Abstract
Disorders of sex development in the human population range in severity from mild genital defects to gonadal sex reversal. XY female development has been associated with heterozygous mutations in several genes, including SOX9, WT1 and MAP3K1. In contrast, XY sex reversal in mice usually requires complete absence of testis-determining gene products. One exception to this involves T-associated sex reversal (Tas), a phenomenon characterized by the formation of ovotestes or ovaries in XY mice hemizygous for the hairpin-tail (Thp) or T-Orleans (TOrl) deletions on proximal mouse chromosome 17. We recently reported that mice heterozygous for a null allele of Map3k4, which resides in the Thp deletion, exhibit XY ovotestis development and occasional gonadal sex reversal on the sensitized C57BL/6J-YAKR (B6-YAKR) genetic background, reminiscent of the Tas phenotype. However, these experiments did not exclude the possibility that loss of other loci in the Thp deletion, or other effects of the deletion itself, might contribute to Tas. Here, we show that disruption to Sry expression underlies XY gonadal defects in B6-YAKR embryos harbouring the Thp deletion and that a functional Map3k4 bacterial artificial chromosome rescues these abnormalities by re-establishing a normal Sry expression profile. These data demonstrate that Map3k4 haploinsufficiency is the cause of T-associated sex reversal and that levels of this signalling molecule are a major determinant of the expression profile of Sry.
INTRODUCTION
In humans, 46,XY gonadal dysgenesis (46,XY GD) is characterized by abnormal testis determination and is an example of a wider class of abnormalities known as disorders of sex development (DSD). In cases of pure or complete gonadal dysgenesis (CGD), the testes are absent and bilateral streak gonads are observed. Molecular genetic studies of individuals with 46,XY GD and CGD have played a critical role in the identification of human testis-determining genes such as SRY (1), SOX9 (2,3), WT1 (4) and SF1 (5). In all autosomal cases, mutation of one copy of the gene can be sufficient to disrupt testis development in some individuals.
XY gonadal sex reversal in the mouse has been described in cases of homozygous deletion of a number of testis-determining genes, including Sox9 (6–8), Fgf9 (9), Fgfr2 (10,11), Cbx2 (M33) (12,13) and Wt1(+KTS) (14). The C57BL/6J (B6) background is sensitized to disruptions to testis determination due to the relatively delayed expression of testis-determining genes and higher levels of expression of ovary-determining genes and, in most sex-reversing mouse mutants examined, B6 increases the amount of ovarian tissue that forms in mutant XY embryos (15–17). This sensitivity increases still further if an AKR/J-derived Y chromosome (YAKR) is present, resulting in sex reversal even in cases of haploinsufficiency of testis-determining genes, such as Gata4 and Fog2 (18). These latter studies suggest that no inherent species differences exist in sensitivity to disruption to testis determination by altered gene dosage; rather, differing sensitivity to gene dosage in both humans and mice is dependent on genetic background.
A number of additional cases of XY sex reversal in the mouse depend on the presence of a Mus domesticus Y chromosome and the C57BL/6J genetic background: so called B6.YDOM sex reversal (19,20). One variant of this phenomenon, known as T-associated sex reversal (Tas), requires a domesticus Y chromosome from the AKR/J strain (YAKR) in combination with a dominant brachyury (T) mutation, either hairpin-tail (Thp) or T-Orleans (TOrl), on the B6 background (21–23). The latter T mutations are both caused by overlapping deletions of proximal mouse chromosome 17. Analysis of embryonic gonads revealed that XY Thp/+ individuals develop ovotestes or ovaries, the extent of sex reversal depending on the number of backcrosses performed to B6 (21,23). Ovotestes tend to resolve as development proceeds and form hypoplastic testes, permitting the identification of occasional fertile males (23). Studies of XY gonad development in sex-reversed TOrl carriers also reveal delayed and significantly reduced levels of Sry expression (24). Thus, it has been suggested that haploinsufficiency of a gene (or genes) located in a region common to the Thp and TOrl deletions, and required for normal timing and levels of Sry expression, is the cause of Tas. However, alternative explanations have been proposed, including the possibility that delayed growth in embryos simply harbouring chromosomal deletions such as Thp and TOrl might disrupt testis determination, irrespective of the specific gene content of the deletions (25).
We have previously described XY embryonic gonadal sex reversal in embryos lacking Map3k4, which encodes a signalling molecule in the mitogen-activated protein kinase (MAPK) pathway (26). Embryos lacking MAP3K4 exhibit reduced Sry expression at 11.5 days post coitum (dpc) and this results in failure to up-regulate expression of genes functioning in the genetic pathway of testis development, including the key gene, Sox9. These data suggested a role for MAPK signalling in the control of Sry expression, and thus testis determination. Moreover, the report of mutations in the human MAP3K1 gene associated with 46,XY DSD and CGD suggests that MAPK signalling is an important component of human testis determination too (27). More recently, we have described data suggesting that disruption to a number of elements of a GADD45γ/MAP3K4/p38 MAPK signalling pathway in somatic cells of the newly formed mouse gonad can result in a delay in Sry expression that results in XY gonadal sex reversal (28).
Map3k4 maps close to T on the proximal region of mouse chromosome 17 and a genetic test indicated that it resides in the region deleted in Thp (26). Thus, Map3k4 is an outstanding candidate for a gene, haploinsufficiency for which causes T-associated sex reversal. Mice heterozygous for a targeted allele of Map3k4 (Map3k4tm1Flv) on B6-YAKR display testicular hypoplasia and occasional XY female development, similar to that described in Tas mice. Moreover, heterozygous XY embryos frequently developed ovotestes by 14.5 dpc, consistent with a delay and/or reduction in expression of Sry and Sox9, although gene expression in the developing gonads at the sex-determining stage (11.5 dpc) was not tested in affected individuals (26). Thus, loss of a single copy of Map3k4 would be predicted to contribute significantly to the XY sex reversal and ovotestis development observed in Thp/+ C57BL/6J-YAKR mice. However, whether Map3k4 is the only gene in the Thp deletion that predisposes to XY sex reversal when haploinsufficient is unclear from these experiments. That question can only be addressed by determining whether additional functional copies of Map3k4 can rescue the sex-reversal phenotype of Thp/+ C57BL/6J-YAKR mice.
Here, we characterize the Thp deletion in more detail and estimate its minimal size and gene content. We also examine the Thp/+, B6.YAKR sex-reversal phenotype further, showing that sex reversal in this strain is more pronounced than in Map3k4tm1Flv/+, B6.YAKR mice. We show that reduced Sry expression at 11.5 dpc underlies XY gonadal defects in Thp/+ embryos. We then describe an in vivo gain-of-function genetic experiment using a mouse line carrying a functional Map3k4 bacterial artificial chromosome (BAC) transgene. When introduced onto the Thp/+, B6.YAKR genetic background this transgene completely rescues the Tas sex-reversal phenotype by re-establishing a normal Sry expression profile. We conclude from these data that: (i) Map3k4 haploinsufficiency is necessary for T-associated sex reversal, (ii) some interaction between reduced Map3k4 dosage and another genetic deficiency in Thp/+ mice exacerbates the sex-reversal defect in Thp/+ embryos when compared with Map3k4tm1Flv/+ embryos and (iii) Map3k4 is a major determinant of the timing and levels of Sry expression in developing mouse gonads.
