Abstract
At the end of the growth phase, mouse antral follicle oocytes acquire full developmental competence. In the mouse, this event is marked by the transition from the so-called non-surrounded nucleolus (NSN) chromatin configuration into the transcriptionally quiescent surrounded nucleolus (SN) configuration, which is named after a prominent perinucleolar condensed chromatin ring. However, the SN chromatin configuration alone is not sufficient for determining the developmental competence of the SN oocyte. There are additional nuclear and cytoplamic factors involved, while a little is known about the changes occurring in the cytoplasm during the NSN/SN transition. Here, we report functional analysis of maternal ELAVL2 an AU-rich element binding protein. Elavl2 gene encodes an oocyte-specific protein isoform (denoted ELAVL2°), which acts as a translational repressor. ELAVL2° is abundant in fully grown NSN oocytes, is ablated during the NSN/SN transition and remains low during the oocyte-to-embryo transition (OET). ELAVL2° overexpression during meiotic maturation causes errors in chromosome segregation, indicating the significance of naturally reduced ELAVL2° levels in SN oocytes. On the other hand, during oocyte growth, prematurely reduced Elavl2 expression results in lower yields of fully grown and meiotically matured oocytes, suggesting that Elavl2 is necessary for proper oocyte maturation. Moreover, Elavl2 knockdown showed stimulating effects on translation in fully grown oocytes. We propose that ELAVL2 has an ambivalent role in oocytes: it functions as a pleiotropic translational repressor in efficient production of fully grown oocytes, while its disposal during the NSN/SN transition contributes to the acquisition of full developmental competence.
Keywords: ELAVL2, NSN, SN, chromatin, oocyte, ARE
Introduction
During life, cohorts of mammalian oocytes resting in follicles resume growth in response to hormones and other signaling cues while staying arrested at the prophase of the first meiotic division (reviewed in ref. 1). Growing oocytes increase their volume and accumulate factors that support later development. The end of the growth phase is marked by an acquisition of meiotic competence and, subsequently, full developmental competence of fully grown germinal vesicle (GV) oocytes. A common mark of the acquisition of developmental competence is transcriptionally silenced condensed chromatin that in the mouse transits gradually from the non-surrounded nucleolus (NSN) to the surrounded nucleolus (SN) chromatin configuration.2-4 While NSN oocytes show reduced rate of meiotic maturation and developmental arrest at the 2-cell stage, SN oocytes can develop to the blastocyst stage in vitro.5,6 Chromatin remodeling during the NSN/SN transition and meiotic maturation involves global changes in histone acetylation and methylation (as reviewed in ref. 7). Nuclear transplantation experiments suggest that both nuclear and cytoplasmic factors (but not the chromatin remodeling itself) determine the meiotic and developmental competence of fully grown SN oocytes in antral follicles.5 Nuclear factors associated with the NSN/SN transition include nucleophosphin 2 (NPM2), a nuclear factor required for establishment of the SN configuration,8,9 and plu(rC)-binding protein 1 (PCBP1), which is necessary for the maintenance of transcriptionally silent state, and knockdown of which causes higher proportion of the NSN configuration.10 In addition, nuclear factors POU5F1 (OCT-4) and DPPA3 (Stella) become upregulated during the NSN/SN transition and were suggested to contribute to the developmental competence of SN oocytes.11,12 Cytoplasmic factors contributing to the acquisition of developmental competence during the NSN/SN transition are poorly understood. The whole transcriptome analysis of NSN and SN oocytes revealed that oocytes with NSN and SN configurations differ in several metabolic pathways, and that during the NSN/SN transition oocytes accumulate factors implicated in meiotic maturation and early development.13 The NSN/SN transition has been linked with changes in the distribution of cytoplasmic subcortical maternal complex (SCMC)14 and in formation of subcortical aggregates from RNA binding proteins in the subcortical domain.15 Recently, a proteomic study of oocytes showed that FILIA and MATER, components of SCMC, become upregulated during the NSN/SN transition, and that the loss of MATER reduces formation of SN chromatin.16
The absence of mRNA production between the fully grown SN oocyte and zygotic genome activation at the 2-cell stage means that during the OET, gene expression is largely dependent on post-transcriptional regulation of maternal mRNAs. The post-transcriptional control employs cis-acting elements in 3′-untranslated regions (3′UTRs) of maternal mRNAs and trans-acting factors that bind to them. The number, position, and combination of cis-acting elements and the presence of trans-acting factors offer a complex combinatorial system controlling mRNA localization, stability, and translation.17-20 In vertebrate oocytes, the most studied cis-acting 3′UTR element is the cytoplasmic polyadenylation element (CPE) and its binding protein (CPEB) (reviewed in ref. 21). CPEs and CPEBs play a major role in the meiotic maturation that is driven by cytoplasmic polyadenylation and sequential translational activation of dormant maternal mRNAs.17,22,23 Other highly studied cis-acting motifs are AU-rich elements (AREs). In somatic cells, they regulate stability and translation of up to 8% of mammalian mRNAs.24 At least 24 ARE-binding proteins (AUBPs) have been identified (reviewed in ref. 25), and 13 of them were shown to regulate the ARE-mediated mRNA decay or translation (reviewed in ref. 26). Interestingly, AREs can direct mRNA deadenylation without triggering ARE-mediated mRNA decay in Xenopus oocytes, resulting in stabilization of deadenylated transcript until mid-blastula transition.27 The opposing effects of ARE and CPE on maternal mRNA polyadenylation timing in Xenopus oocytes20 suggest that AREs and AUBPs are key modulators, balancing the CPE-mediated polyadenylation and translational activation.
ELAVL (embryonic lethal abnormal vision like) proteins exemplify AUBPs conserved from Drosophila to mammals.28,29 The vertebrate ELAVL family consists of 4 members: the ubiquitously expressed ELAVL1 (HuR) and the neuron-specific members, ELAVL2 (HuB, HelN1), ELAVL3 (HuC), and ELAVL4 (HuD) (reviewed in ref. 30). ELAVL proteins contain 3 RNA recognition motifs (RRMs). The RRM1 and RRM2 are placed in a tandem, followed by a linker region and RRM3. While the 3 RRMs are highly conserved, the N terminus and the linker region define ELAVL family diversity.31 The linker region (also known as a “hinge”) may play a role in nuclear/cytoplasmic shuttling, as demonstrated for the ELAVL1.32 All members of the ELAVL protein family interact with AREs, but also with U-rich or GU-rich regions and regulate mRNA localization, stability, translation, and alternative splicing.30,33,34 ELAVL proteins are pivotal factors during neuronal development, where ELAVL2 is among the first markers of differentiated neurons.28 Elavl2 gene expression has also been detected in vertebrate germ cells, including Xenopus,35 zebrafish,36 cattle,37 and mouse.38 However, ELAVL2 protein function or Elavl2 transcript regulation has been so far described only in Xenopus or zebrafish, respectively. ELAVL2 plays a role in the translational repression and localization of Vg1 mRNA during Xenopus oogenesis.39,40 In zebrafish germ cells, a DAZL protein specifically stabilized the Elavl2 transcript by binding to its 3′UTR.36 Nevertheless, the role of Elavl2 in mammalian oocytes has not been studied yet.
Here, we present a functional study of Elavl2 in female germ cells. Elavl2 was selected based on a bioinformatic survey of maternal AUBPs over-represented in mouse oocytes. Elavl2 encodes multiple splicing variants with different coding potential. In contrast to neuronal cells, oocytes contain mainly short ELAVL2 (denoted ELAVL2°) isoform that acts as a translational repressor and localizes mainly in the cytoplasm. Elavl2 mRNA is stable during meiotic maturation and is downregulated just after fertilization. In contrast, ELAVL2 protein rapidly decreased already at the end of the oocyte growth phase, during the NSN/SN transition. Since ELAVL2° overexpression during meiotic maturation caused errors in chromosome segregation, the natural reduction of ELAVL2 in SN oocytes may have a significant influence on final maturation of pre-ovulated oocytes. Using the transgenic RNAi approach41 to downregulate Elavl2 expression in incompetent oocytes resulted in lower yields of fully grown GV and meiotically competent metaphase II (MII) oocytes. Thus, it seems that ELAVL2 plays an important functional role before meiotic maturation. Moreover, oocytes with Elavl2 knockdown increased their translation rate in contrast to wild-type (WT) fully grown oocytes. Altogether, Elavl2 encodes an abundant oocyte-specific isoform that acts as a pleiotropic translational repressor important for efficient production of fully grown oocytes, while removal of ELAVL2 during the NSN/SN transition seems to contribute to the acquisition of the developmental competence.
Results
Elavl2 encodes an abundant oocyte-specific isoform
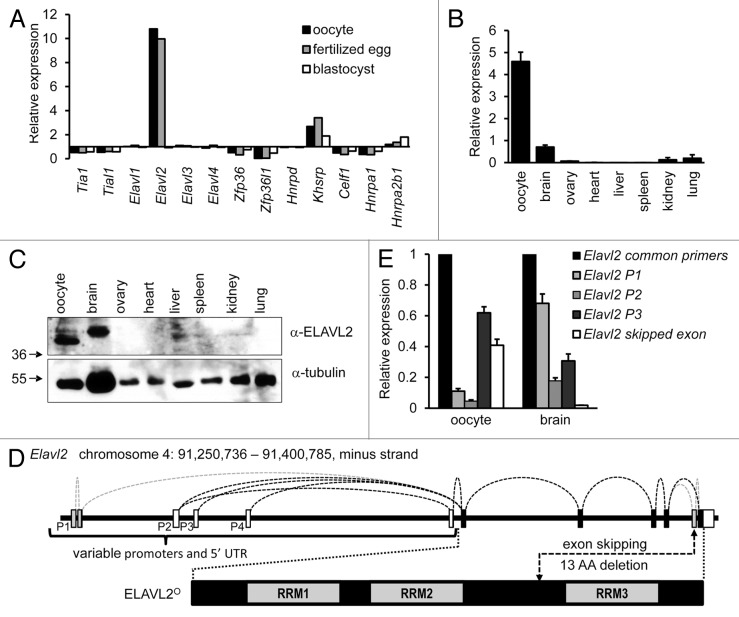
Between the resumption of meiosis and the zygotic genome activation, control of gene expression relies on post-transcriptional regulation of maternal ribonucleoprotein complexes. Since AUBPs contribution to the OET is poorly understood, we searched the BioGPS GNF1M data set42 for maternally expressed AUBPs. We scanned for appropriate candidates whose transcript levels would be enriched during the OET. Elavl2 was found to be the best candidate, because it had the highest relative expression in murine oocytes and fertilized eggs but not in the blastocyst (Fig. 1A). Using primers amplifying a region common for Elavl2 transcript variants annotated in the Ensembl and NCBI databases, we confirmed that Elavl2 mRNA is highly expressed in oocytes relative to a panel of 7 other somatic tissues (Fig. 1B). Consistent with previous results,28 Elavl2 was also readily detectable in the brain (Fig. 1B).
