ABSTRACT
Sperm utilize glycolysis to generate ATP required for motility, and several spermatogenic cell-specific glycolytic isozymes are associated with the fibrous sheath (FS) in the principal piece of the sperm flagellum. We used proteomics and molecular biology approaches to confirm earlier reports that a novel enolase is present in mouse sperm. We then found that a pan-enolase antibody, but not antibodies to ENO2 and ENO3, recognized a protein in the principal piece of the mouse sperm flagellum. Database analyses identified two previously uncharacterized enolase family-like candidate genes, 64306537H0Rik and Gm5506. Northern analysis indicated that 64306537H0Rik (renamed Eno4) was transcribed in testes of mice by Postnatal Day 12. To determine the role of ENO4, we generated mice using embryonic stem cells in which an Eno4 allele was disrupted by a gene trap containing a beta galactosidase (beta-gal) reporter (Eno4+/Gt). Expression of beta-gal occurred in the testis, and male mice homozygous for the gene trap allele (Eno4Gt/Gt) were infertile. Epididymal sperm numbers were 2-fold lower and sperm motility was reduced substantially in Eno4Gt/Gt mice compared to wild-type mice. Sperm from Eno4Gt/Gt mice had a coiled flagellum and a disorganized FS. The Gm5506 gene encodes a protein identical to ENO1 and also is transcribed at a low level in testis. We conclude that ENO4 is required for normal assembly of the FS and provides most of the enolase activity in sperm and that Eno1 and/or Gm5506 may encode a minor portion of the enolase activity in sperm.
Keywords: fibrous sheath, gene expression, glycolysis, spermatid, testis
A novel enolase 4 (ENO4) is required for assembly of the fibrous sheath and is the main source of enolase activity in sperm.
INTRODUCTION
Glycolysis is the primary source of ATP in sperm, and most of the glycolytic enzymes present in sperm have novel structural and/or functional proprieties. Some are products of novel transcript splice variants of genes expressed in other cell types, including hexokinase 1 (HK1S) [1, 2], muscle-type phosphofructokinase (PFKMS) [3], and aldolase (ALDOA_V2) [4]. Others are products of spermatogenic cell-specific genes, including phosphoglycerate kinase 2 (PGK2) [5, 6], lactate dehydrogenase C (LDHC) [7], glyceraldehyde 3-phosphate dehydrogenase, spermatogenic (GAPDHS) [8], aldolase 1A retrogene 1 (ALDOART1), and aldolase 1A retrogene 2 (ALDOART2) [4]. In addition, mammalian sperm were reported to contain atypical forms of the glycolytic enzymes triosephosphate isomerase (TPI) [9], glucose phosphate isomerase (GPI) [10], phosphoglucomutase (PGM) [11], and enolase (ENOS) [12–14]. We have shown that targeted disruption of the Gapdhs, Ldhc, and Pgk2 genes result in reduced levels of ATP in sperm, disruption of sperm motility, and male infertility [15–17], confirming that glycolysis is essential for sperm function in mice [18].
The glycolytic enzymes of sperm localize primarily to the principal piece (PP) of the flagellum. Earlier studies found that some sperm glycolytic enzymes are resistant to detergent extraction, cofractionate with flagellar components, and are present in multienzyme complexes [19–21]. More recent studies have determined that GAPDHS [22], ALDOART1, ALDOA_V2 [4], LDHA, PK [23], and PFKMS [3] are highly resistant to detergent extraction. Of these, GAPDHS [22, 24], LDHA, ALDOA [23], and PK [23, 25] were found to be tightly bound to the fibrous sheath (FS), a novel cytoskeletal structure restricted to the PP of the flagellum. Other glycolytic enzymes present in the PP were readily solubilized by detergents, including HK1S, LDHC, and PGK2 [23]. While HK1S was shown to localize to the PP by tethering to PFKMS, which is bound tightly to glutathione S-transferase, mu 5 (GSTM5) in the FS [3], it remains to be determined how other glycolytic enzymes are restricted to the PP.
The PP occupies over 70% of the length of the mouse sperm flagellum. It is defined by the presence of the FS surrounding the outer dense fibers and axoneme. The FS assembles from distal to proximal and consists of two longitudinal columns interconnected by a network of circumferentially arrayed ribs [26]. The longitudinal columns appear first in round spermatids, and the ribs complete their assembly later in elongating spermatid [27]. The FS was thought originally to provide only mechanical support to modulate flagellar bending and to define the shape of the flagellar beat [28]. It is now known to serve additionally as a scaffold for proteins involved in signal transduction and for the glycolytic enzymes essential for generating the energy required for sperm motility [29].
Enolase (2-phospho-D-glycerate hydrolase; EC 4.2.1.11) catalyzes the conversion of 2-phosphoglycerate to phosphoenolpyruvate, the second of the two high-energy intermediates that generate ATP in the penultimate step of glycolysis. The enolase isozymes in eukaryotes include enolase 1 (α), enolase 2 (γ), and enolase 3 (β), which are encoded by the Eno1, Eno2, and Eno3 genes, respectively [30]. However, a putative sperm-specific enolase (ENOS) was reported in human, ram, and mouse sperm that differed from ENO1, ENO2, and ENO3 by electrophoretic mobility, thermostability, and ability to undergo structural alteration at high temperatures [12]. In addition, enolase was immunolocalized to the flagellum in rat sperm [13] and the PP of human sperm [14, 31]. Furthermore, enolase enzymatic activity was detected in elongating spermatids in mouse sperm [12] and in residual bodies in rat sperm [13].
We used proteomic, bioinformatic, and molecular biology approaches to identify ENO4 as the novel enolase present in mouse sperm. In addition, we generated mice using embryonic stem (ES) cells with an Eno4 allele disrupted by a gene trap (Eno4+/Gt) to define the role of ENO4 in the structure and function of mouse sperm.
MATERIALS AND METHODS
Materials
All reagents were purchased from Sigma-Aldrich (St. Louis, MO) unless indicated otherwise.
Animals
The Crl:CD1 (ICR) (CD-1) and C57BL/6Ncrl (C57BL/6) mice used in these studies were obtained from Charles River Laboratories (Raleigh, NC), and the 129S6SvEvTac (129) mice were obtained from Taconic (Germantown, NY). All animal studies were approved by the NIEHS Institutional Animal Care and Use Committee and carried out according to U.S. Public Health Service guidelines.
The Eno4 gene-trapped ES cell line Eno4Gt(RRF299)Byg (Eno4+/Gt) purchased from the Mutant Mouse Regional Resource Center(http://www.mmrrc.org) was generated using the pGT0Lxf trap vector [32] and ES cells derived from 129 P2/OlaHsd mice. The ES cells were injected into C57BL/6 blastocysts in our laboratory to produce chimeric mice. Male chimeras were bred to C57BL/6 or 129 female mice and offspring genotyped by PCR and Southern blot. The Eno4+/Gt female and male mice were crossed to produce Eno4Gt/Gt mice. To determine the fertility of Eno4Gt/Gt male mice, three Eno4Gt/Gt males were mated with three C57BL/6 or 129 wild-type (WT) females for 1 mo.
Analysis of Testis and Sperm
For histological analysis, testes from WT and Eno4Gt/Gt mice were fixed in Bouin fixative, dehydrated, and embedded in paraffin by standard procedures. Sections (5 μm) were stained with hematoxylin and eosin.
For scanning electron microscopy (SEM), sperm from WT and Eno4Gt/Gt mice were allowed to settle onto L-lysine-coated cover glass (BD Sciences, San Jose, CA) for 30 min at room temperature (RT). Sperm were incubated with PBS with or without 0.1% Triton X-100 for 15 min. After washing with PBS, sperm were fixed in 5% glutaraldehyde in 0.2 M cacodylate buffer for 8 h at 10°C, critical point dried, coated with gold/palladium, and examined in a Supra 25 FESEM (Carl Zeiss, Thornwood, NY) SEM at 20 KV.