RESULTS
Further molecular characterization of the Thp deletion
In order to get a more precise estimate of the gene content of the Thp deletion, we characterized its genomic location in greater detail. We used a strategy based on quantitative polymerase chain reaction (qPCR) to determine the copy number of genes on proximal chromosome 17 in the vicinity of T, Map3k4 and Igf2r (all known to reside in the deletion) in Thp/+ C57BL/6J-YAKR mice and thereby determine the distal limits of the deletion (see Materials and Methods; Supplementary Material, Fig. S1 for more details). The position of known deleted loci allowed us to estimate a minimal size for the deletion of 5.56 Mb. This minimal region (based on Ensembl mouse genome database build GRCm38.p1) is predicted to contain 62 genes. A maximum size of the deletion, based on non-deleted loci, is 6.34 Mb, containing 84 genes. None of the genes within either the calculated maximal or minimal deleted regions, with the exception of Map3k4, has been implicated in testis determination. A more precise delineation of the deletion break points was prevented by regions that contain highly homologous sequences, immediately adjacent to the minimally deleted region (Supplementary Material, Fig. S1). Illegitimate recombination between these repeated regions may offer a mechanistic explanation of the original deletion event itself.
Ovotestis and ovary development in XY Thp/+ and Map3k4tm1Flv/+ embryos at 14.5 dpc
We began our phenotypic analysis of T-associated sex reversal in XY Thp/+ and Map3k4tm1Flv/+ mice by examining embryonic sexual development at 14.5 dpc, a stage at which it is easy to detect the presence of ovotestes (Fig. 1). These have previously been described as a common feature of XY Thp/+ embryos and are thought to be a cause of the hypoplastic testes observed in adult males (21). Thp/+ embryos examined were produced after five generations or more of crossing Thp/+ males to B6, allowing a more appropriate comparison with the embryonic gonadal phenotype of Map3k4tm1Flv/+ heterozygotes, which were congenic on B6.
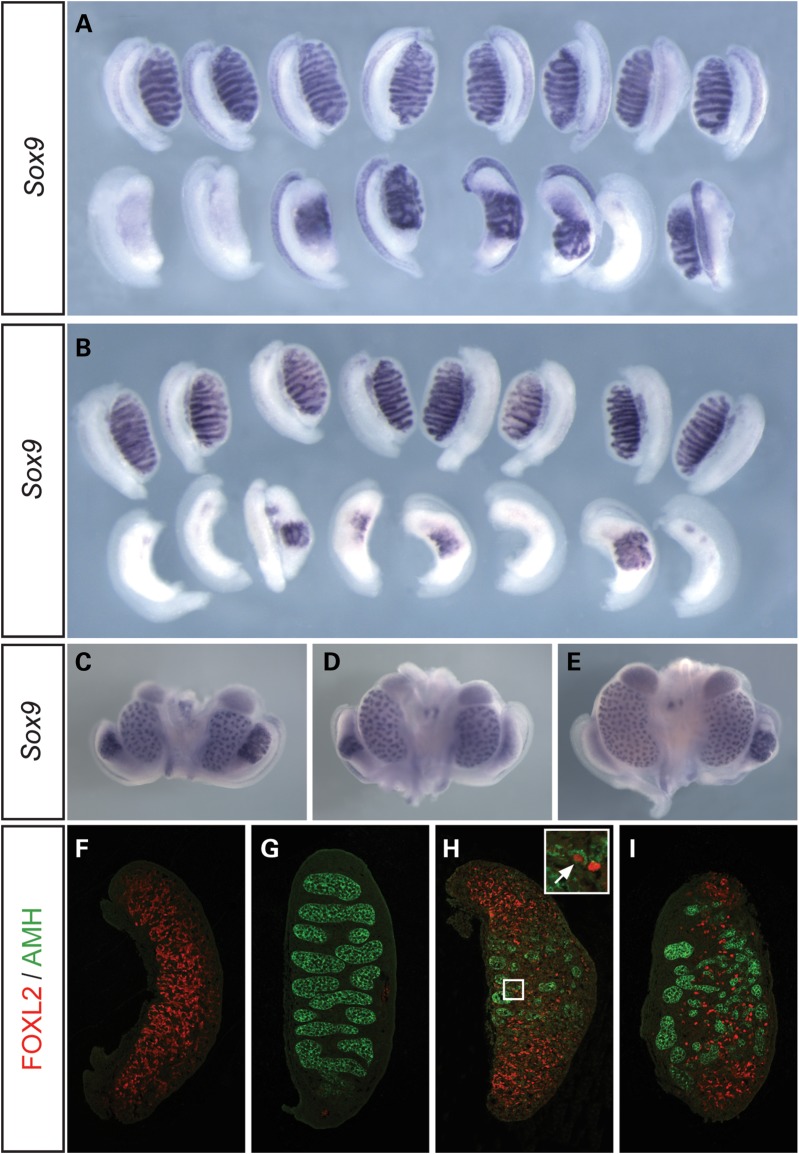
Figure 1.
Abnormalities of testis development in XY Map3k4tm1Flv/+ and XY Thp/+ embryos at 14.5 dpc. (A) WMISH analysis of wild-type (+/+, upper row) and Map3k4tm1Flv/+ (lower row) embryonic gonads on B6.YAKR using a Sox9 probe, showing XY ovary or ovotestis development in Map3k4tm1Flv/+ embryos at 14.5 dpc. (B) Sox9 WMISH analysis of wild-type (upper row) and Thp/+ (lower row) embryonic gonads on B6.YAKR, revealing ovary and ovotestis development in XY Thp/+ embryos with apparent enhanced severity. (C–E) Sox9 WMISH of Map3k4tm1Flv/+ embryonic urogenital organs at 14.5 dpc revealing variable Sox9 expression and gonad morphology even within individual embryos. Embryos may show ovotestes on both sides (C), ovotestis and ovary development on the right and left side, respectively, as in (D), or the converse (E). (F–I) Immunostaining of gonadal tissue sections for FOXL2 (red) and anti-müllerian hormone (AMH) (green) at 14.5 dpc. Images show control (+/+) XX gonad exhibiting only FOXL2 expression (F); control XY gonad exhibiting only AMH expression in testis cords (G); Map3k4tm1Flv/+ gonad with central AMH expression and polar FOXL2 expression (H). Note FOXL2-positive cells in interstitium of the central testicular region and an individual cell positive for both FOXL2 and AMH (inset, white arrow); (I) Thp/+ ovotestis tissue section with cellular distribution similar to (H) .
At 14.5 dpc, analysis of gonadal Sox9 expression by whole mount in situ hybridization (WMISH) revealed varying levels of transcript in both Map3k4tm1Flv/+ and Thp/+ embryos (Fig. 1A and B). Map3k4tm1Flv/+ gonads were frequently ovotestes and exhibited Sox9 expression at high levels only in the centre of the gonad, in association with testis cords, as previously described (26). Dysgenetic testes, with varying numbers of irregular testis cords, were commonly observed. Occasionally, there was greatly reduced Sox9 expression and the gonad had an overtly ovarian appearance (Fig. 1A). Analysis of XY Thp/+ embryos at the same stage revealed a higher frequency of gonads with an overt ovarian morphology and negligible Sox9 expression, in addition to ovotestes (Fig. 1B). Sox9-positive regions of these ovotestes had less well-defined testis cord structures than XY Map3k4tm1Flv/+ equivalents. Interestingly, in addition to variation between individual embryos, variation in the degree of Sox9 expression between gonads from individual XY Map3k4tm1Flv/+ embryos was also common, indicating a non-genetic contribution to the variability in the gonadal phenotypes observed (Fig. 1C–E). The apparent increased frequency and severity of gonadal sex reversal observed in Thp/+ embryos when compared with Map3k4tm1Flv/+ is consistent with our observations of the adult phenotype of these lines. Map3k4tm1Flv/+ and Map3k4byg/+ XY males commonly had reduced testis weights but were infrequently scored as phenotypic females (∼15%, data not shown). In contrast, XY Thp/+ mice exhibited a higher frequency of XY sex reversal (∼40%, data not shown). Morphological abnormalities of the reproductive tracts were common in these adult XY Thp/+ mice, including those scored as males (data not shown).
