Abstract
Post translational modification (PTM) of proteins is ubiquitous and mediates many cellular processes including intracellular localization, protein-protein interactions, enzyme activity, transcriptional regulation and protein stability. While the role of phosphorylation as a key PTM has been well studied, the more evolutionarily conserved acetylation PTM has only recently attracted attention as a key regulator of cellular events. Protein acetylation has been largely studied in the context of its role in histone modification and gene regulation, where histones are modified by histone acetyltransferases (HATs) to promote transcription. However, more recent acetylomic and biochemical studies have revealed that acetylation is mediated by a broader family of protein acetyltransferases (PATs). The recent structure determination of several PATs has provided a wealth of molecular information regarding structural features of PATs, their enzymatic mechanisms, their mode of substrate-specific recognition and their regulatory elements. In this minireview, we will briefly describe what is known about non-histone protein substrates, but mainly focus on a few recent structures of PATs to compare and contrast them with HATs to better understand the molecular basis for protein recognition and modification by this burgeoning family of protein modification enzymes.
Keywords: HATs, PATs, protein acetyltransferases, posttranslational modification enzymes, epigenetics
Introduction
Posttranslational modification (PTM) of proteins provides an essential mechanism for organisms to react to external and internal stimuli, resulting in a myriad of cellular responses [1]. The importance of phosphorylation as a key PTM cannot be understated, as evidenced by the over 500 protein kinases that catalyze this reaction, the even larger number of known substrates, and the diverse array of biological pathways that are regulated by these enzymes [2]. The oncogenic properties of several kinases have also made them attractive pharmacologic targets that continue to be probed with new, more specific and potent enzyme modulators [3].
Protein acetylation has recently emerged as a significant rival to phosphorylation in its biological scope and significance, although our molecular understanding of protein acetylation and the enzymes that catalyze this reaction has lagged behind kinases [4, 5]. Much of our basic understanding has grown out of the investigation of histone acetylation and the histone acetyltransferases (HATs) that mediate this modification [6–8]. The correlation between histone acetylation and gene activation has been known for nearly half a century, but some of the enzymes responsible for these modifications have only been discovered over the past 20 years [9–12], with many other protein acetyltransferase enzymes that are yet to be discovered. To date, many HATs have been identified from yeast to humans, resulting in at least five distinct subfamilies based on sequence and structural divergence: HAT1, Gcn5/PCAF, MYST (MOZ, Ybf2, Sas2, and Tip60), p300/CBP, and Rtt109 [13–22]. With the structure elucidation and biochemical characterization of members of each of these HAT subfamilies, structural and biochemical similarities and differences have been derived. Each of these HAT subfamilies contain a structurally-conserved core region comprised of a three-stranded βsheet and helix on one side of the sheet that participates in AcCoA cofactor binding and templating the respective substrate protein for acetylation. Remarkably, this core region has little to no sequence homology between the different HAT subfamilies. The different HAT subfamilies contain structurally-divergent regions that flank the core region and also use different chemical strategies to acetylate their substrates (reviewed in [5]). These structurally-divergent regions and catalytic strategies appear to participate in HAT-specific activities such as substrate-specific binding and regulation by cofactor proteins (reviewed here, [5]).
While the majority of studies on protein acetylation have focused on histones, many HATs, such as members of the CBP/p300 and MYST families, have been shown to acetylate non-histone proteins, and acetyltransferases have been more recently identified that exclusively acetylate non-histone proteins [23, 24] (herein called PATs), such as the eukaryotic α-tubulin acetyltransferase, αTAT1 [25, 26] and the Mycobacterium tuberculosis acetyl-CoA synthetase acetyltransferase, Rv0998, [27]. In addition, the majority of eukaryotic proteins are modified on their amino termini by a family of N-terminal acetyltransferases (NATs) [28, 29]. Recently, the structure and mechanism of these non-histone acetyltransferases have been characterized, now providing an opportunity to compare the molecular determinants that differentiate these protein acetyltransferases from their HAT relatives. In this mini-review we will briefly summarize the broad range of non-histone substrates of acetylation, but will primarily focus on the structure and mechanism of action of non-histone protein acetyltransferases.
Non-histone acetylation targets
Recent acetylomic studies in several different cell lines reveal that thousands of proteins are acetylated in different cellular compartments to mediate a wide variety of biological processes [30, 31]. While these studies reveal that acetylation maps to many different types of proteins to potentially mediate many different types of acetylation-dependent protein activities, we already appreciate that protein acetylation can affect transcription factor function, protein-protein interaction, protein stability and enzyme activity.
Several proteins involved in gene regulation are modified by acetyltransferases resulting in specific gene regulatory events [32]. A prototypical acetylated transcription factor is the tumor suppressor protein p53, which is acetylated by the p300/CBP HAT on several lysine residues to promote DNA binding and transcriptional activity [33–35]. p53 is also acetylated by the Tip60 HAT to regulate its apoptotic function [36, 37]. Acetylation of p53 is reversible, and deactylation by HDAC1 represses its transcriptional activity [38, 39]. Conversely, acetylation of the Yin Yang 1 (YY1) protein by p300/CBP in two different domains results in decreased DNA binding [40, 41]. Transcription factor acetylation can also have both stimulatory and inhibitory effects dependent on the site of modification. For example, transcription of the interferon-beta gene that is mediated by the HMG-A1 transcription factor can be stimulated by lysine 71 acetylation by the PCAF HAT or inhibited by lysine 65 acetylation by the CBP HAT [42, 43]. Some other transcription factors that are known to be regulated by acetylation include c-MYC, NF-κβ, MyoD, and E2F [44–50]. A more complete review of transcription factor acetylation can be found elsewhere [23, 24].
Other DNA transactions have also been shown to be regulated by protein acetylation. For example, protein acetylation has been shown to play a role in DNA replication through the direct acetylation of the cohesion protein complex that mediates appropriate separation of sister chromatids during mitosis [51]. This acetylation is mediated by the establishment of cohesion 1 and 2 proteins (ECO1 and 2) [52, 53], while the subsequent deacetylation of Smc3 by Hda one similar 1 (Hos1) destabilizes the complex and allows for the separation of sister chromatids during anaphase [54].
