Early seed development in rice is highly sensitive to heat stress and negatively affects seed enlargement because of epigenetic misregulation of endosperm development.
Abstract
Although heat stress reduces seed size in rice (Oryza sativa), little is known about the molecular mechanisms underlying the observed reduction in seed size and yield. To elucidate the mechanistic basis of heat sensitivity and reduced seed size, we imposed a moderate (34°C) and a high (42°C) heat stress treatment on developing rice seeds during the postfertilization stage. Both stress treatments reduced the final seed size. At a cellular level, the moderate heat stress resulted in precocious endosperm cellularization, whereas severe heat-stressed seeds failed to cellularize. Initiation of endosperm cellularization is a critical developmental transition required for normal seed development, and it is controlled by Polycomb Repressive Complex2 (PRC2) in Arabidopsis (Arabidopsis thaliana). We observed that a member of PRC2 called Fertilization-Independent Endosperm1 (OsFIE1) was sensitive to temperature changes, and its expression was negatively correlated with the duration of the syncytial stage during heat stress. Seeds from plants overexpressing OsFIE1 had reduced seed size and exhibited precocious cellularization. The DNA methylation status and a repressive histone modification of OsFIE1 were observed to be temperature sensitive. Our data suggested that the thermal sensitivity of seed enlargement could partly be caused by altered epigenetic regulation of endosperm development during the transition from the syncytial to the cellularized state.
World rice (Oryza sativa) production needs to increase significantly to sustain an increasing population. However, higher average temperatures caused by global warming are predicted to decrease rice yields in many parts of the world, especially Asia. In one study, rice yield was estimated to decrease by 10% for every 1°C rise in minimum growing season temperature (Peng et al., 2004). Comparable yield losses with rising temperatures have been reported for two other major cereal crops: wheat (Triticum spp.) and maize (Zea mays; Wardlaw, 1989; Lobell et al., 2011). Rice, wheat, and maize together are the main sources of calories for most countries (Reynolds et al., 2011). Therefore, it is critical that we understand the agronomic, biological, and economic consequences of high temperature on crop yields.
Heat stress during seed development decreases the seed size in many cereals (Nagato and Ebata, 1960; Hunter et al., 1977; Savin et al., 1996) and when coupled with seed number per unit area, determines seed yield. Seed size is largely contributed by the endosperm, a triploid tissue derived from fusion of the sperm cell with the diploid central cell during the double fertilization event. Endosperm development progresses in distinct developmental stages. After fertilization, the endosperm enters the syncytial stage, where triploid nuclei undergo rapid mitotic divisions without cytokinesis, followed by cellularization and finally, differentiation and maturation (Olsen, 2001; Sabelli and Larkins, 2009b). Duration of the syncytial stage and rate of mitotic divisions during this stage are important determinants of seed size (Mizutani et al., 2010). Successful transition from the syncytial to the cellularization stage is critical for normal seed development (Brown et al., 1996).
In Arabidopsis (Arabidopsis thaliana), the processes controlling early endosperm development and the transition from syncytial to cellularized stage are associated with the Polycomb Repressive Complex2 (PRC2) genes, which includes Fertilization-Independent Endosperm (FIE), Fertilization-Independent Seed2 (FIS2), Medea (MEA), and Multicopy Suppressor of IRA1 (Guitton and Berger, 2005; Baroux et al., 2006; Huh et al., 2007). The PRC2 complex is involved in gene silencing mediated by a repressive histone modification (H3K27me3; Köhler and Villar, 2008). Loss of function of several of these PRC2 genes results in abnormal endosperm development. A notable phenotype observed in Arabidopsis FIS mutants is endosperm overproliferation and seed failure (Kiyosue et al., 1999; Sørensen et al., 2002). Several endosperm-specific MADS-box genes (such as Pheres1, AGAMOUS-LIKE36 (AGL36), and AGL62 among others) are misregulated in seeds that are deficient in PRC2-encoding genes (Kang et al., 2008; Köhler and Villar, 2008; Walia et al., 2009). A loss-of-function mutation in Arabidopsis AGL62 resulted in precocious cellularization and smaller seeds (Kang et al., 2008). Although the function of the PRC2 complex is conserved in cereals such as rice and maize, orthologs of FIS2 and MEA have not been reported (Spillane et al., 2007; Luo et al., 2009). Orthologs of the Arabidopsis FIE gene have been reported in both rice (OsFIE1 and OsFIE2) and maize (ZmFIE1 and ZmFIE2; Springer et al., 2002; Danilevskaya et al., 2003; Luo et al., 2009). OsFIE1 is expressed only in the endosperm, whereas OsFIE2 is expressed in all tissues tested (Luo et al., 2009; Nallamilli et al., 2013). OsFIE1 is an imprinted gene, and its expression is regulated by DNA and H3K9me2 methylation (Luo et al., 2009; Zhang et al., 2012). OsFIE2 has a critical role in normal endosperm development and grain filling (Nallamilli et al., 2013).
Our understanding of the epigenetic regulation of rice seed development has improved significantly (Zemach et al., 2010; Luo et al., 2011; Rodrigues et al., 2013). However, how the epigenetic regulation during seed development is altered during environmental perturbations is not well characterized. Most research efforts in the past have focused on the grain-filling stage (when storage proteins and starch accumulate) under stressful conditions (Yamakawa et al., 2007). However, it is not known if and how an environmental stress that specifically occurs during early seed development impacts seed size in rice. Here, we present evidence that early rice seed development is highly sensitive to heat stress and results in seed size reduction. We suggest a molecular mechanism that involves the rice PRC2 gene OsFIE1 as a potential component involved in regulating seed enlargement under heat stress.
RESULTS
Heat Stress Alters the Duration of Syncytial Stage Endosperm Development
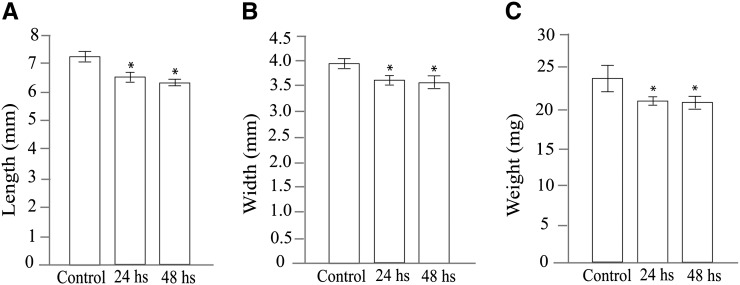
We hypothesized that early endosperm development could be sensitive to high temperature and is a significant factor in reducing seed size and yield. Therefore, our experiments focused on elucidating the effects of heat stress imposed postfertilization at the syncytial stage of endosperm development. The syncytial stage is characterized by a rapid increase in endosperm nuclei number caused by mitotic divisions without formation of individual cells and cell walls and occurs during the first 2 or 3 d after fertilization (DAF; Mizutani et al., 2010; Ishikawa et al., 2011). To observe the specific effects of heat stress on early endosperm development, we imposed a moderate (34°C) and a severe (42°C) heat stress on rice plants at 24 h after fertilization (HAF) for 48 h. A parallel set of rice plants was maintained under optimal conditions (28°C) and used as controls for all experiments. We observed that severe heat stress significantly reduced the size of the developing seeds as well as the final seed size at maturity (Fig. 1; Supplemental Fig. S1). The rapid increase in seed size under control conditions was clearly retarded under severe stress (Fig. 1). We did not notice an obvious difference in seed size between the moderately stressed and control plants during the 2 d of high temperature stress. However, the moderate heat stress of 34°C significantly (P = 0.0001) decreased the size of mature seeds (Fig. 2). The smaller seed size after 24 and 48 h of moderate stress was reflected in reduced seed length, width, and weight at maturity (Fig. 2; Supplemental Fig. S2). In the seeds exposed to severe heat stress, the endosperm failed to fill completely, and the seeds were not viable because they failed to germinate (Supplemental Fig. S1). Notably, our data indicated that even a transient period of moderately high temperature during early seed development can significantly impact seed size. We have identified a previously uncharacterized, to our knowledge, seed development stage, in which high heat sensitivity could be partly responsible for yield losses caused by reduced seed size.