Previous studies of ovotestis development in other genetic models have revealed a fine-grain mixing of ovarian and testicular cell types beneath the coarser testicular and ovarian domains identified morphologically: FOXL2-positive cells have been identified in testicular interstitial regions of B6 XYPOS ovotestes and SOX9- or anti-müllerian hormone (AMH)-positive cells detected in ovarian regions of XX Wt1:Sox9 transgenic ovotestes (29,30). These studies revealed a mutually exclusive pattern of FOXL2 and SOX9 protein expression at the cellular level, reflecting the antagonistic relationship of these molecules and their respective pathways during sex determination. However, a third study reported co-expression of FOXL2 and AMH at the cellular level in ovotestes generated by haploinsufficiency for testis-determining genes in the B6.YAKR gonad (16). We examined Map3k4tm1Flv/+ and Thp/+ ovotestes at 14.5 dpc with antibodies to FOXL2 and AMH in order to examine the distribution of cells committed to the ovarian and testicular fates, respectively (Fig. 1F–I). AMH was detected exclusively in wild-type XY gonads at 14.5 dpc, and FOXL2 exclusively in XX wild-type gonads. In ovotestes from both strains the central, testicular regions exhibited strong AMH expression in testis cords, whilst the gonadal poles showed equally high levels of FOXL2. However, large numbers of FOXL2-positive cells were observed in the testicular region, although these were excluded from well-formed testis cords. AMH-positive cells were not detected in the ovarian poles. Very rarely, an individual cell appeared to express both FOXL2 and AMH (inset, Fig. 1H).
Sox9 and Sry expression is disrupted in Thp/+ and Map3k4tm1Flv/+ embryonic gonads
To determine the molecular basis of ovotestis and ovary development in these mutant lines we began by carefully quantifying expression of Sox9 during the sex-determining period of gonadogenesis, ∼11.25–12.5 dpc, using qRT-PCR. For comparison, we also included samples from B6.YB6 and B6.YAKR wild-type gonads at the same stages. This expression profiling revealed that Sox9 transcript levels rise slowly in B6.YAKR, reaching peak levels ∼25 tail somites (ts) (Fig. 2A). Equivalent levels of Sox9 were observed earlier, at 19–20 ts, in B6.YB6 gonads. This altered profile is consistent with delayed testis cord formation in B6.YAKR gonads, which is associated with transient ovotestis development at 14.5 dpc (24). In Map3k4tm1Flv/+, B6.YAKR gonads, Sox9 levels also remained very low until 18 ts; levels began to rise from ∼19 ts but failed to reach those observed at 25 ts in B6.YAKR controls. In contrast to this, Sox9 levels remained comparatively low in Thp/+ gonads at all stages (Fig. 2A). However, in contrast, strong expression of the ovarian somatic marker Wnt4 was detectable at 18 ts in Thp/+ gonads (Supplementary Material, Fig. S2). Variation in expression levels between individual tissue samples from the same mutant strains was common and likely underlies the phenotypic variability observed at 14.5 dpc. Combining data from multiple samples masks this variation.
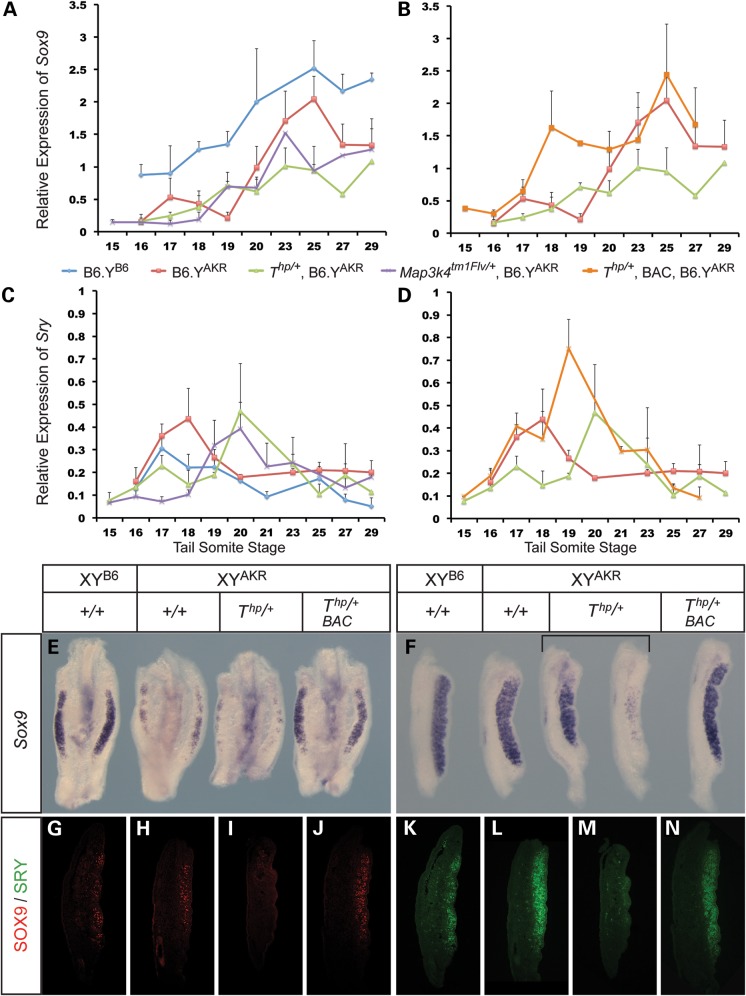
Figure 2.
Expression profiling of Sox9 and Sry expression in XY Map3k4tm1Flv/+, Thp/+ and transgenic Thp/+ embryonic gonads. (A and B) Relative expression levels of Sox9 for different genotypes (indicated by colour key beneath plots) between 15 ts and 29 ts stages. (C and D) Relative expression levels of Sry in gonads from the same genotypic classes and stages as in A and B. Note the delay in peak Sry expression in Map3k4tm1Flv/+ and Thp/+ gonads (C) and the enhanced, early Sry peak in transgenic Thp/+ gonads (D). Error bars indicate standard error mean. (E) Sox9 WMISH analysis at 20 ts in genotypes as indicated. (F) Sox9 WMISH analysis at 27 ts (∼12.5 dpc) in genotypes as indicated. (G–J) Anti-SOX9 antibody immunostaining of control B6.YB6 (G), control B6.YAKR (H), Thp/+ (I) and rescued (BAC transgenic) Thp/+ (J) gonadal tissue sections at 18 ts. (K–N) Anti-SRY antibody immunostaining of the same series of 18 ts gonadal sections as in G–J.
These data suggest a delay in execution of the sex-determining programme in B6.YAKR gonads in comparison to B6.YB6, as evidenced by Sox9 expression, that is exacerbated further in Map3k4tm1Flv/+ and even more so in Thp/+ embryonic gonads. WMISH analyses at 11.75 dpc (20 ts, Fig. 2E) and 12.5 dpc (27 ts, Fig. 2F) using a Sox9 probe corroborated the qRT-PCR data, revealing a reduction or delay in Sox9 expression in Thp/+ that, in some gonads, is associated with spatial restriction of expression or negligible detectable levels by 12.5 dpc (Fig. 2F).
