Abstract
MLL1 is a histone H3Lys4 methyltransferase and forms a complex with WDR5 and other components. It plays important roles in developmental events, transcriptional regulation, and leukemogenesis. MLL1 -fusion proteins resulting from chromosomal translocations are molecular hallmarks of a special type of leukemia, which occurs in over 70% infant leukemia patients and often accompanies poor prognosis. Investigations in the past years on leukemogenesis and the MLL1-WDR5 histone H3Lys4 methyltransferase complex demonstrate that epigenetic regulation is one of the key steps in development and human diseases.
Keywords: MLL1, WDR5, leukemia, epigenetic regulation, signal transduction
Introduction
Leukemia is characterized by an abnormal increase of white blood cells in the blood or bone marrow. Among all types of cancers, the morbidity of leukemia is the highest for the patients below 35 years old. Over 70% of the infant leukemia patients bear a translocation involving chromosome 11, which results in the fusion of the mixed lineage leukemia gene (MLL1) with other genes[1]. MLL1 translocations are also found in approximately 10% of adult acute myeloid leukemia (AML) patients, who are previously treated with topoisomerase II inhibitors for other types of cancers[1]. MLL1 is the human homologue of Saccharomyces cerevisiae (S. cerevisiae ) gene Set1 and Drosophila gene Tor. It encodes an enzyme catalyzing the methylation of histone H3 at lysine 4 (H3Lys4)[1]. Trimethylation of histone H3Lys4 is a hallmark of active gene transcription, and alteration of this process often causes changes in gene expression pattern. MLL1 translocation is also linked to altered transcription of important genes involved in stem cell maintenance and development and, thus, leads to leukemogenesis.
Roles of MLLs in Development and Leukemogenesis
MLL1 and leukemogenesis
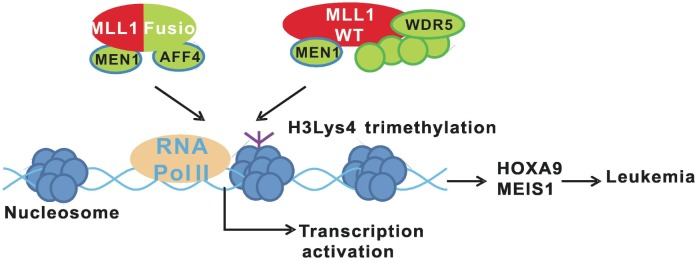
The MLL1 gene was first discovered in leukemia patients in 1991[1]. Its cDNA contains ∼12 kb nucleotides and encodes a peptide over 4000 amino acids in length. In the cell, the premature MLL1 protein is digested by taspase, which results in two peptides: a 300 kDa N-terminal fragment and a 170 kDa C-terminal fragment. The two cleaved peptides form a heterodimer, which is then complexed with other components, including WD repeat domain 5 (WDR5), retinoblastoma binding protein 5 (RBBP5), ash2 (absent, small, or homeotic)-like (ASH2L), dpy-30 homolog (DPY30), multiple endocrine neoplasia I (MEN1), and host cell factor C1 (HCF1)[2]. In some leukemia patients, chromosomal translocation results in fusion of ∼4.2 kb DNA of the MLL1 N-terminal coding region with some other genes[3]. The generation of MLL1 fusion protein is sufficient to induce leukemia, which is demonstrated in animal models[1]. The mechanisms of MLL1 fusion-mediated leukemia have been studied extensively in the past twenty years. Normal MLL1 regulates the expression of homeobox A9 (HOXA9) and me/s homeobox 1 (MEIS1), which are essential for self-renewal of hematopoietic stem cells (HSCs)[4]. After HSC differentiation, the expression of these genes is shut down to prevent tumorigenesis. However, in leukemia patients, MLL1 fusion proteins constitutively activate the expression of HOXA9 and MEIS1 in differentiated cells, leading to leukemia (Figure 1). MLL1 requires a series of binding events to transcribe HOXA9 successfully[5]; its cysteine-x-x-cysteine (CXXC) domain and plant homeodomain (PHD) are required to bind the polymerase-associated factor (PAF) complex and the histone H3Lys4 methylation site, respectively. Both wild-type and fusion MLL1 exist in leukemia cells. Since MLL1 fusion proteins only contain the N-terminal fragment and lack the PHD fingers, they cannot activate HOXA9 expression alone. Wild-type MLL1 has also been shown to be indispensable for HOXA9 expression in leukemia cells[5].
