A genetic mapping population is developed and reveals female control of mating choice in plants.
Abstract
Female control of nonrandom mating has never been genetically established, despite being linked to inbreeding depression and sexual selection. In order to map the loci that control female-mediated nonrandom mating, we constructed a new advanced intercross recombinant inbred line (RIL) population derived from a cross between Arabidopsis (Arabidopsis thaliana) accessions Vancouver (Van-0) and Columbia (Col-0) and mapped quantitative trait loci (QTLs) responsible for nonrandom mating and seed yield traits. We genotyped a population of 490 RILs. A subset of these lines was used to construct an expanded map of 1,061.4 centimorgans with an average interval of 6.7 ± 5.3 centimorgans between markers. QTLs were then mapped for female- and male-mediated nonrandom mating and seed yield traits. To map the genetic loci responsible for female-mediated nonrandom mating and seed yield, we performed mixed pollinations with genetically marked Col-0 pollen and Van-0 pollen on RIL pistils. To map the loci responsible for male-mediated nonrandom mating and seed yield, we performed mixed pollinations with genetically marked Col-0 and RIL pollen on Van-0 pistils. Composite interval mapping of these data identified four QTLs that control female-mediated nonrandom mating and five QTLs that control female-mediated seed yield. We also identified four QTLs that control male-mediated nonrandom mating and three QTLs that control male-mediated seed yield. Epistasis analysis indicates that several of these loci interact. To our knowledge, the results of these experiments represent the first time female-mediated nonrandom mating has been genetically defined.
The process of pollination offers plants the opportunity to selectively breed. For example, in pollinations that include more than one pollen population, pollen often show differential siring ability. This process is called nonrandom mating. Although pollen may fail in pollinations because they are self pollen in an obligate outcrossing plant or pollen from a different species, we focus our studies on differential siring ability of compatible, conspecific mates (Hogenboom, 1973, 1975; Williams et al., 1999; de Nettancourt, 2001; Husband et al., 2002; Wheeler et al., 2009; Meng et al., 2011; Nasrallah, 2011). Nonrandom mating at this level has received intense interest for its potential to avoid inbreeding depression and its potential to be the result of sexual selection (Charnov, 1979; Mulcahy, 1979; Willson, 1979; Queller, 1983; Stephenson and Bertin, 1983; Willson and Burley, 1983; Marshall and Ellstrand, 1986; Charlesworth and Charlesworth, 1987; Mulcahy and Mulcahy, 1987; Cruzan, 1990; Quesada et al., 1993; Snow, 1994; Paschke et al., 2002; Skogsmyr and Lankinen, 2002; Stephenson et al., 2003; Armbruster and Rogers, 2004; Bernasconi et al., 2004; Lankinen and Armbruster, 2007). Despite a long history of theoretical and experimental attention, very little is known about the underlying genetics that govern the process (Carlson et al., 2011).
One challenge in understanding the genetics of nonrandom mating lies in its complexity, potentially involving multiple distinct pathways specific to either female or male tissues. Physiologically, postpollination nonrandom mating may be a result of intrinsic differences in pollen competitive abilities (male-mediated nonrandom mating). A number of experimental strategies have been employed to demonstrate male-mediated control of nonrandom mating. For example, experiments in radish (Raphanus sativus) found that some pollen sire more seeds than others in mixed pollinations across a range of maternal plants, demonstrating consistency of male function (Marshall and Ellstrand, 1986, 1988; Mitchell and Marshall, 1998). More direct measures of male function, such as in vitro and in vivo pollen tube growth rates, verify variation in male function and demonstrable impact on nonrandom mating (Snow and Spira, 1991a, 1991b; Pasonen et al., 1999; Skogsmyr and Lankinen, 1999; Stephenson et al., 2001; Lankinen and Skogsmyr, 2002; Lankinen et al., 2009). Finally, recent work in our laboratory has directly mapped the genetic loci responsible for the control of male-mediated nonrandom mating in Arabidopsis (Arabidopsis thaliana; Carlson et al., 2011).
Alternatively, or concurrently, nonrandom mating can be the result of differential interaction between the female tissue and competing pollen populations or seeds (female-mediated nonrandom mating). Establishing the female role in nonrandom mating has been more challenging, as most study designs involve the deposition of pollen from multiple donors and thus include the confounding variable of pollen competition. Despite this challenge, a number of experimental strategies have been devised to explore the role of the female in nonrandom mating. For example, a number of studies demonstrate that maternal identity influences nonrandom mating patterns (Marshall and Ellstrand, 1986, 1988; Snow and Mazer, 1988; Johnston, 1993; Marshall et al., 2000; Carlson et al., 2009, 2013). Studies have also established that manipulation of watering or nutrient regimes of maternal plants changes the patterns and magnitude of nonrandom mating (Marshall and Diggle, 2001; Shaner and Marshall, 2003; Haileselassie et al., 2005; Marshall et al., 2007). These studies and others implicate the identity and condition of the female in the process of nonrandom mating. Despite a long history of research, genetic control of female-mediated nonrandom mating has never been demonstrated, and the identity of the genes involved remains unexplored.
In previous work, we developed a system in Arabidopsis to assay nonrandom mating and showed its utility for genetically mapping the loci responsible (Carlson et al., 2009, 2011). Pursuing the genetics of nonrandom mating in a largely selfing plant such as Arabidopsis provides both theoretical and practical advantages. First, outcrossing plants carry higher levels of heterozygosity that produce pollen populations that display different phenotypes because of segregating alleles. This complicates genetic analysis. Also, in outcrossing plants that carry genetic load, reproductive success is context dependent. Pollinations with self pollen or pollen from genetically similar plants often lead to poor reproductive outcomes. For example, in mixed pollinations in generally outcrossing self-compatible plants that include self pollen, self pollen often sire a disproportionally low number of seeds (Bateman, 1956; Weller and Ornduff, 1977; Bowman, 1987; Eckert and Barrett, 1994; Jones, 1994; Hauser and Siegismund, 2000; Teixeira et al., 2009), but other findings have been reported (Sork and Schemske, 1992; Johnston, 1993). Thus, in outcrossing plants, gene variants that influence reproductive success, parental relatedness, and segregating heterozygosity all influence reproductive outcomes. Two of these factors are essentially eliminated by studying plant populations that have historically selfed. As outcrossing populations become increasingly self-fertilizing, they both lose heterozygosity, and their genetic load is purged (Lande and Schemske, 1985; Schemske and Lande, 1985; Charlesworth and Charlesworth, 1987; Lande et al., 1994; Byers and Waller, 1999; Crnokrak and Barrett, 2002). This is the case for Arabidopsis, whose tested populations show relatively low levels of heterozygosity and little evidence for the early-acting inbreeding depression that is indicative of genetic load (Bakker et al., 2006; Bomblies et al., 2010; Platt et al., 2010; Carlson et al., 2013). Thus, this system provides an excellent opportunity to identify and explore the genetic variation in differential reproduction that develops or persists in plant populations unrelated to inbreeding depression.
Using this system, we previously identified potential female control of nonrandom mating in mixed pollinations between Vancouver (Van-0) and Columbia (Col-0) accessions of Arabidopsis (Carlson et al., 2009). When Van-0 and genetically marked Col-0 (Col-NPTII) pollen compete on Col-0 pistils, Col-NPTII pollen sire 43% of the progeny, while Van-0 pollen sire 57%. When these pollen compete on Van-0 pistils, Col-NPTII pollen sire 67.5% of the progeny, while Van-0 pollen sire 32.5%. This system offers us, to our knowledge for the first time, the opportunity to genetically define female-mediated nonrandom mating and map the loci responsible.
