Abstract
Small RNAs recognize, bind, and regulate other complementary cellular RNAs. The introduction of small RNAs to eukaryotic cells frequently results in unintended silencing of related, but not identical, RNAs: a process termed off-target gene silencing. Off-target gene silencing is one of the major concerns during the application of small RNA-based technologies for gene discovery and the treatment of human disease. Off-target gene silencing is commonly thought to be due to inherent biochemical limitations of the RNAi machinery. Here we show that following the introduction of exogenous sources of double-stranded RNA, the nuclear RNAi pathway, but not its cytoplasmic counterparts, is the primary source of off-target silencing in Caenorhabditis elegans. In addition, we show that during the normal course of growth and development the nuclear RNAi pathway regulates repetitive gene families. Therefore, we speculate that RNAi off-target effects might not be “mistakes” but rather an intentional and genetically programmed aspect of small RNA-mediated gene silencing, which might allow small RNAs to silence rapidly evolving parasitic nucleic acids. Finally, reducing off-target effects by manipulating the nuclear RNAi pathway in vivo might improve the efficacy of small RNA-based technologies.
Keywords: Nrde, off target, silencing, repetitive sequence
THE introduction of double-stranded RNA (dsRNA) into organisms elicits a process termed RNA interference (RNAi) that suppresses the expression of genes exhibiting sequence homology (Fire et al. 1998). dsRNA molecules are cleaved by the RNase III-like enzyme DICER into ∼22-nt small interfering RNAs (siRNAs), which are then loaded onto Argonaute proteins. siRNA guides the Argonaute silencing complex to complementary nucleic acids to silence gene expression via a multitude of mechanisms, including initiating the degradation of the target mRNA, blocking translation, triggering heterochromatin formation, and inhibiting transcription elongation (reviewed by Ketting 2011; Feng and Guang 2013).
The nuclear RNAi defective (Nrde) pathway is necessary for the silencing of nuclear-localized RNAs in Caenorhabditis elegans. NRDE-3 is an Argonaute protein that associates with secondary siRNAs in the cytoplasm and escorts them to the nucleus (Guang et al. 2008); the siRNA/NRDE-3 complex then binds to complementary nascent transcripts and recruits NRDE-1 and NRDE-2 (Guang et al. 2008, 2010; Burkhart et al. 2011). With the help of NRDE-4, NRDE-1 associates with chromatin at the targeted genomic locus and elicits histone 3 lysine 9 trimethylation (H3K9me3) at the site (Burkhart et al. 2011). The siRNA/NRDE complex, most likely working together with the chromatin modification machinery, pauses RNA polymerase II and inhibits the elongation of transcription (Guang et al. 2010).
Although much is known about the molecular mechanism of Nrde-mediated gene silencing, little is known about the biological roles of the nuclear RNAi pathway. nrde-1/2/4 and hrde-1 (the germline nuclear Ago) mutants produce fewer progeny and exhibit a mortal germline phenotype, suggesting that nuclear RNAi may be important for fertility and germline immortality (Guang et al. 2010; Burkhart et al. 2011; Buckley et al. 2012). In addition, hrde-1 and nrde-2 mutants exhibit high incidence of males (Him) phenotype, suggesting that nuclear RNAi in the germline might promote chromosome segregation during meiosis (Buckley et al. 2012). The nuclear Ago NRDE-3 binds endogenous siRNAs termed the 22G siRNAs. Many genes, such as eri-1, eri-6, eri-7, ergo-1, drh-3, mut-2, mut-7, and rrf-1 are required for the production of a small subset, respectively, of endogenous siRNAs that engage NRDE-3 (Guang et al. 2008; Fischer et al. 2011). RRF-1 is a somatic RNA-dependent RNA polymerase acting upstream but necessary to feed the nuclear RNAi pathway. RRF-1 is guided by the RDE-1/siRNA complex to complementary mRNA targets to generate secondary siRNAs (Pak et al. 2012), which bind to NRDE-3 to be escorted to the nucleus (Guang et al. 2008).
Although near perfect complementarity between siRNAs and their target RNA sequences is required for efficient gene silencing, the exposure of eukaryotes to siRNA frequently results in the unintended silencing of genes exhibiting <100% sequence identity to the trigger siRNA, a phenomenon termed off-target gene silencing (reviewed by Svoboda 2007). siRNA triggers off-target gene silencing in a variety of ways, including sequence-dependent and sequence-independent mechanisms. For example, siRNA is able to mimic microRNA (miRNA) to inhibit translation or elicit the degradation of mRNAs with partial sequence complementarity (Aleman et al. 2007). In addition, high concentrations of siRNA can induce an interferon response or global changes in gene expression by competing with limiting factors involved in the biogenesis and function of the endogenous RNAi pathway (Grimm et al. 2006). However, the lack of a genetically tractable system to study RNAi off-target effects has hindered the understanding of the mechanism of off-target gene silencing.
We conducted a genetic screen in C. elegans to identify the factors that are specifically required for RNAi off-target gene silencing. This screen identified alleles of rrf-1, nrde-2, and nrde-3. Here we show that following exogenous RNAi, the nuclear Ago NRDE-3 associates with off-target siRNAs and that the nuclear RNAi pathway, but not its cytoplasmic counterparts, is the primary source of off-target silencing in C. elegans. In addition, we find that NRDE-3 associates with endo-siRNAs that target repetitive genomic loci and that the nuclear RNAi pathway targets large gene families for silencing. We conclude that nuclear RNAi acts as a major contributor to exogenous and endogenous RNAi off-target effects in C. elegans.
Materials and Methods
Strains
Bristol strain N2 was used as the standard wild-type strain. The Hawaiian strain CB4856 was used for snp-SNP mapping. All strains were incubated at 20°: dpy-13(e458), sqt-3(e2924), eri-1(mg366), ergo-1(gg098), rde-1(ne219), nrde-1(gg088), nrde-2(gg091), nrde-2(ust005), nrde-3(gg066), nrde-3(gg245), rrf-1(pk1417), rrf-1(gg247), ppw-1(pk2505), ppw-2(tm1120), Y49F6A.1(tm1127), sago-1(tm1195), prg-2(tm1094), sur-5::gfp[kuIs54], FLAG::GFP::NRDE-3(ggIs1), MAGO11(−){wago-1(tm1414); wago-2(tm2686); wago-3(tm1120); wago-4(tm1019); wago-5(tm1113); wago-6(tm894); wago-7(tm914); wago-8(tm1195); wago-9(tm1200); wago-10(tm1186); wago-11(tm1127)}.
RNAi
RNAi experiments were performed as described previously (Timmons et al. 2001). Bacteria expressing dsRNA were mostly obtained from the Ahringer RNAi library and were sequenced to verify their identity (Kamath et al. 2003). The lin-15b RNAi clone has been described previously (Guang et al. 2008). The dpy-13 RNAi clone was constructed by PCR with the primers 5′-GGGAAGCTTCGTTCGTTACGGACGTGAC-3′ and 5′-GGGAAGCTTTTAGCGGCGAGTTCCG-3′, inserted into the HindIII site of the L4440 plasmid (a gift from A. Fire), and transformed into the HT115 Escherichia coli strain. To generate 76-bp dsRNA, synthetic oligonucleotides were annealed, phosphorylated by T4 polynucleotide kinase, and inserted into the HindIII site of the L4440 plasmid. The sequences of the primers used to generate each dpy-13 dsRNA expression clone are available upon request.