Protein acetylation can also affect protein-protein interaction. For example, the acetylation of importin-α by the p300 HAT promotes its interaction with importin-β, which results in the transport of the RNA binding protein HuR through the nuclear pore complex, and disruption of this event has been shown to prevent the import of this mRNA-stabilizing protein [55, 56]. α-tubulin represents another extra-nuclear protein that has been known for nearly thirty years to be highly acetylated, but the exact function of this modification has remained inconclusive [57–59]. Acetylated α-tubulin has been generally associated with highly-stable, long-lasting microtubules, with acetylation of microtubules seemingly linked to vesicle trafficking along them [58, 60].
The acetylation of certain enzymes has also been demonstrated to modulate their catalytic activities. This is highlighted by recent studies focusing on the protein acetylome in both prokaryotic and eukaryotic organisms, revealing that a large number of metabolic enzymes involved in glycolysis, gluconeogenesis, the TCA cycle, fatty acid synthesis and degradation were acetylated [61–66]. These studies also revealed that in a number of cases, the acetylation status affected enzyme activity to control the direction of carbon flux in a pathway. For example, in human liver cells, fatty acid production led to the elevated acetylation of the β-oxidation multifunctional enzyme enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase and its increased catalytic activity [63]. Notably, the control of metabolism via acetylation of enzymes includes the inactivation of the mitochondrial acetyl-CoA synthetase, a key enzyme in the production of the AcCoA cofactor of acetyltransferases, via the acetylation of an active site lysine [67–69]. A more complete review on the regulation of metabolism through protein acetylation can be found elsewhere [64].
The majority of eukaryotic proteins are also cotranslationally acetylated on their amino-termini by a family of proteins that have sequence homology to GNAT proteins called N-terminal acetyltransferases (NATs). N-terminal acetylation regulates a wide range of biological processes [70–73]. For example, Hwang et al. demonstrated that N-terminal protein acetylation regulates the N-end rule for protein degradation [73], and Scott et al. demonstrated that N-terminal methionine acetylation of the E2 enzyme, Ubc12, modulates its binding mode to the Dcn1 E3 ligase to promote attachment of the ubiquitin-like protein Nedd8 to its Cul1 protein substrate [71]. The anti-apoptotic protein Bcl-xl has also been shown to promote cell survival through the downregulation of AcCoA biosynthesis and N-amino acetylation [72]. In yet another study, N-terminal acetylation was shown to inhibit protein targeting to the endoplasmic reticulum, thus revealing that the modification also plays a role in protein localization [74]. Taken together, the picture that is emerging is that protein acetylation has as diverse and as important a regulatory role as protein phosphorylation in biology.
Structure and mechanism of non-histone protein acetyltransferase enzymes
Over the last few years we have begun to accumulate biochemical and structural data on the enzymes that mediate non-histone protein acetylation. This now puts us in a position to compare and contrast these enzymes with HATs to better understand the activities and substrate-binding specificities of the broader family of protein acetyltransferases. In this review, we will focus on four non-histone protein acetyltransferases as model systems: the human α-tubulin acetyltransferase, α TAT1 [75], the human NAT, Naa50p [76], the Mycobacterium tuberculosis ACS acetyltranferase, Rv0998 [77], and the Sulfolobus solfataricus ALBA DNA binding protein acetyltransferase, SsPAT [78]. We will specifically compare their overall structures and AcCoA binding pockets, mode of catalysis, substrate selective binding and mode of regulation.
Overall structure and acetyl-CoA binding
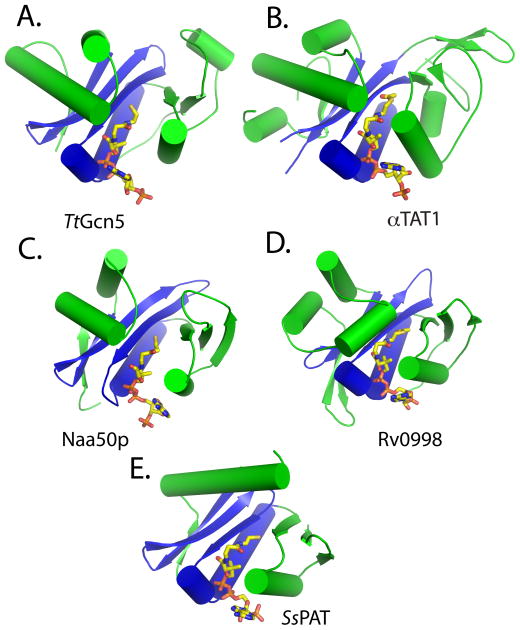
The αTAT1, Naa50p, Rv0998, and SsPAT acetyltransferases are structurally most similar to the Gcn5-related N-acetyltransferase fold (Fig. 1). This observation does not exclude the possibility that other, yet uncharacterized, non-histone acetyltransferases may contain folds similar to the other classes of acetyltransferases. Not surprisingly, each of these proteins contains a β-sheet-helix core region that is structurally conserved among all HATs (colored blue in each panel of Fig. 1), despite the lack of sequence conservation in this region. Also like the different HAT subfamilies, the structurally-conserved core region is flanked by more variable N- and C-terminal segments (colored green in each panel of Fig. 1). Despite the structural variability of the N- and C-terminal segments, each of these proteins contains a binding cavity or groove over the core region that is flanked on opposite sides by helices and loops from the N- and C-terminal segments. The shapes of the respective binding grooves or cavities are different and likely contribute to substrate-specific binding, as will be described in more detail in a later section.
Figure 1. Structure of non-histone protein acetyltransferases.
Cartoon representation of (A) TtGcn5 HAT, (B) αTAT1, (C), Naa50p, (D) activated Rv0998 (only acetyltransferase domain shown), and (E) SsPAT. The conserved acetyltransferase core region is colored blue, and the variable flanking N- and C-terminal segments are colored green. Ac-CoA and CoA are in stick representation and colored according to element: carbon, yellow; nitrogen, blue; oxygen, red.
Like in the HATs, the central core region of the non-histone protein acetyltransferases participates in a large number of the acetyl-CoA binding and stabilizing interactions, with the remainder of the acetyl-CoA interactions being made by two flanking α-helices found in each of the structures. Residues that participate in AcCoA interaction are typically not conserved. Notably, each of the acetyltransferases appears to use CoA, not only as an acetyl donor, but also as a molecule to stabilize the overall fold of the acetyltransferase domain. It is difficult to imagine the folding of these domains in the absence of cofactor and it is indeed not surprising that very few acetyltransferase structures have been reported without bound cofactor or cofactor analog [14, 79].
