Artificial microRNAs and synthetic trans-acting small interfering RNAs produced from new, high-efficiency expression vectors induce reliable gene silencing in Arabidopsis.
Abstract
Artificial microRNAs (amiRNAs) and synthetic trans-acting small interfering RNAs (syn-tasiRNAs) are used for small RNA-based, specific gene silencing or knockdown in plants. Current methods to generate amiRNA or syn-tasiRNA constructs are not well adapted for cost-effective, large-scale production or for multiplexing to specifically suppress multiple targets. Here, we describe simple, fast, and cost-effective methods with high-throughput capability to generate amiRNA and multiplexed syn-tasiRNA constructs for efficient gene silencing in Arabidopsis (Arabidopsis thaliana) and other plant species. amiRNA or syn-tasiRNA inserts resulting from the annealing of two overlapping and partially complementary oligonucleotides are ligated directionally into a zero background BsaI/ccdB-based expression vector. BsaI/ccdB vectors for amiRNA or syn-tasiRNA cloning and expression contain a modified version of Arabidopsis MIR390a or TAS1c precursors, respectively, in which a fragment of the endogenous sequence was substituted by a ccdB cassette flanked by two BsaI sites. Several amiRNA and syn-tasiRNA sequences designed to target one or more endogenous genes were validated in transgenic plants that (1) exhibited the expected phenotypes predicted by loss of target gene function, (2) accumulated high levels of accurately processed amiRNAs or syn-tasiRNAs, and (3) had reduced levels of the corresponding target RNAs.
MicroRNAs (miRNAs) and trans-acting small interfering RNAs (tasiRNAs) are two distinct classes of plant small RNAs that act in posttranscriptional RNA silencing pathways to silence target RNA transcripts with sequence complementarity (Chapman and Carrington, 2007; Martínez de Alba et al., 2013). Target repression can occur through direct endonucleolytic cleavage or through other mechanisms such as target destabilization or translational repression (Huntzinger and Izaurralde, 2011). The miRNAs and tasiRNAs differ in their biogenesis pathways. While miRNAs originate from transcripts with imperfect self-complementary foldback structures that are usually processed by DICER-LIKE1 (DCL1), tasiRNAs are formed through a refined RNA-silencing pathway. TAS transcripts are initially targeted and sliced by a specific miRNA/ARGONAUTE (AGO) complex, and one of the cleavage products is converted to double-stranded RNA (dsRNA) by RNA-DEPENDENT RNA POLYMERASE6 (RDR6). The resulting dsRNA is sequentially processed by DCL4 into 21-nucleotide (nt) small interfering RNA (siRNA) duplexes in register with the miRNA-guided cleavage site (Allen et al., 2005; Dunoyer et al., 2005; Gasciolli et al., 2005; Xie et al., 2005; Yoshikawa et al., 2005; Axtell et al., 2006; Montgomery et al., 2008a, 2008b). For both miRNA and tasiRNA intermediate duplexes, usually one strand is selectively sorted to an AGO protein according to the identity of the 5′ nt or to other sequence/structural elements of the small RNA or the small RNA duplex (Mi et al., 2008; Montgomery et al., 2008a; Takeda et al., 2008; Zhu et al., 2011).
Small RNA-directed gene silencing has been used extensively to selectively regulate plant gene expression. Artificial microRNA (amiRNA), synthetic trans-acting small interfering RNA (syn-tasiRNA), hairpin-based RNA interference, virus-induced gene silencing, and transcriptional silencing methods have been developed (Ossowski et al., 2008; Baykal and Zhang, 2010). Since their initial application (Alvarez et al., 2006; Schwab et al., 2006), amiRNAs produced from different MIRNA precursors have been used to silence reporter genes (Parizotto et al., 2004), endogenous plant genes (Alvarez et al., 2006; Schwab et al., 2006), viruses (Niu et al., 2006), and noncoding RNAs (Eamens et al., 2011). syn-tasiRNAs have been shown to target RNAs in Arabidopsis (Arabidopsis thaliana) when produced from TAS1a (Felippes and Weigel, 2009), TAS1c (de la Luz Gutiérrez-Nava et al., 2008; Montgomery et al., 2008b), and TAS3a (Montgomery et al., 2008a; Felippes and Weigel, 2009) transcripts or from gene fragments fused to an upstream miR173 target site (Felippes et al., 2012). Current methods to generate amiRNA or syn-tasiRNA constructs, however, can be tedious and cost and time ineffective for high-throughput applications.
Here, highly efficient methods for the production of a new generation of plant MIR390a-based amiRNAs and TAS1c-based syn-tasiRNAs are described and validated. The new vectors use positive insert selection and eliminate PCR steps, gel-based DNA purification, restriction digestion, and subcloning of inserts between vectors, making them more suitable for high-throughput libraries. Accurate processing of both MIR390a-based amiRNAs and TAS1c-based syn-tasiRNAs was confirmed by deep sequencing analysis, and target gene silencing was shown through phenotype and molecular analyses.
RESULTS AND DISCUSSION
Selection of Arabidopsis MIR390a Precursor for Direct Cloning of amiRNAs
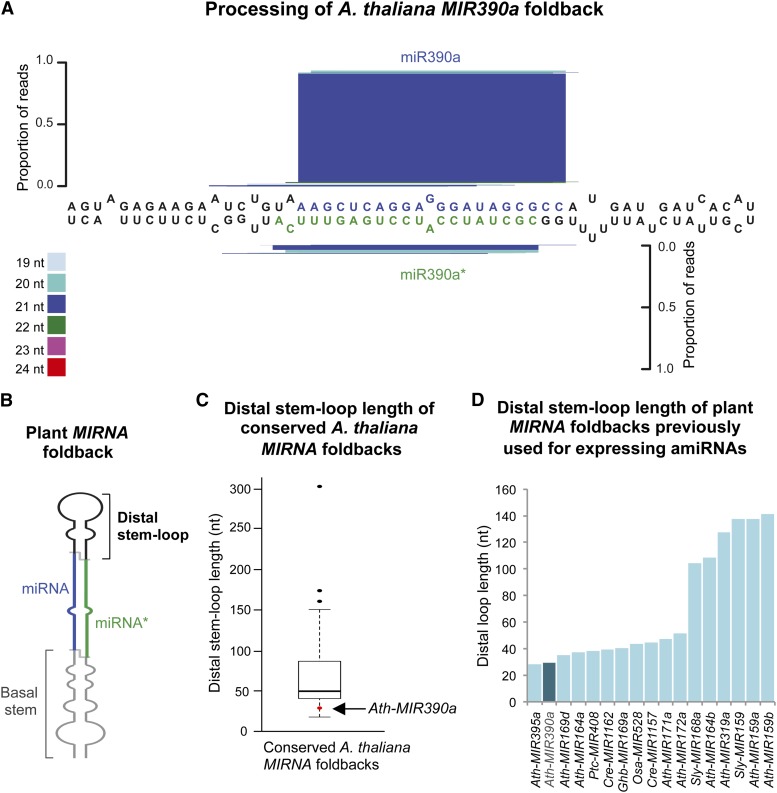
Several properties of the AtMIR390a precursor make it attractive as a backbone to engineer a new generation of amiRNA vectors. First, small RNA library analyses indicate that the AtMIR390a precursor is processed accurately, as the majority of reads mapping to the AtMIR390a foldback correspond to the authentic 21-nt miR390a guide strand (Fig. 1A). Second, as the MIR390 family is deeply conserved in plants (Axtell et al., 2006; Cuperus et al., 2011), AtMIR390a-based amiRNAs are likely to be produced accurately in different plant species. Third, the AtMIR390a precursor was used to express high levels of either 21- or 22-nt amiRNAs of the correct size in Nicotiana benthamiana leaves (Montgomery et al., 2008a; Cuperus et al., 2010; Carbonell et al., 2012), demonstrating that the miR390 duplex sequence provides little or no specific information required for accurate processing. Fourth, the AtMIR390a foldback has a relatively short distal stem loop (31 nt; Fig. 1B) compared with other conserved Arabidopsis MIRNA foldbacks (Fig. 1C), including those used previously for amiRNA expression in plants (Fig. 1D). A short distal stem loop facilitates more cost-effective synthesis of partially complementary oligonucleotides (see next section) that span the entire foldback. And fifth, although authentic miR390a associates preferentially with AGO7, the association of AtMIR390a-based amiRNAs containing a 5′−U or 5′−A can be directed to AGO1 (Montgomery et al., 2008a; Cuperus et al., 2010) or AGO2 (Carbonell et al., 2012), respectively.
Figure 1.
Arabidopsis MIR390a (AtMIR390a) is an accurately processed, conserved MIRNA foldback with a short distal stem loop. A, AtMIR390a foldback processing diagram. miR390a and miR390a* nt are highlighted in blue and green, respectively. The proportion of small RNA reads for the entire foldback is plotted as stacked bar graphs. Small RNAs are color coded by size. B, Diagram of a canonical plant MIRNA foldback (adapted from Cuperus et al., 2011). miRNA guide and amiRNA passenger or star (amiRNA*) strands are highlighted in blue and green, respectively. The distal stem loop and basal stem regions are highlighted in black and gray, respectively. C, Distal stem loop length of Arabidopsis conserved MIRNA foldbacks depicted in a box plot. The distal stem loop length of AtMIR390a is highlighted with the red dot and indicated with the arrow. Outliers are represented with black dots. D, Distal stem loop length of plant MIRNA foldbacks used previously for expressing amiRNAs. The Arabidopsis MIR390a distal stem loop length bar and name are highlighted in dark blue.