Figure 1.
Severe heat stress reduces seed size. A, Images of developing rice seeds at 24, 48, 72, and 96 HAF under control (28°C) conditions. This stage is characterized by rapid increase in seed size. B, Seeds collected from plants exposed to severe heat stress (42°C) imposed at 24 HAF for 2 d show a decrease in the size of developing seeds. Bars = 1 mm.
Figure 2.
Morphometric analysis of mature rice seeds obtained from plants grown under control and moderate heat stress. A, Length. B, Width. C, Weight. Length, width, and weight were significantly (an asterisk indicates P = 0.0001, n = 50) reduced by a short duration of heat stress. The data are presented as means ± sds.
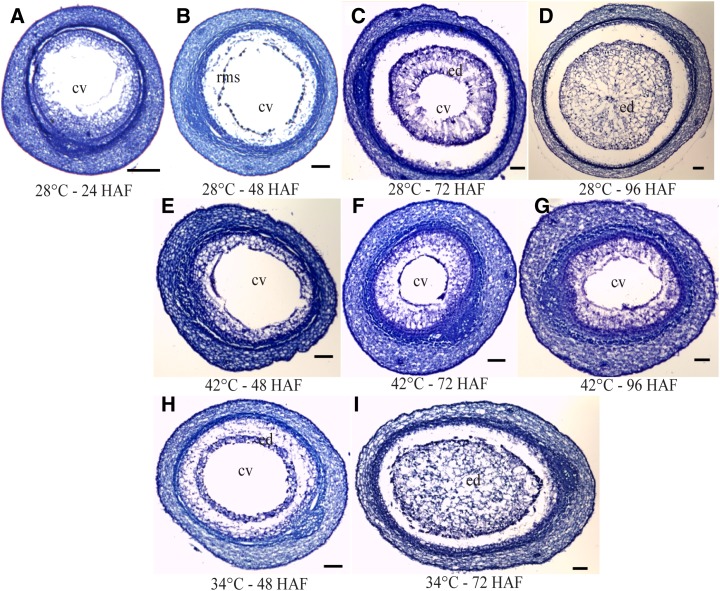
An important event in early endosperm development is the transition from the syncytial stage characterized by the presence of free nuclei to the cellularized stage, when cytokinesis and cell wall formation are observed (Hehenberger et al., 2012). We collected developing seeds from control and moderate and severe heat-stressed plants to document the duration of syncytial stage and the onset of cellularization. Cross sections of rice seeds under control and heat stress conditions are shown in Figure 3. At 48 HAF, rice seeds developed in control conditions were in the syncytial stage (Fig. 3B). Cellularization was initiated between 48 and 72 HAF, and the endosperm was completely cellularized by 96 HAF (Fig. 3, A–D). With moderate heat stress, the developing seeds exhibited precocious cellularization and were completely cellularized by 72 HAF (Fig. 3). In contrast, seeds exposed to 42°C heat stress failed to initiate cellularization at 72 HAF, and the central vacuole was still present at 96 HAF (Fig. 3). A moderate heat stress shortened the duration of the syncytial stage and accelerated the transition to a cellularized endosperm. A severe heat stress, which may not be uncommon in field conditions, resulted in failure of the endosperm to cellularize, resulting in nonviable seeds. We show that the smaller seed size observed during the moderate heat stress is a result of precocious cellularization. Our data suggest that developing rice seeds sense and respond to thermal variations in determining the duration of the endosperm syncytial stage. The molecular basis of this seed-specific response to temperature is not known in plants.
Figure 3.
Histological analysis of early seed development under control and severe and moderate heat stress conditions. Seeds were harvested at 24, 48, 72, and 96 HAF for control and severe stress samples. For moderate stress, seeds were collected at 48- and 72-HAF time points. A to D, Seeds from control condition plants (28°C). Endosperm (ed) cellularization was initiated by 72 HAF and completed by 96 HAF. E to G, Sections were obtained from plants exposed to a severe heat stress of 42°C initiated at 24 HAF for 2 d. Cellularization was delayed under severe heat stress, and the central vacuole (cv) was visible at 96 HAF. H and I, Under moderate heat stress (34°C), seeds exhibited early cellularization, and the endosperm was cellularized by 72 HAF. rms, Radical microtubule system. Bars = 200 µm.
Heat Stress Misregulates OsFIE1 Expression
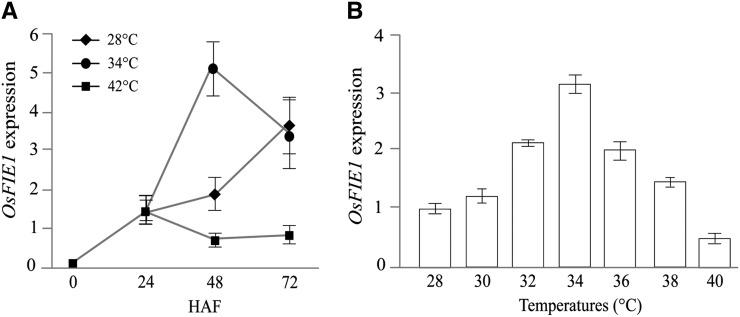
We performed microarray analysis on developing seeds from plants exposed to control (28°C), moderate (34°C), and severe (42°C) heat stress conditions at 48 and 72 HAF. Because moderate and severe heat stress affected endosperm developmental transitions, we mined the data set for genes that are misregulated during heat stress and have a role in endosperm development. A complete list of the all the genes and their expression values under moderate and severe heat stress is provided (Supplemental Data Set S1). We specifically examined the rice PRC2 genes because of their known role in endosperm developmental transition from the syncytial to cellularized state (Fig. 4). Of six PRC2 genes, OsFIE1 was found to be significantly differentially regulated in response to moderate heat stress. The transcript abundance of OsFIE1 was higher in seeds collected from plants exposed to moderate heat stress compared with control samples at 48 HAF. Under severe heat stress, OsFIE1 transcripts did not accumulate at the 48- and 72-HAF time points. To confirm the OsFIE1 expression observed in our microarray data, we performed quantitative reverse transcription PCR (qRT-PCR) assays on rice seeds/ovules before fertilization (0 HAF), and after fertilization at 24, 48, and 72 HAF in control and 34°C and 42°C heat stress conditions. The OsFIE1 transcript begins to accumulate by 48 HAF and increases rapidly by the 72-HAF time point relative to unfertilized ovules (Fig. 5A). At 42°C, heat stress transcript levels of OsFIE1 fail to accumulate by 48 and 72 HAF. In striking contrast, transcript abundance of OsFIE1 was higher at 34°C compared with control seeds at 48 HAF. At 72 HAF, OsFIE1 expression level was not significantly different for 34°C seeds compared with control seeds (Fig. 5A). These data show that, under moderate heat stress, expression of OsFIE1 is higher at 48 HAF, whereas severe heat stress represses OsFIE1, at least at the time points measured in these experiments.
