Abstract
The increasing incidence and mortality associated with advanced stages of melanoma are cause for concern. Few treatment options are available for advanced melanoma and the 5-year survival rate is less than 15%. Targeted therapies may revolutionize melanoma treatment by providing less toxic and more effective strategies. However, maximizing effectiveness requires further understanding of the molecular alterations that drive tumor formation, progression, and maintenance, as well as elucidating the mechanisms of resistance. Several different genetic alterations identified in human melanoma have been recapitulated in mice. This review outlines recent progress made in the development of mouse models of melanoma and summarizes what these findings reveal about the human disease. We begin with a discussion of traditional models and conclude with the recently developed RCAS/TVA somatic cell gene delivery mouse model of melanoma.
Keywords: Melanoma, animal models, RCAS/TVA
The incidence of melanoma has been increasing at an alarming rate over the past 20 years. About 68 130 new cases are expected this year with nearly 8700 resulting in death[1]. Melanoma, the most rapidly increasing malignancy among young people in the United States and the most common cancer for young adults of 25 to 29 years old[2], accounts for the majority of deaths attributed to skin cancer and has a poor prognosis for advanced stages of the disease[3]. Patients with metastatic melanoma have limited treatment options and median survival ranges from 6 to 12 months in clinical trials[4]. Currently, lnterleukin-2 and dacarbazine are two FDA-approved drugs for advanced melanoma, but only a small percentage of patients respond to them[4]. Results from clinical studies with small molecule inhibitors of mutant serine/threonine-protein kinase (BRAF) have been very encouraging and promise to yield a much needed breakthrough in the treatment of tumors with this alteration; however, initial responses are not durable and relapse occurs after a median time of 9 months[5]. Further advances in the management of melanoma require model systems aiding in the understanding of disease behavior and assisting in the development and testing of novel therapeutic strategies. Animal models of melanoma have contributed greatly to the biological understanding of melanoma and also serve as a useful tool for testing potential new therapeutic approaches.
Several animal models of melanoma have been developed, including Xiphophorus fish, guinea pig, opossum, and mouse models [6]. The most relevant and successful model has been the mouse due in part to our broad knowledge of mouse genetics. Numerous types of mouse models have been developed that allow researchers to study different components of the disease. Transplantation models, including xenografts of human tumor tissue into immune-deficient mice and syngeneic mouse melanoma cells, have been used for some time. More recently, the development of genetically engineered mouse models (GEMMs) has expanded our knowledge of gene function and elucidated possible targets for treatment of melanoma. Finally, the development of retroviral-vector delivery systems, such as the RCAS/TVA system, has allowed more rapid assessment of the effect of single or multiple genes on tumor initiation, progression, and maintenance. Each model has advantages and disadvantages. However, using each of these models is necessary to further our understanding of the disease and improve treatment.
Xenograft Transplantation Models
Xenograft models allow the transplantation of human melanoma cells into immune-deficient mice and are useful for studying metastasis, a pattern often retained after xenotransplantation[7]. The major advantage of these models is their ease of implementation and rapidity for results. They have also been useful in determining mutations required for melanocyte transformation and melanoma cell invasion. Chudnovsky et al.[8] expressed several genes implicated in melanogenesis in human melanocytes, integrated these cells into human skin reconstructs, and grafted them onto immune-compromised mice. Their results demonstrated that activation of the Ras pathway, inhibition of the Rb and p53 pathways, and expression of telomerase reverse transcriptase (hTERT) were required to generate invasive melanoma. The ability to study human cells directly is advantageous when considering gene mutation and function, because human skin melanocytes are found throughout the basal layer of the epidermis, whereas mouse melanocytes are predominantly located deeper in hair follicles within the dermis. Mouse melanocytes are only found in the epidermis of hairless areas such as the paws, tails, and ears[6]. Human tissue circumvents this issue when studying the biological transition from radial growth phase (RGP) to vertical growth phase (VGP), which is clinically relevant. A major drawback to the xenograft model is the necessity for immune-compromised mice. However, these mice can accept human immune cells, allowing the interaction between human melanoma cells and specific human immune cells to be assessed[9]. Another disadvantage of the xenograft model is failure to fully replicate the interaction between tumor cells and host stromal cells. This limitation can be overcome by the use of spontaneous melanoma models.
Chemically Induced Models
Spontaneous melanoma is extremely rare in laboratory animals[7]. Chemical carcinogens, such as 7,12-dimethylbenz(a)anthracene (DMBA) and 12-O-tetradecanoylphobol-13-acetate (TPA), have been used to induce melanoma in mice. DMBA is an immuno-suppressing, polycyclic aromatic hydrocarbon[10]. TPA, a phorbol ester, acts as a tumor promoter by activating protein kinase C[11], which in turn phosphorylates some growth factor receptors, including epidermal growth factor receptor[12]. These are often used in combination with other modeling techniques, including ultraviolet (UV) radiation, xenotransplantation, or genetic engineering, to decrease the latency of developing melanoma[13]–[15]. The most obvious disadvantage to chemical induction is the lack of clinical relevance to the human disease. However, an advantage of this model is that the immune system is fully functional and thus, these mice can be used to test immunotherapeutic strategies including vaccines, cytokines, antibodies, or any combination therein. Another potential advantage to this system is that DMBA alone can induce nevi in pigmented mice[16], whose establishment can be used to study mechanism(s) of malignant transformation. To induce melanoma, an initial dose of DMBA (200 to 500 ng) is administered to the skin of mice, and then the same area is treated two to three times per week with TPA (5 µg)[13],[15]. However, the DMBA plus TPA protocol typically yields papillomas and small nevi much more frequently than melanoma. To circumvent this problem, many carcinogen-induced melanomas have been used to develop syngeneic transplantation models.
Syngeneic Transplantation Models
Syngeneic models have been used for more than half a century and include the S91 melanoma in DBA/2 mice[17], Harding-Passey melanoma in BALB/c × DBA/2F1 mice[18] and B16 melanoma in C57BL/6 mice[19],[20]. Recently, a syngeneic model in FVB/n mice has also been developed[21]. These models are useful for addressing basic questions, but because they have a functional immune system, they are most suitable for studying the effects of immunotherapy as a treatment for melanoma. The most widely used syngeneic transplantation melanoma model is B16, which was derived from a chemically induced melanoma arising in a C57BL/6J mouse[20]. While these cells express low levels of major histocompatibility complex class I (MHC I) molecules, they express high levels of melanoma-associated antigens, such as gp100 or tyrosinase related protein 2 (TRP2), which are immunotherapeutic targets[22]. We have previously used this model to demonstrate that the inflammatory killing of normal melanocytes activates a potent T-cell response targeted against a specific subset of self-antigens, but can also lead to the immunoselection of resistant variants[23]. A limitation of most syngeneic melanoma models is a lack of understanding regarding the alterations that drive tumor formation and progression.
