing the sup35-25 (B). Polyclonal antibodies against Sup35 were used.
Table 2.
Sup35 amounts depend on Sfp1 production and on [ISP+] status
| Strains compared | Allele expressed | Relation of Sup35 amounts |
| SFP1[isp−] to sfp1Δ | SUP35 | 2.7 ± 0.03 |
| SFP1[ISP+] to SFP1[isp−] | sup35-25 | 1.5 ± 0.06 |
| SFP1[isp−] to sfp1Δ | sup35-25 | 1.1 ± 0.03 |
Area and density of bands corresponding to Sup35 on protein gel blots was estimated 10 times by ImageJ tool; data are mean ± SEM.
Expression of sup35-25 from a high-copy plasmid in an [isp−] strain causes anti-suppression.
It should be emphasized that strains used in the previous section produced mutant Sup35 encoded by the sup35-25 allele that causes omnipotent nonsense suppression in [isp−] strains.2 Nevertheless, an [ISP+] strain producing increased amounts of mutant protein displayed the non-suppressor phenotype. To examine whether increased amounts of the mutant protein could compensate for defects in translation termination, sup35-25 was expressed from a high copy number plasmid pRSU3-25 in the [isp−] variant 25-25-2V-P3982. Transformation with the original plasmid pRS425 lacking sup35-25, was used as a control. Transformants were selected on SMM-Leu medium. A significant increase in the level of Sup35 in these transformants was confirmed by protein gel blot (Fig. 2A). Phenotypes of 96 transformants were analyzed in both cases. Remarkably, all transformants containing the pRSU3-25 plasmid displayed a non-suppressor phenotype, i.e., His− Lys−, whereas transformants with the original plasmid retained suppression. An example of transformant growth on medium lacking lysine is presented in Figure 2B. These results indicate that an increase in the amount of mutant Sup35 encoded by the sup35-25 allele may compensate for the decrease in termination efficiency caused by the mutation and strongly supports the idea proposed above that the anti-suppression observed in [ISP+] strains may be caused by increased levels of Sup35.
Figure 2.
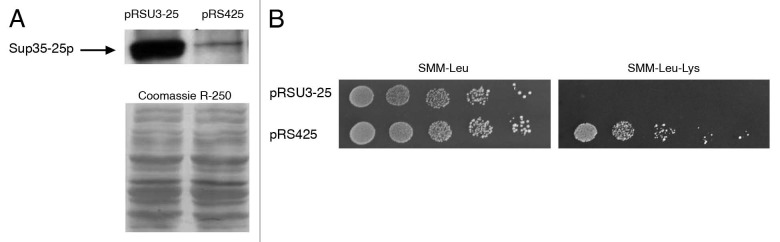
Expression of sup35-25 from a high copy number plasmid increases the amounts of Sup35 and causes anti-suppression of lys2-87 mutation. Detection of Sup35 by protein gel blot in [isp−] strains transformed by pRSU3-25 and pRS425 (A); growth of transformants on SMM-Leu required for maintenance of plasmids and on SMM-Leu-Lys necessary for detection of lys2-87 manifestation (B).
Manifestation of [ISP+] in strains containing wild-type SUP35.
