Abstract
Cancer metabolism has emerged as an important area of research in recent years. Elucidation of the metabolic differences between cancer and normal cells and the underlying mechanisms will not only advance our understanding of fundamental cancer cell biology but also provide an important basis for the development of new therapeutic strategies and novel compounds to selectively eliminate cancer cells by targeting their unique metabolism. This article reviews several important metabolic alterations in cancer cells, with an emphasis on increased aerobic glycolysis (the Warburg effect) and glutamine addiction, and discusses the mechanisms that may contribute to such metabolic changes. In addition, metabolic alterations in cancer stem cells, mitochondrial metabolism and its influence on drug sensitivity, and potential therapeutic strategies and agents that target cancer metabolism are also discussed.
Keywords: Cancer metabolism, the Warburg effect, mitochondria, glycolysis, glutamine, cancer stem cells
Cancer cells have long been known to exhibit profound alterations in their metabolism, as exemplified by the Warburg effect, a phenomenon of cancer cells with elevated aerobic glycolysis[1]–[3]. Reprogramming of the cellular energy metabolism constitutes an emerging hallmark of cancer and may serve as a biochemical basis for new therapeutic intervention[4]. Tremendous efforts in recent years have been devoted to unveiling the underlying mechanisms for metabolic alterations in cancer, thus triggering considerable research interest in mitochondria, bioenergetics, and redox regulation in normal and malignant cells. Both glucose and glutamine are key metabolic substrates in cancer cells and are critical for cancer development, invasion, and metastases [5]–[9]. Glutamine is an abundant amino acid in human plasma and is present at high concentrations in the medium used for in vitro cell culture. Several oncogenes, including c-Myc and Ras, have been identified to promote the expression of metabolic enzymes and regulators that lead to preferential use of glycolysis over mitochondrial oxidative phosphorylation (OXPHOS). Loss of tumor suppressors, such as p53, fumarate hydratase (FH), and succinate dehydrogenase (SDH), also results in significant changes in energy metabolism and may contribute to activation of hypoxia-inducible factor (HIF)-1α-dependent pathways and adaptation to tumor hypoxia[10],[11].
Mitochondria are critical cellular organelles in which many key metabolic pathways converge. Whether metabolic alterations drive tumorigenesis or are a consequence of malignant transformation is still a matter of debate. Mitochondrial dysfunction has been directly linked to alterations in gene expression profiles and significantly affects cancer development. An impaired mitochondrial respiratory chain (MRC) may significantly alter the expression of important genes such as forkhead box O family (FOXO) and apoptosis signal-regulating kinase 1 (ASK-1), leading to changes in cell cycle progression[12]. Mutations in mitochondrial DNA and nuclear DNA-encoded genes can also modulate cancer cell metabolism. Aberrant expression of specific molecules such as the serine/threonine protein kinase AKT and the mammalian target of rapamycin (mTOR) promotes increased glucose metabolism in cancer cells. In fact, AKT alone is sufficient to produce a glycolytic phenotype and glucose dependence in cancers[13]. Furthermore, mTOR, a downstream target of AKT, seems to function as an energy sensor that is sensitive to alterations in nutrients and amino acids[14]–[16] and is deregulated in many cancer types[17]. The tumor suppressor AMP-activated protein kinase (AMPK) is activated in response to stress signals, such as hypoxia and low cellular ATP levels, and regulates various critical cellular processes, including proliferation, cell cycle progression, autophagy, and cellular senescence, through its effects on key molecules such as mTORC1, p53, p27, FOXO3, and others[18],[19]. In addition, metabolic reprogramming is linked to oncogenes such as c-Myc and HIF-1α and tumor suppressors such as p53 that are important players in mediating energetic pathways[20]–[22]. A high dependency on glycolysis in cancers is also associated with altered glucose transporters and glycolytic enzymes such as hexokinase II (HKII) and lactate dehydrogenase (LDH)[23],[24]. Therefore, many of these key molecules that are critical for maintaining cancer metabolism may be considered as potential targets for metabolic intervention in cancer treatment. The following sections provide an overview of several key metabolic alterations in cancers, their potential links to oncogenes and tumor suppressors, and the biochemical and molecular basis for targeting altered metabolism in cancers.
Metabolic Alterations in Cancers
Glycolysis and the Warburg effect
Warburg observed in early 1920s that tumor cells exhibited significant alterations in energy metabolism and mitochondrial respiration compared to normal cells[2],[25]. He showed that cancer cells actively used glycolysis for ATP generation, even in the presence of an abundant supply of oxygen, a phenomenon known as the Warburg effect[1],[25]. Warburg further postulated that the metabolic shift from OXPHOS to glycolysis in neoplastic cells might be due to a respiratory injury (mitochondrial dysfunction) leading to increased aerobic fermentation, a critical event that was considered as the “origin of cancer cells”[1]. Although whether metabolic alterations drive tumorigenesis or are an effect of transformation is still under debate, subsequent studies showed that increased dependence on glycolysis is observed in the majority of tumors and that glycolysis provides ATP as well as the metabolic intermediates essential for cancer cell proliferation and tumor development[8],[26],[27].
Aerobic conversion of glucose to lactate represents a major feature of cancer cell metabolism (Figure 1). The high flux of glycolysis results in an increased output of pyruvate, which may either be converted to lactate by LDH in the cytosol or to acetyl-CoA by pyruvate dehydrogenase (PDH) in the mitochondria. Acetyl-CoA is further metabolized through the Kreb's cycle and the MRC to generate ATP. The tumor hypoxic environment and/or oncogenes such as Ras, Src, and HER2 stabilize HIF-1α, leading to up-regulation of pyruvate dehydrogenase kinase-1 (PDK-1), in turn inactivating PDH and thereby preventing the conversion of pyruvate to acetyl-CoA. Pyruvate is subsequently converted to lactate by LDH with a simultaneous oxidation of nicotinamide adenine dinucleotide (NADH) to NAD+, which is important for the glycolytic reaction at the step catalyzed by glyceraldehyde-3-phosphate dehydrogenase (GAPDH). LDH is up-regulated not only by HIF-1α but also by other oncogenes such as c-Myc, thus ensuring sufficient NAD+ is available for glycolysis in cancer cells.
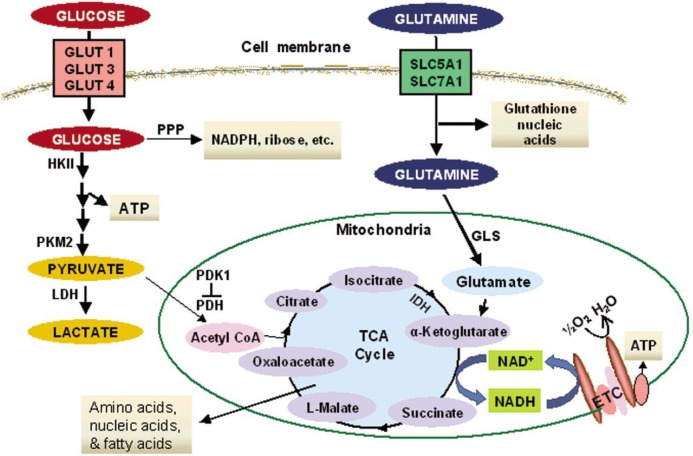
Figure 1. Glucose and glutamine metabolism in cancer cells.
Glucose and glutamine are transported through cell membranes by their respective transporters (GLUT-1, -3 and -4 for glucose; SLC5A1 and SLC7A1 for glutamine). Glucose is metabolized in the cytosol to pyruvate, which can be either converted to lactate or transported into the mitochondria for further catabolism through the tricarboxylic acid (TCA) cycle coupled with respiration through the electron transport chain (ETC). In many cancer cells, glucose is mainly used for the glycolytic pathway, leading to a generation of lactate and important metablic intermediates such as glucose-6-phosphate for the pentose phosphate pathway (PPP) that generates NADPH and ribose for maintaining redox balance and synthesis of nucleic acids. The flow of glucose into mitochondria in the form of pyruvate is relatively low in cancer cells. Glutamine is actively metabolized in cancer cells, both in the cytosol and in the mitochondria, where it is catalyzed by glutaminase to generate glutamate, which is further converted to α-ketoglutarate for utilization through the TCA cycle. Both glycolysis and the TCA cycle provide important metabolic intermediates that serve as substrates for other pathways including the synthesis of nucleic acids, fatty acids, amino acids, and glutathione. The highly active pathways in cancer cells are indicated with bold arrows, whereas the less active metabolic flows are shown with thin arrows. Some of the important enzymes involved in cancer metabolism are indicated. HKII, hexokinase-2; PKM2, pyruvate kinase M2 isoform; LDH, lactate dehydrogenase; PDH, pyruvate dehydrogenase; PDK-1, pyruvate dehydrogenase kinase-1; GLS, glutaminase; IDH, isocitrate dehydrogenase.
The high rate of aerobic glycolysis confers several advantages to cancer cells. In the hypoxic microenvironment of tumor tissues, active glycolysis provides sufficient ATP for tumor cells when mitochondrial OXPHOS is limited. This seems particularly important for actively proliferating cancer cells[1],[28]. Secondly, the metabolic intermediates generated during glycolysis provide important building blocks or precursors for the synthesis of DNA and fatty acids and for redox regulation[29],[30]. For instance, glucose-6-phosphate, the product of the first glycolytic reaction, can be channeled to the pentose phosphate pathway (PPP) to generate ribose-5-phosphate and nicotinamide adenine dinucleotide phosphate (NADPH), which are important for nucleic acid metabolism and redox homeostasis, respectively. NADPH and 3-carbon metabolic intermediates are also important for lipid biosynthesis. Finally, the high levels of lactic acid produced during glycolysis may activate metalloproteinases and matrix remodeling enzymes, thus contributing to cancer invasion and metastasis [29],[31]. It should be noted, however, although most cancer cells are highly active in glycolysis, an increase in mitochondrial OXPHOS has also been observed in some tumors.
Glutamine in cancer metabolism
Another important metabolic substrate and energy source for tumor cells is glutamine[32]. This amino acid is highly abundant in blood, and once it enters the cells, it may serve as an energy source through catabolism or as a building block via anabolism (Figure 1). In addition to its direct involvement in metabolism, glutamine seems able to affect amino acid transport and autophagy through activation of mTOR complex 1 (mTORC1) [33]. Conversion of glutamine to glutamate and its channeling into the tricarboxylic acid (TCA) cycle via α-ketoglutarate seem essential for the oncogenic K-Ras–induced tumor growth in colon cancer cells[5]. Studies using tumor xenograft models have shown that the expression of glutaminase, an enzyme that converts glutamine to glutamate, correlates with tumor growth, and that inhibition of glutaminase in cells led to suppression of tumorigenicity and tumor growth[7],[34]. A metabolic study using nuclear megnetic resonance (NMR) spectroscopic analysis in glioblastoma cells cultured with isotope-labeled glucose and glutamine demonstrated that glutamine supplied the majority of aneplerotic carbon for the TCA cycle in cancer cells[35]. The same study also showed that conversion of glutamate to α-ketoglutarate is the chief source of malate, oxaloacetate, and NADPH for fatty acid biosynthesis in tumors[35]. In fact, active glutaminolysis, the process of glutamine catabolism, is considered as a major metabolic feature of certain tumor cells[36],[37]. Recent studies have shown that glutamine metabolism is regulated by the Oncogene c-Myc, which directly stimulates the expression of the glutamine transporters SLC5A1 and SLC7A1 and indirectly promotes the expression of glutaminase 1 (GLS1) by repressing the expression of miR-23A and miR-23B[38],[39].
Glutamine also contributes to the maintenance of the antioxidant pool in cancer cells. The glutamine-derived malate from the TCA cycle serves as a substrate for malic enzyme 1 to produce NADPH. In glioblastoma cells, the high level of NADPH has been shown to contribute to the active synthesis of glutathione (GSH), which is essential for combating reactive oxygen species (ROS)-induced stress in cancer cells and maintaining redox balance[35]. Interestingly, glutamine also seems to play a role in cancer metastasis. Seyfried et al.[6] recently showed that inhibition of glutamine metabolism in a mouse breast cancer model suppressed tumor metastasis. Although suppression of the mitochondrial glutaminase activity can inhibit oncogenic transformation[40], the exact roles of glutamine in tumorigenesis remain to be elucidated. The role of glutamine metabolism in affecting tumor growth and cancer progression in patients has not also been definitely established. Further research is required to understand the mechanisms underlying the “glutamine addiction” characteristics of cancer cells.
Alterations of metabolic enzymes in cancers
The mitochondrial membrane-bound HKII has been shown to promote the high glycolytic tumor phenotype and to inhibit Bax-induced cytochrome c-mediated apoptosis in HeLa cells[41],[42]. The pyruvate kinase isoform M2 (PKM2) seems to confer a tumorigenic advantage to cancer cells by slowing glycolysis and channeling the glycolytic intermediates to the PPP to produce NADPH and other metabolic substrates[43]. Elevated levels of PKM2 have been observed in various tumor samples[44],[45], consistent with the potential oncogenic role of this enzyme. The co-factor NADPH not only fuels macromolecular biosynthesis but also functions as a major antioxidant in cells. It maintains the redox balance during cell proliferation and is a reducing agent required for the GSH and thioderoxin antioxidant systems that play a major role in quenching R0S[46].
