Abstract
The routine study of human malaria liver-stage biology in vitro is hampered by low infection efficiency of human hepatocellular carcinoma (HCC) lines (<0.1%), poor understanding of steady-state HCC biology, and lack of appropriate tools for trace sample analysis. HC-04 is the only HCC that supports complete development of human malaria parasites. We hypothesized that HCCs are in various intermediate stages of the epithelial-mesenchymal transition (EMT) and HC-04s retain epithelial characteristics that permit infection. We developed a facile analytical approach to test this hypothesis viz. the HC-04 response to hepatocyte growth factor (HGF). We used online two-dimensional liquid chromatography tandem mass spectrometry (2D-LC-MS/MS) to quantify protein expression profiles in HC-04 pre-/post-HGF treatment and validated these results by RT-qPCR and microscopy. We successfully increased protein identification efficiency over offline-2D methods by 12-fold, using less sample material, allowing robust protein quantification. We observed expected up-regulation and down-regulation of EMT protein markers in response to HGF, but also unexpected cellular responses. We also observed that HC-04 is generally more susceptible to HGF-mediated signaling than what was observed for HepG2, a widely used, but poor malaria liver stage-HCC model. Our analytical approach to understanding the basic biology of HC-04 helps us understand the factors that may influence its utility as a model for malaria liver-stage development. We observed that HC-04 treatment with HGF prior to the addition of Plasmodium falciparum sporozoites did not facilitate cell invasion, which suggests unlinking the effect of HGF on malaria liver stage development from hepatocyte invasion. Finally, our 2D-LC-MS/MS approach and broadly applicable experimental strategy should prove useful in the analysis of various hepatocyte-pathogen interactions, tumor progression, and early disease events.
The liver is the target tissue for several microbial pathogens (1). The malaria parasite, Plasmodium, infects the liver as the first stage of infection of a vertebrate host. Once inside hepatocytes, Plasmodium parasites undergo schizogony, developing from invasive sporozoites into hepatic schizonts containing merozoites, which subsequently infect blood cells. The development of asymptomatic pre-erythrocytic liver-stage (LS)1 parasites of the most virulent human malaria parasite, P. falciparum remains poorly studied because of the inherent difficulty of developing in vitro LS models. A clearer understanding of LS biology may lead to the development of interventions that would result in the complete abrogation of disease. Primary hepatocytes and human hepatocellular carcinoma (HCC) cell lines are widespread models for understanding liver biology (2), chronic diseases and microbial pathogenesis (3). Although only a few Plasmodium sporozoites are required for infecting a human (4, 5), infecting hepatocyte cell lines with P. falciparum remains difficult. The use of primary hepatocytes with several Plasmodium species has been successful, but common use is challenging (6, 7). HCCs, such as HepG2 and Huh-7 are expedient for murine malaria LS, but appear to be refractory to infection with P. falciparum (8).
A novel HCC line, HC-04, has been a useful model to examine human LS biology (9, 10), because it is the only HCC that supports complete development of the two most lethal human malaria parasites, P. falciparum and P. vivax, albeit at low infection efficiencies (<0.1% infection) (11). The basic cellular biology of HC-04 is poorly understood and a clearer understanding may lead to enhanced infection efficiencies by P. falciparum and P. vivax, and thus open many avenues for future research into LS biology. We are also interested in examining growth regulation in HC-04 because growth stimulation has been reported to positively influence sporozoite invasion and subsequent LS development in rodent malaria models (12–13). One growth factor in particular, Hepatocyte Growth Factor (HGF), has been implicated in Plasmodium invasion and maturation in hepatocytes in vitro (12, 13); however, its importance in vivo remains controversial (14). Moreover, growth factor stimulation of cultured cells is tightly linked to changes in gene and protein expression that have been described during the epithelial to mesenchymal transition (EMT) (15, 16) in the liver. In the presence of HGF, cells are driven further away from the epithelial state that is representative of hepatocytes in vivo. HCCs are a heterogeneous group of carcinomas themselves, stemming from diverse molecular alterations (15) and thus individual HCC cell lines will likely differ in response to a given EMT-stimulus. We hypothesize that various HCCs that have been used to study LS represent intermediate stages in the EMT spectrum, and thus their intrinsic loss/retention of epithelial characteristics will influence their susceptibility to sporozoite invasion and LS maturation. We further hypothesize that the acute response of HC-04 to HGF can be a direct indication of the relative stage of HC-04 along the EMT spectrum, and that the specific regulation of cellular markers of EMT can help us identify putative “susceptibility factors”. Thus, our primary biological aim is to examine the effect of HGF on HC-04. By shedding light on which epithelial-state protein markers are still expressed in HC-04 in both their naïve and HGF-stimulated state, we will establish a critical foundation for understanding the cellular biology and plasticity of HC-04. In turn, we can directly address the importance of HGF to Plasmodium sporozoite invasion and development within this poorly characterized HCC.
As we will use HC-04 to explore Plasmodium protein trafficking processes and host cell modifications during LS development, we also took the opportunity to pursue our primary technical aim- to develop a sensitive proteomics platform for acquiring quantifiable MS data from an extremely limited sample source. To meet the needs of trace sample analysis with high sensitivity and throughput, the integration of injector, separation column, and stable electrospray for MS on one microfluidic device (the “chip”) has revolutionized HPLC MS (17). Although chip-based HPLC has gained wider use, the separation capacity remains limited by column length and packing materials and one dimensional separation. Offline multi-dimensional (MD) sample fractionation could reduce the sample complexity and improve the orthogonality that is necessary for increasing protein coverage (17, 18). However, offline MD HPLC still suffers from the disadvantages of being time and labor consuming, a higher risk of sample loss or contamination, and requiring a large sample volume/amount, which is unsuitable when protein samples are limiting. To meet our primary technical and biological aims, we developed an online two dimensional (2D) HPLC chip-based method integrating a strong cation exchange (SCX) column. This system used the autosampler to deliver each salt elution buffer for online fractionation of the loaded protein digest to a chip-integrated enrichment column prior to online analytical column separation and mass spectrometer detection.
Here, we report on the application of this system in the direct comparison of label-free (19) protein expression profiles of naïve and HGF-treated HC-04, which we further validated by targeted transcriptomic and immunohistochemical analyses. Taken together, the data revealed an unexpected response profile of HC-04, which suggests a marked propensity of this cell line to maintain certain epithelial characteristics despite induction of EMT.