Figure 1. MLL1 fusion proteins induce leukemia together with the wild-type MLL1. The wild-type MLL1 complex pre-binds to the promoters of oncogenes H0XA9 and MEIS1 and is required for proper transcription activation of these genes. The MLL1 fusion proteins are associated with Menin, AFF4, and other proteins in the cell. The MLL1 fusion complex constantly activates the expression of H0XA9 and MEIS1, leading to induction of leukemia.
Partners of MLL1
The fusion partners of MLL1 are quite diverse. Indeed, more than 60 genes have been found to fuse with the MLL1 N-terminal coding region[3], and in some cases, the N-terminal coding region of the MLL1 gene is duplicated. As all fusions involving MLL1 can eventually cause leukemia, MLL1 was considered to the major determinant of leukemogenesis. This has been demonstrated by the observation that MM artificially fused with the bacterial lacZ gene is still able to induce leukemia in a mouse model [6]. However, compared with fusion proteins that occur naturally in patients, the MLL1-β-galactosidase fusion required a much longer time to induce leukemia, suggesting that the fusion partner genes also influence leukemogenesis. A careful inspection of all the partner genes indicates that the most pathogenic partner genes, such as ALL1-fused gene from chromosome 4 protein (AF4), ALL1-fused gene from chromosome 9 protein (AF9), ENL/MLL fusion (ENL), and eleven- nineteen lysine-rich leukemia gene (ELL)[3] are usually involved in transcription elongation. Further investigations with mouse models have demonstrated that these genes usually require much shorter time to induce leukemia compared with other partners[3].
Recently, Lin et al.[7] purified complexes of several MLL1 fusion proteins from cell extracts and analyzed their associated proteins by mass spectrometry. They found that AF4/FMR2 family, member 4 (AFF4), a component of the ELL-TEFb transcription elongation complex, was commonly associated with many types of MLL1 fusion proteins. Depletion of AFF4 suppressed MLL1-fusion dependent transcription, suggesting an essential role for AFF4 in the function of MLL1 fusions. This discovery links MLL1-dependent leukemogenesis with transcription elongation. It is possible that the acquisition of fusion partners into the MLL1 complex overcomes the requirement for transcription initiation step and results in immediate transcription elongation without upstream signals.
MLL1 homologues and their complex formation
Although MLL1 has been studied extensively in animal models, its biochemical functions were not clear until Set1, its homologue in S. cerevisiae, was characterized as a histone H3Lys4 methyltransferase. Set1 is the only enzyme in budding S. cerevisiae that catalyzes histone H3Lys4 methylation[8]. It is capable of catalyzing mono-, di-, and tri-methylation of histone H3Lys4 in vivo and in vitro. The Set1 complex, also called COMPASS (complex of proteins associated with Set1), contains 6 subun its besides Set1, including Cps60, Cps50, Cps40, Cps35, Cps30, and Cps25.
All components in th e complex are extremely conserved from S. cerevisiae to human. Three homologues exist in Drosophila for S. cerevisiae Set1, including Set1, Trx, and Trl. In mammals, six homologous genes were discovered, including SETD1A and SETD1B for Set1, MLL1/ALL-1/HRX and MLL2/HRX2/WBP7 for Trx, and MLL3/KMT2C and MLL4/ALR for Trl[9](Table 1). The gene products of these mammalian homologues each contain a conserved SET domain, which catalyzes the methylation of histone H3 at lysine 4.
