An Arabidopsis receptor-like kinase negatively regulates defense gene expression and resistance against microbial pathogens and is required for late responses to abscisic acid.
Abstract
Transmembrane receptor-like kinases characterized by the presence of one or more lysin motif (LysM) domains in the extracytoplasmic portion (LysM-containing receptor-like kinases [LYKs]) mediate recognition of symbiotic and pathogenic microorganisms in plants. The Arabidopsis (Arabidopsis thaliana) genome encodes five putative LYKs; among them, AtLYK1/CHITIN ELICITOR RECEPTOR KINASE1 is required for response to chitin and peptidoglycan, and AtLYK4 contributes to chitin perception. More recently, AtLYK3 has been shown to be required for full repression, mediated by Nod factors, of Arabidopsis innate immune responses. In this work, we show that AtLYK3 also negatively regulates basal expression of defense genes and resistance to Botrytis cinerea and Pectobacterium carotovorum infection. Enhanced resistance of atlyk3 mutants requires PHYTOALEXIN-DEFICIENT3, which is crucial for camalexin biosynthesis. The expression of AtLYK3 is strongly repressed by elicitors and fungal infection and is induced by the hormone abscisic acid (ABA), which has a negative impact on resistance against B. cinerea and P. carotovorum. Plants lacking a functional AtLYK3 also show reduced physiological responses to ABA and are partially resistant to ABA-induced inhibition of PHYTOALEXIN-DEFICIENT3 expression. These results indicate that AtLYK3 is important for the cross talk between signaling pathways activated by ABA and pathogens.
Plants must recognize potential pathogens and mount effective defense responses in a timely manner to restrict microbial growth. Molecules conserved among microbes are recognized by plant cells and activate an immune response that contributes to resistance against infections. These molecules are often referred to as microbe-associated molecular patterns (MAMPs), analogous to those that trigger an immune response in animals (Mackey and McFall, 2006). MAMPs are often structural components of the pathogen cell wall or other conserved macromolecules (Boller and Felix, 2009). A prototypical MAMP is flg22, a 22-amino acid peptide derived from the N-amino terminal region of bacterial flagellin that is able to induce defense responses in different plants (Gómez-Gómez et al., 1999; Gómez-Gómez and Boller, 2000). Other well-characterized bacterial MAMPs are peptidoglycans (PGNs; Gust et al., 2007; Erbs et al., 2008), lipopolysaccharides (Newman et al., 2007), and the bacterial elongation factor thermo-unstable (Ef-Tu) and its derived 18-amino acid peptide elf18 (Zipfel et al., 2006), whereas fungal MAMPs include the cell wall polymer chitin (Zhang et al., 2002; Kaku et al., 2006; Wan et al., 2008) and its deacetylated derivative chitosan (Doares et al., 1995; Povero et al., 2011). Molecules released from the plant cell upon damage or pathogen infection can also be perceived and activate defense responses and are indicated as damage-associated molecular patterns (DAMPs; Bianchi, 2007; Boller and Felix, 2009). Well-characterized DAMPs are the oligogalacturonides (OGs), long-chain pectin fragments released from the plant cell wall by microbial polygalacturonases (De Lorenzo and Ferrari, 2002; Federici et al., 2006; Galletti et al., 2009; Ferrari et al., 2013). Defense responses induced by MAMPs and DAMPs are qualitatively similar to those activated during the gene-for-gene resistance (Tao et al., 2003), and plants unable to perceive specific MAMPs often display enhanced susceptibility to virulent microbial strains (Zipfel et al., 2004, 2006; Miya et al., 2007; Wan et al., 2008), indicating that this line of defense contributes to a great extent to basal resistance to pathogens.
Activation of defense responses after MAMP/DAMP perception is at least partly independent of salicylic acid (SA), ethylene, and jasmonic acid (JA), three hormones important for resistance to infections (Glazebrook, 2001). For instance, Arabidopsis (Arabidopsis thaliana) plants impaired in responsiveness to these three signaling molecules are still protected against the fungal pathogen Botrytis cinerea by treatments with OGs or flg22 (Ferrari et al., 2007). Notably, expression of the PHYTOALEXIN-DEFICIENT3 (PAD3) gene is also induced by OGs and flg22 independently of SA, JA, and ethylene signaling (Ferrari et al., 2007). PAD3 is a cytochrome P450 that catalyzes the last biosynthetic step of camalexin (Zhou et al., 1999), a phytoalexin required for both basal and MAMP/DAMP-triggered resistance to B. cinerea (Ferrari et al., 2003, 2007). These data suggest that, in addition to those involving SA, ethylene, and JA, additional signaling pathways regulate plant immunity.
In the past years, significant advances in the understanding of MAMP and DAMP perception and transduction have been made (Boller and Felix, 2009). Recognition of these molecules by plants is often mediated by receptor-like kinases (RLKs), integral plasma membrane proteins containing an extracytoplasmic receptor domain (ectodomain), a single transmembrane domain, and a cytoplasmic protein kinase domain. The first identified plant MAMP receptor is FLAGELLIN-SENSITIVE2 (FLS2), which is responsible for flg22 perception in Arabidopsis (Gómez-Gómez and Boller, 2000). Another well-characterized Arabidopsis MAMP receptor is EF-TU RECEPTOR (EFR), which recognizes elf18 (Zipfel et al., 2006). The ectodomains of FLS2 and EFR contain Leu-rich repeats, which are common structural motifs amenable to protein-protein interaction (Padmanabhan et al., 2009). RLKs that have one or more lysin motifs (LysMs) in their ectodomain (LysM-containing RLKs [LYKs]) also mediate the perception of MAMPs and other microbial signals (Gust et al., 2012; Tanaka et al., 2012). LysM is 42 to 48 amino acids long and was originally identified in bacterial proteins, where it recognizes PGN, a major structural component of the bacterial cell walls composed of alternating β-1,4-linked N-acetylglucosamine (NAG) and N-acetylmuramic acid residues (Bateman and Bycroft, 2000). Structurally, LysM contains two α-helices on one side of a two-stranded antiparallel β-sheet (Bateman and Bycroft, 2000). This motif has been found in most living organisms, except Archaea, though only in plants it is associated to a kinase domain to form LYKs (Zhang et al., 2007). LYKs were first identified in legumes as receptors of the Nod factors, lipochitooligosaccharides secreted by nitrogen-fixing rhizobacteria that are major determinant of host specificity (Limpens et al., 2003; Madsen et al., 2003; Radutoiu et al., 2007). More recently, perception of chitin has also been shown to depend on LYKs, namely LYSM-CONTAINING RECEPTOR-LIKE KINASE1 (AtLYK1)/CHITIN ELICITOR RECEPTOR KINASE1 (AtCERK1) in Arabidopsis (Miya et al., 2007; Wan et al., 2008) and OsCERK1 in rice (Oryza sativa; Shimizu et al., 2010). The structure of chitin, a linear polymer of β-1,4-linked NAG units, is identical to that of the carbohydrate backbone of Nod factors (Limpens et al., 2003). Perception of chitin in rice cells also requires O. sativa chitin elicitor-binding protein (OsCEBiP), a LysM transmembrane receptor-like protein (LYP) lacking a kinase domain (Kaku et al., 2006). Both OsCEBiP and AtLYK1/AtCERK1 can directly bind chitin (Iizasa et al., 2010; Petutschnig et al., 2010). Chitin binding induces the dimerization of AtLYK1/AtCERK1, and this step is essential for downstream signaling (Liu et al., 2012). Another Arabidopsis LYK, AtLYK4, can be pulled down by chitin magnetic beads and eluted by chitooctaose, and null mutants for this protein are partially insensitive to chitin (Wan et al., 2012). AtLYK1/AtCERK1 also mediates perception of PGN, which also requires LYSM DOMAIN GPI-ANCHORED PROTEIN1 (LYM1) and LYM3, two LYPs that physically interact with PGN (Willmann et al., 2011). These data suggest that Arabidopsis LYKs and LYPs may interact to form receptor complexes involved in the perception and transduction of different NAG-containing MAMPs.