Genetically Engineered Models
Molecular analysis of familial and sporadic melanomas has identified several genomic loci implicated in the genesis and progression of melanoma [6],[24],[25] (Table 1), with several of these alterations being reproduced in mice (Table 2)[13],[14],[21],[26]–[50]. Linkage studies in melanoma-prone families have implicated the tumor suppressor locus CDKN2A (cyclin-dependent kinase inhibitor 2A), located at 9p21, as a melanoma susceptibility locus [51]. This locus is functionally inactivated in a significant percentage of sporadic melanoma [24] and encodes two independent protein products, p16INK4a and p14[52],[53]. p16INK4a, also known as multiple tumor suppressor 1 (MTS1) or CDKN2A, is a specific inhibitor of Cyclin D/CDK4 or CDK6 complexes[54]. By inhibiting the kinase activity of CDK4 and CDK6, p16INK4a blocks pRB phosphorylation and prevents G1/S cell cycle progression[52]. P14ARF (p19ARF in mice)[55] stabilizes p53[52], and promotes cell cycle arrest in G1 and G2 in response to oncogenic stimuli[56],[57]. In familial and sporadic melanomas, exon 2, which is common to both p16 and p14, is frequently deleted[24], ablating tumor suppression by the RB and TP53 pathways, possibly explaining why TP53 is found mutated in only a small percentage of human melanoma[58].
Table 1. Summary of putative loci and their involvement in familial or sporadic melanoma.
| Locus: gene | Familial or sporadic | Alteration in familial melanoma | Alteration in sporadic melanoma | Melanoma samples analyzed |
| 9p21: CDKN2A (p16INK4a p14ARF) | Both | Point mutation | Point mutations, deletions, promoter methylation | Cell lines, melanoma, metastases |
| 12q14: CDK4 | Both | Point mutation | Point mutations | Cell lines |
| 1p36 | Familial | Linkage | – | – |
| 6p24: TFAP2A | Both | Linkage | Decreased expression | Cell lines |
| 7p11–13: EGFR | Sporadic | – | Amplification | Cell lines |
| 7q33: MET | Sporadic | – | Amplification | Melanoma |
| 3p21: CTNNB1 (β-catenin) | Sporadic | – | Point mutations | Cell lines |
| 13q14: RB1 | Sporadic | – | Point mutations | Cell lines |
| 17p13: TP53 | Sporadic | – | Point mutations | Cell lines |
| 16q22: CDH1 (E-cadherin) | Sporadic | Decreased expression | Cell lines, melanoma, metastases | |
| 12p13: CDKN1B (p27) | Sporadic | Decreased expression | Melanoma, metastases | |
| 6q | Sporadic | – | LOH and cytogenetic alterations | Cell lines |
| 10q23: PTEN | Sporadic | – | LOH and point mutations | Cell lines, melanoma |
| 11q22–23 | Sporadic | – | LOH | Cell lines |
| 1p13: NRAS | Sporadic | – | Point mutations | Melanoma |
| 8q24: MYC | Sporadic | – | Overexpression | Cell lines, melanoma, & metastases |
| 7q34: BRAF | Sporadic | – | Point mutations | Cell lines, melanoma |
| 2q34: ERBB4 | Sporadic | – | Point mutations | Cell lines, melanoma |
| 16q24: MC1R (melanocortin receptor) | Sporadic | – | Point mutations | Blood cells |
Modified from reference [24]. “–” indicates not applicable.
Table 2. Summary of genetically engineered melanoma mouse models.
| Genetic change | Background | Latency/Penetranee | Promoter | Reference(s) |
| NRasQ61R-IRES-Cre | Ink4a/Arff/f | 8 weeks (median survival)/63% | DCT-TVA | [21] |
| BRafCA/wt Ptenf/f | 10 weeks/100% | Tyr::CreERT2 | [26] | |
| LSL-KRasG12D | 17 weeks/100% | Tyr::CreERT2 | [27] | |
| p53f/f | 31 weeks median/45% | [28] | ||
| p16f/f | 24 weeks median/73% | |||
| p53f/f p16f/f | 9 weeks median/100% | |||
| HRasV12G | Ink4a/Arf−/−, Pten ± | ∼19 weeks/75% | Tyr | [29] |
| tetO::HRasG12V | Ink4a/Arf−/− | 26 weeks/60% | Tyr::rtTA | [30] |
| LSL-KRasG12D, LSL-BRafD549A | 26 weeks/100% | Tyr::CreERT2 | [31] | |
| β-cateninsta, NRasN61K | 27.6 weeks median/85% | Tyr | [32] | |
| NRasQ61K Tyr::CreERT2/p53f/f | 28.5 weeks/100% | Tyr | [33] | |
| HGF/SF | Cdk4R24C/R24C | 12 weeks/100% with DMBA/TPA | MT | [34] |
| p16−/− | < 30 weeks/100% with neonatal UVR | |||
| p19−/− | 15 weeks median/70% with neonatal UVR | [35] | ||
| p16−/− p19−/− | 7 weeks/100% with neonatal UVR | |||
| 30 weeks/50% with DMBA/TPA | [13] | |||
| Ptenf/f ± β-cateninloxe3/wt | Ink4a/Arff/f | 40 weeks/100% | Tyr::CreERT2 | [36] |
| MIP-2 | P16 ± p19 ± | ∼28 weeks median/18.5% | Tyr | [37] |
| BRafV600E | Ink4a/Arf ± | 7–58 weeks median/90%–100% | Tyr | [38] |
| p53± | 15–65 weeks median/30%–100% | |||
| 42–85 weeks/<10% | ||||
| NRasQ61K | Cdk4R24C/R24C | ∼43 weeks/100% | Tyr | [33] |
| Arf−/− | 25% | |||
| p16−/− | 26 weeks median/>90% | [39] | ||
| Ret | 28 weeks/80% with UVR | MT | [40] | |
| Ednrb ± | 70 weeks/∼40% | [41] | ||
| IL6−/− | 65 weeks/47% | [42] | ||
| Xpc−/− | Ink4a/Arf−/− | 50 weeks/∼70% with neonatal UVR | [43] | |
| LSL-BRafV600E | 52 weeks median/64% | Tyr::CreERT2 | [44] | |
| p16−/− | 13 weeks median/80% | Tyr | ||
| HRasG12V | 52 weeks/57% with UVR | [45] | ||
| Cdk4R24C/R24C | 65 weeks/33% and 52 weeks/100% | [14],[46] | ||
| p19−/− | 81 weeks/52% and 50 weeks/50% | [47],[48] | ||
| p16−/− | 89 weeks/33% and 50 weeks/50% | [47],[48] | ||
| p53−/− | 17 weeks median/26% | [48],[49] | ||
| Grm1 | Up to 87 weeks/100% | DCT | [50] |
f/f, flox/flox; CA, cre-activated; UVR, UV radiation; LSL, lox-stop-lox; sta, stable; Tyr, tyrosinase; MT, metallothionein; DCT, dopachrome tautomerase.