Table 1. Complexes of mammalian histone H3Lys4 methyltransferases.
| Histone H3Lys4 Methyltransferases | Yeast Drosophila Human | Set1 |
|||||
| Set1 |
Trx |
Trl |
|||||
| SETD1A | SETD1B | MLL1 | MLL2 | MLL3 | MLL4 | ||
| Subunits In Human Complexes | Shared subunits | WDR5 | |||||
| RBBP5 | |||||||
| ASH2L | |||||||
| DPY30 | |||||||
| Specific subunits | WDR82 | MEN1 | UTX | ||||
| CXXC1 | HCF1 | PTIP | |||||
| PA1 | |||||||
| NCOA6 | |||||||
Set1 is the only histone H3Lys4 methyltransferase in the S. cerevisiae. It has three homologues in Drosophila and six in mammals. The six mammal enzymes have four common subunits, including WDR5, RBBP5, ASH2L, and DPY30. SETD1A and SETD1B are homologues for Drosophila Set1 and their specific subunits are WDR82 and CXXC1. MLL1 and MLL2 are homologues for Drosophila Trx and their specific subunits are MEN1 and HCF1. MLL3 and MLL4 are homologues for Drosophila Trl and their specific subunits are PTIP, PA1, UTX, and NCOA6.
In past years, considerable efforts have been invested to characterize the complexes associated with the six SET1/MLL proteins[9]. Several studies show that the complexes are very similar and all contain several common components, including WDR5, RBBP5, ASH2L, and DPY30 (Table 1). These four proteins, conserved from S. cerevisiae to human, are believed to bind to conserved SET domains in the SET1/MLL proteins. Each complex contains specific subunits, suggesting that the biological functions of the six SET1/MLL proteins are different (Figure 2). This notion is supported by studies with animal models. Deletion of MM gene in mice caused embryonic lethality at 10.5 days [10], whereas deletion of MII2 gene led to embryonic lethality at 11.5 days[11], suggesting that both MM and MII2 have important and non-redundant roles in development. Biochemical studies indicated that complexes involving the gene products of SETD1A and SETD1B, SET1A and SET1B, respectively, are the most robust trimethylating enzymes both in vitro and in vivo[12]. Knockdown of SETD1A and SETD1B led to a severe reduction of global H3Lys4 trimethylation but not mono- or dimethylation in cells.
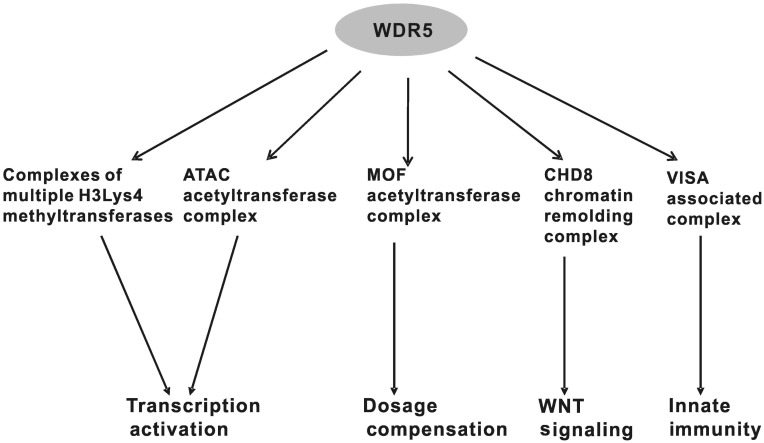
Figure 2. WDR5 serves as an adaptor protein in multiple complexes and related biological processes.
Methylation of histone H3Lys4
The N-terminal tail of histone H3 is covalently modified in many ways, including methylation, acetylation, ubiquitination, phosphorylation, and others[13]. The fourth lysine of histone H3 can be methylated to mono-, di-, and tri-methylated states. Most of the histone H3Lys4 trimethylation occurs near the transcription start site of actively transcribed genes, and the level of methylation is well correlated with the gene transcription level[9]. Many studies investigating histone H3Lys4 methylation have been published, but how histone H3Lys4 methylation regulates gene transcription is still not fully understood. Several proteins have been shown to bind and readout marks of histone H3Lys4 methylation. These include chromodomain helicase DNA binding protein 1 (CHD1), which binds methylated H3Lys4 through its chromo domain, and several other proteins containing PHD fingers, such as inhibitor of growth protein 2 (ING2), inhibitor of growth protein 4 (ING4), inhibitor of growth protein 5 (ING5), recombination activating gene 2 (RAG2), bromodomain PHD finger transcription factor (BPTF), and TBP-associated factor 3 (TAF3)[14]–[22].