Direct Cloning of amiRNA Sequences in AtMIR390a-Based Vectors
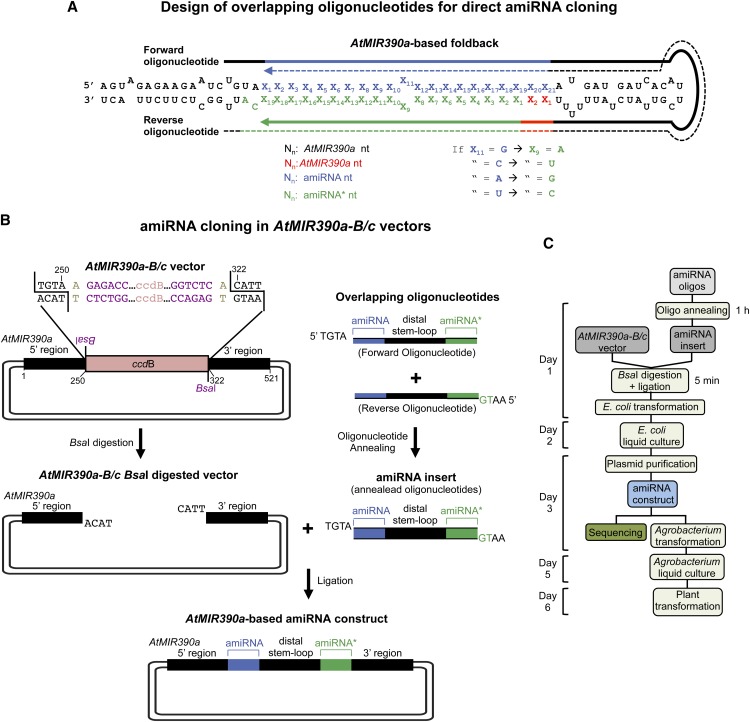
Details of the zero background cloning strategy to generate AtMIR390a-based amiRNA constructs are illustrated in Figure 2A. The amiRNA insert is derived by annealing of two overlapping and partially complementary 75-base oligonucleotides covering the amiRNA/AtMIR390a-distal loop/amiRNA* sequence (Fig. 2A; Supplemental Protocol S1). The design of amiRNA oligonucleotides is described in detail in Supplemental Protocol S1. Forward and reverse oligonucleotides must have 5′-TGTA and 5′-AATG overhangs, respectively, for direct cloning into AtMIR390a-based vectors (see below). This strategy requires no oligonucleotide enzymatic modifications, PCR steps, restriction digestion, or DNA fragment isolation.
Figure 2.
Direct cloning of amiRNAs in vectors containing a modified version of AtMIR390a that includes a ccdB cassette flanked by two BsaI sites (B/c vectors). A, Design of two overlapping oligonucleotides for amiRNA cloning. Sequences covered by the forward and reverse oligonucleotides are represented with solid and dotted lines, respectively. Nt of AtMIR390a foldback, amiRNA guide strand, and amiRNA* strand are in black, blue, and green, respectively. Other AtMIR390a nt that may be modified for preserving authentic AtMIR390a foldback secondary structure are in red. Rules for assigning identity to position 9 of amiRNA* are indicated. B, Diagram of the steps for amiRNA cloning in AtMIR390a-B/c vectors. The amiRNA insert obtained after annealing the two overlapping oligonucleotides has 5′-TGTA and 5′-AATG overhangs and is directly inserted in a directional manner into an AtMIR390a-B/c vector previously linearized with BsaI. Nt of the BsaI sites and those arbitrarily chosen and used as spacers between the BsaI recognition sites and the AtMIR390a sequence are in purple and light brown, respectively. Other details are as described in A. C, Flow chart of the steps from amiRNA construct generation to plant transformation.
A series of AtMIR390a-based cloning vectors were developed and named AtMIR390a-B/c vectors (from AtMIR390a-BsaI/ccdB). They contain a truncated AtMIR390a precursor sequence whose miRNA/distal stem loop/amiRNA* region was replaced by a 1,461-bp DNA cassette including the ccdB gene (Bernard and Couturier, 1992) flanked by two BsaI sites (Fig. 2B; Table I; Supplemental Fig. S1). The BsaI restriction enzyme is a type IIs endonuclease with nonpalindromic recognition sites [GGTCTC(N1/N5)] that are distal from the cleavage sites. Here, BsaI recognition sites are inserted in a configuration that allows both BsaI cleavage sites to be located outside the ccdB cassette (Fig. 2B). After BsaI digestion, AtMIR390a-B/c vectors have 5′-TACA and 5′-CATT ends, which are incompatible. This prevents vector self-ligation and eliminates the need to modify the ends of insert oligonucleotide sequences (Schwab et al., 2006; Molnar et al., 2009). The use of two BsaI sites in this configuration has been adapted from the Golden Gate cloning method (Engler et al., 2008) and was used in other amiRNA cloning methods (Chen et al., 2009; Zhou et al., 2013). BsaI digestion of the B/c vector and subsequent ligation of the amiRNA oligonucleotide insert can be done in separate reactions or combined in a single 5-min reaction (Supplemental Protocol S1). The amiRNA insert is ligated directionally into the BsaI-digested AtMIR390a-B/c vector and introduced into Escherichia coli. Nonlinearized plasmid molecules with no amiRNA insert fail to propagate in E. coli ccdB-sensitive strains, such as DH5α or DH10B. In summary, compared with other amiRNA cloning methods (Schwab et al., 2006; Qu et al., 2007; Chen et al., 2009; Molnar et al., 2009; Wang et al., 2010, 2012; Eamens et al., 2011; Yan et al., 2011; Liang et al., 2012; Zhou et al., 2013), this method is relatively simple, fast, and cost effective (Fig. 2C).
Table I. BsaI/ccdB-based vectors for direct cloning of amiRNAs and syn-tasiRNAs.
CaMV, Cauliflower mosaic virus; nos, nopaline synthase; rbcS, Rubisco small subunit.
| Vector | Small RNA Class | Bacterial Antibiotic Resistance | Plant Antibiotic Resistance | Gateway Use | Backbone | Promoter | Terminator | Plant Species Tested |
|---|---|---|---|---|---|---|---|---|
| pENTR-AtMIR390a-B/c | amiRNA | Kanamycin | – | Donor | pENTR | – | – | – |
| pFK210B-AtMIR390a-B/c | amiRNA | Spectinomycin | BASTA | – | pGreen III | CaMV 35S | rbcS | Arabidopsis |
| pMDC123SB-AtMIR390a-B/c | amiRNA | Kanamycin | BASTA | – | pMDC123 | CaMV 2× 35S | – | Arabidopsis |
| N. benthamiana | ||||||||
| pMDC32B-AtMIR390a-B/c | amiRNA | Kanamycin | Hygromycin | – | pMDC32 | CaMV 2× 35S | nos | Arabidopsis |
| Hygromycin | N. benthamiana | |||||||
| pENTR-AtTAS1c-B/c | syn-tasiRNA | Kanamycin | – | Donor | pENTR | – | – | – |
| pMDC123SB-AtTAS1c-B/c | syn-tasiRNA | Kanamycin | BASTA | – | pMDC123 | CaMV 2× 35S | nos | N. benthamiana |
| Hygromycin | ||||||||
| pMDC32B-AtTAS1c-B/c | syn-tasiRNA | Kanamycin | Hygromycin | – | pMDC32 | CaMV 2× 35S | nos | Arabidopsis |
| Hygromycin | N. benthamiana |
pMDC32B-AtMIR390a-B/c, pMDC123SB-AtMIR390a-B/c, and pFK210B-AtMIR390a-B/c expression vectors were generated for direct cloning of amiRNAs and tested in different plant species (Table I; Supplemental Fig. S1). Each vector contains a unique combination of bacterial and plant antibiotic resistance genes. The direct cloning of amiRNA inserts into plant expression vectors avoids the need for subcloning the amiRNA cassette from an intermediate plasmid to the expression vector (Schwab et al., 2006; Qu et al., 2007; Warthmann et al., 2008; Eamens et al., 2011; Yan et al., 2011). A pENTR-AtMIR390a-B/c Gateway-compatible entry vector was generated for direct cloning of the amiRNA insert and subsequent recombination into a preferred Gateway expression vector containing a promoter, terminator, or other features of choice (Table I; Supplemental Fig. S1).
Comparison of amiRNA Production from AtMIR390a and AtMIR319a Precursors
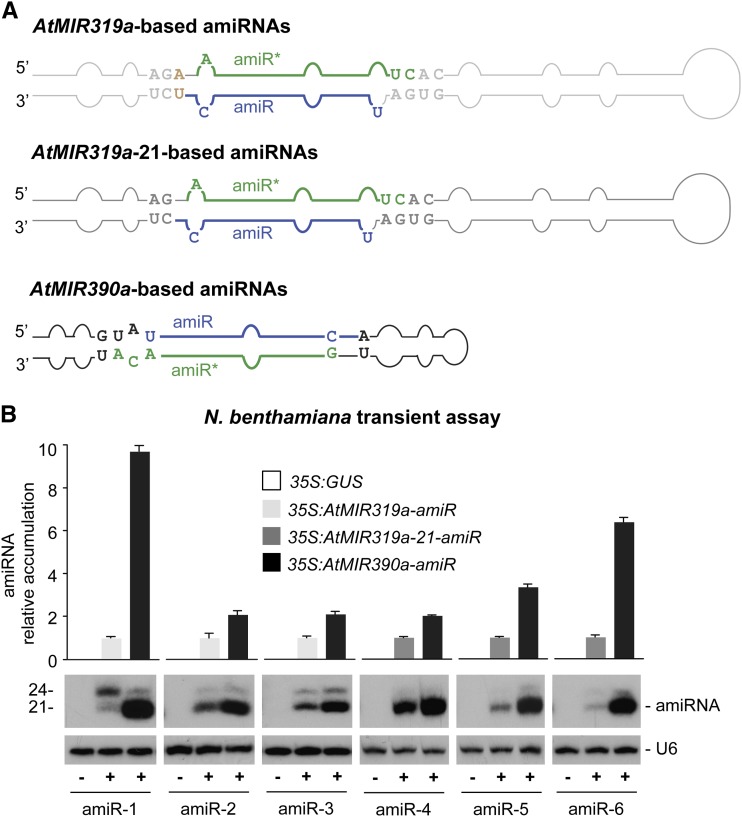
To verify the accumulation in planta of AtMIR390a-derived amiRNAs, six different amiRNA sequences (amiR-1–amiR-6; Supplemental Fig. S2; Supplemental Text S1) were directly cloned into pMDC32B-AtMIR390a-B/c (amiR-2 and amiR-3) or pMDC123SB-AtMIR390a-B/c (amiR-1, amiR-4, amiR-5, and amiR6) and expressed transiently in N. benthamiana leaves. All AtMIR390a-based amiRNAs had a U and C in 5′ to 3′ positions 1 and 19, respectively, of the guide strand. They also contained G, A, C, and A in 5′ to 3′ positions 1, 19, 20, and 21, respectively, of the amiRNA* strand (Fig. 3A; Supplemental Fig. S2). In addition, position 11 of the amiRNA guide strand was kept unpaired with position 9 of amiRNA* to preserve the authentic AtMIR390a base-pairing structure (Fig. 2A).