For transmission electron microscopy (TEM), testes from WT and Eno4Gt/Gt mice were fixed in 2% paraformaldehyde (PFA), 2.5% glutaraldehyde, and 0.2% picric acid in 0.1 M sodium cacodylate buffer (pH 7.4) for 8 h at 4°C and postfixed in 1% osmium tetroxide in the same buffer for 1 h at RT. Samples were dehydrated in ethanol and embedded in Polybed 812 epoxy resin. Sections were poststained with uranyl acetate and lead citrate and examined in a LEO EM-910 (Carl Zeiss) TEM at 80 KV. The SEM and TEM used were in the Microscopy Services Laboratory of the Department of Laboratory Medicine and Pathology, University of North Carolina School of Medicine, Chapel Hill.
Sperm motility parameters were measured using computer-assisted sperm analysis (CASA). Sperm from the cauda epididymis were incubated in M2 medium (EMD Millipore Corp., Billerica, MA) at RT and loaded into assay chambers at 5, 60, and 120 min. Sperm tracks (1.5 sec, 30 frames) were captured at 60 Hz using an HTM-IVOS Sperm Analyzer (Hamilton Thorne Inc., Beverly, MA). The parameters during measurements were minimum contrast, 30; minimum cell size, four pixels; STR threshold, 50.0%; VAP cutoff, 10.0 μm/sec; progressive minimal smoothed path velocity (VAP), 50.0 μm/sec; static head size, 0.13–2.43; static head intensity, 0.10–1.52; and static elongation, 5–80. The concentrations of sperm from the cauda epididymis were determined on samples diluted 1:10 in water and counted using a hemocytometer.
β-Galactosidase Histochemistry
Testes from WT and Eno4Gt/Gt mice were fixed for 2 h in 4% PFA in PBS, followed by immersion in 20% sucrose in PBS for 16 h at 4°C. Frozen sections (10 μm) were washed in PBS; immersed briefly in rinse buffer containing 0.1 M phosphate buffer (pH 7.4), 2 mM MgCl2, 0.01% sodium deoxycholate, and 0.02% NP-40; and stained at 37°C overnight in rinse buffer containing 2 mg/ml X-gal, 5 mM K3Fe (CN)6, and 5 mM K4Fe (CN)6. After staining, the slides were washed in PBS, postfixed in 4% PFA in PBS, and observed using Nomarski interference optics.
Antibody Procedures
An antiserum was prepared to a synthetic peptide corresponding to amino acids (aa) 334–353 of ENO4 (N-CLPPPKQETKKGHNGSKRAQP-COOH) conjugated to keyhole limpet hemocyanin by immunizing New Zealand White rabbits (Covance Inc., Denver, PA). Immunoglobulins were precipitated from the serum by saturating with ammonium sulfate at 4°C for 2 h and collected by centrifugation at 24 600 × g for 30 min at 4°C. The pellet was resuspended in 10 mM Tris buffer (pH 7.5) and dialyzed against 10 mM Tris (pH 7.5) overnight at 4°C. The dialysate was affinity purified using CNBr-4B beads (GE Healthcare Life Sciences, Pittsburgh, PA) as recommended by the supplier.
Enolase was localized in sperm from the cauda epididymis that were fixed in 2% PFA in PBS for 15 min at RT, immersed in 50 mM glycine in PBS for 30 min to block free aldehyde groups, and attached to slides as described [33]. After being permeabilized with ice-cold methanol for 1 min, sperm were immunostained with ENO4 or pan-enolase (sc-15343; Santa Cruz Biotechnology, Santa Cruz, CA) antisera and Alexa 488-tagged antiserum to rabbit IgG (Life Technologies Corp., Grand Island, NY). Images were captured with an Axioplan microscope (Carl Zeiss) fitted with fluorescence optics and an EX16C digital camera (DAGE-MTI, Michigan City, IN). They were recorded as TIFF images, and figures were prepared using Photoshop (Adobe, San Jose, CA).
For immunoblotting, testes from WT or Eno4Gt/Gt mice were homogenized in lysis buffer containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 1% Triton X-100, 0.5% NP-40, and one tablet of proteinase inhibitor cocktail (Roche Applied Sciences, Indianapolis, IN) per 10 ml of lysis buffer. After determined the protein concentration, the samples were added to equal volume of 2× sample buffer (4% SDS, 100 mM Tris-HCl [pH 6.8], 20% glycerol, 2% 2-mercaptoethanol, and 0.001% bromophenol blue).
Samples were separated by electrophoresis on 10% ready gels (Bio-Rad, Hercules, CA). Proteins were transferred from the gel to Immobilon-P PVDF membrane (EMD Millipore). Proteins were detected on membranes using antibodies to pan-enolase (Santa Cruz Biotechnology), ENO2 (GTX73150; GeneTex, Inc., Irvine, CA), ENO3 (611196; BD Biosciences, San Jose, CA), PGAM1/2/4 (EB05028; Everest Biotech Ltd, Oxfordshire, U.K.), AKAP4 [34], anti-rabbit IgG β-galactosidase antibody (55976; MP Biomedicals, Solon, OH), and antiserum to ENO4, using the ECL procedure (GE Healthcare Life Sciences) as recommended by the supplier.
To assay enolase solubility, sperm from CD-1 mice were lysed sequentially with NP-40 and 6M urea as described previously [34]. Sperm were lysed initially in a buffer containing 1% NP-40, 10 mM Hepes (pH 7.5), 50 mM NaCl, 10 mM EDTA, 1% NP-40, 0.2 mM PMFS, and 10 μg/ml aprotinin. After 60 min of incubation on ice, samples were centrifuged at 500 × g for 10 min at 4°C and the supernatants collected (NP-40 lysate). The pellets was washed with 50 mM Tris-HCl (pH 8.2) and suspended in buffer containing 6 M urea, 50 mM Tris-HCl (pH 8.0), 10 mM EDTA, 6 M urea, 0.2 mM DTT, and 10 μg/ml aprotinin. After incubation for 120 min on ice, samples were centrifuged at 12 000 × g for 20 min at 4°C, and the supernatants collected (6 M urea lysate) for immunoblotting.
Sperm lysate for immunoprecipitation was prepared as described [34]. The in vitro translated [35S]-labeled proteins (see below) were incubated with 1 mg sperm lysate for 2 h at 4°C. Antibody (10 μg) was added and incubated for 2 h at 4°C, and then Immunopure immobilized protein G beads (Pierce Biotechnology, Rockford, IL) were added and incubated for 2 h at 4°C on a rotating platform. The beads were washed with TBS, mixed with 1× sample buffer, and heated for 5 min at 86°C. After centrifugation at 5000 × g for 3 min at RT, the supernatants were separated by SDS-PAGE, and the gels were immersed in Amplify (GE Healthcare Life Sciences) for 30 min, vacuum dried, and exposed to Hyperfilm MP (GE Healthcare Life Sciences) at −70°C.
In Vitro Translation
The cDNAs encoding ENO4_V1 (aa 1–572), ENO4-N (aa 1–228), ENO4-C (aa 228–618), and full-length PGAM2 (PGAM-FL) were subcloned into the pcDNA3.1 (+)-His/Myc vector (Life Technologies) for in vitro translation. In vitro translation of ENO4_V1, ENO4-N, ENO4-C, and PGAM2-FL was carried out in the TNT coupled reticulocyte lysate system (Promega Corp., Madison, WI) according to the supplier's instructions. Translation was performed for 2 h at 30°C in a volume of 25 μl containing 20 μCi of [35S] methionine (GE Healthcare Life Sciences). After incubation, the labeled proteins were separated from unincorporated [35S] using MicroSpin G-25 columns (GE Healthcare Life Sciences).
