Abstract
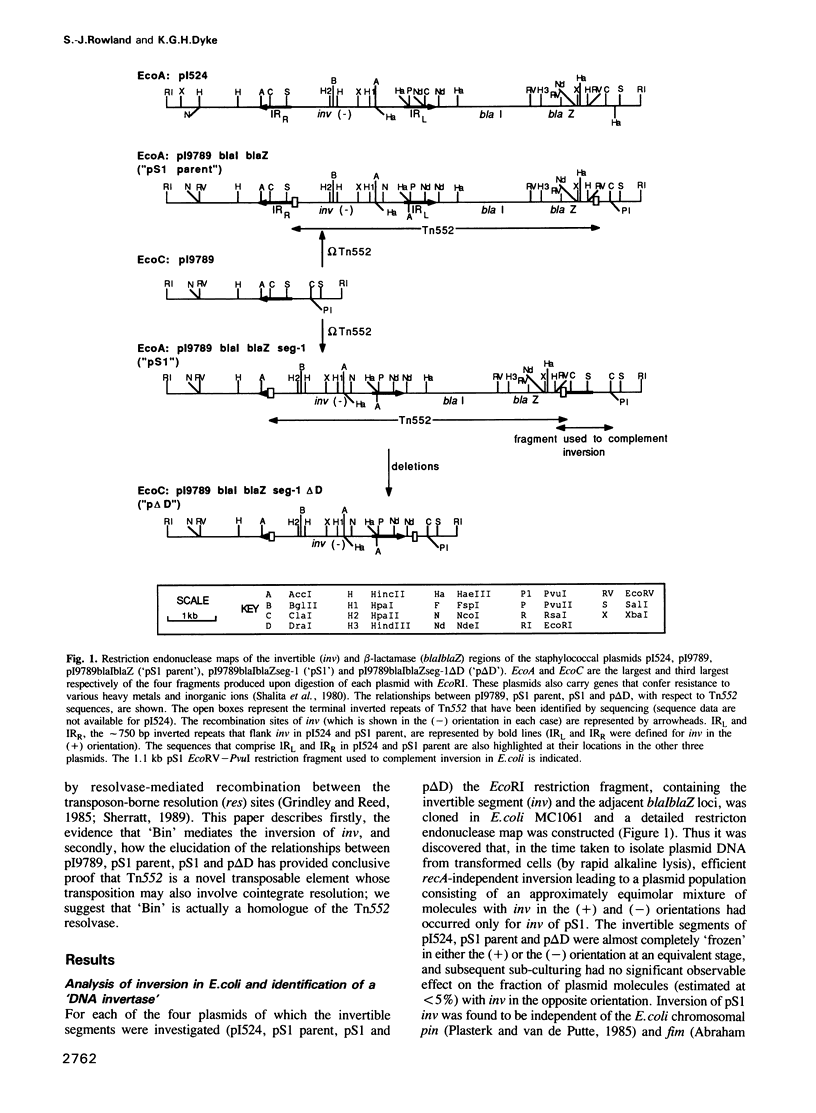
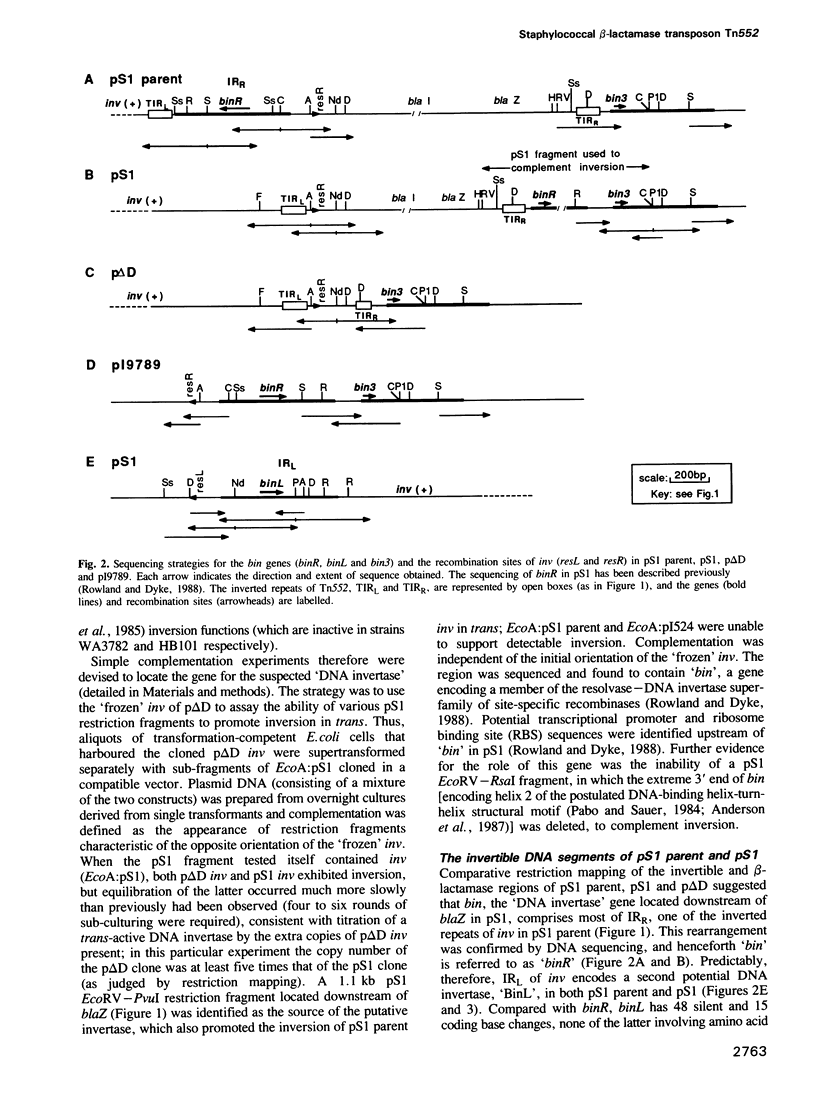
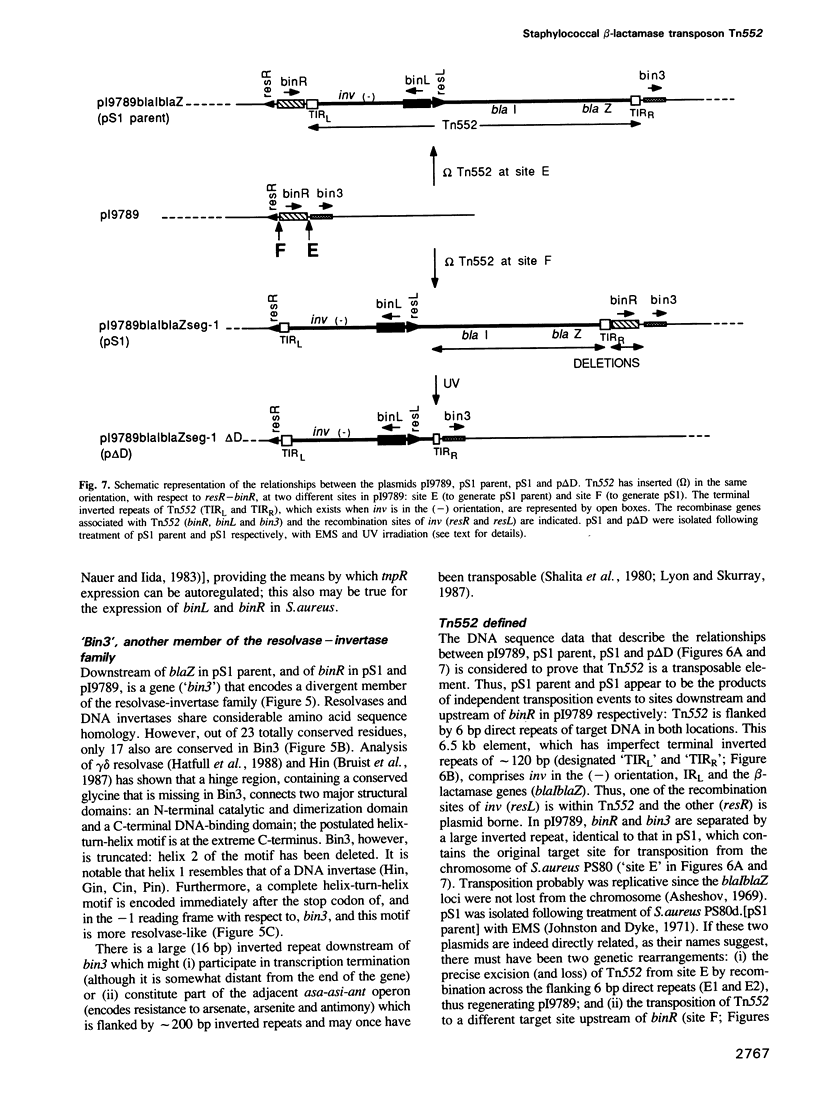
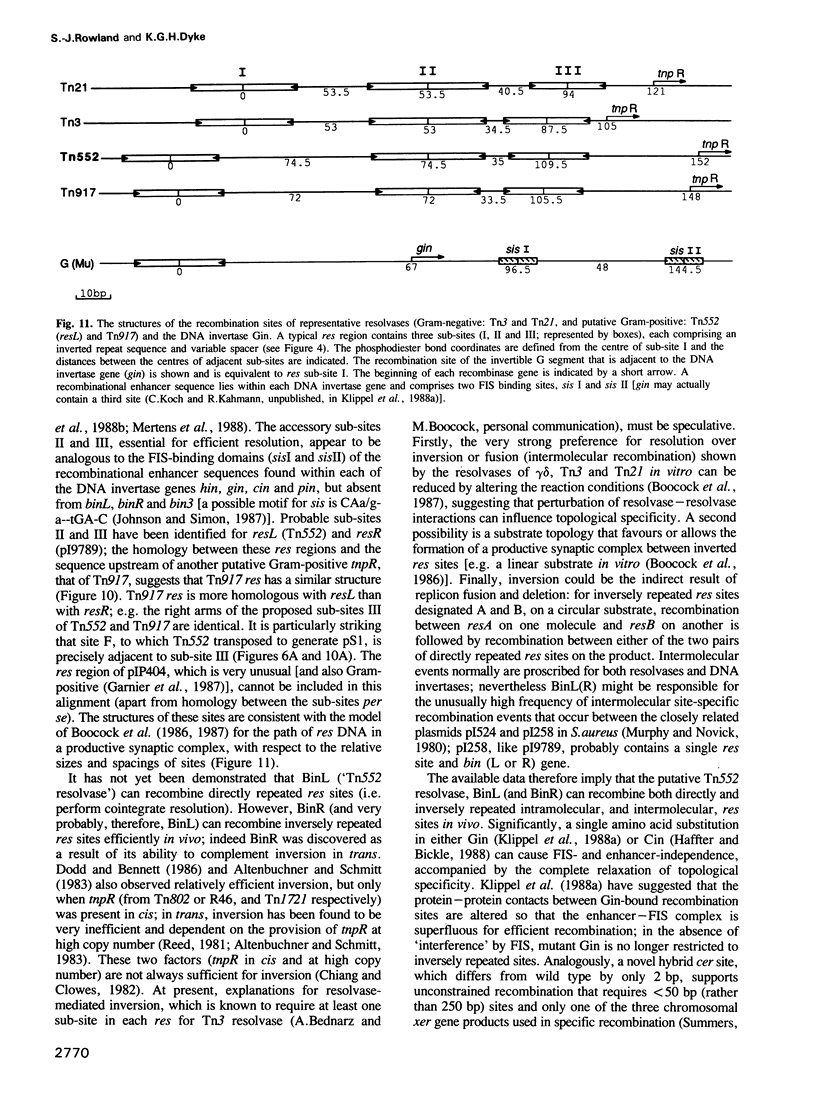
The staphylococcal beta-lactamase transposon Tn552 is a member of a novel group of transposable elements. The organization of genes in Tn552 resembles that of members of the Tn21 sub-group of Tn3 family transposons, which transpose replicatively by cointegrate formation and resolution. Thus, a possible resolution site ('resL') and a resolvase gene (tnpR or 'binL') have been identified. However, consistent with the fact that Tn552 generates 6 bp (rather than 5 bp) flanking direct repeats of target DNA, neither the putative transposase protein, nor the terminal inverted repeats of Tn552 are homologous to those of Tn3 elements. Tn552, like phage Mu and retroelements, is defined by the terminal dinucleotides 5' TG .. CA 3'. A naturally occurring staphylococcal plasmid, pI9789, contains a Tn552-derived resolution system ('resR-binR') that acts as a 'hotspot' for Tn552 transposition; insertion creates a segment of DNA flanked by inversely repeated resolution sites, one (resR) on pI9789 and the other (resL) on Tn552. The putative Tn552 resolvase, the most closely related of known resolvases to the homologous DNA invertases, initially was identified as a DNA invertase ('Bin') as a result of its ability to mediate efficient inversion of this segment in vivo.
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abraham J. M., Freitag C. S., Clements J. R., Eisenstein B. I. An invertible element of DNA controls phase variation of type 1 fimbriae of Escherichia coli. Proc Natl Acad Sci U S A. 1985 Sep;82(17):5724–5727. doi: 10.1073/pnas.82.17.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altenbuchner J., Schmitt R. Transposon Tn1721: site-specific recombination generates deletions and inversions. Mol Gen Genet. 1983;190(2):300–308. doi: 10.1007/BF00330655. [DOI] [PubMed] [Google Scholar]
