The hydrates formed from NADH and NADPH by chemical or enzymatic damage are repaired in plants by highly conserved enzymes that are targeted to multiple compartments.
Abstract
NADH and NADPH undergo spontaneous and enzymatic reactions that produce R and S forms of NAD(P)H hydrates [NAD(P)HX], which are not electron donors and inhibit various dehydrogenases. In bacteria, yeast (Saccharomyces cerevisiae), and mammals, these hydrates are repaired by the tandem action of an ADP- or ATP-dependent dehydratase that converts (S)-NAD(P)HX to NAD(P)H and an epimerase that facilitates interconversion of the R and S forms. Plants have homologs of both enzymes, the epimerase homolog being fused to the vitamin B6 salvage enzyme pyridoxine 5′-phosphate oxidase. Recombinant maize (Zea mays) and Arabidopsis (Arabidopsis thaliana) NAD(P)HX dehydratases (GRMZM5G840928, At5g19150) were able to reconvert (S)-NAD(P)HX to NAD(P)H in an ATP-dependent manner. Recombinant maize and Arabidopsis epimerases (GRMZM2G061988, At5g49970) rapidly interconverted (R)- and (S)-NAD(P)HX, as did a truncated form of the Arabidopsis epimerase lacking the pyridoxine 5′-phosphate oxidase domain. All plant NAD(P)HX dehydratase and epimerase sequences examined had predicted organellar targeting peptides with a potential second start codon whose use would eliminate the targeting peptide. In vitro transcription/translation assays confirmed that both start sites were used. Dual import assays with purified pea (Pisum sativum) chloroplasts and mitochondria, and subcellular localization of GFP fusion constructs in tobacco (Nicotiana tabacum) suspension cells, indicated mitochondrial, plastidial, and cytosolic localization of the Arabidopsis epimerase and dehydratase. Ablation of the Arabidopsis dehydratase gene raised seedling levels of all NADHX forms by 20- to 40-fold, and levels of one NADPHX form by 10- to 30-fold. We conclude that plants have a canonical two-enzyme NAD(P)HX repair system that is directed to three subcellular compartments via the use of alternative translation start sites.
Many metabolites are subject to damaging reactions that occur spontaneously or are mediated by side activities of enzymes (Golubev, 1996; D’Ari and Casadesús, 1998; Tawfik, 2010; Copley, 2012). The products of such chemical or enzymatic damage are essentially useless and can be toxic (Linster et al., 2013). Given its prevalence, metabolite damage has been overlooked to a surprising extent, as have the mechanisms by which cells deal with it (Van Schaftingen et al., 2009; Vinci and Clarke, 2010). However, it is now increasingly recognized that damaged metabolites are ubiquitous, and that they may be reconverted to their undamaged forms by highly conserved, but hitherto unrecognized, repair enzymes (Galperin et al., 2006; Linster et al., 2013).
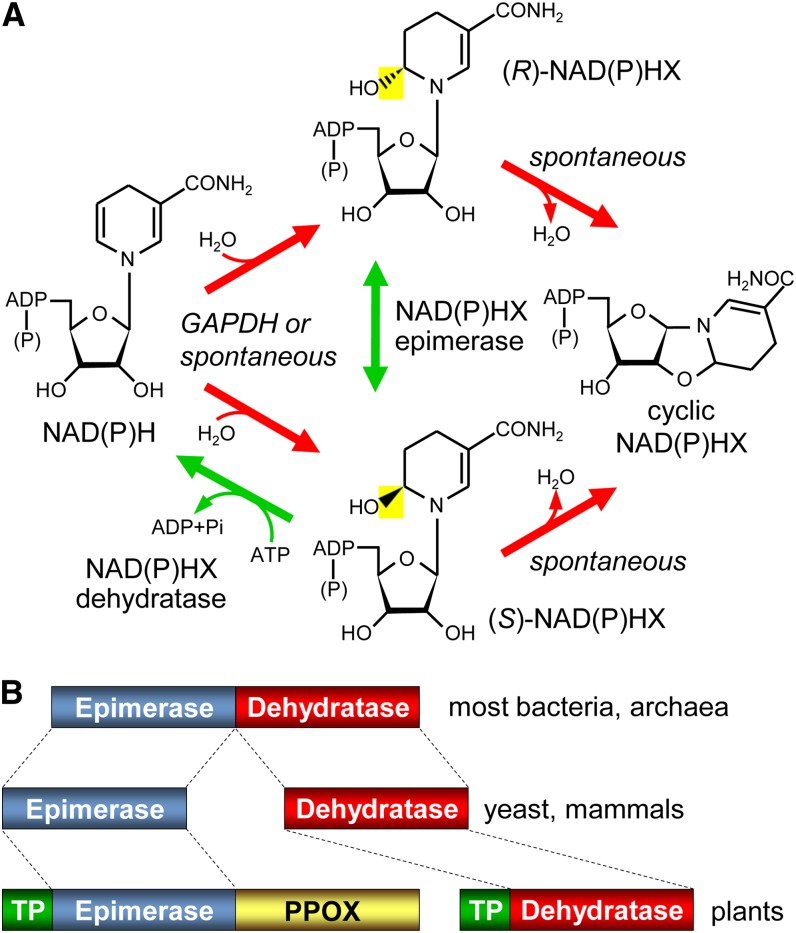
An iconic example of metabolite damage and repair is the formation of NADH and NADPH hydrates, which exist as R and S epimers [(R)- and (S)-NAD(P)HX], and their reconversion to NAD(P)H (Fig. 1A). Classical work in yeast (Saccharomyces cerevisiae) showed that a side reaction of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) converts NADH to NADHX (Rafter et al., 1954; Chaykin et al., 1956; Oppenheimer and Kaplan, 1974), and that NAD(P)H hydration also occurs spontaneously at physiological pH and is promoted by high temperatures (Acheson et al., 1988; Marbaix et al., 2011). NADHX and NADPHX cannot serve as electron donors and inhibit several dehydrogenases (Yoshida and Dave, 1975; Prabhakar et al., 1998), making their removal important. An ATP-dependent yeast dehydratase that achieves this removal by converting NAD(P)HX back to NAD(P)H was discovered soon after the hydration reaction (Meinhart et al., 1956), was shown to be specific for the S epimer (Acheson et al., 1988), and was recently cloned (Marbaix et al., 2011). NAD(P)HX can also spontaneously form cyclic NAD(P)HX (Fig. 1A), for which no repair system is known.
Figure 1.
NAD(P)H damage and repair reactions, and domain structures of the repair enzymes. A, The spontaneous or GAPDH-mediated hydration of NAD(P)H, and the enzymatic epimerization and dehydratase reactions that reconvert the resulting hydrates, (R)- and (S)-NAD(P)HX, to NAD(P)H. NAD(P)H hydrates can also spontaneously cyclize. The bacterial dehydratase reaction is ADP dependent rather than ATP dependent. B, Domain architectures of the NAD(P)HX epimerase and dehydratase enzymes of prokaryotes, yeast, and mammals, and of epimerase and dehydratase homologs in plants. TP, Predicted organellar targeting peptide.
The yeast NAD(P)HX dehydratase was found to have homologs in mammals and prokaryotes (Fig. 1B), and those from mouse and Escherichia coli were confirmed to have (S)-NAD(P)HX dehydratase activity, the latter being ADP dependent rather than ATP dependent (Marbaix et al., 2011). The E. coli dehydratase and its counterparts in almost all other prokaryotes were shown to be fused to an N-terminal domain (Fig. 1B) that was predicted, and then demonstrated, to have NAD(P)HX epimerase activity (Marbaix et al., 2011). This epimerase domain has stand-alone homologs in yeast and mammals that were likewise shown to have NAD(P)HX epimerase activity (Marbaix et al., 2011). Yeast, mammals, and prokaryotes thus share an epimerase and dehydratase that, acting together, can reconvert either epimer of NAD(P)HX to NAD(P)H (Fig. 1A).
It is not known how plants deal with NAD(P)HX, but they have homologs of the dehydratase and epimerase with predicted organellar targeting peptides (Fig. 1B). Because of the centrality of NADH and NADPH in all metabolism, and especially that of NADPH in photosynthesis, we determined whether the plant NAD(P)HX dehydratase and epimerase homologs function in NAD(P)HX repair. We first characterized the biochemical activity and subcellular location of the Arabidopsis (Arabidopsis thaliana) and maize (Zea mays) enzymes, and then investigated the effect on NAD(P)HX levels of ablating the Arabidopsis gene specifying the dehydratase homolog.