Figure 4.
Expression-based heat map of the rice PRC2 genes in response to moderate heat stress. The heat map was generated from the microarray expression values for seed samples collected at 48 and 72 HAF under control and moderate heat stress conditions. The six rice PRC2 members in the cluster are OsFIE1 (Os08g04290), OsFIE2 (Os08g04270), OsSET1 (Os03g19480), OsCLF (Os06g16390), OsEMF2a (Os04g08034), and OsEMF2b (Os09g13630). Log-transformed (base 2) expression values were scaled between −1 and 1. Higher expression values are represented in red, and lower values are represented in blue. CLF, Curly Leaf; EMF, Embryonic flower.
Figure 5.
Gene expression of rice OsFIE1 during early seed development. A, OsFIE1 relative expression under control (28°C) and severe (42°C) and moderate (34°C) stress exhibits transcript abundance shift in response to temperature. Unfertilized ovules sample (0 HAF) was used as the baseline for relative expression. B, Gene expression of OsFIE1 at 48 HAF in seeds developing in a range of temperatures (28°C to –40°C). The 28°C seed sample was used as the baseline for relative expression of OsFIE1. The data are presented as means ± sds.
We next explored the temperature sensitivity of OsFIE1 expression in developing rice seeds grown in temperature regimes that differed by 2°C increments from 28°C to 40°C. All seed samples were collected at 48 HAF, and OsFIE1 transcript abundance was determined. The 28°C sample served as the baseline. We observed that the OsFIE1 transcript level increased with increasing temperature, peaking at 34°C, but declined with additional incremental increases in temperature (Fig. 5B). Inflection of the OsFIE1 expression trajectory at 48 HAF occurred between 32°C and 36°C. Our data suggest that OsFIE1 induction during the course of seed development (Fig. 5B) correlates with the transition from the syncytial to the cellularization stage (Fig. 3). Under control conditions, cellularization is initiated between the 48- and 72-HAF time points, and OsFIE1 transcript accumulates rapidly during this time. However, under a moderate heat stress of 34°C, we observed a shortened duration of the syncytial stage and precocious cellularization, which coincided with higher induction of OsFIE1 expression. We did not detect OsFIE1 expression during severe heat stress. We also did not observe cellularization in severely stressed seeds (Figs. 3, E–G, and 5A). These data suggested that OsFIE1 activity is highly sensitive to thermal variation and can be a component of the thermal-sensing mechanism that regulates early seed development.
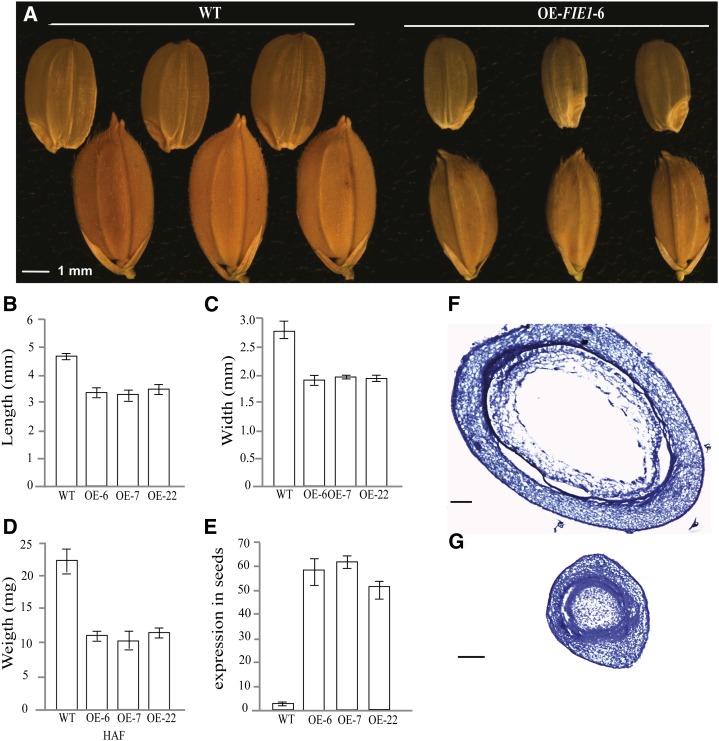
To directly test the effect of OsFIE1 expression on the duration of the endosperm syncytial stage, we generated 21 independent transgenic rice lines that overexpressed OsFIE1 using the maize ubiquitin promoter and characterized early seed development (Fig. 6). Data are presented for three independent lines. Higher expression of OsFIE1 was confirmed at 48 HAF in overexpression seeds from independent transgenic events and compared with wild-type cv Kitaake seeds (Fig. 6E). Plants overexpressing OsFIE1 produced smaller seeds at maturity compared with the wild type (Fig. 6A). Decreased seed size was reflected in reduced length, width, and seed weight (Fig. 6, B–D). We sectioned developing seeds from OsFIE1-overexpressing and wild-type plants at 48 HAF to compare endosperm development. The 48-HAF seeds from overexpressing plants were either completely cellularized or mostly cellularized, whereas cellularization was not observed in wild-type seeds (Fig. 6, F and G). These data suggested that higher expression of OsFIE1 in rice seeds led to precocious cellularization, resulting in smaller seeds. Because reduced seed size and precocious cellularization were also observed in moderately heat-stressed wild-type plants, the increased transcript abundance of OsFIE1 could be a contributing factor in the observed heat stress phenotypes.
Figure 6.
Effect of OsFIE1 overexpression on mature seed size and the duration of the syncytial stage of the endosperm. A, Comparison between mature seed of rice wild type (WT) and three independent transgenic plants overexpressing OsFIE1. B, Length. C, Width. D, Weight. Controls for seed size and weight measurements are seeds obtained from the wild-type cv Kitaake plants grown under the same conditions and times as transgenic lines (n > 30 for all lines). E, Relative gene expression of OsFIE1 in seeds of transgenic lines and the wild type. F, Section from control seeds (28°C) harvested at 48 HAF. G, Sections from OE-OsFIE1 seeds harvested at 48 HAF. Bar in F = 200 µm. Bar in G = 100 µm.