AtLYK1/AtCERK1 and AtLYK4 belong to a family of five Arabidopsis LYK genes, and the function of the other three members (AtLYK2, AtLYK3, and AtLYK5) is currently unclear. Single mutants for these genes, as well as triple atlyk2/atlyk3/atlyk5 mutants, show wild-type expression of marker genes in response to chitin treatment (Wan et al., 2008, 2012), suggesting that they are not involved in the perception of this MAMP. A recent paper shows that AtLYK3 is required for the repression of MAMP-triggered immunity by Nod factors (Liang et al., 2013). Here, we show that AtLYK3 negatively regulates basal resistance to pathogen infection. Furthermore, lack of a functional AtLYK3 increases basal levels of defense gene transcripts and reduces responses to abscisic acid (ABA). These results point to a role of AtLYK3 in the negative cross talk between this hormone and the plant immune response.
RESULTS
Mutants Lacking a Functional AtLYK3 Show Increased Expression of Defense Genes and Enhanced Pathogen Resistance
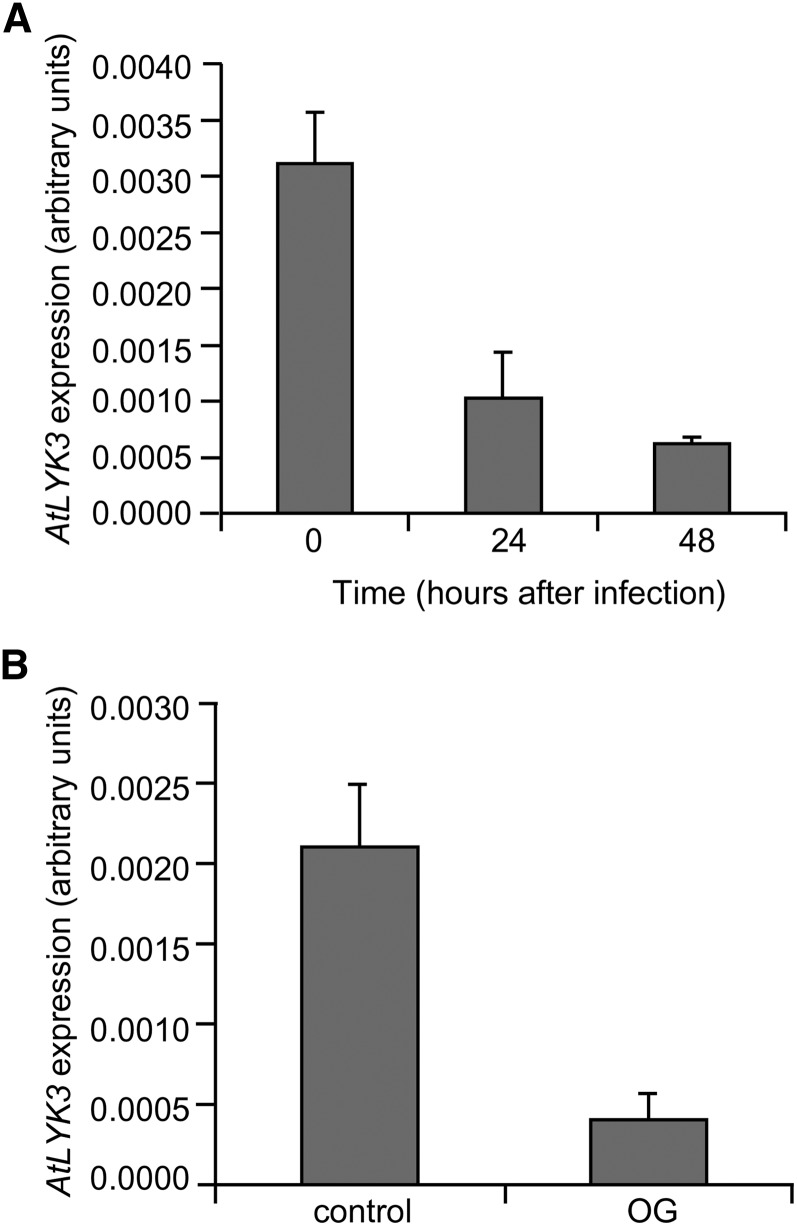
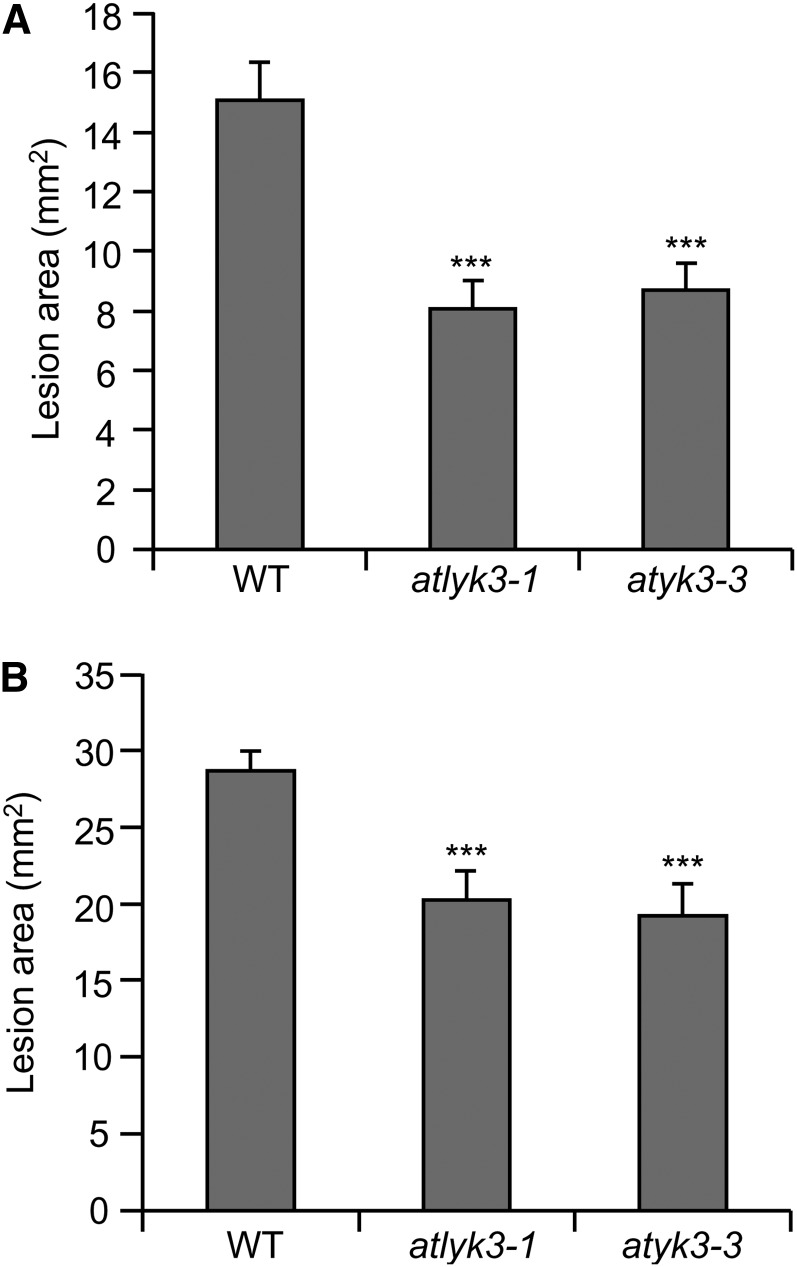
Previous transcript profile analyses indicate that, in contrast to the other members of the Arabidopsis LYK gene family, AtLYK3 (At1g51940) is transcriptionally repressed in response to B. cinerea infection and to treatments with OGs or flg22 (Ferrari et al., 2007; Denoux et al., 2008). A metaanalysis of public microarray data using Genevestigator (Zimmermann et al., 2004) showed that expression of this gene is also repressed by pathogens as diverse as Pseudomonas syringae and Phytophthora infestans and by different elicitors, including chitin and bacterial lipopolysaccharides (Supplemental Fig. S1A). The only exception to this pattern was Alternaria brassicicola infection, which represses all Arabidopsis LYK genes but induces AtLYK3 expression (Supplemental Fig. S1A). Quantitative PCR (qPCR) experiments indicated that AtLYK3 expression was significantly reduced 24 h after inoculation with B. cinerea, when disease symptoms were not yet visible, and was almost completely abolished at 48 h post infection (Fig. 1A). Moreover, we confirmed that expression of AtLYK3 is also significantly repressed in seedlings treated for 3 h with OGs (Fig. 1B) or chitin (Supplemental Fig. S1B). By contrast, AtLYK1/AtCERK1 expression increased after OG treatment, in accordance with the available microarray data (Supplemental Fig. S1C). These data suggest that AtLYK3 has an important role in plant immunity. To investigate this hypothesis, we obtained two homozygous insertional lines for AtLYK3, WiscDsLox318C07 and SALK_140374, which were renamed atlyk3-1 and atlyk3-3, respectively (Supplemental Fig. S2, A–C). AtLYK3 contains 11 predicted exons, and atlyk3-1 and atlyk3-3 were annotated in the SIGNAL database (Alonso et al., 2003) as carrying an insertion in exon 3 and exon 10, respectively. Reverse transcription (RT)-PCR analysis confirmed the transfer DNA (T-DNA) localization in atlyk3-1 and revealed that the insertion in atlyk3-3 was instead in the last exon (Supplemental Fig. S2, D and E). Inoculation of both atlyk3-1 and atlyk3-3 plants with B. cinerea resulted in smaller lesions compared with wild-type plants (Fig. 2A), suggesting that AtLYK3 plays a negative role in defense against fungal infection. Similarly, disease symptoms caused by the necrotrophic bacterial pathogen Pectobacterium carotovorum were significantly reduced in both mutants compared with the parental line (Fig. 2B). These data indicate that AtLYK3 is required for full susceptibility to different necrotrophic pathogens.
Figure 1.
Expression of AtLYK3 is repressed in response to fungal infection and elicitor treatment. Expression of AtLYK3 was analyzed by qPCR in Arabidopsis leaves inoculated for the indicated times with B. cinerea (A) or in liquid-grown seedlings treated for 3 h with water (control) or 100 μg mL–1 OG (B). The UBQ5 gene was used as reference. Bars indicate average expression ± sd of three independent biological replicates.
Figure 2.
Increased resistance to pathogen infection in atlyk3 mutants. Wild-type (WT), atlyk3-1, and atlyk3-3 rosette leaves were inoculated with B. cinerea (A) or with P. carotovorum for 24 h (B), and lesion areas were measured after 24 and 48 h, respectively. Values are means ± se (n ≥ 30). Asterisks indicate statistically significant differences between wild-type and mutant plants, according to Student’s t test (***P ≤ 1 × 10–4).
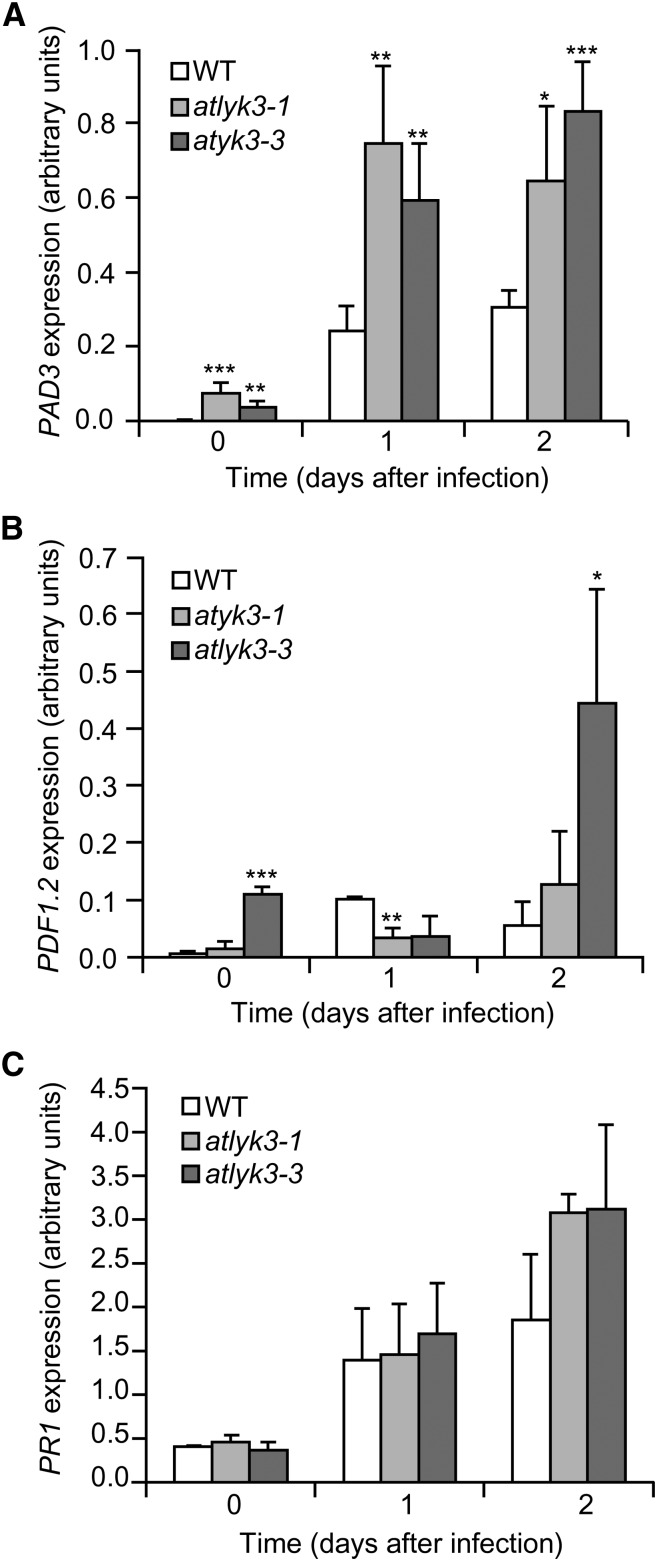
To investigate the cause of the enhanced resistance of plants lacking a functional AtLYK3, we evaluated the expression of the defense-related genes PAD3, PLANT DEFENSIN1.2 (PDF1.2), and PATHOGENESIS-RELATED1 (PR1), which are regulated by different signaling pathways. PAD3 is up-regulated by fungal infection and elicitors independently of SA-, JA-, and ethylene-mediated pathways (Ferrari et al., 2007); PDF1.2 encodes a defensin, and its expression is dependent on JA and ethylene (Penninckx et al., 1996); and PR1 is a marker gene for responses mediated by SA (Maleck et al., 2000). Basal expression of PAD3 was higher in atlyk3-1 and atlyk3-3 than in wild-type leaves (Fig. 3A). After infection with B. cinerea, expression of PAD3 was induced in atlyk3-1 and atlyk3-3 leaves to a much greater extent than in wild-type leaves at both 1 and 2 d post infection (dpi; Fig. 3A), suggesting that lack of AtLYK3 increases both basal and early inducible expression of this gene. Expression of PDF1.2 was similar to the wild type in atlyk3-1 plants, whereas, in atlyk3-3 plants, it was significantly higher at 0 and 2 dpi, but it was induced to a lesser extent at 1 dpi (Fig. 3B), suggesting that this gene may also be affected by AtLYK3, but it probably plays a minor role in the mutant-resistant phenotype. PR1 expression in healthy leaves or after fungal infection did not significantly differ in wild-type and atlyk3 plants at all time points analyzed (Fig. 3C), though a slight increase compared with the wild type, was observed in both mutants at 2 dpi; this suggests that SA-mediated responses are affected to a limited extent by the absence of a functional AtLYK3. Taken together, these data suggest that mutations in AtLYK3 derepress the expression of some defense-related genes, in particular PAD3. Diaminobenzidine staining of uninfected rosette leaves did not reveal a constitutively high basal accumulation of reactive oxygen species (ROS) in atlyk3-1 or atlyk3-3 plants (Supplemental Fig. S3); moreover, after fungal infection, staining was comparable in wild-type and mutant plants, suggesting that ROS production is little affected by AtLYK3 and therefore does not play a major role in the resistant phenotype of the mutants.
Figure 3.
Expression of defense genes in atlyk3 mutants inoculated with B. cinerea. Wild-type (WT; empty bars), atlyk3-1 (light-gray bars), and atlyk3-3 (dark-gray bars) rosette leaves were inoculated with B. cinerea, and total RNA was extracted at the indicated times. Expression of PAD3 (A), PDF1.2 (B), and PR1 (C) was analyzed by qPCR using UBQ5 as reference. Bars indicate average expression ± sd of three independent biological replicates. Asterisks indicate significant differences between the wild type and mutants, according to Student’s t test (*P < 0.05, **P < 0.02, ***P < 0.01).
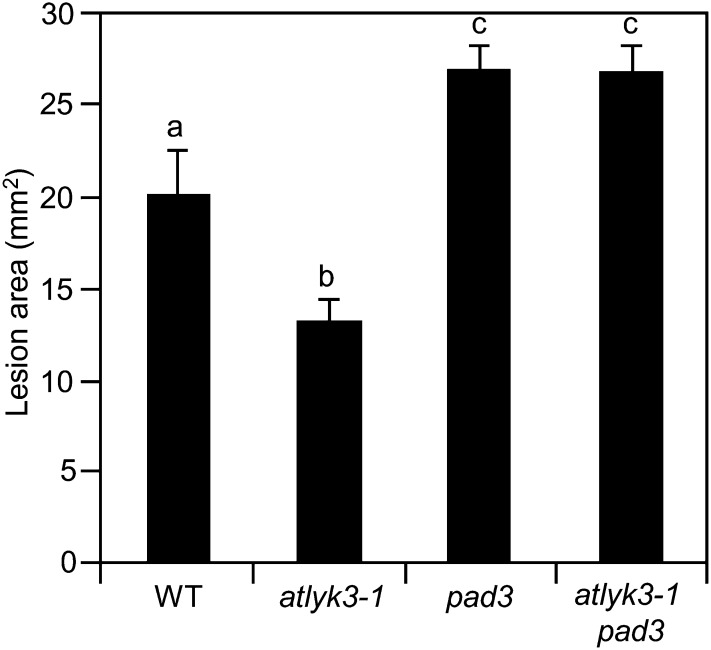
Because PAD3 is important for basal resistance to B. cinerea (Ferrari et al., 2003), we speculated that the elevated expression of this gene in plants mutated in AtLYK3 gives a major contribution to their enhanced resistance to infection. To verify this hypothesis, we generated double mutants by crossing atlyk3-1 with pad3, a line carrying a point mutation in PAD3 that results in a dramatic reduction of expression of this gene and an almost complete lack of camalexin production after B. cinerea infection (Zhou et al., 1999; Ferrari et al., 2003). Homozygous atlyk3-1 pad3 double mutant plants did not accumulate AtLYK3 transcripts and, after B. cinerea infection, displayed very low levels of expression of PAD3 compared with wild-type plants (Supplemental Fig. S4). Notably, double mutants also showed enhanced susceptibility to the fungus, with an average lesion size similar to that observed in the pad3 single mutant (Fig. 4). These results suggest that the enhanced resistance to B. cinerea of atlyk3-1 depends on the increased expression of PAD3 observed in this mutant.