RESULTS
Sequence Features of Plant NAD(P)HX Dehydratase and Epimerase Homologs
Whereas plant NAD(P)HX dehydratase homologs are like those of yeast and mammals in being stand-alone proteins, plant epimerase homologs have an extra C-terminal domain that belongs to the pyridoxine/pyridoxamine phosphate oxidase (PPOX) family (Cluster of Orthologs no. COG0259; Fig. 1B). PPOX activities participate in the salvage of vitamin B6 (Gerdes et al., 2012). Both activities have been demonstrated in vitro for the PPOX domain of the Arabidopsis epimerase-PPOX protein (Sang et al., 2007). Relative to their prokaryotic homologs, plant NAD(P)HX dehydratase and epimerase-PPOX proteins have poorly conserved N-terminal extensions (Supplemental Fig. S1) that are predicted to be mitochondrial and/or plastidial targeting sequences (Supplemental Table S1). Close inspection of these sequences from diverse plants revealed that all have a downstream Met residue that, if used to initiate translation, would remove the targeting signal (Supplemental Fig. S1). The N-terminal regions of plant dehydratase and epimerase-PPOX proteins thus have features suggesting the possibility of triple location in mitochondria, plastids, and cytosol.
Enzymatic Activities of Arabidopsis and Maize Dehydratase and Epimerase Homologs
Arabidopsis and maize dehydratase and epimerase-PPOX coding sequences, each beginning at the second potential start codon, were expressed in E. coli as N-terminally His-tagged proteins, purified by Ni2+-affinity chromatography (Supplemental Fig. S2), and used to assay enzyme activities.
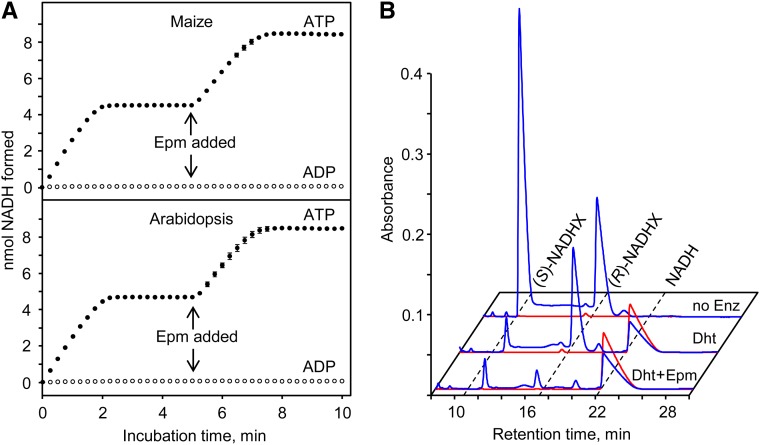
The expected dehydratase and epimerase activities were both tested using a spectrophotometric method in which the conversion of NAD(P)HX to NAD(P)H is monitored (Marbaix et al., 2011). When a purified equilibrium mixture of (S)- and (R)-NADHX (55:45 ratio; Marbaix et al., 2011) was incubated with ATP and Arabidopsis or maize dehydratase, NADH accumulated to a level equivalent to 55% of the total NADHX added to the assay (Fig. 2A), indicating that all (S)-NADHX in the mixture had been converted. No NADH formation was detected when ADP replaced ATP (Fig. 2A). Subsequent addition of the corresponding epimerase-PPOX protein to the reaction caused further accumulation of NADH to a level equal to the total NADHX in the assay (Fig. 2A). This second phase of NADH formation is consistent with epimerase-mediated conversion of (R)-NADHX to (S)-NADHX, followed by dehydration of the latter. Similar results were obtained for Arabidopsis and maize dehydratase and epimerase when a mixture of (S)- and (R)-NADPHX replaced (S)- and (R)-NADHX (not shown).
Figure 2.
In vitro activities of maize and Arabidopsis NAD(P)HX dehydratase and epimerase-PPOX proteins. A, Activities monitored spectrophotometrically at 22°C. Assays contained 25 mm Tris-HCl, pH 8.0, 5 mm KCl, 2 mm MgCl2, 0.1 mg/mL bovine serum albumin, 90 µm NADHX [55% (S)-NADHX, 45% (R)-NADHX], and 1 mm ATP (black circles) or ADP (white circles). Reactions were started by adding 5 µg of maize or Arabidopsis NAD(P)HX dehydratase and monitoring A340. At 5 min, 10 µg of the respective maize or Arabidopsis NAD(P)HX epimerase-PPOX (Epm) was added. Data are means ± se of three replicates; where no error bars appear, they are smaller than the symbols. B, Activities monitored by HPLC. Assays were as above but contained 1 mm NADHX, 5 mm ATP, and 20 µg of maize NAD(P)HX dehydratase (Dht) and epimerase-PPOX as indicated, and were incubated at 30°C for 30 min. A control without enzyme (no Enz) was included; the profile of this sample was the same as a sample given no incubation (not shown). Reactions were analyzed by HPLC, monitoring A279 (blue trace) and A340 nm (red trace). Peak fronts of (S)-NADHX, (R)-NADHX, and NADH are marked by dashed lines. Arabidopsis proteins gave similar results.
To confirm NAD(P)HX dehydration and isomerization directly, enzyme reactions were analyzed by HPLC. Analysis of a mixture of both NADHX epimers before incubation, or after incubation without enzymes, showed the presence of (S)- and (R)-NADHX in a 55:45 ratio (Fig. 2B). Addition of plant dehydratase caused the disappearance of (S)-NADHX and the appearance of an equivalent quantity of NADH, whereas (R)-NADHX was unaffected (Fig. 2B). Adding both plant dehydratase and epimerase caused the disappearance of both NADHX epimers and the appearance of NADH (Fig. 2B). As expected (Marbaix et al., 2011), the dehydratase caused no disappearance of cyclic NAD(P)HX (not shown).
Kinetic characterization of Arabidopsis and maize dehydratases using the spectrophotometric assay gave Km values for (S)-NADHX and (S)-NADPHX of 0.69 to 1.15 μm, catalytic constant (Kcat) values of 0.26 to 0.38 s−1, and Kcat/Km ratios that indicated a mild (less than 2-fold) preference for (S)-NADPHX (Table I). All of these values are near those for the mammalian and bacterial dehydratases (Marbaix et al., 2011).
Table I. Kinetic properties of Arabidopsis and maize NADHX and NADPHX dehydratases.
Activities were determined spectrophotometrically at 22°C, as described in “Materials and Methods.” Values are means ± se of three replicates.
| Species | Substrate | Km | Kcat | Kcat/Km |
|---|---|---|---|---|
| µm | s−1 | mm−1 s−1 | ||
| Arabidopsis | NADHX | 1.15 ± 0.26 | 0.26 ± 0.01 | 262 |
| NADPHX | 0.69 ± 0.26 | 0.34 ± 0.02 | 509 | |
| Maize | NADHX | 0.86 ± 0.21 | 0.30 ± 0.01 | 358 |
| NADPHX | 0.89 ± 0.39 | 0.38 ± 0.03 | 524 |
Together, the above results establish that plant dehydratases catalyze ATP-dependent conversion of (S)-NAD(P)HX to NAD(P)H, that the epimerases facilitate conversion of (R)-NAD(P)HX to (S)-NAD(P)HX, and that their kinetic properties are consistent with their playing these roles in vivo.
