Abstract
We consider genomic imputation for low-coverage genotyping-by-sequencing data with high levels of missing data. We compensate for this loss of information by utilizing family relationships in multiparental experimental crosses. This nearly quadruples the number of usable markers when applied to a large rice Multiparent Advanced Generation InterCross (MAGIC) study.
Keywords: GBS, MPRIL, HMM, MAGIC
WHILE genotyping-by-sequencing (GBS) technology has made dense genotyping cost effective for a wide variety of species, the often high levels of missing data can result in a large loss of information (Elshire et al. 2011). Imputation is possible for human populations with reference panels of high-coverage genotypes (International Hapmap Consortium 2003), but such panels are rarely available for plant species, making it difficult if not impossible to apply standard software.
The popularity of GBS makes the development of efficient imputation approaches a priority even for species lacking the resources of human populations. In the contexts of genomic selection and map construction, Rutkoski et al. (2013) and Ward et al. (2013) have considered imputation approaches for species without reference genomes. Here we consider imputation under the further difficulty caused by multiparental experimental crosses.
Multiparental experimental cross designs are becoming increasingly common in plant studies, as they offer greater diversity than traditional biparental designs do with less complexity than genome-wide association panels (Cavanagh et al. 2008; McMullen et al. 2009; Kover et al. 2009). The limited set of founders offers a ready-made “reference panel” for imputation of genotypes, with approaches designed either for unrelated populations (Browning and Browning 2009; Howie et al. 2009; Li et al. 2010) or for inbred lines (Mott et al. 2000; Huang and George 2011).
For many imputation approaches, however, high-quality founder genotypes are essential. Markers with missing founder genotypes must be discarded, which can result in a large loss of data if both founders and progeny are genotyped using low-coverage GBS. Here we present an approach to imputing founder genotypes in these populations, which allows recovery of a large proportion of markers. Once founder genotypes have been imputed, we assess the efficacy of a population-oriented approach (BEAGLE; Browning and Browning 2009) against a family-oriented approach (R/mpMap; Huang and George 2011) in imputing progeny genotypes. Further, we compare both approaches to the general purpose alternative of weighted k nearest-neighbor imputation (Schwender 2012). We apply our strategy to an eight-parent rice Multiparent Advanced Generation InterCross (MAGIC) population and demonstrate the potential gain from imputation.
Materials and Methods
Our approach has been implemented in R (R Core Team 2013) and is available as the function “mpimpute” in R/mpMap (http://github.com/behuang/mpMap). The procedure for imputation of genotypes is outlined in Figure 1. We first fit a hidden Markov model (HMM), which allows missing founder genotypes to calculate founder allele probabilities in the progeny (Broman et al. 2003). Next, for each missing founder value, we audit the observed progeny genotypes among lines inheriting an allele with high probability from the missing founder and impute the most likely value. Finally, we reestimate the founder allele probabilities using the imputed founder genotypes. For each missing progeny value, we combine the founder probabilities across observed genotypes and impute the most likely value. In the latter two steps, we can set thresholds on the most likely genotype probability, so that values will not be imputed if insufficiently probable. Alternately, progeny genotypes can be output as the expected allelic value, which may be preferable in downstream analyses such as association mapping (Zaykin et al. 2002).
Figure 1.
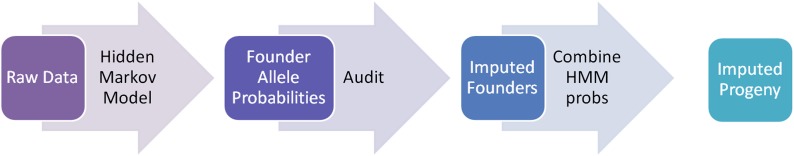
Diagram of process for imputing missing genotypes. We first construct a hidden Markov model (HMM) based on the progeny genotypes to estimate founder haplotype probabilities across the genome. Then at each position, for each missing founder, we audit genotypes among progeny inheriting that founder haplotype. The most common genotype inherited in those progeny is imputed in the founders. The imputed founders are then used as a reference panel to impute missing progeny data by summing HMM probabilities for possible alleles.