Figure 1. Characterization of Elavl2 expression in oocytes and somatic tissues. (A) Elavl2 transcript level is enriched in oocytes and fertilized eggs relative to somatic tissues. A graph shows an expression ratio of 13 different AUBPs in oocytes, fertilized eggs, blastocysts, and median expression values in somatic tissues calculated from the BioGPS GNF1M.gcrma data set42 providing normalized gene expression data for a comprehensive set of tissues and cell types. (B) RT-PCR analysis of Elavl2 mRNA expression in several tissues using Elavl2 common primers detecting all known Elavl2 splicing variants. Data represent the mean Elavl2 expression ± s.e.m. relative to Hprt1 (n = 2). (C) ELAVL2 expression in different tissues. Twenty micrograms of protein lysates from different tissues and 200 oocytes were loaded per a lane and the blot was probed with ELAVL2 antibody. Tubulin was used as a loading control. The fastest migrating ELAVL2 isoform, which is the most abundant in oocytes, was denoted ELAVL2°. (D) A scheme of the mouse oocyte-enriched ELAVL2 isoform (ELAVL2°) and key differences in alternative Elavl2 splicing variants. Mouse Elavl2 gene is located on chromosome 4 (RRM, RNA binding domain; P1–P4, promoter 1–4; UTR, untranslated region; AA, amino acid). (E) RT-PCR analysis of Elavl2 splicing variants expression in the mouse brain and oocytes. Data represent the mean ± s.e.m. normalized to Hprt1 (n = 3). A signal from Elavl2 common primers was set to one. Elavl2 P1, P2, or P3 primer pairs detect Elavl2 transcripts starting from the promoter 1, 2, or 3, respectively, with or without second-to-last exon deletion. Elavl2 skipped exon primers stands for the Elavl2 splicing variants missing second-to-last exon here denoted Elavl2°. (n, independent experiment performed in triplicates)
In accordance with the Elavl2 transcript expression pattern, ELAVL2 protein was detected in oocytes and the brain but not in other organs (Fig. 1C). The intensity of the ELAVL2 band from 200 fully grown GV oocytes was comparable to that from 20 μg of a brain lysate, indicating that ELAVL2 is indeed highly abundant in oocytes. Western blot revealed the presence of additional faster-migrating ELAVL2 isoforms in oocytes compared with the brain tissue. The neuronal isoform described earlier28,43 is 373 amino acids long (Refseq NP_997568.1 [ELAV-like protein 2 isoform 1]) and has a predicted molecular weight ~41 kDa. In contrast, the main ELAVL2 isoform expressed in oocytes has lower MW (Fig. 1C). Based on its dominant appearance in oocytes, we will refer to the dominant, fast-migrating maternal ELAVL2 isoform as ELAVL2° hereafter.
Elavl2 transcripts show variable 5′ untranslated regions, first coding exons and alternative skipping of the second-to-last exon.28 To understand the origin of the ELAVL2° isoform, we explored existing Refseq and ENSEMBL transcriptome annotations and confronted them with next generation sequencing data44 and RT-PCR analysis. In the oocyte, Elavl2 apparently employs 3 different promoters (P2, P3, P4 in Fig.1D), producing 4 alternatively spliced 5′ UTRs (Fig. 1A and D). Three of these variants are spliced with the first coding exon, which carries the most downstream initiation codon (denoted ATG3 in Fig. 1A). The last 5′UTR variant is alternatively spliced with an additional exon upstream of the exon carrying ATG3. This extra exon carries another potential initiation codon (denoted ATG2 in Fig. 1A), producing the coding sequence longer by 29 codons than transcript variants employing ATG3. In addition, the second-to-last exon skipping appears highly frequent in the oocyte (Fig. 1A and D). The exon skipping causes truncation of the “hinge” region between the RRM2 and RRM3 domains by 13 amino acids (Fig. 1D).
Considering apparent molecular weights of dominant and minor ELAVL2 isoforms observed by western blotting (Fig. 1A and C), we conclude that the ELAVL2° isoform translation most likely initiates from ATG3, and the transcript lacks the second-to-last exon (Fig. 1A and D). Such combination would produce a 347-amino-acids-long protein of a predicted molecular weight ~38 kDa, which is consistent with western blot data (Fig. 1A and C). Results are also in agreement with RT-PCR analysis of different Elavl2 splicing variants in oocytes and brain tissue (Fig. 1E) where it seems that the resulting oocyte-specific Elavl2 transcript starts from promoter P3 and misses the second-to-last exon. The RT-PCR data also correlate with Elavl2 cDNA clones obtained from mouse oocytes (data not shown).
Taken together, an AUBP survey identified Elavl2 as an interesting candidate for a maternally expressed AUBPs contributing to the OET. Subsequent expression analysis revealed that Elavl2 produces a highly abundant oocyte-specific isoform ELAVL2°, which differs from the main neuronal ELAVL2 isoform by a shorter N terminus and a 13-amino acid deletion in the “hinge” region.
ELAVL2° is downregulated during the NSN/SN transition
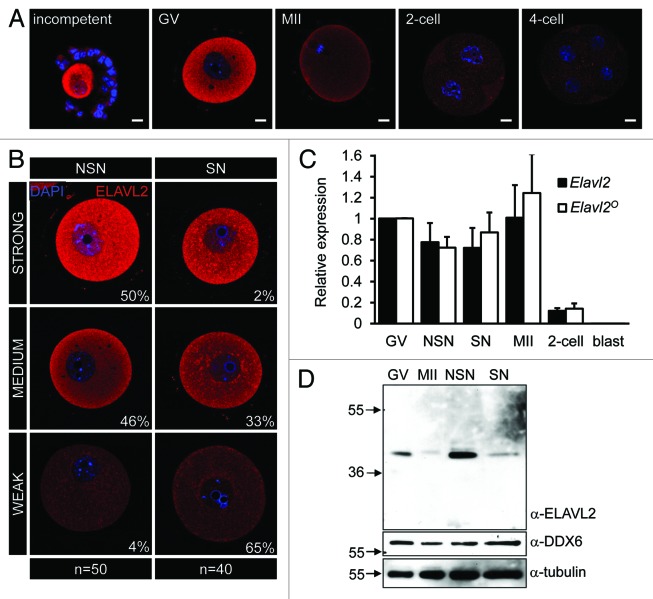
To examine ELAVL2 expression during the OET, we performed immunofluorescent staining and analyzed images by confocal microscopy. We observed robust cytoplasmic staining in meiotically incompetent and fully grown GV oocytes, while the signal was strongly reduced in MII eggs and early embryos (Fig. 2A). In contrast to bovine oocytes,37 we did not observe nuclear ELAVL2 signal. Interestingly, immunofluorescent staining of fully grown GV oocytes showed variable ELAVL2 levels, suggesting that ELAVL2 downregulation was initiated prior to and not by resumption of meiosis, which is a well-known trigger of changes in mRNA stability and translation. To gain more insights into the basis of the ELAVL2 expression variability, we compared levels of ELAVL2 signal in fully grown NSN and SN oocytes. While the majority (96%) of NSN oocytes showed medium-to-strong ELAVL2 staining, the majority (97%) of SN oocytes had weak-to-medium staining (Fig. 2B), indicating that ELAVL2 level is gradually downregulated in fully grown GV oocytes during the NSN/SN transition. Furthermore, ELAVL2 downregulation is apparently initiated at the NSN stage, since reduced ELAVL2 levels were found in approximately half of NSN oocytes (Fig. 2B).
Figure 2. Unique ELAVL2 expression in oocytes. (A) ELAVL2 expression in oocytes and early embryos. Confocal images of incompetent, fully grown GV and MII oocytes, 2-cell and 4-cell embryos after staining with ELAVL2 antibody (red color). DNA staining (DAPI) is shown in blue. The ELAVL2 signal decreases in MII oocyte stage. Scale bar = 10 μm. (B) Confocal images of ELAVL2 expression in fully grown oocytes at non-surrounded and surrounded nucleolus (NSN and SN) stages. Signal intensity was defined using rank-sorted densitometric analysis of 50 NSN and 40 SN oocytes by ImageJ software. While SNS oocytes typically have strong-to-medium ELAVL2 signal intensity, SN oocytes show weak-to-medium intensity suggesting that ELAVL2 is destabilized during the NSN/SN transition. DNA staining (DAPI) is shown in blue. Scale bar = 10 μm. (C) Expression profile of all (Elavl2) and oocyte-specific (Elavl2°) transcript variants in GV, NSN, SN, and MII oocytes, 2-cell embryos and blastocysts (blast). Data were normalized to Hprt1 and represent the mean ± s.e.m from 3 independent RT-PCR experiments. The average Elavl2 and Elavl2° expression were set one in GV oocytes. (D) ELAVL2 downregulation during the NSN/SN transition. One hundred and fifty oocytes from GV, MII, NSN, and SN stages were loaded per a lane. Western blot was probed with ELAVL2 antibody and loading control antibodies (α-tubulin and α-DDX6).
ELAVL2° protein expression is uncoupled from Elavl2 mRNA levels, since RT-PCR analysis revealed that Elavl2 mRNA remains stable from the NSN stage until fertilization. We obtained the same result for both the total Elavl2 mRNA pool and the Elavl2° transcript lacking the second-to-last exon (Fig. 2C). This indicates that ELAVL2 protein is either selectively post-translationally destabilized, or its transcript is not efficiently translated during the NSN/SN transition.