Given the reported role for SRY in regulation of Sox9 expression in the mouse (31), and the significance attributed to reduction of, and delay in, Sry expression in models of B6.YDOM sex reversal (32,33), we next examined the expression profile of Sry in both mutant strains and controls by qRT-PCR at multiple developmental stages. In wild-type B6.YAKR gonads, Sry levels rose to a peak at 18 ts before slowly declining (Fig. 2C). Interestingly, a similar profile was observed in B6.YB6 gonads but the SryB6 allele peaked earlier (at 17 ts) and did not reach the levels of SryAKR. This reduced expression of SryB6 compared with SryAKR in the B6 genetic background has been previously reported (33,34). Moreover, SryB6 expression dropped to lower levels than observed for SryAKR, which remained higher throughout the time-course. In mutant Map3k4tm1Flv/+ and Thp/+ gonads, Sry rose slowly and did not reach levels equivalent to peak expression in wild-type B6.YAKR gonads until ∼20 ts; levels at 18 ts in both mutant strains were significantly reduced in comparison to B6.YAKR wild-type controls (Fig. 2C). Profiles of SryAKR expression in Thp/+ and Map3k4tm1Flv/+ gonads did not differ substantially, although levels rose slightly earlier in Map3k4tm1Flv/+ gonads, resulting in significantly higher levels in this strain at 19 ts. This may account for an apparently reduced severity of sex-reversal phenotype in the Map3k4tm1Flv/+ Tas model. As in the case for Sox9, considerable variation between individual samples was observed. Overall, these data indicate a delay in Sry expression reaching peak levels in Map3k4tm1Flv/+ and Thp/+ gonads on B6.YAKR, likely accounting for defects in testis determination in these strains.
Functional Map3k4 BAC transgenes rescue defects in Thp/+ gonads
Map3k4 haploinsufficiency results in many of the gonadal abnormalities associated with Tas, although with an apparent reduced frequency and severity of sex reversal. In order to test whether Map3k4 haploinsufficiency is absolutely required for Tas, or whether loss of other loci in the Thp deletion is also sufficient to generate a sex reversal phenotype, we performed a gain-of-function experiment utilizing a previously reported Map3k4 BAC transgenic line, c06 (28) (Fig. 3), and lines harbouring an independent Map3k4 BAC, k10 (data not shown). Each BAC contained the complete Map3k4 transcriptional unit and varying amounts of 5′ and 3′ flanking DNA (for details see Materials and Methods). No other complete transcriptional units are predicted to reside within either BAC clone.
Figure 3.
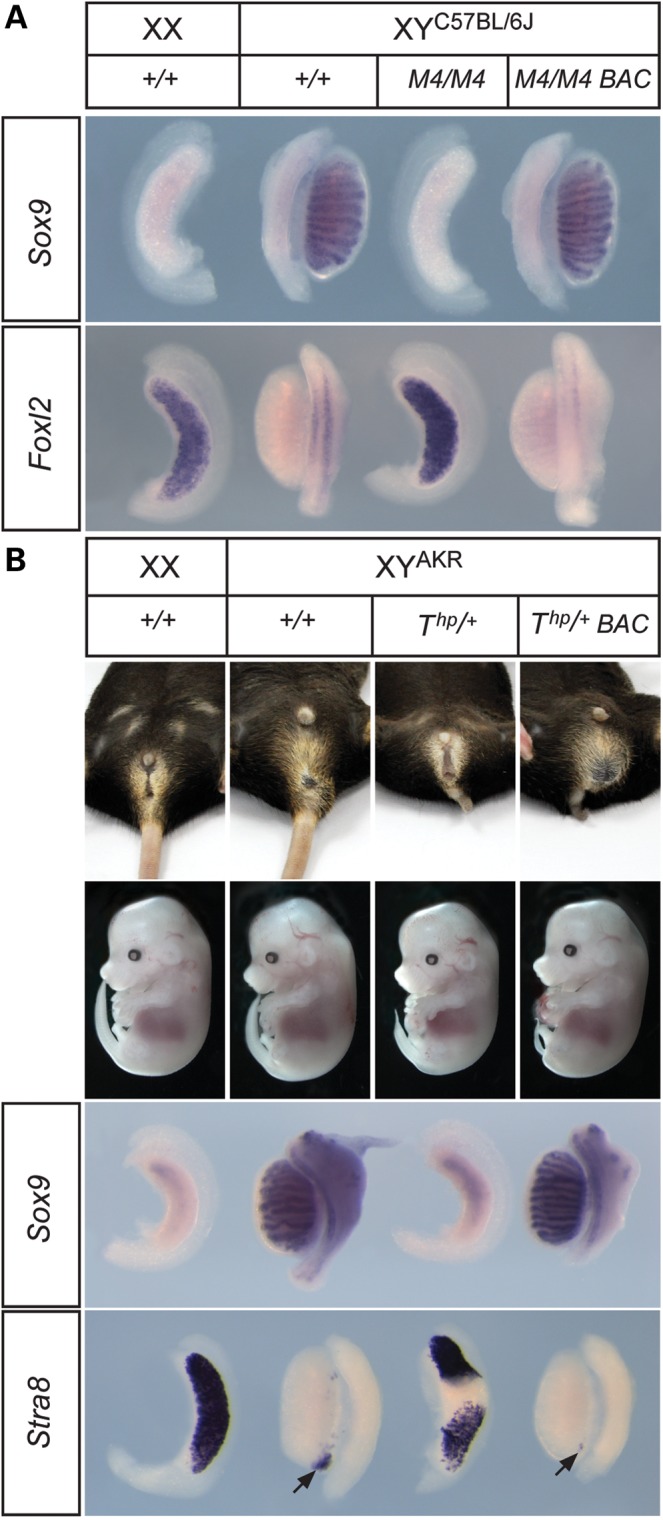
A functional Map3k4 BAC transgene rescues abnormalities of XY gonad development in Thp/+ embryos. (A) WMISH analysis with Sox9 (upper row) and Foxl2 (lower row) of gonads at 14.5 dpc in control (+/+) and Map3k4-deficient (M4/M4) and transgenic Map3k4-deficient (M4/M4, BAC) embryos, revealing rescue of XY gonadal sex reversal in the latter class. (B) External genitalia of adult mice with genotypes indicated (upper row) and gross morphology of 14.5 dpc embryos with genotypes indicated (beneath). Note abnormal tails in Thp/+ animals. Lower panels show WMISH analyses with Sox9 (upper) and Stra8 (lower) of embryonic gonads at 14.5 dpc in genotypes indicated, revealing rescue of gonadal sex reversal in transgenic XY Thp/+ embryos. Arrows indicate clusters of Stra8-positive cells of varying sizes at the poles of the gonads.
To test for functionality of the BACs in these lines, we generated embryos homozygous for the Map3k4tm1Flv targeted knockout that also carried a BAC transgene. XY embryos homozygous for Map3k4tm1Flv exhibit gonadal sex reversal, containing ovaries at 14.5 dpc (Fig. 3A). However, when the c06 and k10 (data not shown) BAC transgenes were present, Map3k4tm1Flv homozygous XY embryos (n = 3) developed testes with a normal morphology. WMISH with Sox9 also revealed that Sertoli cell differentiation had occurred normally and examination of Foxl2 expression demonstrated absence of ovarian tissue (Fig. 3A). Thus, we conclude that Map3k4 expressed from either BAC transgene was sufficient to rescue the sex reversal phenotype in these homozygous mutant embryos. Prior qRT-PCR analysis of Map3k4 expression levels in transgenic and wild-type gonads at 11.5 dpc had revealed that expression levels were ∼5-fold higher in transgenic (line c06) XY gonads at 11.5 dpc (28).
