Abstract
Pancreatic adenocarcinoma and hepatocellular carcinoma are devastating human malignancies that are characterized by poor prognosis, late onset, and a lack of known biomarkers. New diagnostic and therapeutic molecular targets are desperately needed to develop novel and effective treatment strategies. MicroRNAs (miRNAs) are an emerging class of molecules with roles in various cellular processes, including growth, survival, and apoptosis. Most importantly, aberrant expression of miRNAs has been implicated in cancer pathogenesis. miRNA expression profiles of pancreatic adenocarcinoma and hepatocellular carcinoma indicate selective overexpression of oncogenic miRNAs and down-regulation of tumor suppressive miRNAs in these cancers. This review summarizes results from key studies conducted to characterize the miRNA expression profiles of pancreatic adenocarcinoma and hepatocellular carcinoma and describes the potential mechanisms by which some oncogenic or tumor suppressive miRNAs act. Furthermore, this review outlines novel therapeutic strategies for targeting miRNAs.
Keywords: MicroRNA, pancreatic adenocarcinoma, hepatocellular carcinoma
MicroRNAs (miRNAs) are an abundant class of endogenous 20 to 22 nucleotide (nt) non-coding RNA molecules that post-transcriptionally modulate target gene expression[1]. Although miRNAs constitute only 1% to 3% of the human genome, they are estimated to control approximately 30% of all coding genes in the human genome[2]–[4]. At present, over 1000 miRNAs have been identified in humans according to miRBase[5]. It is well documented that miRNAs generally possess multiple downstream target mRNAs, with some individual miRNAs controlling over 100 mRNAs[6]. Therefore, miRNAs exhibit widespread regulatory activity in virtually all important cellular processes, including development, survival, proliferation, and apoptosis, and have thus been implicated in diverse diseases such as cancer, immune disorders, and viral infections[1].
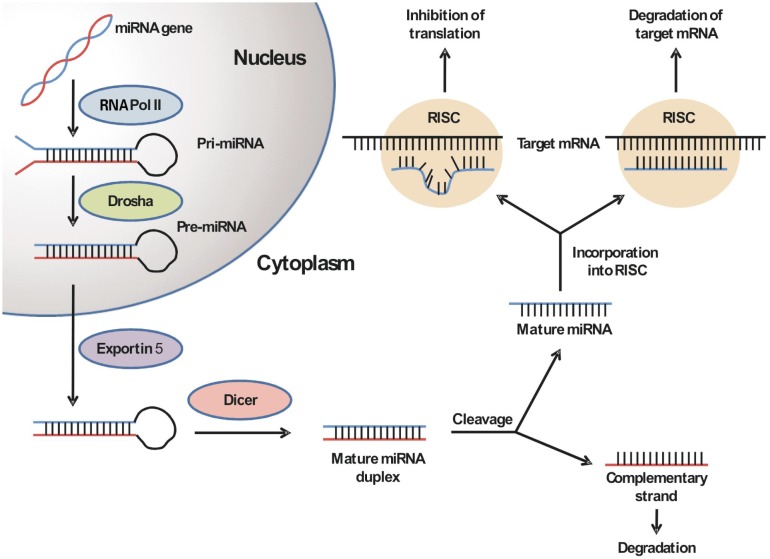
miRNA biogenesis involves a multistep process that is initiated by the nuclear processing of a long primary transcript (pri-miRNA) into 70 to 100 nt hairpin precursor miRNAs (pre-miRNAs) by the ribonuclease Drosha/DGCR8[7]. The pre-miRNA is then translocated to the cytoplasm by exportin-5 in a RanGTP-dependent fashion, where it is further cleaved by the ribonuclease Dicer into a mature miRNA duplex. Subsequent incorporation of the mature miRNA duplex into the RNA-induced silencing complex (RISC) leads to the degradation of the duplex into a single-stranded, mature miRNA. Within the RISC, the mature miRNA can bind to its target mRNAs by complementary base pairing at its 3′ untranslated region. The degree of complementarity between the miRNA and the target mRNA determines whether the miRNA will inhibit translation or, less frequently, induce degradation of the target mRNA (Figure 1)[8],[9].
Figure 1. The biogenesis of microRNAs (miRNAs).
The miRNA gene is initially transcribed into a primary miRNA (pri-miRNA) by RNA polymerase II in the nucleus. The pri-miRNA is then processed by the RNase III Drosha into the 70 to 100 nt hairpin precursor miRNA (pre-miRNA). Exportin-5 translocates the pre-miRNA into the cytoplasm, where it is again cleaved by the ribonuclease Dicer into a mature miRNA duplex. The duplex binds to the RNA-induced silencing complex (RISC), and the complementary miRNA strand is released and degraded. In the RISC, the mature single-stranded miRNA binds to its target mRNA and either induces its degradation or inhibits its translation, depending on the level of binding complementarity.
Emerging evidence suggests a key role for miRNAs in promoting Carcinogenesis. Genome-wide studies have shown that more than 50% of human miRNA genes locate at fragile genomic sites associated with cancer. In addition, abnormal miRNA expression levels have recently been reported in many human cancers, including pancreatic cancer, prostate cancer, thyroid cancer, melanoma, ovarian cancer, colon cancer, and breast cancer[10]–[17].
miRNAs that affect cancer development can act either as oncogenes or tumor suppressors. Oncogenic miRNAs are generally overexpressed in tumors, whereas tumor suppressor miRNAs are generally down-regulated[18]. The first identified miRNAs with tumor suppressor activity are miR-15 and miR-16, which are found to be deleted and/or down-regulated in approximately 70% of chronic lymphocytic leukemia cases [19],[20]. Other miRNAs with known tumor suppressor functions are miR-29 and lethal-7 (let-7) in lung cancer, miR-10b, miR-125b, and miR-145 in breast cancer, and miR-34 in ovarian and colon cancers [17],[21]–[24]. Conversely, miRNAs with tumor-promoting activities include miR-21 in breast cancer and glioblastoma, miR-155 in many types of cancers, and miR-373 and miR-520c as metastasis-promoting miRNAs in breast cancer[11],[16],[17],[25]–[27].
Pancreatic ductal adenocarcinoma (PDAC), the most common type of pancreatic cancer, is one of the most formidable human malignancies, with the annual mortality nearly equaling the annual incidence. As the fourth leading cause of cancer death in the USA, PDAC is characterized by extremely poor prognosis, late onset, a lack of effective diagnostic markers, and high invasiveness and metastatic potential. Less than 20% of PDAC patients present with resectable and potentially curable disease, and the 5-year survival rate is the lowest among all cancers at <5%[28]–[30].
Similarly, hepatocellular carcinoma (HCC), the most prevalent form of liver cancer, is one of the most common and lethal tumors, accounting for ∼600 000 deaths per year worldwide[31],[32]. Approximately half of these deaths are estimated to occur in China[33]. As in PDAC, the lethality of HCC stems from a lack of reliable diagnostic biomarkers, enhanced chemoresistance mechanisms, and underlying liver diseases including cirrhosis and hepatitis[34]. Although resection and transplantation are the best therapeutic options, only 5% to 15% of patients are eligible for surgical intervention[35].