To obtain further insight into the discrepancy between ELAVL2 staining and mRNA analysis, we analyzed ELAVL2 expression in oocytes using western blot. The experiment confirmed strong reduction of the dominant ELAVL2° isoform in SN oocytes compared with NSN oocytes (Fig. 2D). Consistently, ELAVL2° levels in a mixed population of GV oocytes were somewhere between levels observed in NSN and SN oocytes. The lowest ELAVL2° expression was observed in MII eggs (Fig. 2D). Thus, ELAVL2 is the first identified cytoplasmic protein whose level dynamically changes during the NSN/SN transition.
ELAVL2° has properties of a translational repressor
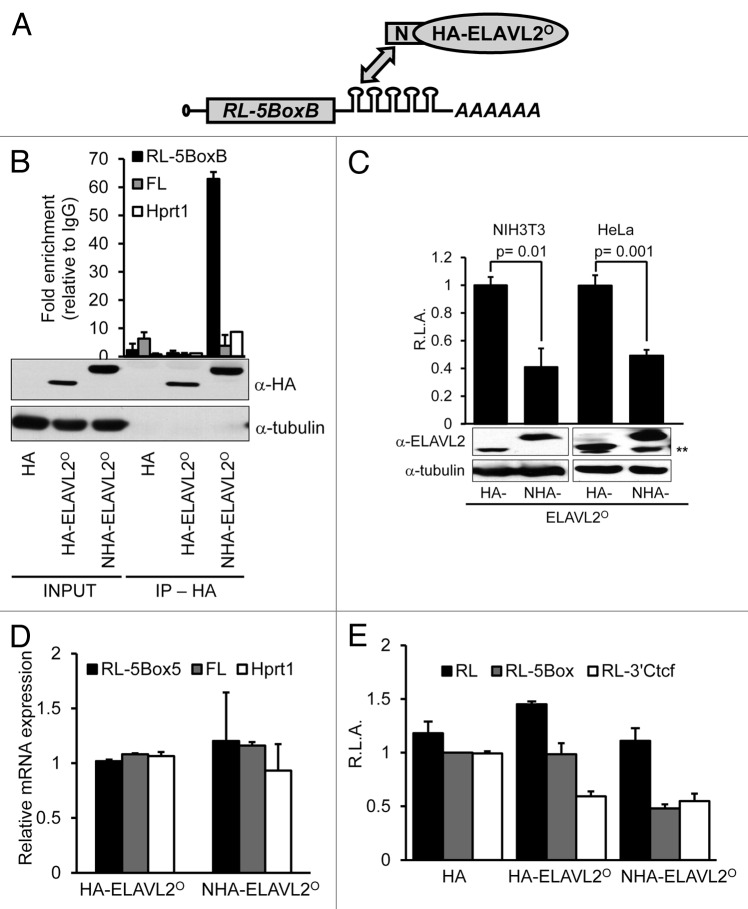
Since the oocyte model has its limits in biochemical studies, we examined functional properties of ELAVL2° isoform in a tethering assay. The tethering assay tests whether binding of a specific protein to a 3′UTR has an effect on mRNA stability or translation. We constructed Renilla luciferase (RL) reporter transcript carrying 5 BoxB hairpins (RL-5BoxB) in the 3′-UTR, that specifically bind the lambda phage λN peptide fused to a candidate protein45 (Fig. 3A). We either expressed ELAVL2° N-terminally fused with a hemaglutinin (HA) tag (HA-ELAVL2°) or to λN peptide-HA tag (NHA-ELAVL2°).
Figure 3. Ectopically-expressed ELAVL2° acts as a translational repressor (A) Tethering assay principle. A Renilla luciferase reporter carrying 5 BoxB binding sites (RL-5BoxB) interacts with the λN-peptide fused to a tested protein. (B) NHA-ELAVL2° specifically interacts with RL-5BoxB. After NIH 3T3 cell transfection with constructs expressing RL-5BoxB, FL, HA tag, and indicated ELAVL2° proteins, lysates were immunoprecipitated with IgG or α-HA antibodies. mRNA was recovered from pellets and selected transcripts were detected by RT-PCR. The data are normalized to the IgG background (mean ± s.e.m., n = 2). Expression levels (INPUT) and immunoprecipitation efficiency (IP, HA) of tagged ELAVL2° proteins were determined by western blot using α-HA antibody. Tubulin served as a loading control. (C) Tethering assay showing 50% reduction of RL-5BoxB signal in presence of NHA-ELAVL2°. HeLa and NIH 3T3 cells were co-transfected with RL-5BoxB, non-targeted firefly luciferase (FL), and HA-tagged ELAVL2° without (HA-ELAVL2°) or with (NHA-ELAVL2°) λN-peptide. The graph represents relative Renilla luciferase activity (R.L.A.) normalized to FL (mean ± s.e.m, n = 4). Expression of tagged ELAVL2° proteins was monitored by western blot (shown below graphs). Tubulin served as a loading control. **Marks non-specific bands in HeLa cell extract. (D) RT-PCR analysis of RL-5BoxB, FL, and Hprt1 mRNAs in cells expressing differently tagged ELAVL2° proteins. Data represent relative mRNA expression normalized to globin mRNA (mean ± s.e.m, n = 2). (F) Luciferase assays in HeLa cells expressing RL, RL-5BoxB or RL-3′Ctcf reporters, non-targeted FL reporter, and HA tag alone, or indicated ELAVL2° proteins. The graph presents relative Renilla activity normalized to FL (R.L.A., mean ± s.e.m, n = 2). The average RL-5BoxB expression in the presence of the HA tag was set to one (n, independent experiment performed in triplicates).
We transiently co-transfected HeLa or NIH3T3 cells with the RL reporter and the ELAVL2°-expressing vector and analyzed the expression by western blot or immunofluorescent analysis (Figs. 2A and 3B). Specific binding of NHA-ELAVL2° to the RL-5BoxB reporter was confirmed by RNA immunoprecipitation (Fig. 3B), which showed that NHA-ELAVL2° binds RL-5BoxB but not transcripts lacking BoxB hairpins, such as firefly luciferase (FL) or Hprt1 mRNAs. Likewise, HA tag alone and HA-ELAVL2° does not bind RL-5BoxB, FL or Hprt1 transcripts (Fig. 3B).
The specific ELAVL2° effect on the RL-5BoxB expression was inferred from comparison of luciferase activities in cells transfected with plasmids expressing ELAVL2° tagged without (HA-ELAVL2°) and with (NHA-ELAVL2°) the N-peptide. The co-transfected FL reporter, which binds neither ELAVL2° protein constructs, served as a control of transfection efficiency. In the luciferase assay, NHA-ELAVL2° acted as a repressor and reduced RL-5BoxB expression by approximately 50% relative to untethered HA-ELAVL2° in both mouse NIH3T3 and human HeLa cells (Fig. 3C). Western blot of transfected cells revealed that the protein level of NHA-ELAVL2° was higher than that of HA-ELAVL2° (Fig. 3C). To confirm that the inhibitory effect was not simply generated by higher level of NHA-ELAVL2° expression, we repeated the assay with lower amounts of the NHA- ELAVL2°-expressing vector. We still observed, ~50% reduction of RL-5BoxB expression when 4 times less of the NHA-ELAVL2°-expressing vector (relative to HA-ELAVL2°-expressing vector) was used (Fig. 2A and B).
The ELAVL2°-inhibitory effect on the RL reporter may be mediated by translational repression and/or mRNA degradation. Therefore, we analyzed steady-state levels of RL-5BoxB transcripts in transfected cells using RT-PCR. Since the RL-5BoxB transcript did not decrease upon NHA-ELAVL2° co-transfection (Fig. 3D), we concluded that the ELAVL2°-mediated repression occurs at the translational level.
To further examine the repressive ability of ELAVL2°, we generated an additional RL reporter carrying Ctcf 3′UTR (RL-3′Ctcf), which contains 2 ELAVL2 consensus binding sites and has been identified as an ELAVL2 target.33 This reporter should be inhibited by ELAVL2° directly and independently of the N-peptide. Indeed, in transiently transfected HeLa cells, HA-ELAVL2° repressed the RL-3′Ctcf reporter to the same extent as NHA-ELAVL2°-repressed RL-5BoxB reporter in previous tethering experiments (Fig. 3E). Taken together, the tethering assay results suggest that ELAVL2° acts as a translational repressor.
High levels of ELAVL2° interfere with meiotic maturation
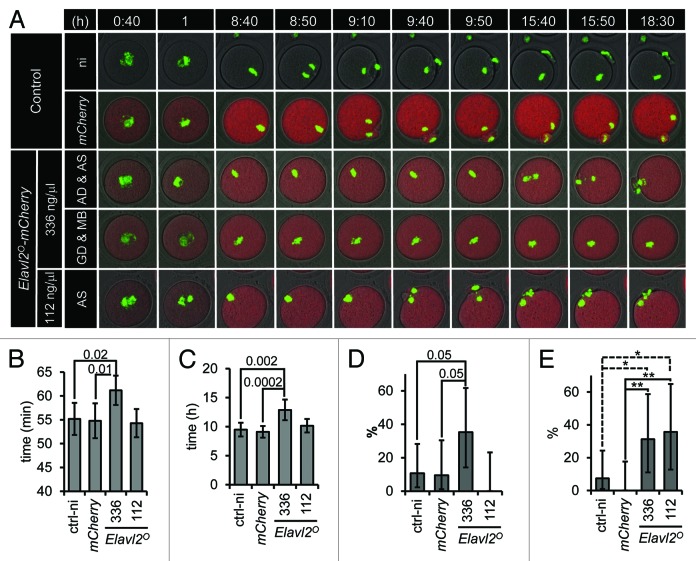
ELALVL2 downregulation during the NSN/SN transition raised the question of whether reduced ELAVL2° level is important for subsequent development. Therefore, we artificially raised ELAVL2° level during meiotic maturation by microinjecting recombinant Elavl2°-mCherry mRNA into fully grown GV oocytes and observed maturing microinjected oocytes by time-lapse confocal microscopy. We used oocytes from H2B-GFP mice allowing us to monitor expression of microinjected recombinant mRNA and chromosome dynamics at the same time. We cultured microinjected GV oocytes for 2 h in the presence of milrinone to allow ELAVL2° accumulate prior to meiotic resumption. After washing milrinone out, oocytes were left to mature for 18.5 h. The images were captured every 10 min. Representative images are depicted in the Figure 4A, a movie made from all acquired images is provided in the supplementary material.