Heat Stress Alters the Methylation Status of OsFIE1
The OsFIE1 locus is marked by DNA methylation at the 5′ end of the coding region and histone methylation (Zhang et al., 2012). We wished to determine if the DNA methylation in this region was affected by moderate heat stress. We studied the methylation status of the OsFIE1 locus in developing seeds at the two time points (48 and 72 HAF) using plants grown in control and moderate heat stress conditions. Under moderate heat stress, OsFIE1 expression was higher at 48 HAF relative to control plants. We used McrBC endonuclease digestion to assay the methylation status of the OsFIE1 locus. McrBC digests between two methylated cytosines (either AmC or GmC). We used a primer set to cover a 5′ region in the coding sequence of OsFIE1 (+246 to +817), which was previously reported to be methylated, and another primer set as a control to amplify a nonmethylated OsFIE1 region (+2,931 to +3,679; Fig. 7A; Zhang et al., 2012). Our results from the McrBC assay suggested that the moderate heat stress treatment decreased DNA methylation level of OsFIE1 at 48 and 72 HAF relative to control seeds (Fig. 7B). Methylation was lowest in the 7-DAF positive control sample compared with 48- and 72-HAF control and moderately stressed seeds. Based on our experiments and publically available microarray data set, transcript abundance of OsFIE1 begins to increase around 48 HAF and continues to increase at least until 9 DAF. No band was detected for the leaf negative control sample, where OsFIE1 is methylated and not expressed.
Figure 7.
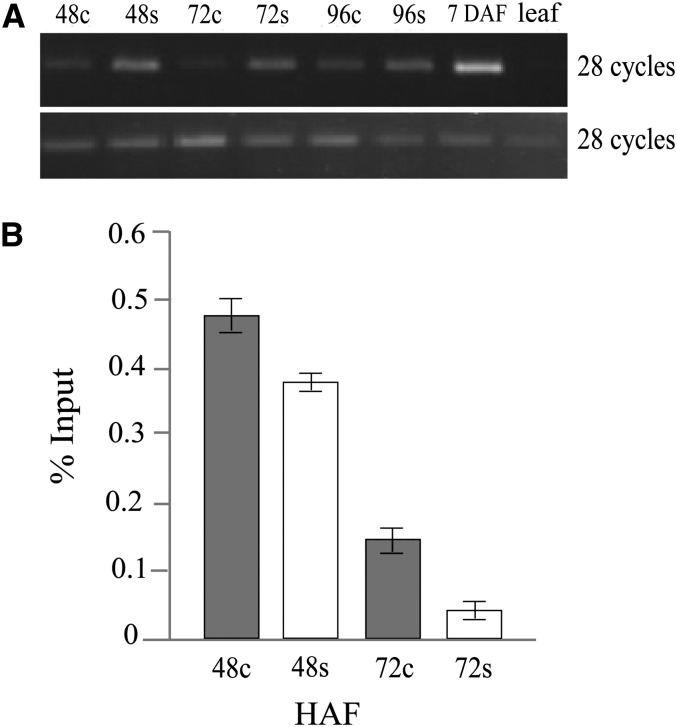
DNA and histone methylation analysis of OsFIE1. A, McrBC-based assay for DNA methylation level of OsFIE1 during early seed development under control and moderate heat-stressed (34°C) conditions at 48, 72, and 96 HAF; 7 DAF (OsFIE1 highly expressed) and leaf tissue (OsFIE1 not expressed) are positive and negative controls, respectively. McrBC digests methylated DNA. Top is from the primer pair targeting known methylated sites, and in the bottom, primers are controls from an unmethylated region of OsFIE1. B, ChIP analyses of OsFIE1 under control (28°C; gray bars) and moderate stress (34°C; white bars) for enrichment of silencing histone modification H3K9me2. The level of H3K9me2 is expressed as the percentage of input DNA used for the ChIP assays. Values are means ± sds of three biological replicates.
We next quantified the change in the DNA methylation status of OsFIE1 during moderate heat stress and in control conditions at 48 and 72 HAF using bisulfite sequencing. We observed that both CG and CHG (where H = A, T, or C) methylation levels were reduced during heat stress at 48 HAF by 8.8% and 6.6%, respectively. At 72 HAF, whereas the CG methylation level decreased by 22% during heat stress, the methylation level in the CHG context increased by 25%. The increase in CHG methylation at 72 HAF is inconsistent with the McrBC digestion assay. However, the qRT-PCR data indicated that the expression of OsFIE1 was down-regulated under moderate heat stress at 72 HAF compared with 48-HAF heat-stressed seeds (Fig. 5A).
DNA methylation is not the only epigenetic modification associated with OsFIE1 in rice. Reduction in dimethylation on H3K9 has also been associated with increased OsFIE1 expression (Zhang et al., 2012). Therefore, we compared H3K9 dimethylation levels of OsFIE1 in seeds developing under moderate heat stress with controls at 48 and 72 HAF using chromatin immunoprecipitation (ChIP)-quantitative PCR assays. These data showed in both control and heat conditions that the level of H3K9me2 associated with OsFIE1 was reduced in 72-HAF seeds compared with 48-HAF seeds. Notably, the reduction in H3K9me2 marks was greater in the heat-stressed samples at both time points. The difference was more dramatic between the 72-HAF seeds from control and moderately heat-stressed plants. At 72 HAF, the H3K9me2 level in heat-stressed samples was roughly one-fourth that observed in seeds collected from control plants (Fig. 7B). Collectively, our methylation experiments suggest that both DNA and H3K9 methylation of OsFIE1 exhibit temperature sensitivity. However, the correlation between transcript abundance and decreased methylation was observed for the 48-HAF time point only.