Figure 4.
Enhanced resistance against B. cinerea of atlyk3-1 plants is dependent on PAD3. Wild-type (WT), atlyk3-1, pad3-1, and atlyk3-1 pad3 rosette leaves were inoculated with B. cinerea, and lesion areas were measured 48 h after inoculation. Values are means ± se (n ≥ 10). Different letters indicate statistically significant differences, according to one-way ANOVA followed by Tukey’s honestly significant difference test (P ≤ 0.01). This experiment was repeated twice with similar results.
AtLYK3 Affects Responses Regulated by ABA
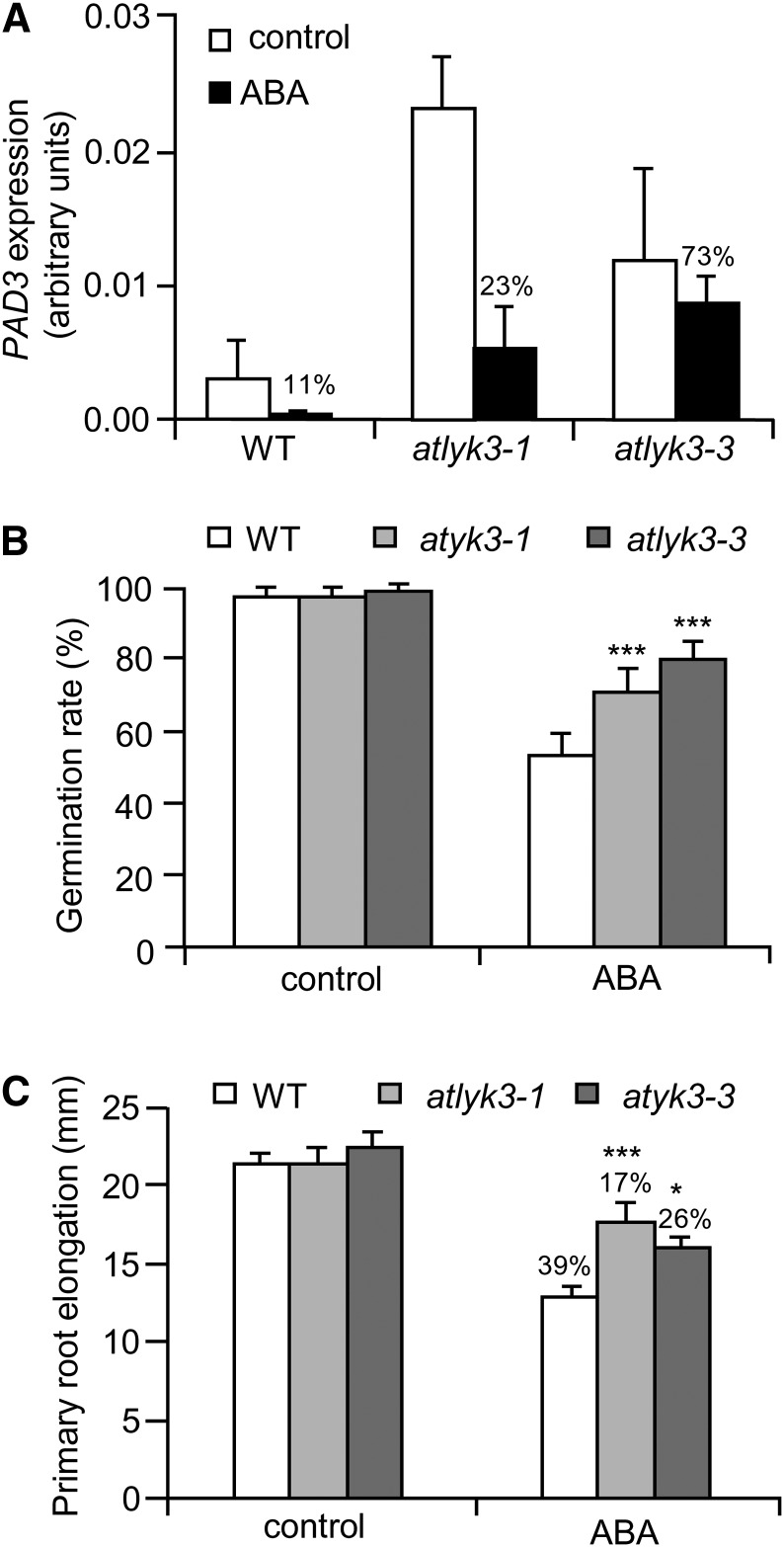
Previous reports indicate that ABA plays a negative role in defense against B. cinerea and that mutants impaired in the biosynthesis or in the transduction of this hormone display increased resistance to this fungus (Audenaert et al., 2002a; Asselbergh et al., 2007). Furthermore, it is known that pretreatments with ABA repress flg22-induced expression of several defense-related genes (Clay et al., 2009), also pointing to ABA as a negative regulator of defense. Therefore, we investigated whether increased resistance of atlyk3 mutants is due to a defective response to ABA. We found that, whereas a treatment of 24 h with 10 µm ABA decreased PAD3 transcripts to almost undetectable levels in wild-type seedlings, the same treatment was less effective in atlyk3-1 and atlyk3-3 (Fig. 5A). To corroborate the observation that responses to ABA are altered in the absence of a functional AtLYK3, other typical responses to this hormone were analyzed. The germination rate in the presence of ABA was significantly higher for atlyk3-1 and atlyk3-3 than for wild-type seeds (Fig. 5B), and inhibition of primary root growth by this hormone was also reduced in both mutants (Fig. 5C). We also analyzed the ability of atlyk3 mutants to cope with salt stress, which is largely dependent on ABA (Koornneef et al., 1998). Both atlyk3-1 and atlyk3-3 seedlings displayed a significant reduction of primary root elongation in the presence of 75 mm NaCl, a concentration that did not affect wild-type seedlings (Supplemental Fig. S5). Notably, sensitivity to salt of atlyk3 mutants was similar to that of aba2-3, a mutant impaired in ABA biosynthesis as a consequence of a mutation in a gene encoding the enzyme that converts xanthoxin to abscisic aldehyde (Laby et al., 2000; González-Guzmán et al., 2002). Altered physiological responses to ABA in plants carrying mutations in AtLYK3 were not dependent on the enhanced expression of PAD3 because mutations in this gene did not affect ABA responsiveness both in the wild type and in atlyk3-1 background (Supplemental Fig. S6).
Figure 5.
AtLYK3 is required for full response to ABA. A, Wild-type (WT), atlyk3-1, and atlyk3-3 liquid-grown seedlings were treated for 24 h with 0.01% MeOH (control, white bars) or 10 μm ABA (black bars). Total RNA was extracted, and expression of PAD3 was analyzed by qPCR, using UBQ5 as reference. Bars indicate average expression ± sd of three independent biological replicates. For each genotype, the percentage of expression in ABA-treated samples, with respect to the control, is indicated above bars. B, Wild-type, atlyk3-1, and atlyk3-3 seeds were stratified for 3 d at 4°C and sown on solid medium supplemented with 0.05% MeOH (control) or 5 μm ABA. Germination rate was determined after 3 d. Bars indicate average percentage of germinated seeds in three independent experiments ± sd (n > 30 for each experiment). C, Wild-type, atlyk3-1, and atlyk3-3 seedlings were germinated on solid medium, and after 4 d, they were transferred to plates containing solid medium supplemented with 0.05% MeOH (control) or 2.5 μm ABA. Elongation of the primary root was measured after 2 d. Bars indicate the average elongation of roots from seedlings grown on ABA or on control plates ± sd (n > 10). The percentage of reduction of root elongation, with respect to the controls, is indicated above bars. Asterisks indicate significant differences between the wild type and mutants treated with the same conditions, according to Student’s t test (***P < 0.01).
Because our results indicate that AtLYK3 is required for full late responses to ABA, we examined whether earlier responses to this hormone are also affected by the loss of this gene. Wild-type, atlyk3-1, and atlyk3-3 seedlings were treated for 4 h with a low dose (1 µm) of the hormone, and the expression of two well-characterized ABA-induced marker genes, i.e. RAB GTPASE HOMOLOG B18 (RAB18; Lång and Palva, 1992) and ABA INSENSITIVE1 (ABI1; Meyer et al., 1994), was analyzed. We did not observe significant differences in the levels of expression of both genes in wild-type and mutant plants (Supplemental Fig. S7, A and B). Moreover, transcript levels for PYRABACTIN RESISTANCE1 (PYR1) and PYR1-LIKE4 (PYL4), which encode two ABA receptors whose expression is repressed by the hormone (Ma et al., 2009; Park et al., 2009), were similar in untreated wild-type, atlyk3-1, and atlyk3-3 seedlings and, after ABA treatment, decreased in a similar way in all three genotypes (Supplemental Fig. S7, C and D). Thus, mutations in AtLYK3 appear to affect only a set of late, ABA-dependent responses but not perception or early responses to this hormone. Enhanced resistance to B. cinerea observed in Arabidopsis mutants impaired in ABA biosynthesis or transduction has been linked to increased cuticle permeability (Bessire et al., 2007; Chassot et al., 2007; Curvers et al., 2010; L’Haridon et al., 2011). However, we could not observe an altered cuticle permeability in atlyk3-1 or atlyk3-3 rosette leaves, as determined by measuring the rate of chlorophyll extraction with ethanol, whereas, as expected, permeability of aba2-3 leaves was significantly increased (Supplemental Fig. S8). We therefore conclude that the enhanced resistance observed in atlyk3 mutants is not due to an altered cuticle permeability.
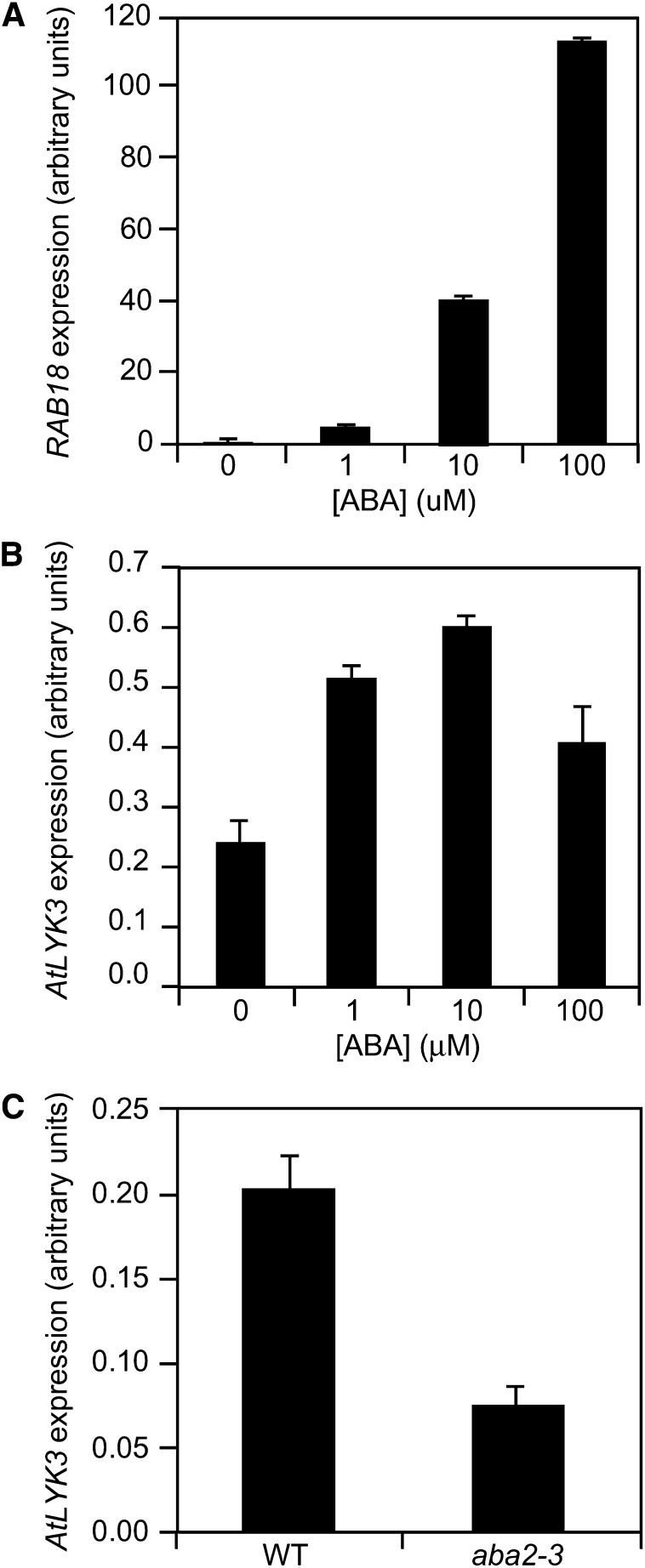
Because our results point to AtLYK3 as a player in the late signal transduction cascade triggered by ABA, we analyzed whether AtLYK3 expression in wild-type plants is also regulated by this hormone; as a control, we measured transcript levels for RAB18. In wild-type seedlings treated for 24 h with different ABA concentrations (0, 1, 10, and 100 µm), expression of RAB18 increased, as expected, in a dose-dependent manner (Fig. 6A), indicating that the treatments were effective. Expression of AtLYK3 also increased after treatment with ABA; however, induction was maximal at a dose of 10 µm and decreased at 100 µm (Fig. 6B), suggesting that expression of this gene is subject to negative feedback at high doses of the hormone. Moreover, basal AtLYK3 expression levels were significantly reduced in aba2-3 seedlings (Fig. 6C). These results indicate that AtLYK3 expression is positively regulated by ABA.