In order to genetically map female control of nonrandom mating, we constructed a new advanced intercross recombinant inbred line (RIL) mapping population derived from a cross between Van-0 and Col-0 accessions of Arabidopsis. RILs are powerful tools that allow high-resolution genetic mapping of loci that direct complex traits. Each RIL contains chromosomes that are defined homozygous patchworks of parental DNA, in this case Van-0 and Col-0. By phenotyping these lines, we can statistically associate nonrandom mating and seed yield phenotypes with chromosomal regions. We chose these two accessions because (1) our previous experiments predict clear female control of nonrandom mating and (2) we have previously mapped male-mediated nonrandom mating controls using a Col-4/Landsberg mapping population (a population that does not display female control of nonrandom mating; Carlson et al., 2011). Thus, this new population provides us the opportunity to map loci that control female nonrandom mating and investigate the degree of conservation of loci that affect male-mediated nonrandom mating. We use this new mapping population to perform quantitative trait locus (QTL) mapping and identify multiple loci that direct both female- and male-mediated control of nonrandom mating and seed yield traits.
RESULTS
Marker Analysis and Map Construction
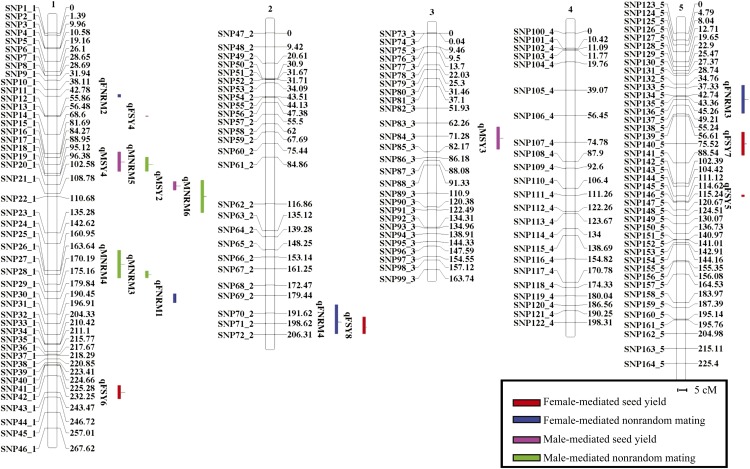
The initial advanced intercross RIL population was generated by four generations of intercrossing followed by seven generations of self pollinations. A total of 490 Van-0 × Col-0 RILs were deposited in the stock center (CS37809). Segregation bias was observed in the Van-0 × Col-0 lines by assaying marker pairs for their fit to Mendelian ratios for independent assortment, particularly with markers on chromosome 5 due to a high level (up to 75.1%) of Van-0 alleles in this region (data not shown). We selected 82 RILs for use in this study to remove allelic bias in our mapping population, as this can complicate mapping and future characterization of loci. Although a few single-nucleotide polymorphism (SNP) markers retained a bias in this set (most notably SNP5 on chromosome 1, SNP116 on chromosome 4, and SNP157 on chromosome 5), elimination of these markers had no effect on subsequent trait mapping. From this set of 82 RILs, a linkage map was generated (Fig. 1). This map contains 32 ± 10 markers per chromosome and an average distance between the markers of 6.7 ± 5.3 centimorgans (cM; Table I). We detected no residual heterozygosity in this subset.
Figure 1.
Van-0 × Col-0 linkage map with analyzed QTLs. Depicted are the five Arabidopsis chromosomes with linkage values indicated for the SNP markers used in this study. Loci for the indicated traits are shown to the right of each chromosome by a colored bar representing the 2-LOD confidence interval of the QTL.
Table I. Features of the map generated with the Van-0 × Col-0 RIL population.
Map distances were calculated with the Mapmaker/QTL 3.0 program. The kb per cM estimates were generated by a linear regression of the map to the known bp positions of the SNPs from the physical map.
Variation in Phenotypes in Female-Mediated Nonrandom Mating and Seed Yield
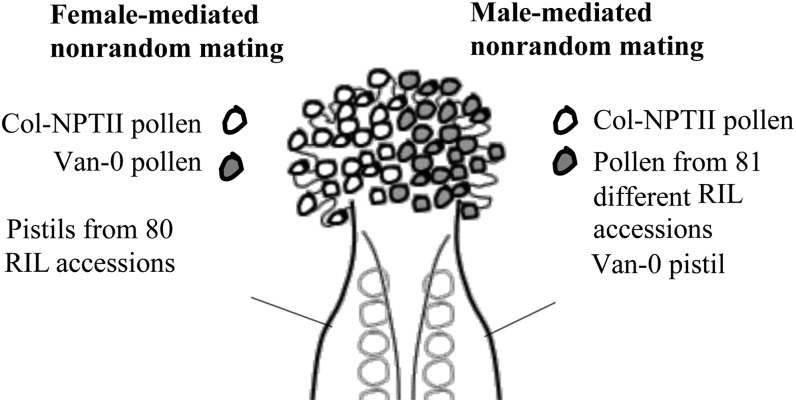
Crossing the RILs separately as mothers and fathers provides independent assays for female and male controls in fertilization (Fig. 2). To map female-mediated nonrandom mating, we performed mixed pollinations between Col-NPTII pollen and Van-0 pollen on virgin RIL pistils. By using parental pollen competitors on different RIL pistils, we expect that differential siring success will be the result of differing maternal genotypes, detectable as QTLs.
Figure 2.
Nonrandom mating experimental design. To map loci that control female-mediated nonrandom mating, pollen from Van-0 and Col-NPTII strains are placed in competition on RIL pistils. Thus, differences in siring success can be attributed to maternal genotypes, detectable as QTLs. To map loci that control male-mediated nonrandom mating, pollen from Col-NPTII is placed in competition with RIL pollen on Van-0 pistils. Thus, differences in siring success can be attributed to paternal genotypes, detectable as QTLs.
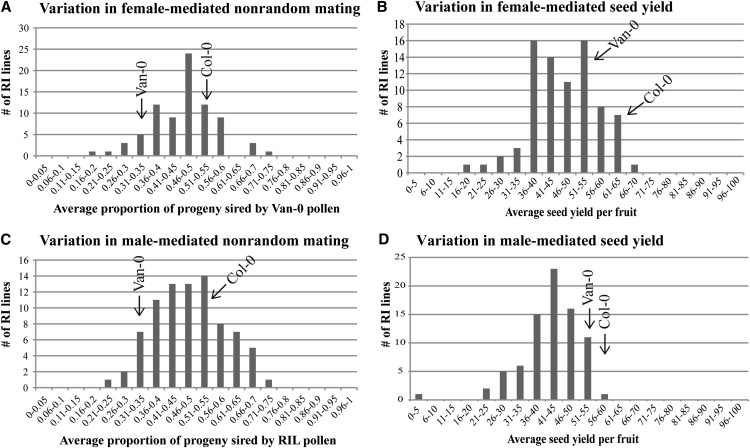
A total of 684 mixed pollinations were performed using pistils from 80 different RILs (8.6 ± 1.7 mixed pollinations per RIL). We paternity tested the resulting 32,309 seeds to calculate the proportion of seeds sired by Van-0. Because of the nature of the data we collected, we were also able to analyze the number of live seeds (seed yield) from each pollination. The RIL pistils display a wide array of phenotypic variation for both female-mediated nonrandom mating (the proportion of seeds sired by Van-0 pollen) and seed yield traits (Fig. 3, A and B). In nonrandom mating and seed yield, RILs displayed transgressive phenotypes that were both higher and lower than their respective parental lines. The average proportion of seeds sired by Van-0 ranged from 0.209 to 0.720. The average number of seeds per fruit ranged from 16.8 to 69.7.
Figure 3.