EXPERIMENTAL PROCEDURES
Cell Culture
HC-04 and HepG2 cells were grown in Iscove's Modified Dulbecco's Medium (IMDM) (Invitrogen, Carlsbad, CA) supplemented with 5% heat-inactivated fetal bovine serum (Corning Life Sciences, Tewksbury, MA), and Penicillin/streptomycin (Invitrogen) in a humidified chamber at 5% CO2. Cells were seeded into 175 cm2 (for proteomics) or 24 well plates (for qPCR), vacuum plasma treated tissue culture vessels at 28,000 cells per cm2, and grown for 4 days with medium changed on the second, and third day prior to addition of HGF. For HGF treated samples, at 3 days post seeding concentrated HGF (Invitrogen) in PBS was added to a final concentration of 50 ng/ml (20), or an equal volume of PBS was added to the control group. Cells were then grown for an additional 24 h prior to harvesting. Before harvesting, cells were washed twice, rapidly, in cold PBS. For RNA extraction cells were collected at 24 h post HGF addition by the direct addition of TriZol reagent (Invitrogen). For protein analysis, cells were scraped from the tissue culture vessel, pelleted at 800 × g, and then re-suspended and washed twice in cold PBS. For immunohistochemistry, cells were fixed and stained following our standard protocol (see Supplemental Methods). Three biological replicates were collected for both treated and control groups.
Extraction and Digestion of Soluble and Membrane Protein Fractions
Soluble proteins were extracted using 5 mm phosphate buffer containing 0.5 mm PMSF, 1 mm EDTA, and 1 mm protease inhibitors mixture (Sigma) and 4 cycles of freezing and thawing. The membrane fraction was collected from the cell pellets following centrifugation of the supernatant at 20,000 × g for 5 min at 4 °C, solubilization in SDST-lysis buffer (4% SDS (w/v), 100 mm Tris/HCl, 0.1 m DTT, pH 7.6), and boiling at 95 °C for 5 min. We performed soluble protein digestion using RapiGest SF as described (21), and the membrane protein fraction was digested according to a Filter-Aided Sample Preparation (FASP) protocol (22) using a 10 kDa molecular weight cutoff filter (EMD Millipore, Billerica, MA). Acidified tryptic peptides from soluble proteins and FASP approaches were desalted using an HPLC C18 column on an Agilent 1200 HPLC system (Agilent Technologies, Santa Clara, CA), and peptide amounts recovered from FASP were compared with each other. Concentration was then estimated by comparison with soluble samples whose concentrations, as determined by BCA, were known.
RNA Extraction and cDNA Synthesis
RNA was extracted from cells in 24 well culture dishes by direct application of TriZOL reagent. After a standard TriZOL RNA extraction protocol was followed, samples were DNase treated and further purified using an RNeasy kit (Qiagen, Gaithersburg, MD). Eluate RNA concentration was determined using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Rockville, MD). For first strand cDNA synthesis, 1 μg of template RNA was processed using a Revertaid First Strand cDNA synthesis Kit (Thermo Fisher Scientific) under standard conditions. Standard SYBR-based qPCR protocols were then followed to measure mRNA levels of the genes in Table I (see supplementary Methods).
Table I. Genes in naïve and hepatocyte growth factor (HGF) treated HC-04 cells whose observed protein regulation was either expected or unexpected based on established evidence regarding the epithelial-mesenchymal transition of hepatocytes following acute exposure to HGF. Proteins were either present (•) or absent in HC-04 in naïve or +HGF treatments. The mean spectral count indicates the fold-increase in protein above the baseline background across three biological replicate experiments (shown in supplemental Table S4). RT-qPCR values are linear changes versus naïve control.
| Group | Naïve | +HGF | Accession no. | Protein name (Reference) | Mr (kDa) | Spectral count | RT-qPCR HGF/naïve |
|
|---|---|---|---|---|---|---|---|---|
| HC-04 | HepG2 | |||||||
| EXPECTED | • | GLRX5_HUMAN | Glutaredoxin-related protein 5, mitochondrial (39) | 17 | 3.37 | 0.79 | 1.02 | |
| • | ST2A1_HUMAN | Bile salt sulfotransferase (40) | 34 | 2.94 | 1.0 | 0.92 | ||
| • | ZO1_HUMAN | Tight junction protein ZO-1 (1, 15) | 195 | 2.91 | 0.92 | 1.14 | ||
| • | RRAS_HUMAN | Ras-related protein R-Ras (41) | 23 | 2.06 | 0.88 | 1.17 | ||
| • | RAGP1_HUMAN | Ran GTPase-activating protein 1 (42) | 64 | 5.81 | 1.17 | 1.28 | ||
| • | SODM_HUMAN | Superoxide dismutase [Mn2+], mitochondrial (43) | 25 | 4.97 | 1.32 | 1.0 | ||
| • | DPP3_HUMAN | Dipeptidyl peptidase 3 (44) | 83 | 4.77 | 1.53 | 1.0 | ||
| • | PLOD2_HUMAN | Procollagen-lysine,2-oxoglutarate 5-dioxygenase 2 (45) | 85 | 3.13 | 1.0 | 1.42 | ||
| UNEXPECTED | • | IF2B2_HUMAN | Insulin-like growth factor 2 mRNA-binding protein 2 (32) | 66 | 6.30 | 0.83 | 1.0 | |
| • | SAE1_HUMAN | SUMO-activating enzyme subunit 1 (26) | 38 | 5.46 | 1.10 | 1.17 | ||
| • | SIAS_HUMAN | Sialic acid synthase (27) | 40 | 5.04 | 0.61 | 1.11 | ||
| • | SSA27_HUMAN | Sjoegren syndrome/scleroderma autoantigen 1 (46) | 21 | 2.93 | 0.94 | 1.02 | ||
| • | PLST_HUMAN | Plastin-3 (29) | 71 | 2.09 | 1.31 | 1.06 | ||
| • | EIF3F_HUMAN | Eukaryotic translation initiation factor 3 subunit F (30) | 38 | 5.96 | 1.63 | 0.94 | ||
| • | ZYX_HUMAN | Zyxin (33) | 61 | 2.95 | 1.0 | 0.87 | ||
| • | HEM3_HUMAN | Porphobilinogen deaminase (34, 35) | 39 | 1.98 | 1.0 | 1.10 | ||
Online 2D LC-MS/MS
To construct the online 2D methods, we integrated one SCX column (150 μm i.d. * 2 cm length, PolySULFOETHYL ATM, 5 μm 300 A, PolyLC INC) into an Agilent LC-MS system comprised of a 1200 LC system coupled to a 6520 QTOF via an HPLC Chip Cube interface. For the online SCX fractionation, in the first dimension peptides were loaded into the SCX column at 1.8 μl/min and the peptides were eluted using the autosampler by injecting 6 μl of each increasing salt concentration (0, 15, 30, 45, 60, 120, 160, and 300 mm NaCl in 2% ACN/0.1% FA; followed by one injection of 500 mm NaCl in 2% ACN/0.1% FA to wash the column). The salt elution was captured by a C18 enrichment column integrated in the Agilent Polaris-HR-Chip-3C18 chip (360 nL, 180 Å C18 trap with a 75 μm i.d., 150 mm length, 180 Å C18 analytical column). In the second dimension, with valve switched and RPLC gradient started, the peptides were eluted from the enrichment column and separated by a C18 analytical column. Elution of peptides from the analytical column was performed using a gradient starting at 97% A (A: 99.9% water, 0.1% FA) at 300 nL/min. The mobile phase was 3–10% B for 4 min, 10–35% B for 56 min, 35–99% for 2 min, and maintained at 99% B (B: 90% ACN, 9.9% water, 0.1% FA) for 6 min, followed by re-equilibration of column with 3% B for 10 min. Data dependent (autoMS2) MS acquisition was performed by an Agilent 6520 QTOF at 2 GHz. Precursor MS spectra were acquired from m/z 315 to 1700 and the top 4 peaks were selected for MS/MS analysis. Product scans were acquired from m/z 50 to 1700 at a scan rate of 1.5 spectra per second. A medium isolation width (∼4 amu) was used, and a collision energy of slope 3.6 V/100 Da with a 2.9 V offset was applied for fragmentation. A dynamic exclusion list was applied with precursors excluded for 0.50 min after two MS/MS spectrum was acquired.