Figure 6.
Expression of AtLYK3 is regulated by ABA. A and B, Wild-type (WT) seedlings were treated for 24 h with 0.1% MeOH (control) or ABA at the indicated concentrations, and expression of RAB18 (A) and AtLYK3 (B) was analyzed by qPCR, using UBQ5 as reference. C, Expression of AtLYK3 was analyzed as in A in untreated wild-type and aba2-3 seedlings. Bars indicate average expression ± sd of three technical replicates. These experiments were repeated at least three times with similar results.
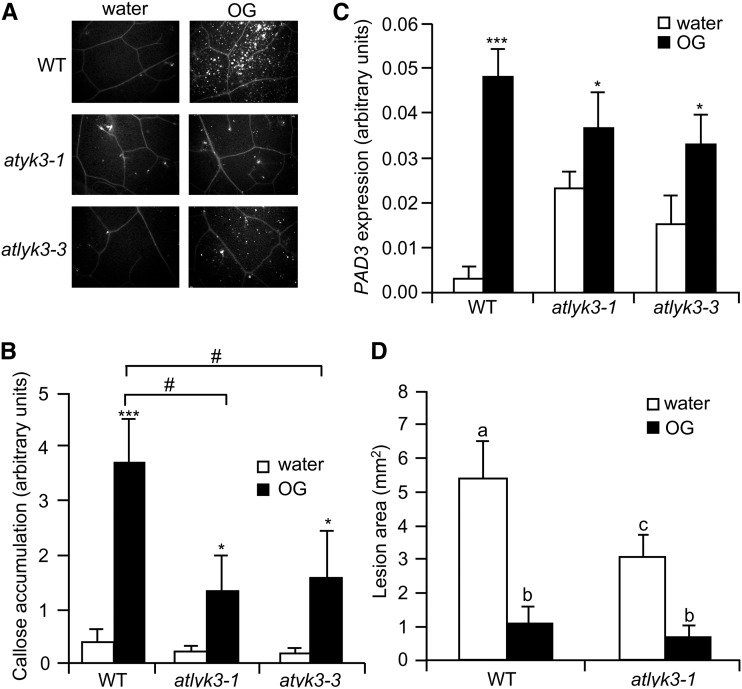
To investigate whether AtLYK3 affects responses to exogenously applied elicitors, we infiltrated rosette leaves of wild-type, atlyk3-1, and atlyk3-3 plants with water or OGs, and after 24 h, we stained them with aniline blue for callose deposits. UV fluorescence microscopy revealed that after treatment with the elicitor, both mutants displayed reduced staining with respect to the wild type (Fig. 7, A and B). A similar reduced accumulation of callose was observed in atlyk3-1 and atlyk3-3 leaves infiltrated with the bacterial MAMP flg22 (Supplemental Fig. S9A). This indicates that their resistance to pathogens is not due to enhanced callose deposition; furthermore, our data suggest that AtLYK3 is required for full activation of this defense response. To determine whether other elicitor-induced responses are impaired in atlyk3 mutants, we analyzed the expression of PAD3 in control- and OG-treated seedlings. As previously reported (Ferrari et al., 2007), PAD3 expression significantly increased in wild-type seedlings treated with the elicitor (Fig. 7C). PAD3 transcript levels in untreated atlyk3-1 and atlyk3-3 seedlings were, as already observed, higher than in the wild type and, in response to OGs, further increased to values comparable to those observed in OG-treated wild-type seedlings (Fig. 7C). Levels of PAD3 transcripts accumulating in response to flg22 were also similar in wild-type and mutant seedlings (Supplemental Fig. S9B). Finally, we tested whether OG-induced resistance to B. cinerea is affected in the absence of a functional AtLYK3. Wild-type and atlyk3-1 plants were sprayed with water or OGs, and after 24 h, leaves were inoculated with the fungus. As expected, water-treated atlyk3-1 plants developed smaller lesions than the wild type (Fig. 7D). Notably, OG pretreatments effectively increased resistance to similar levels in all tested genotypes (Fig. 7D). Taken together, these data indicate that pathogen resistance and increased PAD3 expression in atlyk3 mutants is not due to an enhanced response to elicitors.
Figure 7.
Responses to OGs in atlyk3 mutants. A and B, Rosette leaves of adult wild-type (WT), atlyk3-1, and atlyk3-3 plants were infiltrated with water or 100 μg mL–1 OG. Leaves were harvested after 24 h, and callose deposits were stained with aniline blue. A, Representative pictures of treated leaves obtained with a fluorescence microscope. B, Quantification of callose accumulation. Bars represent average fluorescence ± sd of at least six different samples. Asterisks indicate significant differences between water- and OG-treated samples, according to Student’s t test (*P < 0.05, ***P < 0.01). #, Significant difference between wild-type and mutant leaves treated with OGs, according to Student’s t test (P < 0.05). This experiment was repeated twice with similar results. C, Expression of PAD3 was analyzed by qPCR in wild-type, atlyk3-1, and atlyk3-3 liquid-grown seedlings treated for 3 h with water or 100 μg mL–1 OGs. The UBQ5 gene was used as reference. Bars indicate average expression ± sd of three independent biological replicates. Asterisks indicate significant differences between water- and OG-treated samples, according to Student’s t test (*P < 0.05, ***P < 0.01). D, Four-week-old wild-type and atlyk3-1 plants were sprayed with water or 200 μg mL–1 OGs, and after 24 h, they were inoculated with B. cinerea. Lesion size was measured at 48 h post infection. Bars indicate average lesion area ± se of at least 12 lesions. Different letters indicate statistically significant differences, according to one-way ANOVA followed by Tukey’s honestly significant difference test (P ≤ 0.01). This experiment was repeated three times with similar results.
An AtLYK3-GFP Fusion Is Localized in the Plasma Membrane and Complements atlyk3 Phenotypes
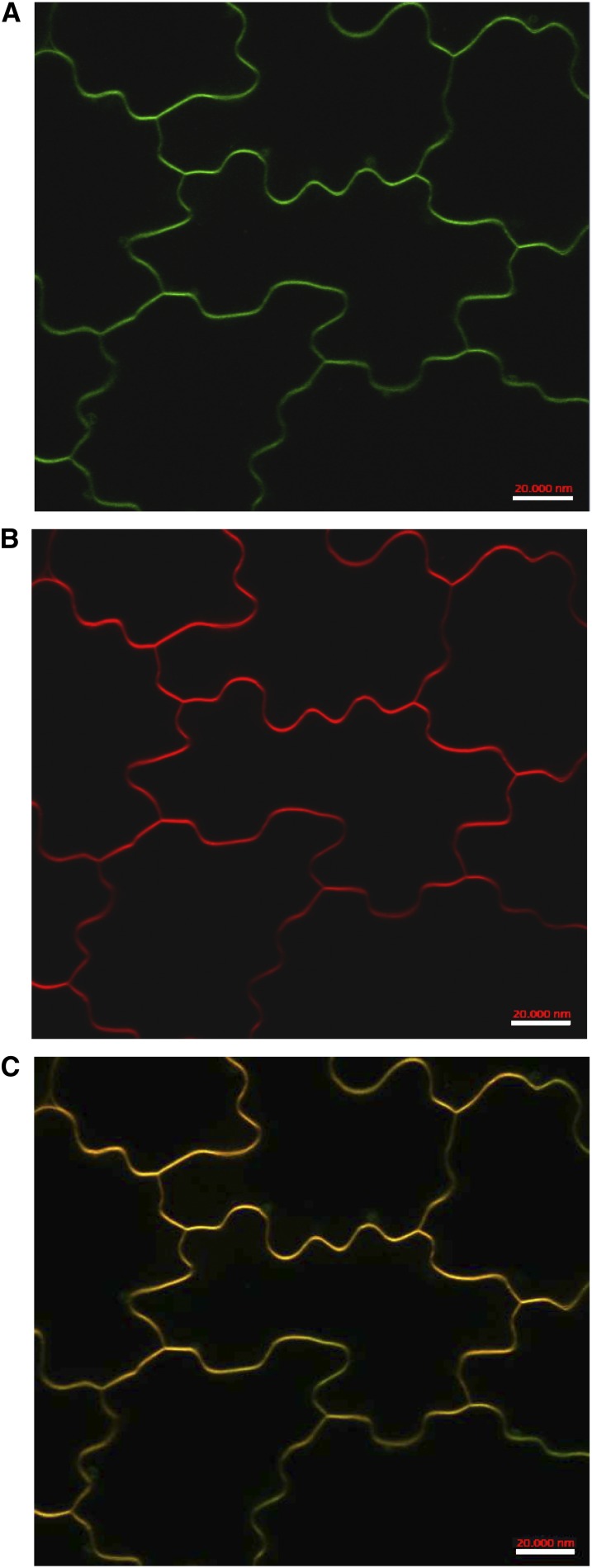
All LYK proteins so far characterized are localized in the plasma membrane, where they act as bona fide receptors or coreceptors of MAMPs or other molecules released by plant-interacting microbes. To determine the subcellular localization of AtLYK3, we generated transgenic Arabidopsis plants expressing a translational fusion of the protein with the GFP under the control of the constitutive promoter Cauliflower mosaic virus (CaMV) 35S. Confocal laser scanning microscopy analyses of cotyledons of primary transformants revealed a strong green fluorescence in correspondence of the plasma membrane, as confirmed by colocalization experiments with the plasma membrane-specific dye FM4-64 (Brandizzi et al., 2004; Fig. 8) and by plasmolysis experiments (Supplemental Fig. S10). We therefore concluded that AtLYK3 is likely localized mainly on the plasma membrane.
Figure 8.
AtLYK3-GFP is localized in the plasma membrane. Ten-day-old transgenic seedlings expressing AtLYK3-GFP under the control of the CaMV 35S promoter were stained with the plasma membrane-specific dye FM4-64, and images of epidermal cells were taken by confocal laser scanning microscopy. A, GFP. B, FM4-64. C, Merge of A and B. Bars = 20 μm. Images are representative of five independent transgenic lines.
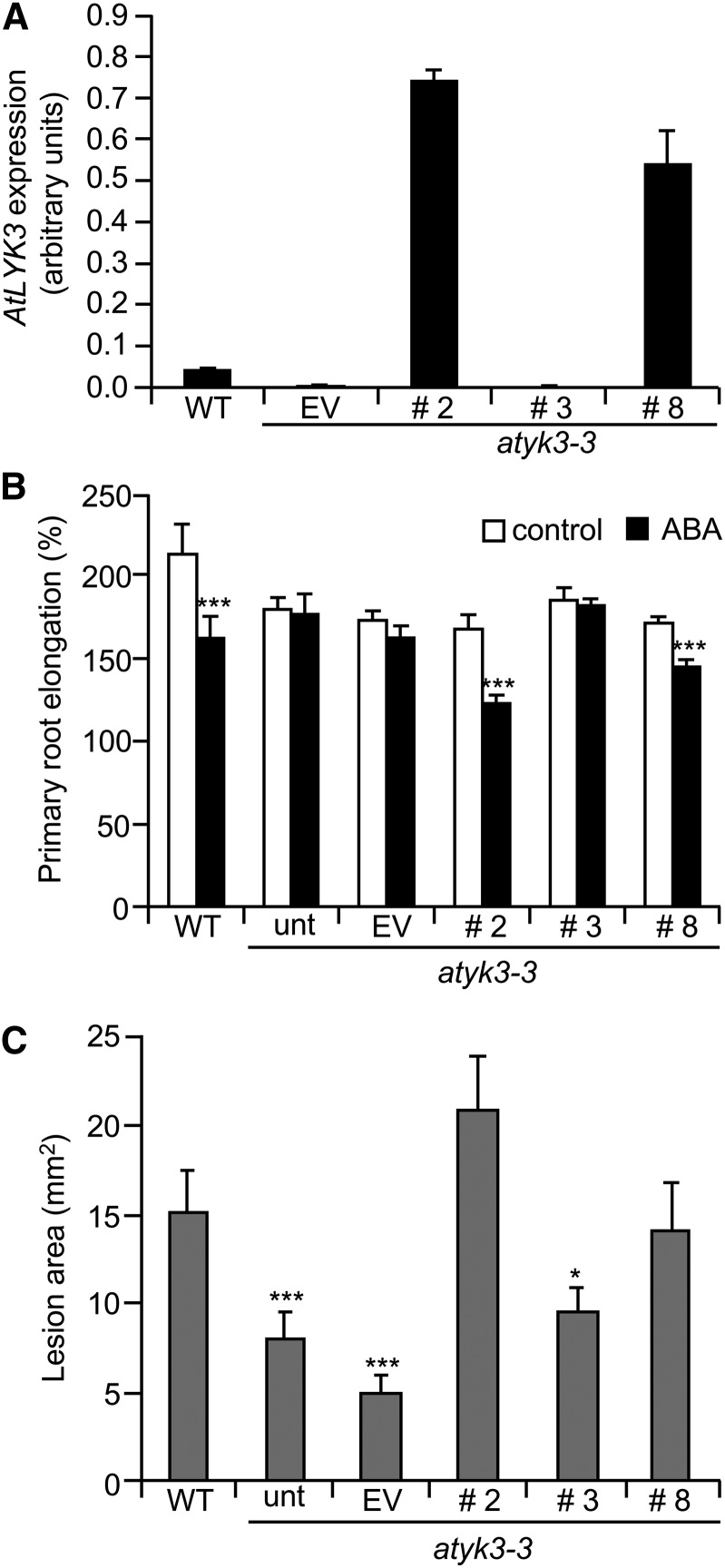
To verify that the AtLYK3-GFP fusion protein was functional, we expressed it in the atlyk3-3 background and determined if it was able to complement the mutant phenotypes. Expression of the transgene was determined by qPCR in leaves of three independent transgenic lines (2, 3, and 8) and, as a control, atlyk3-3 plants transformed with the empty vector (EV). The latter line showed a low but detectable expression of AtLYK3, as expected for atlyk3-3 plants (Fig. 9A). Two lines transformed with AtLYK3-GFP (lines 2 and 8) showed a segregation ratio of 3:1 for their resistance to phosphinothricin, and herbicide-resistant plants expressed AtLYK3 at high levels, whereas line 3, which had a segregation ratio of 15:1 and therefore is likely to carry at least two copies of the transgene, did not show any detectable expression of AtLYK3 (Fig. 9A), possibly as a consequence of silencing of both the transgene and the endogenous copy of the gene. These lines were first tested for root sensitivity to inhibition by exogenous ABA. As shown in Figure 9B, like untransformed atlyk3-3 seedlings, both EV and line 3 were almost completely insensitive to 5 µm ABA, in terms of primary root growth; by contrast, like the wild type, lines 2 and 8 displayed a significant reduction of root elongation in the presence of ABA compared with control-grown seedlings. B. cinerea susceptibility was also determined in the parental and transgenic lines. As expected, lesion size in untransformed atlyk3-3 plants was significantly reduced compared with the wild type; a similar degree of resistance was also observed in the EV line as well as in the silenced line (Fig. 9C). By contrast, lesion size in leaves of lines 2 and 8 were not significantly different from those in the wild type, indicating that AtLYK3-GFP restored susceptibility to the fungus. Taken together, these data suggest that the AtLYK3-GFP fusion protein is functional and able to complement both ABA insensitivity and increased resistance to infection in the atlyk3-3 mutant.