To generate GFP dsRNAs with increasing mismatches, the GFP sequence was amplified from pPD95.79 (a gift from A. Fire) by PCR under error-prone conditions with 4 mM MgCl2 and Taq polymerase. The PCR process was repeated 14 times, and the PCR products were cloned into L4440 and sequenced sequentially. The 10% and 20% mismatch levels were achieved at round 8 and round 14, respectively.
RNAi involving a dilution series was performed by mixing overnight cultures of bacteria expressing dsRNA with HT115 control bacteria. HT115 is an RNase III mutant bacterial strain that was modified to express T7 RNA polymerase from an IPTG-inducible promoter, which does not express either empty vector or dsRNA.
Images were collected using Zeiss Imager D1 and Leica DM2500 microscopes.
Genetic screening
To identify the factors specifically required for off-target gene silencing, we screened for cellular factors required for the dpy-13 dsRNA-induced silencing of off-target RNAs but dispensable for canonical RNAi silencing (see Supporting Information, Figure S2). dpy-13 RNAi results in a superdumpy phenotype in eri-1(mg366); dpy-13(e458) animals. Thus, eri-1(mg366); dpy-13(e458) animals were mutagenized by ethyl methanesulfonate (EMS), and the F2 progeny worms were grown on E. coli expressing dpy-13 dsRNA. Suppressors of the superdumpy phenotype were selected and tested with pos-1 and lir-1 RNAi to exclude the Rde and Nrde genes (Guang et al. 2008). Animals that suppressed the dpy-13 RNAi-induced superdumpy phenotype but still responded to pos-1 and lir-1 RNAi were selected for further analysis. Nine mutants were isolated from this genetic screening, three of which were identified by snp-SNP mapping and sequencing.
Quantitative RT-PCR
RNAs were isolated from embryos using a dounce homogenizer (pestle B) in TRIzol solution followed by purification with an RNeasy kit including on-column DNase I digestion (Qiagen). cDNAs were generated from RNAs using the iScript cDNA Synthesis kit (Bio-Rad) according to the vendor’s protocol. qPCR was performed using an MyIQ2 machine (Bio-Rad) with iQ SYBR Green Supermix (Bio-Rad). The primers for pre-mRNA analysis that were used in qRT-PCR are listed in Table S7. Independent primer pairs were also used in qRT-PCR and gave similar results (data not shown). eft-3 mRNA was used as an internal control for sample normalization. The data analysis was performed using a ΔΔCT approach.
Isolation and deep sequencing of NRDE-3-associated RNAs
NRDE-3-associated siRNAs were isolated from embryo lysates as previously described (Figure S5) (Guang et al. 2008). Briefly, the animals were sonicated in lysis buffer [20 mM Tris-HCl (pH 7.5), 200 mM NaCl, 2.5 mM MgCl2, and 0.5% NP-40]; the lysate was precleared with protein G-agarose beads (Roche), and incubated with anti-FLAG M2 agarose beads (Sigma). The beads were washed extensively, and FLAG::GFP::NRDE-3 and associated RNA were eluted with 100 μg/ml 3× FLAG peptide (Sigma). The eluates were incubated with TRIzol reagent (Invitrogen), followed by isopropanol precipitation. The precipitates were treated with calf intestinal alkaline phosphatase (CIAP, Invitrogen), reextracted with TRIzol, and treated with T4 polynucleotide kinase (T4 PNK, New England Biolabs) in the presence of 1 mM ATP.
The NRDE-3-associated siRNAs were cloned and deep sequenced using an Illumina platform, according to the manufacturer’s instructions, by the Beijing Genomics Institute (BGI Shenzhen). sRNAs ranging from 20 to 30 nt were gel purified and ligated to a 3′ adaptor (5′-pUCGUAUGCCGUCUUCUGCUUGidT-3′; p, phosphate; idT, inverted deoxythymidine) and a 5′ adaptor (5′-GUUCAGAGUUCUACAGUCCGACGAUC-3′). The ligation products were gel purified, reverse transcribed, and amplified using Illumina’s sRNA primer set (5′-CAAGCAGAAGACG GCATACGA-3′; 5′-AATGATACGGCGACCACCGA-3′). The samples were sequenced using an Illumina Hiseq platform.
The Illumina-generated raw reads were first filtered to remove adaptors, low quality tags, as well as contaminants to get clean reads at BGI Shenzhen. Clean reads ranging from 20 to 30 nt were mapped to the C. elegans genome and the transcriptome assembly WS229, respectively, using Bowtie2 with default parameters (Langmead and Salzberg 2012). The number of reads targeting each gene was counted by custom Perl scripts and displayed by IGV (Thorvaldsdottir et al. 2013).
AMA-1 (RNA polymerase II) ChIP-seq datasets were retrieved from GSM677643 of the modENCODE project, which identified the binding region of AMA-1 in L4 wild-type animals (Gerstein et al. 2010). The targets of WAGO-1, CSR-1, and eri-6/7 had been published previously (Claycomb et al. 2009; Gu et al. 2009; Fischer et al. 2011).
Results
RNAi triggers off-target gene silencing in C. elegans
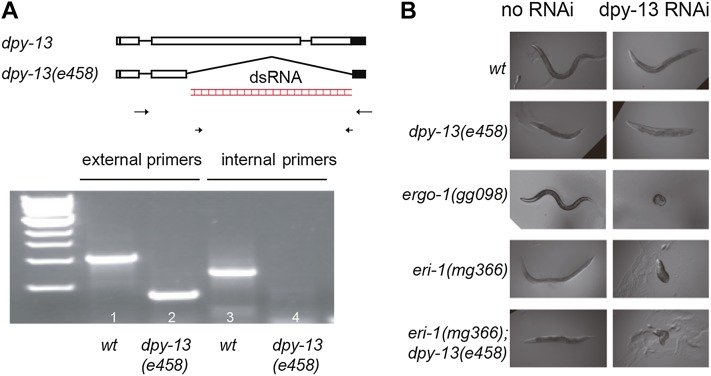
dpy-13 is a collagen gene that belongs to a large family of >100 members that share high sequence similarity in C. elegans. dpy-13(e458) lacks most of the dpy-13 coding region and is likely to be a null mutant (Figure 1A) (von Mende et al. 1988); dpy-13(e458) animals exhibit a dumpy phenotype with a length that is approximately half that of wild-type N2 animals (Figure 1B). Exposure of wild-type animals to dsRNA targeting dpy-13 resulted in the expected dumpy phenotype, consistent with dpy-13 dsRNA silencing dpy-13. Mutant backgrounds have been identified that enhance RNAi silencing. For instance, mutation of the eri-1, rrf-3, eri-6, eri-7, and ergo-1 genes results in an enhanced RNAi (Eri) phenotype (Kennedy et al. 2004; Duchaine et al. 2006; Fischer et al. 2011). Strikingly, RNAi of dpy-13 in ergo-1(gg098) and eri-1(mg366) animals elicited a worm-ball-like phenotype that is more severe (superdumpy) than that of the dpy-13 null phenotype. Furthermore, RNAi of dpy-13 in eri-1(mg366); dpy-13(e458) animals also induced the superdumpy phenotype. Note, the region of dpy-13 targeted by dpy-13 dsRNA in this experiment is missing in the eri-1(mg366); dpy-13(e458) strain, indicating that the superdumpy phenotype is not simply the result of highly efficient silencing of dpy-13 in enhanced RNAi backgrounds.