Mechanism of catalysis
One of the surprising findings of comparing the different HAT subfamilies is that they use different chemical strategies to carry out the same chemical reaction of acetylation [5]. This likely derives from the relative chemical simplicity of transferring an acetyl group from a CoA thioester to an amine of a lysine side-chain, which likely allows these enzymes to employ chemical strategies that best suit their respective biochemical activities. This catalytic diversity within the HATs appears to extend to the non-histone protein acetyltransferases.
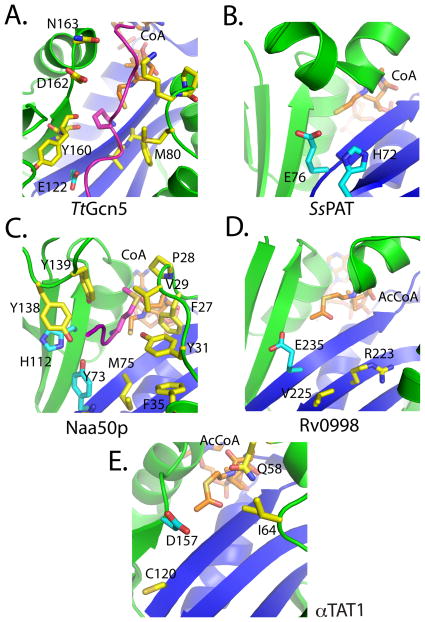
The Gcn5/PCAF and Gcn5-related N-acetyltransferases have been very well characterized structurally and enzymatically and shown to utilize a conserved glutamate residue to function as a general base through a bi-bi ternary complex mechanism [79–81] (Fig 2A). In this mechanism, both the acetyl-CoA and the substrate protein must be bound to the enzymes before catalysis can occur, and the turnover number for Gcn5 (kcat) is very efficient at ~210 min−1 [82]. Interestingly, although SsPAT also contains a glutamate residue (E76) at the equivalent position in 3-D space, it does not utilize this residue exclusively as a general base to facilitate deprotonation, but rather a number of residues (Y38, E42, E43, D53, H72, E76) act as a proton wire to shuttle the proton from the active site (Fig 2B) [78]. The rate of reaction by SsPATs is significantly slower than Gcn5, kcat= 2 min−1, but we note that this is the in vitro measured value at 75 °C for a thermophilic organism, so the naturally-occurring rate may be different. Studies on Naa50p also suggest that catalysis does not rely on one specific residue, with a tyrosine and histidine (Y73 and H112) fulfilling the roles of the general base (Fig 2C) [76]. This reaction appears to occur slightly more efficiently than SsPAT (kcat= ~7 min−1 for Naa50p), but still not as efficiently as Gcn5.
Fig 2. Catalytic pockets of non-histone protein acetyltransferases.
Cartoon representation of acetyltransferases highlighting key catalytic residues colored in cyan and other important residues in yellow. Active site pockets are shown for (A) TtGcn5 with protein contact residues highlighted in yellow, (B) SsPAT, (C) Naa50p with protein contact residues highlighted in yellow, (D) Rv0998, with mutationally sensitive residues in the active site highlighted in yellow and (E) αTAT1 with mutationally sensitive residues in the active site highlighted in yellow. The coloring scheme of the catalytic core and flanking domains is the same as Figure 1.
Through the structural analysis of Rv0998, it was observed that E235 was positioned in a location similar to the conserved glutamate of Gcn5 (Fig 2D), and when mutated to an alanine, catalysis was ablated [77]. However, a more thorough enzymatic analysis has yet to be completed, so it is possible that the catalytic mechanism of Rv0998 may be more complicated.
The simultaneous reporting of the structure of the αTAT1/acetyl-CoA complex revealed structures with nearly identical overall folds [75, 83]. However, the investigators’ interpretations of the catalytic properties of αTAT1 were different. Previous work had suggested that a conserved aspartate (D157), not a glutamate, acted as a general base for catalysis (Fig 2E). Friedmann et al. showed that mutation of this residue resulted in a catalytically-defective enzyme. Furthermore, mutation of this residue to a glutamate (D157E) failed to rescue activity, suggesting that the bond distances in the catalytic pocket are crucial for effective catalysis. Also identified in this study was a cysteine residue (C120) in close proximity to the catalytic pocket. Mutation of this residue to either an alanine or serine also resulted in a catalytically-defective enzyme, thus suggesting a role for this residue in catalysis as well. To probe whether αTAT1 employed a bi-bi ternary complex mechanism like the Gcn5 HAT or a ping-pong catalytic mechanism that might employ this cysteine residue to form an acetyl-enzyme intermediate, as is the case with the MYST HAT proteins [84], Friedmann at al. performed bisubstrate kinetics under steady state conditions. This revealed that the mechanism proceeded through a bi-bi ternary complex mechanism. Based on this data, Friedmann et al. proposed that αTAT1 uses D157 and C120 as general bases for catalysis. The kinetic analysis performed here also supported the previous claim of a very inefficient enzyme (kcat= ~.12 min−1). This observation may suggest why acetylated tubulin is only found in long-lived microtubules, as dynamic microtubules are not stable long enough for αTAT1 to act upon them. Contrary to the observations of Friedmann et al., Taschner et al. showed that D157 and a nearby glutamine residue (Q58) were both mutationally sensitive for acetylation activity (Fig. 1E), suggesting that these two residues may cooperate for general base catalysis [83]. While further studies of α TAT1 will be required to address this discrepancy, it appears that α TAT1 uses yet another chemical strategy for substrate acetylation, again highlighting the diversity of chemical strategies used by acetylation enzymes.