Frequency distributions in RIL populations of nonrandom mating and seed yield traits. A, Variation in female-mediated nonrandom mating in mixed pollinations with Col-NPTII and Van-0 pollen on RIL pistils. Arrows indicate the proportion of seeds sired by Van-0 pollen in mixed pollinations with Col-NPTII on either Van-0 or Col-0 pistils (parental phenotypes). B, Variation in female-mediated seed yield in mixed pollinations with Van-0 and Col-NPTII pollen on RIL pistils. Arrows indicate seed yield in mixed pollinations with Col-NPTII and Van-0 pollen on either Van-0 or Col-0 pistils (parental phenotypes). C, Variation in male-mediated nonrandom mating in mixed pollinations with Col-NPTII and RIL pollen on Van-0 pistils. Arrows indicate the proportion of seeds sired by either Van-0 or Col-0 pollen in mixed pollinations with Col-NPTII pollen on Van-0 pistils (parental phenotypes). D, Variation in male-mediated seed yield in mixed pollinations. Arrows indicate the seed yield in mixed pollinations with Col-NPTII pollen and either Van-0 or Col-0 pollen on Van-0 pistils (parental phenotypes).
We calculated broad-sense heritability (H2) using the genetic (VG) and error (VE) variance components of a random-effects ANOVA, calculated as H2 = VG/(VG + VE) (Table II). The broad-sense heritability for female-mediated nonrandom mating is 0.21. The broad-sense heritability of female-mediated seed yield is 0.41. Spearman analysis indicates no correlation between these two traits (r2 = 0.004, n = 684, P = 0.723).
Table II. Components of variance and broad-sense heritability.
Components of variance were estimated from an ANOVA of mean square (MS) values, where the dependent variable was paternal proportions or seed yield, and the random factor was RIL identity. The F ratio (MSTrait/MSError) and resulting P values are indicated.
| Trait | No. | Mean | sd | Genetic Variance | Error Variance | Broad-Sense Heritability | F Ratio | P |
|---|---|---|---|---|---|---|---|---|
| Female-mediated nonrandom mating | 684 | |||||||
| Paternal proportions of Van-0 pollen | 0.466 | 0.174 | 0.00653 | 0.0241 | 0.213 | 3.23 | <0.0001 | |
| Seed yield | 47.2 | 14.7 | 59.8 | 84.7 | 0.414 | 7.28 | <0.0001 | |
| Male-mediated nonrandom mating | 744 | |||||||
| Paternal proportions of RIL pollen | 0.489 | 0.182 | 0.00924 | 0.0245 | 0.274 | 4.37 | <0.0001 | |
| Seed yield | 42.5 | 12.7 | 51.7 | 114 | 0.312 | 5.04 | <0.0001 |
Variation in Phenotypes in Male-Mediated Nonrandom Mating and Seed Yield
To map male-mediated nonrandom mating, we performed mixed pollinations between Col-NPTII pollen and RIL pollen on virgin Van-0 pistils (Fig. 2). By using Van-0 maternal pistils and Col-NPTII pollen in mixed pollinations, we expect that differential siring success will be the result of differing RIL pollen genotypes, detectable as QTLs.
We analyzed 744 mixed pollinations from 81 RILs (9.2 ± 1.7 per RIL). We assayed the paternity of the resulting 31,625 seeds to determine the proportion of seeds sired by the RIL pollen. Similar to female-mediated traits, the use of RIL pollen resulted in transgressive phenotypes for male-mediated nonrandom mating and seed yield phenotypes (Fig. 3, C and D). The proportion of seeds sired by RIL pollen in mixed pollinations ranged from 0.221 to 0.738. The average number of seeds per fruit ranged from 3.3 to 57.3.
The broad-sense heritability of male-mediated nonrandom mating was estimated to be 0.274 (Table II). The broad-sense heritability of male-mediated seed yield was 0.312. There was no significant correlation between these two traits (r2 = −0.018, n = 744, P = 0.296; Spearman).
Mapping of Female-Mediated Nonrandom Mating Reveals Epistatic Interactions
We used the above data to perform composite interval mapping and identified four QTLs that direct female-mediated nonrandom mating (qFNRM1–qFNRM4; Fig. 1; Table III). qFNRM1 is present on chromosome 1 and results in a roughly 3% decrease in Van-0 progeny in mixed pollinations when the Van-0 allele is present. It accounts for 3.4% of the variance of the trait. qFNRM3 is present on chromosome 5 and results in a roughly 3% increase in Van-0 progeny when the Van-0 allele is present. It accounts for roughly 7.2% of the variance of the trait. qFNRM4 is present on chromosome 2 and results in a 2.5% decrease in Van-0 progeny when the Van-0 allele is present. It accounts for 3.5% of the variance for the trait. Finally, qFNRM2 is present on chromosome 1 and has a smaller effect on mixed pollinations. Although this last peak was detected in our composite interval mapping, it was not significant by general linear model (GLM), possibly due to its strong epistatic interaction with qFNRM3 (see below).
Table III. QTLs detected for female- and male-mediated nonrandom mating and seed yield traits.
Associated Marker, Position refers to the marker and calculated position (cM) closest to the QTL peak. CI refers to the 2-LOD confidence interval in cM. Variance refers to the percentage of variance explained by the QTL. Additive Effect indicates the change in trait value associated with Van-0 genotypes at the indicated QTLs. Significant LOD scores were determined by calculating P values associated with the maximal LOD scores obtained by 10,000 permutations of the mapping data. Variance components were estimated by restricted maximum likelihood. Asterisks indicate QTLs that did not have a statistically significant effect on trait values by GLM.
| Trait | Chromosome | Associated Marker, Position | CI | LOD | Additive Effect | Variance |
|---|---|---|---|---|---|---|
| % | ||||||
| qFNRM1 | 1 | SNP29_1, 179.85 | 163.65–189.85 | 7.48 | −2.65% | 3.4 |
| qFNRM2 | 1 | SNP10_1, 42.62 | 40.62–42.62 | 4.65 | 1.10% | 1.8* |
| qFNRM3 | 5 | SNP136_5, 45.27 | 34.77–54.22 | 4.11 | 2.98% | 7.2 |
| qFNRM4 | 2 | SNP71_2, 197.63 | 181.45–205.13 | 4.00 | −2.46% | 3.5 |
| Epistasis | qFNRM2*qFNRM3 | −2.55% | 3.3 | |||
| qFSY4 | 1 | SNP12_1, 56.37 | 55.87–56.37 | 10.04 | −2.56 seeds | 9.30 |
| qFSY5 | 5 | SNP144_5, 111.13 | 111.13–112.63 | 9.55 | 3.24 seeds | 15.20 |
| qFSY6 | 1 | SNP43_1, 243.48 | 236.26–245.48 | 5.66 | −3.77 seeds | 21.80 |
| qFSY7 | 5 | SNP140_5, 75.53 | 66.12–82.03 | 5.06 | −3.04 seeds | 12.80 |
| qFSY8 | 2 | SNP71_2, 201.13 | 192.13–206.13 | 4.07 | −1.49* | |
| qMNRM3 | 1 | SNP26_1, 163.65 | 161.96–166.65 | 10.84 | −4.62% | 12.0 |
| qMNRM4 | 1 | SNP24_1, 156.63 | 149.63–168.65 | 8.14 | −0.83% | 2.35 |
| qMNRM5 | 1 | SNP17_1, 88.96 | 84.28–93.96 | 5.89 | 2.57% | 2.51* |
| qMNRM6 | 1 | SNP22_1, 110.69 | 102.59–124.69 | 4.21 | −2.64% | 2.70* |
| Epistasis | qMNRM3*qMNRM4 | 2.40% | 5.0 | |||
| qMSY2 | 1 | SNP20_1, 103.59 | 102.59–108.59 | 5.32 | 2.22 seeds | 11.90 |
| qMSY3 | 3 | SNP84_3, 72.79 | 63.77–79.29 | 4.91 | 1.43 seeds | 6.80 |
| qMSY4 | 1 | SNP16_1, 87.28 | 78.11–91.96 | 4.22 | −2.02 seeds | 9.10 |
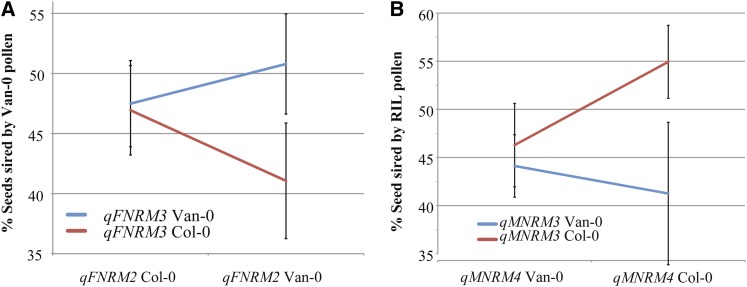
Interestingly, composite interval mapping detected qFNRM2 as having an influence on nonrandom mating, yet the trait means of RILs carrying Van-0 or Col-0 alleles at this locus were similar (proportion of seeds sired by Van-0 pollen across RIL pistils was 47 ± 12 when Van-0 was at qFNRM2 and 47 ± 7 when Col-0 was at qFNRM2, respectively). Since there does not seem to be a simple additive effect of this trait, this suggests an epistatic effect between qFNRM2 and other loci. Indeed, the additive effect of qFNRM3 is completely dependent on Van-0 genotypes at the qFNRM2 locus (Fig. 4A). When qFNRM2 is present as the Col-0 allele, qFNRM3 has little effect on nonrandom mating patterns. In contrast, when qFNRM2 is present as the Van-0 allele, a Van-0 allele at qFNRM3 results in a higher proportion of seeds sired by Van-0 pollen (Table III).