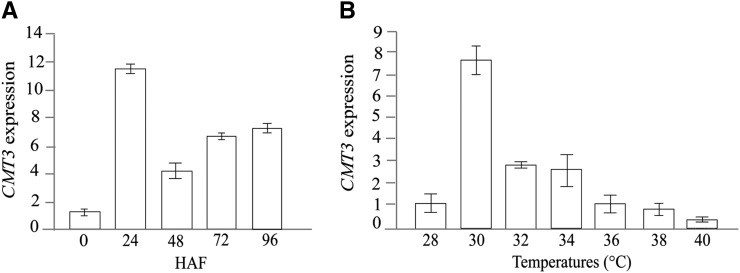
In view of the observed temperature responsiveness of DNA and histone methylation changes at the OsFIE1 locus, it is notable that the transcript abundance of CHROMOMETHYLASE3 (CMT3) was repressed by the moderate heat stress at 48 HAF based on microarray data. CMT3 is a plant-specific methyltransferase required for maintenance of CHG methylation in Arabidopsis and maize (Lindroth et al., 2001; He et al., 2011). We confirmed the differential expression of CMT3 with qRT-PCR assays and found that CMT3 expression decreased by 3.3-fold in moderately heat-stressed seeds at 48 HAF compared with control seeds. No significant difference was observed at 72 HAF between the control and the moderately stressed seeds in the microarray and qRT-PCR experiments. During severe heat stress, CMT3 expression was strongly down-regulated at both 48 and 72 HAF (Supplemental Data Set S1). Under control conditions, the expression of CMT3 was highest at 24 HAF but dropped significantly as the seed development progressed past 48 HAF, leading into cellularization (Fig. 8A). However, under moderate heat stress, the expression of CMT3 was down-regulated at 48 HAF compared with the control seeds (Fig. 8A). We tested the temperature sensitivity of CMT3 expression in the seeds collected from the earlier described temperature gradient experiment, and the transcript abundance of CMT3 increased about 7-fold in 30°C seeds at 48 HAF relative to control seeds (28°C) but dropped significantly with additional increases in temperature (Fig. 8B).
Figure 8.
Expression of rice CMT3 in developing seeds. A, CMT3 expression was measured during early seed development. The gene expression levels are relative to the 0-HAF sample; 24 and 48 HAF correspond to syncytial stages of endosperm development when the nuclei divide rapidly before cellularization. B, Expression of rice CMT3 at the 48-HAF time point in seeds developing under a range of temperatures with 2°C increments. The data are presented as means ± sds from three independent biological replicates.
Seed-Specific Type I MADS-Box Genes Exhibit Temperature Sensitivity
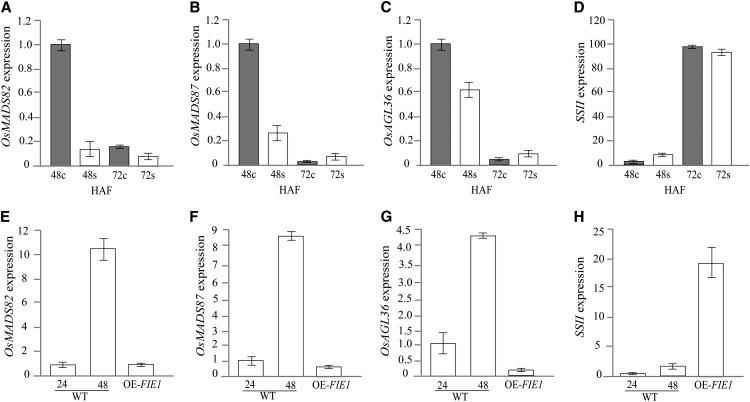
Our results suggest that an early increase in OsFIE1 expression during the course of seed development under moderate heat stress coincides with a shortened duration of the syncytial stage. In this context, we found the repression of three type I MADS-box genes from moderate heat-stressed seeds to be noteworthy. The expression of OsMADS82 and OsMADS87 is induced during the syncytial stage of endosperm development and repressed with initiation of cellularization (Ishikawa et al., 2011). Both of these genes are putative orthologs of the Arabidopsis endosperm-specific gene Pheres1, which is regulated by the PRC2 complex in Arabidopsis (Köhler and Villar, 2008; Ishikawa et al., 2011). Under moderate heat stress, the expression of OsMADS82 and OsMADS87 was repressed at 48 HAF compared with controls, consistent with the observed early cellularization in these seeds (Fig. 9, A and B). The rice ortholog of the Arabidopsis AGL36 (Os11g30220) gene was also repressed by moderate heat stress at 48 HAF (Fig. 9C). AGL36 expression in Arabidopsis is seed-specific and regulated by both PRC2 and DNA methylation (Shirzadi et al., 2011). In contrast to the MADS-box genes, the transcript abundance of a STARCH SYNTHASE II (SSII) gene was induced 6-fold under moderate heat stress at 48 HAF (Fig. 9D). It has been shown that SSII induction coincides with endosperm cellularization in rice (Ishikawa et al., 2011). These data indicate that three of the rice type I MADS-box genes with syncytial stage-specific expression are repressed during moderate heat stress, further supporting precocious cellularization in heat-stressed seeds.
Figure 9.
Gene expression of syncytial stage-specific MADS-box genes (OsMADS82, OsMADS87, and AGL36) and cellularization stage-specific SSII. A to D, Gene expression levels for syncytial and cellularization stage-specific genes in control and moderately heat-stressed seeds at 48 HAF. The stress was imposed at 24 HAF. E to H, Gene expression was measured in wild-type (WT) seeds and OsFIE1 overexpression seeds. Wild-type seeds were sampled at 24 and 48 HAF to compare with the 48-HAF OE-OsFIE1 seeds for measuring expression of MADs-box genes and ssII. The data are presented as means ± sds.
We measured transcript abundance of SSII, a cellularization stage-specific gene, and syncytial stage-associated genes AGL36, OsMADS82, and OsMADS87 in developing seeds at 48 HAF from OsFIE1 overexpression plants and compared them with the wild type. The expression of SSII in transgenic seeds overexpressing OsFIE1 was higher compared with wild-type seeds, consistent with precocious cellularization (Fig. 9H). However, the expression of type I MADS-box genes AGL36, OsMADS82, and OsMADS87 was strongly repressed in the overexpression of OsFIE1 seeds, suggesting a shortened duration of the syncytial stage in seeds overexpressing OsFIE1 (Fig. 9, F and G).
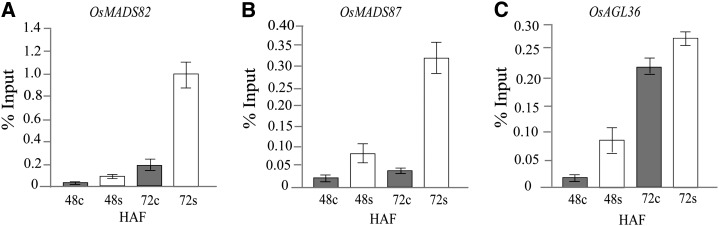
Because Arabidopsis orthologs of the type I MADS-box genes (AGL36 and Pheres1) are regulated by the PRC2 complex through H3K27me3, we tested the hypothesis that expression of rice AGL36, OsMADS82, and OsMADS87 was differentially regulated by H3K27me3 under heat stress compared with control conditions (Köhler et al., 2003; Shirzadi et al., 2011). Using ChIP-quantitative PCR assays with seeds collected at 48 and 72 HAF that were subjected to control and moderate heat stress, we observed that the H3K27me3 silencing mark is associated with OsMADS82, OsMADS87, and AGL36 (Fig. 10). All three genes had increased enrichment for H3K27me3 marks under heat stress relative to control seeds. OsMADS82 and OsMADS87 exhibited a more striking differential enrichment at 72 HAF relative to control seeds (Fig. 10, A and B). Level of enrichment for OsMADS82 and OsMADS87 is relatively low for seeds derived from 48 HAF under heat stress. H3K27me3 enrichment of AGL36 is higher under heat stress at 48 HAF compared with control seeds (Fig. 10C). Collectively, these data suggest that the observed early repression of syncytial stage-specific MADS-box genes under heat stress correlates with increased deposition of H3K27me3 silencing marks by the PRC2 complex compared with seeds developing under optimal conditions.