Gain-of-function mutations in isocitrate dehydrogenase I and II (IDH1 and IDH2) have been identified in several types of human cancers, especially in certain gliomas and acute myelogenous leukemia[47]–[49]. While the wild-type IDH1 and IDH2 catalyze the conversion of isocitrate to α-ketoglutarate, mutated IDH (IDH1R132, IDH2R172, R140) produces 2-hydroxyglutarate (2-HG), a metabolite that seems to have oncogenic properties due to its ability to affect DNA methylation[50]–[52]. 2-HG, which is present at low concentrations in normal cells, may reach abnormally high concentrations in glioma samples and the serum of AML patients[49],[51]. It has been reported to decrease OXPHOS, thus potentiating the cellular glycolysis capacity. IDH1 overexpression has been observed in over 95% of advanced gliomas, suggesting that it might have prognostic value in gliomas[53]. Nevertheless, further study is needed to understand why IDH mutations have preferential distribution in certain cancers, as well as to specifically evaluate the mechanistic role of these mutations in cancer development and test its value as a potential therapeutic target.
Further, deactivating mutations in FH and SDH, first identified in head and neck paragangliomas, have also been reported in hereditary leiomyomatosis, renal cell carcinoma, and certain gastrointestinal tumors[54]–[56]. The succinate dehydrogenase subunit D (SDHD) protein is one of the four subunits composing the SDH complex (complex II). Mutations in SDHD and FH alter the function of mitochondrial complex II, which affects the electron flow involving flavine adenine dinucleotide (FADH2). These mutations seem to compromise mitochondrial respiratory function and may contribute to the Warburg effect. Loss of FH and SDH activity leads to accumulation of their substrates fumarate and succinate, possibly interfering with the function of the α-ketoglutarate-dependent dioxygenases, including prolyl hydroxylases and HIF-1α, which catalyze a host of biochemical reactions[11],[29],[57],[58].
Metabolic alterations in cancer stem cells
The study of cancer stem cells, a rare subpopulation of malignant cells with long-term self-renewal capacity and high tumorigenic potential, has become an important and intensive research area in recent years. Owing to their special biological properties, cancer stem cells are thought to play key roles in tumor initiation, resistance to chemotherapy and radiotherapy, and disease recurrence[59]. Although extensive information on the biological properties of cancer stem cells, specifically with respect to self-renewal, tumorigenesis, maintenance of sternness, differentiation, and regulatory mechanisms, has been generated during the past decade, our understanding of the metabolic properties of cancer stem cells is rather limited. This is due in part to limited availability of proper cancer stem cell models for metabolic study. The low number of cancer stem cells that can be isolated from primary tumors also limit the scope of metabolic study in this subpopulation of cells.
Notably, certain regulatory pathways known to play important roles in governing cell metabolism and energy sensing have been shown to be involved in maintaining the self-renewal property of cancer stem cells. For instance, the tumor suppressor PTEN negatively regulates the AKT pathway, thus affecting cellular uptake and use of glucose. PTEN blockage has been shown to increase self-renewal capacity and promote clonogenicity of glioblastoma stem cells[60]. Interestingly, the tumor suppressor p53 is also involved in metabolic regulation through its effects on mitochondrial respiratory activity[22]. Inhibition of p53 increased the self-renewal capacity of glioma stem cells, whereas activation of p53 promoted cell differentiation[61]. The energy censor mTOR has been shown to play an essential role in maintaining the hematopoietic stem cells at undifferentiated stage by down-regulating mitochondrial respiration[62]. The transcription factor HIFs are often up-regulated in cancer cells and have been shown to promote the maintenance of cancer stem cells in an undifferentiated state[63],[64]. Interestingly, glioma stem cells seem to be confined in the tissue niches in the brain where the oxygen concentration is restricted[65]. Such a hypoxic microenvironment would favor the stabilization of HIF-1α.
Aldehyde dehydrogenase 1 (ALDHI) is a metabolic enzyme involved in detoxification of certain toxic compounds and in the metabolism of biomolecules, such as the conversion of vitamin A (retinol) to retinoic acid. ALDHI is often expressed in cancer stem cells and has been used as a functional marker for isolating cancer stem cells[66]. The retinoic acid signaling is believed to be important in determining the cell fate. ALDHI has been used as a marker in detection of breast cancer stem cells [67]–[69] and, more recently, in lung cancer[70]. In hematopoetic stem cells, inhibition of ALDHI activity leads to an increase in the population[71]. Interestingly, the PI3K/AKT pathway seems to play a major role in maintaining the ALDH1–positive population of breast cancer[72].
In several studies, the metabolic function in cancer stem cells has been directly assessed. Zhou et al. [73] reported that the stem-like glioma cells with enriched CD133+ cells showed increased tumorigenic capacity, low mitochondrial respiration, and high glycolytic activity. Furthermore, these metabolic properties seemed to confer cancer stem cells the ability to survive and proliferate in a hypoxic microenvironment and render them highly sensitive to glycolytic inhibition[73]. Consistently, a recent study in human pluripotent stem cells and their differentiated counterparts showed that the stem cells expressed high levels of glycolytic enzymes and relied mostly on glycolysis to meet their energy demands[74]. In hypoxia-adapted Bcr-Abl(+) cells that exhibited stem cell-like characteristics, the expression of glyoxalase-I, an enzyme that detoxifies methylglyoxal (a toxic metabolite of glycolysis) was high, and these cells were much more sensitive to glyoxalase-l inhibitors[75]. These studies together suggest that cancer stem cells may have unique metabolic properties that can serve as potential targets for anticancer therapy. As cancer stem cells are more resistant to conventional therapeutic agents and play a major role in therapeutic failure and cancer recurrence, the development of novel agents that effectively kill cancer stem cells based on their unique metabolic properties will have major therapeutic implications.
Metabolic Alterations and Drug Resistance
Defects in the mitochondrial respiration and mutations in mitochondrial DNA (mtDNA) have been implicated to affect drug sensitivity. Several groups have used respiration-deficient ρ0 cells to investigate the role of mitochondrial respiration in drug resistance. Singh et al.[76] showed that the ρ0 cells were resistant to photodynamic therapy and adriamycin but remained sensitive to DNA alkylating agents and γ-radiation. Cai et al.[77] reported that although staurosporine treatment induced release of cytochrome c and activation of caspases in both ρ0 and ρ+ cells, the redox homeostasis remained unchanged in ρ0 cells upon induction of apoptosis, suggesting that apoptosis and bioenergetics are two separate events. In another study using SK-Hep1 hepatoma cells, deprivation of mtDNA resulted in ρ0 cells with elevated expression of the antioxidant enzymes manganese superoxide dismutase and glutathione peroxidase. These cells were relatively resistant to H2O2 and ROS-inducing agents such as doxorubicin, paraquat, and menadione, suggesting a significant role of mitochondrial respiration in drug sensitivity[78]. Furthermore, respiration-defective ρ0 prostate cancer cells were resistant to the cytotoxic effects of the synthetic retinoid 4HPR, implying that OXPHOS may be critical to mediate the effects of 4HPR in transformed human prostate cells[79]. Thus, a defect in mitochondrial respiration function may positively or negatively impact drug sensitivity, depending on the mechanisms of actions of the compounds. Ironically, the mtDNA-depleted HeLa cells (EB8 cells) lost their ability to form colonies in the anchorage-independent colony formation assay and to form tumors in nude mice, although both features were restored upon introduction of normal mtDNA into the cells[80]. It appears that a complete loss of mitochondrial respiration may compromise the cell's ability to survive and proliferate in the in vivo tissue environment.
Apart from mtDNA mutations and respiratory dysfunction, ROS and cellular redox homeostasis also play a critical role in determining and modulating cellular sensitivity to drugs. ROS can cause DNA mutations that may contribute to the development of a drug-resistant phenotype. However, an increase in ROS beyond a certain cellular threshold may lead to cell death through various mechanisms, such as activation of the JNK/p38-mediated signaling pathway, direct damage to the mitochondrial membrane, and release of cytochrome c[81]–[84]. interestingly, malignant cells in advanced stage cancers may develop resistance to ROS stress by up-regulation of their antioxidant capacity. For instance, resistance to arsenic trioxide in leukemia cells has been correlated with high cellular GSH levels and elevated expression of superoxide dismutase 1[85],[86]. Similarly, resistance to paclitaxel and cisplatin has been associated with increased antioxidant levels[87]–[89]. Thus, abrogation of the cancer cell antioxidant defense mechanism, such as depletion of GSH using pharmacological agents, seems to be an effective strategy to overcome such drug resistance[90],[91]. For drug resistance associated with mitochondrial respiration defects and increased glycolysis, the use of glycolytic inhibitors is an effective strategy[92].
Mechanisms Underlying Metabolic Alterations
After his initial observation of increased glycolysis in cancer cells[2], Warburg later attributed this metabolic alteration to “respiratory injury” and considered this as the main cause of cancer[1]. This theory was immediately challenged by Weinhouse, who contended that mitochondrial respiration was functioning in cancer cells and that active glycolysis and OXPHOS together would result in a generation of excess ATP, thus creating a metabolic imbalance in cancer cells[93]. More recently, Moreno-Sanchez et al.[94] suggested that cancer cells are heterogeneous in their energy metabolic profiles, with some primarily using glycolysis whereas others use OXPHOS. Since mitochondria are the powerhouses of the cell and serve as the converging points of multiple metabolic pathways, mitochondrial dysfunction has long been suspected to play a key role in metabolic alterations in cancers. Interestingly, a recent study by Compton et al.[95] demonstrated that mitochondrial respiratory state can profoundly affect p53 expression and, thus, might play a role in tumorigenesis. Alterations in cancer cell metabolism have been attributed to dysfunctional mitochondria resulting in part from mtDNA mutations, and metabolic reprogramming may be linked to oncogenes and tumor suppressors that either affect mitochondrial function or regulate important molecules involved in the energetic pathways.
Mitochondria mutations in cancers
A major role of mitochondria is to generate energy in the form of ATP through OXPHOS, which is carried out through the electron transport chain (ETC) associated with the mitochondrial inner membranes[96]. The ETC consists of four respiratory complexes (I–IV), each with multiple protein components or subunits. Electrons are carried through these complexes to molecular oxygen with a simultaneous pumping of protons across the inner membranes to generate an electrochemical gradient, which is then used as the energy source to drive ATP synthesis at complex V. Each ETC complex contains subunits that are encoded by both mtDNA and nuclear DNA, with an exception of complex II, whose subunits are all encoded by nuclear DNA. Mammalian mitochondria contain a genome of 16.5 kb of double-stranded circular DNA encompassing 37 genes, of which 13 are components of respiratory chain complexes[96]–[98]. The lack of histone protection as well as the close physical proximity of the mitochondrial genome to the source of ROS generation makes mtDNA highly prone to mutations[99].
Accumulating evidence suggests that mtDNA mutations occur at higher frequency in cancer cells than in normal cells, perhaps due in part to the increased ROS generation in the cancer cell mitochondria[100]. Because each cell harbors numerous mitochondria with multiple copies of mtDNA, mutations at mtDNA can be heteroplasmic (variably mutated mtDNA copies co-exist in the same cells) or homoplasmic (all mtDNA copies carry the same mutations). For the heteroplasmic mutations to have a significant effect on the cells, it is estimated that such mutations should be predominant and reach a minimal threshold of 60% of the mitochondrial copies[101],[102]. However, due to in vivo selection of mutations that confer advantage for cell survival and proliferation, the heteroplasmic mutations with functional consequences may eventually become homoplasmic or eliminated (disadvantage mutations). This may explain why the majority of mutations detected in human cancers are homoplasmic[103],[104]. Figure 2 shows a summary of mtDNA mutations in the coding region of respiratory chain components detected in various types of cancer tissues. Mutations can occur in any part of mtDNA, although there may be certain “hot spots” where mutations are more concentrated. Many mutations have also been detected in the non-coding region (D-loop), where the origin of replication and the transcription promoter sequences are located. Mutations of D-loop do not directly affect the structure and function of any particular protein encoded by mtDNA but may affect mtDNA replication and transcription.
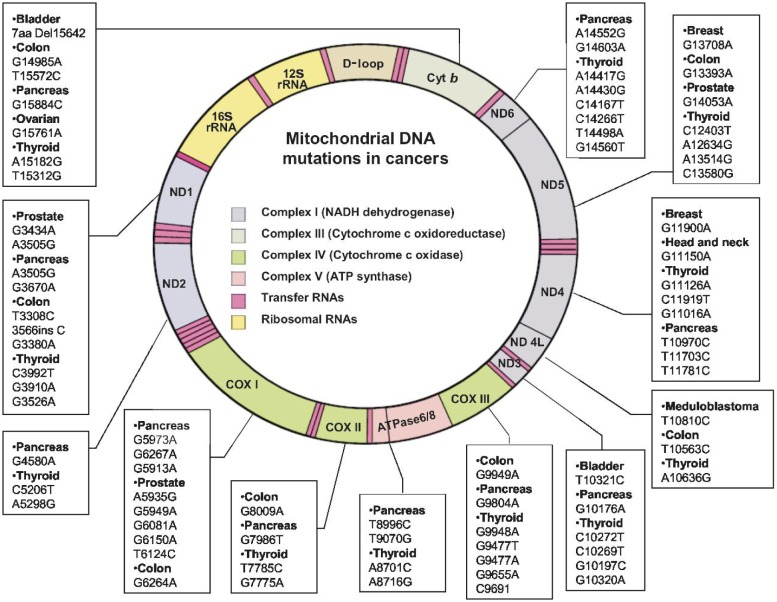
Figure 2. Human mitochondrial DNA (mtDNA) map and mutations in cancers related to metabolism.