Figure 1.
Off-target silencing occurs in enhanced RNAi animals. (A) Gene structure of dpy-13. Recessive dpy-13(e458) is a 723-bp deletion, which removed 71% of the protein coding region and the translation stop codon. All of the conserved Gly-X-Y repeats, which are essential for the function of collagens, are deleted. wt, wild-type. (B) Feeding RNAi targeting the dpy-13(e458)-deleted sequence induced a worm-ball-like superdumpy phenotype in eri animals. L1 animals were fed dpy-13 dsRNA.
We asked if the superdumpy phenotype exhibited by Eri animals was caused by off-target gene silencing. We first mapped the minimal sequence in dpy-13 sufficient to induce a superdumpy phenotype by exposing eri-1(mg366) and eri-1(mg366); dpy-13(e458) animals to portions of the original dpy-13 dsRNA. The 76-nt fragment dpy-13g, which targets the 3′ end of the dpy-13 mRNA, was sufficient to induce a superdumpy phenotype (Figure S1). The same 76-nt region derived from other collagen genes, including sqt-3, col-43, col-93, and col-94, also induced a superdumpy-like phenotype (Table S1). dpy-13g does not contain a single 20+ nt sequence that perfectly matches another C. elegans gene, suggesting that dpy-13g-mediated superdumpy phenotype is due to off-target gene silencing. To test this idea, we introduced mutations into dpy-13g and found that these variants of dpy-13g were also able to trigger off-target silencing (Figure S1). These data indicate that the superdumpy phenotype elicited by dpy-13 dsRNAs is caused by RNAi off-target effects. Below, we show that superdumpy is triggered by the off-target silencing of other C. elegans collagen genes by dpy-13 dsRNA (see below).
Nuclear RNAi contributes to off-target silencing
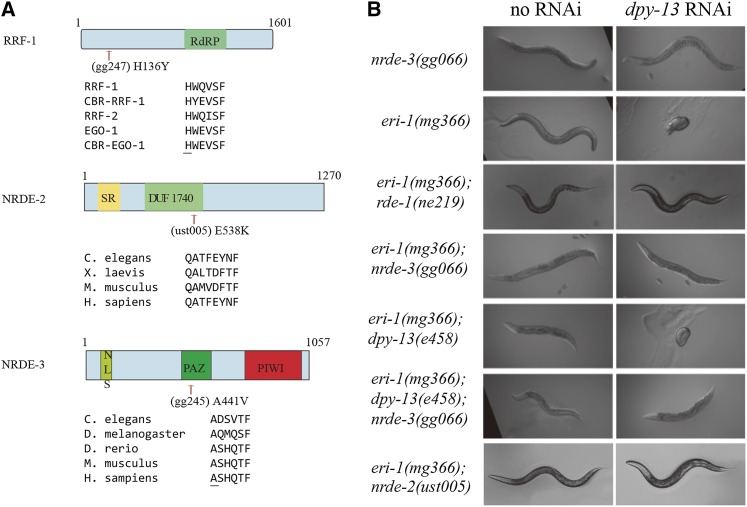
To identify the factors required for off-target gene silencing, we mutagenized eri-1(mg366); dpy-13(e458) animals and screened for mutants that failed to exhibit off-target silencing in response to dpy-13 dsRNA, but retained the ability to initiate RNAi against perfectly complementary cellular RNAs (Figure S2). Nine mutants were isolated that did not exhibit the dpy-13 RNAi-mediated superdumpy phenotype (off target), but were still fully responsive to pos-1 and unc-15 RNAi (on target). We mapped three of our nine mutant alleles by single-nucleotide polymorphism (SNP) mapping and found them to be rrf-1(H136Y), nrde-2(E538K), and nrde-3(A441V) (Figure 2A, Table 1). rrf-1, nrde-2, and nrde-3 are components of the C. elegans nuclear RNAi pathway, which are required for silencing of nuclear RNAs by dsRNA (Guang et al. 2008; Burkhart et al. 2011). The identification of rrf-1, nrde-2, and nrde-3 in our screen suggests that dsRNAs and siRNAs may guide RRF-1 to off-target mRNAs (partial sequence complementarity to siRNA) to direct unintended secondary siRNA synthesis and, consequently, nuclear RNAi at off-target sites within nuclei. The following results suggest that the alleles of rrf-1, nrde-2, and nrde-3 identified in our screen are hypomorphic alleles. Polycistronic RNAs in C. elegans are trans-spliced to monocistronic messages within nuclei. Therefore, targeting one gene in a polycistron with dsRNA and quantifying silencing of other genes encoded in the polycistron allows for quantification of nuclear RNAi. Two such polycistrons are lin-15a/lin-15b and lir-1/lin-26. Null alleles of rrf-1, nrde-2, and nrde-3 abolish nuclear RNAi at both of these polycistronic RNAs (Guang et al. 2008). The alleles of rrf-1, nrde-2, and nrde-3 identified in this screen, however, were defective for nuclear RNAi at the lin-15a/lin-15b operon, but not at the lir-1/lin-26 operon, suggesting that these alleles may be hypomorphic alleles of these genes. Consistent with this idea, we tested putative null alleles of rrf-1, nrde-2, and nrde-3 and found that these mutations also abolished off-target silencing by dpy-13 dsRNA (Figure 2B). For the remainder of this manuscript, we use the reference (null) alleles of rrf-1, nrde-2, and nrde-3 to explore the role of these nuclear RNAi factors in off-target gene silencing.
Figure 2.
(A) Genetic screening identified the alleles of rrf-1, nrde-2, and nrde-3 that are required for off-target gene silencing. eri-1(mg366); dpy-13(e458) animals were chemically mutagenized and screened for mutants that suppress off-target RNAi effect but are still sensitive to pos-1 and lir-1 RNAi. The mutants were mapped via the snp-SNP technique and sequenced. Underlining indicates the mutated amino acids. RRF-1, RRF-2, and EGO-1 are RNA-dependent RNA polymerases. CBR-EGO-1 and CBR-RRF-1 are C. briggsae genes. (B) Nuclear RNAi is required for dpy-13 dsRNA-induced off-target silencing. L1 animals of the indicated genotypes were fed dpy-13 dsRNA, and images were obtained 3 days later. RDE-1 is a primary Argonaute protein that is a prerequisite for feeding RNAi (Steiner et al. 2009). dpy-13 RNAi failed to produce any visible silencing effects in eri-1(mg366); rde-1(ne219) animals.