CHD1 is a common subunit in both the SAGA (Spt-Ada-Gcn5 Acetyltransferase) and SLIK (SAGA-like) histone acetyltransferase complexes[14]. It contains two chromo domains, one of which is able to recognize and bind the dimethylated form of histone H3Lys4. CHD1 may bridge histone H3Lys4 dimethylation with histone acetylation and the subsequent gene transcription steps[14]. BPTF is a subunit of nucleosome remodeling factor (NURF), an ISWI-containing ATP-dependent chromatin-remodeling complex[20],[21]. Its PHD finger binds trimethylated histone H3Lys4. BPTF deficiency causes the abnormal expression of hox genes in Xenopus embryos, which mimics the WDR5 loss-of-function phenotypes. The basal transcription factor TFIID can also recognize histone H3Lys4 trimethylation by its TAF3 subunit, which also contains a PHD finger[23]. The aforementioned observations suggest that histone H3Lys4 di- and trimethylation marks crosstalk with active gene transcription. Interestingly, one member of the ING protein family, ING2, was shown to bind trimethylated histone H3Lys4 on actively transcribed genes and then repress transcription after DNA damage[15]. This suggests that histone H3Lys4 trimethylation serves as a marker for actively transcribed genes, and that the recognition of this marker can lead to different results, including transcription activation or repression.
Besides transcriptional regulation, histone H3Lys4 methylation also plays important roles in chromatin recombination[19]. RAG2 is an essential component of the RAG1/2 V (D)J recombinase, which mediates antigen-receptor gene assembly during B-cell development. It contains a PHD finger that specifically recognizes histone H3 trimethylated at lysine 4. In vivo, RAG2 mutations affecting the PHD finger impaired V (D)J recombination. Reducing the level of histone H3Lys4 trimethylation also led to a decrease of V (D)J recombination[19]. Furthermore, a genome-wide study of the localization of histone H3Lys4 trimethylation in meiotic S. cerevisiae showed that the level of histone H3Lys4 trimethylation was constantly high at sites of programmed double-strand breaks (DSBs) that initiate interhomologue recombination[24]. When the only histone H3Lys4 methyltransferase in S. cerevisiae, Set1, was deleted, the rate of DSB at these sites was severely reduced. These observations suggest that H3Lys4 methylation plays an important role in chromatin recombination during meiosis.
MLL1 in transcription regulation
In the past twenty years, the mechanism by which MLL1 fusion proteins regulate leukemogenesis has been heavily investigated. In contrast, only very few studies have explored the physiological functions of MLL1. Because histone H3Lys4 methylation has a global regulatory function in gene transcription, the six mammalian methyltransferases, which include MLL1, should have much more important roles in development and other physiological events. Gene knockout mouse models also support this idea. Mll1+/− mice are small at birth and have retarded growth[10]. Though the maturation of myeloid and lymphoid cell seems normal, B-cells are consistently reduced in number[10]. These observations suggest that MLL1 plays roles in multiple stages in the development.
An early ChIP-on-chip assay indicated that MLL1 often co-localized with RNA polymerase II and that histone H3Lys4 trimethylation occurred on the promoter region of actively transcribed genes[25], therefore prompting the conclusion that MLL1 is a common transcription factor essential for gene transcription. However, other studies reached different conclusions because the embryonic fibroblasts of Mll1 knockout mice grew normally, and gene expression patterns in deficient cells were not significantly altered[26],[27]. Furthermore, MLL1 has also been reported to interact with RNA polymerase II and regulate the expression of only a subset of actively transcribed targets in vivo[26]. Genome-wide ChlP-on-chip assays indicated that histone H3Lys4 trimethylation was reduced on only ∼5% of the actively transcribed genes in MLL1 knockout cells in comparison with the wild-type counterparts[27]. A similar result was also observed for global gene expression profile. Why MLL1 protein binds to a major fraction of gene promoters but with limited functions is still unknown at this moment.