Figure 4. ELAVL2° overexpression affects meiotic maturation. (A) Representative time-lapse images of maturing oocytes. Oocytes from H2B-GFP (CD1 strain) mice were used to visualize chromatin (green color). Non-injected (ni) or injected oocytes with mCherry or Elavl2°-mCherry mRNAs were let to mature and confocal images (starting 40 min after milrinone removal) were acquired every 10 min for 18 h. Two Elavl2°-mCherry mRNAs concentration were used: high, 336 ng/μl (336); low, 112 ng/μl (112). First 2 image sequences show controls, the third and fourth ones depict anaphase delay (AD) with abnormal chromosomal segregation (AS) and germinal vesicle brakedown delay (GD) together with metaphase I block (MB) when high concentration of Elavl2° was used, respectively. The last line shows abnormal chromosomal segregation (AS) without anaphase delay when low amount of Elavl2° was injected. All image sequences represent maximum projection of H2B-GFP channel, single confocal sections of Elavl2°-mCherry and bright field channels. Totally 27 control non-injected, 23 control mCherry, 17 Elavl2°-mCherry (336 ng/ul), and 14 Elavl2°-mCherry (112 ng/ul) oocytes were imaged and analyzed, respectively. (B) Elavl2°-mCherry-injected in high concentration (336) into oocytes caused slightly but significantly delayed GVBD (mean value ± 95% confidence interval (ci), P = 0.01, t test). (C) Anaphase I (defined as the time between the GVBD and the first visible sign of chromosome segregation) is delayed in oocytes with a high but not a low level of Elavl2°-mCherry mRNA (mean values ± 95% ci, P = 0.0002, t test). (D) In contrast to 10% of mCherry-injected oocytes, 35% of Elavl2°-mCherry-injected oocytes stopped development at metaphase I (mean value ± 95% asymmetric ci, P = 0.05, Chi-Square). (E) Approximately 31 and 35% of high and low Elavl2°-mCherry-injected oocytes respectively exhibit problems with chromosome segregation (high: mean value ± 95% asymmetric ci, P = 0.008, Chi-Square; low: mean value ± 95% asymmetric ci, P = 0.005, Chi-Square).
Strong ELAVL2° overexpression had a negative impact on meiotic maturation when compared with controls expressing mCHERRY alone (Fig. 4A). Oocytes microinjected with mCherry mRNA were indistinguishable from non-injected control oocytes (Fig. 4B–E). Oocytes overexpressing ELAVL2° underwent slight but significant delay in GVBD (Fig. 4B) and strong delay in the anaphase I onset (Fig. 4C) when compared with mCHERRY-expressing oocytes. A significant number of ELAVL2°-overexpressing oocytes (35%, P = 0.0527) were arrested at metaphase I in comparison to mCHERRY control (Fig. 4D). Yet, 3 times lower amount of microinjected Elavl2 mRNA did not induce these phenotypes, suggesting that they are induced by ELAVL2 overexpression above a threshold. However, increased chromosome segregation errors (31% and 35%, P = 0.0085, and P = 0047, high and low dose, respectively) were observed with both Elavl2° mRNA doses (Fig. 4E). This suggests that low levels of ELAVL2° during meiotic maturation might be significant and functionally important for the developmental competence of fully grown GV oocytes.
Premature Elavl2 knockdown leads to a fewer ovulating oocytes
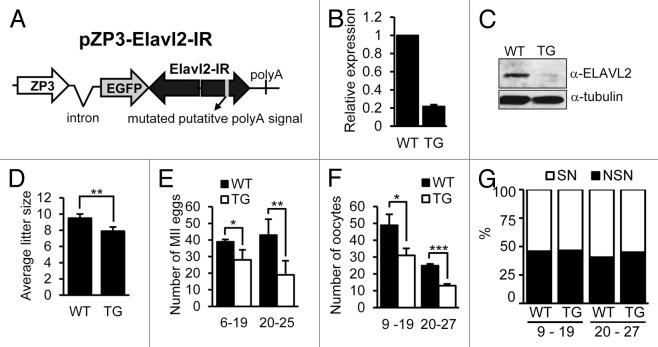
To assess effects of premature Elavl2 downregulation during oocyte development, we created a transgenic RNAi model. We used an expression vector with long double-stranded RNA (dsRNA) under the ZP3 promoter, which induces RNAi-mediated sequence-specific mRNA knockdown solely in oocytes (reviewed in ref. 46). dsRNA produced by the pZP3-Elavl2-IR transgene (Fig. 5A) was directed against 4 coding exons to target all Elavl2 splicing variants. Thirty-nine mice were obtained upon pronuclear microinjection and embryo transfer; 8 mice were positive for the pZP3-Elavl2-IR transgene and were further mated. Five founders produced transgene-positive F1 progeny and gave rise to 5 lines, which were further characterized (denoted after founder animal tg1601, tg1629, tg1632, tg1635, tg1639). The strongest Elavl2 mRNA knockdown was observed in tg1629 animals (~80%, Fig. 5B), whereas mRNA in tg1632 and tg1639 was eliminated only by 60% and 50%, respectively (Fig. 3A). At the protein level, we observed strong ELAVL2° knockdown only in oocytes from the tg1629 line (Figs. 3A, B, and 5C). Therefore, all subsequent experiments were performed using the line tg1629.
Figure 5. Initial characterization of Elavl2 knockdown in transgenic mice oocytes. (A) Design of the transgenic RNAi construct. A putative polyA signal was mutated in the inverted repeat sequence (pZP3-Elavl2-IR). (B) Elavl2 mRNA downregulation in fully grown GV oocytes from transgenic (TG) mice. Data from 4 independent RT-PCR experiments represent the mean Elavl2 expression ± s.e.m. normalized to Hprt1. Relative Elavl2 expression in wild-type (WT) oocytes was set to one. (C) Western blot of ELAVL2 protein expression in fully grown GV oocytes from WT and TG mice. One hundred and fifty oocytes were loaded per lanes. (D) An average litter size of WT and TG females below 20-weeks-of-age. Each mean value (± s.e.m) represents an average of 18 litters. (E) The average number of MII oocytes obtained from superovulated WT (n = 10) and TG (n = 10) mice younger and older than 20 weeks. (F) The average number of fully grown GV oocytes recovered from WT (n = 15) and TG (n = 23) mice younger and older than 20 weeks. (G) The ratio of NSN and SN oocytes obtained from WT (n = 13) and TG (n = 23) females is similar for all animals. One-tailed t test was used for statistical analysis. P < 0.05, 0.01, and 0.001 were denoted by 1, 2, and 3 asterisks, respectively.
Initial studies of the phenotype in transgenic (TG) animals showed that these females were fertile but had slightly smaller average litter size than their WT littermates (7.9 and 9.5 newborns, respectively, P value < 0.01; Fig. 5D). When MII eggs were isolated from superovulated TG animals, we observed up to 50% reduced yield compared with WT littermates (Fig. 5E). The same effect was monitored when fully grown GV oocytes were isolated from non-superovulated animals. The yield of fully grown GV oocytes from TG animals at 9–19 and 20–27-weeks-of-age was ~40% and ~50% lower than from WT littermates, respectively (Fig. 5F).
Since ELAVL2 expression was naturally strongly and rapidly reduced during the NSN/SN transition, we tested whether the NSN/SN chromatin configuration distribution differed in oocytes from WT and TG females. Nevertheless, we found that upon Elavl2 knockdown the NSN/SN ratio did change (Fig. 5G). Thus, these data demonstrated that ELAVL2 reduction did not involve the NSN/SN chromatin remodeling. As ELAVL2 downregulation starts apparently at the NSN stage (Fig. 2B), it indicates that ELAVL2 functions in parallel independently of the chromatin configuration changes.
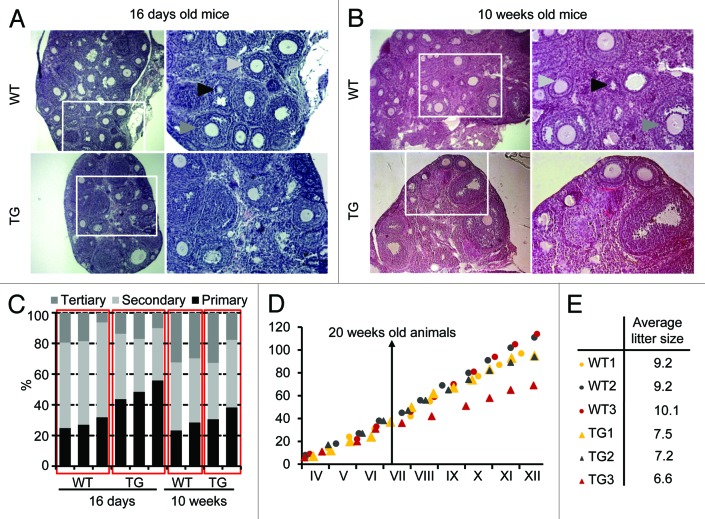
We did not pinpoint the trigger of the lower yield of fully grown GV and MII oocytes yet. Overall morphology of TG ovaries from both young and old mice appeared normal (Fig. 6A and B). Detailed analysis of follicles in histological sections showed increased abundance of primary follicles relative to secondary and antral (tertiary) ones in 16-d-old TG mice (Fig. 6C). However, this difference disappeared in 10-week-old mice. Furthermore, since the absolute number of secondary and tertiary follicles per ovary did not differ between TG and WT ovaries in respect to the lower yield of ovulating oocytes, now the significance of higher number of primary follicles is unclear.
Figure 6. Ovarian histology and fertility of Elavl2-knockdown mice. Hematoxylin and eosin staining of ovarian sections from (A) 16-day- and (B) 10-week-old mice. Black, light gray, and dark gray arrows point on primary, secondary and tertiary (antral) oocytes, respectively. (C) Frequency of primary, secondary and tertiary (antral) oocytes in WT and TG ovaries. (D) Long-term breeding performance. Three pairs of littermates were continuously mated for 9 mo. The y-axis shows the cumulative litter size upon each litter. The x-axis represents calendar months. (E) The average litter size of WT and TG mice during 9 months of breeding.