We then generated mice transgenic for the c06 BAC transgene that also carried the Thp deletion on B6.YAKR. In contrast to the XY gonadal abnormalities usually associated with Thp on this genetic background, adult transgenic XY Thp/+ mice were all fertile males with overtly normal morphology of the reproductive organs (data not shown) and external genitalia (Fig. 3B). We then generated embryos in the control and rescued genotypic classes. Examination of XY embryonic gonads at 14.5 dpc also confirmed that the presence of the c06 BAC transgene suppressed ovotestis development in XY Thp/+ embryos and thus rescued the Tas phenotype. Marker gene expression in gonads from rescued embryos revealed prominent Sox9 expression in testis cords at 14.5 dpc and absence of Stra8, a marker of meiotic germ cells that is ovary-specific at this stage (Fig. 3B). Equivalent data were generated using the k10 BAC transgene (data not shown). From these data we conclude that haploinsufficiency for Map3k4 is key to development of the Tas phenotype.
To examine the molecular basis of rescue of the Tas phenotype during the sex-determining period we performed qRT-PCR profiling of Sry and Sox9 in Thp/+ embryos harbouring the BAC transgene. Examination of Sry expression in transgenic Thp/+, B6.YAKR gonads revealed a positive shift in its profile in comparison to non-transgenic controls, with levels at 17 ts already comparable to the peak of expression observed in Thp/+ gonads at 20 ts (Fig. 2D). Moreover, levels continued to rise to a peak at 19 ts in transgenic Thp/+ gonads, greater even than those observed in control, wild-type B6.YAKR gonads, before dropping quickly to negligible levels by 27–29 ts. Sox9 expression profiles in transgenic gonads also revealed early, robust expression, in contrast to Thp/+ gonads (Fig. 2B). Indeed, levels comparable to those found in non-transgenic, wild-type B6.YAKR controls at 23–25 ts were observed by 18 ts. This early surge of Sox9 expression in rescued transgenic gonads, in addition to accounting for rescue of the Thp/+ phenotype, might account for an apparent acceleration in testis development in these samples when compared with wild-type B6.YAKR controls, as evidenced by the reduced expression of Stra8 at the gonadal poles at 14.5 dpc (Fig. 3B). WMISH analysis also confirmed the rescue of robust Sox9 expression in transgenic Thp/+ gonads at 19 ts (Fig. 2E) and 12.5 dpc (Fig. 2F). Moreover, immunostaining of sectioned gonads with an anti-SOX9 antibody revealed increased numbers of SOX9-positive cells in transgenic XY Thp/+ gonads at ∼18 ts in comparison to non-transgenic gonads from Thp/+ littermates (Fig. 2G–J). Finally, detection of SRY protein at the same stage also revealed large numbers of SRY-positive cells in rescued gonads when compared with Thp/+ equivalents (Fig. 2K–N). Thus, we conclude that transgenic expression of Map3k4 rescues the Tas phenotype by re-establishing early, robust Sry and Sox9 expression profiles that are disrupted in Thp/+ embryos.
DISCUSSION
T-associated sex reversal (Tas) in the mouse was first described before the identification of Sry as the mammalian testis-determining gene (21). Subsequent studies on one particular Tas model phenotype, B6 TOrl/+ XYAKR, suggested severely reduced Sry expression as a possible cause of sex reversal (24). This, along with the development of normal testes in B6 TOrl/+ XYAKR mice when a Mus musculus-derived Sry transgene is present, indicates that the SryAKR allele is deficient on the B6 TOrl/+ genetic background due to abnormalities in the regulation of its expression. By inference, given the overlapping nature of the TOrl and Thp deletions, a similar mechanistic explanation was thought to account for the Thp/+ version of Tas.
Here, we confirm that Sry expression is disrupted in B6 Thp/+ XYAKR embryos. Moreover, we demonstrate that MAP3K4 function is central to a mechanistic account of Tas by showing that Map3k4 haploinsufficiency on B6.YAKR also results in disrupted Sry expression in a manner similar to that observed in Thp/+ gonads and that expression of Map3k4 from a BAC transgene is sufficient to rescue Tas in the mouse. XY transgenic Thp/+ gonads show no signs of ovotestis or ovary development. This rescue is the result of early and robust Sry expression in transgenic gonads, in contrast to deletion carriers that up-regulate Sry expression later, with predicted detrimental consequences for the activation of Sox9 expression and testis determination. Whilst, we cannot formally exclude a direct, positive effect of the Map3k4 BAC transgene on testis-determining genes downstream of Sry, such as Sox9, the normal sexual development and fertility of XX transgenic females does not support such a model. Our previous studies on testis determination suggest that MAP3K4 signalling in gonadal somatic cells activates p38 MAPK and results in phosphorylation of GATA4, a transcription factor known to regulate expression of Sry (28). Thus, transgenic expression of Map3k4 is presumed to rescue defects in this signalling pathway in XY Thp/+ gonads. We cannot formally exclude, however, other positive impacts of MAP3K4 on Sry expression or function.
The rescue of complete and partial XY gonadal sex reversal by the Map3k4 transgene establishes Map3k4 as the key gene in the Thp deletion in the aetiology of T-associated sex reversal (Tas). How, therefore, do we account for the apparent increased frequency and severity of XY gonadal sex reversal in Thp/+ carriers when compared with that observed in embryos heterozygous for a Map3k4 null allele alone? In both cases, the copy number of Map3k4 is the same. Three possible explanations are: (i) haploinsufficiency for another locus (or loci) in the Thp deletion, exacerbating the disruption to XY gonad development; (ii) disruption to a testis-determining gene outside, but close to, the Thp deletion in the form of a position effect or (iii) the presence of the deletion itself contributing to disruption to testis determination. All of these potential explanations rely on a form of genetic interaction that is asymmetrically dependent on Map3k4 haploinsufficiency, since the proposed genetic deficits do not disrupt testis determination in the presence of the transgene. We cannot exclude any of these possibilities. With respect to the first possibility, our novel data concerning the extent of the Thp deletion indicate that it contains no other genes implicated in testis determination. However, further investigation may reveal a novel testis-determining function encoded by the deleted region. In the case of the second possibility, we have recently reported the role of p38α and p38β MAPK in mouse testis determination (28). p38α (Mapk14) and p38δ (Mapk13) map to proximal mouse chromosome 17, but we have shown that neither resides within the Thp deletion region and their expression levels are not significantly disrupted in XY Thp/+ gonads at 11.5 dpc (Supplementary Material, Fig. S3). With respect to the third explanation, the mere presence of hemizygous deletions in the mouse, quite apart from their actual gene content, has previously been invoked as an explanation of sex-reversal phenomena such as Tas, due to possible effects on embryonic growth (25). Here, we show that any contribution that the Thp deletion itself makes to Tas, independently of specific gene dosage alterations, is conditional on the haploinsufficiency of Map3k4.
The re-establishment of early, robust Sry expression at 17 ts in transgenic Thp/+ B6.YAKR gonads, and the increased levels attained when compared with wild-type XYAKR controls, in addition to explaining the phenotypic rescue itself, might account for a small, but detectable, acceleration of testis development in these embryos when compared with B6.YAKR wild-type controls (compare, e.g. testis cord morphology and Stra8 expression pattern in gonads shown in Fig. 3B). Transgenic gonads have an appearance and Stra8 expression profile more reminiscent of B6 gonads at the same stage, rather than B6 XYAKR. This phenomenon suggests that MAP3K4 activity may be a limiting factor in the degree of up-regulation of Sry expression on the B6.YAKR background. This observed transgenic rescue is also not apparently hampered by SRY protein isoform differences between the SryB6 and SryAKR alleles, which, along with altered Sry expression, have been implicated in the delay in B6 XYAKR testis determination (32). While the disruption to Sry expression in Thp/+ and Map3k4tm1Flv/+ gonads is relatively subtle, it is well established that only minor alterations to the timing or levels of Sry expression can radically alter phenotypic consequences: ovary, ovotestes or dysgenetic testes can all arise depending on subtle changes to Sry expression control (17,33,35–37).