Figure 4.
Epistatic effects of female- and male-mediated nonrandom mating loci. A, Mean of means for the proportion of seeds sired by Van-0 pollen in mixed pollinations of Col-NPTII and Van-0 pollen on RIL pistils. Van-0 and Col-0 genotypes for RILs at qFNRM3 (SNP136_5; blue and red lines, respectively) are indicated with qFNRM2 genotypes in the background (SNP10_1; x axis). Error bars represent 2 se. B, Mean of means for the proportion of seeds sired by RIL pollen in mixed pollinations of Col-NPTII and RIL pollen on RIL pistils. Van-0 and Col-0 genotypes for RILs at qMNRM3 (SNP26_1; blue and red lines, respectively) are indicated with qMNRM4 genotypes in the background (SNP24_1; x axis). Error bars represent 2 se.
Mapping of Male-Mediated Nonrandom Mating Also Reveals Epistatic Interactions
When we analyzed the proportion of seeds sired by RIL pollen in mixed pollinations on Van-0 mothers, four QTLs were identified on chromosome 1 (Fig. 1) for male-mediated nonrandom mating. Since two QTLs using this assay on a different RIL population have already been identified, we adopted the convention of numbering these QTLs consecutively (Carlson et al., 2011). Thus, the four QTLs we identify in this report are labeled qMNRM3 to qMNRM6. qMNRM3 had the largest effect, accounting for 12% of variance of the trait. Because qMNRM3 and qMNRM4 both decrease the proportion of seeds sired by RIL pollen when Van-0 alleles are present and they overlap in position on chromosome 1, we initially considered that they may represent a single locus. However, ANOVA indicated that there is a strong effect of qMNRM3 that is independent of qMNRM4 but masked by Van-0 genotypes (Fig. 4B). When a RIL contains the Col-0 allele at qMNRM3, the Col-0 genotype of qMNRM4 increases the proportion of seeds by 8.7%. In short, containing the Col-0 allele at both these loci results in higher proportions of seeds sired. qMNRM5 and qMNRM6 were not significant via GLM.
Mapping of Seed Yield Traits
Five QTLs were identified for female-mediated seed yield from an analysis of seed yield from mixed pollinations between Col-NPTII and Van-0 pollen on RIL pistils (qFSY4–qFSY8, numbered consecutively after previously identified QTLs; Fig. 1; Table III; Carlson et al., 2011). qFSY4, qFSY6, and qFSY7 all decrease seed yield when the Van-0 allele is present by 2.56, 3.77, and 3.04 seeds per pollination, on average (Fig. 1; Table III). This accounts for 9.3%, 21.8%, and 12.8% of the variance of this trait. qFSY5 increases seed yield when the Van-0 allele is present by an average of 3.24 seeds per pollination, accounting for 15.2% of variance in this trait. qFSY8 showed no significant additive effect and thus was not included in GLM.
Three QTLs were identified as influencing male-mediated seed yield using RIL pollen in mixed pollinations (qMSY2–qMSY4, numbered consecutively after previously identified QTLs; Carlson et al., 2011). When qMSY2 is present as the Van-0 allele in pollen used in mixed pollinations, it results in an average increase of 2.22 seeds per pollination. This accounts for 11.9% of the variance in this trait. qMSY3, when present as the Van-0 allele, results in an increase of 1.43 seeds per pollination, accounting for 6.8% of variance in this trait. Finally, qMSY4, when present as the Van-0 allele, decreases seed yield by 2.02 seeds per pollination, accounting for 9.1% of variance in this trait.
DISCUSSION
The Van-0/Col-0 RIL Population
In order to map predicted controls of female-mediated nonrandom mating in Van-0 and Col-0 accessions, we constructed an expanded linkage map from a new advanced intercross RIL population generated from an initial cross between Van-0 and Col-0 accessions. The map was constructed with a total of 164 SNPs, 76 of which were identified for this study. The relative length of the chromosomes in cM is larger than has been seen in maps with other populations (Lister and Dean, 1993; Alonso-Blanco et al., 1999; Loudet et al., 2002; Clerkx et al., 2004). We note an unusually large amount of segregation distortion in our population, especially on chromosome 5. Chromosome 5 distortions have also been seen to a lesser extent in other populations (Loudet et al., 2002; el-Lithy et al., 2006; Törjék et al., 2006). The basis for segregation distortion is unclear, partly because it could be the result of a number of different phenomena, including postzygotic selection, pollen-pistil interactions, gamete competition, epistatic interactions, and/or seed abortion (Hormaza and Herrero, 1992; Li et al., 1997; Lord and Russell, 2002; Lu et al., 2002). It is of interest that the phenotypes that we test in this paper, nonrandom mating and seed yield, both could result in segregation distortion and that three major effect QTLs reside in the distorted regions (qFNRM3, qFSY7, and qFSY5). We previously noted the colocalization of areas of segregation distortion in RILs and loci that affected male-mediated nonrandom mating and seed yield in our study of Columbia-4 (Col-4)/Landsberg erecta (Ler-0) RILs, further implicating these phenotypes as a potential cause of segregation distortion (Carlson et al., 2011). Segregation distortion can lead to a RIL population with unequal sampling of alleles, which can complicate both the mapping of traits and the subsequent characterization of QTLs. To avoid this, we selected a subset of 82 lines to perform experiments for this report. All materials generated in this paper are available from the Arabidopsis Biological Resource Center (ABRC).