Figure 10.
Silencing histone modification (H3K27me3) levels of syncytial stage-specific type I MADS-box genes under moderate heat stress. The rice MADS-box genes assayed are OsMADS82 (A), OsMADS87 (B), and AGL36 (C). Level of H3K27me3 marks is presented as a percentage of input DNA.
DISCUSSION
Our study suggests that heat stress imposes limitations on endosperm enlargement and thus, yield, possibly because of altered epigenetic regulation during early seed development. The sensitivity of endosperm development to drought stress has been shown in maize through transcriptome studies (Yu and Setter, 2003; Kakumanu et al., 2012). A commonality observed by these studies was the down-regulation of genes involved in cell cycle and cell division, suggesting that endosperm proliferation and endoreduplication are impacted by drought stress (Sabelli and Larkins, 2009a; Kakumanu et al., 2012). However, the sensitivity of the syncytial stage endosperm to abiotic stresses and the underlying cellular and molecular mechanisms have remained unexplored in cereals. Our work shows that the duration of the syncytial stage of endosperm development is highly sensitive to heat stress, which leads to earlier cellularization and reduced seed enlargement. Furthermore, our data suggest that OsFIE1 could be a key regulator of the duration of the syncytial stage.
We propose a mechanism that involves the differential regulation of OsFIE1 and a set of seed-specific MADS-box genes in response to heat stress during the syncytial stage of endosperm development. We observed lower levels of DNA methylation associated with OsFIE1 under moderate heat stress at 48 and 72 HAF compared with control plants (Fig. 7A). A similar observation was made for H3K9me3 marks for both time points (Fig. 7B). However, the expression of OsFIE1 was only higher at 48 HAF but not at 72 HAF under moderate heat stress compared with control conditions (Fig. 5A). This lack of correlation between DNA and histone methylation levels and transcript abundance at 72 HAF suggests that OsFIE1 regulation during seed development could involve yet-to-be-discovered component(s) in addition to DNA and H3K9 methylation levels. In another report, it was noted that, in the revertants of the OsFIE1 epi-allele line, OsFIE1 expression was silenced, but the DNA methylation status was not restored to the levels observed in wild-type plants (Zhang et al., 2012). Collectively, our experiments suggest that both DNA methylation and H3K9 methylation levels associated with OsFIE1 are sensitive to moderate heat stress and may be important (if not the only) components involved in regulating OsFIE1 when developing seeds are exposed to a moderate heat stress.
We observed that CMT3 was down-regulated in response to moderate heat stress at 48 HAF but not at 72 HAF. The expression of CMT3 under control condition is highest at 24 HAF, when the endosperm is in the syncytial stage with rapidly dividing nuclei that presumably require active maintenance of DNA methylation. It is conceivable that the decrease in CMT3 activity at 48 HAF coincides with precocious initiation of cellularization of the nuclei under moderate heat stress, which results in a phase of relatively slower nuclear divisions around 48 HAF. Alternatively, it could be that moderate heat stress reduces the number of nuclei at syncytial stage, hence requiring lower CMT3 levels. The possibility of CMT3-mediated DNA methylation being an upstream regulator of OsFIE1 needs to be explored in future experiments.
Expression of two genes, AGL62 and THOUSAND-GRAIN Weight6, has been shown to accelerate the transition from the syncytial stage to cellularization in Arabidopsis and rice, respectively (Kang et al., 2008; Ishimaru et al., 2013). In both instances, the observed precocious cellularization resulted in smaller seeds. We provide evidence that OsFIE1, a putative PRC2 member, could play a role in initiation of cellularization and seed size regulation. We provide evidence that the rice AGL36 and the orthologs of Pheres1, OsMADS82, and OsMADS87 are epigenetically repressed by the PRC2. Preliminary data from ongoing experiments suggest that overexpression of OsMADS87 directly regulates seed size. Future experiments will focus on elucidating the function of these MADS-box genes. In the context of improving tolerance to heat stress, it will be useful to mine rice germplasm for allelic variants of these MADS-box genes.
We provide a molecular framework to further examine the impact of epigenetic misregulation on decreasing sink capacity and endosperm size under heat stress. The window of early seed development is highly sensitive to temperature fluctuations, and it can be a potential developmental stage that could be targeted for mitigating yield losses caused by transient spikes in temperature that are common in rice fields (Peng et al., 2004; Welch et al., 2010). From an ecological perspective, OsFIE1 and other uncharacterized regulators may be part of the thermal sensory mechanism that triggers an escape response in plants, accelerating development when conditions become unfavorable. Epigenetic regulation of endosperm development might have a key role in balancing seed size versus number of viable seeds produced by the plant.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Seeds of rice (Oryza sativa ‘Kitaake’) were germinated in water under dark conditions. Seedlings were transplanted to soil and grown under controlled conditions (27°C–30°C) in the greenhouse until approaching anthesis. Plants were moved from the greenhouse to reach-in growth chambers (I-36VL; Percival Incubators) 4 to 5 d before panicle emergence with a 16-h-light/8-h-dark cycle (300–400 µmol photons m−2 s−1) at 28°C for control condition and 34°C and 42°C for moderate and severe heat stress, respectively, and a relative humidity of 75% to 80%. For the temperature gradient (28°C–40°C) experiment, the heat stress treatment was applied using multiple growth chambers in two batches. The temperature of the individual growth chambers was also monitored independently using a thermometer placed in each chamber. Unfertilized ovules were marked early in the morning and then closely observed for the fertilization event. Closed spikelets with anthers outside were then marked as fertilized by midmorning. Heat stress was imposed 24 HAF. Only a few spikelets were selected from each plant. After the heat stress treatment, some of the plants were transported back to the greenhouse and grown under control conditions until maturity for final seed size measurements. For all seed size measurements, the control and heat-stressed seeds were obtained from at least seven plants for each replicate and three or four independent biological replicates. The same procedure was used for collecting seed material for expression analysis, chromatin assays, and sectioning experiments. All these assays were performed on the whole seeds, which includes the embryo, endosperm, and seed coat.
OsFIE1 Cloning and Transformation
Rice FIE1 (Os08g04290) full length was PCR amplified using specific primers. OsFIE1 PCR product was cloned into pENTR/D-TOPO (Invitrogen) entry vector and then transferred to the final Ubi-FIE1 destination vector using LR clonase recombination (Invitrogen). The final construct was transformed to Agrobacterium tumafaciens strain EHA105. Rice transformation was performed as described by Cheng et al. (1997). Hygromycin was used to screen transformed resistant callus. Transgenic plants were regenerated from resistant callus. We characterized three independent transformation lines for OsFIE1 overexpression. In total, 21 events were generated over a course of several cycles of transformations. The seed phenotype was consistent among the independent transformation events. The overexpression was confirmed using qRT-PCR in leaves and early stage seeds.