Offline Peptide Fractionation
Lyophilized FASP digested peptides from naïve HC-04 cell samples was re-dissolved and fractionated by the 3100 OFFGEL fractionator (Agilent Technologies). The fractionator and OFFGEL kit (pH 3–10; 12-well format) were prepared according to the manufacturer's protocols. Twelve fractions were collected from the fractionator and desalted using C18, dried, reconstituted in 2% ACN/0.1% FA loading buffer, and subjected to HPLC-Chip MS analysis. The HPLC gradient and Mass Spectrometer conditions were the same as those used for online second dimensional analysis.
Database Searching and Label Free Quantification Analysis
All the LC-MS/MS raw data were converted to Mascot generic Format (.mgf) by Agilent MassHunter Qualitative Analysis B.04.00. Mascot version 2.4.1 was used to search the SwissProt human 2012 protein FASTA sequence database (20,234 sequences) for peptide sequence assignments using the following parameters: precursor ion mass tolerance of 50 ppm and a fragment ion mass tolerance of 0.2 daltons. Peptides were searched using fully tryptic cleavage constraints and up to two internal cleavages sites were allowed for tryptic digestion. Fixed modifications consisted of carbamidomethylation of cysteine. Variable modifications considered were oxidation of methionine residues. The Mascot searched results were exported as .DAT format and then imported into the Scaffold software (Version 4.0.4, Proteome Software) for curation, label-free quantification analysis, and visualization. Scaffold's normalized spectral counting was employed to compare relative protein abundance between naïve HC-04 and HGF-treated HC-04 cell samples in each experiment as the basis for normalization of the spectral counts for all other LC-MS/MS data in that experiment. Scaffold calculates the spectrum count quantitative value by normalizing spectral counts across an experiment. The process of calculating normalized spectral counts is as follows: (a) Scaffold takes the sum of all the Total Spectrum Counts for each MS sample; (b)The sums are then scaled to the same level; and (c) Scaffold then applies the scaling factor for each sample to each protein group to produce an output with a normalized quantitative value. Overall, protein false discovery rates of less than 1% and peptide false discovery rates of less than 1% were obtained with Scaffold filters, and each protein has ≥ 2 unique peptides. Our data meets all the standards regarding the Minimum Information About a Proteomics Experiment (MIAPE), and data have been deposited to the ProteomeXchange Consortium (http://www.proteomexchange.org) via the PRIDE partner repository (23) with the dataset identifier PXD000682 and DOI 10.6019/PXD000682.
Immunohistochemistry
Cells grown on glass coverslips were fixed in either 100% acetone or 4% paraformaldehyde. (Samples were blocked with 5% fetal bovine serum in PBS (FBS-PBS), washed and incubated in 5 μg/ml rabbit, or non-immune IgG in FBS-PBS for 1 h at RT. After washing, the antibodies were detected with either 2 μg/ml goat α-rabbit, or α-mouse Alexa Fluor 594 secondary antibodies (Invitrogen), mounted on slides using Aqua-Poly/Mount (Polysciences Inc., Warrington, PA) and fluorescence images were acquired with a Nikon 90i microscope (Nikon Corp., Tokyo, Japan) and then analyzed with Volocity software (Perkin Elmer).
Real-time Quantitative PCR
For qPCR reactions, ∼100 ng of template cDNA was used per well. The housekeeping genes, UBC, B2M, RPL13A, ACTB, and GAPDH were used for between sample normalization in this study. Primer sequences were obtained from the Harvard PrimerBank [Spandidos] or RTPrimerDB and primers were produced by Integrated DNA Technologies (Coralville, Iowa). Quantas PerfeCTa master mix with low ROX (Gaithersburg, MD) was used for reaction preparation. An Illumina Eco qPCR device was used for running qPCR reactions and thermal cycling was performed per standard SYBR protocol: 10 min at 95 °C, followed by 40 cycles of 95 °C for 10 s, 58 °C for 60 s with intensity capture at the end of each 58 °C cycle. Commercial genomic DNA specific primer sets (BioRad, Hercules, CA; supplemental Methods) and RT-negative control samples were run alongside each reaction to ensure cDNA quality. Primer sequences are shown in supplemental Table S5.
Plasmodium falciparum Sporozoite HC-04 Invasion Assay
HC-04 cells were grown in equal volumes DMEM (Invitrogen) and Ham's F-12 nutrient mix (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (Corning Life Sciences; Tewksbury, MA), 15 mm HEPES (Sigma Aldrich), 1.5g/L sodium bicarbonate (Sigma Aldrich), and Penicillin/streptomycin (Invitrogen; Carlsbad, CA) in a humidified chamber at 5% CO2. Cells were seeded in 24-well plates at 50,000 cells per well and grown for 30 h in the presence or absence of 50 ng/ml HGF (Invitrogen). P. falciparum NF54 sporozoites were isolated from salivary glands of Anopheles stephensi mosquitoes and added to cells at a multiplicity of infection of 1 in fresh media with a continued presence or absence of HGF. Immunohistochemistry was preformed 24 h after sporozoite addition using an inside-outside stain of circumsporozoite protein with 2A10 monoclonal antibody before and after HC-04 cell permeabilization, followed by quantification of internalized sporozoites by microscopy. For quantification, the number of internalized sporozoites and the number of HC-04 cells were counted across 19 fields under the 40× objective on a Nikon E800 microscope. The percentage of HC-04 cells infected was calculated by division of the number of internalized sporozoites by the number of HC-04 cells (between 2000–4000 cells), followed by multiplication by 100.