Clearly, there is a pressing need to elucidate the mechanisms underlying PDAC and HCC pathogenesis and to develop novel and effective diagnostic and therapeutic techniques to improve clinical outcomes in patients with PDAC or HCC. Understanding the roles played by miRNAs in these cancers may lead to enhanced therapeutic strategies and improved patient outcomes.
The present review summarizes the current advances in miRNA research, with a specific focus on the roles of miRNAs in PDAC and HCC. Potential therapeutic implications of miRNAs in these cancers will also be discussed. Such information will be highly relevant to researchers and clinicians who wish to acquire an up-to-date understanding of this emerging field and elucidate or identify possible targets for investigation involving miRNAs as important underlying mediators of human cancers.
Roles of miRNAs in Pancreatic Adenocarcinoma
Current miRNA profiling studies in PDAC
Recent evidence has shown that aberrant miRNA expression contributes to Carcinogenesis in the pancreas, and that pancreatic cancer tissues exhibit unique miRNA expression profiles compared to normal pancreatic and chronic pancreatitis cells and tissues[11]. Among the most commonly dysregulated miRNAs in PDAC are miR-21, miR-155, miR-221, miR-216, and miR-217. In recent years, several high-throughput miRNA profiling studies have characterized a variety of miRNA expression patterns in PDAC using different techniques such as miRNA microarray and quantitative RT-PCR, and these studies have identified numerous specific miRNAs as being significantly up- or down-regulated in PDAC (Table 1).
Table 1. MicroRNA profiling studies in pancreatic adenocarcinoma.
| Reference | Study specifications | Up-regulated miRNAs | Down-regulated miRNAs |
| Lee 2007[11] | RT-PCR assay testing for 222 miRNAs in 28 PDAC tissues and 15 adjacent benign tissues, 4 CP tissues, 6 normal pancreas tissues, and 9 PDAC cell lines | miR-221, miR-424, miR-301, miR-100, miR-376a, miR-125b-1, miR-21, miR-16-1, miR-181a,c, miR-92-1, miR15b, miR-155, let-7f-1, miR-212, miR-107, miR-24-1,2, let-7d | miR-345, miR-142P, miR-139 |
| Zhang 2009[36] | RT-PCR assay testing for 95 miRNAs in 5 PDAC tissues and adjacent benign tissues, 3 PDAC cell lines, 1 CP tissue | miR-196a, miR-190, miR-186, miR-221, miR-222, miR-200b, miR-15b, miR-95 | None found |
| Kent 2009[37] | Microarray testing for 321 miRNAs in 21 PDAC cell lines and 2 non-transformed human pancreatic ductal epithelial cell lines | miR-200a, miR-29b, miR-182, miR-141, miR-222, miR-191, miR-15a, miR-361, miR-17-3p, miR-200b, miR-26b, miR-155, miR-16, miR-96, miR-30a-5p, miR-7, miR-429, miR-30c, miR-30b, miR-345, miR-31, miR-193b, miR-30d, miR-26a, miR-134, miR-151 | let-7c, let-7b, miR-99b, miR-204, miR-125a, miR-143, let-7e, miR-34a, miR-145, miR-199a, miR424, miR-199a* |
| Bloomston 2007[38] | Microarray testing for 326 human miRNAs in 65 PDAC tissues and adjacent benign tissues, 42 CP tissues and adjacent benign tissues | miR-221, miR-181a, miR-155, miR-210, miR-213, miR-181b, miR-222, miR-181b-2, miR-21, miR-181b-1, miR-181c, miR-220, miR-181d, miR-223, miR-100-1/2, miR-125a, miR-143, miR-10a, miR-146, miR-99, miR-100, miR-199a-1,miR-10b, miR-199a-2, miR-10b, miR-199a-2, miR-107, miR-103-2, miR-125b-1, miR-205, miR-23b, miR-23a | miR-148a, miR-148b, miR-375 |
| Szafranska 2007[39] | Microarray testing for 377 miRNAs in 33 PDAC tissues and adjacent benign tissues | miR-205, miR-143, miR-145, miR-146a, miR-196b, miR-93, miR-31, miR-210, miR-196a, miR-18a, miR-203, miR-150, miR-155, miR-221, miR-222, miR-223, miR-224 | miR-29c, miR-216, miR-217, miR-375, miR-148a, miR-96, miR-148b, miR-141, miR-130b |
Results of studies attempting to determine the miRNA expression signature of pancreatic adenocarcinoma. PDAC, pancreatic ductal adenocarcinoma; CP, chronic pancreatitis.
Lee et al.[11] used real-time PCR to define the miRNA signature of PDAC by comparing human PDAC specimens, paired benign tumor tissues, normal pancreas, chronic pancreatitis, and 9 PDAC cell lines. One hundred pre-miRNAs were found to be differentially expressed in PDAC, with the majority being overexpressed. The top twenty aberrantly expressed miRNAs, miR-221, miR-424, miR-301, miR-100, miR-376a, miR-125b-1, miR-21, miR-16-1, miR-181a,c, miR-92-1, miR-15b, miR-155, let-7f-1, miR-212, miR-107, miR-24-1,2, let-7d, miR-345, miR-142-P, and miR-139, were proposed to characterize PDAC[11]. Zhang et al. [36] performed a similar study evaluating the expression levels of 95 miRNAs in PDAC cell lines and tissue specimens by real-time RT-PCR. Eight miRNAs, miR-196a, miR-190, miR-186, miR-221, miR-222, miR-200b, miR-15b, and miR-95, were found to be significantly up-regulated in most of the PDAC tissues and cells lines, with the magnitudes of increase ranging from 3- to 2018-fold.
In a global miRNA analysis of 21 human pancreatic adenocarcinoma cell lines, the most extensive PDAC cell analysis to date, Kent et al.[37] found 39 miRNAs to be differentially expressed by at least a factor of two or more compared to control pancreatic ductal cell lines. Bloomston et al.[38] similarly assessed miRNA expression levels in 65 human PDAC tissue samples and 42 benign adjacent pancreatic tissue and chronic pancreatitis specimens by miRNA microarray. In comparison to benign pancreatic tissue, 21 miRNAs were found to be significantly up-regulated and 4 down-regulated in PDAC tissues. Similarly, 15 miRNAs were found to be overexpressed and 8 down-regulated in PDAC tissues in comparison to chronic pancreatitis. In another study comparing miRNA expression levels in normal pancreas, chronic pancreatitis, PDAC tissues, PDAC-derived cell lines, as well as a reference set of 33 human tissues, Szafranksa et al.[39] found 26 miRNAs to be prominently down-regulated in PDAC. Additionally, they identified 2 miRNAs, miR-216 and miR-217, as highly expressed in normal pancreas but only minimally expressed in PDAC tissues and cell lines. miR-196a and miR-196b were found to be absent in normal and chronic pancreatitis tissues but significantly up-regulated in PDAC tissues.