Figure 3.
Comparative analysis of the accumulation of several amiRNAs produced from AtMIR319a, AtMIR319a-21, and AtMIR390a foldbacks. A, Diagrams of AtMIR319a, AtMIR319a-21, and AtMIR390a foldbacks. Nt corresponding to the miRNA guide strand are in blue, and nucleotides of the miRNA* strand are in green. Other nt from the AtMIR319a, AtMIR319a-21, and AtMIR390a foldbacks are in light gray, dark gray, and black, respectively, except those nt that were added in the AtMIR319a configuration, which are in light brown. Shapes of the AtMIR319a, AtMIR319a-21, and AtMIR390a foldbacks are in light gray, dark gray, and black, respectively. B, Accumulation of several amiRNAs expressed from the AtMIR319a, AtMIR319a-21, or AtMIR390a foldbacks in N. benthamiana leaves. The graph at top shows mean (n = 3) relative amiRNA levels + sd when expressed from the AtMIR319a (light gray; amiRNA level = 1.0), AtMIR319a-21 (dark gray; amiRNA level = 1), or AtMIR390a (black) foldback. Only one blot from three biological replicates is shown. The U6 RNA blot is shown as a loading control.
For comparative purposes, the same six amiRNA sequences were also expressed from AtMIR319a precursor, which has been most widely used to express amiRNAs in plants (Schwab et al., 2006). In this case, amiRNAs were cloned into pMDC32B-AtMIR319a-B/c (amiR-2 and amiR-3) or pMDC123SB-AtMIR319a-B/c (amiR-1, amiR-4, amiR-5, and amiR6; Fig. 3A; Supplemental Fig. S2), following the protocols used previously (Schwab et al., 2006). In the original AtMIR319a-based cloning configuration, a 20-bp sequence in AtMIR319a was replaced by a 21-bp sequence (Schwab et al., 2006), because it was initially thought that miR319a was only 20 bases long (Palatnik et al., 2003; Sunkar and Zhu, 2004). Later analyses, however, revealed that miR319a is predominantly a 21-mer, like the majority of plant miRNAs (Rajagopalan et al., 2006; Fahlgren et al., 2007). Consequently, the AtMIR319a foldbacks in the original AtMIR319a-based configuration had a 1-bp elongated basal stem that did not seem to affect foldback processing (Schwab et al., 2006). Here, amiR-1, amiR-2, and amiR-3 were cloned in the original 20-mer configuration (AtMIR319a; Schwab et al., 2006) and amiR-4, amiR-5, and amiR-6 were cloned in the more recent 21-mer configuration (AtMIR319a-21; http://wmd3.weigelworld.org), where the authentic 21-nt sequence of endogenous miR319a is replaced by the 21-nt sequence of amiRNA, preserving the foldback structure of authentic AtMIR319a (Fig. 3A; Supplemental Fig. S2). All AtMIR319a- and AtMIR319a-21-based amiRNAs had U and C in positions 1 and 19, respectively, in the amiRNA guide and A, U, U, and C in positions 1, 19, 20, and 21, respectively, of amiRNA*. Position 12 of amiRNA* was kept unpaired with position 8 of the guide strand to preserve the authentic AtMIR319a base-pairing structure. Note that an extra A-U base pair is found in AtMIR319a-based foldbacks due to the AtMIR319a original 20-mer configuration (Fig. 3A; Supplemental Fig. S2).
In transient expression assays using N. benthamiana, each of the six amiRNAs derived from the AtMIR390a foldbacks accumulated predominantly as 21-nt species, suggesting that the amiRNA foldbacks were likely processed accurately. In each case, the amiRNA from the AtMIR390a foldbacks accumulated to significantly higher levels than did the corresponding amiRNA from the AtMIR319a and AtMIR319a-21 foldbacks (P ≤ 0.02 for all pairwise Student’s t test comparisons; Fig. 3B). The basis for these differences in accumulation levels was not explored further. However, it is suggested that the more noncanonical loop-to-base processing mechanism for the AtMIR319a foldback (Addo-Quaye et al., 2009; Bologna et al., 2009, 2013) may be relatively less efficient than the canonical base-to-loop processing pathway for the AtMIR390a foldback.
Functionality of AtMIR390a-Based amiRNAs in Arabidopsis
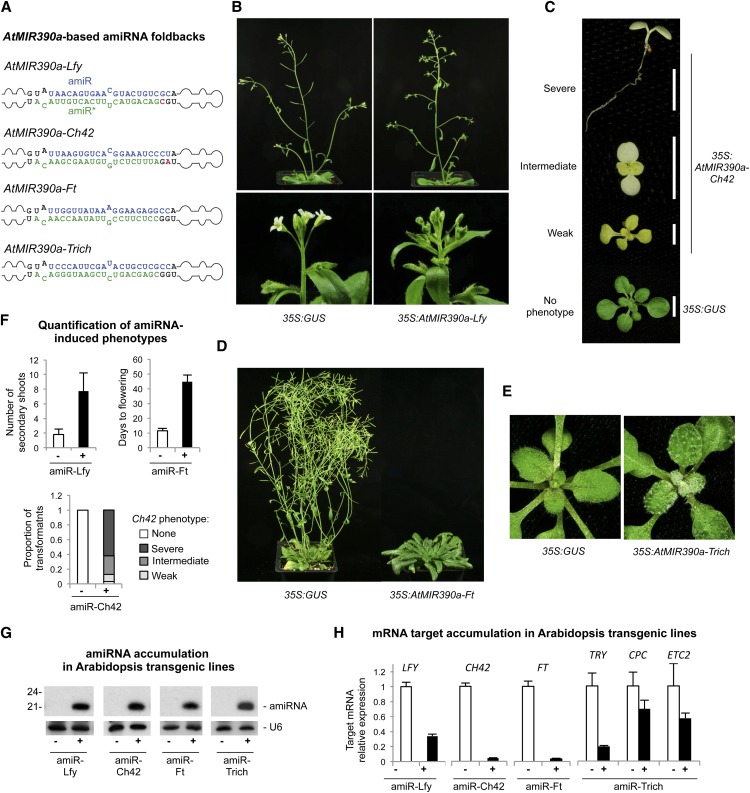
To test the functionality of AtMIR390a-based amiRNAs in repressing target transcripts, four different amiRNA constructs (Fig. 4A) were introduced into Arabidopsis Columbia-0 (Col-0) plants. The small RNA sequences were shown previously to repress gene expression when expressed as amiRNAs from an AtMIR319a-based foldback (Schwab et al., 2006; Liang et al., 2012) or from a syn-tasiRNA construct (Felippes and Weigel, 2009). In particular, amiR-Ft, amiR-Lfy, and amiR-Ch42 each targeted a single gene transcript (LEAFY [LFY], CHLORINA42 [CH42], and FLOWERING LOCUS T [FT], respectively), and amiR-Trich targeted three MYB transcripts (TRIPTYCHON [TRY], CAPRICE [CPC], and ENHANCER OF TRIPTYCHON AND CAPRICE2 [ETC2]; Supplemental Fig. S3). Plant phenotypes, amiRNA accumulation, mapping of amiRNA reads in the corresponding AtMIR390a foldback, and target mRNA accumulation were measured in Arabidopsis T1 transgenic lines.
Figure 4.
Functionality of AtMIR390a-based amiRNAs in Arabidopsis Col-0 T1 transgenic plants. A, AtMIR390a-based foldbacks containing Lfy-, Ch42-, Ft-, and Trich-amiRNAs. Nt corresponding to the miRNA guide and miRNA* strands are in blue and green, respectively; nt from the AtMIR390a foldback are in black, except those that were modified to preserve authentic AtMIR390a foldback secondary structure, which are in red. B to E show representative images of Arabidopsis Col-0 T1 transgenic plants expressing amiRNAs from the AtMIR390a foldback. B, Adult plants expressing 35S:GUS control (left) and 35S:AtMIR390a-Lfy with increased number of secondary shoots (top right) and leaf-like organs instead of flowers (bottom right). C, Ten-day-old seedlings expressing 35S:AtMIR390a-Ch42 and showing bleaching phenotypes. D, Adult control plant (35S:GUS) and 35S:AtMIR390a-Ft plant with a delayed flowering phenotype. E, Fifteen-day-old control seedling (35S:GUS) and a seedling expressing 35S:AtMIR390a-Trich with increased number of trichomes. F, Quantification of amiRNA-induced phenotypes in plants expressing amiR-Lfy (top left), amiR-Ft (top right), and amiR-Ch42 (bottom). G, Accumulation of amiRNAs in Arabidopsis transgenic plants. One blot from three biological replicates is shown. Each biological replicate is a pool of at least eight independent plants. The U6 RNA blot is shown as a loading control. H, Mean relative level + se of Arabidopsis LFY, CH42, FT, TRY, CPC, and ETC2 mRNAs after normalization to ACTIN2 (ACT2), CAP-BINDING PROTEIN20 (CBP20), SAND, and POLYUBIQUITIN10 (UBQ10), as determined by quantitative RT-PCR (RT-qPCR) (35S:GUS = 1.0 in all comparisons).