RESULTS
Evaluation of the Online 2D SCX-Polaris Reversed Phase (RP) Liquid Chromatography Chip System
Accurate protein identification and quantification of complex proteomes requires the development and application of methods capable of capturing the dynamic protein range. Agilent's HPLC-Chip Cube system (16, 24) is limited to the separation power of a single dimension of RPLC. We married a SCX column (as the first dimensional HPLC column) with the Chip Cube system (second dimensional column), to enable sample loading prior to Chip-HPLC separation (Fig. 1). To evaluate this 2D system, peptide digests from 2 μg of a HC-04-FASP processed sample (naïve group) was loaded onto the SCX column via the autosampler and the peptides were then eluted to the C18 enrichment column of the Polaris Chip followed by C18 analytical column separation and MS detection. For comparison, we also loaded the same sample directly into the Polaris Chip. The results following three continuous runs are shown in supplemental Fig. S1. Solid reproducibility and overlapping protein identifications for 1D and 2D systems was achieved (supplemental Fig. S2A–S2C). Both systems achieved similar protein identifications (> 93% overlap) and notably, almost no peptide was lost during the SCX column step.
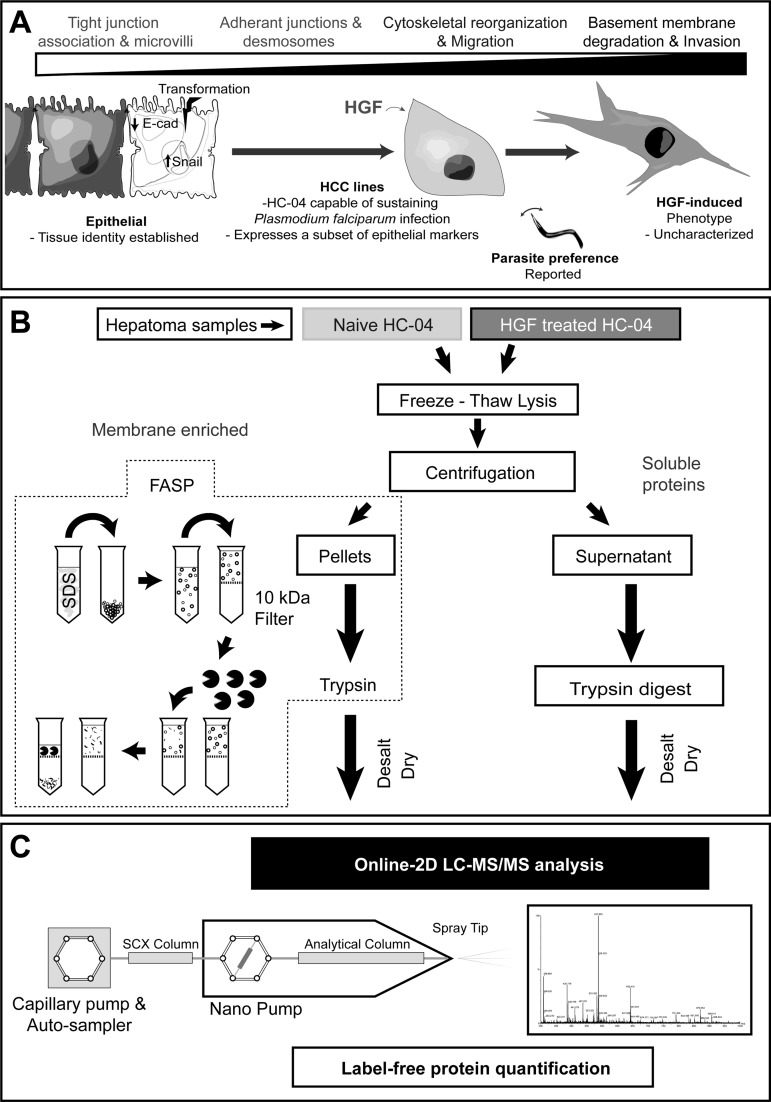
Fig. 1.
Overview of the strategy and the proteomics analytical platform for label-free quantification of hepatocyte proteins. A, Overview of the epithelial-mesenchymal transition (EMT) in the context of our questions. The EMT is a process where epithelial cells lose their established tissue identity (cell polarity and cell-cell adhesion) and is important during development, wound healing, carcinogenesis and the development of cell lines. Here, we assess the effect of HGF on an HCC line in relation to EMT and possible effects on invasion by the malaria parasite. B, Protein sample preparation. Following freezing-thaw lysis of samples a membrane enriched fraction was pelleted by centrifugation prior to FASP digestion. The remaining soluble protein fraction was then processed by standard trypsin digestion. Each sample was then desalted, dried and dissolved in loading buffer prior to online-two dimensional (2D) LC-MS/MS analysis. C, Overview of online-2D LC-MS/MS platform. Our platform was constructed by integrating an SCX column into a chip-cube based RPLC system as the first dimensional separation step. All LC-MS/MS data was searched using Mascot prior to Scaffold for curation and label free quantitation analysis.
Online 2D-LC-MS/MS Platform Permitted Robust Identification and Quantification of HC-04 Proteins Over Offline Approaches
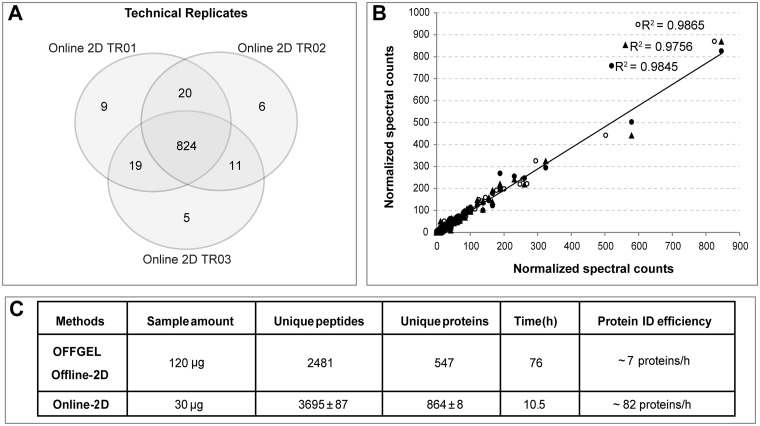
We further evaluated the orthogonality of the 2D system by using seven stepwise salt elution steps to elute the peptide from the SCX column into the Polaris Chip prior to MS detection. We identified 864 ± 8 proteins with two unique peptides in a single 2D run, and noted that > 92% of the total proteins was identified across all three runs, demonstrating marked reproducibility (Fig. 2A, supplemental Table S1A). As expected, given the high degree of reproducibility, we also observed very good linear fit curves across all three runs, demonstrating that the 2D approach is well-suited for quantitative analysis (Fig. 2B).