Oncogenic miRNAs in PDAC
miRNAs can act either as oncogenes or tumor suppressors, depending on their target mRNAs. Several oncogenic miRNAs that are significantly up-regulated in PDAC tumors and cell lines (Table 2) have been identified in the previously mentioned profiling studies, in addition to other functional characterization studies. Among the most commonly identified oncogenic miRNAs in PDAC is miR-155, which has been associated with decreased survival in PDAC patients[11],[38]–[40]. Gironella et al.[41] identified tumor protein 53-induced nuclear protein 1 (TP53INP1), a proapoptotic stress-induced p53 target gene, as a key target of miR-155. This suggests that miR-155 promotes pancreatic Carcinogenesis by down-regulating a key tumor suppressor, TP53INP1.
Table 2. Dysregulated microRNAs, their functions, and possible molecular targets in pancreatic adenocarcinoma.
| MicroRNA(s) | Role | Molecular target(s) | Biological functlon(s) | Reference(s) |
| miR-15a | Tumor suppressor | WNT3A, FGF7 | Increase proliferation and growth; Inhibit apoptosis | Zhang 2010[51] |
| miR-27a | Oncogene | Spry2 | Increase growth, migration, and colony formation in vitro | Ma 2010[49] |
| miR-34a | Tumor suppressor | p53 | Inhibit growth and proliferation | Kent 2009[37] |
| miR-96 | Tumor suppressor | K-Ras/Akt | Decrease Invasion, migration, and growth | Yu 2010[52] |
| miR-132 | Oncogene | PRB/E2F | Increase proliferation | Park 2011[48] |
| miR-155 | Oncogene | TP53INP1 | Decrease apoptosis | Gironella 2007[41] |
| miR-194, miR-200b, miR-220c, miR-429 | Oncogene | EP300 | Increase growth and metastasis | Mees 2010[45] |
| miR-212 | Oncogene | pRB/E2F | Increase proliferation | Park 2011[48] |
| miR-214 | Oncogene | ING4 | Reduce sensitivity to GEM-induced apoptosis; increase growth and angiogenesis | Zhang 2010[51] |
| miR-301a | Oncogene | NK-κB-repressing factor | Increase growth | Lu 2011[50] |
| miR-375 | Tumor suppressor | PDK1/Akt, 14-3-3zeta | Decrease growth; increase apoptosis | Tsukamoto 2010[53] |
| miR-421, miR-483-3p | Oncogene | DCP4/Smad4 | Increase proliferation and colony formation in vitro | Hao 2010[47], Hao 2011[46] |
The E1A-binding protein p300 (EP300) gene encodes for a histone acetyltransferase that modulates transcription via chromatin remodeling, and it has been implicated in the regulation of cell cycle arrest, growth and differentiation signaling, and tumorigenesis[42]–[44]. A recent study showed that certain miRNAs targeting EP300, such as miR-194, miR-200b, miR-220c, and miR-429, were up-regulated in highly metastatic PDAC cell lines as compared to low or nonmetastatic PDAC cell lines, and that EP300 mRNA expression was correspondingly decreased in highly metastatic cell lines[45]. Since EP300 is a widely recognized tumor and metastasis suppressor, these results suggest that these miRNAs may stimulate oncogenesis by reducing the expression of EP300.
DPC4/Smad4 encodes a key messenger in the TGF-β signaling pathway, which is a significant inhibitor of cell growth. In a recent study, miR-421 was found to be up-regulated in human PDAC tissues, whereas DPC4/Smad4 was down-regulated. Ectopic expression of miR-421 decreased levels of DPC4/Smad4 protein, and promoted proliferation and colony formation in vitro, suggesting that miR-421 promotes oncogenesis by acting on DPC4/Smad4[46]. A similar study showed that up-regulated miR-483-3p also targets DPC4/Smad4 in PDAC tissues[47].
Retinoblastoma protein (pRB) is a widely recognized tumor suppressor. Its anti-tumor activity is partly attributable to its binding to and inactivation of E2F proteins. miR-132 and miR-212 are up-regulated in PDAC tissues, and overexpression of these miRNAs reduced pRB expression, increased expression of several E2F target genes, and increased cell proliferation[48]. These results indicate that these miRNAs promote oncogenesis by reducing pRB, thus increasing expression of E2F gene products.
Ma et al.[49] recently reported that inhibition of miR-27a in PDAC cells reduced growth, colony formation, migration, and also down-regulated Spry2, a negative feedback regulator of multiple receptor tyrosine kinases that are active in several cancer types. These data suggest that miR-27a may have an oncogenic effect via down-regulating Spry2. miR-301a was recently shown to down-regulate NF-κB-repressing factor (NKRF) in PDAC, subsequently increasing NF-κB activation. NF-κB then activates the transcription of miR-301a, initiating an oncogenic positive feedback loop. Inhibition of miR-301a or overexpression of NKRF reversed this process and slowed tumor growth, suggesting that miR-301a contributes to oncogenesis by activating NF-κB[50]. Zhang et al.[51] found that inducing overexpression of miR-214 in BxPC-3 pancreatic cancer cells significantly reduced sensitivity to gemcitabine, the current first-line treatment for advanced PDAC. Furthermore, miR-214 was found to potentially inhibit the activity of the tumor suppressor, inhibitor of growth protein 4 (ING4). These data suggest that miR-214 may play a role in the drug resistance and tumorigenesis of PDAC.
Tumor suppressor miRNAs in PDAC
Some miRNAs have also been found to suppress tumorigenesis in PDAC (Table 2). Kent et al.[37] found that miR-34a, which is associated with the p53 pathway, may act as a tumor suppressor. Increasing expression of miR-34a, whose expression is commonly decreased in PDAC, inhibited the growth of Panc-1 and MIA PaCa-2 cell lines, suggesting an anti-proliferative effect for miR-34a via the activation of p53. Meanwhile, miR-96, an miRNA down-regulated in PDAC[39], has been shown to directly target the K-Ras Oncogene. Ectopic expression of miR-96 increased cell apoptosis and reduced migration, proliferation, and invasion of PDAC cell lines. These effects were attributable to the inhibition of the K-Ras/Akt pathway, and indicate that miR-96 functions as a tumor suppressor via regulating K-Ras[52].
Another well-documented tumor suppressor in PDAC is miR-375 [38],[39], which may exert its proapoptotic effect by targeting the caspase-mediated apoptotic pathway. Overexpression of miR-375 has been shown to inhibit PDK1 expression, thereby down-regulating the PDK1/Akt survival pathway. Moreover, it is possible that miR-375 also acts by down-regulating the anti-apoptotic gene 14-3-3zeta[53].