The data obtained earlier showed that [ISP+] status provided a selective advantage over [isp−] strains containing specific sup35 mutations in terms of a higher growth rate and increased resistance to antibiotics that inhibit translation.1,2 To address the question of whether [ISP+] specifically influences cells with unimpaired translation termination, we needed to produce an [ISP+] strain expressing wild-type SUP35. While it is possible to cytoduce the [ISP+] from the sup35 mutant into the initial strain expressing SUP35 as was done in our earlier studies,2 this approach is complicated, since the effects of [ISP+] are not manifested phenotypically in the absence of specific sup35 mutations. As a result, identification of [ISP+]-containing cytoductants is possible only by genetic analysis of individual clones. In addition, the efficiency of [ISP+] cytoduction is about 17%, which is much lower than that of other yeast prions and is likely due to the predominantly nuclear location of [ISP+].1 Therefore, we used another approach and constructed strains where the manifestations of the recessive sup35-25 allele were complemented by wild-type SUP35. To this aim, the [ISP+] strain 25-25-2V-P3982 containing the sup35-25 allele was transformed with a pRSU1 plasmid bearing wild-type SUP35. Simultaneously, the [isp−] and sfp1Δ derivatives of this strain were transformed with the same plasmid. Twenty transformants were selected on SMM-Leu medium for each of the recipient strains. As expected, all transformants of the [ISP+] strain retained the same non-suppressor (His−Lys−) phenotype of the recipient. To control the [ISP+] retention in these transformants, the phenotype of the clones obtained in mitotic progeny of the transformants after pRSU1 loss on YPD medium was examined. All 48 clones obtained from three transformants (16 clones from each) displayed the non-suppressor phenotype (not shown). Thus, [ISP+] is stably maintained in cells containing the wild-type SUP35 allele.
Transformants of the [isp−] strain also displayed the non-suppressor phenotype due to complementation of sup35-25 effects, but after loss of the SUP35 bearing plasmid on YPD, all of them displayed suppression of his7-1 and lys2-87 (not shown). As such, transformants of both [ISP+] and [isp−] strains did not differ phenotypically, but instead differed in their [ISP+] status. Interestingly, transformants of the sfp1Δ strain retained the suppressor phenotype despite their wild-type SUP35 expression, even though the suppression efficiency was significantly decreased compared with the recipient strain (not shown). Retention of nonsense suppression in these transformants is most likely caused by decreases in the amount of Sup35 due to SFP1 deletion.
Transformants of [ISP+], [isp−] and sfp1Δ strains were compared by their growth rate in SMM-Leu medium, which is necessary for pRSU1 maintenance. Figure 3 shows that the growth rates of [ISP+] and [isp−] strains transformed by pRSU1 were not significantly different, but transformants of the sfp1Δ strain grew much more slowly.
Figure 3.
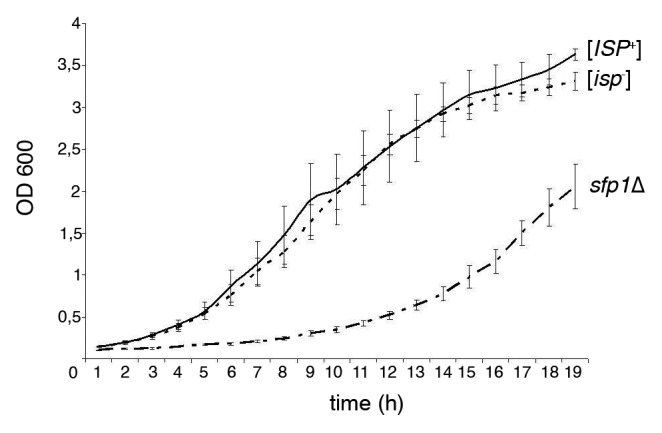
The [ISP+] and [isp−] strains transformed by pRSU1 do not differ by their growth rates; transformants of sfp1Δ strain grow essentially slower. SMM-Leu required for pRSU1 maintenance was used. The means for five independent clones of each strain are presented; error bars indicate SEM.
SFP1 is known to be one of the key genes that controls cell size in yeast; therefore, cells lacking Sfp1p have a reduced size.11–15 At the same time, [ISP+] strain cells are significantly larger than sfp1Δ cells and are slightly larger than [isp−] cells.1 Here we compared transformants of [ISP+], [isp−] and sfp1Δ strains in terms of cellular area (see Materials and Methods) and discovered that in spite of the presence of SUP35, the same pattern was retained: transformants of the sfp1Δ strain had the smallest cell size (18.0 ± 0.56 µm2), and the [ISP+] cells were larger than [isp−] cells. Interestingly, the distinction between transformants of [ISP+] and [isp−] was rather significant: 29.0 ± 0.81 µm2 and 23.1 ± 0.67 µm2, respectively.