Protein substrate recognition
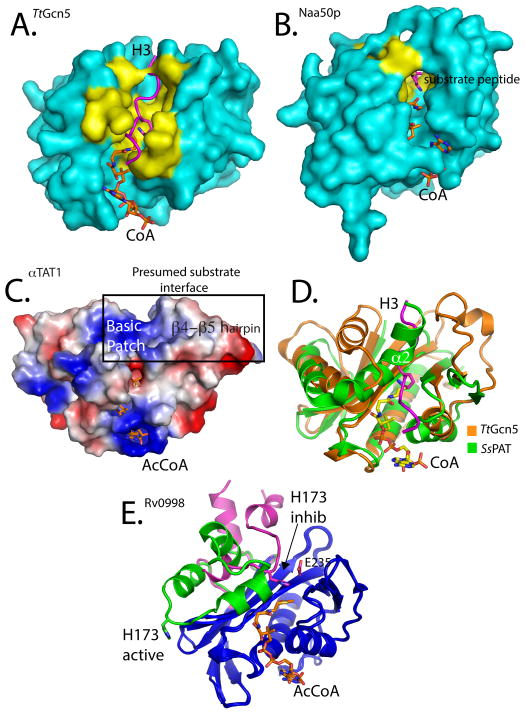
The structure of Gcn5 bound to an 11-residue histone H3 peptide centered on a cognate H3K14 lysine substrate provided the first atomic view of how a HAT recognizes its cognate protein substrate for acetylation [80]. This structure and subsequent structures of Gcn5/PCAF with longer histone or non-histone peptide substrates revealed that protein substrate binds across an enzyme groove formed by the core on the bottom and flanked on opposite sides by the N- and C-terminal segments (Figs. 2A and 3A) [82]. Residues in the α2 and α4 helices of the N- and C-terminal segments, respectively, mediate the majority of interactions with peptide. Interestingly, few substrate side chains participate in interactions with the enzyme, and the majority of interactions are made to the substrate backbone. This may explain the ability of Gcn5 to acetylate diverse histone and non-histone lysine residues.
Figure 3. Substrate specificity determinants and regulation of non-histone protein acetyltransferases.
(A) Surface representation of the TtGcn5/CoA/H3 complex. The H3 peptide is represented as a magenta ribbon. Residues described in the text and shown in Figure 2 to interact with the substrate peptide are colored yellow. (B) Surface representation of the Naa50p/CoA/substrate peptide complex also highlighting the peptide substrate binding residues in yellow. (C) Electrostatic surface potential mapping of the αTAT1/AcCoA complex, with surfaces colored according to charge potential (blue-positive, red-negative, white-neutral). The mutationally sensitive and presumed substrate interface is highlighted, emphasizing the large basic patch and location of the β4-β5 hairpin. (D) Carbon alpha alignment of TtGcn5 (orange) and SsPAT (green). The superimposed H3 peptide substrate from the TtGcn5 structure shown in magenta. The α2 helix of SsPAT that overlaps with the H3 peptide-binding site is highlighted. (E) Cartoon representation carbon alpha alignment of the Rv0998 acetyltransferase in the inhibited (magenta) and activated (green) forms. Structurally similar regions are colored blue. The large domain conformational change that results in the movement of residue H173 is highlighted.
Although the overall structure of Naa50p superimposes well with Gcn5 with a root mean square deviation of Cα atoms of 4.1 Å, the Naa50p protein substrate-binding site shows significant differences that appear to correlate with its ability to acetylate an N-terminal amino group instead of a lysine side chain amino group [76]. Specifically, a long loop in the C-terminal segment occupies one end of what would be the peptide-binding groove of Gcn5. This makes the substrate binding region of Naa50p much more “closed” than Gcn5, having a width of only ~9Å, instead of ~ 17Å in Gcn5. This appears to prevent protein segments from lying across Naa50p to insert lysine side chain residues. Instead, the narrower width of the Naa50p binding cleft creates a “tunnel” that allows the amino-terminal end of the cognate substrate to extend into the enzyme active site in an orientation that is roughly perpendicular to the protein substrate for Gcn5 (Fig 3B). In addition, a hydrophobic pocket that is created by the α1-α2 helices and the β6-β7 strands of the N- and C-terminal segments that surround the active site, accommodate the Met-Val sequence that is located at the peptide N-terminus. These residues are conserved among Naa50p enzymes, but not the other classes of N-terminal acetyltransferases, and explain why Naa50p has such defined substrate specificity. Structural characterization of the other NATs will likely reveal similar overall active site architectures to accommodate N-amino protein substrates, but differences in the substrate active site will likely specify their respective N-terminal cognate sequences.
The lack of structural data on other PATs bound to their respective cognate substrates precludes a detailed analysis of the determinants of substrate specificity. However, the available data on the unliganded enzymes does make possible some predictions that are worth noting. αTAT1 displays a strict specificity for α-tubulin, which may be mediated in part by a large positively-charged surface near the presumed substrate binding cleft (Fig. 3C) [75, 83]. This positively-charged surface potential, which is flanked by small apolar and acidic residues, would seemingly prohibit histone binding and may help coordinate α-tubulin K40 recognition,. The αTAT1 structure revealed structural elements flanking the conserved core that when mutated affect catalysis and a wide substrate binding cleft of ~20Å. Additionally, mutation of a novel β-hairpin structure located in the C-terminal segment was shown to have debilitating or activating effects on catalysis in vitro and tubulin acetylation in cells (Fig. 3C) [75], which further validates the importance of these structural elements for substrate recognition by αTAT1.
The structure of SsPAT did not include a protein substrate peptide, but nonetheless suggested important components for substrate recognition. Similar to Naa50p, SsPAT displayed a very narrowed binding cleft of 8–10Å. However, the SsPAT binding cleft is constricted because of an unusual bent helix (α2) that sits in the space that would be occupied by the peptide substrate as it is bound to Gcn5 (Fig 3D, green). The α2 helix of SsPAT appears to play a role in substrate recognition, as mutation of residues in this helix or residues that contact it affected catalysis [78]. It was proposed that this helix is dynamic and participates in substrate binding, although the mechanism for this is still not clear [78]. Taken together, like the HATs, the non-histone protein acetyltransferases appear to exploit divergent regions N- and C-terminal to the structurally-conserved core region for substrate-specific recognition.
Regulatory mechanisms
The structure of Rv0998 does not reveal the manner by which the protein substrate binds to the enzyme, but it does, however, reveal how an acetyltransferase might respond to allosteric signals. In this particular case, the acetyltransferase domain of Rv0998 is fused to a cAMP binding domain. The crystal structures of the autoinhibited and cAMP-activated Rv0998 reveals the mode of cAMP-mediated activation of the acetyltransferase [77]. Specifically, in the absence of cAMP, a portion of the acetyltransferase domain referred to as the “lid” lies over the top of the catalytic glutamate, with a histidine (H173) from the loop positioned to mimic the cognate lysine substrate (Fig 3E, pink). Upon binding of the cAMP to the regulatory domain over 30Å away from the active site, a series of conformational changes occur that results in significant movement of the “lid” to a position that allows the substrate to gain access to the active site (Fig 3E, green). Many other acetyltransferases, such at the NATs, are also regulated by the binding of other protein domains, typically from other polypeptide chains, and it will be interesting to learn if they are regulated by similar or distinct mechanisms to Rv0998.