The reduced number of fully grown GV oocytes and ovulated MII eggs isolated from TG ovaries, prompted us to test whether the normal litter size would decline with age. A similar phenomenon has been observed in transgenic RNAi-mediated knockdown of CPEB.18 Therefore, we continuously mated 3 TG females and their 3 WT female littermates for 9 months while monitoring their cumulative litter sizes. Consistent with reduced litter size observed in the pZP3-Elavl2-IR mice (Fig. 5D), long-term breeding showed that the TG females produced less progeny and had smaller average litter sizes than their WT littermates (Fig. 6D and E). In addition, the litter size of TG mice has declined with age. While WT mice maintained approximately the same litter size during the experiment (4–19 weeks: 9.6 and 20+ weeks: 9.5), the litter size of TG mice went down from 8.3 in 4–19-weeks-old mice to 6.3 in mice older than 20 weeks. However, we did not observe an age-associated strong reduction of fertility as in the CPEB case. Thus, consistent with ovarian histology, the Elavl2 knockdown does not strongly reduce the pool of resting ovarian oocytes.
Enhanced protein synthesis in oocytes depleted of ELAVL2
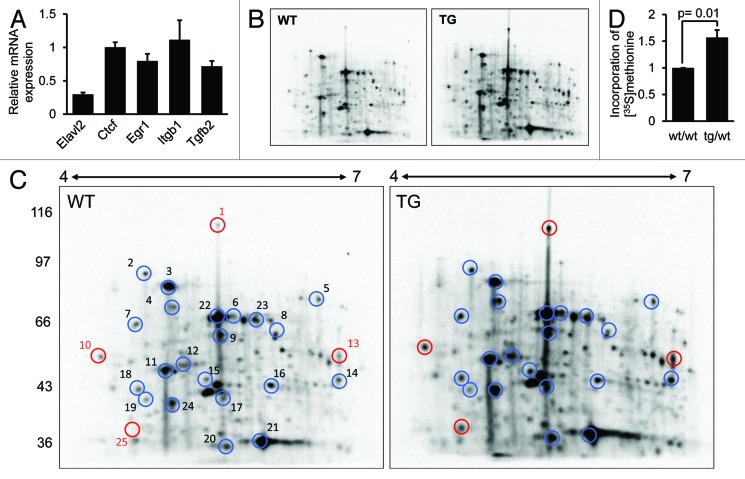
As mentioned above, tethering assays showed that ELAVL2° inhibits translation but does not destabilize cognate mRNAs. Moreover, in agreement with unchanged relative RL-5BoxB mRNA expression in the presence or absence of λN peptide-fused ELAVL2° (Fig. 3E), we found that mRNA stability of 4 previously identified ELAVL2 targets were not influenced by Elavl2 knockdown in TG oocytes (Fig. 7A).33 Therefore, we used our transgenic RNAi model and examined protein translation in fully grown GV oocytes using metabolic labeling with [35S]methionine and 2-dimensional (2D) gel electrophoresis. Approximately 200 spots were recognized in each 2D gel autoradiogram. The overall pattern of radiolabeled protein distribution for WT and TG oocytes was similar (Fig. 7B). However, we observed a higher incorporation of [35S]methionine into proteins in TG oocytes where most of the spots showed a similarly increased signal intensity (average difference 1.9×, blue circles) relative to WT oocytes (Fig. 7C). Correspondingly, the rate of protein synthesis measured by the incorporation of [35S]methionine into TCA-precipitable material was enhanced in TG compared with WT oocytes (mean value from 3 independent experiments:1.57× ± 0.14 s.d.) (Fig. 7D). These results suggest that in mouse oocytes Elavl2 knockdown causes global increase in protein synthesis. This is likely an indirect effect of ELAVL2 downregulation, since ELAVL2 should selectively bind U- or GU-rich regions,30,34,43,47,48 or ARE motifs that are found in approximately 5% of mouse transcripts.24 Accordingly, only several spots showed higher [35S]methionine incorporation than the 1.9× average difference (Fig. 7C, red circles; or Table 1). These spots could represent proteins encoded by transcripts directly regulated by ELAVL2. Notably, we did not observe new prominent spots in Elavl2-knockdown oocytes, indicating that ELAVL2 reduces translation of maternal transcripts during oocyte growth rather than completely blocking it.
Figure 7.Elavl2 knockdown enhances translation in fully grown GV oocytes. (A) The mRNA stability of 4 previously identified ELAVL2 targets stays unchanged in TG vs. WT oocytes. The graph represents relative mRNA expression normalized to Hprt1 mRNA measured by RT-PCR (mean ± s.e.m, n = 2). Elavl2 mRNA is a control of knockdown efficiency. (B) Images of 2-dimensional (2D) electrophoresis. (C) The graph represents total [35S]methionine incorporation measured by trichloracetic acid (TCA) precipitation (mean ± s.e.m, n = 2, t test). (D) 2D electrophoresis shows [35S]methinone incorporation into proteins in WT and TG fully grown GV oocytes. The intensity ratio between the total pixel volumes of the entire gel of TG vs. WT oocytes is 1.9 and corresponds to the average increase of spot intensity in the TG sample. All 25 selected spots are presented in a Table 1 and those with relative intensity change in TG sample higher than 2.85 (1.5× above the average difference) are highlighted in red. These spots represent putative primary targets of ELAVL2. The experiment was been performed twice with a similar result.
Table 1.
| Spot number | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TG/WT | 7.8 | 2.0 | 1.7 | 2.1 | 1.5 | 1.9 | 2.1 | 0.8 | 2.2 | 2.9 | 2.1 | 1.9 | 3.4 | 2.4 | 2.0 | 1.1 | 2.3 | 1.9 | 1.9 | 2.2 | 1.5 | 1.4 | 1.4 | 2.1 | 4.1 |
The table with 25 selected spots from 2D electrophoresis showing the [35S]methinone incorporation into proteins in TG vs. WT fully grown GV oocytes (relative intensity change).
Discussion
While evaluating expression of AUBPs during the OET, Elavl2 emerged as an outstanding candidate for functional analysis, because it showed the most enriched transcript levels in the oocyte relative to somatic tissues. Although, ELAVL2 was originally described as a neuronal protein,28,43 several studies suggest conserved Elavl2 expression in vertebrate germ cells.35-38 We are first to show existence of ELAVL2°, the major ELAVL2 isoform expressed in oocytes. ELAVL2° is shorter than the major neuronal ELAVL2 isoform, localizes mostly in the cytoplasm, does not alter stability of its target mRNAs, acts as a translational repressor, and it is strongly downregulated during the NSN/SN transition.
Elavl2 splicing is complex. We have identified 4 alternative different 5′ and 2 alternative 3′ transcript ends, which have the potential to encode 4 different proteins in mouse oocytes (Fig. 1A and C). The shortest of these protein variants seems to correspond to the ELAVL2° isoform. This conclusion is consistent with western blot data, sequence tag frequencies observed in high-throughput sequencing,44 RT-PCR, and mRNA cloning results. ELAVL2° isoform can be produced from 3 different transcripts differing in the transcription start site and the first non-coding exon. Due to the second-to-last exon skipping, ELAVL2° carries a truncation of the functionally important “hinge” region.31,32 Several studies of ELAVL proteins indicate that the hinge affects nuclear–cytoplasmic shuttling, alternative splicing, protein–protein interaction (mostly oligomerization), and/or RNA-binding specificity due to a changed distance between the ARE-binding sequence (RRM2) and the RNA-protein complex stabilizing domain (RRM3).30,32,49-51 Effects of the hinge truncation on the cellular localization, protein, and RNA interaction of ELAVL2° are currently under investigation.
While highly related ELAVL proteins, such as the mostly studied ELAVL1, are mainly connected with a translational activation,52,53 ELAVL2° exhibits features of a translational repressor. This is likely not an artifact, although proteins that do not naturally regulate translation and/or RNA stability may show some activity in the tethering assay. For example, it has been shown that LacZ tethering positively affects the tethered reporter by apparently increasing its mRNA stability.45 However, additional data minimize the possibility that the translational repression mediated by ELAVL2° is the tethering assay artifact. First, ELAVL proteins are well-established direct RNA binders and post-transcriptional regulators.54 Second, we observed that the translational repression was independent on the N-peptide/BoxB tethering system. The luciferase reporter carrying Ctcf 3′ UTR with 2 known ELAVL2 binding sites33 was repressed by the non-tethered ELAVL2° isoform (Fig. 3E). Third, the tethering assay results are consistent with Elavl2-knockdown data performed in oocytes where we detected stable steady-state levels of known ELAVL2 mRNA targets (Fig. 7A) and higher protein synthesis (Fig. 7C). Finally, our results are consistent with a study of Xenopus ELAVL2 homolog, ElrB, showing that it plays a role in the translational repression of Vg1 mRNA during early oogenesis.39 ELAVL2 appears to influence the translation in 2 ways: (1) stronger selective repression of primary targets, which is a direct effect of ELAVL2 binding, and (2) weaker global reduction of translation. Whether the global effect on translation is a direct ELAVL2 function or it is a secondary effect of translational repression of a primary target remains to be determined. Interestingly, our preliminary data indicate that reduced ELAVL2 levels anti-correlate with higher translation in SN oocytes (Fig. 8), where increased translation in SN oocytes is reminiscent of the Elavl2-knockdown effect on translation. Furthermore, globally increased translation during NSN/SN transition represents an interesting research direction for understanding cytoplasmic changes and formation of full developmental competence in antral SN oocytes.
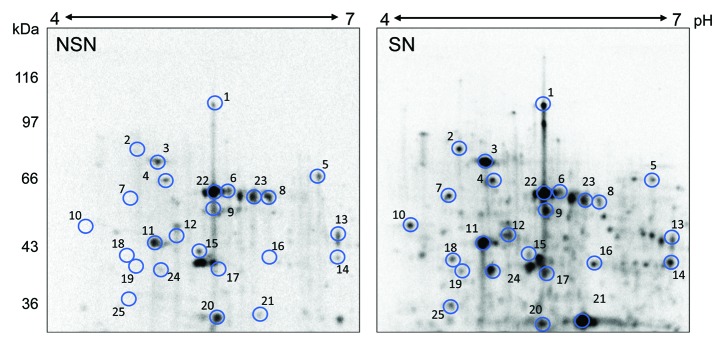
Figure 8.Elavl2 knockdown enhances translation in fully grown GV oocytes. 2D electrophoresis shows [35S]methinone incorporation into proteins in NSN and SN fully grown GV oocytes. The intensity SN/NSN ratio between the total pixel volumes of the entire gel is 6.7. The experiment was been performed twice with a similar result. Selected spots match those analyzed in Figure 7.