Next, the antibiotic sensitivity of the transformants was compared, with cells transformed by empty plasmid acting as a control. As mentioned above, compared with [isp−] and sfp1Δ strains, the initial [ISP+] strain demonstrated an increased resistance to the translation inhibitors paromomycin and cycloheximide.1 Since SFP1 deletion is known to increase the sensitivity of yeast cells to the macrolide rapamycin, an inhibitor of the TORC1 kinase complex,16,17 the effects of Sfp1 prionization on the rapamycin sensitivity of the transformants were also examined.
The data presented in Table 3 show that transformants of the sfp1Δ strain are the most sensitive to all antibiotics used. Notably, SUP35 containing transformants of sfp1Δ and [isp−] strains are more resistant relative to strains expressing only sup35-25. At the same time, SUP35 containing transformants of the [ISP+] strain are more sensitive to all three drugs than strains expressing the sup35-25 and transformants of the [isp−] strain. It should also be stressed that the [ISP+] strain transformed by pRS315 is more sensitive to rapamycin than the [isp−] strain transformed by the same plasmid.
Table 3.
Characterization of drug sensitivity of [ISP+], [isp−] and sfp1Δ strains transformed by pRSU1 and pRS315 plasmids
| Recipient strain | ||||||
| Drug | [ISP+] | [isp−] | sfp1Δ | |||
| pRSU1 | pRS315 | pRSU1 | pRS315 | pRSU1 | pRS315 | |
| Paromomycin | 15.1 ± 0.71 | 11.0 ± 0.11 | 13.8 ± 0.17 | 16.8 ± 0.39 | 17.2 ± 0.13 | 22.7 ± 0.24 |
| Cycloheximide | 26.9 ± 0.33 | 24.0 ± 0.31 | 24.4 ± 0.14 | 25.5 ± 0.39 | 29.2 ± 0.26 | 32.3 ± 0.35 |
| Rapamycin | 21.1 ± 0.20 | 19.2 ± 0.16 | 20.0 ± 0.16 | 18.2 ± 0.21 | 22.7 ± 0.29 | 26.3 ± 0.25 |
The average value of inhibition zones (mm) determined by ImageJ is presented. For each case, n = 50; data are mean ± SEM.
Thus, the [ISP+] effect may be beneficial in specific genetic backgrounds and under particular growth conditions, for example, when the compensation of translation termination defects is needed to ensure survival. In contrast, some effects of [ISP+] are not related to translation, for instance, compensation of the termination defect by SUP35 expression does not abolish the influence of [ISP+] on cell size and does not decrease the sensitivity of [ISP+] strains to rapamycin.
Discussion
The prion form of the transcriptional regulator Sfp1 known as [ISP+] has properties of a typical yeast prion as well as several other unique features (see Introduction). While the consequences of protein prionization and deletion of its host gene are usually similar, [ISP+] strains differ in phenotype from both [isp−] strains and strains containing the SFP1 deletion. [ISP+] strains containing nonsense mutations and specific sup35 mutations display a non-suppressor phenotype, which is in contrast to isogenic sfp1Δ strains, in which the manifestations of nonsense mutations are suppressed.1 This difference might be explained by variations in the level of SUP35 expression of the strains examined, which is consistent with data from transcriptome analysis indicating that Sfp1 regulates SUP35 transcription.11,13 Indeed, our results from qRT-PCR analysis of wild-type and mutant SUP35 transcription and protein gel blotting to determine Sup35 protein levels support this conclusion. The level of SUP35 expression in the sfp1Δ strain is decreased relative to strains containing SFP1. At the same time, the [ISP+] strain expressed SUP35 more efficiently than did the sfp1Δ and [isp−] strains. As to Sfp1 level, our data obtained earlier indicate that Sfp1 production is not increased in [ISP+] cells expressing SFP1 from the native promoter relatively to [isp−] cells.1
Thus, the anti-suppression observed in the [ISP+] strain correlates with increased amounts of Sup35. It should be stressed, however, that compensation for the translation termination defect is achieved in this case by the increased production of mutant Sup35. Such a mechanism of anti-suppression received additional support from results of sup35-25 expression from a high copy number plasmid.