Twenty-three of 67 transgenic lines containing the 35S:AtMIR390a-Lfy construct showed morphological defects like lfy mutants (Schultz and Haughn, 1991; Weigel et al., 1992; Schwab et al., 2006; Supplemental Table S1), including obvious floral defects with leaf-like organs (Fig. 4B) and significantly increased numbers of secondary inflorescence shoots (P < 0.01, two-sample Student’s t test; Fig. 4F). Ninety-eight of 101 transgenic lines containing the 35S:AtMIR390a-Ch42 construct were smaller than controls and had pale or bleached leaves and cotyledons (Fig. 4C; Supplemental Table S1) as expected due to defective chlorophyll biosynthesis with a loss of Ch42 magnesium chelatase (Koncz et al., 1990; Felippes and Weigel, 2009). Sixty-three of these plants had a severe bleached phenotype with a lack of visible true leaves at 14 d after plating (Fig. 4, C and F; Supplemental Table S1). Each of the 34 transformants containing 35S:AtMIR390a-Ft was significantly delayed in flowering time compared with control plants not expressing the amiRNA (P < 0.01, two-sample Student’s t test; Fig. 4D; Supplemental Table S1), as observed previously in small RNA knockdown lines (Schwab et al., 2006; Liang et al., 2012) and ft mutants (Koornneef et al., 1991). Finally, 52 out of 53 lines containing 35S:AtMIR390a-Trich had increased numbers of trichomes in rosette leaves; 15 lines had highly clustered trichomes on leaf blades like try cpc double mutants (Schellmann et al., 2002) or other amiR-Trich overexpressor transgenic lines (Schwab et al., 2006; Liang et al., 2012; Fig. 4E; Supplemental Table S1). Each of the MIR390a-based amiRNAs, therefore, conferred a high proportion of expected target-knockdown phenotypes in transgenic plants.
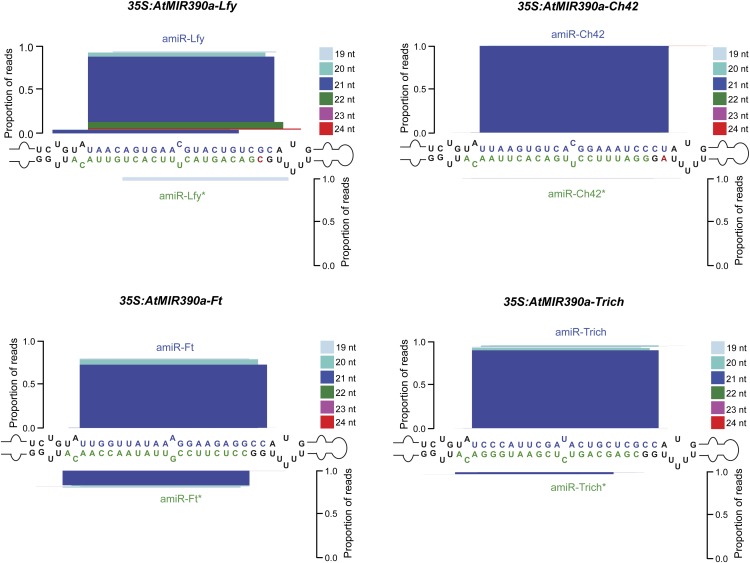
The accumulation of all four amiRNAs was confirmed by RNA-blot analysis in T1 transgenic lines showing amiRNA-induced phenotypes (Fig. 4G). In all cases, amiRNAs accumulated as a single species of 21 nt (Fig. 4G), suggesting that AtMIR390a-based amiRNAs were precisely processed. To more accurately assess the processing and accumulation of the amiRNA populations, small RNA libraries from samples containing each of the AtMIR390a-based constructs were prepared. In each case, the majority of reads from the AtMIR390a foldback corresponded to correctly processed 21-nt amiRNA, while reads from the amiRNA* strands were always relatively underrepresented (Fig. 5). It is possible that amiRNA* strands with an AGO-nonpreferred 5′ nt (5′−C for amiR-Ft* and amiR-Trich* and 5′−G for amiR-Lfy* and amiRCh42*) were actually produced but were less stable. The library read data support the rational design strategy to place an AGO-nonpreferred 5′ nt (such as 5′−G) at the 5′ end of the amiRNA* to avoid competition with the amiRNA guide strand for AGO loading. Combined with previous data (Cuperus et al., 2010), AtMIR390a-based foldbacks can be rationally designed to produce accurately processed amiRNAs of 21 or 22 nt, the latter of which can be used to trigger tasiRNA biosynthesis.
Figure 5.
Mapping of amiRNA reads from AtMIR390a-based foldbacks expressed in Arabidopsis Col-0 T1 transgenic plants. Analysis was performed for amiRNA and amiRNA* reads in plants expressing amiR-Ft (top left), amiR-Lfy (top right), amiR-Ch42 (bottom left), and amiR-Trich (bottom right). amiRNA guide and amiRNA* strands are highlighted in blue and green, respectively. Nt from the AtMIR390a foldback are in black, except those that were modified to preserve authentic AtMIR390a foldback secondary structure, which are in red. The proportion of small RNA reads is plotted as stacked bar graphs. Small RNAs are color coded by size.
The accumulation of amiRNA target mRNAs in Arabidopsis transgenic lines was analyzed by quantitative reverse transcription (RT)-PCR assay. The expression of all target mRNAs was significantly reduced compared with control plants (P < 0.02 for all pairwise Student’s t test comparisons; Fig. 4H) when the specific amiRNA was expressed.
Direct Cloning of syn-tasiRNAs in AtTAS1c-Based Constructs
A new generation of functional syn-tasiRNA vectors based on a modified TAS1c gene was produced with the potential to multiplex syn-tasiRNA sequences at DCL4-processing positions 3′D3[+] and 3′D4[+] of the AtTAS1c transcript (Montgomery et al., 2008b). The design of AtTAS1c-based syn-tasiRNA constructs expressing two syn-tasiRNAs is shown in Figure 6A.
Figure 6.
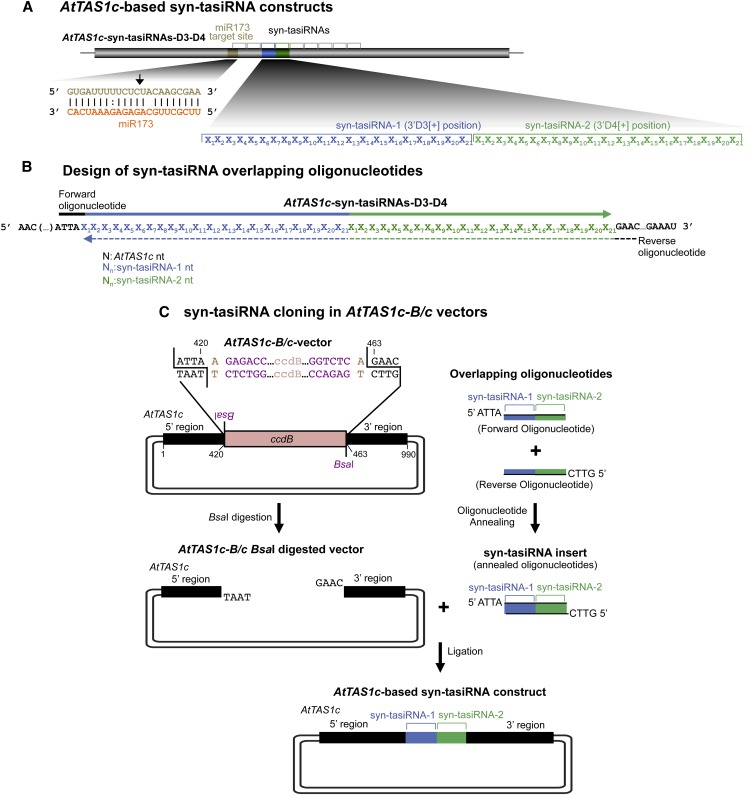
Direct cloning of syn-tasiRNAs in vectors containing a modified version of AtTAS1c with a ccdB cassette flanked by two BsaI sites (B/c vectors). A, Diagram of AtTAS1c-based syn-tasiRNA constructs. tasiRNA production is initiated by miR173-guided cleavage of the AtTAS1c transcript. syn-tasiRNA-1 and syn-tasiRNA-2 are generated from positions 3′D3[+] and 3′D4[+] of the AtTAS1c transcript, respectively. Nt of AtTAS1c, miR173, syn-tasiRNA-1, and syn-tasiRNA-2 are in black, orange, blue, and green, respectively. B, Design of two overlapping oligonucleotides for syn-tasiRNA cloning. The sequences covered by the forward and reverse oligonucleotides are represented with solid and dotted lines, respectively. C, Diagram of the steps for syn-tasiRNA cloning in AtTAS1c-B/c vectors. The syn-tasiRNA insert obtained after annealing the two overlapping oligonucleotides has 5′-ATTA and 5′-GTTC overhangs and is directly inserted into the BsaI-linearized AtTAS1c-B/c vector. Nt of the BsaI sites and arbitrary nt used as spacers between the BsaI recognition site and the AtMIR390a sequence are in purple and light brown, respectively. Other details are as in A.
syn-tasiRNA vector construction is similar to that described for the amiRNA constructs (Fig. 6C). Briefly, two overlapping and partially complementary oligonucleotides containing syn-tasiRNA sequences are designed (for details, see Fig. 6B; Supplemental Protocol S1). The sequence of syn-tasiRNA-1 can be identical to or different from the sequence of syn-tasiRNA-2. Theoretically, more than two syn-tasiRNA sequences can be introduced in the modified AtTAS1c, with such design being more attractive if multiple and unrelated sequences have to be targeted from the same syn-tasiRNA construct. The syn-tasiRNA insert results from the annealing of two 46-nt-long oligonucleotides and will have 5′-ATTA and 5′-GTTC overhangs. No PCR, restriction enzyme digestion, or gel purification steps are required to obtain the syn-tasiRNA insert. Several AtTAS1c-based cloning vectors were developed and named AtTAS1c-B/c vectors (from AtTAS1c-BsaI/ccdB; Table I; Supplemental Fig. S4). These contain a truncated AtTAS1c sequence with the 3′D3[+]-3′D4[+] region replaced by the 1,461-bp ccdB cassette flanked by two BsaI sites in the orientation that allows both BsaI recognition sites to be located outside of the AtTAS1c sequence (Fig. 6C). Annealed oligonucleotides are directly ligated into the linearized AtTAS1c-B/c expression vector in a directional manner (Fig. 6C). Subcloning is only required if the syn-tasiRNA insert is inserted in the Gateway entry vector pENTR-AtTAS1c-B/c, which allows recombination with the AtTAS1c-syn-tasiRNA cassette to the Gateway expression vector of choice (Table I; Supplemental Fig. S4). Compared with other syn-tasiRNA cloning methods (de la Luz Gutiérrez-Nava et al., 2008; Montgomery et al., 2008a; Felippes and Weigel, 2009), this method is relatively fast, efficient, and cost effective.