Several reports have demonstrated that the autoacetylation of HATs also plays important regulatory functions. In particular, the Rtt109, p300/CBP, and MYST HAT families have all been shown to require autoacetylation for maximal acetyltranferase activity [14, 85–88]. Interestingly, each of these HAT families is regulated by autoacetylation in different ways. A lysine-rich loop in the p300 protein is proposed to occlude the active site, but is displaced when it is autoacetylated, which allows for binding and acetylation of the cognate substrate [87]. Several reports have shown that the MYST acetyltransferases are autoacetylated [89, 90], and Yuan et al. showed that a strictly-conserved buried acetyllysine residue in the active site makes essential stabilizing contacts with neighboring residues and facilitates substrate binding and catalysis [14]. Rtt109 also contains a strictly-conserved buried acetyllysine [88, 91, 92], but this residue is about 8Å away from the active site and has been shown to lower the KM for AcCoA binding and increase the catalytic rate [93], although the mechanism for this is unclear.
A study by Kalebic et al. identified four lysine residues in αTAT1 that were autoacetylated, and this autoacetylation was demonstrated to increase the catalytic activity of the enzyme towards its microtubule substrate [94]. The exact mechanism by which this operates is not clear, as two of the lysines are distant from the catalytic domain, and the other two are at the edge of the catalytic domain and not resolved in the crystal structure. Nonetheless, it appears that the regulation of acetyltransferase activity by autoacetylation may extend beyond the HATs to the broader family of protein acetyltransferases.
Conclusions and perspectives
The study of acetyltransferases has come a long way from the first observation of acetylated histones and the subsequent structure determination of the Gcn5 HAT. To date, we have identified a handful of HATs and we now know that they fall into subfamilies that contain a structurally-conserved core region, more variable N- and C-terminal segments and divergent catalytic strategies that participate in substrate-specific acetylation and HAT-specific biochemical properties. We also now know that HAT activity is often modulated by their interaction with other protein domains and/or proteins and by autoacetylation. Perhaps not surprisingly, it appears that PATs follow the same rules. Future studies on these proteins clearly need to focus on better understanding their mode of substrate specific activities and their regulation by other protein domains, protein subunits, autoacetylation and possibly other post-translational modifications.
A remarkable feature of acetyltransferases is that they lack strong sequence homology. This fact, together with recent acetylomic studies showing that thousands of proteins in different cellular compartments and with diverse biological functions are acetylated, suggests that other PATs exist that are yet to be identified. Indeed, the PATs that have been characterized to date have been identified through their limited sequence homology to the GNAT proteins. It is therefore likely that other PATs with homology to the MYST or p300/CBP exist, and perhaps still new subfamilies of PATs are yet to be discovered. One thing that is becoming clear is that PATs are fascinating biochemical machines and we are only beginning to uncover their mechanism of action. On the biological side, an argument can be made that PATs are playing as important a role as protein kinases in signal transduction pathways. Together with the correlation of altered histone and protein acetylation in various human pathologies [95, 96], we propose that PATs may also become important drug targets as well. Indeed, the divergent catalytic strategies that are used by PATs should facilitate the development of PAT-specific inhibitors for use in therapy.
Abbreviations
- HAT
histone acetyltransferase
- PAT
protein acetyltransferase
References
- 1.Walsh C. Posttranslational Modification Of Proteins: Expanding Nature’s Inventory. Roberts and Company Publishers; 2006. [Google Scholar]
- 2.Cohen P. The origins of protein phosphorylation. Nature Cell Biol. 2002;4:E127–30. doi: 10.1038/ncb0502-e127. [DOI] [PubMed] [Google Scholar]
- 3.Cohen P. Protein kinases--the major drug targets of the twenty-first century? Nature Rev Drug Dis. 2002;1:309–15. doi: 10.1038/nrd773. [DOI] [PubMed] [Google Scholar]
- 4.Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 2000;19:1176–9. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yuan H, Marmorstein R. Histone acetyltransferases: Rising ancient counterparts to protein kinases. Biopolymers. 2013;99:98–111. doi: 10.1002/bip.22128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eberharter A, Becker PB. Histone acetylation: a switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep. 2002;3:224–9. doi: 10.1093/embo-reports/kvf053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kimura A, Matsubara K, Horikoshi M. A decade of histone acetylation: marking eukaryotic chromosomes with specific codes. J Biochem. 2005;138:647–62. doi: 10.1093/jb/mvi184. [DOI] [PubMed] [Google Scholar]
- 8.Yang XJ, Seto E. HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene. 2007;26:5310–8. doi: 10.1038/sj.onc.1210599. [DOI] [PubMed] [Google Scholar]
- 9.Allfrey VG, Faulkner R, Mirsky AE. Acetylation and Methylation of Histones and Their Possible Role in the Regulation of Rna Synthesis. Proc Natl Acad Sci USA. 1964;51:786–94. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–51. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 11.Kleff S, Andrulis ED, Anderson CW, Sternglanz R. Identification of a gene encoding a yeast histone H4 acetyltransferase. J Biol Chem. 1995;270:24674–7. doi: 10.1074/jbc.270.42.24674. [DOI] [PubMed] [Google Scholar]
- 12.Ruiz-Garcia AB, Sendra R, Galiana M, Pamblanco M, Perez-Ortin JE, Tordera V. HAT1 and HAT2 proteins are components of a yeast nuclear histone acetyltransferase enzyme specific for free histone H4. J Biol Chem. 1998;273:12599–605. doi: 10.1074/jbc.273.20.12599. [DOI] [PubMed] [Google Scholar]