Activated RAS oncogenes, which constitutively stimulate mitogen-activated protein kinase (MAPK) signaling[59], have been detected in approximately 20% of human melanomas[60]. Recently, mutations in BRAF, which activate MAPK signaling, have also been found in a high percentage (> 65%) of malignant melanoma[61]. With mutually exclusive mutations taking place in RAS and BRAF[61], the MAPK signaling pathway is constitutively activated in over 85% of malignant melanoma cases, indicating the importance of the MAPK pathway in melanomagenesis.
A role for HRas and NRas in melanoma formation was confirmed by tyrosinase-driven expression of mutant HRasG12V and NRasQ61K in the melanocytes of Ink4a/Arf-deficient mice, resulting in the formation of melanoma in 50% or 90% of the mice, respectively, by 6 months [30],[39]. An inducible HRasG12V melanoma mouse model null for the tumor suppressors Ink4a/Arf also showed the importance of HRasG12V expression in the maintenance of melanoma[62]. Inducible tyrosinase-driven expression of BRafV600E in mouse melanocytes leads to benign melanocytic hyperplasia[26],[38] and tumor formation in about 50% of the mice with a median latency of 12 months[44]. Loss of Ink4a or Ink4a/Arf decreased the latency and increased the penetrance of tumor formation in this model[38],[44]. BRafV600E expression in combination with conditional Pten gene silencing in melanocytes leads to metastatic melanoma with a median latency of 50 days[26]. This model was also used to test the therapeutic efficacy of combined MEK and mTOR inhibition using PD0325901 and rapamycin, respectively. The mice treated with either agent had stable disease, whereas the mice treated with the combination therapy demonstrated significant tumor regression[26].
Exposure to UV radiation is thought to be a causal event in at least 80% of the malignant melanoma cases[63],[64]. Epidemiological data suggest that malignant melanoma results from an exposure to intense UV light, especially during childhood[65]. Experiments in mice corroborate these findings. Exposure to UV radiation in neonatal hepatocyte growth factor/scatter factor (HGF/SF)–transgenic mice induced melanoma in 50% of the mice by 1 year[66], with the loss of Ink4a/Arf significantly promoting melanomagenesis, as 50% of the mice developed melanoma by 50 days[35]. In transgenic mice expressing HGF, the location of mouse melanocytes more closely resembles that of human skin as they are aberrantly distributed throughout the epidermis, including the dermoepidermal junction[67]. Thus, this model is well suited to elucidate the role of environmental damage versus genetic predisposition.
Gene knockout and transgenic technology has facilitated the development of mouse strains that can be used as model systems to assess tumor development and treatment. However, despite the great use of such models, there are several limitations. Most human tumors contain multiple genetic changes required for tumor initiation, progression, and metastasis[68]. To accurately model a particular human tumor, many mouse strains generally have to be interbred in a costly and labor intensive process. In addition, many model systems are based on animals that express oncogenes under the control of tissue-specific promoters and/or contain germline inactivating mutations in tumor suppressor genes. In many cases, these alterations have deleterious effects on development and reproductive fitness[69], making it very difficult to obtain animals carrying the desired combination of genotypes. Moreover, tumors in other tissues can potentially arise. For example, tyrosinase-driven expression of activated HRas or NRas in the melanocytes of Ink4a/Arf-deficient mice results in the formation of melanoma in 50% or 90% of the mice, respectively, by 6 months[30],[39],[62]. However, nearly 70% of these mice develop lymphomas and sarcomas by 4 months due to the germline deficiency of Ink4a/Arf[54]. Although these models have shown the importance of Ras in melanoma formation and maintenance, they are not ideal to study metastatic melanoma because most of the animals expire rapidly and it is difficult to discern if tumors found in other tissues are of primary or metastatic origin.
The development of inducible gene expression systems has helped to overcome some of the aforementioned limitations[62],[70]. By expressing the gene in a temporally restricted manner, inducible systems avoid the deleterious effects of the Oncogene on development and fitness. Embryonic lethality associated with the germline deficiency of many tumor suppressor genes can be overcome by the cre/lox system, allowing temporal and spatial control of tumor suppressor gene inactivation[71]–[73]. Before the development of this technology, analysis of tumor formation in mice with inactivating mutations was limited to mice heterozygous for the specific mutation [74] or chimeric mice partially derived from embryonic stem cells homozygous for a particular mutation [75]. Although these conditional approaches overcome the adverse effects on development, they still require the generation of multiple transgenic or conditionally-inactivated strains to accurately model tumors. In addition, the induction techniques tend to activate a host of melanocytes simultaneously rather than individual cells, which differs from the natural occurrence of human disease.
RCAS/TVA Mouse Models
A retrovirally-based gene transfer method has been designed to overcome many of the aforementioned limitations. This system uses a viral vector, RCAS, derived from the avian leukosis virus (ALV)[76]. The receptor for subgroup A-ALV is encoded by the tv-a gene and is normally expressed in avian cells as a transmembranous (tva950) or glycosylphosphatidylinositol (GPI)-membrane linked form (tva800) as a result of alternative splicing[77],[78]. Expression of either form in cells resistant to infection by ALV has been shown to confer susceptibility to infection by RCAS[77]–[79]. In addition, transgenic mice expressing tva950 or tva800 have been generated and are susceptible to RCAS infection in vivo[79]–[81].
Several features of the RCAS/TVA retroviral vector system offer unique advantages, particularly the ability to deliver genes in an efficient and stable manner. Infection with RCAS results in the stable integration of the viral DNA into the genome of replicating cells. In avian cells, the viral vector is replication-competent and can deliver experimental genes of reasonably useful size (up to about 3 kb). Defective vectors, which lack the envelope gene, can deliver genes up to about 4 kb when the envelope protein is supplied in trans[82]. High titer viral vector stocks can be generated in the DF-1 cell line, derived from immortalized chicken fibroblasts [83],[84]. In replicating mammalian cells expressing TVA, the viral vector can stably integrate into the DNA and express the inserted experimental gene at high levels, but the virus is replication-defective since viral RNA and proteins are inefficiently produced[79]. Therefore, the viral vectors cannot spread in the target animals. In addition, since the envelope protein is poorly produced, there is no interference to superinfection. Theoretically, there is no limit to the number of experimental genes that can be introduced. The ability of TVA-expressing mammalian cells to be infected by multiple ALV-derived viruses allows efficient modeling of human melanoma because multiple oncogenic alterations can be introduced into the same cell or animal without the expense associated with mating multiple strains of mice.