Phenotypic variation, even between individuals that are essentially isogenic, is a feature of the Map3k4tm1Flv/+ B6.YAKR gonadal phenotype. Crucially, asymmetric gonad morphology within individual embryos is observed, ruling out a residual genetic contribution to this variability, at least in these individuals. This variability is likely related to the reduction of MAP3K4 functionality to near-threshold levels, and the consequence of this for levels and timing of Sry expression during gonadogenesis. Developmental stochasticity is a research area receiving increased attention (38). A number of features of stochastic systems may be relevant in this context. Firstly, strong fluctuations can be a consequence of low numbers in any system. It is known that SRY-positive cells arise in the centre of the developing gonad at ∼10.75 dpc in low numbers (39). These first, potentially stochastic, events in gonadal SRY expression are likely to result in subsequent recruitment of further cells to the testis-determining (pre-Sertoli cell) pathway. In other systems, gene regulatory networks act to buffer stochastic variability in gene expression, and loss of particular genes can produce pronounced phenotypic variation, including variable penetrance, that reveals underlying stochasticity of gene expression (40,41). Loss of a single copy of Map3k4, on an already sensitized genetic background, appears to exacerbate inherent stochasticity in the testis-determining mechanism, although the precise events governed by this stochasticity are unknown. Phenotypic plasticity and stochastic noise have also been linked to epigenetic mechanisms (42). The significance of epigenetic regulation of the Y chromosome, in particular the Sry locus, for testis determination has recently been underlined by a report of XY gonadal sex reversal in mice lacking the histone demethylase, JMJD1A (43). Finally, these observations in mice suggest that stochasticity, in addition to variation in genetic background, may underlie some of the phenotypic variability between individuals with 46,XY pure or CGD caused by mutation of autosomal or X-linked human genes functioning in early steps of testis determination (27).
MATERIALS AND METHODS
Mouse mutants utilized and genotyping
All animal experimentation was approved by the Animal Welfare and Ethical Review Body at MRC, Harwell, and mice used in this study were bred with licensed approval from the UK Home Office (PPL 30/2877). Mice harbouring a targeted null mutation of Map3k4 (Map3k4tm1Flv) have been previously described (44,45). Map3k4tm1Flv/+ mice were maintained on the C57BL/6J background and genotyped as previously described (26). Thp/+ mice were also maintained on C57BL/6J, and carriers identified by the distinctive shortened or kinked tail. This scoring method was verified by the use of a copy-number assay. Real-time PCR using Taqman chemistry permitted a copy number call of the brachyury gene (T), which is within the Thp deletion. Amplification of T was normalized relative to the endogenous control Dot1L. The ABI 7500 fast SDS software was used to determine the relative copy number of T. This approach was also employed to test other genes for residency within the deleted region. Primer sequences for all loci tested are shown in Supplementary Material, Table S1.
In experiments using the BAC transgene, the presence of the BAC was confirmed by amplification from the chloramphenicol resistance gene in the backbone of the BACe3.6 vector using the following primer pair: 5′-GCGTGTTACGGTGAAAACCT-3′ and 5′-GGGCACCAATAACTGCCTTA-3′.
Adult mice and embryos were sexed by a PCR assay that simultaneously amplifies the Ube1y1 and Ube1x genes, using the following primer pair: 5′-TGGATGGTGTGGCCAATG-3′ and 5′-CACCTGCACGTTGCCCTT-3′ (46). The presence of the YAKR chromosome was confirmed by a PCR assay as previously described (26).
BAC transgenesis
Identification of BAC clones: NOD/MrkTac BAC clones bQ279c06 (c06) and bQ285k10 (k10) were sourced from the Centre for Applied Genomics, Toronto, Canada. c06 comprised 178.7 kb of DNA including 78.7 kb of DNA 5′ and 8.8 kb 3′ of the Map3k4 transcriptional unit. Clone k10 was 139.7 kb in length, including 30.3 kb 5′ and 9.4 kb 3′ of Map3k4. No other complete transcriptional units are predicted to reside in these BACs. The NOD/MrkTac mouse BAC library end sequences were mapped to the NCBIM37 mouse assembly using SSAHA alignment of the reads. No amino acid differences exist between the NOD/MrkTac- and C57BL/6J-encoded versions of MAP3K4. BAC DNA was prepared and injected into C57BL/6J 1-cell embryos to produce transgenic founders as described in (47).
Generation of embryos and expression analyses
Noon on the day of the copulatory plug was counted as 0.5 dpc. Embryos were staged accurately based on the number of tail somites or limb and gonad morphology. WMISH analysis of embryonic tissues was performed as previously described (48,49). Probes for Sox9 (50), Sry (51), Wnt4 and Stra8 (26,52) have been previously described. A 540 bp Foxl2 probe was generated using primers 5′-AGAACGTGTCTGGTCGCTCT-3′ and 5′-GATCCGGGGAAATTTGTTTT-3′.
Quantitative RT-PCR
Total RNA was extracted using RNeasy plus micro kit (Qiagen) from gonads separated from the mesonephros. RT was carried out with 150 ng of total RNA using the high-capacity cDNA RT kit (Applied Biosystem). qRT–PCR was performed with Fast SYBR Green Master Mix (Life technologies) on a 7500 Fast Real-Time PCR system (Applied Biosystem). RNA expression levels were normalized to those of Hrpt1 (endogenous control) using the ΔΔCt method. At least three samples for each genotype were analysed. Primer sequences are shown in Supplementary Material, Table S1.
Immunohistochemistry
Antibodies to the following proteins were utilized in this study: SOX9 (Millipore, #AB5535); AMH (Santa Cruz, #sc28912); FOXL2 (a kind gift from Dagmar Wilhelm and Peter Koopman). A polyclonal anti-SRY antibody was generated by first expressing a poly-histidine-tagged recombinant protein containing amino acids 82–395 of mouse SRY (UniProt accession no. Q05738) in Escherichia coli BL21 (DE3). Protein was purified using TALON resin (Clontech), mixed sufficiently with Freund's complete adjuvant to give a suspension and then injected intradermally into guinea pig (female, Hartley). Anti-SRY antibody was affinity-purified from serum using antigen. Immunostaining was performed on sectioned, paraffin wax-embedded tissue using the above primary antibodies (1:100) and Alexa Fluor 594 (FOXL2, SOX9) or 488 (AMH, SRY) conjugated secondary antibodies (1:200). Images were captured using a Zeiss 710 multiphoton microscope.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the United Kingdom Medical Research Council by Core funding to A.G. at the Mammalian Genetics Unit, Harwell (MC_A390_5RX50). Funding to pay the Open Access publication charges for this article was provided by the Medical Research Council.