Fig. 2.
Evaluation of the online-2D LC-MS/MS platform. A, Overlap of identification of three continuous technical runs. B, Normalized spectral counts of total identified proteins from same three runs. R2 values are derived from sample-to-sample comparison for each protein. C, Comparison of the performance of our online-2D system versus a commercial OFFGEL peptide fractionation Offline-2D system.
For a comparison, we performed a popular peptide offline fractionation approach using the OFFGEL system (16), which reduces peptide complexity based on peptide isoelectric point (pI). We fractionated 120 μg of a HC-04 FASP digested sample into 12 fractions prior to Polaris Chip LC-MS/MS analysis using the same RPLC gradient from our 2D approach. We identified 547 protein groups by OFFGEL, with a total analysis time of ∼73 h including 48 h focusing, 7 h desalting and drying, and 18 h of HPLC-MS/MS analysis (supplemental Table S1B). However, with a 30 μg sample, it took only 10.5 h to identify more proteins with the online 2D system; clearly demonstrating ∼12-fold greater protein identification efficiency (Fig. 2C).
Proteomic Profile Changes Following HGF Treatment
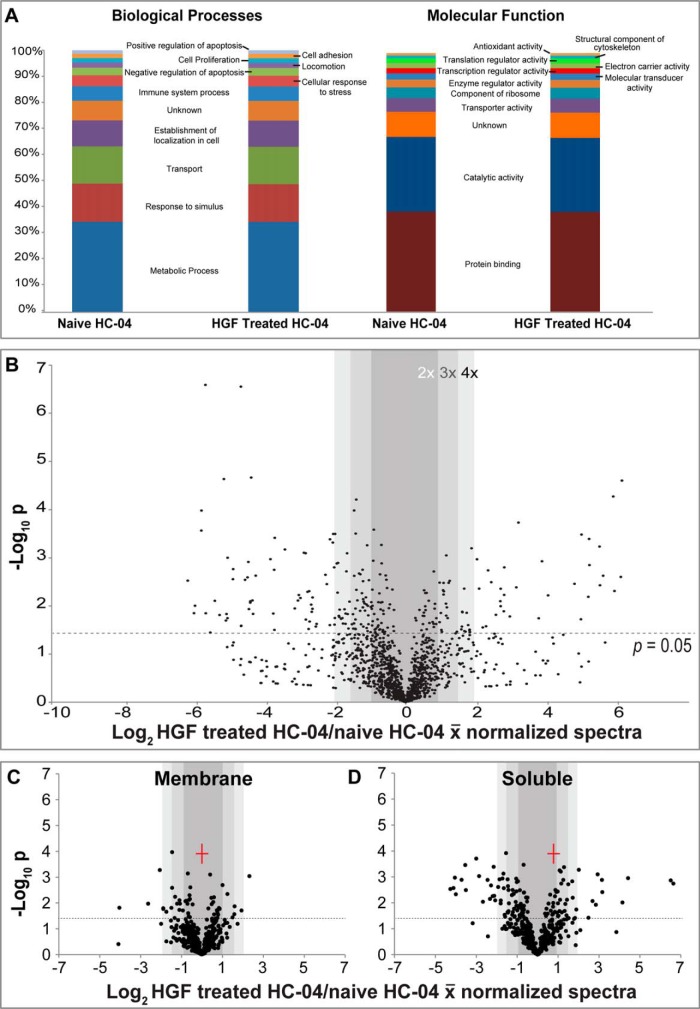
We applied online 2D methods to profile protein expression in naïve and HGF treated HC-04 (supplemental Table S2). A total of 1316 and 1283 protein groups were identified in HC-04-naive samples and HGF-treated samples, respectively. Approximately 92% of the identified proteins from both samples overlapped (supplemental Fig. S2A). To acquire comprehensive proteome information, we also performed soluble and membrane protein extraction prior to online 2D analysis (supplemental Table S3). Using the spectral count values (supplemental Table S2), 829 proteins appeared to be down-regulated and 512 proteins up-regulated following HGF treatment. Of the 829 proteins that were quantifiable, 477 were down-regulated by at least 1.5-fold and 320 proteins were down-regulated at least twofold. In the up-regulated proteins, 212 of the 512 proteins were up-regulated >1.5-fold and 124 proteins exhibited twofold up-regulation.
We used Gene Ontology classifications to parse the data and determine larger trends. Using pre-determined GO filters, we found that the identity of the proteins detected did not differ significantly following HGF treatment (Fig. 3A). However, there were marked changes in protein quantity between several important groups (Fig. 3B–3D). First, the proportion of soluble proteins that significantly changed was greater than the proportion identified in the membrane fractions (Fig. 3C–3D). This may partly be because of membrane proteins being notoriously difficult to extract and thus quantify. This outcome might also be connected to the large number of mitochondrial proteins found in any membrane fraction and that not all proteins in the membrane fraction are bona fide membrane proteins (supplemental Table S3). We observed that several GO Biological Processes (BP) pathways including cell proliferation (supplemental Fig. S4A), cellular response to stress (supplemental Fig. S4B); and negative regulation of apoptosis (data not shown) were all up-regulated in response to HGF treatment. These pathways are generally expected to increase following HGF treatment and could be important for Plasmodium-hepatocyte interactions (13, 15). Using GO Molecular Function (MF) terms we found slight increases in several categories of interest including regulation of transcription (supplemental Fig. S4C) and translation activities (supplemental Fig. S4D).
Fig. 3.
Quantitative proteomic analysis of HC-04 following HGF treatment using the online-2D LC-MS/MS platform revealed significant up-regulation of proteins according to cellular compartment. A, Stacked bar graphs of select Gene Ontology (GO) terms associated with our identified proteins. Abundance in functional categories is very similar across each group in this qualitative analysis. B, Volcano plot of quantifiable proteome comparing HC-04 protein levels before and after HGF treatment. C-D, Volcano plots of different quantifiable subsets of proteins based on cell compartment as determined by fractionation into membrane (C) and soluble (D) protein samples.