Enolase Assay
Sperm from the cauda epididymis of CD-1 mice were lysed in 100 μl of PBS containing 1% Triton X-100, left on ice for 20 min, and centrifuged 12 000 rpm for 5 min at 4°C. The pellet was resuspended in the same volume. Sperm from the cauda epididymis of WT C57BL/6 and Eno4Gt/Gt mice were suspended in buffer containing 2 mM MgSO4, 50 mM imidazol-HCl (pH 6.8), and proteinase inhibitor cocktail (Roche Applied Sciences) and sonicated. Enolase activity was measured as described [35]. The reaction solution containing 1 mM D-(+)-2 phosphoglyceric acid (Toronto Research Chemicals, Inc., North York, ON), 2 mM MgSO4, 0.4 M KCl, and 50 mM imidazol-HCl (pH 6.8) was mixed with 50 μl of sample and enzymatic activity measured at 240-nm wavelength with a UV spectrometer (Life Technologies). Protein concentrations of each sample were measured with Bradford reagent (Bio-Rad).
ATP Assay
Sperm from the cauda epididymis were suspended in 50 μl of PBS after being incubated in M2 medium (EMD Millipore) for 5 min at RT and washed in PBS by centrifugation at 900 × g for 3 min. The sperm pellets were then suspended in 450 μl of preheated extraction buffer (4 mM EDTA/0.1 mM Tris-HCl, pH 7.75) and incubated at 100°C for 2 min. After centrifugation at 9000 × g for 2 min, the supernatant was collected and ATP measured in duplicate 50-μl aliquots using the ATP Bioluminescence Assay Kit CLS II (Roche Applied Sciences). Protein concentrations of the supernatants were measured by spectrophotometry using Bradford reagent (Bio-Rad).
Northern Blot
Total RNA was extracted from various tissues and whole testes of 10- to 35-day-old CD-1 mice or from testes of adult Eno4Gt/Gt and WT mice using TRI-ZOL reagent (Life Technologies). The RNAs were separated by electrophoresis in a 1% agarose gel containing 0.66 M formaldehyde and transferred to Hybond N nylon membrane (GE Healthcare Life Sciences) in 10× saline-sodium citrate (SSC). [32P]-labeled probes were generated using a random primed labeling kit (Roche Applied Sciences) with an Eno4 cDNA (1155 base pairs [bp]) as template. Ribosomal protein 7 (Rpl7) was used as an internal control. Hybridization was performed in Hybrisol I (Chemicon, Purchase, NY) at 42°C overnight. The membrane was washed with 2× SSC containing 0.1% SDS at RT for 10 min, with 0.1× SSC containing 0.1% SDS at 50°C for 10 min, and subjected to autoradiography.
RACE Method
The 5′RACE was performed using a Marathon-ready testis cDNA library and a BD Advantage 2 PCR kit (Clontech, Mountain View, CA) according to the supplier's directions. Adaptor primer 1 (AP1) from the kit and an Eno4 primer (primer 1: 5′-GGA GGT AGA AGG TGG AGT TG-3′), a Gm5506 primer (primer 1: 5′-AAG GAG AAG TAC GGG AAG GAC GCC AC-3′), or a Pgam2 primer (primer 1: 5′-ACG CAC CAC GGG CAC CCA CAT TT-3′) were used for the first PCR amplification. The first PCR amplification products were amplified with adapter primer 2 (AP2) from the kit and an Eno4 primer (primer 2: 5′-TAG AAG GTG GAG TTG AGC AG-3′), a Gm5506 primer (primer 2: 5′-GAG AAG TAC GGG AAG GAC GCC ACC AA-3′), or a Pgam2 primer (primer 2: 5′-GCC CGC TTC AGC ACC GAC GTG TAG CAG A-3′). RACE products were ligated into pGEM-T vector (Promega) for sequencing.
Laser Capture Microdissection
Frozen serial sections (6–9 μm thick) from testes of adult male CD-1 mice were placed on PET foil slides, the slides were fixed with fresh 75% ethanol in DE-PC-treated water and stained with 1% Crystal Violet acetate, and then >1500 cells (pachytene spermatocytes or late spermatids) were harvested within 20 min. The MMI Cellcut Plus (Molecular Machines & Industries Inc., Haslett, MI) laser capture microdissection (LCM) instrument was used for the LCM. Laser power parameters were optimized for the thinnest cut (1 μm or less) with laser speed at 15% and laser power at 53%. Laser focus was optimized for the 8-μm stained sections. As a positive control, a whole testis section was lysed at the end of each 20-min session to check for RNA quality. RNA from the collecting cells and whole testis section were extracted using the Picopure RNA isolation kit (Molecular Devices, Sunnyvale, CA).
Conventional RT-PCR
To determine which of the enolase gene family members are expressed in mouse testis, total RNA was extracted from testis using Trizol reagent according to the manufacturer's instructions (Life Technologies). Total RNA (1 μg) was treated with RNase-free DNase I and reverse transcribed using oligo dT primers. The PCR was carried out using primer pairs shown in Supplemental Table S1 (all Supplemental Data are available online at www.biolreprod.org). The 20-μl reaction mixture was processed using an initial denaturation step at 95°C for 2 min, followed by 35–50 amplification cycles (95°C for 30 sec, 60 or 63°C for 30 sec, and 72°C for 30 sec). Products were separated on 1.2% agarose gels.
Quantitative Real-Time RT-PCR
Total RNA was extracted using Trizol (Life Technologies) from whole testes of 10- to 35-day-old mice and from cells isolated using LCM. The cDNA was synthesized with reverse transcriptase (Life Technologies). Specific primer pairs for Eno4 and Gapdh are shown in Supplemental Table S2. The quantitative real-time RT-PCR (qPCR) analyses were performed using SYBR Green PCR Master Mix reagents (Life Technologies) and the ABI PRISM 7900HT Sequence Detection System (Life Technologies) according to the manufacturer's protocols. The cDNA served as a template in a 50-μl reaction mixture and was processed using an initial denaturation step at 95°C for 10 min, followed by 45 amplification cycles (95°C for 10 sec and 60°C for 1 min) and one dissociation stage cycle (95°C for 15 sec, 60°C for 15 sec, and 95°C for 15 sec). The relative steady-state transcript levels were calculated using cycle time (Ct) values and the following equation: relative quantity = 2−ΔΔCt [36]. The expression levels were normalized using Gapdh as an internal control for each sample. The relative ratios (folds) of transcript levels in each sample were calculated using the lysates of whole testes section values as one. The qPCR reactions were performed in triplicate with each the samples.
Southern Blot
Southern analysis was performed using genomic DNA extracted from testis with lysis buffer containing 60 mM Tris-HCl (pH 8.0), 100 mM EDTA, 0.5% SDS, and 500 μg/ml proteinase K (Life Technologies) at 56°C overnight. The genomic DNA was digested with HindIII, separated on a 0.85% agarose (w/v) gel, and transferred to Hybond-N nylon membrane (GE Healthcare Life Sciences). A 714-bp probe, corresponding to a region of the intron between exons 1 and 2, was amplified with primers: forward (5′GCC CTT CTC CGT TGT TGA 3′)/reverse (5′CTT CTG AGC GCT GTG CCT AC 3′). An 824-bp probe, corresponding to the β-galactosidase-coding region in the gene trap vector (GenBank no. AB255647), was amplified with primers: forward (5′AAG ATC AGG ATA TGT GGC GG 3′)/reverse (5′GAT ACA GCG CGT CGT GAT TA 3′). The probes were labeled by [32P]-dCTP (PerkinElmer, Waltham, MA) using a random primed labeling kit (Roche Applied Sciences).
Statistical Analysis
Statistical analyses were performed using the Student t-test (two sample, assuming unequal variances) calculating the means and SD.