Wingless-type MMTV integration site family, member 3A (WNT3A) is part of the Wnt/β-catenin pathway, and stimulates survival and proliferation pathways via phosphorylation of ERK and Akt. Similarly, fibroblast growth factor-7 (FGF-7) is involved in a variety of biological processes relating to survival. Zhang et al.[51] showed that miR-15a, which is down-regulated in PDAC tissues, directly targets WNT3A and FGF-7. These data indicate that miR-15a may serve as a tumor suppressor by modulating expression of the survival pathways related to WNT3A and FGF-7.
Roles of miRNA in Hepatocellular Carcinoma
Current miRNA profiling studies in HCC
As with PDAC, many miRNA profiling studies have been recently conducted to identify a characteristic miRNA signature for H CC (Table 3). HCC is a very heterogeneous disease with many subtypes, including diseases with viral, non-viral, or cirrhotic etiologies, cancer stem cell-like or mature hepatocyte-like features, malignant versus benign phenotypes, and metastatic versus non-metastatic phenotypes. These subtypes add significant complexity to miRNA profiling studies targeting HCC[54].
Table 3. MicroRNA profiling studies in hepatocellular carcinoma.
| Reference | Study specifications | Up-regulated miRNAs | Down-regulated miRNAs |
| Murakami 2006[55] | Microarray testing for 180 mature and 206 precursor miRNAs in 24 HCC tissues and 22 adjacent non-tumor tissues, 9 CH samples | pre-miR-18, miR-18, miR-224 | miR-125a, miR-195a, miR-199a, miR-199a*, miR-200a |
| Budhu 2008[56] | Microarray testing for unspecified number of miRNAs in 29 metastatic HCC tissues and 102 non-metastatic HCC tissues | miR-185, miR207, miR-219-1, miR-338 | let-7g, miR-1-2, miR-9-2, miR-15a, miR-19a, miR-30a, miR-30c-1,miR-30e, miR-34a, miR-122a, miR-124a-2, miR-125b-2, miR-126, miR-148a, miR-148b, miR-194 |
| Wang 2008[57] | RT-PCR assay testing for 157 miRNAs in 19 HCC tissues and adjacent benign tissues | miR-9, miR-9*, miR-21, miR-25, miR-137, miR-151, miR-155, miR-182, miR-182*, miR-183, miR-186, miR-216, miR-221, miR-222, miR-224, miR-301, | miR-139, miR-145, miR-214 |
| Huang 2007[58] | Microarray testing for 331 miRNAs in 10 HCC tissues and adjacent benign tissues from patients without viral hepatitis |
miR-324-5p, miR-374 let-7a, let-7b, let-7c, let-7d, let-7e, let-7g, let-7i, miR-21, miR-22, miR-98, miR-126, miR-126-3p, miR-195, miR-352 |
miR-235 |
Results of studies attempting to determine the miRNA expression signature of hepatocellular carcinoma. HCC, hepatocellular carcinoma; CH, chronic hepatitis.
Murakami et al.[55] performed the first comprehensive miRNA profiling study in 24 HCC tissues and 22 adjacent non-HCC tissues and found 3 miRNAs, miR-18, pre-miR-18, and miR-224, to be up-regulated in the HCC samples and 5 miRNAs, miR-199a*, miR-199a, miR-195, miR-200a, and miR-125a, to be down-regulated. In a larger study of HCC and adjacent non-HCC specimens from 241 HCC patients, Budhu et al.[56] outlined a 20-miRNA profile of metastatic HCC capable of distinguishing primary HCC tissues and non-metastiatic solitary tumors. Four of these, miR-338, miR-219-1, miR-207, and miR-185, were up-regulated, whereas 16 were down-regulated, with the most down-regulated being miR-34a, miR-148a, and miR-124a-2.
Three similar studies in HCC and adjacent non-HCC specimens found varying results. Wang et al.[57] found 19 up-regulated and 3 down-regulated miRNAs in HCC. Consistent with the study by Murakami et al., miR-199a, miR-200a, miR-125a, and miR-224 were all found to be dysregulated in HCC. Huang et al.[58] found 15 up-regulated and 1 down-regulated miRNAs with little similarity to past studies, and found 11 up-regulated and 23 down-regulated miRNAs in another study[59]. Recently, a profiling study involving tissue samples and cell lines revealed a set of 12 miRNAs that characterize HCC[60]. Among these were miR-21, miR-221/222, miR-34a, and let-7c, which have been identified in other similar studies as being highly dysregulated in HCC.
Viral-associated tumors are a common subtype of HCCs. In an analysis of 52 HCC tumors from patients infected with hepatitis C virus, Varnholt et al.[61] found 29 differentially expressed miRNAs compared to normal liver, but minimal overlap was found between this study and others. A similar study compared miRNA expression profiles of HCC tissues from hepatitis B patients and hepatitis C patients. Two types of differentially expressed miRNAs were identified as either associated with the type of hepatitis (n = 19) or disease stage (n = 31).
Approximately 80% of HCCs develop in cirrhotic liver[62]. In a study focusing on cirrhotic HCC and cirrhotic non-HCC tissues, Gramantieri et al.[63] found 35 differentially expressed miRNAs including members of the let-7 family, miR-221 and miR-145, which had previously been shown to be dysregulated in other cancers. A more recent study found similar results, establishing a signature of 40 miRNAs that could accurately distinguish HCC from adjacent cirrhotic tissues[64].
Hepatocellular stem cells (HSCs) give rise to the proliferative and self-renewal potential of liver tumors. In a global miRNA microarray comparing human HSCs, all members of the let-7 and miR-181 families were expressed >4.0 folds in HSCs compared to HepG2 HCC cells and normal liver stem cells. Another study by Ji et al. [65] identified a 20-miRNA signature differentiating HSC-like HCC from mature hepatocyte-like HCC (MH-HCC).
Oncogenic miRNAs in HCC
Several miRNAs that may serve as oncogenes have been found to be overexpressed in HCC tissues and cell lines (Table 4). One of the most commonly identified up-regulated miRNAs in HCC is miR-21[60], which may promote oncogenesis by repressing expression of the tumor suppressor phosphate and tensin homolog (PTEN)[66],[67]. PTEN has been shown to be a significant contributor to HCC pathogenesis, and suppression of PTEN in murine hepatocytes led to HCC development[68]. Inhibition of miR-21 in HCC cells increased PTEN expression and decreased tumor proliferation, migration, and invasion, suggesting that up-regulation of miR-21 in HCC can contribute to HCC pathogenesis by suppressing PTEN and PTEN-dependent pathways.