The human mitochondrial genome Is a 16.5 kb double-stranded circular DNA, which contains 37 genes: 13 polypeptides that are components of the electron transport complexes (ETC), 22 tRNAs, and 2 ribosomal RNAs. Mutations at the 13 ETC-encoding genes may cause alterations of the respiratory chain activity and thus affect metabolism. Each box linked to a particular mitochondrial gene region contains a partial list of mutations that have been identified in that specific mtDNA in human cancers, with the specific cancer types indicated. The numbers represent the locations of the mutated bases (C, cytosine; G, guanine; T, thymine; A, Adenine). Note that the two adjacent genes for ATPase6 and ATPase8 are shown in a single box.
Extensive mtDNA sequencing in breast cancer conducted by Tan et al.[105] revealed that 74% of patient samples had mutations at 27 different sites, most of which were situated in the D-loop region. Parella et al.[106] detected mtDNA mutations in 61% of fine-needle aspirates of breast cancer tissues, with the majority of mutations again located in the D-loop region. Single base substitutions and deletions seem to be the most common mutations found in breast cancer mtDNA. In a colorectal cancer study, Polyak et al.[107] reported the occurrence of mutations in colon cancer cell lines as well as the original tumors from which the cells were derived. The mutations occurred in the coding regions including the NADH dehydrogenase 1 (ND1), ND4L, ND5, cytochrome b, and 12S and 16S RNA genes. Mutations in the D-loop region in colorectal cancer have also been reported[108]. Another interesting finding was the presence of mutations in the normal colonic crypt stem cells, suggesting that mutations may occur before the development of colon cancer and contribute to Carcinogenesis, although this functional role has not been experimentally demonstrated[109]. In gastric cancer, the D-loop region harbored mutations in 48% of the carcinoma samples sequen ced by Wu et al.[110]. Additionally, mtDNA mutations have been detected in ovarian[111], hepatocellular[112],[113], pancreatic[114], prostate[115], thyroid[116], brain[117], and lung cancers[118].
Role of oncogenes and tumor suppressor genes in alterations of cancer cell metabolism
Emerging evidence suggests that metabolic deregulations play a crucial role in supporting cancer cell survival, proliferation, and tumor growth and that certain oncogenes may orchestrate the activation of enzymes that are important for the glycolytic pathway and thus contribute to the Warburg effect. On the other hand, certain metabolic pathways may lead to production of metabolites that promote silencing of tumor suppressors and activation of certain oncogenes.
Akt
The serine/threonine kinase AKT, a downstream effector of the insulin signaling, plays a major role in cell survival and proliferation and is an important promoter of glysolysis[119]. In the majority of tumor cells, Akt is hyperactive and seems to drive addiction to glucose metabolism for survival and proliferation[13]. The Akt glycolytic enhancer role is driven by stimulation of glucose uptake in response to insulin[13]. Akt up-regulates GLUT1 by enhancing its expression at transcription level. Moreover the AKT signaling enhances glucose metabolism within the cell by stimulating the association of HKII with the mitochondrial outer membrane, thus positioning it to phosphorylate glucose to glucose-6-phosphate for use in glycolysis and other metabolic pathways[120]. Akt's ability to promote metabolism via glycolysis is also partly due to induced expression of glycolytic enzymes through the mTOR and HIF-1 signaling[16],[121].
mTOR
mTOR is an energy censor that promotes nutrient uptake and glycolysis. Deregulation of mTOR pathway is often found in cancer cells[17]. Under nutrient deprivation, mTOR forms a stable complex with raptor, leading to an inhibition of mTOR kinase activity[122]. Akt regulates mTOR through the inactivation of the protein tuberous sclerosis-2 (TSC2). TSC2 and TSC1 associate and form a complex that suppresses mTOR activity[123]. Activated AKT phosphorylates TSC2, thus affecting its interaction with the small G protein Rheb, which when loaded with GTP, activates mTORC1[122],[124]. The activation of mTORC1 increases glycolysis by increasing the translation of the mRNA encoding HIF-1α, thus stimulating the expression of glycolytic enzymes [125]. Importantly, mTOR is regulated by liver kinase B1 (LKB1) and AMPK[126]. The mTOR energy sensor function is conferred in part by AMPK, a heterotrimeric serine/threonine kinase consisting of a catalytic subumit (α) and two regulatory subunits (β and γ)[127]. Nutrient depravation causes cellular stress, leading to an increase of the AMP/ATP ratio, which promotes the binding of AMP to the γ-regulatory subunit and triggers AMPK activation[128]. AMPK activation in turn induces TSC2 phosphorylation, leading to suppression of mTOR activity and inhibition of protein translation. The serine/threonine kinase LKB1 activates AMPK under low ATP conditions[129]. Therefore, the LKB1/AMPK/mTOR axis serves as an important energy sensor that links energy status and the metabolic signaling pathways[130].
HIF-1
Hypoxia inducible factor-1, a heterodimer protein complex that consists of HIF-1α and HIF-1β, has been implicated in the induction of key genes involved in cell proliferation, oxygen and nutrient delivery, and anaerobic energy metabolism[131]. HIF-1 regulation is achieved by protein degradation of the HIF-1α subunit in response to oxygen levels in the microenvironment. Oxygen promotes prolyl hydroxylation, which stimulates the association of HIF-1α with the von Hippel-Linau (VHL) tumor suppressor, thereby targeting HIF-1α for ubiquitination and proteosomal degradation. Hypoxia suppresses prolyl hydroxylation through a process involving mitochondria-generated ROS[132],[133]. As such, HIF-1 is considered as the master sensor that orchestrates cellular responses to changes in oxygen homeostasis. In fact, HIF-1 has been found to be highly expressed within hypoxic tumors and at low levels within normal tissue[134].
HIF-1 up-regulates 9 of the 10 genes involved in glycolysis and, thus, plays a key role in switching glucose metabolism from OXPHOS to glycolysis when cells are in a hypoxic environment[134],[135]. For instance, HIF promotes glucose uptake by up-regulating the expression of the GLUT1 and glucose metabolism by up-regulating hexokinase, the enzyme responsible for catalyzing the first glycolytic reaction. HIF-1 also promotes the expression of LDH-A at the transcription level[136],[137]. Furthermore, HIF-1 enhances glycolysis by preventing the use of pyruvate by the mitochondria through the up-regulation of PDK1, which phosphorylates PDH1 and inhibits its ability to convert pyruvate to acetyl-CoA as the fuel for the TCA cycle[138]. In addition to response to environmental oxygen fluctuation, HIF-1 can also be regulated by molecules such as FH and SDH. Loss of function mutations of FH and SH lead to the accumulation of the TCA cycle intermediates furmate and succinate, resulting in inhibition of the α-ketoglutarate–dependent prolyl hydroxylase, thereby stabilizing HIF-1[139].
c-Myc
c-Myc is an Oncogene that affects many cellular functions including energy metabolism [20]. Myc overexpression is estimated to be associated with at least 40% of all cancers[140]. Similar to HIF-1, c-Myc promotes glycolysis mainly through up-regulation of glycolytic molecules including GLUT1, HKII, phosphofructokinase (PFK), enolase, and LDH[21]. It is important to note that c-Myc also plays a major role in promoting glutamine use in cancer cells through up-regulating the expression of glutamine transporters SLC5A1 and SLC7A1 and enhancing the expression of GLS1 by repressing the expression of miR-23A and miR-23B, thus releasing the suppressive effect of these miRNAs on GLS1 expression[38],[39].
p53
The tumor suppressor p53 is a stress sensor and cell cycle checkpoint regulator in mammalian cells that plays essential roles in cell cycle regulation, apoptosis, and genome stability[141]. The role of p53 in energy metabolism was recognized by Matoba et al.[22], who found that loss of p53 diminishes the synthesis of cytochrome c oxidase (SCO2), a nuclear DNA-encoding gene whose product is necessary for the assembly of mitochondrial respiratory complex IV. Thus, this study directly links p53 to OXPHOS. The loss of p53 shifted the cellular ATP production from mitochondrial respiration to glycolysis. Moreover, p53 protein represses the expression of the GLUT1 and GLUT4 transporters. Thus, the loss of p53 enhances the expression of glucose transporters and further promotes glycolysis. Interestingly, Benssad et al.[21] identified a p53 target gene known as TP53-induced glycolysis and apoptosis regulator (TIGAR), which is a potent glycolysis regulator. Expression of TIGAR decreases the intracellular level of fructose-2,6-bis-phosphate, which otherwise suppresses glycolysis by shunting glucose to the PPP[142]. In addition, p53 also affects energy metabolism by regulating AMPK, mTOR, PTEN, and IGF-binding protein-3[143].
Ras
Ras mutations are found in approximately 30% of all human cancers[144] and are important in promoting cancer initiation and progression[145]. K-Ras, the most commonly mutated oncogenic Ras in pancreatic cancer, has been shown to affect the shape and function of the mitochondria during fibroblast transformation[145]. Further studies showed that fibroblasts transformed by activated K-Ras attenuated OXPHOS by suppressing the activity of respiratory complex I, with a corresponding decrease in the expression level of complex I proteins[147]. Similarly, H-Ras transformed mouse fibroblasts exhibited low mitochondrial respiration, an increased dependency on glycolysis, a sensitivity to glycolytic inhibitors, and an insensitivity to OXPHOS inhibitors[148]. However, a recent study showed that K-Ras may affect the synthesis of the mitochondrial Phospholipid cardiolipin and that the absence of K-Ras led to increased respiration[148], suggesting that the effect of K-Ras on mitochondrial respiration is likely complex and requires further study.
Therap eutic approaches targeting cancer cell metabolism
Although metabolic alterations in cancer cells are highly complex and the detailed underlying mechanisms remain to be elucidated, the unique metabolic profiles in cancer cells may serve as a biochemical basis for developing new therapeutic strategies to effectively kill the malignant cells with high selectivity. For cancer cells that are highly dependent on glycolysis, inhibiting glycolysis seems to be a logical therapeutic strategy. Similarly, interference with glutamine metabolism may significantly impact cancer cells that depend on glutamine for survival and proliferation. Due to the metabolic adaptability of cancer cells, it may be necessary to combine agents that target different metabolic pathways to achieve high therapeutic activity. The following sections describe several therapeutic strategies that target various metabolic pathways important for cancer cells. The representative compounds are listed in Table 1.
Table 1. Therapeutic strategies to target cancer metabolism and relevant agents.
| Therapeutic approach | Metabolic target | Agent | References |
| Inhibition of glycolysis | HK | 2-Deoxyglucose, 3-bromopyruvate Lonidamine | [153]–[158], [217], [218] |
| HK-VDAC complex | Methyl jasmonate | [120], [160]–[162] | |
| LDH & lactate transport | Oxamate, shRNAs, α-cyano-4-hydroxy cinnamic acid | [3], [171], [173], [174] | |
| PDK | Dichloroacetate | [170], [175], [176] | |
| Glucose transporter | Phloretin | [177] | |
| Phosphofructokinase | PFKFB3-3(3-pyridinyl)-1-(4-pyridinyl)-2-p, 3PO, Clotrimazole | [178], [179], [219] | |
| Pyruvate Kinase | CAP-232/TLN-232 | [217] | |
| Interfering glutamine metabolism | Glutamine (analogs) | 6-Diazo-5-oxo-L-norleucine, azaserine, acivicin | [182]–[184], [191] |
| Glutamine | L-Asparaginase, phenylbutyrate, | [181], [185], [186] | |
| Glutamine transport | L-γ-glutamyl-p-nitroanilide (GPNA), | [33], [192] | |
| Glutaminase | Compound 968, 6-diazo-5oxo-norleucine | [40], [193] | |
| Transaminase | Amino-oxyacetic acid | [33], [195], [196] | |
| Targeting energy sensors & regulators | AMPK | Metformin thiazolidinediones, AICAR | [199]–[202], [220] |
| HDAC | Romidepsin, SAHA | [205], [206] | |
| HIF-1 | Romidepsin, | [206] | |
| AKT | NSC 644221, | [189] | |
| PI3K | RX-0047 | [190] | |
| mTOR | Genistein, celecoxib, perifosine, GST-anti-Akt1-MTS | [207], [208] | |
| LY294002, wortmannin | [209] | ||
| Rapamycin, Temsirolimus (CCI-779), Everolimus (RAD-001) | [211]–[213] |
HK, hexokinase; VDAC, voltage-dependent anion channel; LDH, lactate dehydrogenase; PDK, pyruvate dehydrogenase kinase; AMPK, AMP-activated protein kinase; HDAC, histone deacetylase; SAHA, suberoylanilide hydroxamic acid; AICAR, 5-aminoimidazole-4-carboxamide ribonucleotide; HIF-1: hypoxia-inducible factor-1; mTOR: mammalian target of rapamycin.
Inhibition of the glycolytic pathway
HKII has been shown to be elevated in cancers and possibly up-regulated by HIF-1 [150]–[152]. 2-Deoxyglucose (2-DG), a non-metabolically active glucose analog that inhibits HKII and represses tumor growth, is currently in clinical trials for cancer treatment[153],[154]. 2-DG preferentially kills cancer cell exhibiting mitochondrial defects or under hypoxia[155]–[157]. Combination of 2-DG with cisplatin has been shown to increase cytotoxicity through oxidative stress in human head and neck cancer cells[153]. Increased cytotoxic effects were also observed when 2-DG was combined with adriamycin and paclitaxel in mice bearing human osteosarcoma and in the MV522 non–small cell lung cancer xenograft model[158].