Supplementary Material
ACKNOWLEDGEMENTS
We thank staff of the Mary Lyon Centre (MLC) for animal husbandry support, in particular Jackie Harrison and Lee Kent in Ward 5. We thank Peter Koopman and Dagmar Wilhelm for kindly providing anti-FOXL2 antibody. We thank Kevin Glover for help with photography and Steve Thomas for assistance with figure production, Jim Humphreys and Dave Shipston in the necropsy facility of the MLC for support with tissue collection, staff of the MLC histology facility for sectioning and staining, staff of the GEMS facility for genotyping and sequencing and Martin Fray and his staff in the FESA Core for rederivations and the transgenic facility for pronuclear injections.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Berta P., Hawkins J.R., Sinclair A.H., Taylor A., Griffiths B.L., Goodfellow P.N., Fellous M. Genetic evidence equating SRY and the testis-determining factor. Nature. 1990;348:448–450. doi: 10.1038/348448A0. [DOI] [PubMed] [Google Scholar]
- 2.Wagner T., Wirth J., Meyer J., Zabel B., Held M., Zimmer J., Pasantes J., Bricarelli F.D., Keutel J., Hustert E., et al. Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell. 1994;79:1111–1120. doi: 10.1016/0092-8674(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 3.Foster J.W., Dominguez-Steglich M.A., Guioli S., Kwok C., Weller P.A., Stevanovic M., Weissenbach J., Mansour S., Young I.D., Goodfellow P.N., et al. Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature. 1994;372:525–530. doi: 10.1038/372525a0. [DOI] [PubMed] [Google Scholar]
- 4.Barbaux S., Niaudet P., Gubler M.C., Grunfeld J.P., Jaubert F., Kuttenn F., Fekete C.N., Soulerreau-Therville N., Thibaud E., Fellous M., et al. Donor splice-site mutations in WT1 are responsible for Frasier syndrome. Nat. Genet. 1997;17:467–470. doi: 10.1038/ng1297-467. [DOI] [PubMed] [Google Scholar]
- 5.Achermann J.C., Ito M., Hindmarsh P.C., Jameson J.L. A mutation in the gene encoding steroidogenic factor-1 causes XY sex reversal and adrenal failure in humans. Nat. Genet. 1999;22:125–126. doi: 10.1038/9629. [DOI] [PubMed] [Google Scholar]
- 6.Chaboissier M.C., Kobayashi A., Vidal V.I., Lutzkendorf S., van de Kant H.J., Wegner M., de Rooij D.G., Behringer R.R., Schedl A. Functional analysis of Sox8 and Sox9 during sex determination in the mouse. Development. 2004;131:1891–1901. doi: 10.1242/dev.01087. [DOI] [PubMed] [Google Scholar]
- 7.Barrionuevo F., Bagheri-Fam S., Klattig J., Kist R., Taketo M.M., Englert C., Scherer G. Homozygous inactivation of Sox9 causes complete XY sex reversal in mice. Biol. Reprod. 2006;74:195–201. doi: 10.1095/biolreprod.105.045930. [DOI] [PubMed] [Google Scholar]
- 8.Lavery R., Lardenois A., Ranc-Jianmotamedi F., Pauper E., Gregoire E.P., Vigier C., Moreilhon C., Primig M., Chaboissier M.C. XY Sox9 embryonic loss-of-function mouse mutants show complete sex reversal and produce partially fertile XY oocytes. Dev. Biol. 2011;354:111–122. doi: 10.1016/j.ydbio.2011.03.029. [DOI] [PubMed] [Google Scholar]
- 9.Colvin J.S., Green R.P., Schmahl J., Capel B., Ornitz D.M. Male-to-female sex reversal in mice lacking fibroblast growth factor 9. Cell. 2001;104:875–889. doi: 10.1016/s0092-8674(01)00284-7. [DOI] [PubMed] [Google Scholar]
- 10.Kim Y., Bingham N., Sekido R., Parker K.L., Lovell-Badge R., Capel B. Fibroblast growth factor receptor 2 regulates proliferation and Sertoli differentiation during male sex determination. Proc. Natl. Acad. Sci. U S A. 2007;104:16558–16563. doi: 10.1073/pnas.0702581104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bagheri-Fam S., Sim H., Bernard P., Jayakody I., Taketo M.M., Scherer G., Harley V.R. Loss of Fgfr2 leads to partial XY sex reversal. Dev. Biol. 2008;314:71–83. doi: 10.1016/j.ydbio.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 12.Katoh-Fukui Y., Tsuchiya R., Shiroishi T., Nakahara Y., Hashimoto N., Noguchi K., Higashinakagawa T. Male-to-female sex reversal in M33 mutant mice. Nature. 1998;393:688–692. doi: 10.1038/31482. [DOI] [PubMed] [Google Scholar]
- 13.Katoh-Fukui Y., Miyabayashi K., Komatsu T., Owaki A., Baba T., Shima Y., Kidokoro T., Kanai Y., Schedl A., Wilhelm D., et al. Cbx2, a polycomb group gene, is required for Sry gene expression in mice. Endocrinology. 2012;153:913–924. doi: 10.1210/en.2011-1055. [DOI] [PubMed] [Google Scholar]
- 14.Hammes A., Guo J.K., Lutsch G., Leheste J.R., Landrock D., Ziegler U., Gubler M.C., Schedl A. Two splice variants of the Wilms’ tumor 1 gene have distinct functions during sex determination and nephron formation. Cell. 2001;106:319–329. doi: 10.1016/s0092-8674(01)00453-6. [DOI] [PubMed] [Google Scholar]
- 15.Munger S.C., Aylor D.L., Syed H.A., Magwene P.M., Threadgill D.W., Capel B. Elucidation of the transcription network governing mammalian sex determination by exploiting strain-specific susceptibility to sex reversal. Genes Dev. 2009;23:2521–2536. doi: 10.1101/gad.1835809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Correa S.M., Washburn L.L., Kahlon R.S., Musson M.C., Bouma G.J., Eicher E.M., Albrecht K.H. Sex reversal in C57BL/6J XY mice caused by increased expression of ovarian genes and insufficient activation of the testis determining pathway. PLoS Genet. 2012;8:e1002569. doi: 10.1371/journal.pgen.1002569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warr N., Greenfield A. The molecular and cellular basis of gonadal sex reversal in mice and humans. WIREs Dev. Biol. 2012;1:559–577. doi: 10.1002/wdev.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bouma G.J., Washburn L.L., Albrecht K.H., Eicher E.M. Correct dosage of Fog2 and Gata4 transcription factors is critical for fetal testis development in mice. Proc. Natl. Acad. Sci. U S A. 2007;104:14994–14999. doi: 10.1073/pnas.0701677104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eicher E.M., Washburn L.L., Whitney J.B., Morrow K.E. Mus poschiavinus Y chromosome in the C57BL/6J murine genome causes sex reversal. Science. 1982;217:535–537. doi: 10.1126/science.7089579. [DOI] [PubMed] [Google Scholar]
- 20.Albrecht K.H., Eicher E.M. DNA sequence analysis of Sry alleles (subgenus Mus) implicates misregulation as the cause of C57BL/6J-Y(POS) sex reversal and defines the SRY functional unit. Genetics. 1997;147:1267–1277. doi: 10.1093/genetics/147.3.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Washburn L.L., Eicher E.M. Sex reversal in XY mice caused by dominant mutation on chromosome 17. Nature. 1983;303:338–340. doi: 10.1038/303338a0. [DOI] [PubMed] [Google Scholar]
- 22.Washburn L.L., Eicher E.M. Normal testis determination in the mouse depends on genetic interaction of locus on chromosome 17 and the Y chromosome. Genetics. 1989;123:173–179. doi: 10.1093/genetics/123.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Washburn L.L., Lee B.K., Eicher E.M. Inheritance of T-associated sex reversal in mice. Genet. Res. 1990;56:185–191. doi: 10.1017/s001667230003528x. [DOI] [PubMed] [Google Scholar]
- 24.Washburn L.L., Albrecht K.H., Eicher E.M. C57BL/6J-T-associated sex reversal in mice is caused by reduced expression of a Mus domesticus Sry allele. Genetics. 2001;158:1675–1681. doi: 10.1093/genetics/158.4.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cattanach B.M. Sex-reversed mice and sex determination. Ann. N. Y. Acad. Sci. 1987;513:27–39. doi: 10.1111/j.1749-6632.1987.tb24996.x. [DOI] [PubMed] [Google Scholar]