The Identification of Female-Mediated Nonrandom Mating
One reason nonrandom mating in plants has received intense interest is because it may result from sexual selection, either by male competition between pollen grains or female choice by maternal tissue (e.g. the “good genes” hypothesis of sexual selection theory; Charnov, 1979; Mulcahy, 1979; Willson, 1979; Stephenson and Bertin, 1983; Willson and Burley, 1983; Marshall and Ellstrand, 1986; Snow, 1994; Skogsmyr and Lankinen, 2000; Pasonen et al., 2001). Unlike in animals, where the phenomenon is well established, the prospect of sexual selection remains controversial in plants for a number of reasons (Charlesworth et al., 1987; Lyons et al., 1989; Grant, 1995; Winsor et al., 2000). As noted by Marshall et al. (2007), two problems in particular are: (1) the extent to which nonrandom mating is a result of inbreeding depression and the degree of relatedness of the pollen donors with the maternal plants and (2) the difficulty of distinguishing pollen competition from female-mediated nonrandom mating. In this report, we use a plant system that does not display appreciable inbreeding depression to demonstrate, to our knowledge for the first time, unambiguous female genetic control of nonrandom mating.
In plants that routinely outcross, it has long been established that fertilization success in mixed pollinations is context dependent, with genetically related pollen often performing poorly. This is likely the result of deleterious recessive alleles (genetic load) pairing and negatively affecting embryo viability and/or the result of mate discrimination that evolves as a result of inbreeding depression (Bateman, 1956; Weller and Ornduff, 1977; Eckert and Barrett, 1994; Jones, 1994; Waser et al., 1995; Hauser and Siegismund, 2000; Teixeira et al., 2009). This aspect of nonrandom mating is less pronounced in predominantly selfing plants, likely because of reduced genetic load. In Arabidopsis, self pollen does not perform noticeably worse, and in some cases it performs better, than outcross pollen, and there is no correlation between reproductive success and genetic distances between pollen and maternal plants (Carlson et al., 2013). Thus, predominantly selfing plants provide the opportunity to examine aspects of nonrandom mating separable from the effects of inbreeding depression.
The second issue in understanding nonrandom mating patterns is the difficulty of distinguishing between pollen competition and female-mediated nonrandom mating. Interpreting studies in female-mediated nonrandom mating is nontrivial, as study designs require the deposition of multiple pollen populations and thus include the confounding variable of pollen competition. A major advantage of a genetic approach to this issue is that female and male roles in nonrandom mating may be separately mapped, unambiguously identifying gender roles in differential mating. In a previous report, we identified potential female control of nonrandom mating in mixed pollinations between the Van-0 and Col-0 accessions (Carlson et al., 2009). In order to define the genetics of nonrandom mating between these two accessions, we constructed a new mapping population developed from an initial mating between Van-0 and Col-0. By performing mixed pollinations using Col-NPTII and Van-0 pollen on RIL pistils, we identified four QTLs that direct female-mediated nonrandom mating. This makes it clear that, even in a predominantly selfing system, the genetics of the maternal plant do influence what pollen are most successful in mixed pollinations. This is an important first step in identifying the genes that vary between populations that influence female control of nonrandom mating. Unfortunately, because there are few maternal genes known in the literature that affect the dynamics of pollen germination and tube growth, and none of these localized to QTLs, the construction of near isogenic lines coupled with fine-mapping will likely be needed to identify the responsible genes.
Comparison of QTL Positions with Previously Identified QTLs
The mapping of QTLs at similar positions for the same trait in different mapping populations raises the possibility that identical genes or alleles are causative in different populations. One reason we pursued mapping male-mediated nonrandom mating traits in Van-0/Col-0 RILs was to compare with the results of our previous mapping of male-mediated nonrandom mating in Col-4/Ler-0 RILs (Carlson et al., 2011). In both mapping experiments, we identified male-mediated nonrandom mating loci: two using the Col-4/Ler-0 RIL population (qMNRM1 and qMNRM2) and four using the Van-0/Col-0 RIL mapping population (qMRNM3–qMRNM6). Interestingly, even though both RIL populations were constructed with the Col parental accession, the loci that affect male-mediated nonrandom mating in the two populations do not overlap, indicating a variety of genes in different populations of Arabidopsis that affect the dynamics of nonrandom mating. This may be because the male loci we isolated from these two populations could be affecting nonrandom mating in mechanistically distinct ways.
For example, it is possible for male-mediated nonrandom mating to be influenced only by genes in the male (the paternal sporophyte, or male gametophyte). In essence, some pollen may be more competitive than others, regardless of the condition or genotype of the female where the competition takes place. This seems to be the case in Col-NPTII and Ler-0 mixed pollinations, where the identity of the female has minimal effect on the results of the competitions (Carlson et al., 2009, 2011). A single-sex effect is also possible in female-mediated nonrandom mating, in cases when female tissue might distinguish between pollen populations from paternal plants grown in very different environmental conditions. For example, it might be possible for female tissue to distinguish between pollen grown in drought conditions versus nondrought conditions, regardless of the genotype of the pollen. But for pollen from plants grown in very similar conditions, it is likely that female tissue influences pollen success based on pollen genotype. The Van-0/Col-0 RIL population reveals a sizeable female effect. And since the pollen for these mixed pollinations came from plants grown in identical environmental conditions, we hypothesize that some male nonrandom mating loci may be the mechanism that distinguishes sires during the female-mediated nonrandom mating. Thus, the mechanism of male-mediated nonrandom mating in the two RIL populations may be quite different.
Finally, relatively few studies have genetically analyzed the basis for seed yield (seeds per fruit) in Arabidopsis (Alonso-Blanco et al., 2003; Barth et al., 2003; House et al., 2010; Li et al., 2010), and only one study has mapped the female and male contributions to these traits separately (Carlson et al., 2011). Like male-mediated nonrandom mating, the loci that affect female- and male-mediated seed yield do not overlap with those previously identified (qFSY1–qFSY3 and qMSY1) in a study involving the Col-4/Ler-0 population (Carlson et al., 2011).
CONCLUSION
Nonrandom mating has received theoretical and experimental attention for its role as a consequence of inbreeding depression and sexual selection as well as for its role in reinforcing reproductive barriers and speciation. Female control of this system has been hypothesized as necessary for sexual selection via female choice as well as choice mechanisms related to inbreeding, but it has never been genetically established. Here, we use a largely selfing species to demonstrate that, even in the absence of the influence of strong inbreeding depression, the genetics of the maternal plant differentially influence the success of pollen populations. We also identify loci directing male-mediated nonrandom mating and female- and male-mediated seed yield traits. To do so, we generated a new population of advanced intercross RILs between Col-0 and Van-0 accessions. Further studies focused on the identification and analysis of allelic diversity, coupled with the dissection of the physiological mechanisms of nonrandom mating, will provide new insight into this widespread yet little understood phenomenon.
MATERIALS AND METHODS
Plant Growth
Arabidopsis (Arabidopsis thaliana) plants were treated in the following way. Seeds were imbibed and cold treated at 4°C for 3 d to break dormancy and promote uniform germination. For plants that require an overwinter to flower, plants were grown for 1 month before treating at 4°C for 6 weeks. Plants were grown in 4.5-inch pots with 10 plants per pot in Percival growth chambers (Percival Scientific). Plants were grown in Shultz premium potting soil, watered every second day, and fertilized (18-18-21 [for percentage of nitrogen, phosphorus, and potassium, respectively]) twice per week. Plants were subjected to 12 h of 130 μE fluorescent lighting at 22°C.
Generation of RILs
Seeds for accessions Col-0 (accession no. CS22625) and Van-0 (accession no. CS22627) were obtained from the ABRC (http://abrc.osu.edu/). The initial RIL population was generated by intercrossing 300 F2 lines (150 hybrids). Two seeds from each cross were planted, and the intercross cycle was repeated for a total of four generations. Subsequently, eight seeds were propagated from each intercross (1,200 lines), and these were self pollinated for seven additional generations to create inbred lines with loss due to sterility and/or disease. A total of 490 Van-0 × Col-0 RIL lines were deposited in the stock center in 2006 (CS37809) and are formally described here. The lines and marker data (see below) are available through the ABRC. A subset of 82 lines was selected for mapping to avoid segregation distortion (see below).