3-Bromopyruvate (3-BrPA), another inhibitor of HKII, has been shown to be effective in killing lymphoma and colon cancer cells as well as other cancer cells with mitochondrial defects[92]. Consistent with its ability to inhibit glycolysis, 3-BrPA is particularly effective against cancer cells under hypoxia, a condition where cells become highly dependent on glycolysis. Interestingly, Chen et al.[159] reported that 3-BrPA triggered leukemia cell death by disrupting the association between HKII and apoptosis-inducing factor in the mitochondria. Similarly, the plant-derived methyl jasmonate or the HKII amino terminus-derived peptide TAT-HK can also disrupt the association between hexokinase and the voltage-dependent anion channel (VDAC), a phenomenon observed more in cancer cells compared to normal counterparts[120],[160],[161]. In a recent study, methyl jasmonate was found to activate the PI3K/Akt pathway in sarcoma cell lines, whereas its combination with 2-deoxy-D- glucose to inhibit glycolysis resulted in a synergistic cytotoxic effect[162].
Lonidamine, a derivative of indazole-3-carboxylic acid, was shown to inhibit tumor growth through inhibition of HKII, depletion of ATP, reduction of oxygen consumption, and lactate production[163]–[165]. Combination of lonidamine with chemotherapeutic drugs such as doxorubicin and cisplatin have been tested in clinical trials in breast cancer, ovarian cancer, glioblastoma, and non–small cell lung cancer[166]–[169]. In addition to its applications in cancer treatment, lonidamine was developed by Threshold Pharmaceuticals as TH-070 to treat benign prostatic hyperplasia (BPH). Unfortunately, clinical trials of TH-070 in patients with BPH did not show a significant therapeutic effect, and the development of TH-070 was discontinued in 2006. It should be noted, however, that the ability of lonidamine to inhibit glycolysis and its potent effect against cancer cells suggest that this compound may still have potential value in cancer treatment, especially for the highly glycolytic cancer types.
A logical approach to target glycolysis is to inhibit LDH, which is an NADH-dependent enzyme that converts pyruvate to lactate and has been shown to be elevated in certain cancer cells[3],[170]. Inhibition of LDH has been demonstrated by use of oxamate, resulting in reduced glycolytic rate, decreased glucose uptake, and suppressed growth of HeLa cells[171]. Combination of LDH inhibition with doxorubicin resulted in enhanced ATP depletion and overall cyto toxicity[172]. Additionally, α-cyano-4-hyroxycinnamic acid is an agent that inhibits lactate transport and, thus, can affect lactate metabolism and potentially interfere with glycolysis in cancer cells[173],[174].
Another strategy for inhibiting the glycolytic process is targeting PDK. PDK phosphorylates PDH, resulting in inhibition of PDH activity[170]. Dichloroacetate inhibits PDK and indirectly increases production of acetyl-CoA, which can then enter the TCA cycle, thereby switching cellular energy metabolism from glycolysis to mitochondrial OXPHOS. This compound has been previously used in treating lactic acidosis and seems to be well tolerated in young children[175]. Bonnet et al. [176] showed that dichloroacetate reduced tumor cell proliferation and increased apoptosis by activating the expression of the K+ (Kv1.5) channel through nuclear factor of activated T cells and by reducing the mitochondrial membrane potential in tumor cells. The therapeutic activity of dichloroacetate for cancer remains to be evaluated in clinical trials.
Additionally, as glycolysis is dependent on glucose uptake, inhibiting glucose transporters is an alternate method to repress active glycolysis often observed in cancer cells. Phloretin is a glucose transporter inhibitor reported to induce apoptosis and overcome drug resistance in hypoxic conditions[177]. Moreover, since PFK is an important regulatory enzyme in glycolysis, it is considered a high-impact target for antitumor drugs[178]. PFKFB3-3(3-pyridinyl)-1-(4-pyridinyl)-2-propen-1-one(3PO), a small-molecule inhibitor, was shown to inhibit PFK activity, reduce glucose uptake, and suppress tumor growth in several human malignant hematopoietic and adenocarcinoma cell lines[179].
Although many cancer cells are highly dependent on the glycolytic pathway, certain tumor cells, including those of the lung, mammary glands, skin, and cervix, are active in OXPHOS[94]. Therefore, it must be noted that the metabolic profiles of tumor cells are dependent on the cancer type, microenvironment, differentiation, and cancer stage. Further, inhibiting glycolysis may not be a universal strategy to target cancer metabolism.
Inhibition of glutamine metabolism
The dependency of certain cancer cells on exogenous glutamine for survival has been referred to as “glutamine addiction”, which renders cancer cells highly sensitive to glutamine starvation[180],[181]. Early studies have shown that glutamine analogues, such as 6-diazo-5-oxo-L-norleucine (L-DON), azaserine, and acivicin, exhibited potential anticancer activity, but the efforts to develop these compounds as therapeutic drugs were hampered due in part to potential neurotoxicity, gastrointestinal toxicity, and myelosuppression[182]–[184]. Interestingly, a recent study showed that inhibition of glutamine metabol ism using L-DON suppressed cancer metastasis in a mouse model of breast cancer[6].
Another approach to inhibit glutamine metabolism is to lower the glutamine levels in blood using agents such as L-asparaginase and phenylbutyrate. L-asparaginase, an enzyme that hydrolyzes asparagine to aspartic acid, has been used successfully in treating pediatric acute lymphoblastic leukemia[185], as these particular leukemia cells are unable to synthesize asparagines. L-asparaginase can also hydrolyze glutamine to glutamic acid and ammonia, thereby depleting glutamine in the blood[181],[186]. Phenylbutyrate, also known as Buphenyl (Ucyclyd Pharma) or Ammonaps (Swedish Ophan International), is an FDA-approved agent for the treatment of hyperammonemia. This compound is metabolized in the human body to phenylacetate, which is then conjugated with glutamine to form phenylacetylglutamine for excretion by the kidneys. As such, phenylbutyrate is able to deplete plasma glutamine levels. Phenylacetate has also been used in patients with various cancers and other medical conditions[187]–[189].
Glutamine transporters such as SLC1A5 (ASCT2) and SLC1A7 are up-regulated in cancers, making them attractive targets for the suppression of glutamine uptake[38],[39],[190]. Recent studies also suggest that glutamine levels affect activation of mTORC[191]. IL-γ-glutamyl-p-nitroanilide (GPNA) is a SLC1A5 inhibitor shown to inhibit glutamine uptake and attenuate glutamine-dependant mTOR activation, leading to induction of autophagy in cancer cells[33],[192].
Cancer cells display increased activity in glutaminase, an enzyme responsible for hydrolyzing glutamine to glutamate and ammonia. This enzyme seems to play an important role in energy metabolism and survival of certain cancer cells, thus providing a therapeutic window through its inhibition[193]. Human B-cell lymphoma and prostate cancer cells show increased glutaminase expression, and the Oncogene c-Myc plays a major role in promoting glutaminase expression through down-regulation of miR-23[39]. Similarly, breast cancer cell line MDA-MB231 showed a higher expression of glutaminase when compared to normal mammary epithelial cells[40]. A small molecule known as compound 968 has been shown to inhibit glutaminase and suppress oncogenic transformation [40]. Conversion of glutamine to α-ketoglutarate as a metabolic intermediate for the TCA cycle was demonstrated as essential for activating various pathways involving K-Ras that promote tumorgenesis. As the main route through which glutamine enters the TCA cycle is transamination, the transaminase inhibitor amino-oxyacetic acid (AOA) has been suggested as potential therapeutic agent for cancer[194]. AOA exerts a cytotoxic effect on glutamine- dependent Myc-amplified glioblastoma cells and inhibits breast cancer cell growth in a mouse xenograft model[33],[195]. This compound has also been shown to sensitize melanoma cells to TRAIL-mediated killing by interfering with glutamine metabolism[196].
Targeting the energy sensors and regulators
AMPK activators & HDAC inhibitors
AMPK, an evolutionarily conserved enzyme that plays a pivotal role in maintaining cellular energy homeostasis[197], is activated by an elevated ratio of AMP/ATP under various stress conditions, such as hypoxia, glucose deprivation, and oxidative stress. As described earlier, activated AMPK has an effect on various molecules that are involved in cancer-related cellular processes. A recent study examined the role of AMPK in breast cancer progression and showed that reduced expression of AMPK in breast cancer specimens was inversely correlated with axillary node metastasis and histological grade, suggesting an important role of AMPK signaling in cancer progression and metastasis[198]. These observations also imply that activation of AMPK might have therapeutic potential in cancer treatment. Biguanides and thiazolidinediones, two classes of compounds used in treating diabetes, can activate AMPK. A clinical study that involved treating diabetic breast cancer patients with or without the AMPK activator metformin (biguadine) showed that patients in the metformin treatment arm exhibited complete pathologic responses[199]. The antitumor effect of troglitazone (thiazolidinedione) has been demonstrated in various cancer cells involving activation of PPAR-γ[200],[201]. Similarly, 5-aminoimidazole-4-carboxamide ribonucleoside reversed the sensitivity of Akt-expressing glioblastoma cells to glucose deprivation by activation of AMPK[202]. Another metabolic therapeutic approach involves the use of histone deacetylase (HDAC) inhibitors. AMPK activation promotes the phosphorylation of mitochondrial acetyl-CoA carboxylase, leading to fatty acid and cholesterol synthesis, while acetyl-CoA is necessary for histone acetylation[203]. Therefore, epigenetic regulation can be modulated by metabolic factors with therapeutic potential in cancer[204]. The HDAC inhibitor suberoylanilide hydroxamic acid was found to induce oxidative stress, autophagy, and apoptosis in imatinib-resistant chronic myelogenous leukemia[205]. Romidepsin, another HDAC inhibitor, is in clinical phase II trials for the treatment of refractory myeloma and has been shown to inhibit the HIF-1 pathway[206].
Inhibition of PI3K/AKT/mTOR axis
The PI3K/AKT/mTOR signaling pathway plays a key role in energy metabolism and regulates cell growth and proliferation in a variety of tumor cells, thereby serving as a critical target for cancer therapy. Genistein, celecoxib, and perifosine are pharmacologic agents that target Akt and have been tested for treating prostate cancer[207]. A cell permeable Akt antibody that blocks the catalytic site of AKT is effective in triggering cell death in various cancer cell lines[208]. The PI3K inhibitors LY294002 and wortmannin and other derivatives have been investigated in various types of cancer [209]. The mTOR pathway can activate the transcription factor HIF-1 and enhance angiogenesis and glycolysis while down-regulating OXPHOS, rendering tumor cells more glycolytic and aggressive[210]. Rapamycin has been shown to have an anticancer effect, and other mTOR inhibitors such as temsirolimus (CCI-779) and everolimus (RAD-001) are currently in clinical trials for cancer treatment [211]–[213]. Inhibition of mTOR leads to profound change in cellular metabolism and attenuation of protein synthesis.
Targeting other regulators of cancer metabolism
The Myc Oncogene contributes to cancer development by encoding a transcription factor c-Myc, which plays a key role in cellular metabolism to Carcinogenesis[20]. Myc affects metabolism through its ability to regulate various genes involved in glucose and glutamine metabolism, including GLUT1, HKII, PFKM, enolase, LDH-A, and glutaminase. Similarly, HIF-1 also plays a major role in sensing changes in microenvironment and promotes glycolysis through regulation of glycolytic enzymes. Various strategies are currently being explored to target these important regulatory molecules and their downstream effectors for potential use in cancer treatment[214]–[220].
Concluding Remarks
An increase in aerobic glycolysis, first reported by Otto Warburg more than 80 years ago, represents the most prominent metabolic alteration observed in cancer cells. Other important metabolic changes, such as glutamine addiction and active OXPHOS in certain tumor cells, were subsequently discovered. The differences between cancer cells and normal cells in their energy metabolism provide a biochemical basis for developing new therapeutic strategies to preferentially kill cancer cells with selectivity using metabolism-targeted compounds. Inhibiting glycolysis and interfering glutamine metabolism are two possible metabolic intervention approaches. Additionally, emerging metabolic profiling of cancer stem cells will also provide new therapeutic opportunities. Mitochondrial dysfunctions, activation of oncogenes, loss of tumor suppressors, and tumor tissue microenvironment conditions are known to profoundly affect the metabolism of cancer cells, rendering heterogeneous metabolic profiles among different cancer types. As such, it is important to determine the specific metabolic alterations in each particular cancer type so that effective cancer type-specific metabolic intervention strategies can be developed. Furthermore, a combination of conventional chemotherapeutic agents and metabolic modulators may enhance therapeutic activity and should be further evaluated. It should be cautioned, however, that as energy metabolism and its regulatory machinery are evolutionarily conserved and shared by various normal cells, metabolic inhibitors are likely to affect normal cells to various degrees. Despite a number of pharmacologic agents that show promising results in preclinical studies, whether the metabolic alterations in cancer cells can be targeted efficiently and safely in the clinic has yet to be determined. Thus, it is vitally important to identify the metabolic steps that are altered and critical in cancer cells but dispensable in normal cells as the valid therapeutic targets, which will enable the development of effective agents to improve cancer treatment outcomes. A comprehensive understanding of the metabolic differences between cancer and normal cells through detail investigation will pave the way to achieve this goal.