We tested this approach first through simulation. We generated 1000 data sets using R/mpMap for each of 16 different scenarios, varying marker density, sample size, and percentage of missing data (script in supporting information, File S1, and example data in File S2). Each data set consisted of 200 or 400 lines from an eight-parent MAGIC population with one funnel selfed for six generations. A single chromosome of length 100 cM was simulated with markers evenly spaced every 0.5 or 1 cM. Individuals were simulated with percentage missing data in the progeny, %MISS, and this value squared in the founders. This corresponds to two replicates of each founder being genotyped, with missing data occurring randomly in each sample. In general, we expect that in GBS data the missing markers may be different for each progeny and founder sample, and hence we generate the missing data randomly across the genome for each line.
We applied the imputation approach to a real data set derived from an eight-parent MAGIC rice population. Full details of the population design and genotyping can be found in Bandillo et al. (2013). In brief, 8 parent and 178 S4 lines (selfed for four generations; pedigree in File S3) were genotyped using genotyping-by-sequencing (Elshire et al. 2011) with position aligned to Os-Nipponbare-Reference-IRGSP-1.0 (IRGSP-1.0). The average depth of coverage was 4.8 reads per site, with ∼300,000 markers called per sample initially. Heterozygotes were treated as missing data. This was done due to less certainty in the accuracy of heterozygote calls; as S4 lines are close to fixed, heterozygotes may represent incorrect calls.
As a preprocessing step, we removed markers across the genome with (a) >60% missing data in the progeny, (b) no alleles observed in the parents, and (c) complete data in the parents but only a single allele observed. These filtering criteria allow the removal of regions of the genome for which there may be low confidence in the alignment of reads or that do not vary in the population. In the remaining 37,240 markers (File S4), the average spacing was 0.04 cM (9955 bp), and the largest gap was 2 cM (502,775 bp). However, in markers with complete founder genotypes prior to imputation, the average spacing was 0.16 cM (39,398 bp), and the largest gap was 7.5 cM (1,875,124 bp).
Results and Discussion
Simulations demonstrated generally good performance of the method, with accuracy increasing with marker density and sample size. Table 1 summarizes our results for a variety of scenarios. Founder genotypes were nearly always fully imputed accurately (columns %F, %FC). The five nearest neighbor markers based on simple matching coefficient distance were also used for imputation (%FK, %K) but had much poorer performance.
Table 1. Simulation results averaged across 1000 replicates of eight-parent MAGIC populations.
| M | N | %MISS | %F0 | %F | %FC | %FK | %K | %B | %M |
|---|---|---|---|---|---|---|---|---|---|
| 101 | 200 | 30 | 46.9 | 100 | 100 | 86.6 | 79.8 | 93.7 | 96.3 |
| 101 | 200 | 40 | 24.5 | 100 | 100 | 85.4 | 78.8 | 93.0 | 95.5 |
| 101 | 200 | 50 | 9.8 | 100 | 99.6 | 83.9 | 77.5 | 92.0 | 94.8 |
| 101 | 200 | 60 | 3.5 | 99.9 | 98.3 | 81.6 | 75.7 | 88.7 | 93.2 |
| 101 | 400 | 30 | 47.3 | 100 | 100 | 88.4 | 80.3 | 94.3 | 96.3 |
| 101 | 400 | 40 | 24.9 | 100 | 100 | 87.4 | 79.4 | 93.8 | 95.5 |
| 101 | 400 | 50 | 10.1 | 100 | 100 | 86.2 | 78.2 | 92.6 | 94.8 |
| 101 | 400 | 60 | 3.6 | 100 | 99.7 | 84.3 | 76.7 | 90.7 | 93.5 |
| 201 | 200 | 30 | 47.1 | 100 | 100 | 90.7 | 83.5 | 96.7 | 98.3 |
| 201 | 200 | 40 | 24.8 | 100 | 100 | 89.5 | 82.3 | 96.3 | 98.0 |