Sectioning
Rice plants were cultured as described earlier, and plants were transferred to growth chambers for heat stress or control treatments. Three independent experiments were performed for collecting seeds for sectioning. The images of seed sections in Figure 3 are representative of our observation across the replicates. Rice developing grains were harvested at different times points (24, 48, 72, and 96 HAF) and fixed in 3.7% (v/v) formaldehyde, 5% (v/v) acetic acid, and 50% (v/v) ethanol at 4°C overnight. Samples were dehydrated through a graded ethanol series and infiltrated with xylene. Seed samples were embedded in paraffin (Fisher Scientific), sectioned at 8 µm, and stained with 0.1% toluidine blue. Sections were observed and photographed using a bright-field microscope (Leica DM-2500).
Real-Time PCR
Total RNA was extracted from leaves and developing seeds at different times points (24, 48, 72, and 96 HAF) using Trizol and treated with DNase I (Qiagen) according to the manufacturer's instruction. One microgram total RNA was used for complementary DNA synthesis using the SuperScript VILO cDNA Synthesis Kit (Invitrogen). Real-time PCR analysis was conducted using a Lightcycler 480 Real-Time PCR System (Roche) with the SYBR Green Master Mix (Bio-Rad). The rice proteasome gene (Os03g63430) was used as the endogenous control. The relative expressions of specific genes were quantitated using the 2−ΔΔCt method (Livak and Schmittgen, 2001). For all real-time assays, we used three to five independent biological replicates and at least two technical replicates for each biological replicate. Primer sequence information is provided in Supplemental Table S1.
McrBC-PCR
Genomic DNA from rice seeds under control and heat treatment was isolated using the cetyltrimethylammonium bromide method. For McrBC-PCR analysis of the OsFIE1 gene, 0.5 μg of Genomic DNA was digested with 20 units of McrBC restriction endonuclease (New England Biolabs) in a 50-µL reaction mix overnight. For PCR amplification, 100 ng of McrBC-digested DNAs was used, and the products were separated on 1% agarose gels.
Bisulfite Sequencing Analysis
Genomic DNA from rice developing seeds was obtained at 48 and 72 HAF from plants under control and moderate heat stress using the cetyltrimethylammonium bromide method. Five hundred grams of genomic DNA from each sample was treated with sodium bisulfite using the EZ DNA Methylation Kit (Zymo Research) according to the manufacturer’s protocol. The treated DNA was resuspended in 15 µL of distilled water, and 4 µL of this solution was then used as the template in a 25-µL PCR reaction. Primer sequence is provided in Supplemental Table S1. PCR products were purified and cloned into pGEM-T Easy Vector (Promega) and sequenced. Sequencing data were analyzed with Kismeth software (Gruntman et al., 2008). For each sample, at least 15 clones were sequenced for this analysis.
ChIP
Chromatin was extracted from endosperm following the protocol described previously (Zhang et al., 2012) with minor modifications. Antibodies used for ChIP were anti-H3K9me2 (07-441; Millipore) and anti-H3K27me3 (07-449; Millipore). DNA was immunoprecipitated and then used for real-time PCR.
Supplemental Data
The following materials are available in the online version of the article.
Supplemental Figure S1. Comparison between mature control seeds (28°C) and mature severe heat-stressed seeds at 42°C (mature stressed seeds were unviable).
Supplemental Figure S2. Comparison between control seeds (28°C) and moderate heat stress of 34°C for 24 and 48 h imposed 24 h after fertilization.
Supplemental Table S1. Sequences for primers used in the experiments.
Supplemental Data Set S1. Gene expression values of rice genes in developing seeds under control and heat-stressed conditions.
Supplementary Material
Glossary
- ChIP
chromatin immunoprecipitation
- DAF
days after fertilization
- HAF
hours after fertilization
- qRT
quantitative reverse transcription
Footnotes
This work was supported by the National Science Foundation (award no. 1121648 to D.W. and H.W.).
The online version of this article contains Web-only data.
References
- Baroux C, Gagliardini V, Page DR, Grossniklaus U. (2006) Dynamic regulatory interactions of Polycomb group genes: MEDEA autoregulation is required for imprinted gene expression in Arabidopsis. Genes Dev 20: 1081–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RC, Lemmon BE, Olsen OA. (1996) Development of the endosperm in rice (Oryza sativa L): cellularization. J Plant Res 109: 301–313 [Google Scholar]
- Cheng X, Sardana RK, Altosar I. (1997) Rice transformation by Agrobacterium infection: recombinant proteins from plants: production and isolation of clinically useful compounds. Methods Biotechnol 3: 1–9 [Google Scholar]
- Danilevskaya ON, Hermon P, Hantke S, Muszynski MG, Kollipara K, Ananiev EV. (2003) Duplicated fie genes in maize: expression pattern and imprinting suggest distinct functions. Plant Cell 15: 425–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guitton AE, Berger F. (2005) Loss of function of MULTICOPY SUPPRESSOR OF IRA 1 produces nonviable parthenogenetic embryos in Arabidopsis. Curr Biol 15: 750–754 [DOI] [PubMed] [Google Scholar]
- Gruntman E, Qi Y, Slotkin RK, Roeder T, Martienssen RA, Sachidanandam R. (2008) Kismeth: analyzer of plant methylation states through bisulfite sequencing. BMC Bioinformatics 9: 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XJ, Chen T, Zhu JK. (2011) Regulation and function of DNA methylation in plants and animals. Cell Res 21: 442–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehenberger E, Kradolfer D, Köhler C. (2012) Endosperm cellularization defines an important developmental transition for embryo development. Development 139: 2031–2039 [DOI] [PubMed] [Google Scholar]
- Huh JH, Bauer MJ, Hsieh TF, Fischer R. (2007) Endosperm gene imprinting and seed development. Curr Opin Genet Dev 17: 480–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter RB, Tollenaar M, Breuer CM. (1977) Effects of photoperiod and temperature on vegetative and reproductive growth of a maize (Zea-Mays) hybrid. Can J Plant Sci 57: 1127–1133 [Google Scholar]
- Ishikawa R, Ohnishi T, Kinoshita Y, Eiguchi M, Kurata N, Kinoshita T. (2011) Rice interspecies hybrids show precocious or delayed developmental transitions in the endosperm without change to the rate of syncytial nuclear division. Plant J 65: 798–806 [DOI] [PubMed] [Google Scholar]
- Ishimaru K, Hirotsu N, Madoka Y, Murakami N, Hara N, Onodera H, Kashiwagi T, Ujiie K, Shimizu B, Onishi A, et al. (2013) Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat Genet 45: 707–711 [DOI] [PubMed] [Google Scholar]