Table 4. Dysregulated miRNAs, their functions, and possible molecular targets in hepatocellular carcinoma.
| MicroRNA(s) | Role | Molecular target(s) | Biological function(s) | Reference(s) |
| let-7 family | Tumor suppressor | RAS, HMGA2, Collagen type 1 alpha2 | Inhibit growth, migration, and proliferation | Johnson 2007[22], Lee 2007[76], Ji 2010[65] |
| miR-17-92 cluster | Oncogene | Bin, PTEN, CTGF, Tsp1, E2F1/2/3 | Increase growth, proliferation, and angiogenesis; decrease apoptosis | Olive 2010[73] |
| miR-18a | Oncogene | Estrogen Receptor α | Increase proliferation; block protective effect of estrogen | Liu 2009[75] |
| miR-21 | Oncogene | PTEN | Increase proliferation, migration, and invasion | Buscaglia 2011[66] |
| miR-122 | Tumor suppressor | Cyclin G1, Bcl-w | Increase apoptosis; inhibit growth | Gramantieri 2007[63], Lin 2008[78] |
| miR-199a/a* | Tumor suppressor | MET, ERK2, mTOR, CD44 | Inhibit proliferation, migration, and invasion | Kim 2008[79], Henry 2010[80], Fornari 2008[81] |
| miR-221/222 | Oncogene | p27Kip1,CDKN1C/p57, DDIT4/mT0R, PPP2R2A/Akt | Increase growth and proliferation | Ie Sage 2007[70], Fornari 2008[81] |
miR-221 and miR-222, neighbors in human genomes, share a high degree of homology and are commonly overexpressed in HCC [60],[63],[69]. miR-221 and miR-222 have been shown to stimulate tumor growth by inhibiting expression of p27Kip1, a cell cycle inhibitor and tumor suppressor[70]. Other proven targets of miR-221 are the Cyclin-dependent kinase inhibitor (CDKN1C/p57) and DNA damage-inducible transcript 4 (DDIT4), a modulator of the mammalian target of rapamycin (mTOR) pathway[60],[63]. Wong et al.[64] also showed that miR-222 may further promote oncogenesis by enhancing Akt signaling, possibly through the protein phosphatase 2, regulatory subunit B, alpha (PPP2R2A). PPP2R2A regulates protein phosphatase 2 α PPP2A, a key serine/threonine phosphatase that controls Akt signaling[71].
The miRNA-17-92 cluster comprises 7 miRNAs, miR-17-5p, miR-17-3p, miR-18, miR-19a, miR-19b, miR-20a, and miR-92[72], many of which have been found to be commonly overexpressed in HCC[55],[73]. This cluster may have several key molecular targets including p21, which regulates proliferation; Bim and PTEN, regulators of apoptosis; connective tissue growth factor (CTGF) and thrombospondin 1 (Tsp1), regulators of angiogenesis; and E2F1/2/3, transcription activators that stimulate cell cycle progression [73]. Taken as a whole, these data suggest that miR-17-92 promotes oncogenesis in HCC through the modulation of several key pathways related to growth and survival.
HCC occurs more frequently in men than in women, and this pattern may be partly attributable to the protective function of estrogen in women[74]. Interestingly, the differential expression levels of certain oncogenic miRNAs between men and women may help explain this gender disparity. Liu et al.[75] found that miR-18a, an miRNA within the miR-17-92 cluster, is significantly up-regulated in male HCC tissues compared to female tissues. Moreover, they found that increased miR-18a levels in women correlated with decreased expression of ESR1 gene, which encodes estrogen receptor α (ERα) protein, and that miR-18a directly targets ERα by binding to ERα mRNA at its 3′ untranslated region (UTR). These data suggest that increased levels of miR-18a in men may subsequently decrease levels of ERα, thus blocking the documented protective effect of estrogen and stimulating HCC pathogenesis.
Tumor suppressor miRNAs in HCC
Members of the let-7 miRNA family were found to be among the commonly down-regulated miRNAs in HCC (Table 4)[60],[63],[64]. This miRNA family is known to regulate RAS, so decreased expression of let-7 may simultaneously increase expression of RAS, suggesting a mechanism by which decreased levels of tumor suppressor let-7 miRNAs promote hepatocarcinogenesis. Additional targets of let-7 have been proposed, including the high-mobility group AT-hook 2 HMGA2 Oncogene and collagen type I alpha 2, indicating that let-7 may act as a tumor suppressor by suppressing multiple oncogenic signaling pathways and metastatic factors[65],[76],[77].
miR-122 accounts for ∼70% of the entire miRNA population in adult liver[78] and is commonly identified as another down-regulated tumor suppressor in HCC[56],[64],[76],[78]. Several studies have identified different molecular targets of miR-122, including cyclin G1[63] and the anti-apoptotic gene Bcl-w[78].
miR-199a/a* has emerged as another possible tumor suppressor in HCC[55],[64]. Several target genes of miR-199a/a* have been proposed. One of these is the MET Oncogene and its downstream effector extracellular regulated kinase-2 (ERK2), which promote proliferation, motility, and invasiveness[79]. Other possible targets include mTOR protein and CD44[80],[81].
Therapeutic Strategies Targeting miRNA
Because PDAC and HCC have a poor prognoses that have not improved significantly for decades, new diagnostic methods and therapeutic modalities are desperately needed. Relevant targetable miRNAs may either be up-regulated oncogenes or down-regulated tumor suppressors. In the case of the former, direct targeting of oncogenic miRNAs using anti-miRNA molecules, such as anti-miRNA oligonucleotides, antago-miRNAs, DNAzymes, ribozymes, and antisense molecules, may down-regulate or silence the miRNA and its oncogenic effects. Conversely, in the case of down-regulated tumor suppressor miRNAs, miRNA replacement therapies such as miRNA mimics may be used to restore the tumor suppressor functions of the original down-regulated miRNAs[82].
For example, in a study attempting to increase levels of tumor suppressor miR-26a in a mouse liver cancer model, administration of a single miR-26a-expressing adenovirus increased miR-26a expression and led to significant inhibition of proliferation, induction of tumor-specific apoptosis, and protection from disease[83]. In an attempt to decrease levels of oncogenic miRNAs, Pineau et al.[60] recently observed that the introduction of 2 antagomiRs to reduce levels of miR-221 and miR-222 in liver cancer cell lines led to decreased cell growth. These data suggest that the introduction or knockdown of a single tumor suppressor or oncogenic miRNA, respectively, can have a significant impact on a disease such as HCC.