An RCAS retroviral vector containing the gene for Cre recombinase has also been generated to allow targeted inactivation of tumor suppressor genes flanked by loxP recombination sites[85]. Although it will still be necessary to breed mice containing the specific allele flanked by loxP sites with mice that express TVA in the desired cell type, this system eliminates the need to express Cre from a tissue-specific promoter and allows a more selective inactivation of the tumor suppressor gene. In addition, a knock-in mouse line was recently generated that allows expression of TVA in a Cre-recombinase-dependent manner. This strain greatly enhances this system by making it easier to generate mice that express TVA in a tissue-specific fashion[86].
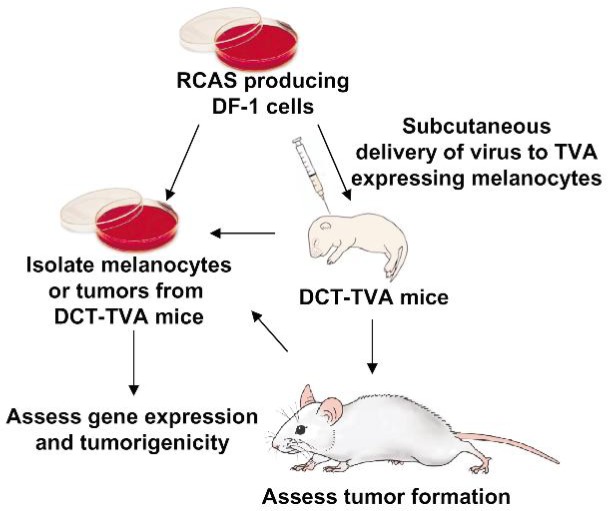
The RCAS family of retroviral vectors has been successfully used in the development of several cancer models in mice[87],[88]. In these models, tumors evolve from gene mutations in developmentally normal somatic cells in the context of an unaltered microenvironment, thereby closely mimicking the human disease. Using this system, newly identified genes can be rapidly validated for their roles in tumor formation, progression, maintenance, and resistance to therapy. To model human melanoma using the RCAS/TVA system (Figure 1), transgenic mice expressing TVA from the dopachrome tautomerase (DCT) promoter, also known as tyrosinase-related protein 2 (TRP2), were generated[89]. This promoter was chosen to drive the expression of the viral receptor, since this gene is expressed early in melanocyte development when the cells are mitotically active[89]–[92]. Because a significant percentage of familial and sporadic melanomas have mutations that functionally inactivate INK4a and ARF, DCT-TVA mice were crossed to Ink4a/Arflox/lox mice to generate DCT-TVA–Ink4a/Arflox/lox mice. As proof-of-principle, newborn mice were injected subcutaneously with RCAS viruses containing Cre-recombinase and NRASQ61R. Whereas no tumors were detected in TVA-negative mice, melanomas were visible in DCT-TVA–Ink4a/Arflox/lox mice as early as 3 weeks. Within 12 weeks, more than one-third of DCT-TVA–Ink4a/Arflox/lox mice developed melanoma histologically similar to the human disease. Delivery of a virus in which NRASQ61R and Cre expression was linked by an internal ribosomal entry site (IRES) resulted in tumor formation in more than two-thirds of TVA-positive mice. Short-term cultures from the primary tumors were established and these cells were syngeneic with the DCT-TVA–Ink4a/Arflox/lox strain, forming tumors in 100% of recipient mice. Passage of these cells in vivo resulted in the development of spontaneous metastases[21].
Figure 1. Diagram of the RCAS/TVA melanoma mouse model and associated procedures. Expression of the TVA viral receptor is driven by the DCT promoter, which is expressed early in melanocyte development when cells are mitotically active. DCT-TVA mice are crossed with Ink4a/Arflox/lox mice to generate DCT-TVA–Ink4a/Arflox/lox mice. TVA negative mice are used as a control. Newborn mice are injected subcutaneously with viral producing cells, which are cleared by the host immune system within 1 week. Animals are monitored for tumor development, which is first observed after a latency of about 4 weeks. Melanocytes and tumors can be isolated and established in culture for further analysis. Images were produced by MediaLab at the Department of Biochemistry, University of Wisconsin at Madison.
Cancer progression is dynamic and depends on interaction between the tumor and its microenvironment. In this model, only a few cells are modified and thus, cells surrounding the tumor are normal. Because the mechanisms by which the microenvironment facilitates tumor progression are not well understood, this model is ideal for studying the interaction between the tumor and the surrounding stroma as well as further defining the role the microenvironment plays in melanoma progression and metastasis. It has also become evident that the order of genetic changes is critical to the successful formation and progression of the tumors. Genes can easily be delivered sequentially in this model to further study the natural evolution of this disease.
Most cancer models using the RCAS/TVA system have analyzed gain-of-function phenotypes by delivering and overexpressing a particular gene of interest. As others have showed that retroviral vectors can be used to stably express short hairpin RNA (shRNA) under the control of an RNA pol III promoter[93]–[95] to reduce target gene expression through RNA interference (RNAi), it was a natural progression to apply this technology to the RCAS family of vectors to take advantage of this replication-competent retroviral system[96]. For this approach, we used the RCAN vector such that the inserted shRNA could be expressed under the control of the human H1 promoter and not the viral long terminal repeat (LTR) promoter and enhancer. Human melanoma cells were engineered to express TVA to allow cell-specific targeted infection by the retroviral vector. We demonstrated that viral-mediated delivery of shRNA specifically reduces target gene expression in melanoma cells in vitro. Recently, it has also been demonstrated that these retroviral vectors can also be used to deliver specific micro RNA (miRNA) sequences[97]. This approach extends the use of the RCAS/TVA system to include loss-of-function analyses of specific genes.
We have further modified this model system to allow the regulation of gene expression post-delivery using the tetracycline (tet)-regulated system [98]. A tet-responsive element (TRE) was inserted into the RCAN viral vector to drive the expression of the inserted gene. Expression from the TRE requires the presence of a tetracycline transcriptional activator (tTA) such as Tet-off, or a reverse tTA (rtTA) such as Tet-on. In the context of Tet-on, the Tet-responsive gene is only expressed in the presence of doxycycline (Dox); in the context of Tet-off, the Tet-responsive gene is repressed in the presence of Dox[99]. Using this approach, we demonstrated that the suppression of NRas causes melanoma regression induced by NRas and Cre in DCT-TVA–Ink4a/Arflox/lox mice (unpublished data). This data confirms the importance of Ras signaling for melanoma maintenance in this context. We hope to use this model to further delineate mechanisms of resistance to Ras inhibition such that rational combination therapies can be developed.