- Anderson J. E., Ptashne M., Harrison S. C. Structure of the repressor-operator complex of bacteriophage 434. 1987 Apr 30-May 6Nature. 326(6116):846–852. doi: 10.1038/326846a0. [DOI] [PubMed] [Google Scholar]
- Asheshov E. H. The genetics of penicillinase production in Staphylococcus aureus strain PS80. J Gen Microbiol. 1969 Dec;59(3):289–301. doi: 10.1099/00221287-59-3-289. [DOI] [PubMed] [Google Scholar]
- Benjamin H. W., Cozzarelli N. R. Isolation and characterization of the Tn3 resolvase synaptic intermediate. EMBO J. 1988 Jun;7(6):1897–1905. doi: 10.1002/j.1460-2075.1988.tb03023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boocock M. R., Brown J. L., Sherratt D. J. Structural and catalytic properties of specific complexes between Tn3 resolvase and the recombination site res. Biochem Soc Trans. 1986 Apr;14(2):214–216. doi: 10.1042/bst0140214a. [DOI] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Bruist M. F., Horvath S. J., Hood L. E., Steitz T. A., Simon M. I. Synthesis of a site-specific DNA-binding peptide. Science. 1987 Feb 13;235(4790):777–780. doi: 10.1126/science.3027895. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang S. J., Clowes R. C. Recombination between two TnA transposon sequences oriented as inverse repeats is found less frequently than between direct repeats. Mol Gen Genet. 1982;185(1):169–175. doi: 10.1007/BF00333809. [DOI] [PubMed] [Google Scholar]
- Dodd H. M., Bennett P. M. Location of the site-specific recombination system of R46: a function necessary for plasmid maintenance. J Gen Microbiol. 1986 Apr;132(4):1009–1020. doi: 10.1099/00221287-132-4-1009. [DOI] [PubMed] [Google Scholar]