References
- 1.Warburg O. On the origin of cancer cells [J] Science. 1956;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 2.Warburg O, Wind F, Negelein E. The metabolism of tumors in the body [J] J Gen Physiol. 1927;8(6):519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fantin VR, Berardi MJ, Scorrano L, et al. A novel mitochondriotoxic small molecule that selectively inhibits tumor cell growth [J] Cancer Cell. 2002;2(1):29–42. doi: 10.1016/s1535-6108(02)00082-x. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation [J] Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Weinberg F, Hamanaka R, Wheaton WW, et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity [J] Proc Natl Acad Sci USA. 2010;107(19):8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shelton LM, Huysentruyt LC, Seyfried TN. Glutamine targeting inhibits systemic metastasis in the VM-M3 murine tumor model [J] Int J Cancer. 2010;127(10):2478–2485. doi: 10.1002/ijc.25431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aledo JC, Segura JA, Medina MA, et al. Phosphate-activated glutaminase expression during tumor development [J] FEBS Lett. 1994;341(1):39–42. doi: 10.1016/0014-5793(94)80236-x. [DOI] [PubMed] [Google Scholar]
- 8.Brand K. Glutamine and glucose metabolism during thymocyte proliferation. Pathways of glutamine and glutamate metabolism [J] Biochem J. 1985;228(2):353–361. doi: 10.1042/bj2280353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Medina RA, Owen GI. Glucose transporters: expression, regulation and cancer [J] Biol Res. 2002;35(1):9–26. doi: 10.4067/s0716-97602002000100004. [DOI] [PubMed] [Google Scholar]
- 10.Semenza GL. HIF-1: upstream and downstream of cancer metabolism [J] Curr Opin Genet Dev. 2010;20(1):51–56. doi: 10.1016/j.gde.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pollard PJ, Briere JJ, Alam NA, et al. Accumulation of Krebs cycle intermediates and over-expression of HIF1alpha in tumours which result from germline FH and SDH mutations [J] Hum Mol Genet. 2005;14(15):2231–2239. doi: 10.1093/hmg/ddi227. [DOI] [PubMed] [Google Scholar]
- 12.Owusu-Ansah E, Yavari A, Mandai S, et al. Distinct mitochondrial retrograde signals control the G1-S cell cycle checkpoint. Nat Genet. 2008;40(3):356–361. doi: 10.1038/ng.2007.50. [DOI] [PubMed] [Google Scholar]
- 13.Elstrom RL, Bauer DE, Buzzai M, et al. Akt stimulates aerobic glycolysis in cancer cells [J] Cancer Res. 2004;64(11):3892–3899. doi: 10.1158/0008-5472.CAN-03-2904. [DOI] [PubMed] [Google Scholar]
- 14.Hara K, Yonezawa K, Weng QP, et al. Amino acid sufficiency and mTOR regulate p70 S6 kinase and elF-4E BP1 through a common effector mechanism [J] J Biol Chem. 1998;273(23):14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- 15.Hara K, Maruki Y, Long X, et al. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action [J] Cell. 2002;110(2):177–189. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- 16.Hay N. The Akt-mTOR tango and its relevance to cancer [J] Cancer Cell. 2005;8(3):179–183. doi: 10.1016/j.ccr.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 17.Chapuis N, Tamburini J, Green AS, et al. Perspectives on inhibiting mTOR as a future treatment strategy for hematological malignancies [J] Leukemia. 2010;24(10):1686–1699. doi: 10.1038/leu.2010.170. [DOI] [PubMed] [Google Scholar]
- 18.Ruderman N, Prentki M. AMP kinase and malonyl-CoA: targets for therapy of the metabolic syndrome [J] Nat Rev Drug Discov. 2004;3(4):340–351. doi: 10.1038/nrd1344. [DOI] [PubMed] [Google Scholar]
- 19.Luo Z, Saha AK, Xiang X, et al. AMPK, the metabolic syndrome and cancer [J] Trends Pharmacol Sci. 2005;26(2):69–76. doi: 10.1016/j.tips.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 20.Dang CV, Le A, Gao P. MYC-induced cancer cell energy metabolism and therapeutic opportunities [J] Clin Cancer Res. 2009;15(21):6479–6483. doi: 10.1158/1078-0432.CCR-09-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bensaad K, Tsuruta A, Selak MA, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis [J] Cell. 2006;126(1):107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 22.Matoba S, Kang JG, Patino WD, et al. p53 regulates mitochondrial respiration [J] Science. 2006;312(5780):1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 23.Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer's Achilles' heel [J] Cancer Cell. 2008;13(6):472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 24.Bustamante E, Pedersen PL. High aerobic glycolysis of rat hepatoma cells in culture: role of mitochondrial hexokinase [J] Proc Natl Acad Sci USA. 1977;74(9):3735–3739. doi: 10.1073/pnas.74.9.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warburg O. On respiratory impairment in cancer cells [J] Science. 1956;124(3215):269–270. [PubMed] [Google Scholar]
- 26.Roos D, Loos JA. Changes in the carbohydrate metabolism of mitogenically stimulated human peripheral lymphocytes. II. Relative importance of glycolysis and oxidative phosphorylation on phytohaemagglutinin stimulation [J] Exp Cell Res. 1973;77(1):127–135. doi: 10.1016/0014-4827(73)90561-2. [DOI] [PubMed] [Google Scholar]
- 27.Wang T, Marquardt C, Foker J. Aerobic glycolysis during lymphocyte proliferation [J] Nature. 1976;261(5562):702–705. doi: 10.1038/261702a0. [DOI] [PubMed] [Google Scholar]
- 28.Guppy M, Greiner E, Brand K. The role of the Crabtree effect and an endogenous fuel in the energy metabolism of resting and proliferating thymocytes [J] Eur J Biochem. 1993;212(1):95–99. doi: 10.1111/j.1432-1033.1993.tb17637.x. [DOI] [PubMed] [Google Scholar]
- 29.Berardi MJ, Fantin VR. Survival of the fittest: metabolic adaptations in cancer [J] Curr Opin Genet Dev. 2011;21(1):59–66. doi: 10.1016/j.gde.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 30.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, et al. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation [J] Cell Metab. 2008;7(1):11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 31.Izumi H, Torigoe T, Ishiguchi H, et al. Cellular pH regulators: potentially promising molecular targets for cancer chemotherapy [J] Cancer Treat Rev. 2003;29(6):541–549. doi: 10.1016/s0305-7372(03)00106-3. [DOI] [PubMed] [Google Scholar]
- 32.Dang CV. Rethinking the Warburg effect with Myc micromanaging glutamine metabolism [J] Cancer Res. 2010;70(3):859–862. doi: 10.1158/0008-5472.CAN-09-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicklin P, Bergman P, Zhang B, et al. Bidirectional transport of amino acids regulates mTOR and autophagy [J] Cell. 2009;136(3):521–534. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lobo C, Ruiz-Bellido MA, Aledo JC, et al. Inhibition of glutaminase expression by antisense mRNA decreases growth and tumourigenicity of tumour cells [J] Biochem J. 2000;348(Pt 2):257–261. [PMC free article] [PubMed] [Google Scholar]
- 35.DeBerardinis RJ, Mancuso A, Daikhin E, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis [J] Proc Natl Acad Sci USA. 2007;104(49):19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mazurek S, Boschek CB, Hugo F, et al. Pyruvate kinase type M2 and its role in tumor growth and spreading [J] Semin Cancer Biol. 2005;15(4):300–308. doi: 10.1016/j.semcancer.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 37.Deberardinis RJ, Sayed N, Ditsworth D, et al. Brick by brick: metabolism and tumor cell growth [J] Curr Opin Genet Dev. 2008;18(1):54–61. doi: 10.1016/j.gde.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wise DR, DeBerardinis RJ, Mancuso A, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction [J] Proc Natl Acad Sci USA. 2008;105(48):18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao P, Tchernyshyov I, Chang TC, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism [J] Nature. 2009;458(7239):762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang JB, Erickson JW, Fuji R, et al. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation [J] Cancer Cell. 2010;18(3):207–219. doi: 10.1016/j.ccr.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pastorino JG, Shulga N, Hoek JB. Mitochondrial binding of hexokinase II inhibits Bax-induced cytochrome c release and apoptosis [J] J Biol Chem. 2002;277(9):7610–7618. doi: 10.1074/jbc.M109950200. [DOI] [PubMed] [Google Scholar]
- 42.Pedersen PL, Mathupala S, Rempel A, et al. Mitochondrial bound type II hexokinase: a key player in the growth and survival of many cancers and an ideal prospect for therapeutic intervention [J] Biochim Biophys Acta. 2002;1555(1–3):14–20. doi: 10.1016/s0005-2728(02)00248-7. [DOI] [PubMed] [Google Scholar]
- 43.Christofk HR, Vander Heiden MG, Harris MH, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth [J] Nature. 2008;452(7184):230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 44.Schneider J, Neu K, Grimm H, et al. Tumor M2-pyruvate kinase in lung cancer patients: immunohistochemical detection and disease monitoring [J] Anticancer Res. 2002;22(1A):311–318. [PubMed] [Google Scholar]
- 45.Luftner D, Mesterharm J, Akrivakis C, et al. Tumor type M2 pyruvate kinase expression in advanced breast cancer [J] Anticancer Res. 2000;20(6D):5077–5082. [PubMed] [Google Scholar]
- 46.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism [J] Nat Rev Cancer. 2011;11(2):85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 47.Mardis ER, Ding L, Dooling DJ, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome [J] N Engl J Med. 2009;361(11):1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas [J] N Engl J Med. 2009;360(8):765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gross S, Cairns RA, Minden MD, et al. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations [J] J Exp Med. 2010;207(2):339–344. doi: 10.1084/jem.20092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dang L, White DW, Gross S, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate [J] Nature. 2009;462(7274):739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ward PS, Patel J, Wise DR, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate [J] Cancer Cell. 2010;17(3):225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Figueroa ME, Abdel-Wahab O, Lu C, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation [J] Cancer Cell. 2010;18(6):553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Horbinski C, Kofler J, Kelly LM, et al. Diagnostic use of IDH1/2 mutation analysis in routine clinical testing of formalin-fixed, paraffin-embedded glioma tissues [J] J Neuropathol Exp Neurol. 2009;68(12):1319–1325. doi: 10.1097/NEN.0b013e3181c391be. [DOI] [PubMed] [Google Scholar]
- 54.Gill AJ, Chou A, Vilain R, et al. Immunohistochemistry for SDHB divides gastrointestinal stromal tumors (GISTs) into 2 distinct types [J] Am J Surg Pathol. 2010;34(5):636–644. doi: 10.1097/PAS.0b013e3181d6150d. [DOI] [PubMed] [Google Scholar]
- 55.Tomlinson IP, Alam NA, Rowan AJ, et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer [J] Nat Genet. 2002;30(4):406–410. doi: 10.1038/ng849. [DOI] [PubMed] [Google Scholar]
- 56.Baysal BE, Ferrell RE, Willett-Brozick JE, et al. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma [J] Science. 2000;287(5454):848–851. doi: 10.1126/science.287.5454.848. [DOI] [PubMed] [Google Scholar]
- 57.Selak MA, Armour SM, MacKenzie ED, et al. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase [J] Cancer Cell. 2005;7(1):77–85. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 58.Loenarz C, Schofield CJ. Expanding chemical biology of 2-oxoglutarate oxygenases [J] Nat Chem Biol. 2008;4(3):152–156. doi: 10.1038/nchembio0308-152. [DOI] [PubMed] [Google Scholar]
- 59.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions [J] Nat Rev Cancer. 2008;8(10):755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 60.Fan X, Khaki L, Zhu TS, et al. NOTCH pathway blockade depletes CD133-positive glioblastoma cells and inhibits growth of tumor neurospheres and xenografts [J] Stem Cells. 2010;28(1):5–16. doi: 10.1002/stem.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng H, Ying H, Yan H, et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation [J] Nature. 2008;455(7216):1129–1133. doi: 10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen C, Liu Y, Liu R, et al. TSC-mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species [J] J Exp Med. 2008;205(10):2397–2408. doi: 10.1084/jem.20081297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Z, Bao S, Wu Q, et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells [J] Cancer Cell. 2009;15(6):501–513. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Soeda A, Park M, Lee D, et al. Hypoxia promotes expansion of the CD133-positive glioma stem cells through activation of HIF-1alpha [J] Oncogene. 2009;28(45):3949–3959. doi: 10.1038/onc.2009.252. [DOI] [PubMed] [Google Scholar]