Functionality of AtTAS1c-Based syn-tasiRNAs in Arabidopsis
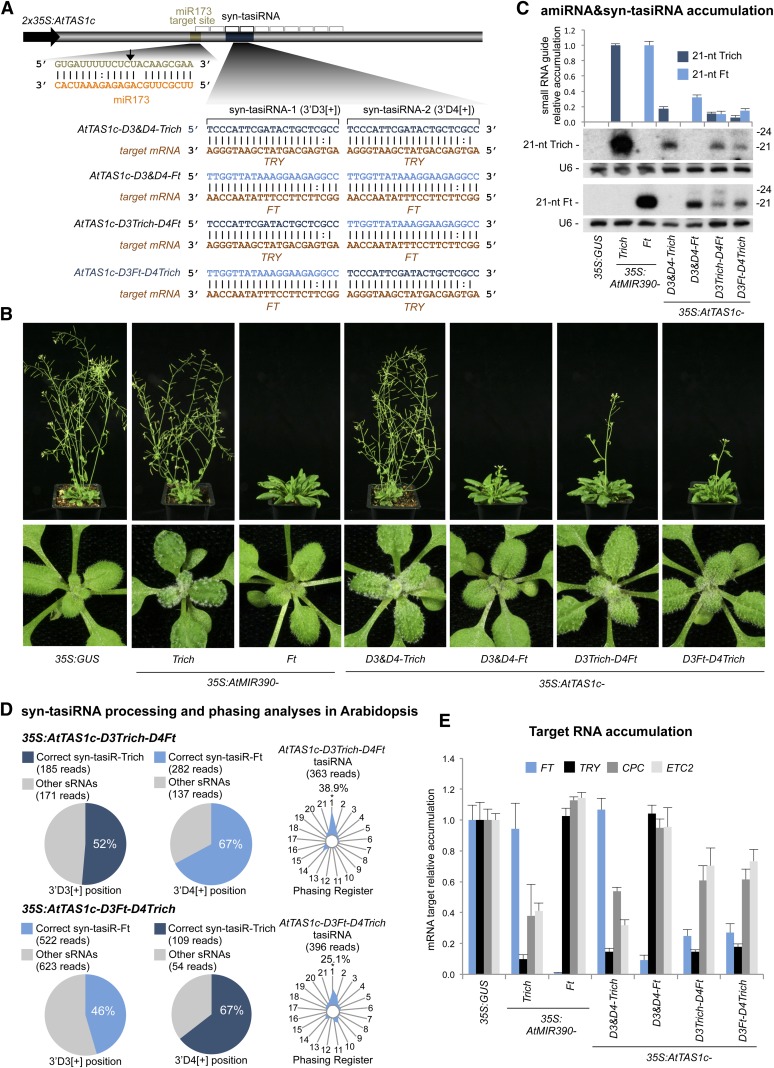
To test the functionality of single and multiplexed AtTAS1c-based syn-tasiRNAs and to compare the efficacy of the syn-tasiRNAs with amiRNA, several syn-tasiRNA constructs were generated and introduced into Arabidopsis Col-0 plants (Fig. 7). These constructs expressed either a syn-tasiRNA targeting FT (syn-tasiR-Ft) and/or a syn-tasiRNA targeting TRY/CPC/ETC2 (syn-tasiR-Trich) in single (35S:AtTAS1c-D3&D4Ft and 35S:AtTAS1c-D3&D4Trich) or dual (35S:AtTAS1c-D3Trich-D4Ft and 35S:AtTAS1c-D3Ft-D4Trich) configurations (Fig. 7A; Supplemental Fig. S5; Supplemental Text S2). For comparative purposes, transgenic lines expressing 35S:AtMIR390a-Ft and 35S:AtMIR390a-Trich, as well as the 35S:GUS control construct, were generated in parallel. The small RNAs produced in each pair of syn-tasiRNA and amiRNA vectors were identical. Plant phenotypes, syn-tasiRNA and amiRNA accumulation, processing and phasing analyses of AtTAS1c-based syn-tasiRNA, and target mRNA accumulation were analyzed in Arabidopsis T1 transgenic lines (Fig. 7; Supplemental Figs. S6–S9; Supplemental Table S2). Plant phenotypes were also analyzed in T2 transgenic lines to confirm the stability of expression (Supplemental Table S3).
Figure 7.
Functionality of AtTAS1c-based syn-tasiRNAs in Arabidopsis Col-0 T1 transgenic plants. A, Organization of syn-tasiRNA constructs. The arrow indicates the miR173-guided cleavage site. tasiRNA positions 3′D1[+] to 3′D10[+] are indicated by brackets, with positions 3′D3[+] and 3′D4[+] highlighted in black. B, Representative images of Arabidopsis Col-0 transgenic lines expressing amiRNA or syn-tasiRNA constructs. C, Accumulation of amiRNAs and syn-tasiRNAs in Arabidopsis transgenic plants. The graph at top shows mean (n = 3) relative Trich 21-mer (dark blue) and Ft 21-mer (light blue) levels + sd (35S:AtMIR390a-Trich and 35S:AtMIR390a-Ft lanes = 1.0 for Trich 21-mer and Ft 21-mer, respectively). One blot from three biological replicates is shown. Each biological replicate is a pool of at least six independent plants. The U6 RNA blot is shown as a loading control. D, syn-tasiRNA processing and phasing analyses in Arabidopsis Col-0 transgenic lines expressing syn-tasiRNAs (35S:AtTAS1c-D3Trich-D4Ft and 35S:AtTAS1c-D3Ft-D4Trich). Analyses of syn-tasiR-Trich, syn-tasiR-Ft, and AtTAS1c-derived siRNA sequences by high-throughput sequencing are shown. Pie charts show percentages of 19- to 24-nt reads; radar plots show percentages of 21-nt reads corresponding to each of the 21 registers from AtTAS1c transcripts, with position 1 designated as immediately after the miR173-guided cleavage site. E, Mean relative levels + se of FT, TRY, CPC, and ETC2 mRNAs after normalization to ACT2, CPB20, SAND, and UBQ10, as determined by RT-qPCR (35S:GUS = 1.0).
Seventy-three percent and 62% of the transformants expressing the dual-configuration syn-tasiRNA constructs 35S:AtTAS1c-D3Ft-D4Trich and 35S:AtTAS1c-D3Trich-D4Ft, respectively, showed both Trich and Ft loss-of-function phenotypes (Supplemental Table S2), which were characterized by increased clustering of trichomes in rosette leaves and a delay in flowering time compared with the 35S:GUS transformants (Fig. 7B). Plants expressing 35S:AtTAS1c-D3&D4Trich or 35S:AtMIR390a-Trich constructs showed clear Trich phenotypes in 82% or 92% of lines, respectively. In contrast with amiR-Trich overexpressors, none of the syn-tasiRNA-Trich constructs triggered the double try cpc phenotype (Supplemental Table S2). Transformants expressing the 35S:AtTAS1c-D3Ft-D4Trich and 35S:AtTAS1c-D3Trich-D4Ft constructs had significant delays in flowering time compared with control lines expressing the 35S:GUS, 35S:AtMIR390a-Trich, or 35S:AtTAS1c-D3&D4Trich constructs (P < 0.01 for all pairwise Student’s t test comparisons), although the 35S:AtMIR390a-Ft amiRNA lines showed the strongest delay in flowering (P < 0.001, two-sample Student’s t test; Fig. 7B; Supplemental Fig. S6; Supplemental Table S2). The trichome phenotypes were maintained in the Arabidopsis T2 progeny expressing 35S:AtMIR390a-Trich, 35S:AtTAS1c-D3&D4-Trich, 35S:AtTAS1c-D3Trich-D4Ft, and 35S:AtTAS1c-D3Ft-D4Trich constructs (Supplemental Table S3).
Next, the accumulation of syn-tasiR-Trich and syn-tasiR-Ft was compared with the accumulation of amiR-Trich and amiR-Ft and analyzed by RNA-blot assays using T1 transgenic plants showing obvious syn-tasiRNA- or amiRNA-induced phenotypes (Fig. 7C). In all cases, syn-tasiRNA accumulated to high levels and as a single band at 21 nt (Fig. 7C), suggesting that the processing of AtTAS1c-based constructs was accurate. When two copies of either syn-tasiR-Ft or syn-tasiR-Trich were expressed from a single construct, the corresponding RNAs accumulated to higher levels compared with when they were expressed in the dual syn-tasiRNA configuration containing only single copies of each RNA (Fig. 7C). Interestingly, amiR-Ft and amiR-Trich accumulated to higher levels than did any of the corresponding syn-tasiRNAs (Fig. 7C). It is possible that one or more factors in the AtTAS1c-dependent tasiRNA-generating pathway is limiting relative to the ubiquitous miRNA biogenesis factors. It is also possible that RDR6-dependent TAS1c dsRNAs may be processed by DCL4 from both ends, resulting in the production of tasiRNAs in two registers (Rajeswaran et al., 2012) and limiting the accumulation of accurately processed syn-tasiRNAs from positions D3[+] and D4[+].