It is evident that the effect of Sup35 would be specific and dependent on the nature of protein damage and/or its level of production. Since the interaction between eRF3(Sup35) and eRF1(Sup45) is known to be required for translation termination,18–20 a mutation that decreases the affinity of Sup35 for Sup45 may be compensated for by increased amounts of Sup35 that could enhance the probability of the protein interaction. In contrast, if a sup35 mutation influences the Sup45 interaction by affecting ribosome binding or stop codon recognition, increased amounts of Sup35 would not rescue this defect. Allele specificity of anti-suppression caused by [ISP+] was revealed in our earlier work whereby amino acid replacements caused by two sup35 mutations that were anti-suppressed in [ISP+] cells were found to be located close to one another (positions 363 and 378) and probably affect the same functional domain of the Sup35.2 This is in contrast to mutations insensitive to [ISP+], which occurred at positions 413 and 575.
Increased levels of SUP35 expression in [ISP+] cells in comparison to sfp1Δ and [isp−] cells implies that a positive effect of Sfp1 on SUP35 transcription is not only retained, but is even enhanced, despite the prion conversion of the protein. So, [ISP+] represents an obvious example of a functionally active prion form of a protein. The possibility of protein function retention after prion switching, in this example, preservation of enzymatic activity, was shown for a fusion protein combining the prion domain of Ure2 and the bacterial ribonuclease barnase.21 Another example is also related to Ure2, which retains its glutathione peroxidase activity after switch to prion form.22 The retention of enzymatic activity by the prion form of the Ure2 was explained by the small size of substrate molecules, which can reach active sites of the protein that remain available after prion filament formation.
Sfp1 is a transcription factor that contains several zinc finger motifs, which should facilitate DNA binding. However, it is difficult to imagine that DNA-binding activity of Sfp1 might be enhanced after prion switch. In reality, some data indicate that Sfp1 may affect its target genes indirectly by mediating the activity of other proteins.23 For instance, Sfp1p prionization may be followed by the binding and sequestration of a potential, as yet uncharacterized, SUP35 repressor protein due to its inclusion in prion aggregates formed by Sfp1, similar to that shown for Sup45 inclusion into [PSI+] aggregates.18 Another possibility is induction of prion conversion of this tentative repressor by [ISP+], akin to [PSI+] induction by [PIN+]; (reviewed in refs. 24 and 25). Both mechanisms would result in enhanced SUP35 transcription and explain the difference in the effects of Sfp1 prionization and SFP1 deletion. Indeed, among proteins interacting with Sfp1, there are several transcriptional factors and components of chromatin remodeling complexes (see the-biogrid.org/31660).
An additional aspect of [ISP+] effects is the advantage of [ISP+] strains over [isp−] strains, which was shown in our earlier work.1,2 This advantage is manifested by the increased growth rates and drug resistance of [ISP+] strains. To determine whether this advantage is specific to [ISP+] and whether [ISP+] confers this benefit by a mechanism other than rescue of defects in translation termination caused by certain sup35 mutations, we compared [ISP+] and [isp−] strains that did not differ from one another by their nonsense suppressor phenotype since they were both transformed by the plasmid bearing wild-type SUP35. In this situation, the growth rates of [ISP+] and [isp−] strains did not differ, and the [ISP+] strain was more sensitive to paromomycin and cycloheximide than was the [isp−] strain, indicating that in certain backgrounds, the effect of [ISP+] may even be harmful.
Sfp1 is a phosphoprotein phosphorylated by TORC1, the rapamycin sensitive protein kinase complex involved in regulating many cellular processes, in particular, protein synthesis, ribosome biogenesis and cell growth; (reviewed in ref. 16, 17 and 26). Phosphorylation of Sfp1 by TORC1 determines Sfp1 localization and, consequently, its function. We found that both the sup35-25 containing [ISP+] strain and its transformants containing the wild-type SUP35 allele were more rapamycin sensitive than [isp−] strains. An exact interpretation of the molecular mechanisms responsible for these effects is complicated by the multiple consequences of TORC1 inhibition by rapamycin. However, it is evident that prion switch of Sfp1 increases sensitivity to this drug.