Epimerase Activity of Arabidopsis Epimerase-PPOX Does Not Require the PPOX Domain
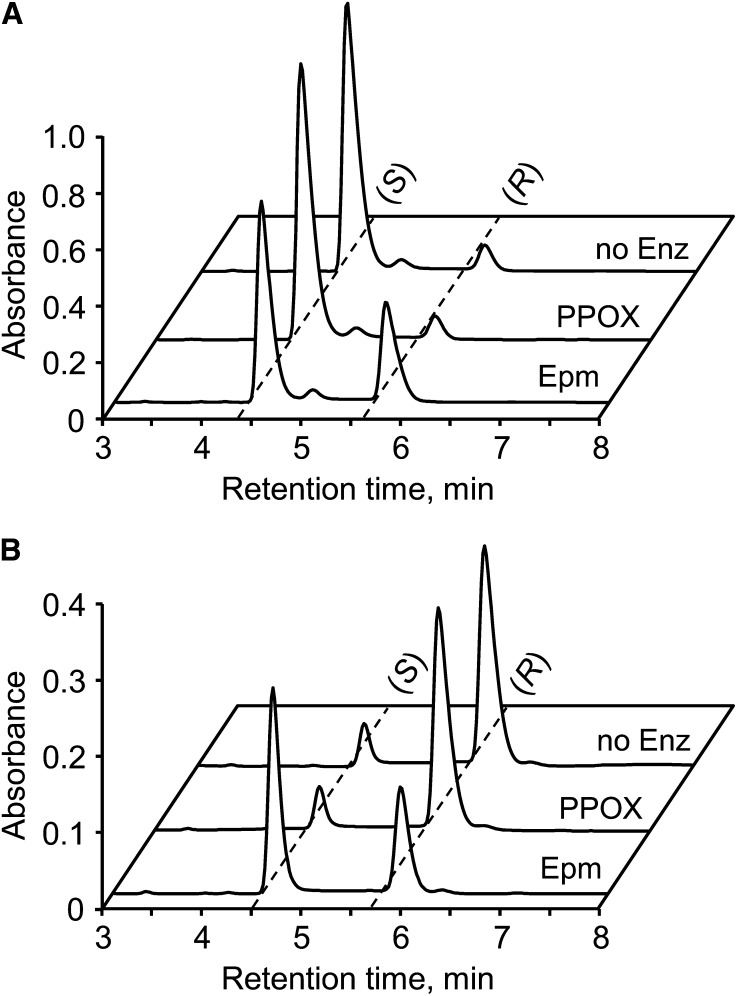
To test whether the epimerase domain is solely responsible for NAD(P)HX epimerase activity, the separate domains of the Arabidopsis epimerase-PPOX fusion were expressed in E. coli as N-terminally His-tagged proteins and purified (Supplemental Fig. S2). When the recombinant domains were incubated with partially purified (S)- or (R)-NADHX, adding the PPOX domain did not result in interconversion of (S) and (R) epimers, but adding the epimerase domain caused rapid conversion of (S)- to (R)-NADHX and vice versa until the (S)- to (R)-NADHX ratio reached the equilibrium value of 55:45 (Fig. 3). This result confirms that epimerase activity resides in the epimerase domain alone, and complements the finding that PPOX activity resides in the PPOX domain (Sang et al., 2007).
Figure 3.
Epimerase activity of the epimerase domain, but not the PPOX domain, of the Arabidopsis NAD(P)HX epimerase-PPOX protein. Assays contained 25 mm Tris-HCl, pH 9.0, 1 mm KCl, NADHX preparations enriched in (S)-NADHX (1 mm; A) or (R)-NADHX (0.5 mm; B), and 10 µg of the separate epimerase (Epm) or PPOX domains. Incubation was at 30°C for 10 min. Controls without enzyme (no Enz) were included. Reactions were analyzed by HPLC, monitoring A279. Peak fronts of (S)- and (R)-NADHX are marked by dashed lines.
Subcellular Localization of Dehydratase and Epimerase-PPOX
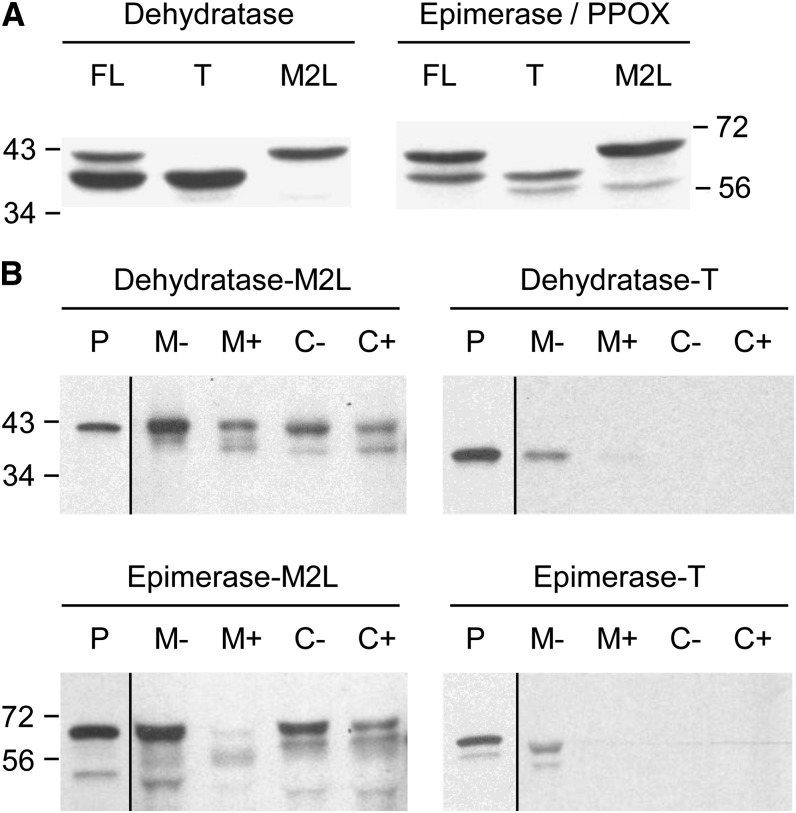
We first determined which of the potential start sites in the dehydratase and epimerase/PPOX are functional in vitro, using Arabidopsis complementary DNAs (cDNAs) and a coupled transcription-wheat germ (Triticum aestivum) translation system. Native full-length cDNAs of both genes gave two translation products, whereas cDNAs whose second predicted start codon was replaced by Leu (M2L) gave only the larger product and cDNAs truncated at the second start codon (T) gave only the smaller one (Fig. 4A). These results establish that the dehydratase and epimerase both have alternative start sites, at least in vitro.
Figure 4.
The in vitro translation products of Arabidopsis NAD(P)HX dehydratase and NAD(P)HX epimerase-PPOX, and their import into purified pea mitochondria and chloroplasts. A, Analysis of the products of in vitro transcription-translation. cDNAs that were full length (FL) or truncated to begin at the second predicted start Met (T), or with the second predicted start Met changed to Leu (M2L) were transcribed and translated in a wheat germ system containing [3H]Leu. The translation products were resolved by SDS-PAGE and visualized by fluorography. Positions of molecular mass markers (in kilodaltons) are indicated. The engineered (T and M2L) epimerase-PPOX constructs both give an additional minor translation product smaller than either of the others; this may reflect translation initiation at a third cryptic start site unmasked by the engineering. B, Dual import assays with pea chloroplasts and mitochondria. Dehydratase and epimerase-PPOX full-length sequences with the second Met changed to Leu (M2L), or truncated at the second Met (T), were translated in vitro as above. The translation products were incubated for 20 min in the light with mixed mitochondria (M) and chloroplasts (C), which were then reisolated using a Percoll gradient, without (−) or with (+) prior thermolysin treatment to remove adsorbed proteins. Proteins were separated by SDS-PAGE and visualized by fluorography. Samples were loaded next to an aliquot of the translation product (P).
To test whether the N-terminal regions of the full-length dehydratase and epimerase-PPOX are plastid or mitochondrial targeting peptides, we performed dual import assays with mixed pea (Pisum sativum) chloroplasts and mitochondria (Rudhe et al., 2002) using the full-length M2L and truncated T forms of the Arabidopsis proteins. The wild-type full-length forms of both proteins were not tested because, as noted above, both of these sequences gave two translation products, which could potentially yield ambiguous results. As shown in Figure 4B, the full-length M2L dehydratase was imported and processed by both organelles, but the truncated form was not. The full-length M2L epimerase-PPOX was imported and processed by chloroplasts, and to a lesser extent by mitochondria, but the truncated T form did not enter either organelle. These results indicate that both proteins have the potential for dual targeting to plastids and mitochondria, as bioinformatically predicted (Supplemental Table S1).