Table 1. Identification of rrf-1, nrde-2, and nrde-3 alleles required for RNAi.
| Genotype | RNAi | |||||
|---|---|---|---|---|---|---|
| pos-1 | lir-1 | lin-15b | unc-15 | dpy-13 | no RNAi | |
| no hatch | lethal | F1 muv | unc | dumpy | ||
| N2 | ++++ | − | − | ++++ | + | − |
| nrde-3(gg066) | ++++ | − | − | +++ | − | − |
| nrde-2(E538K) | ++++ | − | − | ++++ | − | − |
| eri-1(mg366) | ++++ | ++++ | ++++ | ++++ | ++++ | − |
| eri-1(mg366); rde-1(ne219) | − | − | − | − | − | − |
| ergo-1(gg098) | ++++ | ++++ | ++++ | ++++ | ++++ | − |
| eri-1(mg366); nrde-3(gg066)* | ++++ | − | − | +++ | + | − |
| eri-1(mg366); rrf-1(H136Y) | ++++ | ++++ | − | +++ | + | − |
| eri-1(mg366); nrde-2(E538K) | ++++ | +++ | − | +++ | + | − |
| eri-1(mg366); nrde-3(A441V) | ++++ | ++++ | − | +++ | + | − |
| {eri-1(mg366); MAGO12} wago-1(tm1414); wago-2(tm2686); wago-3(tm1120); wago-4(tm1019); wago-5(tm1113); wago-6(tm894); wago-7(tm914); wago-8(tm1195); wago-9(tm1200); wago-10(tm1186); wago-11(tm1127); nrde-3(tm1116); eri-1(mg366) | − | − | − | ++ | − | − |
| {eri-1(mg366); MAGO11(−); nrde-3(+)} | − | − | − | ++ | + | − |
nrde-3(gg066) is the reference allele.
+ means sensitive to RNAi; − means resistant to RNAi.
Nuclear RNAi is required to couple cosilencing between dpy-13 and sqt-3
siRNA can cause off-target gene silencing through sequence-dependent and sequence-independent mechanisms (Svoboda 2007). To begin to ask if dpy-13 dsRNA-induced superdumpy is caused by sequence dependent or independent mechanisms, we asked if the nuclear Ago NRDE-3, which is required for off-target silencing, associates with off-target siRNAs after dpy-13 dsRNA treatment. We fed eri-1(mg366); dpy-13(e458); FLAG::GFP::NRDE-3 animals with dpy-13 dsRNA (dsRNA targets dpy-13 deleted region). NRDE-3 localizes to the cytoplasm when not bound to siRNA and to nuclei when bound to siRNA (Guang et al. 2008). We observed that dpy-13 dsRNA induced the nuclear accumulation of FLAG::GFP::NRDE-3 in animals lacking the dpy-13 gene (Figure 3A), suggesting that dpy-13 dsRNA targeting the deleted region of dpy-13 induced the generation of off-target siRNAs that associate with NRDE-3. We sequenced small RNAs that associate with NRDE-3 before and after dpy-13 dsRNA treatment. NRDE-3 associated with few endogenous small RNAs targeting collagens in wild-type animals in the absence of dpy-13 dsRNA treatment (Figure 3B, Table S2). Following dpy-13 dsRNA treatment, however, NRDE-3 associated with abundant siRNAs targeting multiple collagen genes. The identification of off-target siRNAs bound to NRDE-3 is consistent with the idea that superdumpy is caused by dpy-13 RNAi-mediated off-target silencing of other related collagen genes. Interestingly, of all the collagen genes the sqt-3 gene was targeted by the most off-target siRNAs, suggesting that sqt-3 may be a major off-target of dpy-13 RNAi (Figure 3B). sqt-3 is a cuticle collagen essential for viability and body morphology, and the molecule is likely to form higher order structures with other cuticle collagens (Novelli et al. 2006). Two lines of evidence support the idea that sqt-3 is a major off-target of dpy-13 RNAi. First, we found that dpy-13(e458); sqt-3(e2924) double mutant animals exhibited a superdumpy-like phenotype (Figure 3C, Figure S3). Second, exposure of eri-1(mg366) and eri-1(mg366); dpy-13(e458) animals to dsRNA, targeting either dpy-13 or sqt-3, elicited superdumpy phenotypes (Figure 3D). eri-1(mg366); nrde-3(gg066) animals were only mildly sensitive to dsRNA, targeting either dpy-13 or sqt-3. However, eri-1(mg366); nrde-3(gg066); dpy-13(e458) animal displayed the superdumpy-like phenotype after sqt-3 RNAi, but not dpy-13 RNAi, suggesting that NRDE-3 likely couples the cosilencing of these two genes (Figure 3D). We conclude that the off-target effects of dpy-13 RNAi are the result of sequence-dependent off-target silencing of related collagen genes.
Figure 3.
Nuclear RNAi couples cosilencing between sqt-3 and dpy-13. (A) Off-target RNAi elicits the nuclear accumulation of NRDE-3. L4 animals were fed bacteria expressing dpy-13 dsRNA, and images of embryos were obtained 2 days later. Arrows indicate nuclei. (B) Feeding dpy-13 dsRNA leads to small RNAi targeting of other collagens. NRDE-3-associated small RNAs were immunoprecipitated from embryo lysates with an anti-FLAG antibody and subjected to deep sequencing. Reads of the small RNAs targeting collagens were compared. (C) A hypostasis interaction between dpy-13 and sqt-3. dpy-13(e458); sqt-3(e2924) double mutant leads to a superdumpy-like phenotype. (D) NRDE-3 is required to couple cosilencing between sqt-3 and dpy-13. L1 animals were fed dsRNAs targeting dpy-13 or sqt-3. The phenotypes were scored 3 days later. 0, no dumpy; 4, superdumpy. The experiments were performed in triplicate (N = 3).