Figure 9.
AtLYK3-GFP complements atlyk3-3 phenotypes. A, atlyk3-3 plants were transformed with a construct for the expression of AtLYK3-GFP under the control of the 35S promoter or with the EV. Expression of AtLYK3 in 10-d-old seedlings of the wild type (WT), three independent AtLYK3-GFP lines (numbers 2, 3, and 8), and one EV line was determined by qPCR, using UBQ5 as control. Bars indicate average expression ± sd of three technical replicates. B, Wild-type and atlyk3-3 untransformed (unt) or transgenic lines were germinated on solid medium and, after 6 d, were transferred to plates supplemented with 0.05% MeOH (white bars) or 5 μm ABA (black bars). For each seedling, primary root length was measured before transfer and at 3 d. Bars indicate the average relative root elongation ± se (n > 10). Asterisks indicate significant differences between control- and ABA-grown seedlings, according to Student’s t test (***P < 0.01). C, Leaves from wild-type and untransformed and transgenic atlyk3-3 4-week-old plants were inoculated with B. cinerea. Lesion area was determined 72 h after infection. Values are means ± se (n ≥ 12). Asterisks indicate statistically significant differences between wild-type and mutant plants, according to Student’s t test (*P ≤ 0.05, ***P ≤ 0.01). This experiment was repeated twice with similar results.
DISCUSSION
ABA is involved in several developmental processes, such as seed dormancy, root geotropism, and stomata opening, and its role in the response to abiotic stresses, such as high salt, drought, and low temperature, is well known (Wasilewska et al., 2008). However, only recently, the importance of this hormone as a regulator of defense responses against pathogens has emerged (Mauch-Mani and Mauch, 2005; Asselbergh et al., 2008b; Bari and Jones, 2009; Cao et al., 2011). It was observed that ABA increases susceptibility in some plant-pathogen interactions (Audenaert et al., 2002b; Asselbergh et al., 2007, 2008a; de Torres-Zabala et al., 2007; Curvers et al., 2010), while it promotes resistance in others (Ton and Mauch-Mani, 2004; Adie et al., 2007). It is therefore likely that different components of the ABA signaling pathways, as well as downstream elements regulated by this hormone, may exert a negative or positive influence on the different layers of defense against bacteria, fungi, and pathogens. The molecular and biochemical mechanisms linking ABA perception to the regulation of specific defense responses are however still poorly understood.
In this paper, we have demonstrated that the Arabidopsis RLK AtLYK3 plays an important role in the cross talk between pathogen resistance and physiological responses mediated by ABA. This conclusion is supported by the following lines of evidence: (1) plants lacking a functional AtLYK3 show enhanced expression of defense genes and increased resistance to B. cinerea and P. carotovorum infection; (2) AtLYK3 expression is repressed in response to infection and elicitor treatment, increases after treatment with exogenous ABA, and is reduced in the aba2-3 mutant, which is impaired in ABA biosynthesis; and (3) atlyk3 mutants show reduced sensitivity to ABA in two different bioassays (inhibition of seed germination and of primary root elongation) and a decreased ABA-induced repression of PAD3 expression; furthermore, atlyk3 mutants show increased sensitivity to salt stress, similarly to mutants impaired in ABA synthesis or transduction (Koornneef et al., 1998). Taken together, these results indicate that AtLYK3 acts both as a positive regulator of ABA signaling and as a negative regulator of defense responses, in particular those effective against the fungal necrotroph B. cinerea. Interestingly, in contrast to B. cinerea, A. brassicicola infection appears to increase AtLYK3 expression (Supplemental Fig. S1A). This is consistent with the previous observation that ABA promotes resistance to A. brassicicola (Ton and Mauch-Mani, 2004).
The contribution of ABA to plant susceptibility to B. cinerea has been previously demonstrated in sitiens tomato (Solanum lycopersicum) plants, which are defective for the enzyme that converts the ABA-aldehyde in ABA and are more resistant to this pathogen (Audenaert et al., 2002a). Also, Arabidopsis plants defective in ABA biosynthesis or signaling exhibit an increased resistance to B. cinerea (Adie et al., 2007). More recently, Arabidopsis mutants impaired in ABA biosynthesis or perception/transduction have been shown to have increased resistance to another necrotrophic fungus, Plectosphaerella cucumerina, as well as constitutive expression of defense responses (Sánchez-Vallet et al., 2012). However, the mechanisms linking a reduced ABA accumulation or perception/transduction to resistance to fungal infection are not fully elucidated. In both tomato and Arabidopsis, resistance to B. cinerea in ABA-deficient mutants is accompanied by an increase in the accumulation of ROS during the early stages of penetration and has been linked to an increased cuticle permeability compared with the wild type (Asselbergh et al., 2007; Curvers et al., 2010; L’Haridon et al., 2011). Alterations of cuticle that lead to increased permeability, as in the case of transgenic plants expressing a fungal cutinase or mutants for the biosynthesis of cutin, greatly enhance resistance to B. cinerea (Chassot et al., 2007; L’Haridon et al., 2011), likely due to a faster diffusion of elicitors or of antimicrobial substances. However, we could not observe a significant increase in cuticle permeability in atlyk3-1 or atlyk3-3 leaves compared with wild-type plants. Furthermore, atlyk3 mutant leaves did not constitutively accumulate high levels of ROS, and accumulation of ROS upon infection was similar in wild type and mutant plants.
Our results suggest that the absence of a functional AtLYK3 rather affects the signaling pathways that normally repress defense responses and that these pathways are regulated by ABA. Several lines of evidence indicate that this hormone negatively regulates the immune response. ABA suppresses SA-dependent signaling in tomato, and this suppression is required for the elevated resistance to B. cinerea of sitiens plants (Audenaert et al., 2002a). Moreover, ABA pretreatments strongly reduce the expression of several early flg22-induced genes (Clay et al., 2009), and under specific growth conditions, ABA is able to prevent flg22-induced deposition of callose in Arabidopsis cotyledons (Luna et al., 2011). We have shown that PAD3 expression is significantly repressed by ABA and that this repression is at least partially dependent on AtLYK3. Because PAD3 is a major determinant of resistance to B. cinerea (Ferrari et al., 2003) and is required for the resistant phenotype of atlyk3-1 plants (this work), it is likely that repression of PAD3 expression, mediated by AtLYK3, contributes to the increased susceptibility to this fungus caused by ABA. Endogenous ABA, which negatively regulates resistance to B. cinerea, positively regulates AtLYK3 expression, as shown by the analysis of the aba2-3 mutant. It must be noted that publicly available microarray experiments, such as those in the Genevestigator database (Zimmermann et al., 2004), do not show an induction of AtLYK3 expression in response to ABA. We ascribe this apparent discrepancy to the fact that our experiments were performed treating seedlings with ABA for 24 h, instead of a few hours as in previous reports. This is consistent with our results that indicate that early ABA responses are unaffected in atlyk3 mutants. On the other hand, the reduced expression of AtLYK3 in aba2-3 supports our conclusion that ABA levels positively regulate this gene. These observations suggest that AtLYK3 modulates long-term responses to ABA, such as growth inhibition or repression of defense responses, but is not necessary for ABA perception or early downstream signaling events and that its expression is differently regulated by short- or long-term treatments with ABA.
It is noteworthy that atlyk3 mutants show increased resistance also against P. carotovorum, a bacterial necrotroph. It was previously shown that ABA deficiency leads to a faster and stronger activation of defense responses in tomato plants inoculated with the closely related pathogen Erwinia chrysanthemi (Asselbergh et al., 2008a). Furthermore, Arabidopsis plants overexpressing EARLY TO DEHYDRATION RESPONSIVE15, a small protein of unknown function essential in several stress responses, show increased resistance against P. carotovorum and reduced sensitivity to ABA (Kariola et al., 2006). Defense responses that are negatively regulated by components of the ABA signaling pathway therefore appear to be effective against a wide range of necrotrophic pathogens. Activation of the ABA signaling pathway, either by increasing its biosynthesis in the host cells or through microbial production, may be used by different pathogens as a strategy to favor infection (Cao et al., 2011). For example, the capability of B. cinerea of synthesizing ABA (Siewers et al., 2004) may contribute to suppress host defenses, possibly by increasing AtLYK3 expression. It is noteworthy that atlyk3 mutants show reduced accumulation of callose after infiltration with OGs (this work). This is not in contrast with the observed resistance to B. cinerea, because infection by this pathogen is not affected by callose levels (Galletti et al., 2008). ABA has been previously proposed to be necessary for callose biosynthesis after pathogen or elicitor recognition, at least under some environmental condition (Ton and Mauch-Mani, 2004; Luna et al., 2011); the reduced callose deposition that we observed in the absence of a functional AtLYK3 may therefore be due to the reduced sensitivity to ABA of atlyk3 mutants. An alternative possibility is that atlyk3 mutants have reduced responsiveness to MAMPs/DAMPs. However, our results indicate that loss of AtLYK3 does not affect other responses to elicitors. Pretreatments with OGs are equally effective in promoting resistance to B. cinerea in atlyk3 and wild-type plants. Moreover, in atlyk3, OG-induced expression of PAD3 is similar to that observed in the wild type, even if its basal expression is increased. The latter observation suggests that transduction and/or regulatory elements that mediate up-regulation of PAD3 expression in response to elicitors are partially activated in the mutant in the absence of stress stimuli to become fully activated upon elicitation like in the wild type. AtLYK3 may therefore play a role in preventing the inappropriate expression of defense responses in normal conditions.
Taken together, these results suggest that AtLYK3 is required for full elicitor-induced expression of defense responses that are positively regulated by ABA, such as callose deposition. On the other hand, AtLYK3 appears to maintain the low basal expression of other defense responses, such as PAD3 expression, that are negatively regulated by ABA, with no major role, however, in their elicitor-induced expression. A corollary of this is that ABA is unlikely to contribute to the regulation of PAD3 expression by elicitors. The elevated basal defenses may be sufficient to slow down pathogen growth and explain the reduced susceptibility of atlyk3 mutants; in addition, the relief of the ABA- and AtLYK3-mediated repression of the expression of PAD3 and other defense responses may contribute to the reduced susceptibility. Additional work is however required to verify this hypothesis.
The fine balance between ABA-promoted inhibition of defenses, in part mediated by AtLYK3, and MAMP/DAMP-induced innate immune responses may therefore contribute to determine the final outcome of a pathogen infection: when this balance is tipped off, for instance with treatments with exogenous elicitors or exogenous ABA, plant resistance appears to increase or decrease, respectively. It has been recently reported that AtLYK3 is required for Nod factor-induced suppression of flg22-triggered defense responses, including production of ROS and phosphorylation of mitogen-activated protein kinases (Liang et al., 2013). Because Arabidopsis is not infected by rhizobia (Remy et al., 1994), the authors suggested that AtLYK3-mediated suppression of defenses may be one of the first steps in the evolution of the recognition mechanisms for these symbiotic bacteria (Liang et al., 2013). In this paper, we show that AtLYK3 does suppress basal defenses effective against at least some fungal and bacterial pathogens and may therefore have evolved to avoid unchecked expression of defense responses in the absence of a potential pathogen. Whether ABA is also involved in the Nod-induced repression of Arabidopsis innate immunity still needs to be investigated.