Morphological Changes Support Our Proteomics Results While Transcriptomics Reveal Moderate Discordance Between Transcriptomic and Proteomic Regulation of EMT Cellular Markers
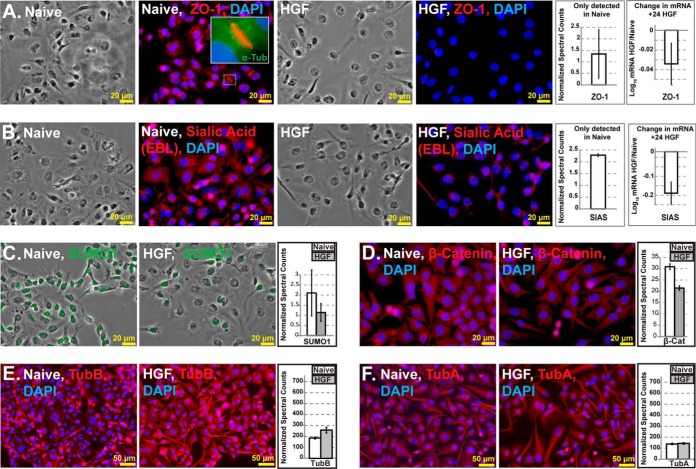
We anticipated that several proteins will be immediately excluded from relative quantifications because of the complete absence of peptide spectra in one comparator group. We interrogated this protein list separately (supplemental Table S4) and further validated the protein quantification by immunofluorescence microscopy and transcriptomic profiling. The antibody staining of EMT markers largely supported our predictions and proteomics results (Fig. 4). Tight junction protein ZO-1 was found to localize at cell-cell interfaces in naïve cells but was found to decrease sharply following HGF treatment (Fig. 4A). This result is in agreement with our proteomics results and supports our hypothesis that naïve HC-04 maintains certain epithelial characteristics. Likewise, staining by the Sambucus nigra lectin (EBL), which binds terminal sialic acids on glycoconjugates demonstrated indirectly that sialic acid synthase (SIAS) was less active following HGF treatment (Fig. 4B). Relative mRNA levels for both ZO-1 and SIAS were found to decrease slightly, but consistently, suggesting that the HGF-directed changes were largely effected post-transcriptionally. The multifunctional proteins SUMO1 (25) and β-catenin (a primary implicated marker for EMT, 15) were both found to decrease slightly in staining intensity by immunohistochemistry, further corroborating our protein quantification. SUMO1 (Fig. 4C) was found to localize primarily in the nuclei while β -catenin was seen throughout the cytoplasm (Fig. 4D). Taken together, the SUMO1 data provide indirect support for our observed down-regulation of SUMO1 activating enzyme 1 (SAE1) protein expression (Table I). SAE1 is co-regulated with SUMO1 (26) and we would expect the observed coordinated down-regulation in protein expression level. Cytoskeletal proteins were also assayed and found to be consistent with the proteomics results wherein β-tubulin staining intensity decreased following HGF treatment (Fig. 4E–4F). We did not observe E-cadherin at FDR<1%, presumably because of the low abundance of E-cadherin in these cell. However, it was important to demonstrate that EMT stimulation was achieved in HC-04. We performed quantitative Western blots using anti-E-cadherin antibodies and observed the reduction in staining intensity of E-cadherin as compared with the β-actin loading control by Western blot (supplemental Fig. 4SE) and quantitative analyses of the blot using densitometry (supplemental Fig. S4F). The staining intensity was poor, further suggesting low protein abundance in cell lysates. We further confirmed the relative reduction of E-cadherin using normalized spectral counts at FDR<5% (data not shown). However, N-cadherin was not detected by Western blot or bioinformatic analyses at FDR<5% (data not shown) Together, these data confirmed that 50 ng/ml HGF stimulated EMT in HC-04 in our studies, however, unlike what we had observed for HepG2, the response to HGF was tempered and did not drive the expression of N-cadherin.
Fig. 4.
Immunohistochemistry analyses supported our predictions and proteomics results whereas transcriptomics results suggested discordance between transcript and protein expression levels at 24 h post HGF treatment. A, Tight junction protein ZO-1 localized at cell-cell interfaces in naïve cells and decreased in abundance following HGF treatment. B, Less staining with lectin (EBL), which binds sialic acids (produced by SIAS) was found in HC-04 following HGF treatment. C, SUMO1 and D, β-catenin both decreased slightly following HGF treatment with SUMO1 localizing primarily in the nuclei, whereas β -catenin was seen throughout the cytoplasm and cell surface. E–F, Cytoskeletal protein β-tubulin (E) staining intensity changed slightly with HGF treatment, whereas α-tubulin (F) localization remained stable.
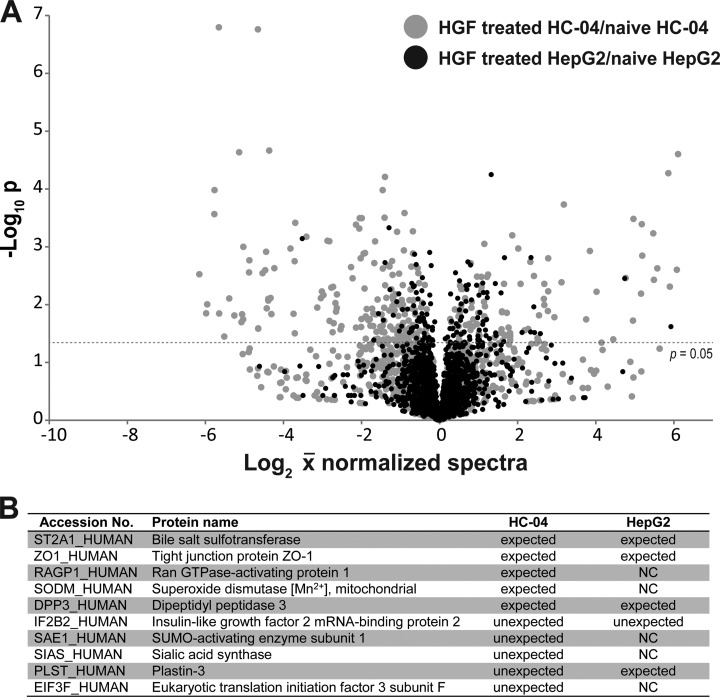
Further RT-qPCR analyses of the 16 genes listed in Table I revealed that at 24 h post-HGF treatment many of the relative mRNA levels are generally not indicative of the changes in protein levels. In total, the expression of 11 of 17 genes did correlate with a positive or negative change in protein level, but changes in mRNA levels were normally several-fold lower than those seen at the protein level. Of the remainder, four genes were found to not change and two showed changes in the opposite direction of that seen in our proteomics results. Treatment of HGF in HepG2 cells produced a similar range of qPCR results, with all genes showing less than two-fold changes and three showing no change (Table I, supplemental Fig. S5, and supplemental Tables S5 and S6). Thus, as expected, the global proteomic change in HepG2 following the same HGF treatment was less dramatic than what we observed in HC-04 (Fig. 5A). Furthermore, approximately half of the EMT transition markers for HC-04 (Table I) were not identified in HepG2, suggesting the more extreme mesenchymal condition of this cell line. Moreover, we observed fewer unexpected changes in the EMT markers assessed for HC-04 (Fig. 5B and Table I). Because HGF treatment still promoted a general transition toward a mesenchymal state (albeit less so than HepG2), we predicted that pre- and co-treatment of HC-04s will not necessarily improve P. falciparum sporozoite invasion. In fact, we observed reduced levels of sporozoites that are physically inside HC-04 at 24 h post exposure to HGF (Fig. 6), but that this difference is not statistically significant (p = 0.1409, Student's t test, α = 0.05).