- 65.Xie Z. Brain tumor stem cells [J] Neurochem Res. 2009;34(12):2055–2066. doi: 10.1007/s11064-009-0079-5. [DOI] [PubMed] [Google Scholar]
- 66.Vasiliou V, Nebert DW. Analysis and update of the human aldehyde dehydrogenase (ALDH) gene family [J] Hum Genomics. 2005;2(2):138–143. doi: 10.1186/1479-7364-2-2-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morimoto K, Kim SJ, Tanei T, et al. Stem cell marker aldehyde dehydrogenase 1–positive breast cancers are characterized by negative estrogen receptor, positive human epidermal growth factor receptor type 2, and high Ki67 expression [J] Cancer Sci. 2009;100(6):1062–1068. doi: 10.1111/j.1349-7006.2009.01151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hwang-Verslues WW, Kuo WH, Chang PH, et al. Multiple lineages of human breast cancer stem/progenitor cells identified by profiling with stem cell markers [J] PLoS One. 2009;4(12):e8377. doi: 10.1371/journal.pone.0008377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Charafe-Jauffret E, Ginestier C, Iovino F, et al. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature [J] Cancer Res. 2009;69(4):1302–1313. doi: 10.1158/0008-5472.CAN-08-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sullivan JP, Spinola M, Dodge M, et al. Aldehyde dehydrogenase activity selects for lung adenocarcinoma stem cells dependent on notch signaling [J] Cancer Res. 2010;70(23):9937–9948. doi: 10.1158/0008-5472.CAN-10-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chute JP, Muramoto GG, Whitesides J, et al. Inhibition of aldehyde dehydrogenase and retinoid signaling induces the expansion of human hematopoietic stem cells [J] Proc Natl Acad Sci USA. 2006;103(31):11707–11712. doi: 10.1073/pnas.0603806103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Korkaya H, Paulson A, Charafe-Jauffret E, et al. Regulation of mammary stem/progenitor cells by PTEN/Akt/beta-catenin signaling [J] PLoS Biol. 2009;7(6):e1000121. doi: 10.1371/journal.pbio.1000121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou Y. Inhibition of glycolysis: a novel strategy to overcome drug resistance in CD133+ tumor initiating cells under hypoxic microenvironment [M] Proc Am Assoc Cancer Res. 2007 [Google Scholar]
- 74.Varum S, Rodrigues AS, Moura MB, et al. Energy metabolism in human pluripotent stem cells and their differentiated counterparts [J] PLoS One. 2011;6(6):e20914. doi: 10.1371/journal.pone.0020914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Takeuchi M, Kimura S, Kuroda J, et al. Glyoxalase-I is a novel target against Bcr-Abl+ leukemic cells acquiring stem-like characteristics in a hypoxic environment [J] Cell Death Differ. 2010;17(7):1211–1220. doi: 10.1038/cdd.2010.6. [DOI] [PubMed] [Google Scholar]
- 76.Singh KK, Russell J, Sigala B, et al. Mitochondrial DNA determines the cellular response to cancer therapeutic agents [J] Oncogene. 1999;18(48):6641–6646. doi: 10.1038/sj.onc.1203056. [DOI] [PubMed] [Google Scholar]
- 77.Cai J, Wallace DC, Zhivotovsky B, et al. Separation of cytochrome c-dependent Caspase activation from thiol-disulfide redox change in cells lacking mitochondrial DNA [J] Free Radic Biol Med. 2000;29(3–4):334–342. doi: 10.1016/s0891-5849(00)00312-9. [DOI] [PubMed] [Google Scholar]
- 78.Park SY, Chang I, Kim JY, et al. Resistance of mitochondrial DNA-depleted cells against cell death: role of mitochondrial superoxide dismutase [J] J Biol Chem. 2004;279(9):7512–7520. doi: 10.1074/jbc.M307677200. [DOI] [PubMed] [Google Scholar]
- 79.Hail N, Jr, Chen P, Kepa JJ. Selective apoptosis induction by the cancer chemopreventive agent N-(4-hydroxyphenyl) retinamide is achieved by modulating mitochondrial bioenergetics in premalignant and malignant human prostate epithelial cells [J] Apoptosis. 2009;14(7):849–863. doi: 10.1007/s10495-009-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hayashi J, Takemitsu M, Nonaka I. Recovery of the missing tumorigenicity in mitochondrial DNA-less HeLa cells by introduction of mitochondrial DNA from normal human cells [J] Somat Cell Mol Genet. 1992;18(2):123–129. doi: 10.1007/BF01233159. [DOI] [PubMed] [Google Scholar]
- 81.Kong Q, Beel JA, Lillehei KO. A threshold concept for cancer therapy [J] Med Hypotheses. 2000;55(1):29–35. doi: 10.1054/mehy.1999.0982. [DOI] [PubMed] [Google Scholar]
- 82.Zamzami N, Marchetti P, Castedo M, et al. Sequential reduction of mitochondrial transmembrane potential and generation of reactive oxygen species in early programmed cell death [J] J Exp Med. 1995;182(2):367–377. doi: 10.1084/jem.182.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Davis W, Jr, Ronai Z, Tew KD. Cellular thiols and reactive oxygen species in drug-induced apoptosis [J] J Pharmacol Exp Ther. 2001;296(1):1–6. [PubMed] [Google Scholar]
- 84.Benhar M, Dalyot I, Engelberg D, et al. Enhanced ROS production in oncogenically transformed cells potentiates c-Jun N-terminal kinase and p38 mitogen-activated protein kinase activation and sensitization to genotoxic stress [J] Mol Cell Biol. 2001;21(20):6913–6926. doi: 10.1128/MCB.21.20.6913-6926.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhou P, Kalakonda N, Comenzo RL. Changes in gene expression profiles of multiple myeloma cells induced by arsenic trioxide (ATO): possible mechanisms to explain ATO resistance in vivo [J] Br J Haematol. 2005;128(5):636–644. doi: 10.1111/j.1365-2141.2005.05369.x. [DOI] [PubMed] [Google Scholar]
- 86.Hour TC, Huang CY, Lin CC, et al. Characterization of molecular events in a series of bladder urothelial carcinoma cell lines with progressive resistance to arsenic trioxide [J] Anticancer Drugs. 2004;15(8):779–785. doi: 10.1097/00001813-200409000-00007. [DOI] [PubMed] [Google Scholar]
- 87.Hoshida Y, Moriyama M, Otsuka M, et al. Gene expressions associated with chemosensitivity in human hepatoma cells [J] Hepatogastroenterology. 2007;54(74):489–492. [PubMed] [Google Scholar]
- 88.Ramanathan B, Jan KY, Chen CH, et al. Resistance to paclitaxel is proportional to cellular total antioxidant capacity [J] Cancer Res. 2005;65(18):8455–8460. doi: 10.1158/0008-5472.CAN-05-1162. [DOI] [PubMed] [Google Scholar]
- 89.Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? [J] Nat Rev Drug Discov. 2009;8(7):579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- 90.Trachootham D, Zhou Y, Zhang H, et al. Selective killing of oncogenically transformed cells through a ROS-mediated mechanism by beta-phenylethyl isothiocyanate [J] Cancer Cell. 2006;10(3):241–252. doi: 10.1016/j.ccr.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 91.Trachootham D, Zhang H, Zhang W, et al. Effective elimination of fludarabine-resistant CLL cells by PEITC through a redox-mediated mechanism [J] Blood. 2008;112(5):1912–1922. doi: 10.1182/blood-2008-04-149815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xu RH, Pelicano H, Zhou Y, et al. Inhibition of glycolysis in cancer cells: a novel strategy to overcome drug resistance associated with mitochondrial respiratory defect and hypoxia [J] Cancer Res. 2005;65(2):613–621. [PubMed] [Google Scholar]
- 93.Weinhouse S. On respiratory impairment in cancer cells [J] Science. 1956;124(3215):267–269. doi: 10.1126/science.124.3215.267. [DOI] [PubMed] [Google Scholar]
- 94.Moreno-Sanchez R, Rodriguez-Enriquez S, Marin-Hernandez A, et al. Energy metabolism in tumor cells [J] FEBS J. 2007;274(6):1393–1418. doi: 10.1111/j.1742-4658.2007.05686.x. [DOI] [PubMed] [Google Scholar]
- 95.Compton S, Kim C, Griner NB, et al. Mitochondrial dysfunction impairs tumor suppressor p53 expression/function [J] J Biol Chem. 2010;286(23):20297–20312. doi: 10.1074/jbc.M110.163063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mitchell P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism [J] Nature. 1961;191:144–148. doi: 10.1038/191144a0. [DOI] [PubMed] [Google Scholar]
- 97.Anderson S, Bankier AT, Barrell BG, et al. Sequence and organization of the human mitochondrial genome [J] Nature. 1981;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 98.Attardi G. Animal mitochondrial DNA: an extreme example of genetic economy [J] Int Rev Cytol. 1985;93:93–145. doi: 10.1016/s0074-7696(08)61373-x. [DOI] [PubMed] [Google Scholar]
- 99.Pelicano H, Carney D, Huang P. ROS stress in cancer cells and therapeutic implications [J] Drug Resist Updat. 2004;7(2):97–110. doi: 10.1016/j.drup.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 100.Carew JS, Huang P. Mitochondrial defects in cancer [J] Mol Cancer. 2002;1:9. doi: 10.1186/1476-4598-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Attardi G, Yoneda M, Chomyn A. Complementation and segregation behavior of disease-causing mitochondrial DNA mutations in cellular model systems [J] Biochim Biophys Acta. 1995;1271(1):241–248. doi: 10.1016/0925-4439(95)00034-2. [DOI] [PubMed] [Google Scholar]
- 102.Hayashi J, Ohta S, Kikuchi A, et al. Introduction of disease-related mitochondrial DNA deletions into HeLa cells lacking mitochondrial DNA results in mitochondrial dysfunction [J] Proc Natl Acad Sci USA. 1991;88(23):10614–10618. doi: 10.1073/pnas.88.23.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fliss MS, Usadel H, Caballero OL, et al. Facile detection of mitochondrial DNA mutations in tumors and bodily fluids [J] Science. 2000;287(5460):2017–2019. doi: 10.1126/science.287.5460.2017. [DOI] [PubMed] [Google Scholar]
- 104.Zhou S, Kassauei K, Cutler DJ, et al. An oligonucleotide microarray for high-throughput sequencing of the mitochondrial genome [J] J Mol Diagn. 2006;8(4):476–482. doi: 10.2353/jmoldx.2006.060008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tan DJ, Bai RK, Wong LJ. Comprehensive scanning of somatic mitochondrial DNA mutations in breast cancer [J] Cancer Res. 2002;62(4):972–976. [PubMed] [Google Scholar]
- 106.Parrella P, Xiao Y, Fliss M, et al. Detection of mitochondrial DNA mutations in primary breast cancer and fine-needle aspirates [J] Cancer Res. 2001;61(20):7623–7626. [PubMed] [Google Scholar]
- 107.Polyak K, Li Y, Zhu H, et al. Somatic mutations of the mitochondrial genome in human colorectal tumours [J] Nat Genet. 1998;20(3):291–293. doi: 10.1038/3108. [DOI] [PubMed] [Google Scholar]
- 108.Alonso A, Martin P, Albarran C, et al. Detection of somatic mutations in the mitochondrial DNA control region of colorectal and gastric tumors by heteroduplex and single-strand conformation analysis [J] Electrophoresis. 1997;18(5):682–685. doi: 10.1002/elps.1150180504. [DOI] [PubMed] [Google Scholar]
- 109.Taylor RW, Barron MJ, Borthwick GM, et al. Mitochondrial DNA mutations in human colonic crypt stem cells [J] J Clin Invest. 2003;112(9):1351–1360. doi: 10.1172/JCI19435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wu CW, Yin PH, Hung WY, et al. Mitochondrial DNA mutations and mitochondrial DNA depletion in gastric cancer [J] Genes Chromosomes Cancer. 2005;44(1):19–28. doi: 10.1002/gcc.20213. [DOI] [PubMed] [Google Scholar]
- 111.Liu VW, Shi HH, Cheung AN, et al. High incidence of somatic mitochondrial DNA mutations in human ovarian carcinomas [J] Cancer Res. 2001;61(16):5998–6001. [PubMed] [Google Scholar]
- 112.Nishikawa M, Nishiguchi S, Shiomi S, et al. Somatic mutation of mitochondrial DNA in cancerous and noncancerous liver tissue in individuals with hepatocellular carcinoma [J] Cancer Res. 2001;61(5):1843–1845. [PubMed] [Google Scholar]
- 113.Wheelhouse NM, Lai PB, Wigmore SJ, et al. Mitochondrial D-loop mutations and deletion profiles of cancerous and noncancerous liver tissue in hepatitis B virus-infected liver [J] Br J Cancer. 2005;92(7):1268–1272. doi: 10.1038/sj.bjc.6602496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jones JB, Song JJ, Hempen PM, et al. Detection of mitochondrial DNA mutations in pancreatic cancer offers a “mass”-ive advantage over detection of nuclear DNA mutations [J] Cancer Res. 2001;61(4):1299–1304. [PubMed] [Google Scholar]