Nuclear RNAi is required for off-target silencing induced by gfp dsRNA
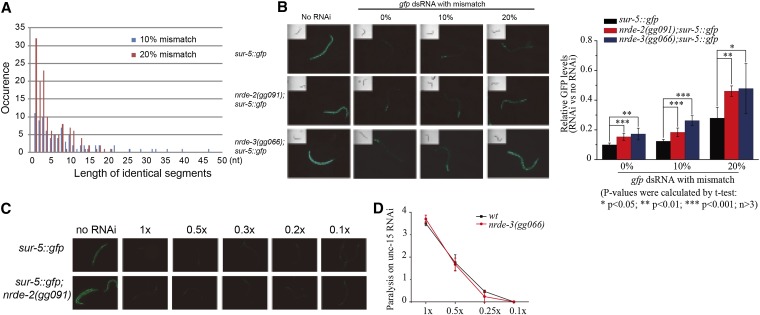
Two lines of evidence argue that the role of nuclear RNAi in off-target silencing is not limited to the collagen genes. First, we subjected animals to RNAi targeting 196 genes (168 operon genes and 28 multigene families), respectively, and asked if the phenotypes induced by these various RNAi treatments depended on the nuclear RNAi pathway. Most RNAi treatments had similar effects in both nuclear RNAi competent and nuclear RNAi defective animals (Table S3). Interestingly, many RNAi treatments that targeted multigene families such as the histone genes, homeobox genes, and G-protein-coupled receptor (GPCR) genes were strongly dependent on a functional nuclear RNAi pathway; animals lacking a functional nuclear RNAi pathway exhibited dramatically reduced phenotypes than animals with a functional RNAi pathway (Table S3, Figure S4). We speculate that the reason that nuclear RNAi is preferentially required for generating phenotypes following RNAi targeting multigene families is because these phenotypes are actually due to off-target silencing of other members of these large gene families. Second, we generated plasmids that express gfp dsRNA harboring randomly generated mutations introduced using error-prone PCR (Figure 4A). Animals expressing the sur-5::gfp transgene were fed wild-type, 10%, or 20% mismatched gfp dsRNA (Figure 4B). Wild-type gfp dsRNA silenced GFP expression in wild-type animals and nrde mutants efficiently, whereas the silencing effect of dsRNA with 10% mismatch was weakened in the nrde animals. gfp dsRNA with 20% mismatch efficiently silenced GFP expression in wild-type animals but showed diminished silencing in nrde mutants, suggesting that the nuclear RNAi machinery may be important for off-target silencing of the gfp gene. We note, however, that imperfectly complementary gfp dsRNA did not work as well as perfectly complementary dsRNA in either wild-type or nrde mutant animals, suggesting that the nuclear RNAi machinery is required for both on- and off-target gene silencing. To exclude the possibility of dose-response effects of the mismatched gfp in nrde mutants, we measured the silencing efficacy of a gfp dsRNA dilution series. We found that wild-type gfp dsRNA diluted 10-fold induced silencing in sur-5::gfp and nrde-2(gg091); sur-5::gfp animals in a similar fashion as the undiluted dsRNA (Figure 4C). Similar responses were also observed when wild-type and nrde-3(gg066) animals were fed serially diluted unc-15 dsRNA (Figure 4D). Therefore, the off-target silencing deficiency of nrde mutants is unlikely due to a dose-response effect of mismatched dsRNA, but, rather, may be an inherent characteristic of the nuclear RNAi pathway. Together, these data suggest that the nuclear RNAi machinery is required for off-target silencing following RNAi targeting many, if not all, genes in C. elegans.
Figure 4.
Nuclear RNAi is required for gfp dsRNA-induced off-target silencing. (A) Sequence comparison of mutagenized gfp dsRNAs that were generated by error-prone PCR. At the 20% mismatch level, one 21-nt segment is identical in sequence to wild-type GFP. (B) Left: L1 animals expressing sur-5::gfp were fed gfp dsRNAs with 0, 10, or 20% sequence mismatch. Right: GFP levels were quantified by ImageJ and normalized to control animals. (C) L1 animals expressing sur-5::gfp were fed a diluted series of gfp dsRNA. The dilution series was performed by mixing overnight cultures of bacteria expressing dsRNA with HT115 control bacteria. (D) L1 animals were fed a diluted series of unc-15 dsRNA, and their phenotypes were scored 3 days later.
Nuclear RNAi is a primary source of off-target silencing in C. elegans
We identified components of the C. elegans nuclear RNAi pathway in our genetic screen for RNAi off-target factors. One of these nuclear RNAi factors was the Ago NRDE-3. C. elegans expresses 12 secondary Argonaute proteins, including NRDE-3 (Yigit et al. 2006). We tested whether off-target gene silencing specifically requires the nuclear RNAi pathway by examining the role of other secondary Argonaute mutants, including ppw-1, ppw-2, Y49F6A.1, sago-1, and prg-2, in off-target gene silencing. We found that, unlike NRDE-3, these four secondary Argonautes were not required for off-target silencing (Table 2). In addition, the duodecuple mutant strain MAGO12 harbors deletion alleles in all 12 secondary Argonaute genes and is resistant to feeding RNAi (Table 1) (Gu et al. 2009). We crossed eri-1(mg366); MAGO12(−) with eri-1(mg366) and isolated eight independent F2 animals that were eri-1(mg366) but suppressed dpy-13 RNAi-induced off-target silencing. We genotyped the 12 Argonaute genes in these eight lines (Table S4). nrde-3(tm1116), but not the other 11 wago deletion alleles, was consistently linked to superdumpy suppression. These data suggest that NRDE-3 is the major secondary Argonaute that is responsible for the off-target silencing effects of dpy-13 dsRNA. We introduced a wild-type copy of nrde-3 into the MAGO12 strain to generate eri-1(mg366); MAGO11(−); nrde-3(+). We found that this strain was moderately defective for dpy-13-induced off-target silencing (Table 1). Thus, unknown combinations of other secondary Argonaute proteins may also contribute redundantly to RNAi off-target effects. We conclude that the nuclear Ago NRDE-3 is the primary, but not the sole, Ago-mediating off-target silencing. These results are consistent with the idea that nuclear RNAi is the major contributor of off-target silencing in C. elegans.
Table 2. Off-target gene silencing specifically requires Nrde pathway.
| Genotype | dpy-13 RNAi |
|---|---|
| dumpy | |
| eri-1(mg366) | ++++ |
| eri-1(mg366); rde-1(ne219) | − |
| eri-1(mg366); nrde-1(gg088) | + |
| eri-1(mg366); nrde-2(gg091) | + |
| eri-1(mg366); nrde-3(gg066) | + |
| eri-1(mg366); ppw-1(pk2505) | ++++ |
| eri-1(mg366); ppw-2(tm1120) | ++++ |
| eri-1(mg366); Y49F6A.1(tm1127) | ++++ |
| eri-1(mg366); sago-1(tm1195) | ++++ |
| eri-1(mg366); prg-2(tm1094) | ++++ |
| eri-1(mg366); MAGO11(−); nrde-3(+) {wago-1(tm1414); wago-2(tm2686); wago-3(tm1120); wago-4(tm1019); wago-5(tm1113); wago-6(tm894); wago-7(tm914); wago-8(tm1195); wago-9(tm1200); wago-10(tm1186); wago-11(tm1127); eri-1(mg366)} | + |
+ means sensitive to RNAi; − means resistant to RNAi.