The data obtained here indicate that Sfp1 prionization may provide an advantage to yeast strains under particular conditions, for example, when increased expression of specific genes regulated by Sfp1 can compensate for defects in certain functions. The participation of Sfp1 in the regulation of basic cellular processes emphasizes that this problem is worthy of special study.
Materials and Methods
Plasmids, strains and genetic methods.
In this work we used Saccharomyces serevisiae strains 2V-P3982 (MATα ade1-14 his7-1 lys2-87 ura3Δ leu2-1 thr4-B15 leu2-1) and its sfp1Δ derivative; 25-25-2V-P3982, which is the MATa derivative of 25-2V-P3982 (MATα ade1-14 his7-1 lys2-87 ura3 Δ leu2-1 thr4-B15 leu2-1 sup45-400 sup35-25),1,2 and [ISP+], [isp−] and sfp1Δ variants of this strain.
Plasmids used in this study were centromeric LEU2 based plasmid pRS315 and its derivative pRSU1 bearing the wild-type SUP35; multicopy LEU2 based plasmid pRS425 and its derivative pRSU3-25 containing the sup35-25 allele. Both pRSU1 and pRSU3-25 express SUP35 alleles from its own promoter.2
Yeast were grown at 26°C in rich medium YPD, supplemented minimal medum (SMM), SMM lacking one or more supplements (e.g., SMM-lysine). All media were prepared as described in reference 27. Standard methods of yeast genetics were used.27 Drug sensitivity test was performed as described in reference 13, using paromomycin (1 M), cycloheximide (5 mM) and rapamycin (1 mM); all from “Sigma.”
Cell-size evaluation.
Strains were grown in YPD up to the early stationary phase. Then, cells were placed in a counting chamber (hemocytometer) and examined in transmitted light under a Leica DM LS microscope equipped with FLUOTAR objective 20x/0.40 photoadapter Leica DFC 320 camera. Images were captured with Leica DFC Twain software and processed with Adobe Photoshop CS2. The photographed areas of 50 randomly selected cells of each strain were estimated and processed by ImageJ 1.34s program (rsb.info.nih.gov/ij).
Preparation of yeast cell lysates; electrophoresis and blotting.
Yeast cultures were grown in liquid media to OD600 0.5–0.7. Cells were harvested, washed in water and lyzed by glass beads in buffer A: 30 mM TRIS-HCl, pH 7.4, 150 mM NaCl, 1 mM dithiothreitol and 1% Triton X-100. To prevent proteolytic degradation, 10 mM phenylmethylsulfonyl fluoride and Complete™ protease inhibitor cocktail (Roche Applied Science) were added. Cell debris was removed by centrifugation at 1,500 g for 4 min. Protein gel blot was performed using rabbit polyclonal antibodies against Sup35p (a gift of S.V. Chabelskaya). Staining of gel with Coomassie R-250 was used for normalization of the total protein amount.
Evaluation of Sup35p amounts.
Relative amounts of Sup35p in strains used were quantified by protein gel blots analysis using ImageJ (rsb.info.nih.gov/ij) as described in reference 28.
Quantitative real-time RT-PCR.