- Kakumanu A, Ambavaram MM, Klumas C, Krishnan A, Batlang U, Myers E, Grene R, Pereira A. (2012) Effects of drought on gene expression in maize reproductive and leaf meristem tissue revealed by RNA-Seq. Plant Physiol 160: 846–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang IH, Steffen JG, Portereiko MF, Lloyd A, Drews GN. (2008) The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell 20: 635–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosue T, Ohad N, Yadegari R, Hannon M, Dinneny J, Wells D, Katz A, Margossian L, Harada JJ, Goldberg RB, et al. (1999) Control of fertilization-independent endosperm development by the MEDEA polycomb gene in Arabidopsis. Proc Natl Acad Sci USA 96: 4186–4191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C, Villar CB. (2008) Programming of gene expression by Polycomb group proteins. Trends Cell Biol 18: 236–243 [DOI] [PubMed] [Google Scholar]
- Köhler C, Hennig L, Spillane C, Pien S, Gruissem W, Grossniklaus U. (2003) The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev 17: 1540–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth AM, Cao XF, Jackson JP, Zilberman D, McCallum CM, Henikoff S, Jacobsen SE. (2001) Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292: 2077–2080 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lobell DB, Schlenker W, Costa-Roberts J. (2011) Climate trends and global crop production since 1980. Science 333: 616–620 [DOI] [PubMed] [Google Scholar]
- Luo M, Platten D, Chaudhury A, Peacock WJ, Dennis ES. (2009) Expression, imprinting, and evolution of rice homologs of the polycomb group genes. Mol Plant 2: 711–723 [DOI] [PubMed] [Google Scholar]
- Luo M, Taylor JM, Spriggs A, Zhang HY, Wu XJ, Russell S, Singh M, Koltunow A. (2011) A genome-wide survey of imprinted genes in rice seeds reveals imprinting primarily occurs in the endosperm. PLoS Genet 7: e1002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani M, Naganuma T, Tsutsumi K, Saitoh Y. (2010) The syncytium-specific expression of the Orysa;KRP3 CDK inhibitor: implication of its involvement in the cell cycle control in the rice (Oryza sativa L.) syncytial endosperm. J Exp Bot 61: 791–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagato K, Ebata M. (1960) Effects of temperature in the ripening periods upon the development and qualities of lowland rice kernels [in Japanese, English summary]. Proc Crop Sci Soc Japan 28: 275–278 [Google Scholar]
- Nallamilli BR, Zhang J, Mujahid H, Malone BM, Bridges SM, Peng Z. (2013) Polycomb group gene OsFIE2 regulates rice (Oryza sativa) seed development and grain filling via a mechanism distinct from Arabidopsis. PLoS Genet 9: e1003322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen OA. (2001) Endosperm development: cellularization and cell fate specification. Annu Rev Plant Physiol Plant Mol Biol 52: 233–267 [DOI] [PubMed] [Google Scholar]
- Peng S, Huang J, Sheehy JE, Laza RC, Visperas RM, Zhong X, Centeno GS, Khush GS, Cassman KG. (2004) Rice yields decline with higher night temperature from global warming. Proc Natl Acad Sci USA 101: 9971–9975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds M, Bonnett D, Chapman SC, Furbank RT, Manès Y, Mather DE, Parry MAJ. (2011) Raising yield potential of wheat. I. Overview of a consortium approach and breeding strategies. J Exp Bot 62: 439–452 [DOI] [PubMed] [Google Scholar]
- Rodrigues JA, Ruan R, Nishimura T, Sharma MK, Sharma R, Ronald PC, Fischer RL, Zilberman D. (2013) Imprinted expression of genes and small RNA is associated with localized hypomethylation of the maternal genome in rice endosperm. Proc Natl Acad Sci USA 110: 7934–7939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabelli PA, Larkins BA. (2009a) The contribution of cell cycle regulation to endosperm development. Sex Plant Reprod 22: 207–219 [DOI] [PubMed] [Google Scholar]
- Sabelli PA, Larkins BA. (2009b) The development of endosperm in grasses. Plant Physiol 149: 14–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savin R, Stone PJ, Nicolas ME. (1996) Responses of grain growth and malting quality of barley to short periods of high temperature in field studies using portable chambers. Aust J Agric Res 47: 465–477 [Google Scholar]
- Shirzadi R, Andersen ED, Bjerkan KN, Gloeckle BM, Heese M, Ungru A, Winge P, Koncz C, Aalen RB, Schnittger A, et al. (2011) Genome-wide transcript profiling of endosperm without paternal contribution identifies parent-of-origin-dependent regulation of AGAMOUS-LIKE36. PLoS Genet 7: e1001303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen MB, Mayer U, Lukowitz W, Robert H, Chambrier P, Jürgens G, Somerville C, Lepiniec L, Berger F. (2002) Cellularisation in the endosperm of Arabidopsis thaliana is coupled to mitosis and shares multiple components with cytokinesis. Development 129: 5567–5576 [DOI] [PubMed] [Google Scholar]
- Spillane C, Schmid KJ, Laoueillé-Duprat S, Pien S, Escobar-Restrepo JM, Baroux C, Gagliardini V, Page DR, Wolfe KH, Grossniklaus U. (2007) Positive darwinian selection at the imprinted MEDEA locus in plants. Nature 448: 349–352 [DOI] [PubMed] [Google Scholar]
- Springer NM, Danilevskaya ON, Hermon P, Helentjaris TG, Phillips RL, Kaeppler HF, Kaeppler SM. (2002) Sequence relationships, conserved domains, and expression patterns for maize homologs of the polycomb group genes E(z), esc, and E(Pc). Plant Physiol 128: 1332–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walia H, Josefsson C, Dilkes B, Kirkbride R, Harada J, Comai L. (2009) Dosage-dependent deregulation of an AGAMOUS-LIKE gene cluster contributes to interspecific incompatibility. Curr Biol 19: 1128–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardlaw IF. (1989) Grain-growth and translocation in water-stressed wheat. Agric Biol Environ 4: 14 [Google Scholar]
- Welch JR, Vincent JR, Auffhammer M, Moya PF, Dobermann A, Dawe D. (2010) Rice yields in tropical/subtropical Asia exhibit large but opposing sensitivities to minimum and maximum temperatures. Proc Natl Acad Sci USA 107: 14562–14567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa H, Hirose T, Kuroda M, Yamaguchi T. (2007) Comprehensive expression profiling of rice grain filling-related genes under high temperature using DNA microarray. Plant Physiol 144: 258–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu LX, Setter TL. (2003) Comparative transcriptional profiling of placenta and endosperm in developing maize kernels in response to water deficit. Plant Physiol 131: 568–582; erratum Yu LX, Setter TL. (2003) Plant Physiol 131: 1921–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D. (2010) Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328: 916–919 [DOI] [PubMed] [Google Scholar]
- Zhang L, Cheng Z, Qin R, Qiu Y, Wang JL, Cui X, Gu L, Zhang X, Guo X, Wang D, et al. (2012) Identification and characterization of an epi-allele of FIE1 reveals a regulatory linkage between two epigenetic marks in rice. Plant Cell 24: 4407–4421 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.