Several recent studies indicate that MLL1 has an important role in cell cycle regulation. MLL1 was reported to be recruited to the promoter of E2F by HCF1, a subunit found in MLL1 complex[28]. E2F is a family of transcription factors regulating cell proliferation by activating or repressing gene expression during G1/S phase transition. Thus, MLL1 regulates cell cycle progression by controlling histone H3Lys4 trimethylation of the E2F promoter, which affects its expression level. Furthermore, during M phase, MLL1 complex has been found to associate with condensed chromatin on genes primarily expressed during interphase [29]. MLL1 is hypothesized to bind these genes and regulate their methylation and transcription immediately after M phase exit. Since MLL1 is important for the gene transcription in the cell cycle, its expression level is also tightly regulated. SCFSKP2, an important E3 Ubiquitin ligase for p27 during G1/S transition, was reported to regulate MLL1 degradation at S phase 1[30]. Also, APCCDC20 ubiquitinates MLL1 at late M phase and mediates its stability[30]. The degradation of MLL1 in S and M phases is essential for proper cell cycle progression. Interestingly, some MLL1 fusion proteins are resistant to the E3 ligases mentioned above, suggesting another possible mechanism of tumorigenesis caused by MLL1 fusion proteins.
Functions of WDR5
WDR5 in MLL1 complex
WDR5 is a common subunit of all six mammalian histone H3Lys4 methyltransferases previously mentioned[31]. Its homologue in S. cerevisiae is Cps30. The protein level of Set1 decreases dramatically in the Cps30 knockout S. cerevisiae strain, probably due to the dissociation of the Set1 complex and subsequent protein degradation[32]. WDR5 consists of 334 amino acids and contains seven typical WD40 repeat domains, each approximately 40 amino acids [33]. Structural studies suggest that the WD40 repeats form a seven-bladed propeller fold, with each blade consisting of a four-stranded anti-parallel sheet. This structural property suggests that WDR5 has many exposed surfaces which make it a perfect adaptor to interact with other proteins. In vitro pull-down assays indicate that WDR5 prefers to bind dimethylated histone H3Lys4 peptide[33]. However, the crystal structure of WDR5 complexed with histone H3 peptide does not confirm in vitro results. Quantitative binding analyses demonstrated that the binding of WDR5 with histone H3 N-terminal peptides methylated to four different states varied only modestly. Structural studies of the whole MLL1-WDR5 complex may resolve conflicting observations between in vivo and in vitro experiments.
To make things more complicated, two recent studies suggested a different role of WDR5 in MLL1 complex. The Win motif, consisting of amino acid residues 3762–3767 next to the SET domain in MLL1 protein, was independently discovered to mediate the interaction of MLL1 with WDR5[34],[35]. The crystal structure of the peptide with WDR5 shows that the Win motif, which is the analogue of H3 N-terminal peptide, binds with WDR5 in the central depression of the β-propeller by adopting a 310-helical structure and inserting Arg3765 into the central channel. This discovery led to different models in the two studies. One proposed that the arginine binding site in WDR5 facilitates its association with MLL1. However, the other hypothesized that binding of mono- and dimethylated histone H3Lys4 with WDR5 dissociates it from the MLL1 complex and thus limits the methyltransferase activity. These two different models have yet to be validated experimentally.
Functions of WDR5 beyond histone methylation
Most of the studies of MLL1 and WDR5 focused on the methylation of histone H3 at the Lys4 site and the relationship to leukemogenesis. However, recent reports suggest that the functions of WDR5 are divergent (Figure 2). WDR5 was reported to associate with chromodomain helicase DNA binding protein 8 (CHD8), a chromatin remodeling factor and regulator of WNT signaling pathway[36]. CHD8 can directly interact with β-catenin and regulate the β-catenin-responsive genes. Thus, WDR5 may be involved in chromatin remodeling and the regulation of genes downstream of β-catenin.