DNA Extraction, SNP Genotyping, and Map Construction
Genomic DNA was extracted from the 490 original lines and parental lines using the DNeasy Plant Mini Kit (Qiagen). These 490 lines were genotyped at 149 SNP markers developed for a previous study (Platt et al., 2010). Sixty-one of these markers showed no polymorphism between Van-0 and Col-0 parental lines. An additional 76 SNPs were identified, to generate the 164 total markers used in this study. Genotyping was performed using the Sequenom MassArray system at the University of Chicago Comprehensive Cancer Center DNA Sequencing and Genotyping Facility (http://cancer-seqbase.uchicago.edu/). To avoid segregation distortion in mapping, we selected 94 lines and reobtained them from the ABRC. When we verified their genotypes, we revealed a discrepancy with the initial genotype in 12 lines, which we eliminated from the study. The linkage map for the 82 Col-0/Van-0 RILs was constructed using Mapmaker/EXP version 3.0b (Lander et al., 2009) using the Kosambi mapping function. The final map was rendered with MetaQTL (Veyrieras et al., 2007). This map contains 32 ± 10 markers per chromosome and an average distance between the markers of 6.7 ± 5.3 cM (Table I). There was no detected residual heterozygosity in this subset.
Mixed Pollination Experimental Design
For mixed pollinations, Col-NPTII is a derivative of the Col-0 accession. Col-NPTII contains an intergenic kanamycin resistance marker between At1g28440 and At1g28450. Col-NPTII is an F2 homozygous T-DNA insertion mutant obtained from the SIGnAL project (GenBank accession no. BZ377762; Alonso et al., 2003). The presence of the kanamycin resistance marker allows for easy paternity testing of the progeny and does not change the competitive performance of the pollen grain. This was demonstrated in two ways. First, when Col-NPTII was crossed to Col-0 and the F1 was allowed to self-fertilize, the resulting progeny displayed the expected 3:1 segregation of NPTII-mediated kanamycin resistance (Carlson et al., 2009). This would not be the case if the NPTII gene influenced nonrandom mating phenotypes. Second, our mixed pollinations of Col-NPTII and Col-0 pollen on virgin Col-0 and Col-4 pistils show no statistical difference in the siring success of the two pollen types and do not differ from the expected 1:1 ratio in progeny (Carlson et al., 2009, 2013). This demonstrates both that the NPTII gene does not change the competitive ability of the pollen and that we consistently deliver roughly equal amounts of pollen during mixed pollinations.
Mixed pollinations were performed as described previously on the newly bolted primary inflorescences of maternal plants (Carlson et al., 2009, 2011). We emasculate buds during stages 11 to 12 of development and allow pistils to mature to stage 14 before performing pollinations (Smyth et al., 1990). We harvest anthers from stage 14 flowers and choose two anthers from each paternal plant. We first apply pollen from the fixed pollen competitor accession (Col-NPTII) on half the available surface area of the stigma. We then apply pollen from the competing accession on the remaining surface area. We use saturating amounts of pollen (more than 500). We collect siliques and assay seed paternity by growing seeds on Murashige and Skoog medium containing 50 μg mL−1 kanamycin (Murashige and Skoog, 1962).
Pollinations that yielded no live seed were excluded from subsequent analysis, as were any RILs that failed to yield at least three successful pollinations (defined as a pollination that yields at least one viable seed). A total of 1,611 successful mixed pollinations were performed for this study.
Statistical Analysis and QTL Mapping
Male- and female-mediated nonrandom mating and seed yield data fit a normal distribution. We partitioned phenotypic variance between RILs into genetic (VG) and error (VE) variance components in a random-effects ANOVA using SAS (SAS Institute). This was used to calculate broad-sense heritability (H2) as follows: H2 = VG/(VG + VE). Phenotypic correlations were performed using a Pearson’s correlation in SAS.
Confidence interval mapping was conducted with Zmapqtl (QTL Cartographer version 1.17j; Basten et al., 2002). The reported P value thresholds were calculated using the maximal log of the odds (LOD) scores from 10,000 permutations of trait data. The 2-LOD confidence intervals were derived from the identification of the nearest markers at which the LOD score for a trait dropped 2-LOD below the LOD maximum. QTL additive values and significance were estimated from a GLM including all detected QTL peaks (trait = μ + βmrk1 × Mrk1 + … + βmrkn × MrkN + ε, where μ = the population mean and ε = residual error), with a crossed term added when epistasis was detected (JMP version 8.0.1; SAS Institute). Variance components were estimated with a restricted maximum likelihood method (JMP).
Acknowledgments
We thank Adam Hammond for useful comments and two anonymous reviewers for comments.
Glossary
- Van-0
Vancouver
- Col-0
Columbia
- RIL
recombinant inbred line
- QTL
quantitative trait locus
- SNP
single-nucleotide polymorphism
- cM
centimorgan
- GLM
general linear model
- Ler-0
Landsberg erecta
- ABRC
Arabidopsis Biological Resource Center
- LOD
log of the odds
- Col-NPTII
genetically marked Columbia
- Col-4
Columbia-4
Footnotes
This work was supported by the National Science Foundation (grant no. IOS–1020325 to R.J.S.).
Articles can be viewed online without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen HM, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C, Bentsink L, Hanhart CJ, Blankestijn-de Vries H, Koornneef M. (2003) Analysis of natural allelic variation at seed dormancy loci of Arabidopsis thaliana. Genetics 164: 711–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C, Blankestijn-de Vries H, Hanhart CJ, Koornneef M. (1999) Natural allelic variation at seed size loci in relation to other life history traits of Arabidopsis thaliana. Proc Natl Acad Sci USA 96: 4710–4717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster WS, Rogers DG. (2004) Does pollen competition reduce the cost of inbreeding? Am J Bot 91: 1939–1943 [DOI] [PubMed] [Google Scholar]
- Bakker EG, Stahl EA, Toomajian C, Nordborg M, Kreitman M, Bergelson J. (2006) Distribution of genetic variation within and among local populations of Arabidopsis thaliana over its species range. Mol Ecol 15: 1405–1418 [DOI] [PubMed] [Google Scholar]
- Barth S, Busimi AK, Utz HF, Melchinger AE. (2003) Heterosis for biomass yield and related traits in five hybrids of Arabidopsis thaliana L. Heynh. Heredity (Edinb) 91: 36–42 [DOI] [PubMed] [Google Scholar]
- Basten CJ, Weir BS, Zeng ZB (2002) QTL Cartographer, Version 1.17. Department of Statistics, North Carolina State University, Raleigh [Google Scholar]
- Bateman AJ. (1956) Cryptic self-incompatibility in the wallflower Cherianthus cheiri L. Heredity 10: 257–261 [Google Scholar]
- Bernasconi G, Ashman TL, Birkhead TR, Bishop JDD, Grossniklaus U, Kubli E, Marshall DL, Schmid B, Skogsmyr I, Snook RR, et al. (2004) Evolutionary ecology of the prezygotic stage. Science 303: 971–975 [DOI] [PubMed] [Google Scholar]