To further analyze the processing and phasing of AtTAS1c-based syn-tasiRNA expressed from the dual-configuration constructs (35S:AtTAS1c-D3Trich-D4Ft and 35S:AtTAS1c-D3Ft-D4Trich), small RNA libraries were produced and analyzed. Analysis of 35S:AtTAS1c-D3Trich-D4Ft small RNA libraries confirmed that the syn-tasiRNA transcript yielded predominantly 21-nt syn-tasiR-Trich and syn-tasiR-Ft (51% and 67% of the reads within ±4 nt of 3′D3[+] and 3′D4[+], respectively) and that the corresponding tasiRNAs were in phase with the miR173 cleavage site (Fig. 7D, top; Supplemental Fig. S7, A and B, left). Similarly, 35S:AtTAS1c-D3Ft-D4Trich libraries revealed a high proportion of 21-nt syn-tasiR-Ft and syn-tasiR-Trich (45% and 65% of the reads within ±4 nt of 3′D3[+] and 3′D4[+], respectively) and accurately phased tasiRNAs (Fig. 7D, bottom; Supplemental Fig. S7, A and B, right). In both 35S:AtTAS1c-D3Trich-D4Ft and 35S:AtTAS1c-D3Ft-D4Trich libraries, relatively low levels of incorrectly processed siRNAs that overlap with the D3[+] and D4[+] positions were detected (Supplemental Fig. S7). While these small RNAs differ from the correctly processed forms by only one or a few terminal nt, it is theoretically possible that these could have altered targeting properties. Additionally, analyses of endogenous small RNAs showed that the expression of the syn-tasiRNA constructs, relative to the expression of the 35S:GUS control construct, did not interfere with the processing or accumulation of authentic AtTAS1c tasiRNAs (Supplemental Figs. S8 and S9).
Finally, the accumulation of target mRNAs in the 35S:AtTAS1c-D3Trich-D4Ft and 35S:AtTAS1c-D3Ft-D4Trich transgenic lines was analyzed by quantitative RT-PCR assay (Fig. 7E). The expression of all four target mRNAs (FT, TRY, CPC, and ETC2) was significantly reduced in lines expressing both dual-configuration syn-tasiRNA constructs compared with control plants expressing the 35S:GUS construct (P < 0.02 for all pairwise Student’s t test comparisons; Fig. 7E). However, target mRNA expression was reduced more in lines expressing the single-configuration syn-tasiRNA constructs and decreased even more in lines expressing the corresponding amiRNA (Fig. 7E). Taken together with the results presented above, the extent of target mRNA knockdown and the resultant phenotypes correlates with amiRNA and syn-tasiRNA dosage.
syn-tasiRNA technology was used before to repress single targets in Arabidopsis (de la Luz Gutiérrez-Nava et al., 2008; Montgomery et al., 2008a, 2008b; Felippes and Weigel, 2009). Here, a single AtTAS1c-based construct expressing multiple distinct syn-tasiRNAs triggered the silencing of multiple target transcripts and the resultant knockdown phenotypes. Theoretically, AtTAS1c-based vectors could be designed to produce more than two syn-tasiRNAs to repress a larger number of unrelated targets. Therefore, the syn-tasiRNA approach may be preferred for applications involving specific knockdown of multiple targets.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) Col-0 and Nicotiana benthamiana plants were grown in a chamber under long-day conditions (16-h-light/8-h-dark photoperiod at 200 μmol m−2 s−1) and 22°C constant temperature. Plants were transformed using the floral dip method with Agrobacterium tumefaciens GV3101 strain (Clough and Bent, 1998). Transgenic plants were grown on plates containing Murashige and Skoog medium and BASTA (50 mg mL−1) or hygromycin (50 mg mL−1) for 10 d before being transferred to soil. Plant photographs were taken with a Canon Rebel XT/EOS 350D digital camera and EF-S18-55mm f/3.5-5.6 II or EF-100mm f/2.8 Macro USM lenses.
DNA Constructs
The cassette containing the AtMIR390a sequence lacking the distal stem loop region, and including two BsaI sites, was generated as follows. A first round of PCR was done to amplify AtMIR390a-5′ or AtMIR390a-3′ regions using primers AtMIR390a-F and BsaI-AtMIR390a-5′-R or BsaI-AtMIR390a-3′-F and AtMIR390a-R, respectively. A second round of PCR was done using as template a mixture of the products of the first PCR round and primers AtMIR390a-F and AtMIR390a-R. The PCR product was cloned into pENTR-D-TOPO (Life Technologies) to generate pENTR-AtMIR390a-BsaI. A similar strategy was used to generate pENTR-AtTAS1c-BsaI containing the AtTAS1c cassette for syn-tasiRNA cloning: oligonucleotide pairs AtTAS1c-F/BsaI-AtTAS1c-5′-R and BsaI-AtTAS1c-3′-F/AtTAS1c-R were used for the first round of PCR, and oligonucleotide pair AtTAS1c-F/AtTAS1c-R was used for the second PCR.
A 2× 35S promoter cassette including the Gateway attR sites of pMDC32 (Curtis and Grossniklaus, 2003) was transferred into pMDC123 (Curtis and Grossniklaus, 2003) to make pMDC123S. An undesired BsaI site contained in pMDC32, pMDC123S, and pFK210 (de Felippes and Weigel, 2010) was disrupted to generate pMDC32B, pMDC123SB, and pFK210B, respectively. pMDC32B-AtMIR390a-BsaI, pMDC123SB-AtMIR390BsaI, and pFK210B-AtMIR390a-BsaI intermediate plasmids were obtained by LR recombination using pENTR-AtMIR390a-BsaI as the donor plasmid and pMDC32B, pMDC123SB, and pFK210B as destination vectors, respectively. Similarly, pMDC32B-AtTAS1c-BsaI and pMDC123SB-AtTAS1c-BsaI intermediate plasmids were obtained by LR recombination using pENTR-AtTAS1c-BsaI as the donor plasmid and pMDC32B and pMDC123SB as destination vectors, respectively.
To generate zero background cloning vectors, a ccdB cassette was inserted between the BsaI sites of plasmids containing the AtMIR390a-BsaI or AtTAS1c-BsaI cassettes. ccdB cassettes flanked with BsaI sites and with AtMIR390a- or AtTAS1c-specific sequences were amplified from pFK210 using primers AtMIR390a-B/c-F and AtMIR390a-B/c-R or AtTAS1c-B/c-F and AtTAS1c-Bc-R, respectively, with an overlapping PCR to disrupt an undesired BsaI site from the original ccdB sequence. These modified ccdB cassettes were then inserted between the BsaI sites into pENTR-AtMIR390a-BsaI, pENTR-AtTAS1c-BsaI, pMDC32B-AtMIR390a-BsaI, pMDC32B-AtTAS1c-BsaI, pMDC123SB-AtMIR390-BsaI, pMDC123SB-AtTAS1c-BsaI, and pFK210B-AtMIR390-BsaI to generate pENTR-AtMIR390a-B/c, pENTR-AtTAS1c-B/c, pMDC32B-AtMIR390a-B/c, pMDC32B-AtTAS1c-B/c, pMDC123SB-AtMIR390a-B/c, pMDC123SB-AtTAS1c-B/c, and pFK210B-AtMIR390a-B/c, respectively.
AtMIR319a-based amiRNA constructs (pMDC32-AtMIR319a-amiR-1, pMDC32-AtMIR319a-amiR-2, pMDC32-AtMIR319a-amiR-3, pMDC32-AtMIR319a-21-amiR-4, pMDC32-AtMIR319a-21-amiR-5, and pMDC32-AtMIR319-21-amiR-6) were generated as described previously (Schwab et al., 2006) using the WMD3 tool (http://wmd3.weigelworld.org). The CACC sequence was added to the 5′ end of the PCR fragments for pENTR-D-TOPO cloning (Life Technologies) and to allow LR recombination to pMDC32B or pMDC123SB. amiR-1, amiR-2, and amiR-3 were inserted in the AtMIR319a foldback, while amiR-4, amiR-5, and amiR-6 were inserted in the AtMIR319a-21 foldback.
The rest of the amiRNA and syn-tasiRNA constructs (pMDC32B-AtMIR390a-amiR-1, pMDC32B-AtMIR390a-amiR-2, pMDC32B-AtMIR390a-amiR-3, pMDC32B-AtMIR390a-21-amiR-4, pMDC32B-AtMIR390a-21-amiR-5, pMDC32B-AtMIR390a-amiR-6, pMDC32B-AtMIR390a-Ft, pMDC32B-AtMIR390a-Lfy, pMDC32B-AtMIR390a-Ch42, pMDC32B-AtMIR390a-Trich, pMDC32B-AtTAS1c-D3&D4Ft, pMDC32B-AtTAS1c-D3&D4Trich, pMDC32B-AtTAS1c-D3Trich-D4Ft, and pMDC32B-AtTAS1c-D3Ft-D4Trich) were obtained as described in the next section. The pMDC32-GUS construct was described previously (Montgomery et al., 2008a).
All oligonucleotides used for generating the constructs described above are listed in Supplemental Table S4. The sequences and predicted targets for all the amiRNAs and syn-tasiRNAs used in this study are listed in Supplemental Table S5. The sequences of the amiRNA and syn-tasiRNA vectors are listed in Supplemental Text S3. The following amiRNA and syn-tasiRNA vectors are available from Addgene at http://www.addgene.org/: pENTR-AtMIR390a-B/c (Addgene plasmid 51778), pMDC32B-AtMIR390a-B/c (Addgene plasmid 51776), pMDC123SB-AtMIR390a-B/c (Addgene plasmid 51775), pFK210B-AtMIR390a-B/c (Addgene plasmid 51777), pENTR-AtTAS1c-B/c (Addgene plasmid 51774), pMDC32B-AtTAS1c-B/c (Addgene plasmid 51773), and pMDC123SB-AtTAS1c-B/c (Addgene plasmid 51772).
amiRNA and syn-tasiRNA Oligonucleotide Design and Cloning
Detailed amiRNA and syn-tasiRNA oligonucleotide design and cloning protocols are given in Figures 2 and 6 and in Supplemental Protocol S1. A Web tool to design amiRNA and syn-tasiRNA sequences, together with the corresponding oligonucleotides for cloning into B/c vectors, will be available at http://p-sams.carringtonlab.org. All oligonucleotides used in this study for cloning amiRNA and syn-tasiRNA sequences are listed in Supplemental Table S4.