| 201 | 200 | 50 | 10.0 | 100 | 100 | 87.8 | 80.8 | 95.4 | 97.6 |
| 201 | 200 | 60 | 2.9 | 100 | 99.7 | 85.2 | 78.8 | 91.2 | 96.8 |
| 201 | 400 | 30 | 47.1 | 100 | 100 | 92.1 | 84.1 | 97.0 | 98.3 |
| 201 | 400 | 40 | 24.9 | 100 | 100 | 91.3 | 83.1 | 96.5 | 98.0 |
| 201 | 400 | 50 | 10.0 | 100 | 100 | 90.1 | 81.8 | 96.0 | 97.6 |
| 201 | 400 | 60 | 2.8 | 100 | 100 | 88.2 | 80.0 | 94.3 | 96.8 |
For each scenario we record the percent of markers with complete founder genotypes originally (%F0), the percentage complete after imputation (%F), the percentage of missing founder genotypes correctly imputed (%FC), the percentage correctly imputed based on the five nearest neighbor markers (%FK), the percentage of missing progeny genotypes correctly imputed based on the five nearest neighbor markers (%K), the percentage correctly imputed with BEAGLE (%B), and the percentage correctly imputed with R/mpMap (%M).
Once founder genotypes were imputed, we used them as a reference panel for imputation of progeny genotypes. As there were few errors in founder imputation, errors here are simply due to uncertainty in which the allele was inherited. Increases in marker density had a greater impact on performance than did increases in sample size, since neighboring markers will then more accurately represent those with missing data. BEAGLE and R/mpMap typically both have ∼5% errors in imputation, although the family approach tends to have slightly fewer errors.
In the rice data, we first considered a test example of markers from chromosome 1. From the set of 1130 markers with complete founder genotypes on the chromosome, we simulated 22% missingness and compared the imputed founder genotypes to the true values. We were able to impute the set of 128 markers with complete founder data up to 1092 with 96% of missing values correctly imputed. The lower accuracy (relative to simulation) should be kept in mind during future analyses, ideally by utilizing methods that account for potential genotyping errors. Following this, we imputed markers in both the founders and the progeny for the full data set, with results summarized in Table 2.
Table 2. Results of imputation in rice 8-way MAGIC.
| Chr | M | %F0 | %F | %MISS | G0 | G | D0 | D |
|---|---|---|---|---|---|---|---|---|
| 1 | 4993 | 22.6 | 93.1 | 39.1 | 5.8 | 1.4 | 0.15 | 0.037 |
| 2 | 4054 | 28.1 | 93.3 | 38.5 | 3.2 | 0.9 | 0.13 | 0.038 |
| 3 | 2592 | 30.7 | 97.2 | 36.9 | 5.2 | 1.2 | 0.18 | 0.058 |
| 4 | 4547 | 23.5 | 91.6 | 39.4 | 7.3 | 1.8 | 0.13 | 0.034 |
| 5 | 2257 | 27.9 | 94.2 | 36.5 | 6.0 | 1.9 | 0.19 | 0.056 |
| 6 | 2294 | 27.2 | 95.1 | 37.6 | 7.5 | 1.3 | 0.20 | 0.057 |
| 7 | 2756 | 27.8 | 96.6 | 38.4 | 4.2 | 1.7 | 0.15 | 0.045 |
| 8 | 2466 | 21.0 | 94.0 | 38.1 | 7.2 | 1.8 | 0.22 | 0.049 |
| 9 | 2029 | 26.3 | 90.3 | 36.1 | 5.2 | 2.0 | 0.17 | 0.049 |
| 10 | 2359 | 23.7 | 91.3 | 37.2 | 2.9 | 1.0 | 0.17 | 0.043 |
| 11 | 3356 | 23.0 | 92.3 | 38.8 | 2.7 | 0.8 | 0.15 | 0.037 |
| 12 | 3537 | 23.8 | 85.9 | 37.0 | 4.2 | 1.7 | 0.13 | 0.036 |
| All | 37240 | 25.2 | 92.7 |
Physical map positions were converted to centimorgans using a conversion factor of 1 cM/250 kb. For each chromosome, we report the number of markers postprocessing (M) to be imputed; the proportion of markers with complete founder genotypes initially (%F0); the proportion of markers with complete founder genotypes postimputation (%F); the percentage of missing data among the progeny (%MISS); the maximum gap between markers with complete founder genotypes in centimorgans before and after imputation (G0/G); and the mean distance between markers with complete founder genotypes before and after imputation in centimorgans (D0/D).