Another area of interest involving miRNAs is their use in diagnosis and characterization of tumors. Differential tumor classification by miRNA-based profiling has been shown to be more accurate than mRNA-based profiling in characterizing human cancers and their developmental history, and it is also an accurate predictor of a patient's overall prognosis[84]. This suggests that, from a clinical standpoint, miRNA profiling may serve as an effective diagnostic and prognostic tool.
miRNA delivery methods
Although miRNAs are emerging as viable cancer therapeutics, their efficacy remains largely dependent on the development of effective delivery strategies. In recent studies demonstrating miRNA-induced inhibition of tumor growth in mouse models, delivery of endogenous miRNAs was performed by either direct intratumoral injection or viral vectors. These methods are useful in mouse models but may not be applicable in humans. Non-viral strategies may provide a viable alternative to these current techniques to deliver tumor suppressor miRNAs or antisense miRNA antagomiRNAs[82]. Several non-viral delivery methods have been developed, including lipid encapsulation, complex formation via a variety of liposomes or Cationic polymers, liposomal nanoparticles, and chemical conjugation of miRNAs to peptides, aptamers, or antibodies[82],[85]. An miRNA sponge may provide a novel method of silencing specific miRNAs. These miRNA sponges can be expressed in cells and contain multiple binding sites to a specific miRNA and thereby may have significant potential to achieve long-lasting inhibition of miRNAs[86].
miRNA-based therapies have effectively suppressed tumorigenesis in vitro and in animal models, but many obstacles remain until these strategies can be used in a clinical setting[87]. To distribute miRNA-based therapies to their proper locations within the body, both local and systemic delivery strategies need to be developed and optimized. Additionally, a key question of interest is whether to manipulate a few specific oncogenes or to manipulate a single specific miRNA, which may in turn change the levels of many different genes with diverse cellular functions. Experiments comparing these two strategies will help clinicians and researchers determine which targets are more appropriate for miRNA-based therapy[38]. Another obstacle of miRNA research is the uncertainty of the target gene identification. The predicted targets of a miRNA need to be validated and confirmed through biochemical and functional assays.
Our prospective outlook for miRNAs as potential therapeutic and diagnostic targets is cautiously optimistic. Despite the previously mentioned obstacles, miRNAs possess broad oncogenic and tumor suppressive activities in PDAC and HCC and may represent an opportunity for targeted therapies to mimic or inhibit miRNAs that contribute to tumorigenesis. miRNAs can also be used as a diagnostic tool, and the prognostic potential of miRNAs has also been shown to be more effective than mRNA-based prognosis. Overall, the development of miRNA-based therapeutic, prognostic, and diagnostic strategies may generate improved clinical practices for the management of PDAC and HCC.
Conclusions
Increasing evidence suggests that miRNAs play a key role in Carcinogenesis. As the mRNA targets of specific miRNAs are discovered, the miRNA expression profiles of individual tumors are clarified, and new therapeutic strategies are developed and improved, it is likely that miRNAs will play an increasingly important role in cancer diagnosis and therapy. The reintroduction of down-regulated tumor suppressor miRNAs and the silencing of overexpressed oncogenic miRNAs may have great therapeutic potential in the coming years. In the cases of PDAC and HCC, which are devastating diseases, several key oncogenic and tumor suppressor miRNAs have been identified, and a few of these have confirmed mRNA targets. While more benchwork needs to be performed to further elucidate the roles of specific miRNAs in these cancers, therapeutic studies involving miRNA-based treatment in animal models as well as the development of systemic delivery systems will be required if miRNAs are to be used effectively in the clinical setting.
Acknowledgments
This work was supported in part by the William and Ella Owens Medical Research Foundation (M. Li), and the Vivian L. Smith Department of Neurosurgery at the University of Texas Health Science Center at Houston, Medical School.
References
- 1.Ambros V. The functions of animal microRNAs [J] Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post–transcriptional regulation by microRNAs: are the answers in sight? [J] Nat Rev Genet. 2008;9(2):102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 3.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets [J] Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 4.Song F, Chen K. Single nucleotide polymorphisms among microRNA: big effects on cancer [J] Chin J Cancer. 2011;30(6):381–391. doi: 10.5732/cjc.011.10142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Griffiths-Jones S, Saini HK, van Dongen S, et al. MiRBase: Tools for microRNA genomics [J] Nucleic Acids Res. 2008;36(Database issue):D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs [J] Nature. 2005;433(7027):769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 7.Lee Y, Ahn C, Han J, et al. The nuclear RNase III drosha initiates microRNA processing [J] Nature. 2003;425(6956):415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function [J] Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? [J] Trends in cell biology. 2007;17(3):118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 10.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers [J] Proc Natl Acad Sci USA. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee EJ, Gusev Y, Jiang J, et al. Expression profiling identifies microRNA signature in pancreatic cancer [J] Int J Cancer. 2007;120(5):1046–1054. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi XB, Xue L, Yang J, et al. An androgen-regulated miRNA suppresses Bak1 expression and induces androgen-independent growth of prostate cancer cells [J] Proc Natl Acad Sci USA. 2007;104(50):19983–19988. doi: 10.1073/pnas.0706641104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitomo S, Maesawa C, Ogasawara S, et al. Downregulation of miR-138 is associated with overexpression of human telomerase reverse transcriptase protein in human anaplastic thyroid carcinoma cell lines [J] Cancer Sci. 2008;99(2):280–286. doi: 10.1111/j.1349-7006.2007.00666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felicetti F, Errico MC, Bottero L, et al. The promyelocytic leukemia zinc fingemicroRNA-221/-222 pathway controls melanoma progression through multiple oncogenic mechanisms [J] Cancer Res. 2008;68(8):2745–2754. doi: 10.1158/0008-5472.CAN-07-2538. [DOI] [PubMed] [Google Scholar]
- 15.Yang H, Kong W, He L, et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN [J] Cancer Res. 2008;68(2):425–433. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- 16.Schetter AJ, Leung SY, Sohn JJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma [J] JAMA. 2008;299(4):425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer [J] Cancer Res. 2005;65(16):7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 18.Kent OA, Mendell JT. A small piece in the cancer puzzle: microRNAs as tumor suppressors and oncogenes [J] Oncogene. 2006;25(46):6188–6196. doi: 10.1038/sj.onc.1209913. [DOI] [PubMed] [Google Scholar]