Fig. 5.
Proteomic analysis of HepG2 following HGF treatment compared with the HC-04 response to HGF. We applied online 2D methods to profile protein expression in naïve and HGF treated HepG2 (supplemental Table S7). A total of 1759 and 1757 protein groups were identified in HepG2-naive samples and HGF-treated samples, respectively. A, Volcano plot of the quantifiable proteomes of HepG2 (black points) and HC-04 (gray points) comparing protein levels before and after HGF treatment. These plots demonstrate the more drastic change in HC-04 following HGF treatment, suggesting that HC-04 is at a different point on the EMT spectrum that other common HCC. B, Observed protein regulation in naïve and hepatocyte growth factor (HGF) treated HepG2 cells in comparison to expected or unexpected changes of HC-04 cells following acute exposure to HGF (from Table I). With the exception of IF2B2, all HepG2 examples either changed in an expected manner based on the epithelial-mesenchymal transition, did not change more than 0.1 fold (NC), or were not detected. Data is based on three biological replicate experiments (shown in supplemental Table S7).
Fig. 6.
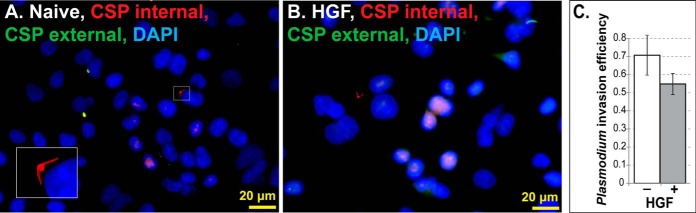
Plasmodium falciparum invasion efficiency is slightly decreased in HC-04 following HGF treatment. A–B, HC-04 cells were grown for 30 h in the absence (A) or presence (B) of HGF before the addition of P. falciparum NF54 sporozoites. After 24 h of incubation with the sporozoites in the continued presence or absence of HGF, inside-outside staining of the sporozoites revealed fewer sporozoites in HC-04 cells treated with HGF compared with those without HGF. A representative microscope field is shown for both treatments. Inset: A sporozoite inside a hepatocyte that is beginning to round-up to form an exoerythrocytic form. Red staining denotes a sporozoite inside an HC-04 cell; green denotes a sporozoite outside; yellow indicates an overlap. DAPI staining of the nuclei appears blue. C, Quantification of inside-outside staining of P. falciparum sporozoites that have invaded a HC-04 cell. Percent invasion efficiency is shown for experimental replicates that received HGF treatment, n = 5, and without HGF (naïve) treatment, n = 8. Error bars indicate the standard error of the mean.
DISCUSSION
A better understanding of hepatocyte biology may lead to the development of high-throughput assays to evaluate novel anti-malarial strategies targeting LS. In vivo, hepatocyte physiological function is tightly linked to the microarchitecture, or zonation, of hepatocytes within each liver lobule. It is theorized that physiological changes in liver zonation are dictated primarily by growth factor regulation (1). We hypothesized that the acute response of HC-04 to HGF would be an indication of its relative stage along the EMT spectrum, and that the specific regulation of the widely implicated EMT cellular markers can help us identify the possible facilitators of LS invasion and development.
We devised a highly orthogonal online 2D-LC-MS/MS system, which lessened sample loss and risk of contamination (as compared with offline approaches) and successfully quantified protein expression levels from a small sample of HC-04 cells pre- and post-HGF treatment. Our results highlight the utility of our online-2D MS/MS system to identify important phenomenon (beyond the malaria paradigm) that would not otherwise be captured by protein identification or transcriptomic profiling alone. Importantly, in support of our hypothesis, the proteomic data and correlative approaches identified several expected markers associated with EMT and paradoxically, markers indicative of an epithelial phenotype following HGF treatment. ZO-1 and β-Catenin expression and localization along the cytoplasmic membrane surface at intercellular tight junctions in many epithelial cells has been shown to strongly correlate with signaling pathways during the EMT (1). As expected, ZO-1 and β-Catenin were down-regulated following HGF treatment. SIAS is involved in the synthesis of sialic acids which are typically found as terminal glycans of cell surface glycoconjugates and hypersialylation has been associated with metastases and liver regeneration (27). However, contrary to predictions, we observed down-regulation of EMT markers such as SIAS (27), RUVBL-1/RUVBL-2 (28), and Plastin-3 (29) with the marked up-regulation of Eukaryotic initiation factor 3 subunit f (30) and Epiplakin (31), which are potentially two unexpected markers for an “epithelial-like” phenotype. We would also expect SAE1 and Insulin-like growth factor 2 mRNA-binding protein 2 to both be up-regulated at the protein level following HGF treatment at the mRNA level (Table I; 26, 32). In the reverse context, Zyxin, which is involved in regulating the disruption of actin membrane linkages at adherens junctions, and thus mediates cell migration during EMT (33), is reduced at the transcript level but is highly up-regulated in terms of protein abundance. Reduced porphobilinogen deaminase (PBGD, HEM3-HUMAN) activity has been shown to be associated with acute hepatic porphyria and HCC (34). However, following HGF stimulation or EMT-induction, no changes in PBGD deaminase transcript has been reported (34). It is therefore unclear if the observed overexpression at the protein level following HGF treatment simply suggests a discordance between transcript and protein levels for this gene, or if the up-regulation is indicative of a counter-response to HGF. It is interesting to speculate that up-regulation of PBGD in HC-04 results in increased enzyme activity, a phenomenon that has been described in C6 glioma cells, wherein overexpression of PBGD resulted in G1 cell cycle attenuation, reduction in synthesis of Vimentin (an EMT marker) and subsequent transformation of C6 cells into a more astrocyte-like morphology (35). It should also be noted that there is growing evidence that expression of the implicated EMT markers are extremely heterogeneous in HCCs (36, 37), and expression is dependent on size and location within tumors, and that HCC cell lines may not be an exception (36). We noted that HC-04 responsiveness to HGF by itself differs from that observed for other HCCs. For example, HGF induces proliferation followed by a cytostatic state in HepG2 cells (38), which is a poor infection model for LS of human malaria. Our data suggests that, at baseline, HGF can still “induce” the markers of proliferation in HepG2 but that the response is less dramatic. Conversely, HC-04s are subject to the influence of growth factors, but the response is tempered by the up-regulation of proteins that are driving the maintenance of residual epithelial characteristics. Taken altogether, HC-04 and HepG2 appear to be at different stages of EMT. HepG2 cells exhibit a more mesenchymal phenotype, whereas HC-04 cells reveal a more hepatocyte-like nature; which can be expected because HC-04 cells were derived spontaneously from a culture of primary human hepatocytes (11).