Quantitative Real-time RT-CR was performed in 96-well plates on a Biorad ICycler (Biorad Laboratories, Inc., Hercules) using the SYBR Green stain that binds double-stranded DNA and ACT1 as a reference gene. The primers used were SUP35qRT_S3 (CCA ACA ACA AGG TAA CAA CAG ATA C) and SUP35qRT_AS3 (GTG GAT TGA ATT GCT GCT GAT AAC). Reactions were performed in a total volume of 25 µL that contained 2.5 µL of cDNA, 0.5 mM of forward and reverse primers, and 12.5 µL of 2x SYBR Green master mix (Biorad). From one (for 2V-P3982 and its sfp1Δ derivative) to three (for [ISP+], [isp−] and sfp1Δ variants of 25-25-2V-P3982) independent clones were used for RNA isolation. Each sample was analyzed three times. A non template control for each primer was included in all real-time plates. Amplifications were performed under the following conditions: 95°C for 3 min; 40 cycles of 95°C for 10 sec, 63°C for 30 sec and 72°C for 30 min; and a final extension at 72°C for 5 min. At the end of the amplification cycle, a melting analysis was conducted to verify the specificity of the reaction. This was performed by heating the amplification products from 60°C to 95°C at 0.5°C/10 sec and monitoring the decrease in fluorescence. The expression levels were described in terms of the cycle threshold value (Ct), which is the number of cycles required to reach a certain fluorescence value (threshold). The threshold values were obtained by using the automated setting of the instrument software (base line subtracted curve fit data) considering the fluorescence when maximum PCR efficiency was achieved. The data expressed as Ct were imported into a Microsoft Excel data sheet for subsequent analysis. The data were analyzed by the comparative CT method of Livak.29
Acknowledgments
The work was supported by the grants from Russian Foundation for Basic Research (11-04-00146-a) and from Russian Ministry of Science and Education (P799). Authors are grateful to A. Petrova for valuable advices concerning real time RT-PCR application, to O. Tarasov for help in statistic processing of the data and to A. Goginashvili for the critical reading of the manuscript. ER and TR contributed equally to this work.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Rogoza T, Goginashvili A, Rodionova S, Ivanov M, Viktorovskaya O, Rubel A, et al. Non-Mendelian determinant [ISP+] in yeast is a nuclear-residing prion form of the global transcriptional regulator Sfp1. Proc Natl Acad Sci USA. 2010;107:10573–10577. doi: 10.1073/pnas.1005949107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Volkov KV, Aksenova AYu, Soom MJ, Osipov KV, Svitin AV, Kurischko C, et al. Novel non-Mendelian determinant involved in the control of translation accuracy in Saccharomyces cerevisiae. Genetics. 2002;160:25–36. doi: 10.1093/genetics/160.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grimminger V, Richter K, Imhof A, Buchner J, Walter S. The prion curing agent guanidinium chloride specifically inhibits ATP hydrolysis by Hsp104. J Biol Chem. 2004;279:7378–7383. doi: 10.1074/jbc.M312403200. [DOI] [PubMed] [Google Scholar]
- 4.Romanova NV, Chernoff YO. Hsp104 and prion propagation. Protein Pept Lett. 2009;16:598–605. doi: 10.2174/092986609788490078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones GW, Tuite MF. Chaperoning prions: the cellular machinery for propagating an infectious protein? Bioessays. 2005;27:823–832. doi: 10.1002/bies.20267. [DOI] [PubMed] [Google Scholar]
- 6.Uptain SM, Lindquist S. Prions as protein-based genetic elements. Annu Rev Microbiol. 2002;56:703–741. doi: 10.1146/annurev.micro.56.013002.100603. [DOI] [PubMed] [Google Scholar]
- 7.Wickner RB, Edskes HK, Ross ED, Pierce MM, Baxa U, Brachmann A, et al. Prion genetics: new rules for a new kind of gene. Annu Rev Genet. 2004;38:681–707. doi: 10.1146/annurev.genet.38.072902.092200. [DOI] [PubMed] [Google Scholar]