- Dodd H. M., Bennett P. M. The R46 site-specific recombination system is a homologue of the Tn3 and gamma delta (Tn1000) cointegrate resolution system. J Gen Microbiol. 1987 Aug;133(8):2031–2039. doi: 10.1099/00221287-133-8-2031. [DOI] [PubMed] [Google Scholar]
- Egner C., Berg D. E. Excision of transposon Tn5 is dependent on the inverted repeats but not on the transposase function of Tn5. Proc Natl Acad Sci U S A. 1981 Jan;78(1):459–463. doi: 10.1073/pnas.78.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fort P., Piggot P. J. Nucleotide sequence of sporulation locus spoIIA in Bacillus subtilis. J Gen Microbiol. 1984 Aug;130(8):2147–2153. doi: 10.1099/00221287-130-8-2147. [DOI] [PubMed] [Google Scholar]
- Garnier T., Saurin W., Cole S. T. Molecular characterization of the resolvase gene, res, carried by a multicopy plasmid from Clostridium perfringens: common evolutionary origin for prokaryotic site-specific recombinases. Mol Microbiol. 1987 Nov;1(3):371–376. doi: 10.1111/j.1365-2958.1987.tb01944.x. [DOI] [PubMed] [Google Scholar]
- Gillespie M. T., Lyon B. R., Skurray R. A. Structural and evolutionary relationships of beta-lactamase transposons from Staphylococcus aureus. J Gen Microbiol. 1988 Nov;134(11):2857–2866. doi: 10.1099/00221287-134-11-2857. [DOI] [PubMed] [Google Scholar]
- Grindley N. D., Reed R. R. Transpositional recombination in prokaryotes. Annu Rev Biochem. 1985;54:863–896. doi: 10.1146/annurev.bi.54.070185.004243. [DOI] [PubMed] [Google Scholar]
- Haffter P., Bickle T. A. Enhancer-independent mutants of the Cin recombinase have a relaxed topological specificity. EMBO J. 1988 Dec 1;7(12):3991–3996. doi: 10.1002/j.1460-2075.1988.tb03287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiestand-Nauer R., Iida S. Sequence of the site-specific recombinase gene cin and of its