RESULTS
Enolase Is Present in Mouse Sperm
Protein kinase A anchoring protein 4 (AKAP4) is a scaffold protein essential for FS integrity and sperm motility [29, 37]. Two-dimensional PAGE of sperm detergent extracts and proteomics analysis were used to identify proteins that were reduced in sperm lacking AKAP4. Protein spots in WT sperm with 1.5-fold or greater optical density than in AKAP4-null sperm were cored from gels and identified using MALDI mass spectrometry (Supplemental Materials and Methods). One of the proteins identified was enolase (Supplemental Fig. S1), and we hypothesized that it was the novel sperm-specific enolase previously reported in mammalian sperm [12–14, 31].
We determined that enolase is localized to the PP of the mouse sperm flagellum by indirect immunofluorescence using a pan-enolase antibody (Fig. 1, A and B). Immunoblotting studies indicated that the majority of the enolase protein and enzymatic activities were present in the insoluble fractions of mouse sperm extracts (Fig. 1, C and D). Although the immunogen used to generate the pan-enolase antibody corresponded to amino acids 1–300 of human ENO1, mouse ENO1, ENO2, and ENO3 contain regions with high identity to the human sequence (95%, 79%, and 82%, respectively). However, ENO2 and ENO3 were not detected in mouse sperm by immunoblotting with antibodies specific to each (Supplemental Fig. S2), strongly suggesting that they are not present in mouse sperm.
FIG. 1.
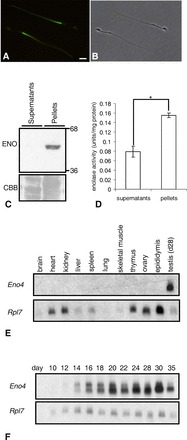
A–C) Enolase in sperm. A) Immunofluorescence localization of enolase by pan-enolase antibody in the PP of the sperm flagellum. B) Phase contrast of same sperm (bar = 8 μm). C) Enolase (ENO) was detected in pellets but not in supernatants of sperm lysates. The same gel stained with Coomassie brilliant blue (CBB) before transfer is shown below to demonstrate protein loading. D) Enolase activity was detected in pellets and at lower levels in supernatants of sperm lysates. Enolase activity in the pellets was significantly higher than in supernatants (*P < 0.05). Data are expressed as means ± SD of three experiments. E and F) Northern blot analysis of Eno4 expression. Northern blot analyses using a probe from the translated region (715-1869 nt) of Eno4 transcript and RNA from various tissues of adult mice (E) or whole testes of Day 10–35 juvenile mice (F). The Rpl7 transcript was used as an internal control.
Eno4 Gene Is Expressed in Mouse Testis
Database searches identified two potential candidate genes for a novel sperm enolase. Sequences present in adult mouse testis EST libraries (UniGene Mm.103154) corresponded to an uncharacterized gene (6430537H07Rik) on chromosome 19. Northern analyses with a probe specific to this gene and RNA from 11 tissues detected a broad ∼4.3-kb band only in adult testis (Fig. 1E). With RNA from testes of mice 10 to 35 days of age, two bands were detected. A ∼6.0-kb band was first seen on Day 12, and an additional ∼4.3-kb band was seen on Day 14 (Fig. 1F). The sequences in this gene encoding the N-terminal 135 amino acid and C-terminal 16 amino acid of the protein were not present in other enolase genes. Based on these and other data contained herein, the 6430537H07Rik gene was renamed Eno4 (MGI: 2441717) by the International Committee on the Standardized Nomenclature for Mice. The alignment of the amino acids of ENO1-4 is shown in Supplemental Figure S3. The identities of mouse ENO1, ENO2, and ENO3 to the mouse ENO4 are 23%, 23%, and 24%, respectively.
Gene tree homology alignments suggest that Eno4 arose during evolution prior to the other three enolase family members (Ensembl ENSGT00550000074560). The Eno4 transcript and two transcript variants expressed in mouse testis were identified using PCR and cDNA sequencing (Fig. 2, A and B; Supplemental Fig. S4A). One transcript variant (Eno4_v1) lacked Eno4 exon 13 (nucleotides 1644–1761), and a second transcript variant (Eno4_v2) encoded an N-terminal truncated protein (Supplemental Fig. S4B). Since the length of the Eno4_v1 variant was about 1.8 kb and the length of the Eno4_v2 variant was about 1.2 kb, the ∼6.0-kb band and the ∼4.3-kb band may be responsible for Eno4_v1 and Eno4_v2 variants, respectively. Both variants had ORFs capable of encoding proteins (Supplemental Fig. S4B). Using PCR with cDNA from germ cells isolated by LCM, the level of Eno4_v1 transcripts was found to be higher in pachytene spermatocytes than in late spermatids, while the level of Eno4_v2 transcripts was higher in late spermatids than in pachytene spermatocytes (Fig. 2C).
FIG. 2.
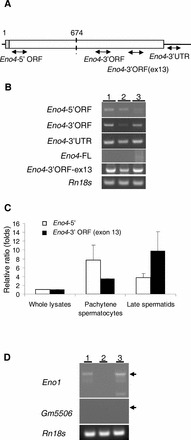
A–C) Expression of Eno1 transcripts and variants. A) The location of primers specific for each of the Eno4 transcripts used in the PCR assays. B) Conventional RT-PCR results with specific primer pairs detecting Eno4 transcript and variants in testes and isolated germ cells. Lanes contained total RNA from testes (lane 1), StaPut-isolated pachytene spermatocytes (lane 2), and StaPut-isolated round spermatids (lane 3). Thirty-five amplification cycles were used for the Eno4 transcript and variants, and the annealing temperature used was 63°C. C) Expression of Eno4 transcripts in pachytene spermatocytes and late spermatids isolated by LCM. Primers specific for each of the Eno4 transcripts was used to assay by qPCR their expression in whole lysates, pachytene spermatocyte, and late spermatids; Eno4-5′, white bars; Eno4-3′ORF (exon 13), black bars. Expression levels were determined as described in Materials and Methods and shown here as ratios (folds) relative to the level on whole lysate, with that level set at one. Data are expressed as means ± SEM. D) Expression of Gm5506 and Eno1 transcripts in mouse testes. Gene specific primers were used to detect Eno1 and Gm5506 using cDNA from testes and isolated germ cells by conventional RT-PCR. The amplification cycles of PCR for Eno1 and Gm5506 were 40 or 50, respectively. The annealing temperature used for Eno1 or Gm5506 was 60°C. Lanes contained total RNA from testes (lane 1), StaPut-isolated pachytene spermatocytes (lane 2), and Sta-Put-isolated round spermatids (lane 3). 18S ribosomal RNA (Rn18s) was used as a control.
Gm5506 Gene Is Expressed in Mouse Testis
The other possible gene identified for sperm enolase was Gm5506 (MGI:3648653) on chromosome 18. It has a nucleotide sequence 99% identical to Eno1 (chromosome 4) and encodes a protein (GenBank LOC433182) 100% identical to ENO1 (MGI: 95393). The Gm5506 gene has two exons, with the ORF restricted to exon 2, while Eno1 has 12 exons, and the 5′ and 3′ flanking sequences of Gm5506 and Eno1 are different, suggesting that Gm5506 is a retrotransposed gene derived from Eno1. Gene tree homology alignments suggest that Gm5506 is present only in mouse (Ensemble ENSMUSG00000059040). In addition, a Gm5506 variant transcript (Gm5506_v1) was identified in testis using PCR and 5′ RACE that contained a sequence corresponding to a region 5′ to Gm5506 on chromosome 18 (Supplemental Fig. S5). PCR results indicated that the Gm5506_v1 transcript level was lower in whole testis than the Eno1 transcript level (Fig. 2D).