ELAVL2 integration with other cytoplasmic factors, such as SCMC components14 or the subcortical domain,15 remains unclear. Given the ELAVL2 expression pattern and proposed role in translational repression, ELAVL2 does not seem to be an integral component of either of the 2 above-mentioned subcortical phenomena, which are highly pronounced in SN oocytes. We show that ELAVL2 downregulation did not affect changes on chromatin, suggesting that ELAVL2 is functionally distinct from the SCMC system, which is involved in the NSN/SN chromatin transition.16 However, since a recent proteomic study showed that FILIA and MATER become upregulated during the NSN/SN transition, it would be interesting to test whether transcripts encoding SCMC components are not regulated by ELAVL2.16
ELAVL2 expression appears functionally important but non-essential during oocyte growth, because premature ELAVL2 downregulation by transgenic RNAi caused a lower yield of fully grown GV oocytes and MII eggs. Transgenic RNAi bears a risk of insertion site effects, which is minimized when the same phenotype is observed in different transgenic lines. Unfortunately, we did not obtain a second transgenic line with a strong ELAVL2 protein depletion. However, it is most likely that the phenotype of tg1629 mice is caused by Elavl2 knockdown. First, TG mice appear normal while phenotypes are associated with cells expressing the transgene. Second, effects observed in TG oocytes are consistent with the role of ELAVL2 in the translational repression (Fig. 7C). Since histology of ovaries from transgenic mice did not show any particular defect in follicular growth, we speculate that there is a stochastic loss of growing oocytes containing low levels of ELAVL2. The premature downregulation of ELAVL2 causes ~50% lower yield of oocytes but smaller reduction in litter size (10–20%). However, even a minor difference in litter sizes could represent a significant selective force for evolving high levels of ELAVL2 in growing and NSN oocytes.
While the NSN/SN transition studies have so far focused on nuclear changes, cytoplasmic changes were also expected to contribute to the acquisition of developmental competence.5 From this perspective, the most revealing observation made in our study is strong ELAVL2° downregulation during the NSN/SN transition followed by low levels of ELAVL2° in fully grown meiotically competent SN oocytes and later stages of the OET. The significance of naturally reduced ELAVL2 levels in fully grown antral SN oocytes remains unclear. Overexpression experiments with ELAVL2° in meiotically maturing oocytes consistently showed increased chromosome segregation errors, while other effects, observed when high amounts of Elavl2° mRNA were microinjected, could be a phenotype of highly overexpressed ELAVL2°. Importantly, reduced ELAVL2 levels were also found in MII by mass-spectroscopy-based proteomic analysis of GV, MII, and fertilized oocytes.55 This makes highly unlikely that our results might reflect some modification on the region recognized by the ELAVL2 antibody used in this publication. The molecular trigger and mechanism for ELAVL2 downregulation remain to be elucidated. Presumably, ELAVL2 is destabilized via ubiquitination and subsequent proteasome-mediated degradation. Active protein degradation is consistent with that fact that Elavl2 mRNA accumulates in polysomal fractions during meiotic maturation of mouse oocytes.56
In conclusion, our data indicate that ELAVL2 has an ambivalent role in oocytes. It functions as a translational repressor during the oocyte growth, while its downregulation during the NSN/SN transition presumably contributes to the acquisition of developmental competence.
Materials and Methods
Plasmid constructs
Construction of plasmids used in this study is described in detail in the Appendix in the Supplementary Materials.
Transgenic mice
Transgenic mice were generated in the transgenic facility of the Institute of Molecular Genetics by injecting linearized pZP3-Elavl2-IR transgenic construct (Fig. 5A) into male pronuclei of C57BL/6 1-cell embryos. Transgene-positive mice (identified by PCR; primer sequences are in the Table 1A) were further mated with BALB/C mice to reduce a possibility of epigenetic silencing of the transgene on the inbred background.
Oocyte isolation and culture
Meiotically incompetent, fully grown GV and MII oocytes were isolated from ovaries of 12-day-old and 8–25-week-old mice, respectively, as described previously.15 To distinguish NSN and SN nucleolus stages, GV oocytes were stained by Hoechst 33342 (1 μg/1ml in M2 medium with 0.2 mM IBMX) for 5 min at 37 °C and 5% CO2 atmosphere and rapidly sorted under a microscope Nikon Eclipse TE300 (Nikon).
RNA isolation and real-time (RT)-PCR
Total RNA from 20 oocytes, tissue biopsies, or cultured cells was isolated using RNAzol (Molecular Research Center, Inc) according to the manufacturer’s protocol. Total RNA from 20 oocytes or 1 μg of total RNA from other samples were reverse transcribed using RevertAid reverse transcriptase (Fermentas) and random hexanucleotides (Fermentas) in 10 µl reactions. The final cDNA was diluted 10 times with water, and a 3-µl aliquot was applied as a template for a 10-µl PCR reaction. RT-PCR was performed on the Mx3000P (Stratagene) machine using Maxima SYBR Green qPCR Master Mix (Fermentas). The RT-PCR was performed in triplicates from 2 or 3 independently obtained biological samples. Relative expression of target mRNAs was normalized to hypoxanthine–guanine phosphoribosyltransferase (Hprt1) housekeeping gene. All primers are listed in the Table 1A.
Cell culture, transfection, and luciferase assay
Human HeLa and mouse NIH3T3 fibroblast cells were maintained in Dulbecco modified Eagle medium (DMEM, Sigma) supplemented with 10% FCS (Sigma), penicillin (100 U/ml, Invitrogen), and streptomycin (100 µg/ml, Invitrogen) at 37 °C and 5% CO2 atmosphere. Transfections and luciferase assays were performed as described previously.57 Cells grown in 24-well plates were transfected with 200 ng of HA- or NHA-tagged vectors expressing ELAVL2° protein, 50 ng of phRL (RL), or phRL-5BoxB (RL-5BoxB) plasmids and 1 ng of firefly (FL) reporter plasmid. After 48 h, cells were washed with PBS and analyzed using the Dual-Luciferase Reporter Assay (Promega).
Histology and immunofluorescent staining
Ovaries were fixed in Bouin fixative for 24 h and processed for paraffin sections. Eight-micrometer sections were stained with hematoxylin and eosin and photographed using a Leica DM 6000B microscope (Leica Microsystems).
Frequency of primary, secondary, and tertiary (antral) oocytes in WT and transgenic (TG) ovaries were determined from the same number of (26) serial sections from each ovary (WT and TG) that were examined under 2 magnifications (20× and 40×). Numbers of the primary follicles (oocyte surrounded with one layer of cuboid granulosa cells), the secondary follicles (oocyte surrounded with 2 or more layers of granulosa cells), and the tertiary (antral) follicles were recorded. Follicles were counted only on sections where an oocyte was present.
Immunofluorescent staining of transfected HeLa cells using rabbit polyclonal α-HuB (ELAVL2; 1:150, H1538, Sigma) and rat monoclonal α-HA (1:200; 3F10, Roche) was performed as described previously.58 Samples were mounted in FluoroMount reagent (SouthernBiotech). Immunofluorescent staining of oocytes with 18033 antibody (human index 18033 [1:500; a kind gift from Marvin J Fritzler]) or α-HuB rabbit polyclonal (1:150; H1538, Sigma) were performed as described previously.15,59 For poly(A) mRNA co-localization, 1 μM tetramethylrhodamine-oligo(dT)18 probe (TAMRA-oligo[dT]18; Sigma) was included in the secondary antibody mixture as described previously.59 Images were obtained using a TSC SP5 confocal microscope (Leica Microsystems).
To calculate relative expression of ELAVL2 from confocal images (Fig. 2B), signal intensity of ELAVL2 was established on a LAS-Life software by defining the same region of interest (ROI) for all tested oocyte (50 NSN and 40 SN GV oocytes from nine mice). Signal intensity was defined using rank-sorted densitometric analysis by ImageJ software. The lowest and highest ELAVL2 signal intensity was set 0 and 1, respectively. The range between 0–0.33 was marked as “weak”, 0.34–0.67 was marked as “medium”, and 0.68–1 was marked as “strong”.
Western blot
Oocytes (150–200) were directly lysed in the SDS loading buffer. Tissue biopsies or cultured cells were lysed with Nonidet P-40 buffer (50 mm TRIS-HCl, pH 8, 120 mm NaCl, 1% Nonidet P-40, 1 mm EDTA, 1:100 protease inhibitor cocktail set III (Calbiochem), 1:50 phosphatase inhibitor cocktail set V [Calbiochem]) for 10 min on ice and centrifuged at 15 000 × g for 15 min at 4 °C to remove cellular debris. Oocyte lysates or 20 μg of total protein/per lane were resolved by SDS-PAGE and transferred to a PVDF membrane that was blocked in 2.5% milk in TBS buffer for 1 h at RT and incubated with primary antibodies (α-HuB (1:1000); mouse monoclonal α-tubulin (1:5000; T6074, Sigma); rabbit polyclonal α-DDX6 (1:5000; A300–461A, Bethyl Laboratories) overnight at 4 °C. The PVDF membranes was then washed 3 times with 0.05% TBS-Tween and incubated with HRP-conjugated secondary antibody (1:10 000; Pierce) for 1 h at RT. Protein bands were visualized with the Supersignal Chemiluminiscence kit (Thermofisher).
mRNA–protein immunoprecipitation (RIP)
Immunoprecipitation of mRNA–protein complexes was performed with small modifications according published protocol.60 Briefly, HA-ELAVL2°, NHA-ELAVL2° transfected HeLa cells were lysed, and 1 µg of rat anti-HA or rabbit anti-IgG was used for immunoprecipitation of 300 μg of proteins. One M urea was omitted from a washing buffer, immunoprecipitated mRNA was eluted by RNAzol reagent and glycogen RNA grade (Fermentas) was used as a carrier instead of yeast tRNA. RNase-free DNase I (Fermentas) was used for DNase treatment before reverse transcription. Reverse transcription was performed using SuperScript III (Invitrogen) and RT-PCR was performed as described above.