AtLYK3 is not the only RLK that acts as a negative regulator of defense responses against pathogens. For instance, Lectin Receptor Kinase V.5 negatively regulates resistance to bacterial pathogens and MAMP-induced stomatal closure (Arnaud et al., 2012; Desclos-Theveniau et al., 2012), and mutants for the PHYTOSULFOKIN RECEPTOR1 gene show enhanced responses triggered by MAMPs (Igarashi et al., 2012). This suggests that plant cells actively maintain basal defenses at low levels in uninfected conditions through a variety of signaling networks that involve surface receptor-like proteins. However, the mechanisms underlying the role of these proteins in defense are still poorly defined. How AtLYK3 modulates defense responses is also currently unknown, though possible mechanisms can be envisioned. AtLYK3, like AtLYK1/AtCERK1, is predicted to possess an intact kinase domain (Tanaka et al., 2012). Because AtLYK3-GFP localizes on the plasma membrane and plant LYKs and LYPs can cooperate to recognize different ligands (Willmann et al., 2011; Wan et al., 2012), likely forming hetero- and homo-oligomers (Shimizu et al., 2010; Liu et al., 2012), it can be speculated that AtLYK3 is part of a multiprotein complex, where it may act as a negative regulator and keep downstream defense responses repressed in the absence of infection. Activation of the innate immune response during pathogen infection may involve a derepression of the complex, through posttranslational modifications and/or through the reduction of AtLYK3 expression, as we have observed in response to B. cinerea or to OGs. In this scenario, ABA contributes to keeping elevated levels of AtLYK3 in uninfected plants and therefore avoids elevated expression of defenses in healthy tissues.
Alternatively, AtLYK3 may be a bona fide receptor of a still unidentified endogenous signal. In contrast to AtCERK1, AtLYK3 possesses a single LysM motif in its ectodomain (Zhang et al., 2007, 2009), which is likely sufficient to bind a tetrasaccharide moiety, such as that of Nod factors, but not sufficient to bindchitooligosaccharides with eliciting activity. It is therefore tempting to speculate that an unidentified short oligosaccharide, likely containing NAG in its structure, can negatively modulate plant defenses through its binding to AtLYK3. Endogenous carbohydrates possessing NAG in their backbone have been previously proposed to play important signaling roles in plants. For instance, fragments of NAG-containing arabinogalactan proteins may be released in carrot (Daucus carota) plants by endogenous chitinases to regulate embryogenesis (van Hengel et al., 2001), and endogenous Nod factor-like lipochitooligosaccharides are able to promote somatic embryogenesis in Norway spruce (Picea abies; Dyachok et al., 2002). Notably, overexpression of a fungal chitinase in Arabidopsis increases resistance to salt stress, and this resistance is mediated by AtCERK1 (Brotman et al., 2012), suggesting that this protein is able to perceive molecules endogenously released by chitinases (de las Mercedes et al., 2006; Kumar et al., 2009). The exact nature of these molecules, and the role of AtLYK3 or other LYKs in their perception, is still not known.
AtLYK3 appears to be important also for physiological responses to ABA, likely acting downstream of the perception of this hormone. This hypothesis is supported by the observation that transcript levels for ABI1, RAB18, PYR1, and PYL4 show similar changes when the wild type and atlyk3 mutants are treated with ABA for a few hours. AtLYK3 may directly act in a branch of the ABA signaling pathway that regulates some late responses to this hormone, including inhibition of seed germination, root growth, and of PAD3 expression. Alternatively, reduction of these responses in atlyk3 mutants may be a consequence of the enhanced activation of defense in these plants. Increasing evidence suggests that activation of plant immune responses negatively regulates ABA signaling. For instance, chemically induced activation of systemic acquired resistance suppresses the expression of ABA biosynthesis-related and ABA-responsive genes in Arabidopsis (Yasuda et al., 2008). The constitutive expresser of PR genes22 lesion mimic mutant, which has as a deletion that fuses two cyclic nucleotide-gated ion channel-encoding genes, exhibits elevated levels of SA, constitutive expression of defense genes, and enhanced resistance to pathogens (Yoshioka et al., 2006). Interestingly, this mutant also shows SA-dependent alterations in ABA signaling (Mosher et al., 2010). Furthermore, the small molecule [5-(3,4-dichlorophenyl)furan-2-yl]-piperidine-1-ylmethanethione rapidly down-regulates ABA-induced responses and stimulates expression of defense-related genes in an SA-independent fashion (Kim et al., 2011). This molecule does not interfere with the earliest events in ABA perception and signaling, like ABA-dependent interaction of PYR1 with ABI1 and activation of sucrose nonfermenting-1-related protein kinase2 (SnRK2), but seems to disrupt cytosolic Ca2+ signaling. Notably, the suppressor of nonexpresser of pr genes1-1, constitutive1 (snc1) mutant, which shows constitutive activation of defense responses and pathogen resistance as a consequence of a point mutation in a gene encoding a Toll-like/Interleukin1 Receptor-Nucleotide Binding-Leu-Rich Repeat protein (Zhang et al., 2003), is also partially insensitive to ABA (Kim et al., 2011). These observations suggest that ABA signaling may be negatively affected by the activation of both SA-dependent and SA-independent defense responses. We did not observe a significant increase of expression of PR1 in healthy and infected atlyk3 mutants; however, we cannot rule out that the effects of AtLYK3 on resistance and on ABA signaling are partly dependent on the SA signaling pathway.
CONCLUSION
Our results indicate that AtLYK3 is a negative regulator of basal Arabidopsis defense responses effective against microbial infections. This protein appears to negatively regulate basal and early pathogen-induced expression of defense genes, in particular PAD3, which is important for the enhanced resistance against B. cinerea observed in the absence of a functional AtLYK3. AtLYK3 is also required for a subset of late responses to ABA, and its expression is positively regulated by this hormone. These observations suggest that AtLYK3 plays an important role in the negative cross talk between the plant innate immunity and the activation of responses mediated by ABA. Our results indicate that AtLYK3 affects a subset of late responses to ABA, including the repression of defenses. This could be explained either assuming that this protein directly affects specific components of the ABA signaling pathway or, alternatively, that unchecked activation of the innate immunity negatively influences physiological responses mediated by this hormone. Future studies will help discriminate between these two hypotheses and determine the biochemical function of AtLYK3.
MATERIALS AND METHODS
Plant Material and Growth Conditions
All Arabidopsis (Arabidopsis thaliana) plants used in this work belong to the ecotype Columbia. Seeds of the T-DNA insertional mutants atlyk3-1 (WiscDsLox318C07) and atlyk3-3 (SALK_140374C) were obtained from the Salk Institute Genomic Analysis Laboratory (Alonso et al., 2003). The aba2-3 mutant (Laby et al., 2000) was obtained from the Nottingham Arabidopsis Stock Centre (University of Nottingham). The pad3 mutant (Glazebrook and Ausubel, 1994) was kindly donated by Frederick M. Ausubel (Massachusetts General Hospital).
Plants were grown on soil (Einheitserde) in a climatic chamber at 22°C and 70% relative humidity with a 16-h-light/8-h-dark cycle (approximately 120 µmol m–2 s–1). For in vitro growth, seeds were surface sterilized and either germinated in 12-well plates (about 10 seeds per well) containing 1 mL of liquid Murashige and Skoog (MS) basal medium (Sigma-Aldrich) supplemented with 0.5% (w/v) Suc, pH 5.5 (Murashige and Skoog, 1962), or on solid medium containing one-half-strength MS basal salts, 1% agar (w/v), and 1% (w/v) Suc, pH 5.7. Plates were incubated at 22°C with a 16-h/8-h light/dark cycle and a light intensity of 120 µmol m–2 s–1.
Mutant Genotyping and Generation of Double Mutants
Genomic DNA was extracted from rosette leaves as previously described (Edwards et al., 1991) and subjected to PCR using primer pairs for the AtLYK3 wild-type allele (RP1 + LP1 for atlyk3-1 and RP3 + LP3 for atlyk3-3) or for the T-DNA insertion (P745 + RP1 for atlyk3-1 and Lba1 + RP3 for atlyk3-3). The sequences of the primers are as follows: Lba1, ATTTTTGTCAACAATGGCCTG; P745, AACGTCCGCAATGTGTTATTAAGTTGTC; RP1, GTTCAGCTTCTTTGTGGTTGC; LP1, CTTTTCAGCATTCCCTTTCG; LP3, TGGGAATCGAAAAAGATTACAAC; and RP3, GGACTTGCAAAACTCGTTGAG. PCR products were separated by agarose gel electrophoresis and visualized by ethidium bromide staining. Plants homozygous for the mutations were propagated and used for all subsequent experiments.
Crosses between atlyk3-1 and pad3-1 plants were made by emasculation of the flowers and manual pollination. All the flowers that were not manually pollinated were removed, as well as the apex of the flowering stem. Homozygous double mutants were identified in the F2 generation by genotyping for the atlyk3-1 allele (as described above) and by RT-PCR analysis for AtLYK3 and PAD3 expression before and after Botrytis cinerea infection (see below).
Elicitor Treatments and Pathogen Infections
OGs (degree of polymerization = 10–15) were obtained as previously described (Galletti et al., 2008). Crab shell chitin (Sigma-Aldrich) was prepared by dissolving 10 mg of the same in 37% (w/w) HCl and adjusting the pH to 5 to 6 with NaOH 12M. Treatments were performed on 10-d-old liquid-grown seedlings. The medium was replaced with a fresh one the day before the experiment, and after 24 h, the elicitor was added to the medium to a final concentration of 100 µg mL–1 OG or 50 µg mL–1 chitin. Seedlings were incubated for 3 h at room temperature. Water was used as control for the OG treatments, whereas a mock solution, obtained by adding to 37% HCl the same amount of NaOH used to adjust the pH of the elicitor stock solution, was used.
All pathogen infections were conducted on rosette leaves of 4-week-old plants grown in soil. B. cinerea growth and inoculation were performed as previously described (Ferrari et al., 2007; Galletti et al., 2008). For elicitor-induced protection experiments, plants were sprayed with water or 200 µg mL–1 OGs 24 h before inoculation as previously described (Ferrari et al., 2007). Pectobacterium carotovorum subsp. carotovorum (strain DSMZ 30169) was obtained by DSMZ GmbH. Bacteria were cultivated in Luria-Bertani (LB) liquid medium (Duchefa Biochemie) for 16 to 18 h at 28°C and 340 rpm. Bacteria were then collected by centrifugation (8,000g for 10 min) and suspended in a 50 mm potassium-phosphate buffer (pH 7.0) at a final optical density at 600 nm of 0.05, corresponding to a concentration of 5 × 107 colony forming units mL–1. Arabidopsis leaves were detached with a scalpel and placed on water-soaked filter paper in petri dishes. Two scratches of about 5 mm were made on the epidermis of the adaxial surface of each leaf, at the sides of the mid rib, using a sterile needle. A droplet of 5 µL of the bacterial suspension was placed on each scratch. Plates were wrapped with transparent plastic film and incubated at the same conditions as the leaves inoculated with B. cinerea. The area of water-soaked lesions was determined 24 h after inoculation.
Analysis of Sensitivity to ABA and Salt Stress, Cuticle Permeability, and Accumulation of ROS
For the analysis of the germination rate, sterilized seeds were stratified for 4 d in the dark at 4°C in sterile water and then distributed in petri dishes containing solid MS medium supplemented with ABA (Sigma-Aldrich) at different concentrations or, as a control, 0.01% (v/v) MeOH. After 2 d of incubation under the conditions described above, the number of seeds that had germinated was determined with the aid of a stereoscope.
To measure the sensitivity of primary roots to ABA, seeds were germinated on solid medium as indicated above. After 4 d, seedlings were transferred under sterile conditions on new plates containing solid MS medium supplemented with 5 µm ABA or, as control, 0.05% (v/v) MeOH. The plates were placed vertically in the growth chamber after marking the position of the root apex of each seedling. After 2 d, the elongation of the primary root was measured with a ruler. For salt stress, seeds were germinated on solid MS and, after 4 d, transferred to control plates or plates that were supplemented with 75 mm NaCl. Elongation of the primary root was measured after 2 d as described above. For the analysis of gene expression in response to ABA, liquid-grown 10-d-old seedlings were treated for the indicated times with ABA or MeOH (as control) and frozen in liquid nitrogen.