For cloning amiRNA or syn-tasiRNA inserts into B/c vectors, 2 μL of each of the two overlapping oligonucleotides (100 μm stock) were annealed in 46 μL of Oligo Annealing buffer (60 mm Tris-HCl, pH 7.5, 500 mm NaCl, 60 mm MgCl2, and 10 mm dithiothreitol) by heating the reaction for 5 min at 94°C and then cooling to 20°C (0.05°C s−1 decrease). The annealed oligonucleotides were diluted in deionized water to a final concentration of 0.3 μm.
A 10-μL digestion-ligation reaction was incubated for 5 min at 37°C, and included 1 μL of the annealed and diluted oligonucleotides (0.3 μm), 50 ng of the corresponding B/c vector, 1 μL of 10× T4 DNA ligase buffer (New England Biolabs), 1 μL of T4 DNA ligase (400 units μL−1; New England Biolabs), and 1 μL of BsaI (10 units μL−1; New England Biolabs). Alternatively, BsaI digestion of the B/c vector and subsequent ligation of the amiRNA oligonucleotide insert can be done in separate reactions. In this case, a 20-μL ligation reaction was incubated for 1 h at room temperature and included 1 μL of the annealed and diluted oligonucleotides (0.3 μm) and 1 μL (75 ng μL−1) of the corresponding B/c vector previously digested with BsaI. In all cases, 1 to 2 μL of the ligation reaction was used to transform an Escherichia coli strain such as DH10B or TOP10 lacking ccdB resistance.
Transient Expression Assays
Transient expression assays in N. benthamiana leaves were done as described (Carbonell et al., 2012) using A. tumefaciens GV3101 strain.
RNA-Blot Assays
Total RNA from Arabidopsis or N. benthamiana was extracted using TRIzol reagent (Life Technologies) as described (Cuperus et al., 2010). RNA-blot assays were done as described (Montgomery et al., 2008b; Cuperus et al., 2010). Oligonucleotides used as probes for small RNA blots are listed in Supplemental Table S4.
Real-Time RT-qPCR
Real-time RT-qPCR was done using the RNA samples that were used for RNA-blot and small RNA library analyses. Two micrograms of DNaseI-treated total RNA was used to produce first-strand complementary DNA using the SuperScript III system (Life Technologies). RT-qPCR was done on optical 96-well plates in the StepOnePlus Real-Time PCR System (Applied Biosystems) using the following program: 20 s at 95°C, followed by 40 cycles of 95°C for 3 s and 60°C for 30 s, with an additional melt curve stage consisting of 15 s at 95°C, 1 min at 60°C, and 15 s at 95°C. The 20-μL reaction mixture contained 10 μL of 2× Fast SYBR Green Master Mix (Applied Biosystems), 2 μL of diluted complementary DNA (1:5), and 300 nm of each gene-specific primer. Primers used for RT-qPCR are listed in Supplemental Table S4. Target mRNA expression levels were calculated relative to four Arabidopsis reference genes (ACT2, CBP20, SAND, and UBQ10) using the delta delta cycle threshold comparative method (Applied Biosystems) of the StepOne Software (version 2.2.2; Applied Biosystems). Three independent biological replicates were analyzed. For each biological replicate, two technical replicates were analyzed by RT-qPCR analysis.
Preparation of Small RNA Libraries
Small RNA libraries were produced using the same RNA samples as used for RNA blots. Fifty to 100 μg of Arabidopsis total RNA was treated as described (Carbonell et al., 2012), but each small RNA library was barcoded at the amplicon PCR step using an indexed 3′ PCR primer (i1, i3, i4, i5, or i9) and the standard 5′ PCR primer (P5; Supplemental Table S6). Libraries were multiplexed and submitted for sequencing using the HiSEquation 2000 sequencer (Illumina).
Small RNA Sequencing Analysis
Sequencing reads were parsed to identify library-specific barcodes and remove the 3′ adaptor sequence and were collapsed to a unique set with read counts. Unique sequences were aligned to a database containing the sequences of AtMIR390a-based amiRNA, AtTAS1c-based syn-tasiRNA, and the control constructs using BOWTIE version 0.12.8 (Langmead et al., 2009) with settings that identified only perfect matches (-f -v 0 -a -S). Small RNA alignments were saved in Sequence Alignment/Map format and were queried using SAMtools version 0.1.19+ (Li et al., 2009). Processing of amiRNA foldbacks and syn-tasiRNA transcripts was assessed by quantifying the proportion of small RNA, by position and size, that mapped within ±4 nt of the 5′ end of the miRNA and miRNA* or DCL4 processing positions 3′D3[+] and 3′D4[+], respectively.
The syn-tasiRNA constructs differ from endogenous AtTAS1c at positions 3′D3 and 3′D4 but are otherwise the same. Therefore, reads for other syn-tasiRNA positions are indistinguishable from endogenous AtTAS1c-derived small RNAs. To assess the phasing of syn-tasiRNA constructs, small RNA reads from libraries generated from plants containing 35S:GUS, 35S:AtTAS1c-D3Trich-D4Ft, or 35S:AtTAS1c-D3Ft-D4Trich were first normalized to account for library size differences (reads per million total sample reads). Next, normalized reads for 21-nt small RNAs that mapped to AtTAS1c in the 35S:GUS plants were subtracted from the corresponding small RNA reads in plants containing syn-tasiRNA constructs to correct for endogenous background tasiRNA expression. Phasing register tables were constructed by calculating the proportion of reads in each register relative to the miR173 cleavage site for all 21-nt positions downstream of the cleavage site.
A summary of high-throughput small RNA sequencing libraries from Arabidopsis transgenic lines is provided in Supplemental Table S6.
Arabidopsis gene and locus identifiers are as follows: ACT2 (AT3G18780), CBP20 (AT5G44200), CH42 (AT4G18480), CPC (AT2G46410), ETC2 (AT2G30420), LFY (AT5G61850), FT (AT1G65480), SAND (AT2G28390), TRY (AT5G53200) and UBQ10 (AT4G05320). The miRBase (http://mirbase.org) locus identifiers of the conserved Arabidopsis MIRNA precursors (Fig. 1C) and of the plant MIRNA precursors used to express amiRNAs (Fig. 1D) are listed in Supplemental Tables S7 and S8, respectively. High-throughput sequencing data from this article can be found in the Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under accession number SRP036134.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. AtMIR390a-B/c vectors for direct cloning of amiRNAs.
Supplemental Figure S2. Diagrams of AtMIR319a, AtMIR319a-21, and AtMIR390a foldbacks used to express several amiRNAs in N. benthamiana.
Supplemental Figure S3. Base pairing of amiRNAs and target mRNAs.
Supplemental Figure S4. AtTAS1c-B/c vectors for direct cloning of syn-tasiRNAs.
Supplemental Figure S5. Organization of syn-tasiRNA constructs.
Supplemental Figure S6. Flowering time analysis of Arabidopsis Col-0 T1 transgenic plants expressing amiRNAs or syn-tasiRNAs.
Supplemental Figure S7. Processing analyses of syn-tasiRNAs expressed in Arabidopsis Col-0 T1 transgenic lines (35S:AtTAS1c-D3Trich-D4Ft and 35S:AtTAS1c-D3Ft-D4Trich).
Supplemental Figure S8. Processing and phasing analyses of endogenous AtTAS1c-tasiRNA in Arabidopsis Col-0 T1 transgenic lines expressing syn-tasiRNAs (35S:AtTAS1c-D3Trich-D4Ft, 35S:AtTAS1c-D3Ft-D4Trich, and 35S:GUS control).
Supplemental Figure S9. Processing analyses of endogenous AtTAS1c-derived siRNAs in Arabidopsis Col-0 T1 transgenic plants expressing syn-tasiRNAs (35S:AtTAS1c-D3Trich-D4Ft, 35S:AtTAS1c-D3Ft-D4Trich, and 35S:GUS control).
Supplemental Table S1. Phenotypic penetrance of amiRNAs expressed in Arabidopsis Col-0 T1 transgenic plants.
Supplemental Table S2. Phenotypic penetrance of amiRNAs or syn-tasiRNAs expressed in Arabidopsis Col-0 T1 transgenic plants.
Supplemental Table S3. Phenotypic penetrance of amiRNAs or syn-tasiRNAs expressed in Arabidopsis Col-0 T2 transgenic plants.
Supplemental Table S4. DNA oligonucleotides used in this study.
Supplemental Table S5. Sequences and predicted targets for all the amiRNAs and syn-tasiRNAs used in this study.
Supplemental Table S6. Summary of high-throughput small RNA libraries from Arabidopsis transgenic lines.
Supplemental Table S7. miRBase locus identifiers of the Arabidopsis conserved MIRNA precursors used in this study.
Supplemental Table S8. miRBase locus identifiers of those plant MIRNA precursors used previously for expressing amiRNAs.
Supplemental Protocol S1. Protocol to design and clone amiRNAs or syn-tasiRNAs in BsaI/ccdB-based vectors containing the AtMIR390a or AtTAS1c precursors, respectively.
Supplemental Text S1. DNA sequences in FASTA format of all MIRNA foldbacks used in this study to express and analyze amiRNAs.
Supplemental Text S2. DNA sequences in FASTA format of all AtTAS1c-based constructs used to express and analyze syn-tasiRNAs.
Supplemental Text S3. DNA sequences of BsaI-ccdB-based vectors used for direct cloning of amiRNAs or syn-tasiRNAs.