- 19.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia [J] Proc Natl Acad Sci USA. 2002;99(24):15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cimmino A, Calin GA, Fabbri M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2 [J] Proc Natl Acad Sci U S A. 2005;102(39):13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fabbri M, Garzon R, Cimmino A, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3a and 3b [J] Proc Natl Acad Sci USA. 2007;104(40):15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson CD, Esquela-Kerscher A, Stefani G, et al. The let-7 microRNA represses cell proliferation pathways in human cells [J] Cancer Res. 2007;67(16):7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 23.Corney DC, Flesken-Nikitin A, Godwin AK, et al. MicroRNA-34b and microRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth [J] Cancer Res. 2007;67(18):8433–8438. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- 24.Tazawa H, Tsuchiya N, Izumiya M, et al. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells [J] Proc Natl Acad Sci USA. 2007;104(39):15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan JA, Krichevsky AM, Kosik KS. MicroRNA21 is an antiapoptotic factor in human glioblastoma cells [J] Cancer Res. 2005;65(14):6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 26.Costinean S, Zanesi N, Pekarsky Y, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice [J] Proc Natl Acad Sci U S A. 2006;103(18):7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang Q, Gumireddy K, Schrier M, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis [J] Nat Cell Biol. 2008;10(2):202–210. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- 28.Jemal A, Siegel R, Xu J, et al. Cancer statistics, 2010 [J] CA Cancer J Clin. 2010;60(5):277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 29.Warshaw AL, Fernandez-del Castillo C. Pancreatic carcinoma [J] N Engl J Med. 1992;326(7):455–455. doi: 10.1056/NEJM199202133260706. [DOI] [PubMed] [Google Scholar]
- 30.Li D, Xie K, Wolff R, et al. Pancreatic cancer [J] Lancet. 2004;363(9414):1049–1057. doi: 10.1016/S0140-6736(04)15841-8. [DOI] [PubMed] [Google Scholar]
- 31.Jemal A, Bray F, Center MM, et al. Global cancer statistics [J] CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 32.Di Bisceglie AM. Issues in screening and surveillance for hepatocellular carcinoma [J] Gastroenterology. 2004;127(5 Suppl 1):S104–S107. doi: 10.1053/j.gastro.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 33.Parkin DM, Pisani P, Ferlay J. Estimates of the worldwide incidence of eighteen major cancers in 1985 [J] Int J Cancer. 1993;54(4):594–606. doi: 10.1002/ijc.2910540413. [DOI] [PubMed] [Google Scholar]
- 34.Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment [J] Nat Rev Cancer. 2006;6(9):674–687. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- 35.El-Serag HB, Marrero JA, Rudolph L, et al. Diagnosis and treatment of hepatocellular carcinoma [J] Gastroenterology. 2008;134(6):1752–1763. doi: 10.1053/j.gastro.2008.02.090. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y, Li M, Wang H, et al. Profiling of 95 microRNAs in pancreatic cancer cell lines and surgical specimens by realtime PCR analysis [J] World J Surg. 2009;33(4):698–709. doi: 10.1007/s00268-008-9833-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kent OA, Mullendore M, Wentzel EA, et al. A resource for analysis of microRNA expression and function in pancreatic ductal adenocarcinoma cells [J] Cancer Biol Ther. 2009;8(21):2013–2024. doi: 10.4161/cbt.8.21.9685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bloomston M, Frankel WL, Petrocca F, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis [J] JAMA. 2007;297(17):1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 39.Szafranska AE, Davison TS, John J, et al. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma [J] Oncogene. 2007;26(30):4442–4452. doi: 10.1038/sj.onc.1210228. [DOI] [PubMed] [Google Scholar]
- 40.Greither T, Grochola LF, Udelnow A, et al. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival [J] Int J Cancer. 2010;126(1):73–80. doi: 10.1002/ijc.24687. [DOI] [PubMed] [Google Scholar]
- 41.Gironella M, Seux M, Xie MJ, et al. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development [J] Proc Natl Acad Sci USA. 2007;104(41):16170–16175. doi: 10.1073/pnas.0703942104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ogryzko W, Schiltz RL, Russanova V, et al. The transcriptional coactivators p300 and CBP are histone acetyltransferases [J] Cell. 1996;87(5):953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 43.Snowden AW, Anderson LA, Webster GA, et al. A novel transcriptional repression domain mediates p21 (WAF1/CIP1) induction of p300 transactivation [J] Mol Cell Biol. 2000;20(8):2676–2686. doi: 10.1128/mcb.20.8.2676-2686.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sahar S, Reddy MA, Wong C, et al. Cooperation of SRC-1 and p300 with NF-kappaB and CREB in angiotensin II-induced IL-6 expression in vascular smooth muscle cells [J] Arterioscler Thromb Vasc Biol. 2007;27(7):1528–1534. doi: 10.1161/ATVBAHA.107.145862. [DOI] [PubMed] [Google Scholar]
- 45.Mees ST, Mardin WA, Wendel C, et al. EP300—a miRNA-regulated metastasis suppressor gene in ductal adenocarcinomas of the pancreas [J] Int J Cancer. 2010;126(1):114–124. doi: 10.1002/ijc.24695. [DOI] [PubMed] [Google Scholar]
- 46.Hao J, Zhang S, Zhou Y, et al. MicroRNA 421 suppresses DPC4/Smad4 in pancreatic cancer [J] Biochem Biophys Res Commun. 2011;406(4):552–557. doi: 10.1016/j.bbrc.2011.02.086. [DOI] [PubMed] [Google Scholar]
- 47.Hao J, Zhang S, Zhou Y, et al. MicroRNA 483-3p suppresses the expression of DPC4/Smad4 in pancreatic cancer [J] FEBS Lett. 2011;585(1):207–213. doi: 10.1016/j.febslet.2010.11.039. [DOI] [PubMed] [Google Scholar]
- 48.Park JK, Henry JC, Jiang J, et al. miR-132 and miR-212 are increased in pancreatic cancer and target the retinoblastoma tumor suppressor [J] Biochem Biophys Res Commun. 2011;406(4):518–523. doi: 10.1016/j.bbrc.2011.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma Y, Yu S, Zhao W, et al. miR-27a regulates the growth, colony formation and migration of pancreatic cancer cells by targeting Sprouty2 [J] Cancer Lett. 2010;298(2):150–158. doi: 10.1016/j.canlet.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 50.Lu Z, Li Y, Takwi A, et al. miR-301a as an NF-κB activator in pancreatic cancer cells [J] EMBO J. 2011;30(1):57–67. doi: 10.1038/emboj.2010.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang XJ, Ye H, Zeng CW, et al. Dysregulation of miR-15a and miR-214 in human pancreatic cancer [J] J Hematol Oncol. 2010;3:46. doi: 10.1186/1756-8722-3-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu S, Lu Z, Liu C, et al. miRNA-96 suppresses KRAS and functions as a tumor suppressor gene in pancreatic cancer [J] Cancer Res. 2010;70(14):6015–6025. doi: 10.1158/0008-5472.CAN-09-4531. [DOI] [PubMed] [Google Scholar]