substrates serving in the inversion of the C segment of bacteriophage P1. EMBO J. 1983;2(10):1733–1740. doi: 10.1002/j.1460-2075.1983.tb01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida S. Bacteriophage P1 carries two related sets of genes determining its host range in the invertible C segment of its genome. Virology. 1984 Apr 30;134(2):421–434. doi: 10.1016/0042-6822(84)90309-x. [DOI] [PubMed] [Google Scholar]
- Johnson R. C., Glasgow A. C., Simon M. I. Spatial relationship of the Fis binding sites for Hin recombinational enhancer activity. Nature. 1987 Oct 1;329(6138):462–465. doi: 10.1038/329462a0. [DOI] [PubMed] [Google Scholar]
- Johnston L. H., Dyke K. G. Stability of penicillinase plasmids in Staphylococcus aureus. J Bacteriol. 1971 Jul;107(1):68–73. doi: 10.1128/jb.107.1.68-73.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamp D., Kardas E., Ritthaler W., Sandulache R., Schmucker R., Stern B. Comparative analysis of invertible DNA in phage genomes. Cold Spring Harb Symp Quant Biol. 1984;49:301–311. doi: 10.1101/sqb.1984.049.01.036. [DOI] [PubMed] [Google Scholar]
- Kanaar R., van de Putte P., Cozzarelli N. R. Gin-mediated DNA inversion: product structure and the mechanism of strand exchange. Proc Natl Acad Sci U S A. 1988 Feb;85(3):752–756. doi: 10.1073/pnas.85.3.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. A., Novick R. P. Terminal nucleotide sequences of Tn551, a transposon specifying erythromycin resistance in Staphylococcus aureus: homology with Tn3. Plasmid. 1980 Sep;4(2):148–154. doi: 10.1016/0147-619x(80)90004-9. [DOI] [PubMed] [Google Scholar]