- Bomblies K, Yant L, Laitinen RA, Kim ST, Hollister JD, Warthmann N, Fitz J, Weigel D. (2010) Local-scale patterns of genetic variability, outcrossing, and spatial structure in natural stands of Arabidopsis thaliana. PLoS Genet 6: e1000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman RN. (1987) Cryptic self-incompatibility and the breeding system of Clarkia unguiculata (Onagraceae). Am J Bot 74: 471–476 [Google Scholar]
- Byers DL, Waller DM. (1999) Do plant populations purge their genetic load? Effects of population size and mating history on inbreeding depression. Annu Rev Ecol Syst 30: 479–513 [Google Scholar]
- Carlson AL, Fitz Gerald JN, Telligman M, Roshanmanesh J, Swanson RJ. (2011) Defining the genetic architecture underlying female- and male-mediated nonrandom mating and seed yield traits in Arabidopsis. Plant Physiol 157: 1956–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson AL, Gong H, Toomajian C, Swanson RJ. (2013) Parental genetic distance and patterns in nonrandom mating and seed yield in predominately selfing Arabidopsis thaliana. Plant Reprod 26: 317–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson AL, Telligman M, Swanson RJ. (2009) Incidence and post-pollination mechanisms of nonrandom mating in Arabidopsis thaliana. Sex Plant Reprod 22: 257–262 [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B. (1987) Inbreeding depression and its evolutionary consequences. Annu Rev Ecol Syst 18: 237–268 [Google Scholar]
- Charlesworth D, Schemske DW, Sork VL (1987) The evolution of plant reproductive characters: sexual versus natural selection. In SC Stearns, ed, Evolution of Sex, Vol 55. Birkhauser, Basel, pp 317–335 [DOI] [PubMed] [Google Scholar]
- Charnov EL. (1979) Simultaneous hermaphroditism and sexual selection. Proc Natl Acad Sci USA 76: 2480–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerkx EJM, El-Lithy ME, Vierling E, Ruys GJ, Blankestijn-De Vries H, Groot SPC, Vreugdenhil D, Koornneef M. (2004) Analysis of natural allelic variation of Arabidopsis seed germination and seed longevity traits between the accessions Landsberg erecta and Shakdara, using a new recombinant inbred line population. Plant Physiol 135: 432–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crnokrak P, Barrett SCH. (2002) Perspective. Purging the genetic load: a review of the experimental evidence. Evolution 56: 2347–2358 [DOI] [PubMed] [Google Scholar]
- Cruzan MB. (1990) Pollen-pollen and pollen-style interactions during pollen-tube growth in Erythronium grandiflorum (Liliaceae). Am J Bot 77: 116–122 [Google Scholar]
- de Nettancourt D (2001) Incompatibility and Incongruity in Wild and Cultivated Plants. Springer, Berlin [Google Scholar]
- Eckert CG, Barrett SCH. (1994) Postpollination mechanisms and the maintenance of outcrossing in self-compatible, tristylous, Decodon verticillatus (Lythraceae). Heredity 72: 396–411 [Google Scholar]
- el-Lithy ME, Bentsink L, Hanhart CJ, Ruys GJ, Rovito DI, Broekhof JLM, van der Poel HJA, van Eijk MJT, Vreugdenhil D, Koornneef M. (2006) New Arabidopsis recombinant inbred line populations genotyped using SNPWave and their use for mapping flowering-time quantitative trait loci. Genetics 172: 1867–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant V. (1995) Sexual selection in plants: pros and cons. Proc Natl Acad Sci USA 92: 1247–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haileselassie T, Mollel M, Skogsmyr I. (2005) Effects of nutrient level on maternal choice and siring success in Cucumis sativus (Cucurbitaceae). Evol Ecol 19: 275–288 [Google Scholar]
- Hauser TP, Siegismund HR. (2000) Inbreeding and outbreeding effects on pollen fitness and zygote survival in Silene nutans (Caryophyllaceae). J Evol Biol 13: 446–454 [Google Scholar]
- Hogenboom NG. (1973) Model for incongruity in intimate partner relationships. Euphytica 22: 219–233 [Google Scholar]
- Hogenboom NG. (1975) Incompatibility and incongruity. 2. Different mechanisms for non-functioning of intimate partner relationships. Proc R Soc Lond B Biol Sci 188: 361–374 [Google Scholar]
- Hormaza JI, Herrero M. (1992) Pollen selection. Theor Appl Genet 83: 663–672 [DOI] [PubMed] [Google Scholar]
- House C, Roth C, Hunt J, Kover PX. (2010) Paternal effects in Arabidopsis indicate that offspring can influence their own size. Proc Biol Sci 277: 2885–2893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husband BC, Schemske DW, Burton TL, Goodwillie C. (2002) Pollen competition as a unilateral reproductive barrier between sympatric diploid and tetraploid Chamerion angustifolium. Proc Biol Sci 269: 2565–2571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston MO. (1993) Tests of 2 hypotheses concerning pollen competition in a self-compatible, long-styled species (Lobelia cardinalis, Lobeliaceae). Am J Bot 80: 1400–1406 [Google Scholar]
- Jones KN. (1994) Nonrandom mating in Clarkia gracilis (Onagraceae): a case of cryptic self-incompatibility. Am J Bot 81: 195–198 [Google Scholar]
- Lande R, Schemske DW. (1985) The evolution of self-fertilization and inbreeding depression in plants. 1. Genetic models. Evolution 39: 24–40 [DOI] [PubMed] [Google Scholar]
- Lande R, Schemske DW, Schultz ST. (1994) High inbreeding depression, selective interference among loci, and the threshold selfing rate for purging recessive lethal mutations. Evolution 48: 965–978 [DOI] [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newberg LA. (2009) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations (vol 1 pg 174, 1987). Genomics 93: 398. [DOI] [PubMed] [Google Scholar]
- Lankinen A, Armbruster WS. (2007) Pollen competition reduces inbreeding depression in Collinsia heterophylla (Plantaginaceae). J Evol Biol 20: 737–749 [DOI] [PubMed] [Google Scholar]
- Lankinen A, Maad J, Armbruster WS. (2009) Pollen-tube growth rates in Collinsia heterophylla (Plantaginaceae): one-donor crosses reveal heritability but no effect on sporophytic-offspring fitness. Ann Bot (Lond) 103: 941–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lankinen A, Skogsmyr I. (2002) Pollen competitive ability: the effect of proportion in two-donor crosses. Evol Ecol Res 4: 687–700 [Google Scholar]
- Li Y, Huang Y, Bergelson J, Nordborg M, Borevitz JO. (2010) Association mapping of local climate-sensitive quantitative trait loci in Arabidopsis thaliana. Proc Natl Acad Sci USA 107: 21199–21204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZK, Pinson SRM, Paterson AH, Park WD, Stansel JW. (1997) Genetics of hybrid sterility and hybrid breakdown in an intersubspecific rice (Oryza sativa L.) population. Genetics 145: 1139–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister C, Dean C. (1993) Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J 4: 745–750 [DOI] [PubMed] [Google Scholar]
- Lord EM, Russell SD. (2002) The mechanisms of pollination and fertilization in plants. Annu Rev Cell Dev Biol 18: 81–105 [DOI] [PubMed] [Google Scholar]
- Loudet O, Chaillou S, Camilleri C, Bouchez D, Daniel-Vedele F. (2002) Bay-0 × Shahdara recombinant inbred line population: a powerful tool for the genetic dissection of complex traits in Arabidopsis. Theor Appl Genet 104: 1173–1184 [DOI] [PubMed] [Google Scholar]
- Lu H, Romero-Severson J, Bernardo R. (2002) Chromosomal regions associated with segregation distortion in maize. Theor Appl Genet 105: 622–628 [DOI] [PubMed] [Google Scholar]
- Lyons EE, Waser NM, Price MV, Antonovics J, Motten AF. (1989) Sources of variation in plant reproductive success and implications for concepts of sexual selection. Am Nat 134: 409–433 [Google Scholar]