- 115.Jessie BC, Sun CQ, Irons HR, et al. Accumulation of mitochondrial DNA deletions in the malignant prostate of patients of different ages [J] Exp Gerontol. 2001;37(1):169–174. doi: 10.1016/s0531-5565(01)00153-x. [DOI] [PubMed] [Google Scholar]
- 116.Maximo V, Soares P, Lima J, et al. Mitochondrial DNA somatic mutations (point mutations and large deletions) and mitochondrial DNA variants in human thyroid pathology: a study with emphasis on Hurthle cell tumors [J] Am J Pathol. 2002;160(5):1857–1865. doi: 10.1016/S0002-9440(10)61132-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Montanini L, Regna-Gladin C, Eoli M, et al. Instability of mitochondrial DNA and MRI and clinical correlations in malignant gliomas [J] J Neurooncol. 2005;74(1):87–89. doi: 10.1007/s11060-004-4036-5. [DOI] [PubMed] [Google Scholar]
- 118.Sanchez-Cespedes M, Parrella P, Nomoto S, et al. Identification of a mononucleotide repeat as a major target for mitochondrial DNA alterations in human tumors [J] Cancer Res. 2001;61(19):7015–7019. [PubMed] [Google Scholar]
- 119.Testa JR, Tsichlis PN. AKT signaling in normal and malignant cells [J] Oncogene. 2005;24(50):7391–7393. doi: 10.1038/sj.onc.1209100. [DOI] [PubMed] [Google Scholar]
- 120.Pastorino JG, Hoek JB, Shulga N. Activation of glycogen synthase kinase 3beta disrupts the binding of hexokinase II to mitochondria by phosphorylate voltage-dependent anion channel and potentiates chemotherapy-induced cytotoxicity [J] Cancer Res. 2005;65(22):10545–10554. doi: 10.1158/0008-5472.CAN-05-1925. [DOI] [PubMed] [Google Scholar]
- 121.Lum JJ, Bui T, Gruber M, et al. The transcription factor HIF-1alpha plays a critical role in the growth factor-dependent regulation of both aerobic and anaerobic glycolysis [J] Genes Dev. 2007;21(9):1037–1049. doi: 10.1101/gad.1529107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kim DH, Sarbassov DD, Ali SM, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery [J] Cell. 2002;110(2):163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 123.Crino PB, Nathanson KL, Henske EP. The tuberous sclerosis complex [J] N Engl J Med. 2006;355(13):1345–1356. doi: 10.1056/NEJMra055323. [DOI] [PubMed] [Google Scholar]
- 124.Inoki K, Li Y, Zhu T, et al. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling [J] Nat Cell Biol. 2002;4(9):648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 125.Duvel K, Yecies JL, Menon S, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1 [J] Mol Cell. 2010;39(2):171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Green AS, Chapuis N, Lacombe C, et al. LKB1/AMPK/mTOR signaling pathway in hematological malignancies: from metabolism to cancer cell biology [J] Cell Cycle. 2011;10(13) doi: 10.4161/cc.10.13.16244. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 127.Carling D, Sanders MJ, Woods A. The regulation of AMP-activated protein kinase by upstream kinases [J] Int J Obes. 2008;32(Suppl 4):S55–S59. doi: 10.1038/ijo.2008.124. [DOI] [PubMed] [Google Scholar]
- 128.Luo Z, Zang M, Guo W. AMPK as a metabolic tumor suppressor: control of metabolism and cell growth [J] Future Oncol. 2010;6(3):457–470. doi: 10.2217/fon.09.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression [J] Nat Rev Cancer. 2009;9(8):563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Scholl C, Gilliland DG, Frohling S. Deregulation of signaling pathways in acute myeloid leukemia [J] Semin Oncol. 2008;35(4):336–345. doi: 10.1053/j.seminoncol.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 131.Patiar S, Harris AL. Role of hypoxia-inducible factor-1alpha as a cancer therapy target [J] Endocr Relat Cancer. 2006;13(Suppl 1):S61–S75. doi: 10.1677/erc.1.01290. [DOI] [PubMed] [Google Scholar]
- 132.Brunelle JK, Bell EL, Quesada NM, et al. Oxygen sensing requires mitochondrial ROS but not oxidative phosphorylation [J] Cell Metab. 2005;1(6):409–414. doi: 10.1016/j.cmet.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 133.Mansfield KD, Guzy RD, Pan Y, et al. Mitochondrial dysfunction resulting from loss of cytochrome c impairs cellular oxygen sensing and hypoxic HIF-alpha activation [J] Cell Metab. 2005;1(6):393–399. doi: 10.1016/j.cmet.2005.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Semenza GL. Targeting HIF-1 for cancer therapy [J] Nat Rev Cancer. 2003;3(10):721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 135.Macheda ML, Rogers S, Best JD. Molecular and cellular regulation of glucose transporter (GLUT) proteins in cancer [J] J Cell Physiol. 2005;202(3):654–662. doi: 10.1002/jcp.20166. [DOI] [PubMed] [Google Scholar]
- 136.Pouyssegur J, Dayan F, Mazure NM. Hypoxia signalling in cancer and approaches to enforce tumour regression [J] Nature. 2006;441(7092):437–443. doi: 10.1038/nature04871. [DOI] [PubMed] [Google Scholar]
- 137.Semenza GL. Hypoxia-inducible factor 1 (HIF-1) pathway [J] Sci STKE. 2007;2007(407):cm8. doi: 10.1126/stke.4072007cm8. [DOI] [PubMed] [Google Scholar]
- 138.Papandreou I, Cairns RA, Fontana L, et al. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption [J] Cell Metab. 2006;3(3):187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 139.Gottlieb E, Tomlinson IP. Mitochondrial tumour suppressors: a genetic and biochemical update [J] Nat Rev Cancer. 2005;5(11):857–866. doi: 10.1038/nrc1737. [DOI] [PubMed] [Google Scholar]
- 140.Wokolorczyk D, Gliniewicz B, Sikorski A, et al. A range of cancers is associated with the rs6983267 marker on chromosome 8 [J] Cancer Res. 2008;68(23):9982–9986. doi: 10.1158/0008-5472.CAN-08-1838. [DOI] [PubMed] [Google Scholar]
- 141.Aylon Y, Oren M. Living with p53, dying of p53 [J] Cell. 2007;130(4):597–600. doi: 10.1016/j.cell.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 142.Chesney J, Mitchell R, Benigni F, et al. An inducible gene product for 6-phosphofructo-2-kinase with an AU-rich instability element: role in tumor cell glycolysis and the Warburg effect [J] Proc Natl Acad Sci USA. 1999;96(6):3047–3052. doi: 10.1073/pnas.96.6.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Feng Z, Levine AJ. The regulation of energy metabolism and the IGF-1/mTOR pathways by the p53 protein [J] Trends Cell Biol. 2010;20(7):427–434. doi: 10.1016/j.tcb.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bos JL. ras oncogenes in human cancer: a review [J] Cancer Res. 1989;49(17):4682–4689. [PubMed] [Google Scholar]
- 145.Friday BB, Adjei AA. K-ras as a target for cancer therapy [J] Biochim Biophys Acta. 2005;1756(2):127–144. doi: 10.1016/j.bbcan.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 146.Chiaradonna F, Gaglio D, Vanoni M, et al. Expression of transforming K-Ras Oncogene affects mitochondrial function and morphology in mouse fibroblasts [J] Biochim Biophys Acta. 2006;1757(9–10):1338–1356. doi: 10.1016/j.bbabio.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 147.Baracca A, Chiaradonna F, Sgarbi G, et al. Mitochondrial Complex I decrease is responsible for bioenergetic dysfunction in K-ras transformed cells [J] Biochim Biophys Acta. 2010;1797(2):314–323. doi: 10.1016/j.bbabio.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 148.Yang D, Wang MT, Tang Y, et al. Impairment of mitochondrial respiration in mouse fibroblasts by oncogenic H-RAS (Q61L) [J] Cancer Biol Ther. 2010;9(2):122–133. doi: 10.4161/cbt.9.2.10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Chun SY, Johnson C, Washburn JG, et al. Oncogenic KRAS modulates mitochondrial metabolism in human colon cancer cells by inducing HIF-1alpha and HIF-2alpha target genes [J] Mol Cancer. 2010;9:293. doi: 10.1186/1476-4598-9-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rose IA, Warms JV. Mitochondrial hexokinase. Release, rebinding, and location [J] J Biol Chem. 1967;242(7):1635–1645. [PubMed] [Google Scholar]
- 151.Mathupala SP, Rempel A, Pedersen PL. Glucose catabolism in cancer cells: identification and characterization of a marked activation response of the type II hexokinase gene to hypoxic conditions [J] J Biol Chem. 2001;276(46):43407–43412. doi: 10.1074/jbc.M108181200. [DOI] [PubMed] [Google Scholar]
- 152.Maher JC, Wangpaichitr M, Savaraj N, et al. Hypoxia-inducible factor-1 confers resistance to the glycolytic inhibitor 2-deoxy-D-glucose [J] Mol Cancer Ther. 2007;6(2):732–741. doi: 10.1158/1535-7163.MCT-06-0407. [DOI] [PubMed] [Google Scholar]
- 153.Simons AL, Ahmad IM, Mattson DM, et al. 2-Deoxy-D-glucose combined with cisplatin enhances cytotoxicity via metabolic oxidative stress in human head and neck cancer cells [J] Cancer Res. 2007;67(7):3364–3370. doi: 10.1158/0008-5472.CAN-06-3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Singh D, Banerji AK, Dwarakanath BS, et al. Optimizing cancer radiotherapy with 2-deoxy-d-glucose dose escalation studies in patients with glioblastoma multiforme [J] Strahlenther Onkol. 2005;181(8):507–514. doi: 10.1007/s00066-005-1320-z. [DOI] [PubMed] [Google Scholar]
- 155.Liu H, Hu YP, Savaraj N, et al. Hypersensitization of tumor cells to glycolytic inhibitors [J] Biochemistry. 2001;40(18):5542–5547. doi: 10.1021/bi002426w. [DOI] [PubMed] [Google Scholar]
- 156.Liu H, Savaraj N, Priebe W, et al. Hypoxia increases tumor cell sensitivity to glycolytic inhibitors: a strategy for solid tumor therapy (Model C) [J] Biochem Pharmacol. 2002;64(12):1745–1751. doi: 10.1016/s0006-2952(02)01456-9. [DOI] [PubMed] [Google Scholar]
- 157.Maher JC, Krishan A, Lampidis TJ. Greater cell cycle inhibition and cytotoxicity induced by 2-deoxy-D-glucose in tumor cells treated under hypoxic vs aerobic conditions [J] Cancer Chemother Pharmacol. 2004;53(2):116–122. doi: 10.1007/s00280-003-0724-7. [DOI] [PubMed] [Google Scholar]
- 158.Maschek G, Savaraj N, Priebe W, et al. 2-deoxy-D-glucose increases the efficacy of adriamycin and paclitaxel in human osteosarcoma and non-small cell lung cancers in vivo [J] Cancer Res. 2004;64(1):31–34. doi: 10.1158/0008-5472.can-03-3294. [DOI] [PubMed] [Google Scholar]
- 159.Chen Z, Zhang H, Lu W, et al. Role of mitochondria-associated hexokinase II in cancer cell death induced by 3-bromopyruvate [J] Biochim Biophys Acta. 2009;1787(5):553–560. doi: 10.1016/j.bbabio.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Chiara F, Castellaro D, Marin O, et al. Hexokinase II detachment from mitochondria triggers apoptosis through the permeability transition pore independent of voltage-dependent anion channels [J] PLoS One. 2008;3(3):e1852. doi: 10.1371/journal.pone.0001852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Goldin N, Arzoine L, Heyfets A, et al. Methyl jasmonate binds to and detaches mitochondria-bound hexokinase [J] Oncogene. 2008;27(34):4636–4643. doi: 10.1038/onc.2008.108. [DOI] [PubMed] [Google Scholar]
- 162.Elia U, Flescher E. PI3K/Akt pathway activation attenuates the cytotoxic effect of methyl jasmonate toward sarcoma cells [J] Neoplasia. 2008;10(11):1303–1313. doi: 10.1593/neo.08636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Floridi A, Paggi MG, Marcante ML, et al. Lonidamine, a selective inhibitor of aerobic glycolysis of murine tumor cells [J] J Natl Cancer Inst. 1981;66(3):497–499. [PubMed] [Google Scholar]
- 164.Floridi A, Bruno T, Miccadei S, et al. Enhancement of doxorubicin content by the antitumor drug lonidamine in resistant Ehrlich ascites tumor cells through modulation of energy metabolism [J] Biochem Pharmacol. 1998;56(7):841–849. doi: 10.1016/s0006-2952(98)00054-9. [DOI] [PubMed] [Google Scholar]
- 165.Floridi A, Paggi MG, D'Atri S, et al. Effect of lonidamine on the energy metabolism of Ehrlich ascites tumor cells [J] Cancer Res. 1981;41(11 Pt 1):4661–4666. [PubMed] [Google Scholar]
- 166.Berruti A, Bitossi R, Gorzegno G, et al. Time to progression in metastatic breast cancer patients treated with epirubicin is not improved by the addition of either cisplatin or lonidamine: final results of a phase III study with a factorial design [J] J Clin Oncol. 2002;20(20):4150–4159. doi: 10.1200/JCO.2002.08.012. [DOI] [PubMed] [Google Scholar]
- 167.De Lena M, Lorusso V, Latorre A, et al. Paclitaxel, cisplatin and lonidamine in advanced ovarian cancer. A phase II study [J] Eur J Cancer. 2001;37(3):364–368. doi: 10.1016/s0959-8049(00)00400-7. [DOI] [PubMed] [Google Scholar]