As with all models, there are limitations to the use of the RCAS/TVA system. The expression of genes greater than 3 kb is unsuccessful, requiring the use of defective vectors that produce lower titers. Target cells must be actively dividing to allow integration into the host genome and although lentiviral vectors can be used to circumvent this limitation, they are less efficient[100]. The site of integration is thought to be random and has the potential to affect the expression of host genes. In addition, the integration site may affect the level of expression of the virally delivered genes. Fortunately, the advantages of this model greatly outnumber the disadvantages and there are numerous alternative strategies available to circumvent the few limitations to this approach.
Conclusions
Although experiments performed in vitro are useful and necessary as a first step to study the effects of gene over-expression or loss and to test potential therapies, it is not possible for in vitro systems to fully recapitulate the complexity of the whole organism and the microenvironment in which tumors develop. Therefore, the development of effective and efficient in vivo models is important to more closely mimic the complex realities of human melanoma and to increase our understanding of the biology of this disease. Increasingly sophisticated models have been developed that not only allow temporal and spatial control of gene expression or loss but also permit the testing of novel anti-melanoma therapies. All the model systems possess unique advantages and disadvantages, necessitating the use of each melanoma model as appropriate. The combined knowledge obtained from the use of each model described above will ultimately bring us closer to developing more effective treatment modalities for patients with advanced melanoma.
References
- 1.Jemal A, Siegel R, Xu J, et al. Cancer statistics, 2010 [J] CA Cancer J Clin. 2010;60(5):277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.SF A, CL K, M K, et al., editors. SEER AYA Monograph. Bethesda, MD: 2007. Seer cancer statistics review; pp. 53–63. [Google Scholar]
- 3.Ahmed I. Malignant melanoma: Prognostic indicators [J] Mayo Clin Proc. 1997;72(4):356–361. doi: 10.4065/72.4.356. [DOI] [PubMed] [Google Scholar]
- 4.Flaherty KT. Narrative review: Braf opens the door for therapeutic advances in melanoma [J] Ann Intern Med. 2010;153(9):587–591. doi: 10.7326/0003-4819-153-9-201011020-00008. [DOI] [PubMed] [Google Scholar]
- 5.Flaherty KT, Puzanov I, Kim KB, et al. Inhibition of mutated, activated braf in metastatic melanoma [J] N Engl J Med. 2010;363(9):809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zaidi MR, Day CP, Merlino G. From uvs to metastases: Modeling melanoma initiation and progression in the mouse [J] J Invest Dermatol. 2008;128(10):2381–2391. doi: 10.1038/jid.2008.177. [DOI] [PubMed] [Google Scholar]
- 7.Rofstad EK, Lyng H. Xenograft model systems for human melanoma [J] Mol Med Today. 1996;2(9):394–403. doi: 10.1016/s1357-4310(96)10035-6. [DOI] [PubMed] [Google Scholar]
- 8.Chudnovsky Y, Adams AE, Robbins PB, et al. Use of human tissue to assess the oncogenic activity of melanoma-associated mutations [J] Nat Genet. 2005;37(7):745–749. doi: 10.1038/ng1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sabzevari H, Reisfeld RA. Human cytotoxic T-cells suppress the growth of spontaneous melanoma metastases in scid/hu mice [J] Cancer Res. 1993;53(20):4933–4937. [PubMed] [Google Scholar]
- 10.Miyata M, Furukawa M, Takahashi K, et al. Mechanism of 7,12-dimethylbenz [a]anthracene-induced immunotoxicity: Role of metabolic activation at the target organ [J] Jpn J Pharmacol. 2001;86(3):302–309. doi: 10.1254/jjp.86.302. [DOI] [PubMed] [Google Scholar]
- 11.Blumberg PM. Protein kinase c as the receptor for the phorbol ester tumor promoters: sixth rhoads memorial award lecture [J] Cancer Res. 1988;48(1):1–8. [PubMed] [Google Scholar]
- 12.Hunter T, Ling N, Cooper JA. Protein kinase c phosphorylation of the egf receptor at a threonine residue close to the cytoplasmic face of the plasma membrane [J] Nature. 1984;311(5985):480–483. doi: 10.1038/311480a0. [DOI] [PubMed] [Google Scholar]
- 13.Tormo D, Ferrer A, Gaffal E, et al. Rapid growth of invasive metastatic melanoma in carcinogen-treated hepatocyte growth factor/scatter factor-transgenic mice carrying an oncogenic cdk4 mutation [J] Am J Pathol. 2006;169(2):665–672. doi: 10.2353/ajpath.2006.060017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chawla R, Procknow JA, Tantravahi RV, et al. Cooperativity of cdk4r24c and ras in melanoma development [J] Cell Cycle. 2010;9(16):3305–3314. doi: 10.4161/cc.9.16.12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inoue-Narita T, Hamada K, Sasaki T, et al. Pten deficiency in melanocytes results in resistance to hair graying and susceptibility to carcinogen-induced melanomagenesis [J] Cancer Res. 2008;68(14):5760–5768. doi: 10.1158/0008-5472.CAN-08-0889. [DOI] [PubMed] [Google Scholar]
- 16.Elmets CA, Yusuf N, Hamza S, et al. Topical application of dimethylbenz [a]anthracene results in the generation of multiple melanocytic nevi in c3h/hen mice [J] Toxicol Appl Pharmacol. 2004;195(3):355–360. doi: 10.1016/j.taap.2003.09.029. [DOI] [PubMed] [Google Scholar]
- 17.Nordlund JJ, Gershon RK. Splenic regulation of the clinical appearance of small tumors [J] J Immunol. 1975;114(5):1486–1490. [PubMed] [Google Scholar]
- 18.Maguire HC., Jr Tumor immunology with particular reference to malignant melanoma [J] Int J Dermatol. 1975;14(1):3–11. doi: 10.1111/j.1365-4362.1975.tb00071.x. [DOI] [PubMed] [Google Scholar]
- 19.Fidler IJ, Darnell JH, Budmen MB. Tumoricidal properties of mouse macrophages activated with mediators from rat lymphocytes stimulated with Concanavalin a [J] Cancer Res. 1976;36(10):3608–3615. [PubMed] [Google Scholar]
- 20.Fidler IJ, Nicolson GL. Organ selectivity for implantation survival and growth of b16 melanoma variant tumor lines [J] J Natl Cancer Inst. 1976;57(5):1199–1202. doi: 10.1093/jnci/57.5.1199. [DOI] [PubMed] [Google Scholar]