[35S] methionine labeling of oocytes and 2D-PAGE experiment
Fully grown GV oocytes were collected into methionine-free DMEM medium (supplemented with 3 mg/ml bovine serum albumin, 0.2 mM isobutylmethylxanthine, and 0.015% glutamine). Oocytes were cultured for 3 h with 1 µCi/µl α-[35S] methionine in methionine-free DMEM medium (supplied as above) at 37 °C in 5% CO2 atmosphere. After labeling, 50 oocytes were washed in PBS/PVA (polyvinyl alcohol) 5 times and transferred into 20 µl of IPG sample buffer (7 M urea, 2 M thiourea, 20 mM Tris, 4% [w/v] CHAPS, 1% [w/v] Triton X-100, 1% [w/v] dithiothreitol, and 0.8% [w/v] Biolyte 3/10). 2D PAGE was performed as described previously.61 2D gels were analyzed by phosphor-imaging using the Quantity One machine and Quantity One software (Biorad). WT, TG, NSN, and SN gels were always developed together abreast on one phosphor-imaging pad and were scanned in one go. Thus, the intensity ratio of entire gels or selected spots could be compared between each other by Quantity One software. TCA precipitation was performed as previously described.62
Microinjection of recombinant RNA and time-lapse confocal microscopy
Recombinant mRNA for microinjection was prepared by in vitro transcription of 500 ng of linearized mCHERRY or ELAVL2°-mCHERRY plasmids using the mMESSAGE kit (Ambion) according to the manufacturer’s instructions. Recombinant mRNA was polyadenylated using ATP-polyA tailing kit (Ambion) and purified by RNeasy mini kit columns (Qiagen). Fully grown GV oocytes for microinjection were obtained from PMSG-primed H2B-GFP mice63 backcrossed to CD1 strain. Oocytes were microinjected with approximately 5 pl of in vitro transcribed mRNA (336 or 112 µg/ml) and cultured for 2 h in OptiMEM medium supplemented with 10% FCS (Invitrogen) containing 2.5 µM milrinone (Sigma) that reversibly blocks maturation of oocytes. After 2 h, oocytes were washed in milrinone-free medium and let to mature for 18.5 h. Leica TSC SP5 (Leica Microsystems) equipped with an HCX PL Apo Lambda Blue 40× 1.25 oil objective was used for time-lapse confocal microscopy. Images(12 z-confocal sections every 7.4 µm, 1024 × 1024 xy pixel resolution, 16 bit depth) were acquired every 10 min for 18 h. Imaging started 40 min after milrinone removal.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Witold Filipowicz (FMI) and Susan Wagner (Institute of Microbiology, AS CR) for material help, Jana Nejepinska, Lenka Sarnova, Inke Beck, Veronika Libova, and Hynek Strnad (Institute of Molecular Genetics, AS CR), and Patricia Jandurova, Irena Deylova, and Stepan Hladky (Institute of Animal Physiology and Genetics, AS CR) for technical assistance and advice. The main support was provided by the Czech Science Foundation grant GACR P305/12/G034 and the Czech Ministry of Education Youth and Sports (MEYS) project KONTAKT II (LH13084). P.S. was supported by GACR P301-11-P081. V.S. was supported by GACR P302–11-P709. R.S. was supported by MEYS project LM2011032. IMG institutional support was provided by RVO 68378050, IAPG institutional support was provided by RVO 67985904. Work at IAPG was also supported by MEYS project ExAM CZ.1.05/2.1.00/03.0124.
Glossary
Abbreviations:
- ELAVL2
embryonic lethal abnormal vision like 2 protein
- NSN
non-surrounded nucleolus
- SN
surrounded nucleolus
- ARE
AU-rich element
- OET
oocyte-to-embryo transition
- GV
germinal vesicle
- CPE
cytoplasmic polyadenylation element
- CPEB
CPE binding protein
- SCMC
subcortical maternal complex
- AUBP
ARE binding protein
- MII
metaphase II
- WT
wild type
- TG
transgenic mouse
- RRM
RNA recognition motif
- RL
Renilla luciferase
- FL
firefly luciferase
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/28107
References
- 1.Edson MA, Nagaraja AK, Matzuk MM. The mammalian ovary from genesis to revelation. Endocr Rev. 2009;30:624–712. doi: 10.1210/er.2009-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tan JH, Wang HL, Sun XS, Liu Y, Sui HS, Zhang J. Chromatin configurations in the germinal vesicle of mammalian oocytes. Mol Hum Reprod. 2009;15:1–9. doi: 10.1093/molehr/gan069. [DOI] [PubMed] [Google Scholar]
- 3.De La Fuente R. Chromatin modifications in the germinal vesicle (GV) of mammalian oocytes. Dev Biol. 2006;292:1–12. doi: 10.1016/j.ydbio.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 4.Mattson BA, Albertini DF. Oogenesis: chromatin and microtubule dynamics during meiotic prophase. Mol Reprod Dev. 1990;25:374–83. doi: 10.1002/mrd.1080250411. [DOI] [PubMed] [Google Scholar]
- 5.Inoue A, Nakajima R, Nagata M, Aoki F. Contribution of the oocyte nucleus and cytoplasm to the determination of meiotic and developmental competence in mice. Hum Reprod. 2008;23:1377–84. doi: 10.1093/humrep/den096. [DOI] [PubMed] [Google Scholar]
- 6.Zuccotti M, Ponce RH, Boiani M, Guizzardi S, Govoni P, Scandroglio R, Garagna S, Redi CA. The analysis of chromatin organisation allows selection of mouse antral oocytes competent for development to blastocyst. Zygote. 2002;10:73–8. doi: 10.1017/S0967199402002101. [DOI] [PubMed] [Google Scholar]
- 7.Gu L, Wang Q, Sun QY. Histone modifications during mammalian oocyte maturation: dynamics, regulation and functions. Cell Cycle. 2010;9:1942–50. doi: 10.4161/cc.9.10.11599. [DOI] [PubMed] [Google Scholar]
- 8.De La Fuente R, Viveiros MM, Burns KH, Adashi EY, Matzuk MM, Eppig JJ. Major chromatin remodeling in the germinal vesicle (GV) of mammalian oocytes is dispensable for global transcriptional silencing but required for centromeric heterochromatin function. Dev Biol. 2004;275:447–58. doi: 10.1016/j.ydbio.2004.08.028. [DOI] [PubMed] [Google Scholar]
- 9.Burns KH, Viveiros MM, Ren Y, Wang P, DeMayo FJ, Frail DE, Eppig JJ, Matzuk MM. Roles of NPM2 in chromatin and nucleolar organization in oocytes and embryos. Science. 2003;300:633–6. doi: 10.1126/science.1081813. [DOI] [PubMed] [Google Scholar]
- 10.Xia M, He H, Wang Y, Liu M, Zhou T, Lin M, Zhou Z, Huo R, Zhou Q, Sha J. PCBP1 is required for maintenance of the transcriptionally silent state in fully grown mouse oocytes. Cell Cycle. 2012;11:2833–42. doi: 10.4161/cc.21169. [DOI] [PubMed] [Google Scholar]
- 11.Zuccotti M, Merico V, Sacchi L, Bellone M, Brink TC, Stefanelli M, Redi CA, Bellazzi R, Adjaye J, Garagna S. Oct-4 regulates the expression of Stella and Foxj2 at the Nanog locus: implications for the developmental competence of mouse oocytes. Hum Reprod. 2009;24:2225–37. doi: 10.1093/humrep/dep191. [DOI] [PubMed] [Google Scholar]
- 12.Zuccotti M, Merico V, Sacchi L, Bellone M, Brink TC, Bellazzi R, Stefanelli M, Redi CA, Garagna S, Adjaye J. Maternal Oct-4 is a potential key regulator of the developmental competence of mouse oocytes. BMC Dev Biol. 2008;8:97. doi: 10.1186/1471-213X-8-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma JY, Li M, Luo YB, Song S, Tian D, Yang J, Zhang B, Hou Y, Schatten H, Liu Z, et al. Maternal factors required for oocyte developmental competence in mice: transcriptome analysis of non-surrounded nucleolus (NSN) and surrounded nucleolus (SN) oocytes. Cell Cycle. 2013;12:1928–38. doi: 10.4161/cc.24991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li L, Baibakov B, Dean J. A subcortical maternal complex essential for preimplantation mouse embryogenesis. Dev Cell. 2008;15:416–25. doi: 10.1016/j.devcel.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flemr M, Ma J, Schultz RM, Svoboda P. P-body loss is concomitant with formation of a messenger RNA storage domain in mouse oocytes. Biol Reprod. 2010;82:1008–17. doi: 10.1095/biolreprod.109.082057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monti M, Zanoni M, Calligaro A, Ko MS, Mauri P, Redi CA. Developmental arrest and mouse antral not-surrounded nucleolus oocytes. Biol Reprod. 2013;88:2. doi: 10.1095/biolreprod.112.103887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piqué M, López JM, Foissac S, Guigó R, Méndez R. A combinatorial code for CPE-mediated translational control. Cell. 2008;132:434–48. doi: 10.1016/j.cell.2007.12.038. [DOI] [PubMed] [Google Scholar]
- 18.Racki WJ, Richter JD. CPEB controls oocyte growth and follicle development in the mouse. Development. 2006;133:4527–37. doi: 10.1242/dev.02651. [DOI] [PubMed] [Google Scholar]
- 19.Kim JH, Richter JD. Measuring CPEB-mediated cytoplasmic polyadenylation-deadenylation in Xenopus laevis oocytes and egg extracts. Methods Enzymol. 2008;448:119–38. doi: 10.1016/S0076-6879(08)02607-4. [DOI] [PubMed] [Google Scholar]
- 20.Belloc E, Méndez R. A deadenylation negative feedback mechanism governs meiotic metaphase arrest. Nature. 2008;452:1017–21. doi: 10.1038/nature06809. [DOI] [PubMed] [Google Scholar]
- 21.Mendez R, Richter JD. Translational control by CPEB: a means to the end. Nat Rev Mol Cell Biol. 2001;2:521–9. doi: 10.1038/35080081. [DOI] [PubMed] [Google Scholar]