- Marshall DL, Avritt JJ, Shaner M, Saunders RL. (2000) Effects of pollen load size and composition on pollen donor performance in wild radish, Raphanus sativus (Brassicaceae). Am J Bot 87: 1619–1627 [PubMed] [Google Scholar]
- Marshall DL, Diggle PK. (2001) Mechanisms of differential pollen donor performance in wild radish, Raphanus sativus (Brassicaceae). Am J Bot 88: 242–257 [PubMed] [Google Scholar]
- Marshall DL, Ellstrand NC. (1986) Sexual selection in Raphanus sativus: experimental data on nonrandom fertilization, maternal choice, and consequences of multiple paternity. Am Nat 127: 446–461 [Google Scholar]
- Marshall DL, Ellstrand NC. (1988) Effective mate choice in wild radish: evidence for selective seed abortion and its mechanism. Am Nat 131: 739–756 [Google Scholar]
- Marshall DL, Reynolds J, Abrahamson NJ, Simpson HL, Barnes MG, Medeiros JS, Walsh S, Oliveras DM, Avritt JJ. (2007) Do differences in plant and flower age change mating patterns and alter offspring fitness in Raphanus sativus (Brassicaceae)? Am J Bot 94: 409–418 [DOI] [PubMed] [Google Scholar]
- Meng XY, Sun PL, Kao TH. (2011) S-RNase-based self-incompatibility in Petunia inflata. Ann Bot (Lond) 108: 637–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell RJ, Marshall DL. (1998) Nonrandom mating and sexual selection in a desert mustard: an experimental approach. Am J Bot 85: 48–55 [PubMed] [Google Scholar]
- Mulcahy DL. (1979) The rise of the angiosperms: a genecological factor. Science 206: 20–23 [DOI] [PubMed] [Google Scholar]
- Mulcahy DL, Mulcahy GB. (1987) The effects of pollen competition. Am Sci 75: 44–50 [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nasrallah JB (2011) Self-incompatibility in the Brassicaceae. In R Schmidt, I Bancroft, eds, Genetics and Genomics of the Brassicaceae, Vol 9. Springer, New York, pp 389–411 [Google Scholar]
- Paschke M, Abs C, Schmid B. (2002) Effects of population size and pollen diversity on reproductive success and offspring size in the narrow endemic Cochlearia bavarica (Brassicaceae). Am J Bot 89: 1250–1259 [DOI] [PubMed] [Google Scholar]
- Pasonen HL, Pulkkinen P, Käpylä M. (2001) Do pollen donors with fastest-growing pollen tubes sire the best offspring in an anemophilous tree, Betula pendula (Betulaceae)? Am J Bot 88: 854–860 [PubMed] [Google Scholar]
- Pasonen HL, Pulkkinen P, Kapyla M, Blom A. (1999) Pollen-tube growth rate and seed-siring success among Betula pendula clones. New Phytol 143: 243–251 [Google Scholar]
- Platt A, Horton M, Huang YS, Li Y, Anastasio AE, Mulyati NW, Agren J, Bossdorf O, Byers D, Donohue K, et al. (2010) The scale of population structure in Arabidopsis thaliana. PLoS Genet 6: e1000843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queller DC. (1983) Sexual selection in a hermaphroditic plant. Nature 305: 706–707 [Google Scholar]
- Quesada M, Winsor JA, Stephenson AG. (1993) Effects of pollen competition on progeny performance in a heterozygous cucurbit. Am Nat 142: 694–706 [DOI] [PubMed] [Google Scholar]
- Schemske DW, Lande R. (1985) The evolution of self-fertilization and inbreeding depression in plants. 2 Empirical observations. Evolution 39: 41–52 [DOI] [PubMed] [Google Scholar]
- Shaner MGM, Marshall DL. (2003) Under how wide a set of conditions will nonrandom mating occur in Raphanus sativus (Brassicaceae)? Am J Bot 90: 1604–1611 [DOI] [PubMed] [Google Scholar]
- Skogsmyr I, Lankinen A. (1999) Selection on pollen competitive ability in relation to stochastic factors influencing pollen deposition. Evol Ecol Res 1: 971–985 [Google Scholar]
- Skogsmyr I, Lankinen A. (2000) Potential selection for female choice in Viola tricolor. Evol Ecol Res 2: 965–979 [Google Scholar]
- Skogsmyr I, Lankinen A. (2002) Sexual selection: an evolutionary force in plants? Biol Rev Camb Philos Soc 77: 537–562 [DOI] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow AA. (1994) Postpollination selection and male fitness in plants. Am Nat (Suppl) 144: S69–S83 [Google Scholar]
- Snow AA, Mazer SJ. (1988) Gametophytic selection in Raphanus raphanistrum: a test for heritable variation in pollen competitive ability. Evolution 42: 1065–1075 [DOI] [PubMed] [Google Scholar]
- Snow AA, Spira TP. (1991a) Differential pollen-tube growth rates and nonrandom fertilization in Hibiscus moscheutos (Malvaceae). Am J Bot 78: 1419–1426 [Google Scholar]
- Snow AA, Spira TP. (1991b) Pollen vigor and the potential for sexual selection in plants. Nature 352: 796–797 [Google Scholar]
- Sork VL, Schemske DW. (1992) Fitness consequences of mixed-donor pollen loads in the annual legume Chamaecrista fasciculata. Am J Bot 79: 508–515 [Google Scholar]
- Stephenson AG, Bertin RI (1983) Mate Competition, Female Choice and Sexual Selection in Plants. Academic Press, Orlando, FL [Google Scholar]
- Stephenson AG, Hayes CN, Johannsson MH, Winsor JA. (2001) The performance of microgametophytes is affected by inbreeding depression and hybrid vigor in the sporophytic generation. Sex Plant Reprod 14: 77–83 [Google Scholar]
- Stephenson AG, Travers SE, Mena-Ali JI, Winsor JA. (2003) Pollen performance before and during the autotrophic-heterotrophic transition of pollen tube growth. Philos Trans R Soc Lond B Biol Sci 358: 1009–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira S, Foerster K, Bernasconi G. (2009) Evidence for inbreeding depression and post-pollination selection against inbreeding in the dioecious plant Silene latifolia. Heredity (Edinb) 102: 101–112 [DOI] [PubMed] [Google Scholar]
- Törjék O, Witucka-Wall H, Meyer RC, von Korff M, Kusterer B, Rautengarten C, Altmann T. (2006) Segregation distortion in Arabidopsis C24/Col-0 and Col-0/C24 recombinant inbred line populations is due to reduced fertility caused by epistatic interaction of two loci. Theor Appl Genet 113: 1551–1561 [DOI] [PubMed] [Google Scholar]
- Veyrieras JB, Goffinet B, Charcosset A. (2007) MetaQTL: a package of new computational methods for the meta-analysis of QTL mapping experiments. BMC Bioinformatics 8: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waser NM, Shaw RG, Price MV. (1995) Seed set and seed mass in Ipomopsis aggregata: variance partitioning and inferences about postpollination selection. Evolution 49: 80–88 [DOI] [PubMed] [Google Scholar]
- Weller SG, Ornduff R. (1977) Cryptic self-incompatibility in Amsinckia grandiflora. Evolution 31: 47–51 [DOI] [PubMed] [Google Scholar]
- Wheeler MJ, de Graaf BHJ, Hadjiosif N, Perry RM, Poulter NS, Osman K, Vatovec S, Harper A, Franklin FCH, Franklin-Tong VE. (2009) Identification of the pollen self-incompatibility determinant in Papaver rhoeas. Nature 459: 992–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JH, Jr, Friedman WE, Arnold ML. (1999) Developmental selection within the angiosperm style: using gamete DNA to visualize interspecific pollen competition. Proc Natl Acad Sci USA 96: 9201–9206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willson MF. (1979) Sexual selection in plants. Am Nat 113: 777–790 [Google Scholar]
- Willson MF, Burley N (1983) Mate Choice in Plants. Princeton University Press, Princeton [Google Scholar]
- Winsor JA, Peretz S, Stephenson AG. (2000) Pollen competition in a natural population of Cucurbita foetidissima (Cucurbitaceae). Am J Bot 87: 527–532 [PubMed] [Google Scholar]