Supplementary Material
Acknowledgments
We thank Goretti Nguyen, Benjamin J. Kelly, and Skyler Mitchell for excellent technical assistance and Dr. Detlef Weigel for pFK210 plasmid.
Glossary
- miRNA
microRNA
- tasiRNA
trans-acting small interfering RNA
- dsRNA
double-stranded RNA
- siRNA
small interfering RNA
- amiRNA
artificial microRNA
- syn-tasiRNA
synthetic trans-acting small interfering RNA
- nt
nucleotide
- Col-0
Columbia-0
- RT
reverse transcription
- RT-qPCR
quantitative real-time reverse transcription-PCR
Footnotes
This work was supported by the National Science Foundation (grant nos. MCB–0956526 and MCB–1231726), the National Institutes of Health (grant no. AI043288), the Japan Society for the Promotion of Science (postdoctoral fellowship to A.T.), and the National Institute of Food and Agriculture (postdoctoral fellowship no. MOW–2012–01361 to N.F.).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Addo-Quaye C, Snyder JA, Park YB, Li YF, Sunkar R, Axtell MJ. (2009) Sliced microRNA targets and precise loop-first processing of MIR319 hairpins revealed by analysis of the Physcomitrella patens degradome. RNA 15: 2112–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC. (2005) MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 [DOI] [PubMed] [Google Scholar]
- Alvarez JP, Pekker I, Goldshmidt A, Blum E, Amsellem Z, Eshed Y. (2006) Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell 18: 1134–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Jan C, Rajagopalan R, Bartel DP. (2006) A two-hit trigger for siRNA biogenesis in plants. Cell 127: 565–577 [DOI] [PubMed] [Google Scholar]
- Baykal U, Zhang Z (2010) Small RNA-mediated gene silencing for plant biotechnology. In AJ Catalano, ed, Gene Silencing: Theory, Techniques and Applications. Nova Science Publishers, Hauppauge, NY, pp 255–269 [Google Scholar]
- Bernard P, Couturier M. (1992) Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. J Mol Biol 226: 735–745 [DOI] [PubMed] [Google Scholar]
- Bologna NG, Mateos JL, Bresso EG, Palatnik JF. (2009) A loop-to-base processing mechanism underlies the biogenesis of plant microRNAs miR319 and miR159. EMBO J 28: 3646–3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bologna NG, Schapire AL, Palatnik JF. (2013) Processing of plant microRNA precursors. Brief Funct Genomics 12: 37–45 [DOI] [PubMed] [Google Scholar]
- Carbonell A, Fahlgren N, Garcia-Ruiz H, Gilbert KB, Montgomery TA, Nguyen T, Cuperus JT, Carrington JC. (2012) Functional analysis of three Arabidopsis ARGONAUTES using slicer-defective mutants. Plant Cell 24: 3613–3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman EJ, Carrington JC. (2007) Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet 8: 884–896 [DOI] [PubMed] [Google Scholar]
- Chen S, Songkumarn P, Liu J, Wang GL. (2009) A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiol 150: 1111–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cuperus JT, Carbonell A, Fahlgren N, Garcia-Ruiz H, Burke RT, Takeda A, Sullivan CM, Gilbert SD, Montgomery TA, Carrington JC. (2010) Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat Struct Mol Biol 17: 997–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus JT, Fahlgren N, Carrington JC. (2011) Evolution and functional diversification of MIRNA genes. Plant Cell 23: 431–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Felippes FF, Weigel D. (2010) Transient assays for the analysis of miRNA processing and function. Methods Mol Biol 592: 255–264 [DOI] [PubMed] [Google Scholar]
- de la Luz Gutiérrez-Nava M, Aukerman MJ, Sakai H, Tingey SV, Williams RW. (2008) Artificial trans-acting siRNAs confer consistent and effective gene silencing. Plant Physiol 147: 543–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer P, Himber C, Voinnet O. (2005) DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat Genet 37: 1356–1360 [DOI] [PubMed] [Google Scholar]
- Eamens AL, Agius C, Smith NA, Waterhouse PM, Wang MB. (2011) Efficient silencing of endogenous microRNAs using artificial microRNAs in Arabidopsis thaliana. Mol Plant 4: 157–170 [DOI] [PubMed] [Google Scholar]
- Engler C, Kandzia R, Marillonnet S. (2008) A one pot, one step, precision cloning method with high throughput capability. PLoS ONE 3: e3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, et al. (2007) High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE 2: e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felippes FF, Wang JW, Weigel D. (2012) MIGS: miRNA-induced gene silencing. Plant J 70: 541–547 [DOI] [PubMed] [Google Scholar]
- Felippes FF, Weigel D. (2009) Triggering the formation of tasiRNAs in Arabidopsis thaliana: the role of microRNA miR173. EMBO Rep 10: 264–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. (2005) Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr Biol 15: 1494–1500 [DOI] [PubMed] [Google Scholar]
- Huntzinger E, Izaurralde E. (2011) Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet 12: 99–110 [DOI] [PubMed] [Google Scholar]
- Koncz C, Mayerhofer R, Koncz-Kalman Z, Nawrath C, Reiss B, Redei GP, Schell J. (1990) Isolation of a gene encoding a novel chloroplast protein by T-DNA tagging in Arabidopsis thaliana. EMBO J 9: 1337–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, van der Veen JH. (1991) A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet 229: 57–66 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, He H, Li Y, Yu D. (2012) A new strategy for construction of artificial miRNA vectors in Arabidopsis. Planta 235: 1421–1429 [DOI] [PubMed] [Google Scholar]
- Martínez de Alba AE, Elvira-Matelot E, Vaucheret H. (2013) Gene silencing in plants: a diversity of pathways. Biochim Biophys Acta 1829: 1300–1308 [DOI] [PubMed] [Google Scholar]
- Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, et al. (2008) Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133: 116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar A, Bassett A, Thuenemann E, Schwach F, Karkare S, Ossowski S, Weigel D, Baulcombe D. (2009) Highly specific gene silencing by artificial microRNAs in the unicellular alga Chlamydomonas reinhardtii. Plant J 58: 165–174 [DOI] [PubMed] [Google Scholar]
- Montgomery TA, Howell MD, Cuperus JT, Li D, Hansen JE, Alexander AL, Chapman EJ, Fahlgren N, Allen E, Carrington JC. (2008a) Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133: 128–141 [DOI] [PubMed] [Google Scholar]
- Montgomery TA, Yoo SJ, Fahlgren N, Gilbert SD, Howell MD, Sullivan CM, Alexander A, Nguyen G, Allen E, Ahn JH, et al. (2008b) AGO1-miR173 complex initiates phased siRNA formation in plants. Proc Natl Acad Sci USA 105: 20055–20062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu QW, Lin SS, Reyes JL, Chen KC, Wu HW, Yeh SD, Chua NH. (2006) Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat Biotechnol 24: 1420–1428 [DOI] [PubMed] [Google Scholar]
- Ossowski S, Schwab R, Weigel D. (2008) Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J 53: 674–690 [DOI] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D. (2003) Control of leaf morphogenesis by microRNAs. Nature 425: 257–263 [DOI] [PubMed] [Google Scholar]
- Parizotto EA, Dunoyer P, Rahm N, Himber C, Voinnet O. (2004) In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev 18: 2237–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu J, Ye J, Fang R. (2007) Artificial microRNA-mediated virus resistance in plants. J Virol 81: 6690–6699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. (2006) A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev 20: 3407–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajeswaran R, Aregger M, Zvereva AS, Borah BK, Gubaeva EG, Pooggin MM. (2012) Sequencing of RDR6-dependent double-stranded RNAs reveals novel features of plant siRNA biogenesis. Nucleic Acids Res 40: 6241–6254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellmann S, Schnittger A, Kirik V, Wada T, Okada K, Beermann A, Thumfahrt J, Jürgens G, Hülskamp M. (2002) TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J 21: 5036–5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz EA, Haughn GW. (1991) LEAFY, a homeotic gene that regulates inflorescence development in Arabidopsis. Plant Cell 3: 771–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Zhu JK. (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16: 2001–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda A, Iwasaki S, Watanabe T, Utsumi M, Watanabe Y. (2008) The mechanism selecting the guide strand from small RNA duplexes is different among argonaute proteins. Plant Cell Physiol 49: 493–500 [DOI] [PubMed] [Google Scholar]
- Wang X, Yang Y, Yu C, Zhou J, Cheng Y, Yan C, Chen J. (2010) A highly efficient method for construction of rice artificial microRNA vectors. Mol Biotechnol 46: 211–218 [DOI] [PubMed] [Google Scholar]
- Wang X, Yang Y, Zhou J, Yu C, Cheng Y, Yan C, Chen J. (2012) Two-step method for constructing Arabidopsis artificial microRNA vectors. Biotechnol Lett 34: 1343–1349 [DOI] [PubMed] [Google Scholar]
- Warthmann N, Chen H, Ossowski S, Weigel D, Hervé P. (2008) Highly specific gene silencing by artificial miRNAs in rice. PLoS ONE 3: e1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM. (1992) LEAFY controls floral meristem identity in Arabidopsis. Cell 69: 843–859 [DOI] [PubMed] [Google Scholar]
- Xie Z, Allen E, Wilken A, Carrington JC. (2005) DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc Natl Acad Sci USA 102: 12984–12989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Zhong X, Jiang S, Zhai C, Ma L. (2011) Improved method for constructing plant amiRNA vectors with blue-white screening and MAGIC. Biotechnol Lett 33: 1683–1688 [DOI] [PubMed] [Google Scholar]
- Yoshikawa M, Peragine A, Park MY, Poethig RS. (2005) A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev 19: 2164–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Yu F, Chen B, Wang X, Yang Y, Cheng Y, Yan C, Chen J. (2013) Universal vectors for constructing artificial microRNAs in plants. Biotechnol Lett 35: 1127–1133 [DOI] [PubMed] [Google Scholar]
- Zhu H, Hu F, Wang R, Zhou X, Sze SH, Liou LW, Barefoot A, Dickman M, Zhang X. (2011) Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell 145: 242–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.