- 13.Marcus GA, Silverman N, Berger SL, Horiuchi J, Guarente L. Functional similarity and physical association between GCN5 and ADA2: putative transcriptional adaptors. EMBO J. 1994;13:4807–15. doi: 10.1002/j.1460-2075.1994.tb06806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan H, Rossetto D, Mellert H, Dang W, Srinivasan M, Johnson J, Hodawadekar S, Ding EC, Speicher K, Abshiru N, Perry R, Wu J, Yang C, Zheng YG, Speicher DW, Thibault P, Verreault A, Johnson FB, Berger SL, Sternglanz R, McMahon SB, Cote J, Marmorstein R. MYST protein acetyltransferase activity requires active site lysine autoacetylation. EMBO J. 2012;31:58–70. doi: 10.1038/emboj.2011.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Candau R, Moore PA, Wang L, Barlev N, Ying CY, Rosen CA, Berger SL. Identification of human proteins functionally conserved with the yeast putative adaptors ADA2 and GCN5. Mol Cell Biol. 1996;16:593–602. doi: 10.1128/mcb.16.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith ER, Belote JM, Schiltz RL, Yang XJ, Moore PA, Berger SL, Nakatani Y, Allis CD. Cloning of Drosophila GCN5: conserved features among metazoan GCN5 family members. Nuc Acids Res. 1998;26:2948–54. doi: 10.1093/nar/26.12.2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith ER, Eisen A, Gu W, Sattah M, Pannuti A, Zhou J, Cook RG, Lucchesi JC, Allis CD. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc Natl Acad Sci USA. 1998;95:3561–5. doi: 10.1073/pnas.95.7.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamamoto T, Horikoshi M. Novel substrate specificity of the histone acetyltransferase activity of HIV-1-Tat interactive protein Tip60. Proc Natl Acad Sci USA. 1997;272:30595–8. doi: 10.1074/jbc.272.49.30595. [DOI] [PubMed] [Google Scholar]
- 19.Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–3. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 20.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–9. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 21.Han J, Zhou H, Horazdovsky B, Zhang K, Xu RM, Zhang Z. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science. 2007;315:653–5. doi: 10.1126/science.1133234. [DOI] [PubMed] [Google Scholar]
- 22.Driscoll R, Hudson A, Jackson SP. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science. 2007;315:649–52. doi: 10.1126/science.1135862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 24.Spange S, Wagner T, Heinzel T, Kramer OH. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. International J Biochem Cell Biol. 2009;41:185–98. doi: 10.1016/j.biocel.2008.08.027. [DOI] [PubMed] [Google Scholar]
- 25.Shida T, Cueva JG, Xu Z, Goodman MB, Nachury MV. The major alpha-tubulin K40 acetyltransferase alphaTAT1 promotes rapid ciliogenesis and efficient mechanosensation. Proc Natl Acad Sci USA. 2010;107:21517–22. doi: 10.1073/pnas.1013728107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akella JS, Wloga D, Kim J, Starostina NG, Lyons-Abbott S, Morrissette NS, Dougan ST, Kipreos ET, Gaertig J. MEC-17 is an alpha-tubulin acetyltransferase. Nature. 2010;467:218–22. doi: 10.1038/nature09324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu H, Hegde SS, Blanchard JS. Reversible acetylation and inactivation of Mycobacterium tuberculosis acetyl-CoA synthetase is dependent on cAMP. Biochemistry. 2011;50:5883–92. doi: 10.1021/bi200156t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown JL, Roberts WK. Evidence that approximately eighty per cent of the soluble proteins from Ehrlich ascites cells are Nalpha-acetylated. J Biol Chem. 1976;251:1009–14. [PubMed] [Google Scholar]
- 29.Arnesen T, Van Damme P, Polevoda B, Helsens K, Evjenth R, Colaert N, Varhaug JE, Vandekerckhove J, Lillehaug JR, Sherman F, Gevaert K. Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc Natl Acad Sci USA. 2009;106:8157–62. doi: 10.1073/pnas.0901931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–40. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 31.Kim GW, Yang XJ. Comprehensive lysine acetylomes emerging from bacteria to humans. TrendsBiochemical Sciences. 2011;36:211–20. doi: 10.1016/j.tibs.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 32.Bannister AJ, Miska EA. Regulation of gene expression by transcription factor acetylation. Cellular Mol Life Sciences : CMLS. 2000;57:1184–92. doi: 10.1007/PL00000758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 34.Li AG, Piluso LG, Cai X, Gadd BJ, Ladurner AG, Liu X. An acetylation switch in p53 mediates holo-TFIID recruitment. Mol Cell. 2007;28:408–21. doi: 10.1016/j.molcel.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 35.Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008;133:612–26. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, McMahon SB. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell. 2006;24:841–51. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang Y, Luo J, Zhang W, Gu W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell. 2006;24:827–39. doi: 10.1016/j.molcel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 38.Murphy M, Ahn J, Walker KK, Hoffman WH, Evans RM, Levine AJ, George DL. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Develop. 1999;13:2490–501. doi: 10.1101/gad.13.19.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo J, Su F, Chen D, Shiloh A, Gu W. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature. 2000;408:377–81. doi: 10.1038/35042612. [DOI] [PubMed] [Google Scholar]
- 40.Lee JS, Galvin KM, See RH, Eckner R, Livingston D, Moran E, Shi Y. Relief of YY1 transcriptional repression by adenovirus E1A is mediated by E1A-associated protein p300. Genes Develop. 1995;9:1188–98. doi: 10.1101/gad.9.10.1188. [DOI] [PubMed] [Google Scholar]
- 41.Yao YL, Yang WM, Seto E. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol Cell Biol. 2001;21:5979–91. doi: 10.1128/MCB.21.17.5979-5991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Munshi N, Merika M, Yie J, Senger K, Chen G, Thanos D. Acetylation of HMG I(Y) by CBP turns off IFN beta expression by disrupting the enhanceosome. Mol Cell. 1998;2:457–67. doi: 10.1016/s1097-2765(00)80145-8. [DOI] [PubMed] [Google Scholar]
- 43.Munshi N, Agalioti T, Lomvardas S, Merika M, Chen G, Thanos D. Coordination of a transcriptional switch by HMGI(Y) acetylation. Science. 2001;293:1133–6. doi: 10.1126/science.293.5532.1133. [DOI] [PubMed] [Google Scholar]