ENO4 Is Required for Male Fertility
To determine the role of ENO4, a gene-trapped ES cell line containing a disrupted Eno4 allele (Eno4+/Gt) was used to generate a strain of mutant mice. The gene trap vector was integrated 300 bp downstream of Eno4 exon 1 (Supplemental Fig. S6A). Southern blot analysis confirmed that the gene trap vector was present in genomic DNA from testes of Eno4+/Gt and Eno4Gt/Gt mice (Supplemental Fig. S6B). The absence of Eno4 transcripts in Eno4Gt/Gt mice confirmed that the Eno4 gene was disrupted (Supplemental Fig. S6C). In addition, ENO4 protein was detected in extracts of sperm from WT mice by immunoblotting but not in sperm from Eno4Gt/Gt mice (Supplemental Fig. S6D).
The histochemical product of β-gal activity was detected in the region of the seminiferous epithelium containing pachytene spermatocytes and spermatids in the testes of Eno4Gt /Gt mice (Fig. 3A) but not in WT mice (Fig. 3B). In addition, an antibody to β-gal detected a band in extracts of testis from Eno4Gt/Gt mice but not in testis from WT mice (Supplemental Fig. S6D). There were no differences in body weight, but testis and epididymis weights were significantly lower in Eno4Gt/Gt mice than in WT mice at 12 wk of age (Table 1). In addition, sperm numbers (Table 1), sperm motility (Supplemental Table S3), enolase activity, and ATP levels (Fig. 3, C and D) were significantly lower in sperm from Eno4Gt/Gt mice than in sperm from WT mice. Male Eno4Gt/Gt mice (n = 3) mated with WT females failed to sire offspring, while male WT littermates (n = 3) were fertile (four litters and 27 offspring in 1 mo), and female Eno4Gt/Gt mice mated with WT males were fertile. These results strongly suggest that ENO4 is expressed in meiotic and postmeiotic germ cells and that ENO4 is required for sperm function and male fertility.
FIG. 3.
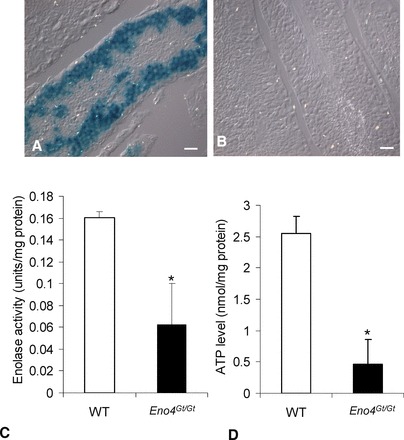
β-galactosidase activity and ATP levels in Eno4Gt/Gt and WT testes. A) β-galactosidase activity was detected in germ cells in testis from Eno4Gt/Gt mice by X-gal staining. B) No β-galactosidase activity was detected in testis from WT mice by X-gal staining. Bars = 25 μm. C) Enolase activity. Sperm were suspended in an imidazole buffer containing 2 mM MgSO4 and 50 mM imidazol-HCl (pH 6.8) and sonicated, and total enolase activity was assayed. Enolase activity in sperm from Eno4Gt/Gt mice was significantly lower than in WT mice (n = 4; *P < 0.05). D) ATP levels. The ATP levels in sperm from Eno4Gt/Gt mice were significantly lower than in WT mice (n = 3; *P < 0.05).
TABLE 1.
Reproductive organ weights and sperm numbers.
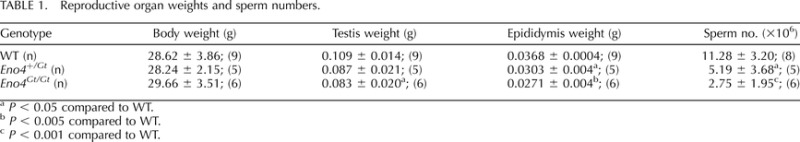
P < 0.05 compared to WT.
P < 0.005 compared to WT.
P < 0.001 compared to WT.
Sperm Morphology Is Abnormal in Eno4Gt/Gt Mice
The majority of sperm from the cauda epididymis of Eno4Gt/Gt mice examined by SEM had obvious morphological defects, frequently including a clubbed flagellum (Fig. 4A). After treating with Triton X-100 to remove the plasma membrane, the middle piece of these sperm was seen to be shortened and the PP components to be wrapped in a coil (Fig. 4B). Other flagellar defects observed include a sharp bend at the midpiece-PP junction, a splayed distal tip, and an abnormally thin PP (Fig. 4C). The heads of most sperm from Eno4Gt/Gt mice appeared comparable to WT sperm (Fig. 4D), but the flagellum usually was abnormal. The midpiece often was incomplete, the cytoskeletal components were disorganized, and the flagellum typically was shortened.
FIG. 4.
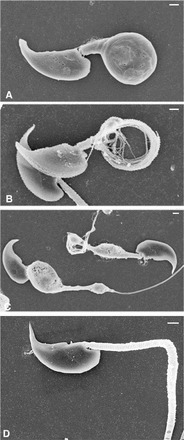
SEM of sperm from Eno4Gt/Gt and WT mice. A and D) Sperm collected from the cauda epididymis and fixed with glutaraldehyde. B and C) Sperm collected from the cauda epididymis extracted with 0.1% Triton X-100 for 5 min before fixation in glutaraldehyde. A–C) Sperm from Eno4Gt/Gt mouse. D) Sperm from WT mouse. Bars = 1 μm.
Most sperm in the cauda epididymis of Eno4Gt/Gt mice examined by TEM had obvious defects in the organization and structure of the PP. The FS ribs were irregular in organization and thickness (Fig. 5A) compared to WT sperm (Fig. 5B), and disorganized aggregates of FS components were seen (Fig. 5C). One or three longitudinal columns often were seen in Eno4Gt/Gt sperm (Fig. 6A), while two FS longitudinal columns occurred invariably in WT sperm (Fig 6C). The typical “9 + 2” microtubule axoneme was disrupted in some flagella, and outer dense fibers in the PP often were displaced and disorganized (Fig. 6B). The annulus, located at the junction between the PP and middle piece of WT sperm (Fig. 5B), often was absent, vestigial, or misplaced in sperm of Eno4Gt/Gt mice (Fig. 6D). These abnormalities were not seen in sperm of Eno4+/Gt mice. CASA analysis showed that sperm motility in Eno4Gt/Gt mice was 2- to 3-fold lower than in WT mice (Supplemental Table S3).
FIG. 5.
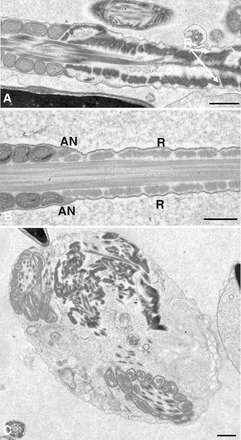
TEM of sperm from Eno4Gt/Gt and WT mice. Sperm collected from cauda epididymis were fixed, sectioned, and examined by TEM. A and C) Sperm from Eno4Gt/Gt mouse. B) Sperm from WT mouse. R, ribs of the FS; AN, annulus. Bars = 0.5 μm.
FIG. 6.
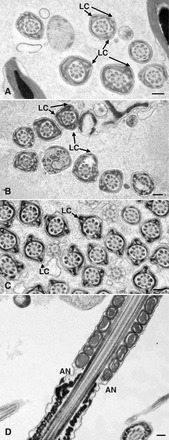
TEM of sperm from Eno4Gt/Gt and WT mice. Sperm collected from cauda epididymis were fixed, sectioned, and examined by TEM. Cross sections (A–C) and sagittal section (D) of sperm flagellum are shown. A, B, and D) Sperm from Eno4Gt/Gt mouse. C) Sperm from WT mouse. LC, longitudinal column; AN, annulus. Bars = 0.2 μm.