WDR5 also plays important roles in histone acetyltransferase complexes and the related physiological functions. The Drosophila homologue of WDR5, Wds, was reported to be one component of ATAC (Ada2a-containing complex) [37], an essential histone acetyltransferase complex in Drosophila. This complex acetylates histone H4Lys16 in Drosophila embryos and facilitates nucleosome sliding along DNA catalyzed by ISWI and SWI-SNF complexes. In human cells, WDR5 was also found to be associated with the human ATAC[38]. Interestingly, many other factors were also reported in the human ATAC, including co-factors of chromatin assembly/remodeling and DNA replication machineries (POLE3/CHRAC17 and POLE4), stress-and TGF-β–activated protein kinase (TAK1/MAP3K7), and MAP3-kinase regulator (MBIP). Furthermore, both in human and Drosophila, WDR5 is a subunit in the MOF complex, which is the key complex regulating X chromosome dosage compensation by acetylating histone H4Lys16[39],[40].
Because WDR5 is an essential component of the histone methylation, acetylation, and chromatin remodeling complexes, WDR5 is believed to serve as an adaptor protein for complex assembly. However, it may also contribute to other physiological phenomena. Recently, it was reported that WDR5 is an important component for assembly or stability of the virus-induced-signaling adapter (VISA)–associated complex, which plays a key role in virus-triggered induction of type I interferons (IFNs) and anti-viral innate immune response[41]. Previous studies have demonstrated that VISA is located at the outer membrane of mitochondria. Interestingly, this study revealed that WDR5 was not only localized in the nucleus as believed before, but also abundantly localized in the cytoplasm. Viral infection caused translocation of WDR5 from the nucleus to the mitochondria-located VISA complex, where it played a role in the assembly and stability of the VISA complex. This study demonstrates for the first time a cytoplasmic function for WDR5, specifically in virus-triggered signaling resulting in induction of type I IFNs (Figure 2)[41].
The Remaining Questions
It has been ∼20 years since the MLL1 gene was discovered and its relationship to leukemia was established. Many details have been elucidated on how MLL1 fusion proteins cause leukemia. However, it is still not clear why MLL1 fusion proteins function differently from their wild-type counterparts. It seems that wild-type MLL1 protein is somehow restricted and not oncogenic in normal cells; however, the MLL1 fusion can overcome these restrictions. Also, it is unclear why MLL1 fusion causes malignancy only in hematopoietic cells but not in other differentiated cells. Identification of the limiting factors in MLL1-activated transcription might help to prevent leukemogenesis or lead to novel therapies for patients with leukemia.
To design specific drugs to treat MLL1-fusion dependent leukemia patients is an ultimate goal of the research. Based on current knowledge, small therapeutic chemicals can be developed following several strategies. The first is to target the enzymatic activity of MLL1. However, since MLL1 may have multiple functions in many other cell types, this strategy could have severe side effects. The second is to target the interaction between the CXXC domain of MLL1 and DNA or between MLL1 and MEN1. However, as wild-type MLL1 also utilizes a similar mechanism, this approach might also have side effects. The third is to target the interaction between MLL1 fusion and the elongation factors, such as AFF4. However, this strategy still needs to be tested.
Despite our growing knowledge about MLL1 fusion proteins and leukemia, very little is known about the MLL1-WDR5 complex. It is highly possible that this complex may have more significant roles than expected. Considering the importance of histone H3Lys4 methylation in global gene transcription regulation, MLL1 may regulate development of many tissues and organs. Studies of conditional knockout mice will be critical to investigating the functions of MLL1 in different tissues. The substrate specificity of the MLL1-WDR5 complex has emerged to be another interesting question. Although the histone lysine methyltransferases, unlike acetyltransferases, seem to have good substrate specificity, some, such as SET7 and SMYD2, have also been reported to have multiple substrates. Does MLL1-WDR5 complex have substrates other than histone H3Lys4? This is very likely as WDR5 interacts with a variety of proteins and even has the ability to translocate to the mitochondria. Undoubtedly, a new picture on the physiological functions of the MLL1-WDR5 complex will soon emerge.