- 44.Patel JH, Du Y, Ard PG, Phillips C, Carella B, Chen CJ, Rakowski C, Chatterjee C, Lieberman PM, Lane WS, Blobel GA, McMahon SB. The c-MYC oncoprotein is a substrate of the acetyltransferases hGCN5/PCAF and TIP60. Mol Cell Biol. 2004;24:10826–34. doi: 10.1128/MCB.24.24.10826-10834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen LF, Mu Y, Greene WC. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-kappaB. EMBO J. 2002;21:6539–48. doi: 10.1093/emboj/cdf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen L, Fischle W, Verdin E, Greene WC. Duration of nuclear NF-kappaB action regulated by reversible acetylation. Science. 2001;293:1653–7. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- 47.Sartorelli V, Puri PL, Hamamori Y, Ogryzko V, Chung G, Nakatani Y, Wang JY, Kedes L. Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol Cell. 1999;4:725–34. doi: 10.1016/s1097-2765(00)80383-4. [DOI] [PubMed] [Google Scholar]
- 48.Polesskaya A, Harel-Bellan A. Acetylation of MyoD by p300 requires more than its histone acetyltransferase domain. J Biol Chem. 2001;276:44502–3. doi: 10.1074/jbc.M106501200. [DOI] [PubMed] [Google Scholar]
- 49.Martinez-Balbas MA, Bauer UM, Nielsen SJ, Brehm A, Kouzarides T. Regulation of E2F1 activity by acetylation. EMBO J. 2000;19:662–71. doi: 10.1093/emboj/19.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marzio G, Wagener C, Gutierrez MI, Cartwright P, Helin K, Giacca M. E2F family members are differentially regulated by reversible acetylation. J Biol Chem. 2000;275:10887–92. doi: 10.1074/jbc.275.15.10887. [DOI] [PubMed] [Google Scholar]
- 51.Unal E, Heidinger-Pauli JM, Kim W, Guacci V, Onn I, Gygi SP, Koshland DE. A molecular determinant for the establishment of sister chromatid cohesion. Science. 2008;321:566–9. doi: 10.1126/science.1157880. [DOI] [PubMed] [Google Scholar]
- 52.Rolef Ben-Shahar T, Heeger S, Lehane C, East P, Flynn H, Skehel M, Uhlmann F. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science. 2008;321:563–6. doi: 10.1126/science.1157774. [DOI] [PubMed] [Google Scholar]
- 53.Zhang J, Shi X, Li Y, Kim BJ, Jia J, Huang Z, Yang T, Fu X, Jung SY, Wang Y, Zhang P, Kim ST, Pan X, Qin J. Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol Cell. 2008;31:143–51. doi: 10.1016/j.molcel.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 54.Borges V, Lehane C, Lopez-Serra L, Flynn H, Skehel M, Rolef Ben-Shahar T, Uhlmann F. Hos1 deacetylates Smc3 to close the cohesin acetylation cycle. Mol Cell. 2010;39:677–88. doi: 10.1016/j.molcel.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 55.Bannister AJ, Miska EA, Gorlich D, Kouzarides T. Acetylation of importin-alpha nuclear import factors by CBP/p300. Current Biol : CB. 2000;10:467–70. doi: 10.1016/s0960-9822(00)00445-0. [DOI] [PubMed] [Google Scholar]
- 56.Wang W, Yang X, Kawai T, Lopez de Silanes I, Mazan-Mamczarz K, Chen P, Chook YM, Quensel C, Kohler M, Gorospe M. AMP-activated protein kinase-regulated phosphorylation and acetylation of importin alpha1: involvement in the nuclear import of RNA-binding protein HuR. J Biol Chem. 2004;279:48376–88. doi: 10.1074/jbc.M409014200. [DOI] [PubMed] [Google Scholar]
- 57.L’Hernault SW, Rosenbaum JL. Chlamydomonas alpha-tubulin is posttranslationally modified by acetylation on the epsilon-amino group of a lysine. Biochemistry. 1985;24:473–8. doi: 10.1021/bi00323a034. [DOI] [PubMed] [Google Scholar]
- 58.Maruta H, Greer K, Rosenbaum JL. The acetylation of alpha-tubulin and its relationship to the assembly and disassembly of microtubules. Journal Cell Biol. 1986;103:571–9. doi: 10.1083/jcb.103.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Piperno G, LeDizet M, Chang XJ. Microtubules containing acetylated alpha-tubulin in mammalian cells in culture. Journal Cell Biol. 1987;104:289–302. doi: 10.1083/jcb.104.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reed NA, Cai D, Blasius TL, Jih GT, Meyhofer E, Gaertig J, Verhey KJ. Microtubule acetylation promotes kinesin-1 binding and transport. Current Biol : CB. 2006;16:2166–72. doi: 10.1016/j.cub.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 61.Chen Y, Zhao W, Yang JS, Cheng Z, Luo H, Lu Z, Tan M, Gu W, Zhao Y. Quantitative acetylome analysis reveals the roles of SIRT1 in regulating diverse substrates and cellular pathways. Mol Cell Proteomics : MCP. 2012;11:1048–62. doi: 10.1074/mcp.M112.019547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–18. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 63.Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, Ning ZB, Zeng R, Xiong Y, Guan KL, Zhao S, Zhao GP. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327:1004–7. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xing S, Poirier Y. The protein acetylome and the regulation of metabolism. Trends Plant Sci. 2012;17:423–30. doi: 10.1016/j.tplants.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 65.Zhang J, Sprung R, Pei J, Tan X, Kim S, Zhu H, Liu CF, Grishin NV, Zhao Y. Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol Cell Proteomics : MCP. 2009;8:215–25. doi: 10.1074/mcp.M800187-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, Li Y, Shi J, An W, Hancock SM, He F, Qin L, Chin J, Yang P, Chen X, Lei Q, Xiong Y, Guan KL. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–4. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci USA. 2006;103:10230–5. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schwer B, Eckersdorff M, Li Y, Silva JC, Fermin D, Kurtev MV, Giallourakis C, Comb MJ, Alt FW, Lombard DB. Calorie restriction alters mitochondrial protein acetylation. Aging Cell. 2009;8:604–6. doi: 10.1111/j.1474-9726.2009.00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Starai VJ, Celic I, Cole RN, Boeke JD, Escalante-Semerena JC. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science. 2002;298:2390–2. doi: 10.1126/science.1077650. [DOI] [PubMed] [Google Scholar]