Testis Morphology in Eno4Gt/Gt Mice
The general structure and organization of the seminiferous epithelium in Eno4Gt/Gt mice was similar to that in WT mice at the histological level, except for the presence of unusual bodies at the luminal surface of stage VII–VIII tubules (Fig. 7A). These bodies were also seen by TEM (Fig. 7B). However, there were not significant differences in the frequency of TUNEL-positive cells in Eno4Gt/Gt and WT mice (Supplemental Fig. S7). At the TEM level, there were no obvious morphological differences in the axoneme and outer dense fibers in the middle piece region of elongating spermatids in Eno4Gt/Gt and WT mice (Fig. 7C). However, abnormalities were seen in the FS in sperm of Eno4Gt/Gt mice, including the presence of one or three longitudinal columns in elongating spermatids (step 9) and disruption of the organization of FS ribs in condensing spermatids (steps 13–16; Fig. 7D).
FIG. 7.
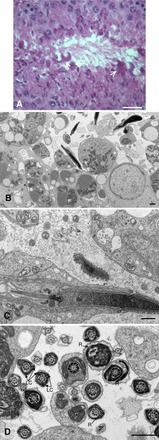
Testis morphology in Eno4Gt/Gt mice. A) Hematoxylin and eosin staining in testis of Eno4Gt/Gt mice. Lobular structures (arrows) were observed along lumen of testis of Eno4Gt/Gt mice. Bar = 25 μm. B, C, and D) Testes were fixed, sectioned, and examined by TEM. Sagittal section (C) and cross section (D) of sperm flagellum are shown. LC, longitudinal column; R, ribs of FS. Bars = 1 μm.
AKAP4 Can Associate with ENO4, Protein Encoded by Gm5506, and Phosphoglycerate Mutase Enzyme
The localization of enolase to the PP region of the flagellum (Fig. 1A) and the reduced level of enolase protein in AKAP4-null sperm compared to WT sperm (Supplemental Fig. S1) suggested that enolase is associated with the FS. When sperm were extracted sequentially with NP-40 and 6M urea, enolase was detected by immunoblotting in the 6 M urea insoluble fraction (Fig. 8), where AKAP4 is located [37]. In addition, coimmunoprecipitation assays using antiserum to AKAP4, testis homogenates, and in vitro synthesized [35S]-ENO4 demonstrated that AKAP4 can associate with the protein encoded by the Eno4_v1 (ENO4_V1) and N-terminal (ENO4-N) regions of ENO4 (Fig. 9B). Although most AKAP4 remains in the insoluble fraction after extraction (Fig. 8), small amounts of AKAP4 apparently remain unincorporated into the FS or are released by the extraction procedure and are available to bind to [35S]-ENO4-C or [35S]-ENO4-V1 and can be precipitated by antiserum to AKAP4 (Fig. 9). Furthermore, the levels of enolase and AKAP4 detected by the pan-enolase and AKAP4 antisera were reduced in Eno4Gt/Gt sperm (Supplemental Fig. S8). The findings that ENO4 was localized to the PP, FS was disrupted in Eno4Gt/Gt sperm, ENO4 was able to associate in vitro with AKAP4, and ENO4 was present in the 6M urea extract suggest that ENO4 and AKAP4 interact in the FS.
FIG. 8.
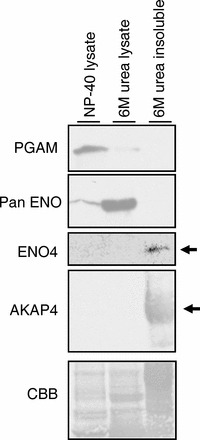
Differences in solubility of PGAM2, enolase, ENO4, and AKAP4. Most of the PGAM2 (detected with PGAM antibody) and some of the enolase (detected with pan-enolase antibody) was detected in the 1% NP-40 sperm lysate. The majority of the enolase was solubilized by 6 M urea. ENO4 and AKAP4 were present in the 6 M urea insoluble sperm fraction.
FIG. 9.
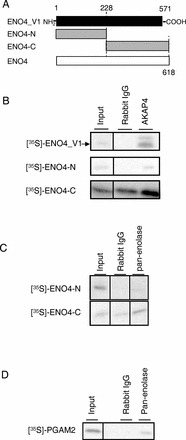
Coimmunoprecipitation assay with ENO4, AKAP4, ENO, and PGAM2. A) Regions of ENO4_V1 (aa 1–571), truncated N-terminal ENO4-N (aa 1–228), and ENO4-C (aa 228–618) used for in vitro synthesized [35S]-labeled probes. B) [35S]-ENO4_V1 and [35S]-ENO4-C were coimmunoprecipitated from sperm lysates with AKAP4 by the antiserum to AKAP4 but not by rabbit IgG. C) [35S]-ENO4-C was coimmunoprecipitated from sperm lysates with ENO1 or GM5506 by the antibody to pan-enolase but not by rabbit IgG. D) [35S]-PGAM2 was coimmunoprecipitated from sperm lysates with ENO1 or GM5506 by the antibody to pan-enolase but not by rabbit IgG.
Because enolase can form homodimers or heterodimers [38, 39], coimmunoprecipitation assays were carried out to determine if heterodimers can form between the proteins encoded by Eno4 with Eno1 or GM5506. Direct interaction between [35S]-ENO4-C and the protein encoded by Eno1 or GM5506 was observed with a coimmunoprecipitation assay using sperm extracts and the pan-enolase antibody (Fig. 9C). In addition, while mouse sperm lack ENO2 and ENO3 (Supplemental Fig. S2), pan-enolase antiserum recognized a band on immunoblots of Eno4Gt/Gt sperm (Supplemental Fig. S8).
The phosphoglycerate mutase (PGAM) enzyme is immediately upstream of enolase in the glycolytic pathway. ESTs for Pgam2 are relatively abundant in spermatocyte and spermatid libraries (UniGene 314657), and expression of Pgam2 in testis was confirmed by PCR and 5′ RACE (data not shown). Direct interaction between [35S]-PGAM2 and the protein encoded by Gm5506 occurred in a coimmunoprecipitation assay using sperm extracts and the pan-enolase antibody (Fig. 9D). Further studies will be necessary to determine if ENO4 and PGAM are associated in the FS.
DISCUSSION
This study confirmed previous proteomic and immunohistochemical studies that an enolase is present in mouse sperm [12, 40, 41]. Enolase was reported to be present in sperm in other species, including human [12–14, 31], and to have novel biochemical and enzymatic properties [12]. We used bioinformatics and molecular biology approaches to identify the previously uncharacterized 6430537H07Rik gene (since renamed Eno4) as a candidate for the novel sperm enolase gene. Mice were generated using ES cells with a gene-trapped Eno4 allele, and male mice homozygous for this mutation (Eno4Gt/Gt) were infertile. The sperm from Eno4Gt/Gt mice had significantly reduced motility, ATP levels, and enolase enzymatic activity as well as a structurally abnormal FS, indicating that Eno4 encodes a novel and essential sperm glycolytic enzyme.
This study indicated that ENO4 is the main enolase in mouse sperm. Low-level enolase activity was present in Eno4Gt/Gt sperm, and a pan-enolase antiserum to a sequence conserved in ENO1, ENO2, and ENO3 recognized a band on immunoblots of Eno4Gt/Gt sperm. However, antisera to ENO2 and ENO3 did not recognize a protein in mouse sperm, suggesting that ENO1 is present in mouse sperm. In addition, a gene trap vector inserted into the first intron of the Eno1 gene was homozygous lethal, but heterozygous males expressed β-gal in the seminiferous epithelium beginning between 16 and 20 days after birth [42]. On the other hand, Gm5506 transcripts are expressed in testis and encode a protein sequence identical to ENO1. This suggests that the ENO1 identified in mouse sperm and testis [40, 41] provides some of the enolase activity in sperm and is the product of Eno1, Gm5506, or both. This is similar to the suggestion that LDHA in the FS [23] provides the low amount of LDH activity in sperm lacking Ldhc [16]. However, additional studies using conditional gene targeting approaches will be necessary to determine if the Gm5506 and/or Eno1 genes encode an enolase with a functional role in mouse sperm.