- 8.Wickner RB, Shewmaker F, Kryndushkin D, Edskes HK. Protein inheritance (prions) based on parallel in-register beta-sheet amyloid structures. Bioessays. 2008;30:955–964. doi: 10.1002/bies.20821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stansfield I, Jones KM, Kushnirov VV, Dagkesamanskaya AR, Poznyakovski AI, Paushkin SV, et al. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 1995;14:4365–4373. doi: 10.1002/j.1460-2075.1995.tb00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhouravleva G, Frolova L, Le Goff X, Le Guellec R, Inge-Vechtomov S, Kisselev L, et al. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 1995;14:4065–4072. doi: 10.1002/j.1460-2075.1995.tb00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jorgensen P, Rupes I, Sharom JR, Schneper L, Broach JR, Tyers M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004;18:2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marion RM, Regev A, Segal E, Barash Y, Koller D, Friedman N, et al. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc Natl Acad Sci USA. 2004;101:14315–14322. doi: 10.1073/pnas.0405353101.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fingerman I, Nagaraj V, Norris D, Vershon AK. Sfp1 plays a key role in yeast ribosome biogenesis. Eukaryot Cell. 2003;2:1061–1068. doi: 10.1128/EC.2.5.1061-8.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jorgensen P, Rupes I, Sharom JR, Schneper L, Broach JR, Tyers M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004;18:2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cipollina C, van den Brink J, Daran-Lapujade P, Pronk JT, Porro D, de Winde JH. Saccharomyces cerevisiae SFP1: at the crossroads of central metabolism and ribosome biogenesis. Microbiology. 2008;154:1686–1699. doi: 10.1099/mic.0.2008/017392-0. [DOI] [PubMed] [Google Scholar]
- 16.Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10:457–468. doi: 10.1016/S1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 17.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 18.Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. Interaction between yeast Sup45p (eRF1) and Sup35p (eRF3) polypeptide chain release factors: implications for prion-dependent regulation. Mol Cell Biol. 1997;17:2798–2805. doi: 10.1128/mcb.17.5.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parsons AB, Brost RL, Ding H, Li Z, Zhang C, Sheikh B, et al. Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat Biotechnol. 2004;22:62–69. doi: 10.1038/nbt919. [DOI] [PubMed] [Google Scholar]
- 20.Fan-Minogue H, Du M, Pisarev AV, Kallmeyer AK, Salas-Marco J, Keeling KM, et al. Distinct eRF3 Requirements Suggest Alternate eRF1 Conformations Mediate Peptide Release During Eukaryotic Translation Termination. Mol Cell. 2008;30:599–609. doi: 10.1016/j.molcel.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baxa U, Speransky V, Steven AC, Wickner RB. Mechanism of inactivation on prion conversion of the Saccharomyces cerevisiae Ure2 protein. Proc Natl Acad Sci USA. 2002;99:5253–5260. doi: 10.1073/pnas.082097899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bai M, Zhou JM, Perrett S. The yeast prion protein Ure2 shows glutathione peroxidase activity in both native and fibrillar forms. J Biol Chem. 2004;279:50025–50030. doi: 10.1074/jbc.M406612200. [DOI] [PubMed] [Google Scholar]
- 23.Gordân R, Hartemink AJ, Bulyk ML. Distinguishing direct versus indirect transcriptional factor-DNA interactions. Genome Res. 2009;19:2090–2100. doi: 10.1101/gr.094144.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN+] Cell. 2001;106:171–182. doi: 10.1016/S0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- 25.Wickner RB, Shewmaker F, Edskes H, Kryndushkin D, Nemecek J, McGlinchey R, et al. Prion amyloid structure explains templating: how proteins can be genes. FEMS Yeast Res. 2010;10:980–991. doi: 10.1111/j.1567-364.2010.00666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lempiäinen H, Shore D. Growth control and ribosome biogenesis. Curr Opin Cell Biol. 2009;21:855–863. doi: 10.1016/j.ceb.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 27.Kaiser C, Michaelis S, Mitchell A. Methods in Yeast Genetics. Plainview NY: Cold Spring Harbor Laboratory Press; 1994. p. 234. [Google Scholar]
- 28.Gassmann M, Grenacher B, Rohde B, Vogel J. Quantifying western blots: pitfalls of densitometry. Electrophoresis. 2009;30:1845–1855. doi: 10.1002/elps.200800720. [DOI] [PubMed] [Google Scholar]
- 29.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]