- Klippel A., Cloppenborg K., Kahmann R. Isolation and characterization of unusual gin mutants. EMBO J. 1988 Dec 1;7(12):3983–3989. doi: 10.1002/j.1460-2075.1988.tb03286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klippel A., Mertens G., Patschinsky T., Kahmann R. The DNA invertase Gin of phage Mu: formation of a covalent complex with DNA via a phosphoserine at amino acid position 9. EMBO J. 1988 Apr;7(4):1229–1237. doi: 10.1002/j.1460-2075.1988.tb02935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon B. R., Skurray R. Antimicrobial resistance of Staphylococcus aureus: genetic basis. Microbiol Rev. 1987 Mar;51(1):88–134. doi: 10.1128/mr.51.1.88-134.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens G., Klippel A., Fuss H., Blöcker H., Frank R., Kahmann R. Site-specific recombination in bacteriophage Mu: characterization of binding sites for the DNA invertase Gin. EMBO J. 1988 Apr;7(4):1219–1227. doi: 10.1002/j.1460-2075.1988.tb02934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E., Novick R. P. Physical mapping of Staphylococcus aureus penicillinase plasmid pI524: characterization of an invertible region. Mol Gen Genet. 1979 Aug;175(1):19–30. doi: 10.1007/BF00267851. [DOI] [PubMed] [Google Scholar]
- Murphy E., Novick R. P. Site-specific recombination between plasmids of Staphylococcus aureus. J Bacteriol. 1980 Jan;141(1):316–326. doi: 10.1128/jb.141.1.316-326.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mézes P. S., Wang W., Yeh E. C., Lampen J. O. Construction of penP delta 1, Bacillus licheniformis 749/C beta-lactamase lacking site for lipoprotein modification. Expression in Escherichia coli and Bacillus subtilis. J Biol Chem. 1983 Sep 25;258(18):11211–11218. [PubMed] [Google Scholar]
- Novick R. P., Bouanchaud D. The problems of drug-resistant pathogenic bacteria. Extrachromosomal nature of drug resistance in Staphylococcus aureus. Ann N Y Acad Sci. 1971 Jun 11;182:279–294. doi: 10.1111/j.1749-6632.1971.tb30664.x. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Plasterk R. H., Van de Putte P. Genetic switches by DNA inversions in prokaryotes. Biochim Biophys Acta. 1984 Jun 16;782(2):111–119. doi: 10.1016/0167-4781(84)90013-7. [DOI] [PubMed] [Google Scholar]