- 21.VanBrocklin MW, Robinson JP, Lastwika KJ, et al. Targeted delivery of nrasq61r and cre-recombinase to post-natal melanocytes induces melanoma in ink4a/arflox/lox mice [J] Pigment Cell Melanoma Res. 2010;23(4):531–541. doi: 10.1111/j.1755-148X.2010.00717.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Becker JC, Houben R, Schrama D, et al. Mouse models for melanoma: a personal perspective [J] Exp Dermatol. 2009;19(2):157–164. doi: 10.1111/j.1600-0625.2009.00986.x. [DOI] [PubMed] [Google Scholar]
- 23.Sanchez-Perez L, Kottke T, Diaz RM, et al. Potent selection of antigen loss variants of b16 melanoma following inflammatory killing of melanocytes in vivo [J] Cancer Res. 2005;65(5):2009–2017. doi: 10.1158/0008-5472.CAN-04-3216. [DOI] [PubMed] [Google Scholar]
- 24.Castellano M, Parmiani G. Genes involved in melanoma: an overview of ink4a and other loci [J] Melanoma Res. 1999;9(5):421–432. [PubMed] [Google Scholar]
- 25.Chin L, Garraway LA, Fisher DE. Malignant melanoma: Genetics and therapeutics in the genomic era [J] Genes Dev. 2006;20(16):2149–2182. doi: 10.1101/gad.1437206. [DOI] [PubMed] [Google Scholar]
- 26.Dankort D, Curley DP, Cartlidge RA, et al. Braf (v600e) cooperates with pten loss to induce metastatic melanoma [J] Nat Genet. 2009;41(5):544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milagre C, Dhomen N, Geyer FC, et al. A mouse model of melanoma driven by oncogenic kras [J] Cancer Res. 2010;70(13):5549–5557. doi: 10.1158/0008-5472.CAN-09-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monahan KB, Rozenberg GI, Krishnamurthy J, et al. Somatic p16(ink4a) loss accelerates melanomagenesis [J] Oncogene. 2010;29(43):5809–5817. doi: 10.1038/onc.2010.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nogueira C, Kim KH, Sung H, et al. Cooperative interactions of pten deficiency and ras activation in melanoma metastasis [J] Oncogene. 2010;29(47):6222–6232. doi: 10.1038/onc.2010.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chin L, Pomerantz J, Polsky D, et al. Cooperative effects of ink4a and ras in melanoma susceptibility in vivo [J] Genes Dev. 1997;11(21):2822–2834. doi: 10.1101/gad.11.21.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heidorn SJ, Milagre C, Whittaker S, et al. Kinase-dead braf and oncogenic ras cooperate to drive tumor progression through craf [J] Cell. 2010;140(2):209–221. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delmas V, Beermann F, Martinozzi S, et al. Beta-catenin induces immortalization of melanocytes by suppressing p16ink4a expression and cooperates with n-ras in melanoma development [J] Genes Dev. 2007;21(22):2923–2935. doi: 10.1101/gad.450107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferguson B, Konrad Muller H, Handoko HY, et al. Differential roles of the prb and arf/p53 pathways in murine naevus and melanoma genesis [J] Pigment Cell Melanoma Res. 2010;23(6):771–780. doi: 10.1111/j.1755-148X.2010.00752.x. [DOI] [PubMed] [Google Scholar]
- 34.Ha L, Ichikawa T, Anver M, et al. Arf functions as a melanoma tumor suppressor by inducing p53-independent senescence [J] Proc Natl Acad Sci USA. 2007;104(26):10968–10973. doi: 10.1073/pnas.0611638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Recio JA, Noonan FP, Takayama H, et al. Ink4a/arf deficiency promotes ultraviolet radiation-induced melanomagenesis [J] Cancer Res. 2002;62(22):6724–6730. [PubMed] [Google Scholar]
- 36.Held MA, Curley DP, Dankort D, et al. Characterization of melanoma cells capable of propagating tumors from a single cell [J] Cancer Res. 2010;70(1):388–397. doi: 10.1158/0008-5472.CAN-09-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang J, Luan J, Yu Y, et al. Induction of melanoma in murine macrophage inflammatory protein 2 transgenic mice heterozygous for inhibitor of kinase/alternate reading frame [J] Cancer Res. 2001;61(22):8150–8157. [PubMed] [Google Scholar]
- 38.Goel VK, Ibrahim N, Jiang G, et al. Melanocytic nevus-like hyperplasia and melanoma in transgenic brafv600e mice [J] Oncogene. 2009;28(23):2289–2298. doi: 10.1038/onc.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ackermann J, Frutschi M, Kaloulis K, et al. Metastasizing melanoma formation caused by expression of activated n- rasq61k on an ink4a-deficient background [J] Cancer Res. 2005;65(10):4005–4011. doi: 10.1158/0008-5472.CAN-04-2970. [DOI] [PubMed] [Google Scholar]
- 40.Kato M, Liu W, Akhand AA, et al. Ultraviolet radiation induces both full activation of ret kinase and malignant melanocytic tumor promotion in rfp-ret-transgenic mice [J] J Invest Dermatol. 2000;115(6):1157–1158. doi: 10.1046/j.1523-1747.2000.0202a-2.x. [DOI] [PubMed] [Google Scholar]
- 41.Kumasaka MY, Yajima I, Hossain K, et al. A novel mouse model for de novo melanoma [J] Cancer Res. 2010;70(1):24–29. doi: 10.1158/0008-5472.CAN-09-2838. [DOI] [PubMed] [Google Scholar]
- 42.von Felbert V, Cordoba F, Weissenberger J, et al. lnterleukin-6 gene ablation in a transgenic mouse model of malignant skin melanoma [J] Am J Pathol. 2005;166(3):831–841. doi: 10.1016/S0002-9440(10)62304-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang G, Curley D, Bosenberg MW, et al. Loss of xeroderma pigmentosum c (xpc) enhances melanoma photocarcinogenesis in ink4a-arf-deficient mice [J] Cancer Res. 2007;67(12):5649–5657. doi: 10.1158/0008-5472.CAN-06-3806. [DOI] [PubMed] [Google Scholar]
- 44.Dhomen N, Reis-Filho JS, da Rocha Dias S, et al. Oncogenic braf induces melanocyte senescence and melanoma in mice [J] Cancer Cell. 2009;15(4):294–303. doi: 10.1016/j.ccr.2009.02.022. [DOI] [PubMed] [Google Scholar]
- 45.Hacker E, Irwin N, Muller HK, et al. Neonatal ultraviolet radiation exposure is critical for malignant melanoma induction in pigmented tpras transgenic mice [J] J Invest Dermatol. 2005;125(5):1074–1077. doi: 10.1111/j.0022-202X.2005.23917.x. [DOI] [PubMed] [Google Scholar]
- 46.Hacker E, Muller HK, Irwin N, et al. Spontaneous and uv radiation-induced multiple metastatic melanomas in cdk4r24c/r24c/tpras mice [J] Cancer Res. 2006;66(6):2946–2952. doi: 10.1158/0008-5472.CAN-05-3196. [DOI] [PubMed] [Google Scholar]