NRDE-3 interacts with 22G-RNAs
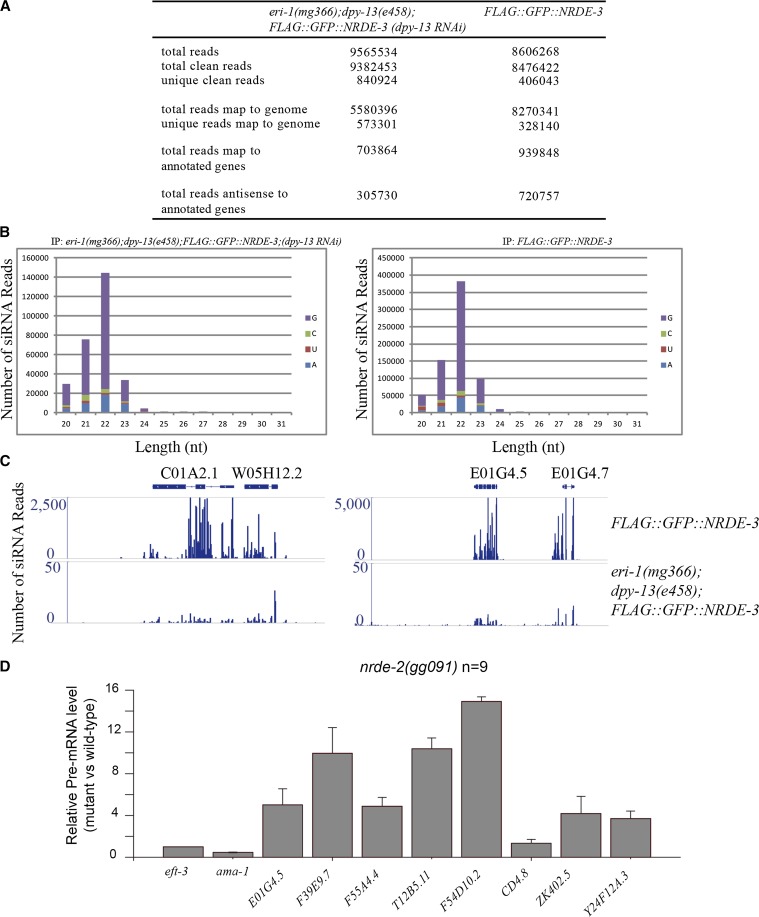
Previously, we sequenced endogenous siRNAs that bind to NRDE-3 on a small scale. To better understand the endogenous roles of the nuclear RNAi pathway, and to ask if nuclear RNAi drives off-target silencing during the normal course of growth and development, we immunoprecipitated NRDE-3 from the FLAG::GFP::NRDE-3 strain and deep sequenced the associated small RNAs from embryo lysates (Figures 5, Figure S5). eri-1(mg366); dpy-13(e458); FLAG::GFP::NRDE-3 fed dpy-13 RNAi was included as a control. We used a small RNA-cloning method compatible with cloning monophosphorylated and triphosphorylated small RNAs (i.e., 22G-RNAs). Small RNAs were isolated, pretreated with calf intestinal alkaline phosphatase to remove phosphate groups, treated with polynucleotide kinase to add a monophosphate group to the 5′ ends, and subjected to deep sequencing using the Illumina platform. Small RNAs were aligned to the C. elegans transcriptome (WS229 assembly) using Bowtie2 software with the default parameters, and the number of small RNAs targeting each gene was counted.
Figure 5.
Deep sequencing indicates that NRDE-3 associates with 22G-RNAs. (A) Summary of the deep-sequenced NRDE-3-associated small RNAs from eri-1(mg366); dpy-13(e458); FLAG::GFP::NRDE-3; dpy-13(RNAi) and FLAG::GFP::NRDE-3 animals. (B) Length and first letter distribution of NRDE-3-associated siRNAs. (C) eri-1 is required for the production of NRDE-3-associated siRNAs. (D) The expression of NRDE-3 targets is misregulated in nrde-2 mutants. Total RNAs were isolated from embryos and subjected to qRT-PCR to detect the pre-mRNA level of the indicated genes. Independent primer sets were applied, and similar results were obtained (data not shown). eft-3 mRNA was used as an internal control for normalization. N = 9.
We first focused on the small RNA reads that were complementary to annotated genes to avoid sense mRNA or sense pre-mRNA sequences that were likely to have co-immunoprecipitated with NRDE-3 (Guang et al. 2008, 2010). An analysis of both the length and first nucleotide distribution of the reads revealed that NRDE-3-associated siRNAs belong to the 22G-RNA class, which is consistent with previous results (Figure 5B; Guang et al. 2008; Zhang et al. 2011). NRDE-3-associated 22G-RNAs were dependent upon eri-1 (Figure 5C). We searched for NRDE-3 target genes by selecting genes that had >10 raw read counts (not normalized) and twofold enrichment (normalized) between FLAG::GFP::NRDE-3 and eri-1(mg366); dpy-13(e458); FLAG::GFP::NRDE-3; (dpy-13 RNAi) animals. Among the set of 178 genes identified as NRDE-3 targets (Table S5), E01G4.5 showed the most siRNA reads, in accordance with our previous result (Guang et al. 2008). Consistently, the expression of NRDE-3 targets was increased in the nrde-2(gg091) mutant, as determined by real-time PCR (Figure 5D).
Nuclear RNAi silences repetitive genomic loci
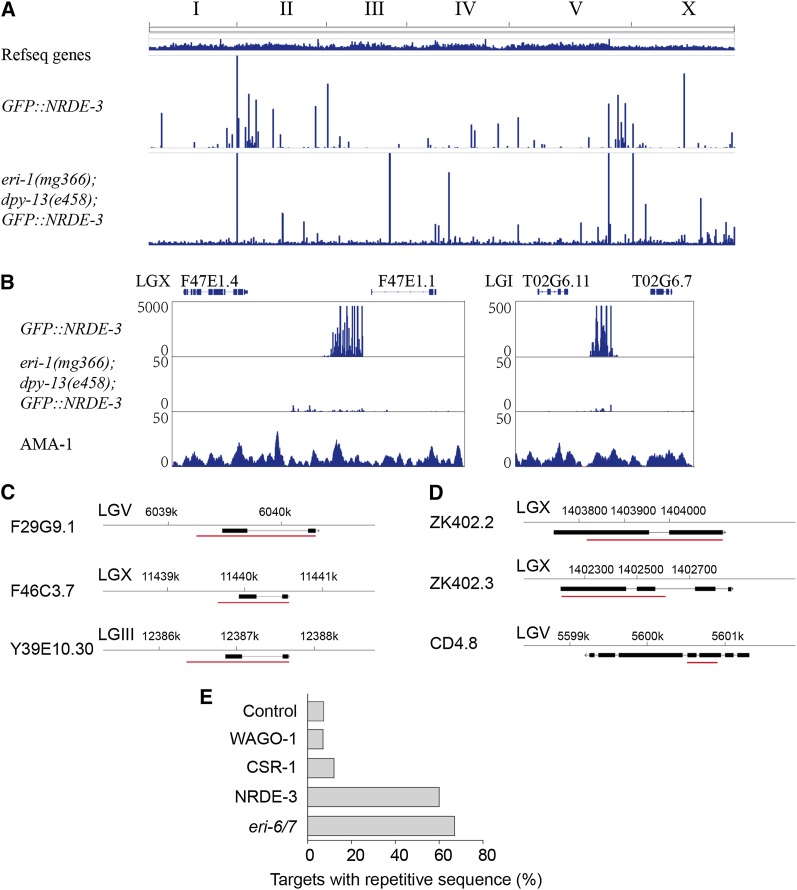
Interestingly, many of the NRDE-3 reads did not map to annotated genes (Figure 6A). We aligned all NRDE-3 associated small RNAs to the C. elegans genome and, surprisingly, found that many small RNAs are derived from intergenic genomic loci (Figure 6, A and B). Despite the lack of annotation, these sequences exist in clusters and are oriented on one strand as if they target expressed transcripts. These loci bind RNA polymerase II to produce nascent transcripts, suggesting that active transcription occurs at these sites (Figure 6B). We speculate that nuclear RNAi may target incompletely annotated transcripts at these genomic loci (Gu et al. 2009).
Figure 6.