- 168.Oudard S, Carpentier A, Banu E, et al. Phase II study of lonidamine and diazepam in the treatment of recurrent glioblastoma multiforme [J] J Neurooncol. 2003;63(1):81–86. doi: 10.1023/a:1023756707900. [DOI] [PubMed] [Google Scholar]
- 169.De Marinis F, Rinaldi M, Ardizzoni A, et al. The role of vindesine and lonidamine in the treatment of elderly patients with advanced non-small cell lung cancer: a phase III randomized FONICAP trial [J] Italian Lung Cancer Task Force Tumori. 1999;85(3):177–182. doi: 10.1177/030089169908500306. [DOI] [PubMed] [Google Scholar]
- 170.Wang F, Ogasawara MA, Huang P. Small mitochondria-targeting molecules as anti-cancer agents [J] Mol Aspects Med. 2010;31(1):75–92. doi: 10.1016/j.mam.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Goldberg EB, Colowick SP. The role of glycolysis in the growth of tumor cells. 3. Lactic dehydrogenase as the site of action of oxamate on the growth of cultured cells [J] J Biol Chem. 1965;240:2786–2790. [PubMed] [Google Scholar]
- 172.Hamilton E, Fennell M, Stafford DM. Modification of tumour glucose metabolism for therapeutic benefit [J] Acta Oncol. 1995;34(3):429–433. doi: 10.3109/02841869509094003. [DOI] [PubMed] [Google Scholar]
- 173.Pedersen PL. The cancer cell's “power plants” as promising therapeutic targets: an overview [J] J Bioenerg Biomembr. 2007;39(1):1–12. doi: 10.1007/s10863-007-9070-5. [DOI] [PubMed] [Google Scholar]
- 174.Mathupala SP, Colen CB, Parajuli P, et al. Lactate and malignant tumors: a therapeutic target at the end stage of glycolysis [J] J Bioenerg Biomembr. 2007;39(1):73–77. doi: 10.1007/s10863-006-9062-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Ohta S. Contribution of somatic mutations in the mitochondrial genome to the development of cancer and tolerance against anticancer drugs [J] Oncogene. 2006;25(34):4768–4776. doi: 10.1038/sj.onc.1209602. [DOI] [PubMed] [Google Scholar]
- 176.Bonnet S, Archer SL, Allalunis-Turner J, et al. A mitochondria-K+ channel axis is suppressed in cancer and its normalization promotes apoptosis and inhibits cancer growth [J] Cancer Cell. 2007;11(1):37–51. doi: 10.1016/j.ccr.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 177.Cao X, Fang L, Gibbs S, et al. Glucose uptake inhibitor sensitizes cancer cells to daunorubicin and overcomes drug resistance in hypoxia [J] Cancer Chemother Pharmacol. 2007;59(4):495–505. doi: 10.1007/s00280-006-0291-9. [DOI] [PubMed] [Google Scholar]
- 178.Chesney J. 6-phosphofructo-2-kinase/fructose-2, 6-bisphosphatase and tumor cell glycolysis [J] Curr Opin Clin Nutr Metab Care. 2006;9(5):535–539. doi: 10.1097/01.mco.0000241661.15514.fb. [DOI] [PubMed] [Google Scholar]
- 179.Clem B, Telang S, Clem A, et al. Small-molecule inhibition of 6-phosphofructo-2-kinase activity suppresses glycolytic flux and tumor growth [J] Mol Cancer Ther. 2008;7(1):110–120. doi: 10.1158/1535-7163.MCT-07-0482. [DOI] [PubMed] [Google Scholar]
- 180.Eagle H. Nutrition needs of mammalian cells in tissue culture [J] Science. 1955;122(3168):501–514. doi: 10.1126/science.122.3168.501. [DOI] [PubMed] [Google Scholar]
- 181.Wu MC, Arimura GK, Yunis AA. Mechanism of sensitivity of cultured pancreatic carcinoma to asparaginase [J] Int J Cancer. 1978;22(6):728–733. doi: 10.1002/ijc.2910220615. [DOI] [PubMed] [Google Scholar]
- 182.Ahluwalia GS, Grem JL, Hao Z, et al. Metabolism and action of amino acid analog anti-cancer agents [J] Pharmacol Ther. 1990;46(2):243–271. doi: 10.1016/0163-7258(90)90094-i. [DOI] [PubMed] [Google Scholar]
- 183.Ovejera AA, Houchens DP, Catane R, et al. Efficacy of 6-diazo-5-oxo-L-norleucine and N-[N-gamma-glutamyl-6-diazo-5-oxo-norleucinyl]-6-diazo-5-oxo-norleucine against experimental tumors in conventional and nude mice [J] Cancer Res. 1979;39(8):3220–3224. [PubMed] [Google Scholar]
- 184.Griffiths M, Keast D, Patrick G, et al. The role of glutamine and glucose analogues in metabolic inhibition of human myeloid leukaemia in vitro [J] Int J Biochem. 1993;25(12):1749–1755. doi: 10.1016/0020-711x(88)90303-5. [DOI] [PubMed] [Google Scholar]
- 185.Narta UK, Kanwar SS, Azmi W. Pharmacological and clinical evaluation of L-asparaginase in the treatment of leukemia [J] Crit Rev Oncol Hematol. 2007;61(3):208–221. doi: 10.1016/j.critrevonc.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 186.Avramis VI, Panosyan EH. Pharmacokinetic/pharmacodynamic relationships of asparaginase formulations: the past, the present and recommendations for the future [J] Clin Pharmacokinet. 2005;44(4):367–393. doi: 10.2165/00003088-200544040-00003. [DOI] [PubMed] [Google Scholar]
- 187.Enns GM, Berry SA, Berry GT, et al. Survival after treatment with phenylacetate and benzoate for urea-cycle disorders [J] N Engl J Med. 2007;356(22):2282–2292. doi: 10.1056/NEJMoa066596. [DOI] [PubMed] [Google Scholar]
- 188.Thibault A, Samid D, Cooper MR, et al. Phase I study of phenylacetate administered twice daily to patients with cancer [J] Cancer. 1995;75(12):2932–2938. doi: 10.1002/1097-0142(19950615)75:12<2932::aid-cncr2820751221>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 189.Mitchell RB, Wagner JE, Karp JE, et al. Syndrome of idiopathic hyperammonemia after high-dose chemotherapy: review of nine cases [J] Am J Med. 1988;85(5):662–667. doi: 10.1016/s0002-9343(88)80239-0. [DOI] [PubMed] [Google Scholar]
- 190.Fuchs BC, Bode BP. Amino acid transporters ASCT2 and LAT1 in cancer: partners in crime? [J] Semin Cancer Biol. 2005;15(4):254–266. doi: 10.1016/j.semcancer.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 191.Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer [J] Trends Biochem Sci. 2010;35(8):427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Esslinger CS, Cybulski KA, Rhoderick JF. Ngamma-aryl glutamine analogues as probes of the ASCT2 neutral amino acid transporter binding site [J] Bioorg Med Chem. 2005;13(4):1111–1118. doi: 10.1016/j.bmc.2004.11.028. [DOI] [PubMed] [Google Scholar]
- 193.Erickson JW, Cerione RA. Glutaminase: a hot spot for regulation of cancer cell metabolism? [J] Oncotarget. 2010;1(8):734–740. doi: 10.18632/oncotarget.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Moreadith RW, Lehninger AL. The pathways of glutamate and glutamine oxidation by tumor cell mitochondria. Role of mitochondrial NAD(P)+-dependent malic enzyme [J] J Biol Chem. 1984;259(10):6215–621. [PubMed] [Google Scholar]
- 195.Thornburg JM, Nelson KK, Clem BF, et al. Targeting aspartate aminotransferase in breast cancer [J] Breast Cancer Res. 2008;10(5):R84. doi: 10.1186/bcr2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Qin JZ, Xin H, Nickoloff BJ. Targeting glutamine metabolism sensitizes melanoma cells to TRAIL-induced death [J] Biochem Biophys Res Commun. 2010;398(1):146–152. doi: 10.1016/j.bbrc.2010.06.057. [DOI] [PubMed] [Google Scholar]
- 197.Luo Z, Zang M, Guo W. AMPK as a metabolic tumor suppressor: control of metabolism and cell growth [J] Future Oncol. 2010;6(3):457–470. doi: 10.2217/fon.09.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Hadad SM, Baker L, Quinlan PR, et al. Histological evaluation of AMPK signalling in primary breast cancer [J] BMC Cancer. 2009;9:307. doi: 10.1186/1471-2407-9-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Jiralerspong S, Palla SL, Giordano SH, et al. Metformin and pathologic complete responses to neoadjuvant chemotherapy in diabetic patients with breast cancer [J] J Clin Oncol. 2009;27(20):3297–3302. doi: 10.1200/JCO.2009.19.6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Sarraf P, Mueller E, Jones D, et al. Differentiation and reversal of malignant changes in colon cancer through PPARgamma [J] Nat Med. 1998;4(9):1045–1052. doi: 10.1038/2030. [DOI] [PubMed] [Google Scholar]
- 201.Lehrke M, Lazar MA. The many faces of PPARgamma [J] Cell. 2005;123(6):993–999. doi: 10.1016/j.cell.2005.11.026. [DOI] [PubMed] [Google Scholar]
- 202.Buzzai M, Bauer DE, Jones RG, et al. The glucose dependence of Akt-transformed cells can be reversed by pharmacologic activation of fatty acid beta-oxidation [J] Oncogene. 2005;24(26):4165–4173. doi: 10.1038/sj.onc.1208622. [DOI] [PubMed] [Google Scholar]
- 203.Motoshima H, Goldstein BJ, Igata M, et al. AMPK and cell proliferation—AMPK as a therapeutic target for atherosclerosis and cancer [J] J Physiol. 2006;574(Pt 1):63–71. doi: 10.1113/jphysiol.2006.108324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Bruserud O, Stapnes C, Tronstad KJ, et al. Protein lysine acetylation in normal and leukaemic haematopoiesis: HDACs as possible therapeutic targets in adult AML [J] Expert Opin Ther Targets. 2006;10(1):51–68. doi: 10.1517/14728222.10.1.51. [DOI] [PubMed] [Google Scholar]
- 205.Carew JS, Nawrocki ST, Kahue CN, et al. Targeting autophagy augments the anticancer activity of the histone deacetylase inhibitor SAHA to overcome Bcr-Abl-mediated drug resistance [J] Blood. 2007;110(1):313–322. doi: 10.1182/blood-2006-10-050260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Stiehl DP, Fath DM, Liang D, et al. Histone deacetylase inhibitors synergize p300 autoacetylation that regulates its transactivation activity and complex formation [J] Cancer Res. 2007;67(5):2256–2264. doi: 10.1158/0008-5472.CAN-06-3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Nelson EC, Evans CP, Mack PC, et al. Inhibition of Akt pathways in the treatment of prostate cancer [J] Prostate Cancer Prostatic Dis. 2007;10(4):331–339. doi: 10.1038/sj.pcan.4500974. [DOI] [PubMed] [Google Scholar]
- 208.Shin I, Edl J, Biswas S, et al. Proapoptotic activity of cell-permeable anti-Akt single-chain antibodies [J] Cancer Res. 2005;65(7):2815–2824. doi: 10.1158/0008-5472.CAN-04-2898. [DOI] [PubMed] [Google Scholar]
- 209.Kong D, Yamori T. Phosphatidylinositol 3–kinase inhibitors: promising drug candidates for cancer therapy [J] Cancer Sci. 2008;99(9):1734–1740. doi: 10.1111/j.1349-7006.2008.00891.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Wu M, Neilson A, Swift AL, et al. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells [J] Am J Physiol Cell Physiol. 2007;292(1):C125–C136. doi: 10.1152/ajpcell.00247.2006. [DOI] [PubMed] [Google Scholar]
- 211.Guertin DA, Sabatini DM. The pharmacology of mTOR inhibition [J] Sci Signal. 2009;2(67):pe24. doi: 10.1126/scisignal.267pe24. [DOI] [PubMed] [Google Scholar]
- 212.Dancey JE. Inhibitors of the mammalian target of rapamycin [J] Expert Opin Investig Drugs. 2005;14(3):313–328. doi: 10.1517/13543784.14.3.313. [DOI] [PubMed] [Google Scholar]
- 213.Shor B, Zhang WG, Toral-Barza L, et al. A new pharmacologic action of CCI-779 involves FKBP12-independent inhibition of mTOR kinase activity and profound repression of global protein synthesis [J] Cancer Res. 2008;68(8):2934–2943. doi: 10.1158/0008-5472.CAN-07-6487. [DOI] [PubMed] [Google Scholar]
- 214.Shim H, Dolde C, Lewis BC, et al. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth [J] Proc Natl Acad Sci USA. 1997;94(13):6658–6663. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Kim JW, Gao P, Liu YC, et al. Hypoxia-inducible factor 1 and dysregulated c-Myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1 [J] Mol Cell Biol. 2007;27(21):7381–7393. doi: 10.1128/MCB.00440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Osthus RC, Shim H, Kim S, et al. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc [J] J Biol Chem. 2000;275(29):21797–21800. doi: 10.1074/jbc.C000023200. [DOI] [PubMed] [Google Scholar]
- 217.Scatena R, Bottoni P, Pontoglio A, et al. Glycolytic enzyme inhibitors in cancer treatment [J] Expert Opin Investig Drugs. 2008;17(10):1533–1545. doi: 10.1517/13543784.17.10.1533. [DOI] [PubMed] [Google Scholar]
- 218.Pelicano H, Martin DS, Xu RH, et al. Glycolysis inhibition for anticancer treatment [J] Oncogene. 2006;25(34):4633–4646. doi: 10.1038/sj.onc.1209597. [DOI] [PubMed] [Google Scholar]
- 219.Zancan P, Rosas AO, Marcondes MC, et al. Clotrimazole inhibits and modulates heterologous association of the key glycolytic enzyme 6-phosphofructo-1–kinase [J] Biochem Pharmacol. 2007;73(10):1520–1527. doi: 10.1016/j.bcp.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 220.Hagland H, Nikolaisen J, Hodneland LI, et al. Targeting mitochondria in the treatment of human cancer: a coordinated attack against cancer cell energy metabolism and signalling [J] Expert Opin Ther Targets. 2007;11(8):1055–1069. doi: 10.1517/14728222.11.8.1055. [DOI] [PubMed] [Google Scholar]