- 53.Tsukamoto Y, Nakada C, Noguchi T, et al. MicroRNA-375 is downregulated in gastric carcinomas and regulates cell survival by targeting PDK1 and 14-3-3zeta [J] Cancer ReS. 2010;70(6):2339–2349. doi: 10.1158/0008-5472.CAN-09-2777. [DOI] [PubMed] [Google Scholar]
- 54.Laurent-Puig P, Legoix P, Bluteau O, et al. Genetic alterations associated with hepatocellular carcinomas define distinct pathways of hepatocarcinogenesis [J] Gastroenterology. 2001;120(7):1763–1773. doi: 10.1053/gast.2001.24798. [DOI] [PubMed] [Google Scholar]
- 55.Murakami Y, Yasuda T, Saigo K, et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues [J] Oncogene. 2006;25(17):2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 56.Budhu A, Jia HL, Forgues M, et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma [J] Hepatology. 2008;47(3):897–907. doi: 10.1002/hep.22160. [DOI] [PubMed] [Google Scholar]
- 57.Wang Y, Lee AT, Ma JZ, et al. Profiling microrna expression in hepatocellular carcinoma reveals microRNA-224 up-regulation and apoptosis inhibitor-5 as a microRNA-224-specific target [J] J Biol Chem. 2008;283(19):13205–13215. doi: 10.1074/jbc.M707629200. [DOI] [PubMed] [Google Scholar]
- 58.Huang YS, Dai Y, Yu XF, et al. Microarray analysis of microRNA expression in hepatocellular carcinoma and non-tumorous tissues without viral hepatitis [J] J Gastroenterol Hepatol. 2008;23(1):87–94. doi: 10.1111/j.1440-1746.2007.05223.x. [DOI] [PubMed] [Google Scholar]
- 59.Chiang CW, Huang Y, Leong KW, et al. PKCalpha mediated induction of miR-101 in human hepatoma HepG2 cells [J] J Biom Sci. 2010;17:35. doi: 10.1186/1423-0127-17-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pineau P, Volinia S, McJunkin K, et al. miR-221 overexpression contributes to liver tumorigenesis [J] Proc Natl Acad Sci U S A. 2010;107(1):264–269. doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Varnholt H, Drebber U, Schulze F, et al. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma [J] Hepatology. 2008;47(4):1223–1232. doi: 10.1002/hep.22158. [DOI] [PubMed] [Google Scholar]
- 62.Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma [J] Lancet. 2003;362(9399):1907–1917. doi: 10.1016/S0140-6736(03)14964-1. [DOI] [PubMed] [Google Scholar]
- 63.Gramantieri L, Ferracin M, Fornari F, et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma [J] Cancer Res. 2007;67(13):6092–6099. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- 64.Wong QW, Ching AK, Chan AW, et al. MiR-222 overexpression confers cell migratory advantages in hepatocellular carcinoma through enhancing akt signaling [J] Clin Cancer Res. 16(3):867–875. doi: 10.1158/1078-0432.CCR-09-1840. [DOI] [PubMed] [Google Scholar]
- 65.Ji J, Zhao L, Budhu A, et al. Let-7g targets collagen type I alpha2 and inhibits cell migration in hepatocellular carcinoma [J] J Hepatol. 2010;52(5):690–697. doi: 10.1016/j.jhep.2009.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Buscaglia LEB, Li Y. Apoptosis and the target genes of miR-21 [J] Chin J Cancer. 2011;30(6):371–380. doi: 10.5732/cjc.011.10132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wei J, Gao W, Zhu C, et al. Identification of plasma miRNA-21 as a biomarker for early detection and chemosensitivity of nonsmall cell lung cancer [J] Chin J Cancer. 2011;30(6):407–414. doi: 10.5732/cjc.010.10522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Horie Y, Ohshima S, Sato W, et al. Hepatocyte-specific Pten deficient mice [J] Nippon Rinsho. 2006;64(6):1033–1042. [in Japanese] [PubMed] [Google Scholar]
- 69.Wong QW, Lung RR, Law PT, et al. MicroRNA-223 is commonly repressed in hepatocellular carcinoma and potentiates expression of stathmini [J] Gastroenterology. 2008 doi: 10.1053/j.gastro.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 70.Ie Sage C, Nagel R, Egan DA, et al. Regulation of the p27 (Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation [J] EMBO J. 2007;26(15):3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cohen PT, Brewis ND, Hughes V, et al. Protein serine/threonine phosphatases; an expanding family [J] FEBS letters. 1990;268(2):355–359. doi: 10.1016/0014-5793(90)81285-v. [DOI] [PubMed] [Google Scholar]
- 72.Shigoka M, Tsuchida A, Matsudo T, et al. Deregulation of miR-92a expression is implicated in hepatocellular carcinoma development [J] Pathol Int. 2010;60(5):351–357. doi: 10.1111/j.1440-1827.2010.02526.x. [DOI] [PubMed] [Google Scholar]
- 73.Olive V, Jiang I, He L. mir-17-92, a cluster of miRNAs in the midst of the cancer network [J] Int J Biochem Cell Biol. 2010;42(8):1348–1354. doi: 10.1016/j.biocel.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Naugler WE, Sakurai T, Kim S, et al. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production [J] Science. 2007;317(5834):121–124. doi: 10.1126/science.1140485. [DOI] [PubMed] [Google Scholar]
- 75.Liu WH, Yeh SH, Lu CC, et al. MicroRNA-18a prevents estrogen receptor-alpha expression, promoting proliferation of hepatocellular carcinoma cells [J] Gastroenterology. 2009;136(2):683–693. doi: 10.1053/j.gastro.2008.10.029. [DOI] [PubMed] [Google Scholar]
- 76.Lee YS, Dutta A. The tumor suppressor microRNA Ief7 represses the HMGA2 Oncogene [J] Genes Dev. 2007;21(9):1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and HMGA2 enhances oncogenic transformation [J] Science. 2007;315(5818):1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lin CJ, Gong HY, Tseng HC, et al. miR-122 targets an antiapoptotic gene, Bcl-w, in human hepatocellular carcinoma cell lines [J] Biochem Biophys Res Commun. 2008;375(3):315–320. doi: 10.1016/j.bbrc.2008.07.154. [DOI] [PubMed] [Google Scholar]
- 79.Kim S, Lee UJ, Kim MN, et al. MicroRNA miR-199a* regulates the MET protooncogene and the downstream extracellular signal-regulated kinase 2 (ERK2) [J] J Biol Chem. 2008;283(26):18158–18166. doi: 10.1074/jbc.M800186200. [DOI] [PubMed] [Google Scholar]
- 80.Henry JC, Park JK, Jiang J, et al. miR-199a-3p targets CD44 and reduces proliferation of CD44 positive hepatocellular carcinoma cell lines [J] Biochem Biophys Res Commun. 2010;403(1):120–125. doi: 10.1016/j.bbrc.2010.10.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fornari F, Milazzo M, Chieco P, et al. MiR-199a-3p regulates mTOR and c-met to influence the doxorubicin sensitivity of human hepatocarcinoma cells [J] Cancer Res. 70(12):5184–5193. doi: 10.1158/0008-5472.CAN-10-0145. [DOI] [PubMed] [Google Scholar]
- 82.Aigner A. Nonviral in vivo delivery of therapeutic small interfering RNAs [J] Curr Opin Mol Ther. 2007;9(4):345–352. [PubMed] [Google Scholar]
- 83.Kota J, Chivukula RR, O'Donnell KA, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model [J] Cell. 2009;137(6):1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers [J] Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 85.Purow B. The elephant in the room: do microRNA-based therapies have a realistic chance of succeeding for brain tumors such as glioblastoma? [J] J Neurooncol. 2010 Nov 17; doi: 10.1007/s11060-010-0449-5. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells [J] Nat Methods. 2007;4(9):721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rupaimoole R, Han H, Lopez-Berestein G, et al. MicroRNA therapeutics: principles, expectations and challenges [J] Chin J Cancer. 2011;30(6):368–370. doi: 10.5732/cjc.011.10186. [DOI] [PMC free article] [PubMed] [Google Scholar]