The permeability of the cuticle of 4-week-old rosette leaves was performed as previously described (Curvers et al., 2010). For visualization of ROS accumulation, leaves were stained with 3,3′-diaminobenzidine (Sigma-Aldrich) as previously described (Ferrari et al., 2008), with the only difference that samples were incubated in the presence of the dye
Gene Expression Analysis
Leaves or seedlings were frozen in liquid nitrogen and homogenized with a MM301 Ball Mill (Retsch) for about 2 min at 30 Hz, and total RNA was extracted with Isol-RNA Lysis Reagent (5′-Prime) according to the manufacturer’s instructions. RNA was treated with RQ1 DNase (Promega), and first-strand complementary DNA (cDNA) was synthesized using ImProm-II reverse transcriptase (Promega). PCR analysis was performed in 30 μL of reaction containing 2 μL of cDNA, 1× reaction buffer, 25 mm MgCl2, 100 µm each deoxynucleotide triphosphate, 0.5 µm of each primer, and 1 unit of Taq DNA Polymerase (RBC-Bioscience). Primer sequences are shown in Supplemental Table S1. Twenty-eight, 30, and 35 cycles were performed for each primer pair to verify linearity of the amplification. PCR products were separated by agarose gel electrophoresis and stained with ethidium bromide.
qPCR analysis was performed using a CFX96 Real-Time System (Bio-Rad). One microliter of cDNA (corresponding to 50 ng of total RNA) was amplified in 30 μL of reaction mix containing 1× Go Taq qPCR Master Mix (Promega) and 0.4 μm of each primer. Expression levels of each gene, relative to the UBIQUITIN5 (UBQ5) gene, were determined using a modification of the Pfaffl method (Pfaffl, 2001) as previously described (Ferrari et al., 2006).
Metaanalysis of publicly available microarray data were performed using the Genevestigator database (https://www.genevestigator.com/gv/plant.jsp; Zimmermann et al., 2004).
Generation of Transgenic Plants and Imaging
A 2,849-bp DNA fragment corresponding to the genomic sequence of AtLYK3 from the start to the last codon of the predicted open reading frame, excluding the stop codon, was amplified with GoTaq DNA Polymerase (Promega) from ecotype Columbia genomic DNA using the following primer pair: L3FB1, GGGGACAAGTTTGTACAAAAAAGCAGGCTCAATGAATCTTACCTTCTACATCTTCTTC; and L3RB2, GGGGACCACTTTGTACAAGAAAGCTGGGTATCTTCCTTGGACTAGACCACTAAAG (the attB sequences, which were introduced for subsequent recombination in Gateway-based plasmids [Karimi et al., 2007a, 2007b], are underlined). The PCR product was cloned into the pDONR221/Zeo vector (Life Technologies) using BP Clonase (Life Technologies), following the manufacturer’s instructions. The resulting plasmid (pENTR-AtLYK3) was introduced in Escherichia coli JM109, and transformants were selected on low-salt LB medium containing 30 µg mL–1 zeocin. After plasmid purification, a three-fragment recombination was performed using LR Clonase II (Life Technologies) to generate a binary vector for the constitutive expression of AtLYK3 translationally fused at the C terminus to GFP. The reaction included, in addition to pENTR-AtLYK3, the Gateway-compatible plasmids pEN-L4-2-R1,0, containing the CaMV 35S promoter sequence; pEN-R2-F-L3,0, containing the GFP sequence; and the binary destination vector pB7m34GW,0, which were all obtained from the Department of Plant Systems Biology of the Vlaams Instituut voor Biotechnologie-Ghent University (http://gateway.psb.ugent.be/). The recombination reaction was then used to transform E. coli OmniMAX 2-T1 electrocompetent cells (Invitrogen), and transformed cells were selected on low-salt LB medium containing 120 µg mL–1 spectinomycin (Duchefa-Biochemie). The obtained plasmid was introduced in Agrobacterium tumefaciens GV3101 by electroporation. Arabidopsis plants were transformed by floral dip (Clough and Bent, 1998), and transgenic seedlings were selected on MS plates containing 20 µg mL–1 phosphinothricin.
For colocalization experiments, detached cotyledons of T1 seedlings were stained for 15 min with 4 mm FM4-64 (Life Technologies; Brandizzi et al., 2004), and images were obtained using a confocal laser scanning microscope (LSM 780 NLO; Zeiss). For plasmolysis experiments, an inverted spinning-disc confocal microscope (CarvX, CrEST) was used. Imaging was performed using CFI Planfluo 40× (Numerical Aperture: 1,4) oil immersion objectives (Nikon) through a 70-μm pinhole disc set at 6,000 rpm. GFP was excited using 473-nm wavelength laser light, while FM4-64 was excited with 532-nm laser light, and both were detected using a cooled CCD camera (CoolSNAP HQ2, Photometrics) through ω band-pass filters (XF100-2 and XF101-2, respectively). The CCD camera, Z-motor, and confocal head were controlled with Metamorph software (Molecular Devices).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At1g51940 (AtLYK3), At3g62250 (UBQ5), At3g26830 (PAD3), At2g14610 (PR1), At5g44420 (PDF1.2), At5g66400 (RAB18), At4g26080 (ABI1), At4g17870 (PYR1), At2g38310 (PYL4), At1g52340 (ABA2), and At3g21630 (AtCERK1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression of Arabidopsis AtLYK genes in response to pathogens and elicitors.
Supplemental Figure S2. Isolation of insertional mutants for AtLYK3.
Supplemental Figure S3. Accumulation of ROS in response to fungal infection is not affected by AtLYK3.
Supplemental Figure S4. Expression of AtLYK3 and PAD3 in atlyk3-1 pad3 double mutants.
Supplemental Figure S5. AtLYK3 is required for resistance to salt stress.
Supplemental Figure S6. ABA-induced physiological responses are independent of PAD3.
Supplemental Figure S7. AtLYK3 is not required for early responses to ABA.
Supplemental Figure S8. Cuticle permeability is unaffected in atlyk3-1 and atlyk3-3 plants.
Supplemental Figure S9. Responses to flg22 in atlyk3 mutants.
Supplemental Figure S10. AtLYK3-GFP localization in plasmolyzed cells.
Supplemental Table S1. Primers for RT-PCR and qPCR used in this work.
Supplementary Material
Glossary
- ABA
abscisic acid
- MAMP
microbe-associated molecular pattern
- PGN
peptidoglycan
- DAMP
damage-associated molecular pattern
- OG
oligogalacturonide
- SA
salicylic acid
- JA
jasmonic acid
- NAG
N-acetylglucosamine
- LYP
lysin motif transmembrane receptor-like protein
- qPCR
quantitative PCR
- RT
reverse transcription
- T-DNA
transfer DNA
- dpi
days post infection
- ROS
reactive oxygen species
- CaMV
Cauliflower mosaic virus
- EV
empty vector
- MS
Murashige and Skoog
- LB
Luria-Bertani
- cDNA
complementary DNA
Footnotes
This work was supported by the Ministero dell’Istruzione, dell’Università e della Ricerca (grant nos. PRIN2009, WTCJL8, and FIRB ERA–PG RBER063SN4 to G.D.L.), the European Research Council (Advanced Grant no. 233083 “FUEL-PATH”), the Istituto Pasteur-Fondazione Cenci-Bolognetti, and the Sapienza Università di Roma (Ricerche Universitarie grant no. C26A10ME4X to G.D.L.).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Adie BA, Pérez-Pérez J, Pérez-Pérez MM, Godoy M, Sánchez-Serrano JJ, Schmelz EA, Solano R. (2007) ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 19: 1665–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Arnaud D, Desclos-Theveniau M, Zimmerli L. (2012) Disease resistance to Pectobacterium carotovorum is negatively modulated by the Arabidopsis Lectin Receptor Kinase LecRK-V.5. Plant Signal Behav 7: 1070–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselbergh B, Achuo AE, Höfte M, Van Gijsegem F. (2008a) Abscisic acid deficiency leads to rapid activation of tomato defence responses upon infection with Erwinia chrysanthemi. Mol Plant Pathol 9: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselbergh B, Curvers K, Franca SC, Audenaert K, Vuylsteke M, Van Breusegem F, Höfte M. (2007) Resistance to Botrytis cinerea in sitiens, an abscisic acid-deficient tomato mutant, involves timely production of hydrogen peroxide and cell wall modifications in the epidermis. Plant Physiol 144: 1863–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselbergh B, De Vleesschauwer D, Höfte M. (2008b) Global switches and fine-tuning-ABA modulates plant pathogen defense. Mol Plant Microbe Interact 21: 709–719 [DOI] [PubMed] [Google Scholar]
- Audenaert K, De Meyer GB, Höfte MM. (2002a) Abscisic acid determines basal susceptibility of tomato to Botrytis cinerea and suppresses salicylic acid-dependent signaling mechanisms. Plant Physiol 128: 491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audenaert K, Pattery T, Cornelis P, Höfte M. (2002b) Induction of systemic resistance to Botrytis cinerea in tomato by Pseudomonas aeruginosa 7NSK2: role of salicylic acid, pyochelin, and pyocyanin. Mol Plant Microbe Interact 15: 1147–1156 [DOI] [PubMed] [Google Scholar]
- Bari R, Jones JD. (2009) Role of plant hormones in plant defence responses. Plant Mol Biol 69: 473–488 [DOI] [PubMed] [Google Scholar]
- Bateman A, Bycroft M. (2000) The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD). J Mol Biol 299: 1113–1119 [DOI] [PubMed] [Google Scholar]
- Bessire M, Chassot C, Jacquat AC, Humphry M, Borel S, Petétot JM, Métraux JP, Nawrath C. (2007) A permeable cuticle in Arabidopsis leads to a strong resistance to Botrytis cinerea. EMBO J 26: 2158–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi ME. (2007) DAMPs, PAMPs and alarmins: all we need to know about danger. J Leukoc Biol 81: 1–5 [DOI] [PubMed] [Google Scholar]
- Boller T, Felix G. (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Brandizzi F, Irons SL, Johansen J, Kotzer A, Neumann U. (2004) GFP is the way to glow: bioimaging of the plant endomembrane system. J Microsc 214: 138–158 [DOI] [PubMed] [Google Scholar]
- Brotman Y, Landau U, Pnini S, Lisec J, Balazadeh S, Mueller-Roeber B, Zilberstein A, Willmitzer L, Chet I, Viterbo A. (2012) The LysM receptor-like kinase LysM RLK1 is required to activate defense and abiotic-stress responses induced by overexpression of fungal chitinases in Arabidopsis plants. Mol Plant 5: 1113–1124 [DOI] [PubMed] [Google Scholar]
- Cao FY, Yoshioka K, Desveaux D. (2011) The roles of ABA in plant-pathogen interactions. J Plant Res 124: 489–499 [DOI] [PubMed] [Google Scholar]
- Chassot C, Nawrath C, Métraux JP. (2007) Cuticular defects lead to full immunity to a major plant pathogen. Plant J 49: 972–980 [DOI] [PubMed] [Google Scholar]
- Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM. (2009) Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323: 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Curvers K, Seifi H, Mouille G, de Rycke R, Asselbergh B, Van Hecke A, Vanderschaeghe D, Höfte H, Callewaert N, Van Breusegem F, et al. (2010) Abscisic acid deficiency causes changes in cuticle permeability and pectin composition that influence tomato resistance to Botrytis cinerea. Plant Physiol 154: 847–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de las Mercedes DM, Pintor-Toro JA, Cubero B. (2006) Transgenic tobacco plants overexpressing chitinases of fungal origin show enhanced resistance to biotic and abiotic stress agents. Plant Physiol 142: 722–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lorenzo G, Ferrari S. (2002) Polygalacturonase-inhibiting proteins in defense against phytopathogenic fungi. Curr Opin Plant Biol 5: 295–299 [DOI] [PubMed] [Google Scholar]
- de Torres-Zabala M, Truman W, Bennett MH, Lafforgue G, Mansfield JW, Rodriguez Egea P, Bögre L, Grant M. (2007) Pseudomonas syringae pv tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO J 26: 1434–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoux C, Galletti R, Mammarella N, Gopalan S, Werck D, De Lorenzo G, Ferrari S, Ausubel FM, Dewdney J. (2008) Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Mol Plant 1: 423–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desclos-Theveniau M, Arnaud D, Huang TY, Lin GJ, Chen WY, Lin YC, Zimmerli L. (2012) The Arabidopsis lectin receptor kinase LecRK-V.5 represses stomatal immunity induced by Pseudomonas syringae pv tomato DC3000. PLoS Pathog 8: e1002513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doares SH, Syrovets T, Weiler EW, Ryan CA. (1995) Oligogalacturonides and chitosan activate plant defensive genes through the octadecanoid pathway. Proc Natl Acad Sci USA 92: 4095–4098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyachok JV, Wiweger M, Kenne L, von Arnold S. (2002) Endogenous Nod-factor-like signal molecules promote early somatic embryo development in Norway spruce. Plant Physiol 128: 523–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K, Johnstone C, Thompson C. (1991) A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res 19: 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbs G, Silipo A, Aslam S, De Castro C, Liparoti V, Flagiello A, Pucci P, Lanzetta R, Parrilli M, Molinaro A, et al. (2008) Peptidoglycan and muropeptides from pathogens Agrobacterium and Xanthomonas elicit plant innate immunity: structure and activity. Chem Biol 15: 438–448 [DOI] [PubMed] [Google Scholar]
- Federici L, Di Matteo A, Fernandez-Recio J, Tsernoglou D, Cervone F. (2006) Polygalacturonase inhibiting proteins: players in plant innate immunity? Trends Plant Sci 11: 65–70 [DOI] [PubMed] [Google Scholar]
- Ferrari S, Galletti R, Denoux C, De Lorenzo G, Ausubel FM, Dewdney J. (2007) Resistance to Botrytis cinerea induced in Arabidopsis by elicitors is independent of salicylic acid, ethylene, or jasmonate signaling but requires PHYTOALEXIN DEFICIENT3. Plant Physiol 144: 367–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S, Galletti R, Pontiggia D, Manfredini C, Lionetti V, Bellincampi D, Cervone F, De Lorenzo G. (2008) Transgenic expression of a fungal endo-polygalacturonase increases plant resistance to pathogens and reduces auxin sensitivity. Plant Physiol 146: 669–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S, Galletti R, Vairo D, Cervone F, De Lorenzo G. (2006) Antisense expression of the Arabidopsis thaliana AtPGIP1 gene reduces polygalacturonase-inhibiting protein accumulation and enhances susceptibility to Botrytis cinerea. Mol Plant Microbe Interact 19: 931–936 [DOI] [PubMed] [Google Scholar]