- 26.Bogani D., Siggers P., Brixey R., Warr N., Beddow S., Edwards J., Williams D., Wilhelm D., Koopman P., Flavell R.A., et al. Loss of mitogen-activated protein kinase kinase kinase 4 (MAP3K4) reveals a requirement for MAPK signalling in mouse sex determination. PLoS Biol. 2009;7:e1000196. doi: 10.1371/journal.pbio.1000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pearlman A., Loke J., Le Caignec C., White S., Chin L., Friedman A., Warr N., Willan J., Brauer D., Farmer C., et al. Mutations in MAP3K1 cause 46,XY disorders of sex development and implicate a common signal transduction pathway in human testis determination. Am. J. Hum. Genet. 2010;87:898–904. doi: 10.1016/j.ajhg.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Warr N., Carre G.A., Siggers P., Faleato J.V., Brixey R., Pope M., Bogani D., Childers M., Wells S., Scudamore C.L., et al. Gadd45gamma and Map3k4 interactions regulate mouse testis determination via p38 MAPK-mediated control of Sry expression. Dev. Cell. 2012;23:1020–1031. doi: 10.1016/j.devcel.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilhelm D., Washburn L.L., Truong V., Fellous M., Eicher E.M., Koopman P. Antagonism of the testis- and ovary-determining pathways during ovotestis development in mice. Mech. Dev. 2009;126:324–336. doi: 10.1016/j.mod.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gregoire E.P., Lavery R., Chassot A.A., Akiyama H., Treier M., Behringer R.R., Chaboissier M.C. Transient development of ovotestes in XX Sox9 transgenic mice. Dev. Biol. 2011;349:65–77. doi: 10.1016/j.ydbio.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sekido R., Lovell-Badge R. Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature. 2008;453:930–934. doi: 10.1038/nature06944. [DOI] [PubMed] [Google Scholar]
- 32.Albrecht K.H., Young M., Washburn L.L., Eicher E.M. Sry expression level and protein isoform differences play a role in abnormal testis development in C57BL/6J mice carrying certain Sry alleles. Genetics. 2003;164:277–288. doi: 10.1093/genetics/164.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bullejos M., Koopman P. Delayed Sry and Sox9 expression in developing mouse gonads underlies B6-Y(DOM) sex reversal. Dev. Biol. 2005;278:473–481. doi: 10.1016/j.ydbio.2004.11.030. [DOI] [PubMed] [Google Scholar]
- 34.Lee C.H., Taketo T. Normal onset, but prolonged expression, of Sry gene in the B6.YDom sex-reversed mouse gonad. Dev. Biol. 1994;165:442–452. doi: 10.1006/dbio.1994.1266. [DOI] [PubMed] [Google Scholar]
- 35.Capel B., Rasberry C., Dyson J., Bishop C.E., Simpson E., Vivian N., Lovell-Badge R., Rastan S., Cattanach B.M. Deletion of Y chromosome sequences located outside the testis determining region can cause XY female sex reversal. Nat. Genet. 1993;5:301–307. doi: 10.1038/ng1193-301. [DOI] [PubMed] [Google Scholar]
- 36.Hiramatsu R., Matoba S., Kanai-Azuma M., Tsunekawa N., Katoh-Fukui Y., Kurohmaru M., Morohashi K., Wilhelm D., Koopman P., Kanai Y. A critical time window of Sry action in gonadal sex determination in mice. Development. 2008;136:129–138. doi: 10.1242/dev.029587. [DOI] [PubMed] [Google Scholar]
- 37.Kashimada K., Koopman P. Sry: the master switch in mammalian sex determination. Development. 2010;137:3921–3930. doi: 10.1242/dev.048983. [DOI] [PubMed] [Google Scholar]
- 38.Oates A.C. What’s all the noise about developmental stochasticity? Development. 2011;138:601–607. doi: 10.1242/dev.059923. [DOI] [PubMed] [Google Scholar]
- 39.Sekido R., Bar I., Narvaez V., Penny G., Lovell-Badge R. SOX9 is up-regulated by the transient expression of SRY specifically in Sertoli cell precursors. Dev. Biol. 2004;274:271–279. doi: 10.1016/j.ydbio.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 40.Raj A., van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raj A., Rifkin S.A., Andersen E., van Oudenaarden A. Variability in gene expression underlies incomplete penetrance. Nature. 2010;463:913–918. doi: 10.1038/nature08781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pujadas E., Feinberg A.P. Regulated noise in the epigenetic landscape of development and disease. Cell. 2012;148:1123–1131. doi: 10.1016/j.cell.2012.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuroki S., Matoba S., Akiyoshi M., Matsumura Y., Miyachi H., Mise N., Abe K., Ogura A., Wilhelm D., Koopman P., et al. Epigenetic regulation of mouse sex determination by the histone demethylase Jmjd1a. Science. 2013;341:1106–1109. doi: 10.1126/science.1239864. [DOI] [PubMed] [Google Scholar]
- 44.Chi H., Lu B., Takekawa M., Davis R.J., Flavell R.A. GADD45beta/GADD45gamma and MEKK4 comprise a genetic pathway mediating STAT4-independent IFNgamma production in T cells. EMBO J. 2004;23:1576–1586. doi: 10.1038/sj.emboj.7600173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chi H., Sarkisian M.R., Rakic P., Flavell R.A. Loss of mitogen-activated protein kinase kinase kinase 4 (MEKK4) results in enhanced apoptosis and defective neural tube development. Proc. Natl. Acad. Sci. U S A. 2005;102:3846–3851. doi: 10.1073/pnas.0500026102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Warr N., Siggers P., Bogani D., Brixey R., Pastorelli L., Yates L., Dean C.H., Wells S., Satoh W., Shimono A., et al. Sfrp1 and Sfrp2 are required for normal male sexual development in mice. Dev. Biol. 2009;326:273–284. doi: 10.1016/j.ydbio.2008.11.023. [DOI] [PubMed] [Google Scholar]
- 47.Gardiner W.J., Teboul L. Overexpression transgenesis in mouse: pronuclear injection. Methods Mol. Biol. 2009;561:111–126. doi: 10.1007/978-1-60327-019-9_8. [DOI] [PubMed] [Google Scholar]
- 48.Grimmond S., Van Hateren N., Siggers P., Arkell R., Larder R., Soares M.B., de Fatima Bonaldo M., Smith L., Tymowska-Lalanne Z., Wells C., et al. Sexually dimorphic expression of protease nexin-1 and Vanin-1 in the developing mouse gonad prior to overt differentiation suggests a role in mammalian sexual development. Hum. Mol. Genet. 2000;9:1553–1560. doi: 10.1093/hmg/9.10.1553. [DOI] [PubMed] [Google Scholar]
- 49.Cox S., Smith L., Bogani D., Cheeseman M., Siggers P., Greenfield A. Sexually dimorphic expression of secreted frizzled-related (SFRP) genes in the developing mouse Mullerian duct. Mol. Reprod. Dev. 2006;73:1008–1016. doi: 10.1002/mrd.20507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wright E., Hargrave M.R., Christiansen J., Cooper L., Kun J., Evans T., Gangadharan G., Greenfield A., Koopman P. The Sry-related gene Sox-9 is expressed during chondrogenesis in mouse embryos. Nat. Genet. 1995;9:15–20. doi: 10.1038/ng0195-15. [DOI] [PubMed] [Google Scholar]
- 51.Bullejos M., Koopman P. Spatially dynamic expression of Sry in mouse genital ridges. Dev. Dyn. 2001;221:201–205. doi: 10.1002/dvdy.1134. [DOI] [PubMed] [Google Scholar]
- 52.Warr N., Bogani D., Siggers P., Brixey R., Tateossian H., Dopplapudi A., Wells S., Cheeseman M., Xia Y., Ostrer H., et al. Minor abnormalities of testis development in mice lacking the gene encoding the MAPK signalling component, MAP3K1. PLoS One. 2011;6:e19572. doi: 10.1371/journal.pone.0019572. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.