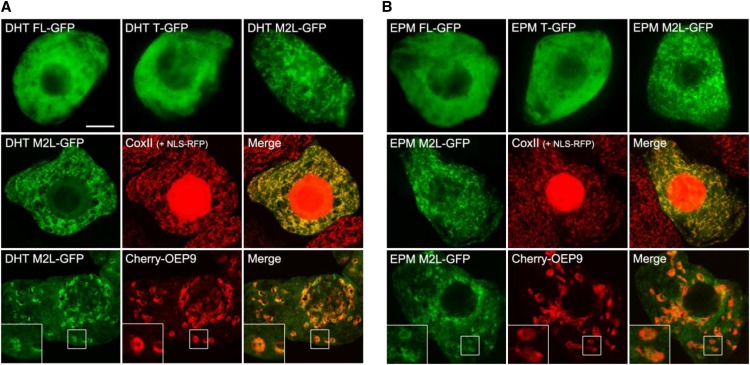
To explore localization in vivo, fluorescence microscopy was used to analyze Arabidopsis dehydratase and epimerase-PPOX C-terminal GFP fusions expressed transiently in tobacco (Nicotiana tabacum) suspension-cultured cells, serving as a well-characterized system for studying protein localization in plant cells (Brandizzi et al., 2003; Miao and Jiang, 2007; Fig. 5). Three types of fusion constructs were examined for each sequence: the wild-type full-length form, which can potentially be translated to yield proteins with or without the putative N-terminal targeting peptide; the truncated T form, which lacks the putative N-terminal targeting peptide; and the full-length M2L form, which forces translation of only the full-length protein, including its putative N-terminal targeting peptide. For both the dehydratase and the epimerase, the wild-type full-length and truncated T forms appeared to be localized primarily in the cytosol, as evidenced by the diffuse fluorescence patterns attributable to these constructs (Fig. 5, top row). By contrast, cells transformed with the full-length M2L versions of both proteins usually exhibited a distinct punctate fluorescence pattern (Fig. 5, top and middle rows) identical to that exhibited by the endogenous mitochondrial marker protein, cytochrome c oxidase subunit II, in the same cells (Fig. 5, middle row). Notably, in a small proportion (approximately 5% to 10%) of cells transformed with the M2L dehydratase or epimerase, the proteins appeared to localize not to mitochondria, but rather to plastids, as well as to cytosol (i.e. both the M2L dehydratase and epimerase proteins in these cells partially colocalized with the coexpressed plastid marker protein, Outer Envelope TA protein9 [OEP9]; Dhanoa et al., 2010; Fig. 5, bottom row). Plastid targeting of the full-length wild-type epimerase-PPOX protein was also reported by Sang et al. (2011). Overall, the in vivo GFP fusion protein localization results indicate that, depending on the translation start site used, the dehydratase and epimerase-PPOX proteins can localize either to the cytosol and/or to mitochondria or plastids. These in vivo results are thus to some extent consistent with the in vitro data (Fig. 4).
Figure 5.
Representative epifluorescence and confocal micrographs of tobacco BY-2 cells transiently expressing GFP fused to the C-terminus of either the native full-length sequence (FL), the truncated sequence beginning at the second predicted start site (T), or the full-length sequence with the second start site Met changed to Leu (M2L) of Arabidopsis NAD(P)HX dehydratase (DHT; A) or Arabidopsis NAD(P)HX epimerase/PPOX (EPM; B). The top rows are epifluorescence images that show, for both dehydratase and epimerase, the localization of the FL and T fusions in the cytosol and the localization of the M2L fusions predominantly to mitochondria. The middle rows are confocal images showing colocalization of the dehydratase and epimerase M2L fusions with endogenous mitochondrial cytochrome c oxidase subunit II. Note that these cells also exhibit red fluorescence in the nucleus because of the coexpressed NLS-RFP fusion protein, which was used as a convenient cell transformation marker. The bottom rows are confocal images showing the colocalization of the dehydratase and epimerase M2L fusions with the plastid (outer envelope) marker protein Cherry-OEP9 that was observed in approximately 5% to 10% of transformed cells. The boxes represent the portion of the cells shown at higher magnification in the insets. NLS-RFP, Nuclear localization signal fused to the red fluorescent protein. Bar = 10 μm in A.
High-throughput proteomics data sets provide additional evidence for multisite localization of the plant epimerase-PPOX protein. Thus, two independent studies detected the Arabidopsis protein in enriched cytosolic fractions (Ito et al., 2011) and in mitochondria (Finkemeier et al., 2011), and unpublished experiments from the Plant Proteome Database (Sun et al., 2009) detected it in the chloroplast stroma. No publicly available proteomics data could be found for the plant dehydratase.
Impact of Ablating Arabidopsis NAD(P)HX Dehydratase on NAD(P)HX Levels
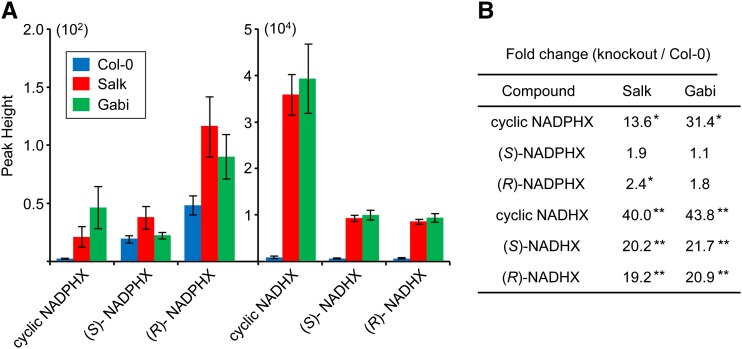
Two potential NAD(P)HX dehydratase knockout lines with transfer DNA (T-DNA) insertions in exons of At5g19150 were identified from the Salk collection (Salk_203790) and the GABI-Kat collection (Gabi_173F11). Homozygous plants were confirmed to lack detectable NAD(P)HX mRNA (Supplemental Fig. S3). Neither NAD(P)HX dehydratase knockout line differed visibly from the wild-type parental line when grown under our standard conditions. Targeted liquid chromatography-mass spectrometry (LC-MS) metabolomics (which measures relative changes in specific metabolites across samples) was used to compare levels of NAD(P)HX and of the corresponding pyrimidine nucleotides in 14-d-old seedlings of each knockout line with those in the wild type. There were 20- to 40-fold increases in (R)- and (S)-NADHX and in cyclic NADHX in both knockouts, as well as large fold changes in the cyclic form of NADPHX (Fig. 6; Supplemental Fig. S4, A–C). No consistent and significant increases or decreases in NAD+, NADH, NADP+, or NADPH were detected in the knockouts relative to the wild type (not shown). It should be noted that although the signals for NADHX forms were two orders of magnitude greater than those for NADPHX forms (Fig. 6A), this does not signify that NADHX is more abundant than NADPHX because signal strength in LC-MS is highly dependent on the behavior of molecules in the mass spectrometer (King et al., 2000). These data confirm that the dehydratase functions in vivo to limit the buildup of NAD(P)HX.
Figure 6.
Relative levels of pyridine nucleotide hydrates in Arabidopsis wild-type and NAD(P)HX dehydratase knockouts. Extracts of 14-d-old Arabidopsis wild-type (Col-0), Salk_203790 (Salk), or Gabi_173F11 (Gabi) seedlings (100 mg) were analyzed by LC-MS. A, Average peak areas of NADPHX forms (×102 scale) and NADHX forms (×104 scale) from six independent samples. B, The fold changes between knockouts and Col-0. *P < 0.05; **P < 0.01. Col-0, Ecotype Columbia 0 of Arabidopsis.
DISCUSSION
Our results provide biochemical and genetic evidence that Arabidopsis and maize NAD(P)HX dehydratase and epimerase homologs function in the coordinated repair of hydrated NAD(P)H. In vitro, the plant dehydratases resembled their mammalian and yeast homologs in being ATP dependent, and had kinetic constants for NADHX and NADPHX similar to those of other NAD(P)HX dehydratases. The plant epimerases, which are fused to PPOX, mediated rapid in vitro interconversion of (R)- and (S)-NAD(P)HX, again like their counterparts in other organisms. Substantiating the operation of the dehydratase in planta, we detected massive accumulations, relative to the wild type, of all forms of NADHX and of the cyclic form of NADPHX in two Arabidopsis dehydratase knockout lines.
Our targeted metabolomics data showing buildup of NAD(P)HX forms in knockout plants appear to be the first demonstration of NAD(P)HX in vivo for any organism. Although the fold accumulations of these hydrates were large, there was no indication that these accumulations were matched by depletions of the NAD(H) or NADP(H) pools. Our data cannot resolve whether the hydrates constituted only a small fraction of the total pools of pyridine nucleotides; however, if this were the case, it would be consistent with the lack of an obvious growth phenotype. We return to this point below.