- 70.Starheim KK, Gevaert K, Arnesen T. Protein N-terminal acetyltransferases: when the start matters. Trends Biochemical Sci. 2012;37:152–61. doi: 10.1016/j.tibs.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 71.Scott DC, Monda JK, Bennett EJ, Harper JW, Schulman BA. N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science. 2011;334:674–8. doi: 10.1126/science.1209307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yi CH, Pan H, Seebacher J, Jang IH, Hyberts SG, Heffron GJ, Vander Heiden MG, Yang R, Li F, Locasale JW, Sharfi H, Zhai B, Rodriguez-Mias R, Luithardt H, Cantley LC, Daley GQ, Asara JM, Gygi SP, Wagner G, Liu CF, Yuan J. Metabolic regulation of protein N-alpha-acetylation by Bcl-xL promotes cell survival. Cell. 2011;146:607–20. doi: 10.1016/j.cell.2011.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hwang CS, Shemorry A, Varshavsky A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science. 2010;327:973–7. doi: 10.1126/science.1183147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Forte GM, Pool MR, Stirling CJ. N-terminal acetylation inhibits protein targeting to the endoplasmic reticulum. PLoS Biol. 2011;9:e1001073. doi: 10.1371/journal.pbio.1001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Friedmann DR, Aguilar A, Fan J, Nachury MV, Marmorstein R. Structure of the alpha-tubulin acetyltransferase, alphaTAT1, and implications for tubulin-specific acetylation. Proc Natl Acad Sci USA. 2012;109:19655–60. doi: 10.1073/pnas.1209357109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liszczak G, Arnesen T, Marmorstein R. Structure of a ternary Naa50p (NAT5/SAN) N-terminal acetyltransferase complex reveals the molecular basis for substrate-specific acetylation. J Biol Chem. 2011;286:37002–10. doi: 10.1074/jbc.M111.282863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee HJ, Lang PT, Fortune SM, Sassetti CM, Alber T. Cyclic AMP regulation of protein lysine acetylation in Mycobacterium tuberculosis. Nature Struct Mol Biol. 2012;19:811–8. doi: 10.1038/nsmb.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brent MM, Iwata A, Carten J, Zhao K, Marmorstein R. Structure and biochemical characterization of protein acetyltransferase from Sulfolobus solfataricus. J Biol Chem. 2009;284:19412–9. doi: 10.1074/jbc.M109.014951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Trievel RC, Rojas JR, Sterner DE, Venkataramani RN, Wang L, Zhou J, Allis CD, Berger SL, Marmorstein R. Crystal structure and mechanism of histone acetylation of the yeast GCN5 transcriptional coactivator. Proc Natl Acad Sci USA. 1999;96:8931–6. doi: 10.1073/pnas.96.16.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rojas JR, Trievel RC, Zhou J, Mo Y, Li X, Berger SL, Allis CD, Marmorstein R. Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide. Nature. 1999;401:93–8. doi: 10.1038/43487. [DOI] [PubMed] [Google Scholar]
- 81.Clements A, Rojas JR, Trievel RC, Wang L, Berger SL, Marmorstein R. Crystal structure of the histone acetyltransferase domain of the human PCAF transcriptional regulator bound to coenzyme A. EMBO J. 1999;18:3521–32. doi: 10.1093/emboj/18.13.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Poux AN, Cebrat M, Kim CM, Cole PA, Marmorstein R. Structure of the GCN5 histone acetyltransferase bound to a bisubstrate inhibitor. Proc Natl Acad Sci USA. 2002;99:14065–70. doi: 10.1073/pnas.222373899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Taschner M, Vetter M, Lorentzen E. Atomic resolution structure of human alpha-tubulin acetyltransferase bound to acetyl-CoA. Proc Natl Acad Sci USA. 2012;109:19649–54. doi: 10.1073/pnas.1209343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yan Y, Harper S, Speicher DW, Marmorstein R. The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate. Nature Struc Biol. 2002;9:862–9. doi: 10.1038/nsb849. [DOI] [PubMed] [Google Scholar]
- 85.Karanam B, Wang L, Wang D, Liu X, Marmorstein R, Cotter R, Cole PA. Multiple roles for acetylation in the interaction of p300 HAT with ATF-2. Biochemistry. 2007;46:8207–16. doi: 10.1021/bi7000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Thompson PR, Wang D, Wang L, Fulco M, Pediconi N, Zhang D, An W, Ge Q, Roeder RG, Wong J, Levrero M, Sartorelli V, Cotter RJ, Cole PA. Regulation of the p300 HAT domain via a novel activation loop. Nature Struc Mol Biol. 2004;11:308–15. doi: 10.1038/nsmb740. [DOI] [PubMed] [Google Scholar]
- 87.Liu X, Wang L, Zhao K, Thompson PR, Hwang Y, Marmorstein R, Cole PA. The structural basis of protein acetylation by the p300/CBP transcriptional coactivator. Nature. 2008;451:846–50. doi: 10.1038/nature06546. [DOI] [PubMed] [Google Scholar]
- 88.Tang Y, Holbert MA, Wurtele H, Meeth K, Rocha W, Gharib M, Jiang E, Thibault P, Verreault A, Cole PA, Marmorstein R. Fungal Rtt109 histone acetyltransferase is an unexpected structural homolog of metazoan p300/CBP. Nature Struc Mol Biol. 2008;15:738–45. doi: 10.1038/nsmb.1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lu L, Li L, Lv X, Wu XS, Liu DP, Liang CC. Modulations of hMOF autoacetylation by SIRT1 regulate hMOF recruitment and activities on the chromatin. Cell Res. 2011;21:1182–95. doi: 10.1038/cr.2011.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sun B, Guo S, Tang Q, Li C, Zeng R, Xiong Z, Zhong C, Ding J. Regulation of the histone acetyltransferase activity of hMOF via autoacetylation of Lys274. Cell Res. 2011;21:1262–6. doi: 10.1038/cr.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lin C, Yuan YA. Structural insights into histone H3 lysine 56 acetylation by Rtt109. Structure. 2008;16:1503–10. doi: 10.1016/j.str.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 92.Stavropoulos P, Nagy V, Blobel G, Hoelz A. Molecular basis for the autoregulation of the protein acetyl transferase Rtt109. Proc Natl Acad Sci USA. 2008;105:12236–41. doi: 10.1073/pnas.0805813105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Albaugh BN, Arnold KM, Lee S, Denu JM. Autoacetylation of the histone acetyltransferase Rtt109. J Biol Chem. 2011;286:24694–701. doi: 10.1074/jbc.M111.251579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kalebic N, Martinez C, Perlas E, Hublitz P, Bilbao-Cortes D, Fiedorczuk K, Andolfo A, Heppenstall PA. The Tubulin Acetyltransferase alphaTAT1 Destabilizes Microtubules Independently of its Acetylation Activity. Mol Cell Biol. 2012 doi: 10.1128/MCB.01044-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fullgrabe J, Kavanagh E, Joseph B. Histone onco-modifications. Oncogene. 2011;30:3391–403. doi: 10.1038/onc.2011.121. [DOI] [PubMed] [Google Scholar]
- 96.Timmermann S, Lehrmann H, Polesskaya A, Harel-Bellan A. Histone acetylation and disease. Cell Mol Life Sci : CMLS. 2001;58:728–36. doi: 10.1007/PL00000896. [DOI] [PMC free article] [PubMed] [Google Scholar]