The approach presented here improves the yield of low-coverage GBS in complex crosses by allowing high-accuracy imputation in both parents and progeny. From simulation we note that the method performs well as long as there is a high density of markers, which is one of the main features of GBS. Increasing filtering thresholds for markers and genotyping several replicates of founders are simple ways to improve the overall data quality.
Our results in real data are slightly poorer than those observed in simulation with similar sample sizes and levels of missingness. Our performance criterion is based on the proportion of genotypes correctly imputed, and we typically manage to recover complete founder genotypes for > 90% of the markers (Table 2, column %F). This may slightly overestimate the performance in imbalanced populations given that the probability of correct imputation depends on allele frequencies. In eight-parent MAGIC populations most markers have minor allele frequencies >10%; however, in different designs with more parents it may be preferable to consider a measure of the genetic variation explained (e.g., correlation between imputed and true results).
A number of factors may explain the differences between real data and simulation. These include lower diversity of haplotypes in the blocks used for imputation, larger gaps between markers, and missingness that is not completely random. The diversity of haplotypes is directly affected by different genetic relationships among the founders. In the rice data, all eight founders are of the indica subtype, with percentage similarity ranging from 61 to 79%. Increased similarity between parents may decrease the accuracy of imputation, as the increased difficulty in discriminating between individual founders will be reflected in lower confidence in the probabilities of inheriting specific alleles.
Gaps between markers and nonrandom missingness may be due to variation in genome alignment of the reads. Indeed, our approach relies on the assumption of correct read alignment; hence it may be desirable to filter regions of the genome where this is a concern. Rice specifically does have substantial genome structure variation; however, past work (Arai-Kichise et al. 2011; Xu et al. 2012) has shown that genomes of different varieties are at least 90% similar to the reference genome. Hence most SNPs from a GBS assay derive from common regions of the reference and the varieties being assayed. In our data gaps between markers typically reflected the sub-centimorgan scale used in simulation; however, a few larger gaps may have affected performance.
Currently, our approach assumes that lines are nearly fully inbred, and heterozygous allele calls are treated as missing data. Hence imputation is not recommended for populations with high levels of residual heterozygosity, as genotypes will not be imputed correctly. However, it is possible to accommodate heteroyzygous genotypes by employing an alternate HMM, which would allow the use of this approach in populations such as heterogeneous stock. Options such as R/DOQTL (http://cgd.jax.org/apps/doqtl/DOQTL.shtml) and reconstruction (http://mus.well.ox.ac.uk/19genomes/magic.html) allow for heterozygosity in the progeny and could be integrated with our approach for imputation in less inbred populations.
Once founder genotypes have been imputed, the final step is to impute the progeny genotypes. R/mpMap is designed specifically for MAGIC populations, and as such it is unsurprising that it has slightly better performance than BEAGLE. However, it will not be applicable to other complex experimental crosses. For this, alternatives such as BEAGLE or HAPPY (Mott et al. 2000) will still provide imputation with low error rates and work well even for large data sets. While founder imputation is not strictly necessary to estimate haplotype probabilities for imputation, we have found that it does reduce the uncertainty in estimates and improves the final results.