The particular prominence of the cyclic forms of NADHX and NADPHX among the accumulated hydrate forms raises the question of whether they were present in vivo or formed from (R)- and (S)-NAD(P)HX during storage at −80°C, extraction, or analysis. Because we have observed that solutions, pH 8.0, of (R)- and (S)-NAD(P)HX standards undergo slow cyclization at −80°C, it is likely that some of the cyclic forms in our samples originated in this way. However, it is also likely that, in the absence of a repair system in the knockout plants, the accumulating (R)- and (S)- NAD(P)HX underwent some cyclization. The latter possibility is supported by the finding that the fold accumulations of the cyclic forms of NADHX and NADPHX were much higher than those of the noncyclic forms (Fig. 6A). Were the cyclic forms to have solely originated postmortem from the noncyclic forms, their fold accumulations would be expected to be the same. If substantial cyclization did take place in vivo in the knockout plants, it emphasizes that the dehydratase must normally intervene promptly to prevent the accumulation of cyclic forms, upon which it cannot act.
The subcellular localization data indicate that alternative translation start sites and dual targeting signals together place plant dehydratases and epimerases in the cytosol, mitochondria, and plastids. Analogous targeting to multiple compartments has been proposed for the mammalian dehydratase and epimerase (Marbaix et al., 2011). Dual targeting peptides that direct proteins to plastids and mitochondria are fairly common in plants (Xu et al., 2013). The use of alternative translation start sites to target proteins to different compartments appears to be less common, but is nevertheless well documented in plants (Chabregas et al., 2003; von Braun et al., 2007; Wamboldt et al., 2009) as well as in mammals and yeast (Porras et al., 2006; Kim et al., 2010). Targeting of the epimerase and dehydratase to multiple compartments is consistent with in situ repair of the spontaneous damage to NAD(P)H that is expected to occur throughout the cell. GAPDH-mediated damage to NAD(P)H is also expected to occur throughout plant cells because of the occurrence of NAD- and NADP-dependent cytosolic and plastidial GAPDH isoforms (Marri et al., 2005; Rius et al., 2006). It should be mentioned, however, that when transiently expressed in tobacco suspension cultures, wild-type full-length forms of both proteins fused to GFP appeared to localize primarily in cytosol, and the full-length M2L forms of both proteins localized either to mitochondria or plastids, but not to both organelles in the same cell (Fig. 5). These results suggest that although both proteins seem to have the ability to localize to the cytosol, mitochondria, and plastids, they are not necessarily targeted to these three intracellular destinations in all cells, implying that their intracellular localizations may vary depending on the cell type, developmental stage, or perhaps in response to particular physiological conditions.
No growth phenotype was observed in Arabidopsis NAD(P)HX dehydratase knockout lines grown in moderate conditions. This seems surprising on two counts. First, epimerase and dehydratase homologs occur in all domains of life and in nearly all organisms, suggesting that their function is ancient and almost universally important. Second, and relatedly, the Arabidopsis dehydratase and epimerase genes are expressed in all organs and under all conditions, as shown by microarray data sets (Steinhauser et al., 2004; Toufighi et al., 2005). However, the lack of an Arabidopsis mutant phenotype is matched by the near absence of growth phenotypes of the yeast dehydratase and epimerase (Breslow et al., 2008; Hillenmeyer et al., 2008) and E. coli dehydratase-epimerase (Nichols et al., 2011) knockouts in standard culture conditions. It remains to be established whether NAD(P)HX dehydratase becomes crucial to plants or other organisms under particular environmental conditions, such as high temperature, which greatly accelerates the spontaneous NAD(P)H hydration rate (Marbaix et al., 2011). The yeast dehydratase and epimerase genes are induced by heat shock (Gasch et al., 2000). However, Arabidopsis dehydratase and epimerase genes are not induced by heat or other stresses (Steinhauser et al., 2004; Toufighi et al., 2005), and the E. coli dehydratase-epimerase knockout shows little impairment of growth at any temperature (Nichols et al., 2011). The lack of marked growth phenotypes of knockout dehydratase mutants in diverse organisms under typical growth conditions implies that there may be spillover disposal routes for NAD(P)HX that limit the magnitude of its accumulation in knockouts (e.g. hydrolysis to ADP-Rib and 6-hydoxynicotinamide derivatives by an nicotinamide mononucleotide nucleosidase-like enzyme; Wagner et al., 1986).
Finally, we demonstrated that the previously mysterious N-terminal domain of plant PPOX proteins (Sang et al., 2007) is NAD(P)HX epimerase, and that this epimerase domain, like the PPOX domain (Sang et al., 2007), does not need its fusion partner for activity. The fusion of the epimerase to PPOX, an enzyme of vitamin B6 salvage, suggests a functional connection between the epimerase and B6 metabolism. Because there seems to be nothing to link the epimerase function in NAD(P)H metabolism with B6, we speculate that the epimerase has a second function that is specifically B6 related.
MATERIALS AND METHODS
Bioinformatics
Protein sequences were taken from GenBank, MaizeSeqence.org, and the JGI Genome Portal, and verified when necessary using ESTs from the GenBank dbEST database. Alignments were made with Multalin (Corpet, 1988). Subcellular localization was predicted using the algorithms given in Supplemental Table S1.
Chemicals
(R)- and (S)-NADHX and NADPHX and their cyclic forms were prepared essentially as described by Acheson et al. (1988). NADH or NADPH (25 mm) in 0.5 m Na phosphate, pH 6.0, was incubated for 30 min at 35°C; the pH was then brought to pH 8.0 with 5 m NaOH. (R)- and (S)-NAD(P)HX (as a mixture or separately) and their cyclic forms were then purified by HPLC on a Polaris C18-A column (150 × 4.6 mm, 3 µm; Agilent) with 10 mm Na phosphate, pH 7.0, as the mobile phase (flow rate, 1 mL/min). Fractions containing NAD(P)HX forms (detected by A279) were collected and adjusted to pH 8.0 with 1 m NaOH. Successive runs were pooled, lyophilized, and redissolved in 10 mm Tris-HCl, pH 8.0. NAD(P)HX concentrations were determined spectrophotometrically (molar extinction coefficient [ε], at 290 nm = 13,500 M−1 cm−1). Solutions were stored at −20°C; upon thawing, a white precipitate appeared (presumably a phosphate salt) that was removed by centrifugation without loss of NAD(P)HX.
cDNAs and Expression Constructs
Supplemental Table S2 lists the PCR primers used. All constructs were sequence verified. Full-length cDNAs for the Arabidopsis (Arabidopsis thaliana) dehydratase (At5g19150), the maize (Zea mays) dehydratase (GRMZM5G840928), and the Arabidopsis epimerase-PPOX (At5g49970) were from the Arabidopsis Biological Resource Center (clone U13060), the Arizona Genomics Institute (clone ZM_BFb0161I05), and the Riken Bio Resource Center (clone RAFL09-46-I120), respectively. A cDNA for the maize epimerase-PPOX (GRMZM2G061988) lacking the first 120 bp of the open reading frame was obtained from the Arizona Genomics Institute (clone ZM_BFc0017D14). The missing 5′ sequence, recoded to conform to Escherichia coli codon usage, was synthesized by GenScript and spliced to the ZM_BFc0017D14 coding sequence as follows. Both sequences were PCR amplified with Phusion High-Fidelity DNA Polymerase (New England Biolabs; primers 2 and 3, and 1 and 4, respectively). The amplicons were purified with the GeneJET Gel Extraction Kit (Thermo Scientific) according to the manufacturer’s recommendations and 10 ng of each was used as a template in a second PCR reaction (primers 3 and 4). The assembled amplicon was purified, digested with NdeI and EcoRI, and ligated into the matching sites of pET28b. Supplemental Figure S5 shows the sequence of the maize epimerase-PPOX recoded 5′ region and the assembled full-length coding sequence.
For expression in E. coli as N-terminally His-tagged proteins, sequences encoding predicted mature dehydratase and epimerase-PPOX (i.e. beginning at the predicted second start Met; Supplemental Fig. S1) were PCR amplified (Arabidopsis dehydratase, primers 5 and 6; Arabidopsis epimerase-PPOX, primers 7 and 8; maize dehydratase, primers 9 and 10; and maize epimerase-PPOX, primers 11 and 4). Amplicons were digested with NdeI and EcoRI and ligated into the matching sites of pET28b.