- 47.Sharpless NE, Kannan K, Xu J, et al. Both products of the mouse ink4a/arf locus suppress melanoma formation in vivo [J] Oncogene. 2003;22(32):5055–5059. doi: 10.1038/sj.onc.1206809. [DOI] [PubMed] [Google Scholar]
- 48.Kannan K, Sharpless NE, Xu J, et al. Components of the rb pathway are critical targets of uv mutagenesis in a murine melanoma model [J] Proc Natl Acad Sci USA. 2003;100(3):1221–1225. doi: 10.1073/pnas.0336397100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bardeesy N, Bastian BC, Hezel A, et al. Dual inactivation of rb and p53 pathways in ras-induced melanomas [J] Mol Cell Biol. 2001;21(6):2144–2153. doi: 10.1128/MCB.21.6.2144-2153.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pollock PM, Cohen-Solal K, Sood R, et al. Melanoma mouse model implicates metabotropic glutamate signaling in melanocytic neoplasia [J] Nat Genet. 2003;34(1):108–112. doi: 10.1038/ng1148. [DOI] [PubMed] [Google Scholar]
- 51.Haluska FG, Hodi FS. Molecular genetics of familial cutaneous melanoma [J] J Clin Oncol. 1998;16(2):670–682. doi: 10.1200/JCO.1998.16.2.670. [DOI] [PubMed] [Google Scholar]
- 52.Chin L, Pomerantz J, DePinho RA. The ink4a/arf tumor suppressor: One gene—two products—two pathways [J] Trends Biochem Sci. 1998;23(8):291–296. doi: 10.1016/s0968-0004(98)01236-5. [DOI] [PubMed] [Google Scholar]
- 53.Roussel MF. The ink4 family of cell cycle inhibitors in cancer [J] Oncogene. 1999;18(38):5311–5317. doi: 10.1038/sj.onc.1202998. [DOI] [PubMed] [Google Scholar]
- 54.Serrano M, Lee H, Chin L, et al. Role of the ink4a locus in tumor suppression and cell mortality [J] Cell. 1996;85(1):27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 55.Quelle DE, Zindy F, Ashmun RA, et al. Alternative reading frames of the ink4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest [J] Cell. 1995;83(6):993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 56.Kamijo T, Zindy F, Roussel MF, et al. Tumor suppression at the mouse ink4a locus mediated by the alternative reading frame product p19arf [J] Cell. 1997;91(5):649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 57.Zhang Y, Xiong Y, Yarbrough WG. Arf promotes mdm2 degradation and stabilizes p53: Arf-ink4a locus deletion impairs both the rb and p53 tumor suppression pathways [J] Cell. 1998;92(6):725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 58.Meier F, Satyamoorthy K, Nesbit M, et al. Molecular events in melanoma development and progression [J] Front Biosci. 1998;3(D):1005–1010. doi: 10.2741/a341. [DOI] [PubMed] [Google Scholar]
- 59.Lewis TS, Shapiro PS, Ahn NG. Signal transduction through map kinase cascades [M] In: Vande Woude GF, Klein G., editors. Advances in cancer research. San Diego: Academic Press; 1998. pp. 49–139. [DOI] [PubMed] [Google Scholar]
- 60.Barbacid M. Ras oncogenes: their role in neoplasia [J] Eur J Clin Invest. 1990;20(3):225–235. doi: 10.1111/j.1365-2362.1990.tb01848.x. [DOI] [PubMed] [Google Scholar]
- 61.Davies H, Bignell GR, Cox C, et al. Mutations of the braf gene in human cancer [J] Nature. 2002;417(6892):949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 62.Chin L, Tarn A, Pomerantz J, et al. Essential role for oncogenic ras in tumour maintenance [J] Nature. 1999;400(6743):468–472. doi: 10.1038/22788. [DOI] [PubMed] [Google Scholar]
- 63.Armstrong BK, Kricker A, English DR. Sun exposure and skin cancer [J] Australas J Dermatol. 1997;38(Suppl 1):S1–S6. doi: 10.1111/j.1440-0960.1997.tb01000.x. [DOI] [PubMed] [Google Scholar]
- 64.MacKie RM. Incidence, risk factors and prevention of melanoma [J] Eur J Cancer. 1998;34(Suppl 3):S3–S6. doi: 10.1016/s0959-8049(98)00003-3. [DOI] [PubMed] [Google Scholar]
- 65.Whiteman DC, Whiteman CA, Green AC. Childhood sun exposure as a risk factor for melanoma: a systematic review of epidemiologic studies [J] Cancer Causes Control. 2001;12(1):69–82. doi: 10.1023/a:1008980919928. [DOI] [PubMed] [Google Scholar]
- 66.Noonan FP, Recio JA, Takayama H, et al. Neonatal sunburn and melanoma in mice [J] Nature. 2001;413(6853):271–272. doi: 10.1038/35095108. [DOI] [PubMed] [Google Scholar]
- 67.Noonan FP, Otsuka T, Bang S, et al. Accelerated ultraviolet radiation-induced Carcinogenesis in hepatocyte growth factor/scatter factor transgenic mice [J] Cancer Res. 2000;60(14):3738–3743. [PubMed] [Google Scholar]
- 68.Fearon E, Vogelstein B. A genetic model for colorectal tumorigenesis [J] Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 69.McClatchey Al, Jacks T. Tumor suppressor mutations in mice: The next generation [J] Curr Opin Genet Dev. 1998;8(3):304–310. doi: 10.1016/s0959-437x(98)80086-5. [DOI] [PubMed] [Google Scholar]
- 70.Bardeesy N, Wong KK, DePinho RA, et al. Animal models of melanoma: Recent advances and future prospects [J] Adv Cancer Res. 2000;79:123–156. doi: 10.1016/s0065-230x(00)79004-x. [DOI] [PubMed] [Google Scholar]
- 71.Furth PA, St Onge L, Boger H, et al. Temporal control of gene expression in transgenic mice by a tetracycline-responsive promoter [J] Proc Natl Acad Sci USA. 1994;91(20):9302–9306. doi: 10.1073/pnas.91.20.9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gossen M, Bonin AL, Freundlieb S, et al. Inducible gene expression systems for higher eukaryotic cells [J] Curr Opin Biotechnol. 1994;5(5):516–520. doi: 10.1016/0958-1669(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 73.Shockett P, Difilippantonio M, Hellman N, et al. A modified tetracycline-regulated system provides autoregulatory, inducible gene expression in cultured cells and transgenic mice [J] Proc Natl Acad Sci USA. 1995;92(14):6522–6526. doi: 10.1073/pnas.92.14.6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Macleod KF, Jacks T. Insights into cancer from transgenic mouse models [J] J Pathol. 1999;187(1):43–60. doi: 10.1002/(SICI)1096-9896(199901)187:1<43::AID-PATH246>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 75.Williams BO, Schmitt EM, Remington L, et al. Extensive contribution of rb-deficient cells to adult chimeric mice with limited histopathological consequences [J] Embo J. 1994;13(18):4251–4259. doi: 10.1002/j.1460-2075.1994.tb06745.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Federspiel M, Hughes S. Retroviral gene delivery [M] In: Emerson C, Sweeney H, editors. Methods in cell biology: Methods in muscle biology. San Diego: Academic Press; 1997. pp. 179–214. [PubMed] [Google Scholar]