- Plasterk R. H., van de Putte P. The invertible P-DNA segment in the chromosome of Escherichia coli. EMBO J. 1985 Jan;4(1):237–242. doi: 10.1002/j.1460-2075.1985.tb02341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R. R. Resolution of cointegrates between transposons gamma delta and Tn3 defines the recombination site. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3428–3432. doi: 10.1073/pnas.78.6.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvo J. J., Grindley N. D. The gamma delta resolvase bends the res site into a recombinogenic complex. EMBO J. 1988 Nov;7(11):3609–3616. doi: 10.1002/j.1460-2075.1988.tb03239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalita Z., Murphy E., Novick R. P. Penicillinase plasmids of Staphylococcus aureus: structural and evolutionary relationships. Plasmid. 1980 May;3(3):291–311. doi: 10.1016/0147-619x(80)90042-6. [DOI] [PubMed] [Google Scholar]
- Summers D. K. Derivatives of ColE1 cer show altered topological specificity in site-specific recombination. EMBO J. 1989 Jan;8(1):309–315. doi: 10.1002/j.1460-2075.1989.tb03378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Weber D. A., Goering R. V. Tn4201, a beta-lactamase transposon in Staphylococcus aureus. Antimicrob Agents Chemother. 1988 Aug;32(8):1164–1169. doi: 10.1128/aac.32.8.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiater L. A., Grindley N. D. Gamma delta transposase and integration host factor bind cooperatively at both ends of gamma delta. EMBO J. 1988 Jun;7(6):1907–1911. doi: 10.1002/j.1460-2075.1988.tb03024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]