For dual import and in vitro transcription/translation assays, full-length and truncated T (beginning at the predicted second start Met) coding sequences of the Arabidopsis proteins were PCR amplified and cloned into pGEM4Z. The primers used were as follows: full-length dehydratase, primers 14 and 16; T dehydratase, primers 15 and 16; full-length epimerase-PPOX, primers 19 and 21; and T epimerase-PPOX, primers 20 and 21). Amplicons were digested with EcoRI and XbaI and ligated into the matching sites of pGEM4Z. Constructs in which the dehydratase or epimerase:PPOX cDNA had their putative second start codon changed Leu (M2L) were created with the QuikChange Site-Directed Mutagenesis Kit (Stratagene), using full-length dehydratase- or epimerase-PPOX-pGEM4Z as templates, and primers 17 and 18, or 22 and 23, respectively.
For transient expression in tobacco (Nicotiana tabacum), Bright Yellow-2 (BY-2) cells of proteins fused C terminally to GFP, Arabidopsis full-length, T, and M2L dehydratase and epimerase-PPOX coding sequences without their stop codons were PCR amplified and cloned into pUC18/NcoI-mGFP (Chiu et al., 1996). Primers were as follows: full-length and M2L dehydratase, primers 24 and 26; T dehydratase, primers 25 and 26; full-length and M2L epimerase-PPOX, primers 27 and 29; and T epimerase-PPOX, primers 28 and 29. Amplicons were digested with BamHI and ligated into the matching site of pUC18/NcoI-mGFP. Restriction analysis was used to identify constructs with the correct insert orientation.
Protein Production and Isolation
E. coli strain BL21 (DE3) RIPL cells harboring each expression plasmid was grown at 37°C in 200 mL of Luria-Bertani medium plus 50 mg/L kanamycin until optical density at 600 nm reached 0.8. Cultures were then cooled to 22°C, supplemented with isopropyl-β-d-thiogalactoside and ethanol (final concentrations 0.5 mm and 4% [v/v], respectively), and incubated for a further 20 h at 22°C. Cells were collected by centrifugation and stored at −80°C. Pellets were resuspended in 7 mL of ice-cold lysis buffer (50 mm potassium phosphate, pH 8.0, 300 mm sodium chloride, 10 mm imidazole) and sonicated (Fisher Scientific Ultrasonic Dismembrator model 150E) using seven 15-s pulses at 70% power, cooling on ice for 2 min between pulses. The resulting lysates were centrifuged (10,000g, 10 min); the supernatant was applied to nickel-nitrilotriacetic acid agarose superflow resin columns (0.75 to 1.5 mL; Qiagen) and proteins were purified using the manufacturer’s protocol. Purified proteins were passed through PD-10 columns (GE Healthcare) equilibrated with 100 mm KCl, 20 mm Tris-HCl, pH 8.0, and 10% (v/v) glycerol, and concentrated to 30 to 120 mg/mL with Amicon Ultra-4 10,000 NMWL centrifugal filters (Millipore). Aliquots (5 to 10 µL) were snap frozen in liquid nitrogen and stored at −80°C. Protein was determined by dye binding (Bradford, 1976).
NAD(P)HX Dehydratase and Epimerase Assays
Enzyme solutions were freshly prepared from aliquots stored at −80°C. Spectrophotometric assays were made at 22°C with a Beckman DU 7400 spectrophotometer, monitoring absorbance at 340 nm (A340) every 15 s. Assays (100 µL total volume) contained 25 mm Tris-HCl, pH 8.0, 5 mm KCl, 2 mm MgCl2, 10 µg bovine serum albumin, 90 µm of a purified mixture of NADHX epimers (55% S, 45% R), and 1 mm of ATP or ADP. Reactions were started by adding 5 µg of Arabidopsis or maize NAD(P)HX dehydratase. At 5 min, 10 µg of Arabidopsis or maize NAD(P)HX epimerase-PPOX was added.
Enzyme assays for analysis by HPLC were as above but contained 1 mm NADHX, 5 mm ATP, and 20 µg of Arabidopsis or maize NAD(P)HX dehydratase and epimerase-PPOX and were incubated at 30°C. Reactions were analyzed by HPLC with a Polaris C18-A column (150 × 4.6 mm, 3 µm) using 10 mm Na phosphate buffer, pH 7.0, at a flow rate of 1 mL/min, and monitoring A279 and A340. The total reaction time before injection was 30 min. For the epimerase assays monitored by HPLC (Fig. 3), preparations enriched in (S)- or (R)-NADHX were obtained by injecting 4 µmol NADHX using the conditions described above and collecting the most concentrated 0.5-mL fraction for each epimer as determined by monitoring A279. To slow spontaneous epimerization, 25 µL of 1 m Tris-HCl, pH 9.0, was promptly added to each fraction, which was then placed on ice. Enriched fractions were used in assays within 1 h of preparation. Assays contained 25 mm Tris-HCl, pH 9.0, 1 mm KCl, 1 mm enriched (S)-NADHX or 0.5 mm enriched (R)-NADHX, and 10 µg of the separate Arabidopsis epimerase or PPOX domains and were incubated at 30°C. Analysis was by HPLC as above but with 3.5% (v/v) methanol added to the mobile phase to allow the NADHX epimers to elute within 10 min. The total reaction time before injection was 10 min.
To determine kinetic constants, the spectrophotometric assay described above was used. The concentration of (S)-NAD(P)HX in equilibrium mixtures was determined enzymatically using NAD(P)HX dehydratase. Assays contained between 1 and 25 µm (S)-NAD(P)HX and were started by adding 1.3 to 1.7 µg of NAD(P)HX dehydratase. A340 was recorded every 5 s for up to 6 min and the initial rate of NAD(P)H formation was calculated. At least three independent data sets consisting of assays at five to seven different substrate concentrations were performed for each treatment. Kinetic constants were obtained by fitting the experimental data to the Michaelis-Menten equation using Prism software (GraphPad Software)
Coupled in Vitro Transcription Translation and Dual Import Assays
In vitro transcription translation was performed using a TNT Coupled Wheat Germ Extract System or, for dual import assays, a TNT Coupled Reticulocyte Lysate System (Promega), according to the manufacturer’s instructions with [3H]Leu (115.8 Ci/mmol; PerkinElmer) as the label. Dual import assays with pea (Pisum sativum) chloroplasts and mitochondria were as described (Rudhe et al., 2002; Frelin et al., 2012), using soybean (Glycine max) alternative oxidase and Arabidopsis YgfZ as positive controls for mitochondrial and chloroplast import, respectively (Frelin et al., 2012). Reaction time was 20 min.
Expression and Visualization of GFP Fusion Proteins in BY-2 Cells
Expression and epifluorescence microscopic analysis of GFP fusion proteins in tobacco (Nicotiana tabacum) BY-2 suspension-cultured cells, as well as immunostaining of BY-2 cells, was performed as previously described (Frelin et al., 2012), except that cells were cobombarded with either pRTL2/NLS-RFP (Dhanoa et al., 2010), which encodes a nuclear localization signal fused to the red fluorescent protein and served as a convenient marker for cotransformed cells, or pRTL2/Cherry-OEP9 (Dhanoa et al., 2010), which encodes the Cherry fluorescent protein fused to the N-terminus of the outer envelope protein of 9 kD (Dhanoa et al., 2010) and also served as a transformation marker as well as a marker for plastids in BY-2 cells. Confocal microscopy of cotransformed BY-2 cells was performed using a Leica DM RBE microscope (Leica Microsystems Inc.) as previously described (Gidda et al., 2011). All confocal microscopy images presented were collected sequentially in double-labeling experiments; single-labeling experiments exhibited no detectable crossover at the settings used for data collections. Primary and secondary antibodies and sources were as follows: rabbit anti-Arabidopsis cytochrome c oxidase subunit II (Frelin et al., 2012) and goat anti-rabbit Rhodamine Red-X (Jackson ImmunoResearch Laboratories). Figures were composed using Adobe Photoshop CS (Adobe Systems). All fluorescence images of cells shown in the figures are representative of >50 independent (transient) cotransformed cells from at least three independent transformation experiments.