Nuclear RNAi targets distinct genomic loci. (A) NRDE-3-associated small RNAs were mapped to the C. elegans genome and displayed using the integrative genomics viewer (IGV). (B) NRDE-3-associated small RNAs target intergenic regions. The left panel indicates X-cluster siRNAs on chromosome X, which has the most NRDE-3-bound small RNA reads (Guang et al. 2008). AMA-1 (RNA polymerase II) ChIP-seq datasets were retrieved from GSM677643 of the modENCODE project to identify the binding region of AMA-1 in L4 wild-type animals (Gerstein et al. 2010). (C and D) The genes targeted by NRDE-3-associated siRNAs were blasted against the genome. Shown are examples of the gene groups that share high sequence similarity with a cutoff of >90% nt identity over a stretch of >200 nt of sequence. The red bars indicate the regions with sequence homology. (E) The repetitiveness of the targets of three secondary Argonaute proteins, including WAGO-1, CSR-1, and NRDE-3, and the targets of eri-6/7 were compared by using a criteria of >90% nt identity in a stretch of >200 nt sequence to one or more of the other targeted genes or genomic loci. A set of randomly selected genes (n = 40) was used as a control.
To analyze the extent of homology between nuclear RNAi target loci, we identified 63 genomic loci that were complementary to small RNA reads (Figure 6A, Table S6). The sequences of these loci were retrieved and aligned to the genome using BLASTn with a cutoff of >90% nt identity over a stretch of >200 nt of sequence. Most of the 63 loci are homologous to another region in the genome (Table S6). We also examined the NRDE-3-targeted genes and found that they are broadly repetitive with each other (Figure 6, C and D). Consistent with this idea, it was recently reported that eri-6/7-targeted genes are repetitive in sequence and that NRDE-3-bound siRNAs depend on eri-6/7 (Fischer et al. 2011). To ask if the repetitiveness of NRDE-3 target loci was unique to NRDE-3 or a general characteristic of small RNA-targeted genes in C. elegans, we compared the repetitiveness of the targets of three secondary Argonaute proteins, including WAGO-1, CSR-1, and NRDE-3 (Figure 6E). As described above, more than 60% of the targets of NRDE-3 were repetitive, yet only 10% of WAGO-1 and CSR-1 targets were repetitive. Within a set of randomly selected genes, ∼7.5% (3 of 40) share this degree of sequence similarity. Thus, NRDE-3-associated 22G siRNAs preferentially target repetitive genomic loci and the repetitiveness of these target loci is particular to NRDE-3-associated siRNAs. We note, however, that this correlation is not perfect: there are very few NRDE-3-associated endogenous siRNAs targeting multigene families such as the collagen genes or other protein coding genes from multigene families and most CSR-1-associated endo-siRNAs target genes are not repetitive (Table S2, Table S5) (Claycomb et al. 2009).
Discussion
Here, we report that components of the C. elegans nuclear RNAi pathway are required for off-target silencing. We also show that members of large gene families are prone to silencing by the nuclear RNAi pathway. Our data suggest that nuclear RNAi, but not its cytoplasmic counterparts, is the primary source of off-target silencing. We also identified endogenous nuclear RNAi targets by deep sequencing and found that NRDE-3 binds to 22G-RNAs and targets distinct repetitive genomic loci. These data revealed unexpected roles for nuclear RNAi in gene silencing.
Why off-target silencing?
Invasive parasitic nucleic acids can evolve rapidly, which results in highly similar and heterogeneous sequences. To protect the host genome, allowing RNAi off-target silencing can enable the host to efficiently recognize, memorize, and defend against these sequences. Consistent with this idea, nuclear RNAi is involved in the inheritance of RNAi silencing (Buckley et al. 2012; Feng and Guang 2013). In addition, many of the NRDE-3 targets are pseudogenes and unlikely express functional proteins (Fischer et al. 2011). C. elegans expresses many pseudogenes that exhibit sequence similarity to other protein coding genes. Pseudogenes can also evolve rapidly and interfere with the normal function of the homologous protein coding genes, therefore requiring an efficient silencing by nuclear RNAi.
Off-target gene silencing is one of the major concerns for the application of RNAi-based technology. Although most people assume that the off-target effects of RNAi are simply due to inherent and unavoidable biochemical limitations of the RNAi machinery, our data suggest that off-target silencing is a genetically programmed aspect of RNAi. Nuclear RNAi machinery is the major contributor to RNAi off-target effects in C. elegans, implying its potential application in clinics. Off-target silencing can be alleviated via the use of low concentrations of siRNA, adding methyl groups, and/or modifying siRNA with locked nucleic acids (LNAs) (Svoboda 2007). Our data suggest that off-target gene silencing could also be reduced by abrogating the nuclear RNAi pathway. As NRDE-2 is a conserved protein that has only one homolog in mammals (Guang et al. 2010), it will be interesting to explore how NRDE-2 is involved in nuclear RNAi and off-target silencing in higher eukaryotes.
RNAi in eri animals induces unintended silencing effects
eri mutant animals respond more robustly than wild-type animals to exogenously provided dsRNA (Kennedy et al. 2004; Duchaine et al. 2006; Fischer et al. 2011). This effect could be partially explained by a competition model in which the exogenous RNAi pathway competes with the endogenous RNAi pathway for limiting factors (Lee et al. 2006; Yigit et al. 2006). Indeed, the overexpression of SAGO-1, SAGO-2, or NRDE-3 enhances feeding RNAi, indicating that some secondary Argonaute proteins could be the limiting factors (Yigit et al. 2006; Zhuang et al. 2013). On the other hand, our data suggest that at least some of the enhanced RNAi effect could arise from off-target silencing, particularly for dsRNAs targeting large gene families.
RRF-1 is required for off-target silencing
Off-target silencing can happen either by imperfect complementation between siRNA with its targets or by synthesizing off-target secondary siRNA in C. elegans. In the latter case, Argonaute-mediated silencing may be a targeted process per se, directed by RdRP-generated off-target secondary siRNAs. The identification of siRNAs that target collagens other than dpy-13 in eri-1(mg366); dpy-13(e458); FLAG::GFP::NRDE-3 after dpy-13 RNAi supports the second model.
It is unknown why nuclear RNAi is special in contributing to off-target silencing. Previous genetic screening to search for nuclear RNAi defective (Nrde) mutants had isolated many rrf-1 alleles, suggesting an inherent connection between rrf-1 and nuclear RNAi (Guang et al. 2008; Burkhart et al. 2011). RRF-1 is required for NRDE-3 to bind both endogenous siRNA and siRNA elicited by feeding RNAi (Guang et al. 2008). However, we have conducted proteomic analysis of NRDE-2 and NRDE-3 but failed to identify RRF-1 interaction with either of them (data not shown). It is possible that RRF-1-synthesized off-target secondary siRNAs can preferentially bind to NRDE-3 and trigger silencing in the nucleus. Alternatively, off-target siRNAs can bind either NRDE-3 or other cytoplasmic Argonautes; yet, nuclear RNAi can be more permissive than cytoplasmic silencing systems. It will be very interesting to sequence and compare siRNAs from all secondary Argonaute proteins after dpy-13 RNAi to answer this question.