References
- 1.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development [J] Nat Rev Cancer. 2007;7(11):823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 2.Ziemin-van der Poel S, McCabe NR, Gill HJ, et al. Identification of a gene, MLL, that spans the breakpoint in 11q23 translocations associated with human leukemias [J] Proc Natl Acad Sci USA. 1991;88(23):10735–10739. doi: 10.1073/pnas.88.23.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin ME, Milne TA, Bloyer S, et al. Dimerization of MLL fusion proteins immortalizes hematopoietic cells [J] Cancer Cell. 2003;4(3):197–207. doi: 10.1016/s1535-6108(03)00214-9. [DOI] [PubMed] [Google Scholar]
- 4.Zeisig BB, Milne T, Garcia-Cuellar MP, et al. Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization [J] Mol Cell Biol. 2004;24(2):617–628. doi: 10.1128/MCB.24.2.617-628.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Milne TA, Kim J, Wang GG, et al. Multiple interactions recruit MLL1 and MLL1 fusion proteins to the HOXA9 locus in leukemogenesis [J] Mol Cell. 2010;38(6):853–863. doi: 10.1016/j.molcel.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dobson CL, Warren AJ, Pannell R, et al. Tumorigenesis in mice with a fusion of the leukaemia Oncogene MII and the bacterial lacZ gene [J] EMBO J. 2000;19(5):843–851. doi: 10.1093/emboj/19.5.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin C, Smith ER, Takahashi H, et al. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia [J] Mol Cell. 2010;37(3):429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller T, Krogan NJ, Dover J, et al. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein [J] Proc Natl Acad Sci USA. 2001;98(23):12902–12907. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation [J] Curr Opin Cell Biol. 2008;20(3):341–348. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu BD, Hess JL, Horning SE, et al. Altered Hox expression and segmental identity in Mil-mutant mice [J] Nature. 1995;378(6556):505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 11.Glaser S, Schaft J, Lubitz S, et al. Multiple epigenetic maintenance factors implicated by the loss of MII2 in mouse development [J] Development. 2006;133(8):1423–1432. doi: 10.1242/dev.02302. [DOI] [PubMed] [Google Scholar]
- 12.Wu M, Wang PF, Lee JS, et al. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS [J] Mol Cell Biol. 2008;28(24):7337–7344. doi: 10.1128/MCB.00976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression [J] Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 14.Pray-Grant MG, Daniel JA, Schieltz D, et al. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation [J] Nature. 2005;433(7024):434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- 15.Shi X, Hong T, Walter KL, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression [J] Nature. 2006;442(7098):96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pena PV, Davrazou F, Shi X, et al. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2 [J] Nature. 2006;442(7098):100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hung T, Binda O, Champagne KS, et al. ING4 mediates crosstalk between histone H3 K4 trimethylation and H3 acetylation to attenuate cellular transformation [J] Mol Cell. 2009;33(2):248–256. doi: 10.1016/j.molcel.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Champagne KS, Saksouk N, Pena PV, et al. The crystal structure of the ING5 PHD finger in complex with an H3K4me3 histone peptide [J] Proteins. 2008;72(4):1371–1376. doi: 10.1002/prot.22140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matthews AG, Kuo AJ, Ramon-Maiques S, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination [J] Nature. 2007;450(7172):1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Ilin S, Wang W, et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF [J] Nature. 2006;442(7098):91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wysocka J, Swigut T, Xiao H, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling [J] Nature. 2006;442(7098):86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 22.Vermeulen M, Mulder KW, Denissov S, et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4 [J] Cell. 2007;131(1):58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 23.Gangloff YG, Pointud JC, Thuault S, et al. The TFIID components human TAF (II)140 and drosophila BIP2 (TAF(II) 155) are novel metazoan homologues of yeast TAF (II)47 containing a histone fold and a PHD finger [J] Mol Cell Biol. 2001;21(15):5109–5121. doi: 10.1128/MCB.21.15.5109-5121.