Isolation and Growth of Arabidopsis Dehydratase Knockout Lines
Two Arabidopsis lines with T-DNA insertions in different exons of the NAD(P)HX dehydratase gene (At5g19150) were identified in the Salk and GABI-Kat collections (Salk_203790 and Gabi_173F11). Seeds from each line and the Columbia-0 wild-type parent were germinated on one-half-strength Murashige and Skoog medium and seedlings were planted in soil and grown in the regime below. Genomic DNA was isolated from young leaves by the method of Edwards et al. (1991). Standard PCR reactions were performed with wild-type allele primers (30 and 31, or 33 and 34) or T-DNA insertion allele primers (32 and 31, or 35 and 34) and amplification products were analyzed by agarose-gel electrophoresis. At least two plants homozygous for the T-DNA insertion allele were identified for each line. The amplicons obtained with the T-DNA insertion allele primers were subcloned into pGEM-T easy (Promega) and sequenced to determine the sites of T-DNA insertion. Total RNA was isolated from homozygous plants using the RNeasy Plant Mini Kit (Qiagen) with on-column DNase treatment. First-strand cDNA was created with the SuperScript III First-Strand Synthesis System (Invitrogen). PCR reactions were performed with primers (36 and 37) designed to amplify a fragment of the NAD(P)HX dehydratase transcript or a fragment of the actin-7 transcript (38 and 39). Amplicons were analyzed by agarose-gel electrophoresis.
Seeds from the wild type and dehydratase knockout Arabidopsis plants grown under identical conditions were used to prepare samples for metabolic analysis. Seeds were surface sterilized, plated on one-half-strength Murashige and Skoog salts with 1% Suc, vernalized for 3 d at 4°C, and placed under fluorescent lights (photosynthetic photon flux 100 to 150 µmol photons m−2 s−1) on a 12-h-light/12-h-dark cycle at 22°C for 14 d prior to collecting 100-mg whole-seedling samples. Samples were frozen in liquid nitrogen and stored at −80°C in 1.5-mL Eppendorf tubes.
Analysis of NADHX, NADPHX, and Their Cyclic Forms
Six 100-mg samples each were analyzed for the wild type and homozygous Salk and GABI-Kat knockout lines. Samples were extracted by adding 400 µL of 50 mm ammonium acetate, pH 7.0, and homogenizing with a SPEX SamplePrep Geno/Grinder 2010 for 2 min at 1,250 rpm with 1.6-mm steel balls. The homogenate was transferred to a fresh 1.5-mL tube; the steel balls were washed with 600 µL of 3:1 acetonitrile/50 mm ammonium acetate, pH 7.0, and the wash was combined with the homogenate. Samples were centrifuged to clear, and the supernatant was transferred to 2-mL tubes containing 900 µL of chloroform. Samples were vortexed for 20 s and centrifuged to separate the phases. The aqueous phase (550 µL) was transferred to a fresh 1.5-mL tube, lyophilized overnight, redissolved in 50 µL of 50 mm ammonium acetate, pH 7.0, and analyzed by LC-MS. Chromatography was performed on an Agilent 1290 Infinity LC System (Agilent Technologies). Samples were maintained at 4°C in the autosampler and 5 µL of sample material was injected onto a Polaris 3 C18-A 150 × 2.0 mm HPLC column kept at ambient temperature. Mobile phase A consisted of 50 mm ammonium acetate in water, pH 7.0, and mobile phase B consisted of acetonitrile. Target metabolites were eluted from the column using the following gradient: 0 to 5 min, 0% B; 5 to 20 min, linear gradient to 5% B; 20 to 25 min, linear gradient to 100% B; and 25 to 26 min, linear gradient to 0% B. Re-equilibration time was 14 min, and flow rate was held at 0.25 mL/min throughout the method. Metabolites were detected with an Agilent 6530 accurate-mass quadrupole time-of-flight mass spectrometer equipped with an Agilent Jet Stream electrospray ionization source. Mass spectrometry data were acquired in negative ionization mode from 100 to 1100 mass-to-charge ratio with a 2 spectra/s scan rate. Mass calibration was maintained by constant infusion of reference ions at 119.036320 and 980.016375 mass-to-charge ratio. Target analytes were identified based on retention time and accurate mass matching to authentic standards.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Amino acid alignment of plant dehydratase and epimerase:PPOX N-terminal sequences.
Supplemental Figure S2. Purification of recombinant proteins used in this study.
Supplemental Figure S3. Confirmation of Arabidopsis dehydratase knockout lines.
Supplemental Figure S4. Metabolomics analysis of NAD(P)HX in Arabidopsis seedlings.
Supplemental Figure S5. Sequence of the maize epimerase:PPOX used in this study.
Supplemental Table S1. Predicted localization of Arabidopsis and maize dehydratase and epimerase:PPOX proteins.
Supplemental Table S2. Oligonucleotide primers used in this study.
Supplementary Material
Acknowledgments
We thank Oceane Frelin and Ken W. Ellens for help with dual import assays, Mike. J. Ziemak for help building expression constructs, Julia Becker and Carole L. Linster for providing the NAD(P)HX extraction method and the liquid chromatography method that allowed NAD(P)HX separation and subsequent detection by mass spectrometry, and Greg Moorhead for providing anti-N-acetylglucosamine kinase antibodies.
Glossary
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- PPOX
pyridoxine/pyridoxamine phosphate oxidase
- Kcat
catalytic constant
- cDNA
complementary DNA
- T-DNA
transfer DNA
- LC-MS
liquid chromatography-mass spectrometry
- BY-2
Bright Yellow-2
Footnotes
This work was supported by the National Science Foundation Plant Genome Research Program (grant no. IOS–1025398 to A.D.H.), the National Sciences and Engineering Research Council (grant no. 217291 to R.T.M.), the National Science Foundation (grant no. MCB–1153491 to O.F.), and an endowment from the C.V. Griffin Sr. Foundation. R.T.M. holds a University of Guelph Research Chair.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Acheson SA, Kirkman HN, Wolfenden R. (1988) Equilibrium of 5,6-hydration of NADH and mechanism of ATP-dependent dehydration. Biochemistry 27: 7371–7375 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brandizzi F, Irons S, Kearns A, Hawes C. (2003) BY-2 cells: culture and transformation for live cell imaging. Curr Protoc Cell Biol Chapter 1: 1.7.1–1.7.16 [DOI] [PubMed] [Google Scholar]
- Breslow DK, Cameron DM, Collins SR, Schuldiner M, Stewart-Ornstein J, Newman HW, Braun S, Madhani HD, Krogan NJ, Weissman JS. (2008) A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat Methods 5: 711–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabregas SM, Luche DD, Van Sluys MA, Menck CF, Silva-Filho MC. (2003) Differential usage of two in-frame translational start codons regulates subcellular localization of Arabidopsis thaliana THI1. J Cell Sci 116: 285–291 [DOI] [PubMed] [Google Scholar]
- Chaykin S, Meinhart JO, Krebs EG. (1956) Isolation and properties of a reduced diphosphopyridine nucleotide derivative. J Biol Chem 220: 811–820 [PubMed] [Google Scholar]
- Chiu W, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J. (1996) Engineered GFP as a vital reporter in plants. Curr Biol 6: 325–330 [DOI] [PubMed] [Google Scholar]
- Copley SD. (2012) Moonlighting is mainstream: paradigm adjustment required. Bioessays 34: 578–588 [DOI] [PubMed] [Google Scholar]
- Corpet F. (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16: 10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ari R, Casadesús J. (1998) Underground metabolism. Bioessays 20: 181–186 [DOI] [PubMed] [Google Scholar]