- Ferrari S, Plotnikova JM, De Lorenzo G, Ausubel FM. (2003) Arabidopsis local resistance to Botrytis cinerea involves salicylic acid and camalexin and requires EDS4 and PAD2, but not SID2, EDS5 or PAD4. Plant J 35: 193–205 [DOI] [PubMed] [Google Scholar]
- Ferrari S, Savatin DV, Sicilia F, Gramegna G, Cervone F, Lorenzo GD. (2013) Oligogalacturonides: plant damage-associated molecular patterns and regulators of growth and development. Front Plant Sci 4: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletti R, De Lorenzo G, Ferrari S. (2009) Host-derived signals activate plant innate immunity. Plant Signal Behav 4: 33–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletti R, Denoux C, Gambetta S, Dewdney J, Ausubel FM, De Lorenzo G, Ferrari S. (2008) The AtrbohD-mediated oxidative burst elicited by oligogalacturonides in Arabidopsis is dispensable for the activation of defense responses effective against Botrytis cinerea. Plant Physiol 148: 1695–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. (2001) Genes controlling expression of defense responses in Arabidopsis—2001 status. Curr Opin Plant Biol 4: 301–308 [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Ausubel FM. (1994) Isolation of phytoalexin-deficient mutants of Arabidopsis thaliana and characterization of their interactions with bacterial pathogens. Proc Natl Acad Sci USA 91: 8955–8959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez L, Boller T. (2000) FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L, Felix G, Boller T. (1999) A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J 18: 277–284 [DOI] [PubMed] [Google Scholar]
- González-Guzmán M, Apostolova N, Bellés JM, Barrero JM, Piqueras P, Ponce MR, Micol JL, Serrano R, Rodríguez PL. (2002) The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell 14: 1833–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gust AA, Biswas R, Lenz HD, Rauhut T, Ranf S, Kemmerling B, Götz F, Glawischnig E, Lee J, Felix G, et al. (2007) Bacteria-derived peptidoglycans constitute pathogen-associated molecular patterns triggering innate immunity in Arabidopsis. J Biol Chem 282: 32338–32348 [DOI] [PubMed] [Google Scholar]
- Gust AA, Willmann R, Desaki Y, Grabherr HM, Nürnberger T. (2012) Plant LysM proteins: modules mediating symbiosis and immunity. Trends Plant Sci 17: 495–502 [DOI] [PubMed] [Google Scholar]
- Igarashi D, Tsuda K, Katagiri F. (2012) The peptide growth factor, phytosulfokine, attenuates pattern-triggered immunity. Plant J 71: 194–204 [DOI] [PubMed] [Google Scholar]
- Iizasa E, Mitsutomi M, Nagano Y. (2010) Direct binding of a plant LysM receptor-like kinase, LysM RLK1/CERK1, to chitin in vitro. J Biol Chem 285: 2996–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaku H, Nishizawa Y, Ishii-Minami N, Akimoto-Tomiyama C, Dohmae N, Takio K, Minami E, Shibuya N. (2006) Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc Natl Acad Sci USA 103: 11086–11091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Bleys A, Vanderhaeghen R, Hilson P. (2007a) Building blocks for plant gene assembly. Plant Physiol 145: 1183–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Depicker A, Hilson P. (2007b) Recombinational cloning with plant gateway vectors. Plant Physiol 145: 1144–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariola T, Brader G, Helenius E, Li J, Heino P, Palva ET. (2006) EARLY RESPONSIVE TO DEHYDRATION 15, a negative regulator of abscisic acid responses in Arabidopsis. Plant Physiol 142: 1559–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Hauser F, Ha T, Xue S, Böhmer M, Nishimura N, Munemasa S, Hubbard K, Peine N, Lee BH, et al. (2011) Chemical genetics reveals negative regulation of abscisic acid signaling by a plant immune response pathway. Curr Biol 21: 990–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Léon-Kloosterziel KM, Schwartz SH, Zeevaart JAD. (1998) The genetic and molecular dissection of abscisic acid biosynthesis and signal transduction in Arabidopsis. Plant Physiol Biochem 36: 83–89 [Google Scholar]
- Kumar V, Parkhi V, Kenerley CM, Rathore KS. (2009) Defense-related gene expression and enzyme activities in transgenic cotton plants expressing an endochitinase gene from Trichoderma virens in response to interaction with Rhizoctonia solani. Planta 230: 277–291 [DOI] [PubMed] [Google Scholar]
- L’Haridon F, Besson-Bard A, Binda M, Serrano M, Abou-Mansour E, Balet F, Schoonbeek HJ, Hess S, Mir R, Léon J, et al. (2011) A permeable cuticle is associated with the release of reactive oxygen species and induction of innate immunity. PLoS Pathog 7: e1002148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laby RJ, Kincaid MS, Kim D, Gibson SI. (2000) The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J 23: 587–596 [DOI] [PubMed] [Google Scholar]
- Lång V, Palva ET. (1992) The expression of a rab-related gene, rab18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.) Heynh. Plant Mol Biol 20: 951–962 [DOI] [PubMed] [Google Scholar]
- Liang Y, Cao Y, Tanaka K, Thibivilliers S, Wan J, Choi J, Kang Ch, Qiu J, Stacey G. (2013) Nonlegumes respond to rhizobial Nod factors by suppressing the innate immune response. Science 341: 1384–1387 [DOI] [PubMed] [Google Scholar]
- Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R. (2003) LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302: 630–633 [DOI] [PubMed] [Google Scholar]
- Liu T, Liu Z, Song C, Hu Y, Han Z, She J, Fan F, Wang J, Jin C, Chang J, et al. (2012) Chitin-induced dimerization activates a plant immune receptor. Science 336: 1160–1164 [DOI] [PubMed] [Google Scholar]
- Luna E, Pastor V, Robert J, Flors V, Mauch-Mani B, Ton J. (2011) Callose deposition: a multifaceted plant defense response. Mol Plant Microbe Interact 24: 183–193 [DOI] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Mackey D, McFall AJ. (2006) MAMPs and MIMPs: proposed classifications for inducers of innate immunity. Mol Microbiol 61: 1365–1371 [DOI] [PubMed] [Google Scholar]
- Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N, et al. (2003) A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425: 637–640 [DOI] [PubMed] [Google Scholar]
- Maleck K, Levine A, Eulgem T, Morgan A, Schmid J, Lawton KA, Dangl JL, Dietrich RA. (2000) The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat Genet 26: 403–410 [DOI] [PubMed] [Google Scholar]
- Mauch-Mani B, Mauch F. (2005) The role of abscisic acid in plant-pathogen interactions. Curr Opin Plant Biol 8: 409–414 [DOI] [PubMed] [Google Scholar]
- Meyer K, Leube MP, Grill E. (1994) A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science 264: 1452–1455 [DOI] [PubMed] [Google Scholar]
- Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, Narusaka Y, Kawakami N, Kaku H, Shibuya N. (2007) CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci USA 104: 19613–19618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher S, Moeder W, Nishimura N, Jikumaru Y, Joo SH, Urquhart W, Klessig DF, Kim SK, Nambara E, Yoshioka K. (2010) The lesion-mimic mutant cpr22 shows alterations in abscisic acid signaling and abscisic acid insensitivity in a salicylic acid-dependent manner. Plant Physiol 152: 1901–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) Revised medium for rapid growth and bioassays with tobacco cultures. Physiol Plant 15: 437–479 [Google Scholar]
- Newman MA, Dow JM, Molinaro A, Parrilli M. (2007) Priming, induction and modulation of plant defence responses by bacterial lipopolysaccharides. J Endotoxin Res 13: 69–84 [DOI] [PubMed] [Google Scholar]
- Padmanabhan M, Cournoyer P, Dinesh-Kumar SP. (2009) The leucine-rich repeat domain in plant innate immunity: a wealth of possibilities. Cell Microbiol 11: 191–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx IA, Eggermont K, Terras FR, Thomma BP, De Samblanx GW, Buchala A, Métraux JP, Manners JM, Broekaert WF. (1996) Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 8: 2309–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petutschnig EK, Jones AM, Serazetdinova L, Lipka U, Lipka V. (2010) The lysin motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. J Biol Chem 285: 28902–28911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povero G, Loreti E, Pucciariello C, Santaniello A, Di Tommaso D, Di Tommaso G, Kapetis D, Zolezzi F, Piaggesi A, Perata P. (2011) Transcript profiling of chitosan-treated Arabidopsis seedlings. J Plant Res 124: 619–629 [DOI] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Jurkiewicz A, Fukai E, Quistgaard EM, Albrektsen AS, James EK, Thirup S, Stougaard J. (2007) LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J 26: 3923–3935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remy W, Taylor TN, Hass H, Kerp H. (1994) Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc Natl Acad Sci USA 91: 11841–11843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Vallet A, López G, Ramos B, Delgado-Cerezo M, Riviere MP, Llorente F, Fernández PV, Miedes E, Estevez JM, Grant M, et al. (2012) Disruption of abscisic acid signaling constitutively activates Arabidopsis resistance to the necrotrophic fungus Plectosphaerella cucumerina. Plant Physiol 160: 2109–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Nakano T, Takamizawa D, Desaki Y, Ishii-Minami N, Nishizawa Y, Minami E, Okada K, Yamane H, Kaku H, et al. (2010) Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J 64: 204–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siewers V, Smedsgaard J, Tudzynski P. (2004) The P450 monooxygenase BcABA1 is essential for abscisic acid biosynthesis in Botrytis cinerea. Appl Environ Microbiol 70: 3868–3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Nguyen CT, Liang Y, Cao Y, Stacey G. (2012) Role of LysM receptors in chitin-triggered plant innate immunity. Plant Signal Behav 8: e22598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Xie Z, Chen W, Glazebrook J, Chang HS, Han B, Zhu T, Zou G, Katagiri F. (2003) Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 15: 317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton J, Mauch-Mani B. (2004) β-Amino-butyric acid-induced resistance against necrotrophic pathogens is based on ABA-dependent priming for callose. Plant J 38: 119–130 [DOI] [PubMed] [Google Scholar]
- van Hengel AJ, Tadesse Z, Immerzeel P, Schols H, van Kammen A, de Vries SC. (2001) N-Acetylglucosamine and glucosamine-containing arabinogalactan proteins control somatic embryogenesis. Plant Physiol 125: 1880–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Tanaka K, Zhang XC, Son GH, Brechenmacher L, Nguyen TH, Stacey G. (2012) LYK4, a lysin motif receptor-like kinase, is important for chitin signaling and plant innate immunity in Arabidopsis. Plant Physiol 160: 396–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Zhang XC, Neece D, Ramonell KM, Clough S, Kim SY, Stacey MG, Stacey G. (2008) A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20: 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasilewska A, Vlad F, Sirichandra C, Redko Y, Jammes F, Valon C, Frei dit Frey N, Leung J. (2008) An update on abscisic acid signaling in plants and more... Mol Plant 1: 198–217 [DOI] [PubMed] [Google Scholar]
- Willmann R, Lajunen HM, Erbs G, Newman MA, Kolb D, Tsuda K, Katagiri F, Fliegmann J, Bono JJ, Cullimore JV, et al. (2011) Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc Natl Acad Sci USA 108: 19824–19829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda M, Ishikawa A, Jikumaru Y, Seki M, Umezawa T, Asami T, Maruyama-Nakashita A, Kudo T, Shinozaki K, Yoshida S, et al. (2008) Antagonistic interaction between systemic acquired resistance and the abscisic acid-mediated abiotic stress response in Arabidopsis. Plant Cell 20: 1678–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka K, Moeder W, Kang HG, Kachroo P, Masmoudi K, Berkowitz G, Klessig DF. (2006) The chimeric Arabidopsis CYCLIC NUCLEOTIDE-GATED ION CHANNEL11/12 activates multiple pathogen resistance responses. Plant Cell 18: 747–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Ramonell K, Somerville S, Stacey G. (2002) Characterization of early, chitin-induced gene expression in Arabidopsis. Mol Plant Microbe Interact 15: 963–970 [DOI] [PubMed] [Google Scholar]
- Zhang XC, Cannon SB, Stacey G. (2009) Evolutionary genomics of LysM genes in land plants. BMC Evol Biol 9: 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XC, Wu X, Findley S, Wan J, Libault M, Nguyen HT, Cannon SB, Stacey G. (2007) Molecular evolution of lysin motif-type receptor-like kinases in plants. Plant Physiol 144: 623–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Goritschnig S, Dong X, Li X. (2003) A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell 15: 2636–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N, Tootle TL, Glazebrook J. (1999) Arabidopsis PAD3, a gene required for camalexin biosynthesis, encodes a putative cytochrome P450 monooxygenase. Plant Cell 11: 2419–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JDG, Boller T, Felix G. (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T. (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.