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borde V, Robine N, Lin W, et al. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites [J] EMBO J. 2009;28(2):99–111. doi: 10.1038/emboj.2008.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guenther MG, Jenner RG, Chevalier B, et al. Global and Hox-specific roles for the MLL1 methyltransferase [J] Proc Natl Acad Sci USA. 2005;102(24):8603–8608. doi: 10.1073/pnas.0503072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milne TA, Dou Y, Martin ME, et al. MLL associates specifically with a subset of transcriptionally active target genes [J] Proc Natl Acad Sci USA. 2005;102(41):14765–14770. doi: 10.1073/pnas.0503630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang P, Lin C, Smith ER, et al. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II [J] Mol Cell Biol. 2009;29(22):6074–6085. doi: 10.1128/MCB.00924-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tyagi S, Chabes AL, Wysocka J, et al. E2F activation of S phase promoters via association with HCF-1 and the MLL family of histone H3K4 methyltransferases [J] Mol Cell. 2007;27(1):107–119. doi: 10.1016/j.molcel.2007.05.030. [DOI] [PubMed] [Google Scholar]
- 29.Blobel GA, Kadauke S, Wang E, et al. A reconfigured pattern of MLL occupancy within mitotic chromatin promotes rapid transcriptional reactivation following mitotic exit [J] Mol Cell. 2009;36(6):970–983. doi: 10.1016/j.molcel.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu H, Cheng EH, Hsieh JJ. Bimodal degradation of MLL by SCFSkp2 and APCCdc20 assures cell cycle execution: a critical regulatory circuit lost in leukemogenic MLL fusions [J] Genes Dev. 2007;21(19):2385–2398. doi: 10.1101/gad.1574507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eissenberg JC, Shilatifard A. Histone H3 lysine 4 (H3K4) methylation in development and differentiation [J] Dev Biol. 2010;339(2):240–249. doi: 10.1016/j.ydbio.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krogan NJ, Dover J, Khorrami S, et al. COMPASS, a histone H3 (Lysine 4) methyltransferase required for telomeric silencing of gene expression [J] J Biol Chem. 2002;277(13):10753–10755. doi: 10.1074/jbc.C200023200. [DOI] [PubMed] [Google Scholar]
- 33.Trievel RC, Shilatifard A. WDR5, a complexed protein [J] Nat Struct Mol Biol. 2009;16(7):678–680. doi: 10.1038/nsmb0709-678. [DOI] [PubMed] [Google Scholar]
- 34.Patel A, Dharmarajan V, Cosgrove MS. Structure of WDR5 bound to mixed lineage leukemia protein-1 peptide [J] J Biol Chem. 2008;283(47):32158–32161. doi: 10.1074/jbc.C800164200. [DOI] [PubMed] [Google Scholar]
- 35.Song JJ, Kingston RE. WDR5 interacts with mixed lineage leukemia (MLL) protein via the histone H3-binding pocket [J] J Biol Chem. 2008;283(50):35258–35264. doi: 10.1074/jbc.M806900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson BA, Tremblay V, Lin G, et al. CHD8 is an ATP-dependent chromatin remodeling factor that regulates beta-catenin target genes [J] Mol Cell Biol. 2008;28(12):3894–3904. doi: 10.1128/MCB.00322-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suganuma T, Gutierrez JL, Li B, et al. ATAC is a double histone acetyltransferase complex that stimulates nucleosome sliding [J] Nat Struct Mol Biol. 2008;15(4):364–372. doi: 10.1038/nsmb.1397. [DOI] [PubMed] [Google Scholar]
- 38.Guelman S, Kozuka K, Mao Y, et al. The double-histone-acetyltransferase complex ATAC is essential for mammalian development [J] Mol Cell Biol. 2009;29(5):1176–1188. doi: 10.1128/MCB.01599-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dou Y, Milne TA, Tackett AJ, et al. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF [J] Cell. 2005;121(6):873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 40.Cai Y, Jin J, Swanson SK, et al. Subunit composition and substrate specificity of a MOF-containing histone acetyltransferase distinct from the male-specific lethal (MSL) complex [J] J Biol Chem. 2010;285(7):4268–4272. doi: 10.1074/jbc.C109.087981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang YY, Liu LJ, Zhong B, et al. WDR5 is essential for assembly of the VISA-associated signaling complex and virus-triggered IRF3 and NF-kappaB activation [J] Proc Natl Acad Sci USA. 2010;107(2):815–820. doi: 10.1073/pnas.0908967107. [DOI] [PMC free article] [PubMed] [Google Scholar]