- Dhanoa PK, Richardson LG, Smith MD, Gidda SK, Henderson MP, Andrews DW, Mullen RT. (2010) Distinct pathways mediate the sorting of tail-anchored proteins to the plastid outer envelope. PLoS ONE 5: e10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K, Johnstone C, Thompson C. (1991) A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res 19: 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkemeier I, Laxa M, Miguet L, Howden AJ, Sweetlove LJ. (2011) Proteins of diverse function and subcellular location are lysine acetylated in Arabidopsis. Plant Physiol 155: 1779–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frelin O, Agrimi G, Laera VL, Castegna A, Richardson LG, Mullen RT, Lerma-Ortiz C, Palmieri F, Hanson AD. (2012) Identification of mitochondrial thiamin diphosphate carriers from Arabidopsis and maize. Funct Integr Genomics 12: 317–326 [DOI] [PubMed] [Google Scholar]
- Galperin MY, Moroz OV, Wilson KS, Murzin AG. (2006) House cleaning, a part of good housekeeping. Mol Microbiol 59: 5–19 [DOI] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11: 4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes S, Lerma-Ortiz C, Frelin O, Seaver SM, Henry CS, de Crécy-Lagard V, Hanson AD. (2012) Plant B vitamin pathways and their compartmentation: a guide for the perplexed. J Exp Bot 63: 5379–5395 [DOI] [PubMed] [Google Scholar]
- Gidda SK, Shockey JM, Falcone M, Kim PK, Rothstein SJ, Andrews DW, Dyer JM, Mullen RT. (2011) Hydrophobic-domain-dependent protein-protein interactions mediate the localization of GPAT enzymes to ER subdomains. Traffic 12: 452–472 [DOI] [PubMed] [Google Scholar]
- Golubev AG. (1996) The other side of metabolism: a review. Biokhimiia 61: 2018–2039 [PubMed] [Google Scholar]
- Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, Proctor M, St Onge RP, Tyers M, Koller D, et al. (2008) The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science 320: 362–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito J, Batth TS, Petzold CJ, Redding-Johanson AM, Mukhopadhyay A, Verboom R, Meyer EH, Millar AH, Heazlewood JL. (2011) Analysis of the Arabidopsis cytosolic proteome highlights subcellular partitioning of central plant metabolism. J Proteome Res 10: 1571–1582 [DOI] [PubMed] [Google Scholar]
- Kim G, Cole NB, Lim JC, Zhao H, Levine RL. (2010) Dual sites of protein initiation control the localization and myristoylation of methionine sulfoxide reductase A. J Biol Chem 285: 18085–18094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King R, Bonfiglio R, Fernandez-Metzler C, Miller-Stein C, Olah T. (2000) Mechanistic investigation of ionization suppression in electrospray ionization. J Am Soc Mass Spectrom 11: 942–950 [DOI] [PubMed] [Google Scholar]
- Linster CL, Van Schaftingen E, Hanson AD. (2013) Metabolite damage and its repair or pre-emption. Nat Chem Biol 9: 72–80 [DOI] [PubMed] [Google Scholar]
- Marbaix AY, Noël G, Detroux AM, Vertommen D, Van Schaftingen E, Linster CL. (2011) Extremely conserved ATP- or ADP-dependent enzymatic system for nicotinamide nucleotide repair. J Biol Chem 286: 41246–41252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marri L, Sparla F, Pupillo P, Trost P. (2005) Co-ordinated gene expression of photosynthetic glyceraldehyde-3-phosphate dehydrogenase, phosphoribulokinase, and CP12 in Arabidopsis thaliana. J Exp Bot 56: 73–80 [DOI] [PubMed] [Google Scholar]
- Meinhart JO, Chaykin S, Krebs EG. (1956) Enzymatic conversion of a reduced diphosphopyridine nucleotide derivative to reduced diphosphopyridine nucleotide. J Biol Chem 220: 821–829 [PubMed] [Google Scholar]
- Miao YS, Jiang LW. (2007) Transient expression of fluorescent fusion proteins in protoplasts of suspension cultured cells. Nat Protoc 2: 2348–2353 [DOI] [PubMed] [Google Scholar]
- Nichols RJ, Sen S, Choo YJ, Beltrao P, Zietek M, Chaba R, Lee S, Kazmierczak KM, Lee KJ, Wong A, et al. (2011) Phenotypic landscape of a bacterial cell. Cell 144: 143–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer NJ, Kaplan NO. (1974) Glyceraldehyde-3-phosphate dehydrogenase catalyzed hydration of the 5-6 double bond of reduced β-nicotinamide adenine dinucleotide (betaNADH). Formation of β-6-hydroxy-1,4,5,6-tetrahydronicotinamide adenine dinucleotide. Biochemistry 13: 4685–4694 [DOI] [PubMed] [Google Scholar]
- Porras P, Padilla CA, Krayl M, Voos W, Bárcena JA. (2006) One single in-frame AUG codon is responsible for a diversity of subcellular localizations of glutaredoxin 2 in Saccharomyces cerevisiae. J Biol Chem 281: 16551–16562 [DOI] [PubMed] [Google Scholar]
- Prabhakar P, Laboy JI, Wang J, Budker T, Din ZZ, Chobanian M, Fahien LA. (1998) Effect of NADH-X on cytosolic glycerol-3-phosphate dehydrogenase. Arch Biochem Biophys 360: 195–205 [DOI] [PubMed] [Google Scholar]
- Rafter GW, Chaykin S, Krebs EG. (1954) The action of glyceraldehyde-3-phosphate dehydrogenase on reduced diphosphopyridine nucleotide. J Biol Chem 208: 799–811 [PubMed] [Google Scholar]
- Rius SP, Casati P, Iglesias AA, Gomez-Casati DF. (2006) Characterization of an Arabidopsis thaliana mutant lacking a cytosolic non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase. Plant Mol Biol 61: 945–957 [DOI] [PubMed] [Google Scholar]
- Rudhe C, Chew O, Whelan J, Glaser E. (2002) A novel in vitro system for simultaneous import of precursor proteins into mitochondria and chloroplasts. Plant J 30: 213–220 [DOI] [PubMed] [Google Scholar]
- Sang Y, Barbosa JM, Wu H, Locy RD, Singh NK. (2007) Identification of a pyridoxine (pyridoxamine) 5′-phosphate oxidase from Arabidopsis thaliana. FEBS Lett 581: 344–348 [DOI] [PubMed] [Google Scholar]
- Sang Y, Locy RD, Goertzen LR, Rashotte AM, Si Y, Kang K, Singh NK. (2011) Expression, in vivo localization and phylogenetic analysis of a pyridoxine 5′-phosphate oxidase in Arabidopsis thaliana. Plant Physiol Biochem 49: 88–95 [DOI] [PubMed] [Google Scholar]
- Steinhauser D, Usadel B, Luedemann A, Thimm O, Kopka J. (2004) CSB.DB: a comprehensive systems-biology database. Bioinformatics 20: 3647–3651 [DOI] [PubMed] [Google Scholar]
- Sun Q, Zybailov B, Majeran W, Friso G, Olinares PD, van Wijk KJ. (2009) PPDB, the Plant Proteomics Database at Cornell. Nucleic Acids Res 37: D969–D974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawfik DS. (2010) Messy biology and the origins of evolutionary innovations. Nat Chem Biol 6: 692–696 [DOI] [PubMed] [Google Scholar]
- Toufighi K, Brady SM, Austin R, Ly E, Provart NJ. (2005) The Botany Array Resource: e-Northerns, Expression Angling, and promoter analyses. Plant J 43: 153–163 [DOI] [PubMed] [Google Scholar]
- Van Schaftingen E, Rzem R, Veiga-da-Cunha M. (2009) L: -2-Hydroxyglutaric aciduria, a disorder of metabolite repair. J Inherit Metab Dis 32: 135–142 [DOI] [PubMed] [Google Scholar]
- Vinci CR, Clarke SG. (2010) Homocysteine methyltransferases Mht1 and Sam4 prevent the accumulation of age-damaged (R,S)-AdoMet in the yeast Saccharomyces cerevisiae. J Biol Chem 285: 20526–20531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Braun SS, Sabetti A, Hanic-Joyce PJ, Gu J, Schleiff E, Joyce PB. (2007) Dual targeting of the tRNA nucleotidyltransferase in plants: not just the signal. J Exp Bot 58: 4083–4093 [DOI] [PubMed] [Google Scholar]
- Wagner R, Feth F, Wagner KG. (1986) The pyridine-nucleotide cycle in tobacco: enzyme activities for the recycling of NAD. Planta 167: 226–232 [DOI] [PubMed] [Google Scholar]
- Wamboldt Y, Mohammed S, Elowsky C, Wittgren C, de Paula WB, Mackenzie SA. (2009) Participation of leaky ribosome scanning in protein dual targeting by alternative translation initiation in higher plants. Plant Cell 21: 157–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Carrie C, Law SR, Murcha MW, Whelan J. (2013) Acquisition, conservation, and loss of dual-targeted proteins in land plants. Plant Physiol 161: 644–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A, Dave V. (1975) Inhibition of NADP-dependent dehydrogenases by modified products of NADPH. Arch Biochem Biophys 169: 298–303 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.