- 22.Richter JD. CPEB: a life in translation. Trends Biochem Sci. 2007;32:279–85. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 23.Mendez R, Barnard D, Richter JD. Differential mRNA translation and meiotic progression require Cdc2-mediated CPEB destruction. EMBO J. 2002;21:1833–44. doi: 10.1093/emboj/21.7.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bakheet T, Williams BR, Khabar KS. ARED 3.0: the large and diverse AU-rich transcriptome. Nucleic Acids Res. 2006;34:D111–4. doi: 10.1093/nar/gkj052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–50. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–26. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 27.Voeltz GK, Steitz JA. AUUUA sequences direct mRNA deadenylation uncoupled from decay during Xenopus early development. Mol Cell Biol. 1998;18:7537–45. doi: 10.1128/mcb.18.12.7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okano HJ, Darnell RB. A hierarchy of Hu RNA binding proteins in developing and adult neurons. J Neurosci. 1997;17:3024–37. doi: 10.1523/JNEUROSCI.17-09-03024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akamatsu W, Okano HJ, Osumi N, Inoue T, Nakamura S, Sakakibara S, Miura M, Matsuo N, Darnell RB, Okano H. Mammalian ELAV-like neuronal RNA-binding proteins HuB and HuC promote neuronal development in both the central and the peripheral nervous systems. Proc Natl Acad Sci U S A. 1999;96:9885–90. doi: 10.1073/pnas.96.17.9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hinman MN, Lou H. Diverse molecular functions of Hu proteins. Cell Mol Life Sci. 2008;65:3168–81. doi: 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakai K, Kitagawa Y, Hirose G. Analysis of the RNA recognition motifs of human neuronal ELAV-like proteins in binding to a cytokine mRNA. Biochem Biophys Res Commun. 1999;256:263–8. doi: 10.1006/bbrc.1999.0282. [DOI] [PubMed] [Google Scholar]
- 32.Fan XC, Steitz JA. HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc Natl Acad Sci U S A. 1998;95:15293–8. doi: 10.1073/pnas.95.26.15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tenenbaum SA, Carson CC, Lager PJ, Keene JD. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc Natl Acad Sci U S A. 2000;97:14085–90. doi: 10.1073/pnas.97.26.14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ince-Dunn G, Okano HJ, Jensen KB, Park WY, Zhong R, Ule J, Mele A, Fak JJ, Yang C, Zhang C, et al. Neuronal Elav-like (Hu) proteins regulate RNA splicing and abundance to control glutamate levels and neuronal excitability. Neuron. 2012;75:1067–80. doi: 10.1016/j.neuron.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Good PJ. The role of elav-like genes, a conserved family encoding RNA-binding proteins, in growth and development. Semin Cell Dev Biol. 1997;8:577–84. doi: 10.1006/scdb.1997.0183. [DOI] [PubMed] [Google Scholar]
- 36.Wiszniak SE, Dredge BK, Jensen KB. HuB (elavl2) mRNA is restricted to the germ cells by post-transcriptional mechanisms including stabilisation of the message by DAZL. PLoS One. 2011;6:e20773. doi: 10.1371/journal.pone.0020773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Calder MD, Madan P, Watson AJ. Bovine oocytes and early embryos express Staufen and ELAVL RNA-binding proteins. Zygote. 2008;16:161–8. doi: 10.1017/S096719940700456X. [DOI] [PubMed] [Google Scholar]
- 38.Calder MD, Watson PH, Watson AJ. Culture medium, gas atmosphere and MAPK inhibition affect regulation of RNA-binding protein targets during mouse preimplantation development. Reproduction. 2011;142:689–98. doi: 10.1530/REP-11-0082. [DOI] [PubMed] [Google Scholar]
- 39.Colegrove-Otero LJ, Devaux A, Standart N. The Xenopus ELAV protein ElrB represses Vg1 mRNA translation during oogenesis. Mol Cell Biol. 2005;25:9028–39. doi: 10.1128/MCB.25.20.9028-9039.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arthur PK, Claussen M, Koch S, Tarbashevich K, Jahn O, Pieler T. Participation of Xenopus Elr-type proteins in vegetal mRNA localization during oogenesis. J Biol Chem. 2009;284:19982–92. doi: 10.1074/jbc.M109.009928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stein P, Svoboda P, Schultz RM. Transgenic RNAi in mouse oocytes: a simple and fast approach to study gene function. Dev Biol. 2003;256:187–93. doi: 10.1016/S0012-1606(02)00122-7. [DOI] [PubMed] [Google Scholar]
- 42.Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, et al. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci U S A. 2002;99:4465–70. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levine TD, Gao F, King PH, Andrews LG, Keene JD. Hel-N1: an autoimmune RNA-binding protein with specificity for 3′ uridylate-rich untranslated regions of growth factor mRNAs. Mol Cell Biol. 1993;13:3494–504. doi: 10.1128/mcb.13.6.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smallwood SA, Tomizawa S, Krueger F, Ruf N, Carli N, Segonds-Pichon A, Sato S, Hata K, Andrews SR, Kelsey G. Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat Genet. 2011;43:811–4. doi: 10.1038/ng.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pillai RS, Artus CG, Filipowicz W. Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. RNA. 2004;10:1518–25. doi: 10.1261/rna.7131604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Svoboda P. RNA silencing in mammalian oocytes and early embryos. Curr Top Microbiol Immunol. 2008;320:225–56. doi: 10.1007/978-3-540-75157-1_11. [DOI] [PubMed] [Google Scholar]
- 47.Barker A, Epis MR, Porter CJ, Hopkins BR, Wilce MC, Wilce JA, Giles KM, Leedman PJ. Sequence requirements for RNA binding by HuR and AUF1. J Biochem. 2012;151:423–37. doi: 10.1093/jb/mvs010. [DOI] [PubMed] [Google Scholar]
- 48.Pascale A, Govoni S. The complex world of post-transcriptional mechanisms: is their deregulation a common link for diseases? Focus on ELAV-like RNA-binding proteins. Cell Mol Life Sci. 2012;69:501–17. doi: 10.1007/s00018-011-0810-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hinman MN, Zhou HL, Sharma A, Lou H. All three RNA recognition motifs and the hinge region of HuC play distinct roles in the regulation of alternative splicing. Nucleic Acids Res. 2013;41:5049–61. doi: 10.1093/nar/gkt166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fialcowitz-White EJ, Brewer BY, Ballin JD, Willis CD, Toth EA, Wilson GM. Specific protein domains mediate cooperative assembly of HuR oligomers on AU-rich mRNA-destabilizing sequences. J Biol Chem. 2007;282:20948–59. doi: 10.1074/jbc.M701751200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Doller A, Schlepckow K, Schwalbe H, Pfeilschifter J, Eberhardt W. Tandem phosphorylation of serines 221 and 318 by protein kinase Cdelta coordinates mRNA binding and nucleocytoplasmic shuttling of HuR. Mol Cell Biol. 2010;30:1397–410. doi: 10.1128/MCB.01373-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jain RG, Andrews LG, McGowan KM, Pekala PH, Keene JD. Ectopic expression of Hel-N1, an RNA-binding protein, increases glucose transporter (GLUT1) expression in 3T3-L1 adipocytes. Mol Cell Biol. 1997;17:954–62. doi: 10.1128/mcb.17.2.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brennan CM, Steitz JA. HuR and mRNA stability. Cell Mol Life Sci. 2001;58:266–77. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simone LE, Keene JD. Mechanisms coordinating ELAV/Hu mRNA regulons. Curr Opin Genet Dev. 2013;23:35–43. doi: 10.1016/j.gde.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang S, Kou Z, Jing Z, Zhang Y, Guo X, Dong M, Wilmut I, Gao S. Proteome of mouse oocytes at different developmental stages. Proc Natl Acad Sci U S A. 2010;107:17639–44. doi: 10.1073/pnas.1013185107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen J, Melton C, Suh N, Oh JS, Horner K, Xie F, Sette C, Blelloch R, Conti M. Genome-wide analysis of translation reveals a critical role for deleted in azoospermia-like (Dazl) at the oocyte-to-zygote transition. Genes Dev. 2011;25:755–66. doi: 10.1101/gad.2028911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nejepinska J, Malik R, Filkowski J, Flemr M, Filipowicz W, Svoboda P. dsRNA expression in the mouse elicits RNAi in oocytes and low adenosine deamination in somatic cells. Nucleic Acids Res. 2012;40:399–413. doi: 10.1093/nar/gkr702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chalupníková K, Lattmann S, Selak N, Iwamoto F, Fujiki Y, Nagamine Y. Recruitment of the RNA helicase RHAU to stress granules via a unique RNA-binding domain. J Biol Chem. 2008;283:35186–98. doi: 10.1074/jbc.M804857200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Flemr M, Svoboda P. Ribonucleoprotein localization in mouse oocytes. Methods. 2011;53:136–41. doi: 10.1016/j.ymeth.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 60.Peritz T, Zeng F, Kannanayakal TJ, Kilk K, Eiríksdóttir E, Langel U, Eberwine J. Immunoprecipitation of mRNA-protein complexes. Nat Protoc. 2006;1:577–80. doi: 10.1038/nprot.2006.82. [DOI] [PubMed] [Google Scholar]
- 61.Vinopal S, Cernohorská M, Sulimenko V, Sulimenko T, Vosecká V, Flemr M, Dráberová E, Dráber P. γ-Tubulin 2 nucleates microtubules and is downregulated in mouse early embryogenesis. PLoS One. 2012;7:e29919. doi: 10.1371/journal.pone.0029919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Poueymirou WT, Schultz RM. Differential effects of activators of cAMP-dependent protein kinase and protein kinase C on cleavage of one-cell mouse embryos and protein synthesis and phosphorylation in one- and two-cell embryos. Dev Biol. 1987;121:489–98. doi: 10.1016/0012-1606(87)90185-0. [DOI] [PubMed] [Google Scholar]
- 63.Hadjantonakis AK, Papaioannou VE. Dynamic in vivo imaging and cell tracking using a histone fluorescent protein fusion in mice. BMC Biotechnol. 2004;4:33. doi: 10.1186/1472-6750-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.