Supplementary Material
Acknowledgments
We thank the Associate Editor and two anonymous reviewers for their helpful comments. Emma Huang is the recipient of an Australian Research Council Discovery Early Career Research Award (DE120101127). Karl W. Broman was supported by National Institutes of Health grant R01 GM074244.
Footnotes
Communicating editor: I. Hoeschele
Literature Cited
- Arai-Kichise Y., Shiwa Y., Nagasaki H., Ebana K., Yoshikawa H., et al. , 2011. Discovery of genome-wide DNA polymorphisms in a landrace cultivar of japonica rice by whole-genome sequencing. Plant Cell Physiol. 52(2): 274–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandillo N., Raghavan C., Muyco P. A., Sevilla M. A. L., Lobina I. T., et al. , 2013. Multi-parent advanced generation inter-cross (MAGIC) populations in rice: progress and potential for genetics research and breeding. Rice 6: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K. W., Wu H., Sen S., Churchill G. A., 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890. [DOI] [PubMed] [Google Scholar]
- Browning B. L., Browning S. R., 2009. A unified approach to genotype imputation and haplotype phase inference for large data sets of trios and unrelated individuals. Am. J. Hum. Genet. 84: 210–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh C., Morell M., Mackay I., Powell W., 2008. From mutations to MAGIC: resources for gene discovery, validation and delivery in crop plants. Curr. Opin. Plant Biol. 11: 215–221. [DOI] [PubMed] [Google Scholar]
- Elshire R. J., Glaubitz J. C., Sun Q., Poland J. A., Kawamoto K., et al. , 2011. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6(5): e19379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B. N., Donnelly P., Marchini J., 2009. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5(6): e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B. E., George A. W., 2011. R/mpMap: a computational platform for the genetic analysis of multi-parent recombinant inbred lines. Bioinformatics 27: 727–729. [DOI] [PubMed] [Google Scholar]
- International HapMap Consortium , 2003. The International HapMap Project. Nature 426: 789–796. [DOI] [PubMed] [Google Scholar]
- Kover P. X., Valdar W., Trakalo J., Scarcelli N., Ehrenreich I. M., et al. , 2009. A multiparent advanced generation inter-cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet. 5(7): e1000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Willer C. J., Ding J., Scheet P., Abecasis G. R., 2010. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 34: 816–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullen M. D., Kresovich S., Villeda H. S., Bradbury P. J., Li H., et al. , 2009. Genetic properties of the maize nested association mapping population. Science 325: 737–740. [DOI] [PubMed] [Google Scholar]
- Mott R., Talbot C. J., Turri M. G., Collins A. C., Flint J., 2000. A new method for fine-mapping quantitative trait loci in outbred animal stocks. Proc. Natl. Acad. Sci. USA 97: 12649–12654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team , 2013. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Rutkoski J. E., Poland J., Jannink J.-L., Sorrells M. E., 2013. Imputation of unordered markers and the impact on genomic selection accuracy. Genes Genomes Genetics 3: 427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwender H., 2012. Imputing missing genotypes with weighted k nearest neighbours. J. Toxicol. Environ. Health A 75: 438–446. [DOI] [PubMed] [Google Scholar]
- Ward J. A., Bhangoo J., Fernandez-Fernandez F., Moore P., Swanson J. D., et al. , 2013. Saturated linkage map construction in Rubus idaeus using genotyping by sequencing and genome-independent imputation. BMC Genomics 14: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Liu X., Ge S., Jensen J. D., Hu F., et al. , 2012. Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat. Biotechnol. 30: 105–111. [DOI] [PubMed] [Google Scholar]
- Zaykin D. V., Westfall P. H., Young S. S., Karnoub M. A., Wagner M. J., et al. , 2002. Testing association of statistically inferred haplotypes with discrete and continuous traits in samples of unrelated individuals. Hum. Hered. 53: 79–91. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