- 77.Bates P, Young J, Varmus H. Receptor for subgroup a rous sarcoma virus is related to the low density lipoprotein receptor [J] Cell. 1993;74:1043–1051. doi: 10.1016/0092-8674(93)90726-7. [DOI] [PubMed] [Google Scholar]
- 78.Young J, Bates P, Varmus H. Isolation of a chicken gene that confers susceptibility to infection by subgroup a avian leukosis and sarcoma viruses [J] Journal of Virology. 1993;67(4):1811–1816. doi: 10.1128/jvi.67.4.1811-1816.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Federspiel MJ, Bates P, Young JA, et al. A system for tissue-specific gene targeting: transgenic mice susceptible to subgroup a avian leukosis virus-based retroviral vectors [J] Proc Natl Acad Sci USA. 1994;91(23):11241–11245. doi: 10.1073/pnas.91.23.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Holland EC, Hively WP, DePinho RA, et al. A constitutively active epidermal growth factor receptor cooperates with disruption of G, cell-cycle arrest pathways to induce glioma-like lesions in mice [J] Genes Dev. 1998;12(23):3675–3685. doi: 10.1101/gad.12.23.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Murphy GJ, Leavitt AD. A model for studying megakaryocyte development and biology [J] Proc Natl Acad Sci USA. 1999;96(6):3065–3070. doi: 10.1073/pnas.96.6.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Boerkoel CF, Federspiel MJ, Salter DW, et al. A new defective retroviral vector system based on the bryan strain of rous sarcoma virus [J] Virology. 1993;195(2):669–679. doi: 10.1006/viro.1993.1418. [DOI] [PubMed] [Google Scholar]
- 83.Himly M, Foster DN, Bottoli I, et al. The df-1 chicken fibroblast cell line: transformation induced by diverse oncogenes and cell death resulting from infection by avian leukosis viruses [J] Virology. 1998;248(2):295–304. doi: 10.1006/viro.1998.9290. [DOI] [PubMed] [Google Scholar]
- 84.Schaefer-Klein J, Givol I, Barsov EV, et al. The ev-o-derived cell line df-1 supports the efficient replication of avian leukosis-sarcoma viruses and vectors [J] Virology. 1998;248(2):305–311. doi: 10.1006/viro.1998.9291. [DOI] [PubMed] [Google Scholar]
- 85.Sauer B. Inducible gene targeting in mice using the cre/lox system [J] Methods. 1998;14(4):381–392. doi: 10.1006/meth.1998.0593. [DOI] [PubMed] [Google Scholar]
- 86.Seidler B, Schmidt A, Mayr U, et al. A cre-loxp-based mouse model for conditional somatic gene expression and knockdown in vivo by using avian retroviral vectors [J] Proc Natl Acad Sci USA. 2008;105(29):10137–10142. doi: 10.1073/pnas.0800487105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fisher GH, Orsulic S, Holland E, et al. Development of a flexible and specific gene delivery system for production of murine tumor models [J] Oncogene. 1999;18(38):5253–5260. doi: 10.1038/sj.onc.1203087. [DOI] [PubMed] [Google Scholar]
- 88.Orsulic S. An rcas-tva-based approach to designer mouse models [J] Mamm Genome. 2002;13(10):543–547. doi: 10.1007/s00335-002-4003-4. [DOI] [PubMed] [Google Scholar]
- 89.Dunn KJ, Williams BO, Li Y, et al. Neural crest-directed gene transfer demonstrates wnt1 role in melanocyte expansion and differentiation during mouse development [J] Proc Natl Acad Sci USA. 2000;97(18):10050–10055. doi: 10.1073/pnas.97.18.10050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dunn KJ, Brady M, Ochsenbauer-Jambor C, et al. Wnt1 and wnt3a promote expansion of melanocytes through distinct modes of action [J] Pigment Cell Res. 2005;18(3):167–180. doi: 10.1111/j.1600-0749.2005.00226.x. [DOI] [PubMed] [Google Scholar]
- 91.Dunn KW, Sandoval RM, Kelly KJ, et al. Functional studies of the kidney of living animals using multicolor two-photon microscopy [J] Am J Physiol Cell Physiol. 2002;283(3):C905–C916. doi: 10.1152/ajpcell.00159.2002. [DOI] [PubMed] [Google Scholar]
- 92.Steel KP, Davidson DR, Jackson IJ. Trp-2/dct, a new early melanoblast marker, shows that steel growth factor (c-kit ligand) is a survival factor [J] Development. 1992;115(4):1111–1119. doi: 10.1242/dev.115.4.1111. [DOI] [PubMed] [Google Scholar]
- 93.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering rnas in mammalian cells [J] Science. 2002;296(5567):550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 94.Heimann MT, Fridman JS, Zilfou JT, et al. An epi-allelic series of p53 hypomorphs created by stable rnai produces distinct tumor phenotypes in vivo [J] Nat Genet. 2003;33(3):396–400. doi: 10.1038/ng1091. [DOI] [PubMed] [Google Scholar]
- 95.Rubinson DA, Dillon CP, Kwiatkowski AV, et al. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by rna interference [J] Nat Genet. 2003;33(3):401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- 96.Bromberg-White JL, Webb CP, Patacsil VS, et al. Delivery of short hairpin rna sequences by using a replication-competent avian retroviral vector [J] J Virol. 2004;78(9):4914–4916. doi: 10.1128/JVI.78.9.4914-4916.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Huse JT, Brennan C, Hambardzumyan D, et al. The pten-regulating microrna mir-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo [J] Genes Dev. 2009;23(11):1327–1337. doi: 10.1101/gad.1777409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Holmen SL, Zylstra CR, Mukherjee A, et al. Essential role of beta-catenin in postnatal bone acquisition [J] J Biol Chem. 2005;280(22):21162–21168. doi: 10.1074/jbc.M501900200. [DOI] [PubMed] [Google Scholar]
- 99.Kistner A, Gossen M, Zimmermann F, et al. Doxycycline- mediated quantitative and tissue-specific control of gene expression in transgenic mice [J] Proc Natl Acad Sci USA. 1996;93(20):10933–10938. doi: 10.1073/pnas.93.20.10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lewis BC, Chinnasamy N, Morgan RA, et al. Development of an avian leukosis-sarcoma virus subgroup a pseudotyped lentiviral vector [J] J Virol. 2001;75(19):9339–9344. doi: 10.1128/JVI.75.19.9339-9344.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]