Nuclear RNAi mediates off-target silencing
We observed that nrde-3, but not ppw-1, ppw-2, Y49F6A.1, sago-1, or prg-2, is phenotypically critical for off-target silencing, suggesting that some of the effects of off-target silencing strongly depend on nuclear RNAi machinery. Alternatively, cytoplasmic Argonaute proteins might be more redundant in C. elegans; therefore, knocking out one cytoplasmic Ago is not sufficient to elicit visible RNAi effects. The eri-1(mg366); MAGO11(−); nrde-3(+) strain is nearly completely RNAi defective in the germline but partially RNAi defective in the soma, and NRDE-3 is only expressed in soma (Guang et al. 2008), suggesting that NRDE-3 likely functions in parallel with cytoplasmic Argonaute proteins to perform silencing. However, the observation that dpy-13 RNAi of eri-1(mg366); MAGO11(−); nrde-3(+) animals does not induce superdumpy and the MAGO12(−) strain is defective in small RNA accumulation (Vasale et al. 2010), suggest that cytoplasmic Argonaute proteins may also act as a whole to stabilize siRNAs and indirectly modulate nuclear RNAi.
NRDE-3 binds to 22G-RNAs
In C. elegans, endogenous siRNAs are mostly categorized according to their length and 5′ nucleotide: 22G-RNAs are predominantly 22 nt in length and contain a triphosphorylated 5′-G. These siRNAs bind to particular subtypes of the 27 C. elegans Argonaute proteins to confer their unique functionality. The majority of endogenous siRNAs are derived from RdRP transcripts and likely undergo additional nuclease-mediated processing. 22G-RNAs are antisense to >50% of annotated genes (Gu et al. 2009). Two major 22G-RNAs exist in C. elegans: those that interact with CSR-1 and those that interact with worm-specific AGO (WAGO) proteins. It has been suggested that the WAGO/22G-RNA system provides surveillance against transposable elements and aberrant transcripts. ERGO-1-dependent 26G-siRNAs were derived from unannotated intergenic regions (Vasale et al. 2010), and NRDE-3 targets repetitive intergenic loci, suggesting that nuclear RNAi suppresses aberrant transcripts at these sites. Further deep sequencing and annotation to identify long intervening noncoding RNAs (lincRNAs) will facilitate an understanding of these intergenic loci in C. elegans.
Supplementary Material
Acknowledgments
We are grateful to Scott Kennedy, Guang Yao, Yupeng Yang, Ge Shan, Qingfa Wu, and Ying Liu and members of the Kennedy and Guang labs for comments. We are grateful to the Caenorhabditis Genetics Center for providing the strains. This work was supported by grants from the Chinese 973 Program (no. 2011CBA01100), the National Natural Science Foundation of China (nos. 31171254 and 31371323), the Fundamental Research Funds for Central Universities (nos. WK2060190018 and WK2070000034), and the research foundation of Chinese Academy of Science, KJZD-EW-L01-2.
Footnotes
Communicating editor: B. Meyer
Literature Cited
- Aleman L. M., Doench J., Sharp P. A., 2007. Comparison of siRNA-induced off-target RNA and protein effects. RNA 13: 385–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley B. A., Burkhart K. B., Gu S. G., Spracklin G., Kershner A., et al. , 2012. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature 489: 447–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart K. B., Guang S., Buckley B. A., Wong L., Bochner A. F., et al. , 2011. A pre-mRNA-associating factor links endogenous siRNAs to chromatin regulation. PLoS Genet. 7: e1002249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb J. M., Batista P. J., Pang K. M., Gu W., Vasale J. J., et al. , 2009. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell 139: 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchaine T. F., Wohlschlegel J. A., Kennedy S., Bei Y., Conte D., Jr, et al. , 2006. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 124: 343–354. [DOI] [PubMed] [Google Scholar]
- Feng X., Guang S., 2013. Small RNAs, RNAi and the inheritance of gene silencing in Caenorhabditis elegans. J. Genet. Genomics 40: 153–160. [DOI] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M. K., Kostas S. A., Driver S. E., et al. , 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. [DOI] [PubMed] [Google Scholar]
- Fischer S. E., Montgomery T. A., Zhang C., Fahlgren N., Breen P. C., et al. , 2011. The ERI-6/7 helicase acts at the first stage of an siRNA amplification pathway that targets recent gene duplications. PLoS Genet. 7: e1002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein M. B., Lu Z. J., Van Nostrand E. L., Cheng C., Arshinoff B. I., et al. , 2010. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330: 1775–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D., Streetz K. L., Jopling C. L., Storm T. A., Pandey K., et al. , 2006. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 441: 537–541. [DOI] [PubMed] [Google Scholar]
- Gu W., Shirayama M., Conte D., Jr, Vasale J., Batista P. J., et al. , 2009. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol. Cell 36: 231–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang S., Bochner A. F., Pavelec D. M., Burkhart K. B., Harding S., et al. , 2008. An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science 321: 537–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang S., Bochner A. F., Burkhart K. B., Burton N., Pavelec D. M., et al. , 2010. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature 465: 1097–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R. S., Fraser A. G., Dong Y., Poulin G., Durbin R., et al. , 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237. [DOI] [PubMed] [Google Scholar]
- Kennedy S., Wang D., Ruvkun G., 2004. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature 427: 645–649. [DOI] [PubMed] [Google Scholar]
- Ketting R. F., 2011. The many faces of RNAi. Dev. Cell 20: 148–161. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R. C., Hammell C. M., Ambros V., 2006. Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans. RNA 12: 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novelli J., Page A. P., Hodgkin J., 2006. The C terminus of collagen SQT-3 has complex and essential functions in nematode collagen assembly. Genetics 172: 2253–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak J., Maniar J. M., Mello C. C., Fire A., 2012. Protection from feed-forward amplification in an amplified RNAi mechanism. Cell 151: 885–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner F. A., Okihara K. L., Hoogstrate S. W., Sijen T., Ketting R. F., 2009. RDE-1 slicer activity is required only for passenger-strand cleavage during RNAi in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 16: 207–211. [DOI] [PubMed] [Google Scholar]
- Svoboda P., 2007. Off-targeting and other non-specific effects of RNAi experiments in mammalian cells. Curr. Opin. Mol. Ther. 9: 248–257. [PubMed] [Google Scholar]
- Thorvaldsdottir H., Robinson J. T., Mesirov J. P., 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14: 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L., Court D. L., Fire A., 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263: 103–112. [DOI] [PubMed] [Google Scholar]
- Vasale J. J., Gu W., Thivierge C., Batista P. J., Claycomb J. M., et al. , 2010. Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. Proc. Natl. Acad. Sci. USA 107: 3582–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mende N., Bird D. M., Albert P. S., Riddle D. L., 1988. dpy-13: a nematode collagen gene that affects body shape. Cell 55: 567–576. [DOI] [PubMed] [Google Scholar]
- Yigit E., Batista P. J., Bei Y., Pang K. M., Chen C. C., et al. , 2006. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 127: 747–757. [DOI] [PubMed] [Google Scholar]
- Zhang C., Montgomery T. A., Gabel H. W., Fischer S. E., Phillips C. M., et al. , 2011. mut-16 and other mutator class genes modulate 22G and 26G siRNA pathways in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 108: 1201–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang J. J., Banse S. A., Hunter C. P., 2013. The nuclear argonaute NRDE-3 contributes to transitive RNAi in Caenorhabditis elegans. Genetics 194: 117–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.