This study showed that ENO4 is highly resistant to detergent extraction, like the FS-associated glycolytic enzymes GAPDHS [15, 22, 23], ALDOA [4, 23], and PK-S [24]. It also showed that ENO4 can associate in vitro with AKAP4, a highly insoluble component of the FS [34, 37, 43], and that ENO4 and the protein encoded by Eno1 or Gm5506 can associate in vitro and with PGAM. This suggests that ENO4 anchors PGAM to the FS. Previous studies demonstrated that HK1S is tethered to the FS through PFKMS [3] and that LDHC is present in a complex containing other ATP-processing proteins [44]. These findings are notable because glycolytic enzymes within sperm have higher enzymatic activities than individual enzymes in vitro [18, 45]. Furthermore, they are consistent with earlier reports that sperm glycolytic enzymes exist in complexes [19–21] and support the concept that such complexes facilitate the efficient production of ATP required for sperm motility.
Targeted disruption of genes encoding the integral FS sperm glycolytic enzymes GAPDHS [15] and PGK2 [17] did not overtly disrupt sperm structure. However, there were irregularities in the number and placement of the longitudinal columns and the organization of the ribs in Eno4Gt/Gt sperm. The longitudinal columns of the FS form before the ribs [27], suggesting that ENO4 is involved in the initial part of FS assembly. While sperm from the cauda epididymis of Eno4Gt/Gt mice often had a clubbed flagellum, this was seen less frequently in testicular sperm. This suggests that flagella lacking ENO4 are prone to damage during passage through the epididymis, possibly due to effects of shear forces on the structurally altered and weakened FS. The major structural protein of the FS, AKAP4, is integrated relatively late into the FS [43], and the FS of sperm lacking AKAP4 is thin [37]. However, AKAP4 is present in sperm from Eno4Gt/Gt mice (Supplemental Fig. S8, right panel), suggesting that ENO4 is not required for integration of AKAP4 into the FS.
Three Eno1 transcript variants and two Gm5506 transcript variants expressed in testis were identified in this study. Transcript variants for Hk1 [1], Pfkm [3], Aldoa [4], and glucose phosphate isomerase 1 (Gpi1) [46] also have been identified in spermatogenic cells. In addition, transcript variants are present in databases for the spermatogenic cell-specific Gapdhs (Ensembl ENSMUSG00000061099) and Ldhc (Ensembl ENSMUG00000030851) genes. Because of the sequence identity of ENO1 and the protein encoded by Gm5506, it is not clear if one or both have a role in sperm glycolysis. It would require using the conditional gene targeting approach for both genes in combination with the disruption of the Eno4 gene to answer this question. However, the relatively low levels of enolase activity and ATP in Eno4Gt/Gt sperm strongly suggest that the Eno1 and Gm5506 genes are not critical for maintenance of mouse sperm glycolysis.
The structure of the Gm5506 gene, the sequence of its encoded protein, and its absence in other species strongly suggest that it is an orthologue of the Eno1 gene that resulted from a recent retrotransposition event in the mouse genome. While retrotransposition has happened frequently during mammalian evolution [47, 48], it is of course only those that occur in germ cells that are heritable and result in new gene family members [4, 49]. Other retrotransposons that encode glycolytic enzymes present in mouse sperm include Pgk2 [5, 6], Aldoart1, and Aldoart2 [4]. While Pgk2 is a relatively ancient gene present in diverse eutherian mammals, Aldoart1 and Aldoart2 evolved more recently and are found only in rodents [4]. In addition to these known transcript variants, the mouse and human genomes contain a substantial number of retrotransposed genes implicated in the evolution of sperm glycolytic enzymes [46]. The current studies provide further emphasis that glycolysis is essential for energy production in mammalian sperm and that a combination of novel genes and variant transcripts are responsible for producing the enzymes involved in this process. Although the Eno4 gene appears to have appeared early in the history of the enolase gene family, variant transcripts and new genes, such as Gm5506, appear in the male germ cell line, and the genes associated with sperm glycolysis continue to evolve.
Supplementary Material
ACKNOWLEDGMENT
The authors thank Linwood Koonce for excellent animal care, Patricia S. Stockton for help with Laser Capture Microdissection procedures, and Liang-Yu Chen and Yukitomo Arao for helpful comments on the manuscript.
Footnotes
Supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (E.M.E., ES070076).
REFERENCES
- Mori C, Nakamura N, Welch JE, Gotoh H, Goulding EH, Fujioka M, Eddy EM. Mouse spermatogenic cell-specific type 1 hexokinase (mHk1-s) transcripts are expressed by alternative splicing from the mHk1 gene and the HK1-S protein is localized mainly in the sperm tail. Mol Reprod Dev 1998; 49: 374–385. [DOI] [PubMed] [Google Scholar]
- Nakamura N, Shibata H, O'Brien DA, Mori C, Eddy EM. Spermatogenic cell-specific type 1 hexokinase is the predominant hexokinase in sperm. Mol Reprod Dev 2008; 75: 632–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Mori C, Eddy EM. Molecular complex of three testis-specific isozymes associated with the mouse sperm fibrous sheath: hexokinase 1, phosphofructokinase M, and glutathione S-transferase mu class 5. Biol Reprod 2010; 82: 504–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemuganti SA, Bell TA, Scarlett DO, Parker CE, Pardo-Manuel de Villena F, O'Brien DA. Three male germline-specific aldolase A isozymes are generated by alternative splicing and retrotransposition. Dev Biol 2007; 309: 18–31. [DOI] [PubMed] [Google Scholar]
- McCarrey JR, Thomas K. Human testis-specific PGK gene lacks introns and possesses characteristics of a processed gene. Nature 1987; 326: 501–505. [DOI] [PubMed] [Google Scholar]
- Boer PH, Adra CN, Lau YF, McBurney MW. The testis-specific phosphoglycerate kinase gene pgk-2 is a recruited retroposon. Mol Cell Biol 1987; 7: 3107–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millan JL, Driscoll CE, LeVan KM, Goldberg E. Epitopes of human testis-specific lactate dehydrogenase deduced from a cDNA sequence. Proc Natl Acad Sci U S A 1987; 84: 5311–5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JE, Schatte EC, O'Brien DA, Eddy EM. Expression of a glyceraldehyde 3-phosphate dehydrogenase gene specific to mouse spermatogenic cells. Biol Reprod 1992; 46: 869–878. [DOI] [PubMed] [Google Scholar]
- Russell DL, Kim KH. Expression of triosephosphate isomerase transcripts in rat testis: evidence for retinol regulation and a novel germ cell transcript. Biol Reprod 1996; 55: 11–18. [DOI] [PubMed] [Google Scholar]
- Buehr M, McLaren A. An electrophoretically detectable modification of glucosephosphate isomerase in mouse spermatozoa. J Reprod Fertil 1984; 63: 169–173. [DOI] [PubMed] [Google Scholar]
- Fundele R, Winking H, Illmensee K, Jagerbauer E-M. Developmental activation of phosphoglycerate mutase-2 in the testis of the mouse. Dev Biol 1987; 124: 562–566. [DOI] [PubMed] [Google Scholar]
- Edwards YH, Grootegoed JA. A sperm-specific enolase. J Reprod Fertil 1983; 68: 305–310. [DOI] [PubMed] [Google Scholar]
- Gitlits VM, Toh BH, Loveland KL, Sentry JW. The glycolytic enzyme enolase is present in sperm tail and displays nucleotide-dependent association with microtubules. Eur J Cell Biol 2000; 79: 104–111. [DOI] [PubMed] [Google Scholar]
- Force A, Viallard JL, Saez F, Grizard G, Boucher D. Electrophoretic characterization of the human sperm-specific enolase at different stages of maturation. J Androl 2004; 25: 824–829. [DOI] [PubMed] [Google Scholar]
- Miki K, Qu W, Goulding EH, Willis WD, Bunch DO, Strader LF, Perreault SD, Eddy EM, O'Brien DA. Glyceraldehyde 3-phosphate dehydrogenase-S, a sperm-specific glycolytic enzyme, is required for sperm motility and male fertility. Proc Natl Acad Sci U S A 2004; 101: 16501–16506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odet F, Duan C, Willis WD, Goulding EH, Kung A, Eddy EM, Goldberg E. Expression of the gene for mouse lactate dehydrogenase C (Ldhc) is required for male fertility. Biol Reprod 2008; 79: 26–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danshina PV, Geyer CB, Dai Q, Goulding EH, Willis WD, Kitto B, McCarrey JR, Eddy EM, O'Brien DA. Phosphoglycerate kinase 2 (PGK2) is essential for sperm function and male fertility. Biol Reprod 2009; 82: 136–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai C, Okuno M. Glycolysis plays a major role for adenosine triphosphate supplementation in mouse sperm flagellar movement. Biol Reprod 2004; 71: 540–547. [DOI] [PubMed] [Google Scholar]
- Harrison RAP. Glycolytic enzymes in mammalian spermatozoa: activities and stabilities of hexokinase and phosphofructokinase in various fractions from sperm homogenates. Biochem J 1971; 124: 741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey BT, Kayne FJ. Energy metabolism of spermatozoa. V. The Embden-Myerhof pathway of glycolysis: activities of pathway enzymes in hypotonically treated rabbit epididymal spermatozoa. Fertil Steril 1975; 26: 1257–1265. [PubMed] [Google Scholar]
- Gillis BA, Tamblyn TA. Association of bovine sperm aldolase with sperm subcellular components. Biol Reprod 1984; 31: 25–35. [DOI] [PubMed] [Google Scholar]
- Bunch DO, Welch JE, Magyar PL, Eddy EM, O'Brien DA. Glyceraldehyde 3-phosphate dehydrogenase-S protein distribution during mouse spermatogenesis. Biol Reprod 1998; 58: 834–841. [DOI] [PubMed] [Google Scholar]
- Krisfalusi M, Miki K, Magyar PL, O'Brien DA. Multiple glycolytic enzymes are tightly bound to the fibrous sheath of mouse spermatozoa. Biol Reprod 2006; 75: 270–278. [DOI] [PubMed] [Google Scholar]
- Westhoff D, Kamp G. Glyceraldehyde 3-phosphate dehydrogenase is bound to the fibrous sheath of mammalian spermatozoa. J Cell Sci 1997; 110: 1821–1829. [DOI] [PubMed] [Google Scholar]
- Feiden S, Stypa H, Wolfrum U, Wegener G, Kamp G. A novel pyruvate kinase (PK-S) from boar spermatozoa is localized at the fibrous sheath and the acrosome. Reproduction 2007; 134: 81–95. [DOI] [PubMed] [Google Scholar]
- Eddy EM. The spermatozoon. : Neill J. (ed.), Knobil and Neill: The Physiology of Reproduction, vol. 1, 3rd ed. San Diego, Elsevier; 2006: 2–54. [Google Scholar]
- Irons MJ, Clermont Y. Kinetics of fibrous sheath formation in the rat spermatid. Am J Anat 1982; 165: 121–130. [DOI] [PubMed] [Google Scholar]
- Fawcett DW. The mammalian spermatozoon. Dev Biol 1975; 44: 394–436. [DOI] [PubMed] [Google Scholar]
- Eddy EM. The scaffold role of the fibrous sheath. : ERS Roldan, Gomendio M. (eds.), Spermatology. SRF Suppl. 65. Nottingham: Nottingham University Press; 2007: 45–62. [PubMed] [Google Scholar]
- Tracy MR, Hedges SB. Evolutionary history of the enolase gene family. Gene 2000; 259: 129–138. [DOI] [PubMed] [Google Scholar]
- Force A, Viallard JL, Grizard G, Boucher D. Enolase isoforms activities in spermatozoa from men with normospermia and abnormospermia. J Androl 2002; 23: 202–210. [PubMed] [Google Scholar]
- Stanford WL, Cohn JB, Cordes SP. Gene-trap mutagenesis: past, present and beyond. Nat Rev Genet 2001; 2: 756–768. [DOI] [PubMed] [Google Scholar]
- Saling PM, Irons G, Waibel R. Mouse sperm antigens that participate in fertilization. I. Inhibition of sperm fusion with the egg plasma membrane using monoclonal antibodies. Biol Reprod 1985; 33: 515–526. [DOI] [PubMed] [Google Scholar]
- Fenderson BA, Toshimori K, Muller CH, Lane TF, Eddy EM. Identification of a protein in the fibrous sheath of the sperm flagellum. Biol Reprod 1988; 38: 345–357. [DOI] [PubMed] [Google Scholar]
- Baranowski T, Wolna E. Enolase from human muscle. Methods Enzymol 1975; 42: 335–338. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2000; 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Miki K, Willis WD, Brown PR, Goulding EH, Fulcher KD, Eddy EM. Targeted disruption of the Akap4 gene causes defects in sperm flagellum and motility. Dev Biol 2002; 248: 331–342. [DOI] [PubMed] [Google Scholar]
- Craig SP, Day IN, Thompson RJ, Craig IW. Localisation of neurone-specific enolase (ENO2) to 12p13. Cytogenet Cell Genet 1990; 54: 71–73. [DOI] [PubMed] [Google Scholar]
- Feo S, Arcuri D, Piddini E, Passantino R, Giallongo A. ENO1 gene product binds to the c-myc promoter and acts as a transcriptional repressor: relationship with Myc promoter-binding protein 1 (MBP-1). FEBS Lett 2000; 473: 47–52. [DOI] [PubMed] [Google Scholar]
- Baker MA, Letherington L, Reeves GM, Aitken RJ. The mouse sperm proteome characterized via IPG strip prefractionation and LC-MS/MS identification. Proteomics 2008; 8: 1720–1730. [DOI] [PubMed] [Google Scholar]
- Asano A, Nelson JL, Zhang S, Travis AJ. Characterization of the proteomes associating with three distinct membrane raft sub-types in murine sperm. Proteomics 2010; 10: 3494–3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couldrey C, Carlton MB, Ferrier J, Colledge WH, Evans MJ. Disruption of murine alpha-enolase by a retroviral gene trap results in early embryonic lethality. Dev Dyn 1998; 212: 284–292. [DOI] [PubMed] [Google Scholar]
- Brown PR, Miki K, Harper DB, Eddy EM. AKAP4 binding proteins in the fibrous sheath of the sperm flagellum. Biol Reprod 2003; 68: 2241–2248. [DOI] [PubMed] [Google Scholar]
- Odet F, Gabel SA, Williams J, London RE, Goldberg E, Eddy EM. Lactate dehydrogenase C and energy metabolism in mouse sperm. Biol Reprod 2011; 85: 556–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai C, Bergkvist M, Nelson JL, Travis AJ. Sequential reactions of surface-tethered glycolytic enzymes. Chem Biol 2009; 16: 1013–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemuganti SA, Pardo-Manuel de Villena F, O'Brien DA. Frequent and recent retrotransposition of orthologous genes plays a role in the evolution of sperm glycolytic enzymes. BMC Genomics 2010; 11 (285): 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H, Koyanagi KO, Imanishi T, Itoh T, Gojobori T. Frequent emergence and functional resurrection of processed pseudogenes in the human and mouse genomes. Gene 2007; 389: 196–203. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Harrison PM, Liu Y, Gerstein M. Millions of years of evolution preserved: a comprehensive catalogue of the process pseudogenes in the human genome. Genome Res 2003; 13: 2541–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinckenbosch N, Depanloup I, Kaessmann H. Evolutionary fate of retroposed gene copies in the human genome. Proc Natl Acad Sci U S A 2006; 103: 3220–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