Successful malaria parasite development in HC-04 may be the result of an amenable intracellular environment or susceptibility to infection, which would result in a larger number of invaded cells. Importantly, HC-04 still responds sufficiently to HGF (i.e. becomes less epithelial) resulting in poorer sporozoite infection efficiency, which corroborates the findings of others (14). Therefore, our data would further argue for unlinking the effect of HGF on LS development from sporozoite invasion; because it is likely that the epithelial character of HC-04, as suggested by our data, more closely resembles the hepatocyte architecture in vivo that is important for presenting the proper cellular context/cellular surface receptors that mediate sporozoite invasion. However, our study focused on invasion and therefore our data is not inconsistent with the reported importance of HGF in LS development and maturation following invasion (12, 13). A direct glycoproteomic analysis of LS permissive and non-permissive HCC cell lines are in progress with the aim of characterizing and validating the repertoire of cell surface molecules that enable sporozoite invasion.
Supplementary Material
Acknowledgments
We thank the Naval Medical Research Center for provision of the original stock of the HC-04 cell line for this study (LP-CRADA-NMRC-11-3857) and the PRoteomics IDEntifications (PRIDE) team) for technical assistance with the first native MzIdentML submission to the data repository.
Footnotes
Author contributions: D.T., J.G.K., P.J.J., J.A.B., and R.R.D. designed research; D.T., J.G.K., R.E.T., and R.R.D. performed research; D.T., R.E.T., and R.R.D. contributed new reagents or analytic tools; D.T., J.G.K., R.E.T., P.J.J., J.A.B., and R.R.D. analyzed data; D.T., J.G.K., R.E.T., P.J.J., J.A.B., and R.R.D. wrote the paper.
* This work was funded by the Human Frontiers Science Program (RGY0073/2012 to J.A.B., R.R.D., and P.J.J.), the Bloomberg Family Foundation and the Johns Hopkins Malaria Research Institute (JHMRI) (to R.R.D.) and the NIH National Center for Research Resources [grant number UL1 RR 025005]. J.A.B. is a QEII Fellow of the Australian Research Council.
 This article contains supplemental Figs. S1 to S5 and Tables S1 to S7.
This article contains supplemental Figs. S1 to S5 and Tables S1 to S7.
Conflict of Interest: The authors declare that no conflicts of interest exist that can be perceived to influence the conduct of the study.
1 The abbreviations used are:
- LS
- liver stages
- HCC
- human hepatocellular carcinoma
- HGF
- hepatocyte growth factor
- EMT
- epithelial-mesenchymal transition
- SCX
- strong cation exchange
- FASP
- Filter-Aided Sample Preparation
- ppm
- parts per million
- FA
- formic acid.
REFERENCES
- 1. Grisham J. W. (2009) Organizational Principles of the Liver, in The Liver: Biology and Pathobiology, Fifth Edition (ed Arias I. M.), John Wiley & Sons, Ltd, Chichester, UK: doi: 10.1002/9780470747919.ch1 [Google Scholar]
- 2. Wilkening S., Stahl F., Bader A. (2003) Comparison of primary human hepatocytes and hepatoma cell line Hepg2 with regard to their biotransformation properties. Drug Metab. Dispos. 31, 1035–1042 [DOI] [PubMed] [Google Scholar]
- 3. Liu L., Cao Y., Chen C., Zhang X., McNabola A., Wilkie D., Wilhelm S., Lynch M., Carter C. (2006) Sorafenib blocks the RAF/MEK/ERK pathway, inhibits tumor angiogenesis, and induces tumor cell apoptosis in hepatocellular carcinoma model PLC/PRF/5. Cancer Res. 66, 11851–11858 [DOI] [PubMed] [Google Scholar]
- 4. Medica D. L., Sinnis P. (2005) Quantitative dynamics of plasmodium yoelii sporozoite transmission by infected anopheline mosquitoes. Infect. Immun. 73, 4363–4369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rosenberg R., Wirtz R. A., Schneider I., Burge R. (1990) An estimation of the number of malaria sporozoites ejected by a feeding mosquito. Trans. R. Soc. Trop. Med. Hyg. 84, 209–212 [DOI] [PubMed] [Google Scholar]
- 6. Mazier D., Landau I., Druilhe P., Miltgen F., Guguen-Guillouzo C., Baccam D., Baxter J., Chigot J., Gentilini M. (1984) Cultivation of the liver forms of plasmodium vivax in human hepatocytes. Nature 307, 367–369 [DOI] [PubMed] [Google Scholar]
- 7. Foley D. A., Kennard J., Vanderberg J. P. (1978) Plasmodium berghei: Infective exoerythrocytic schizonts in primary monolayer cultures of rat liver cells. Exp. Parasitol. 46, 166–178 [DOI] [PubMed] [Google Scholar]
- 8. Prudencio M., Mota M. M., Mendes A. M. (2011) A toolbox to study liver stage malaria. Trends Parasitol. 27, 565–574 [DOI] [PubMed] [Google Scholar]
- 9. Mikolajczak S. A., Sacci J. B., Jr., De La Vega P., Camargo N., VanBuskirk K., Krzych U., Cao J., Jacobs-Lorena M., Cowman A. F., Kappe S. H. (2011) Disruption of the plasmodium falciparum liver-stage antigen-1 locus causes a differentiation defect in late liver-stage parasites. Cell. Microbiol. 13, 1250–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. VanBuskirk K. M., O'Neill M. T., De La Vega P., Maier A. G., Krzych U., Williams J., Dowler M. G., Sacci J. B., Jr., Kangwanrangsan N., Tsuboi T., Kneteman N. M., Heppner D. G., Jr., Murdock B. A., Mikolajczak S. A., Aly A. S., Cowman A. F., Kappe S. H. (2009) Preerythrocytic, live-attenuated plasmodium falciparum vaccine candidates by design. Proc. Natl. Acad. Sci. U.S.A. 106, 13004–13009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sattabongkot J., Yimamnuaychoke N., Leelaudomlipi S., Rasameesoraj M., Jenwithisuk R., Coleman R. E., Udomsangpetch R., Cui L., Brewer T. G. (2006) Establishment of a human hepatocyte line that supports in vitro development of the exo-erythrocytic stages of the malaria parasites plasmodium falciparum and P. vivax. Am. J. Trop. Med. Hyg. 74, 708–715 [PubMed] [Google Scholar]
- 12. Leiriao P., Albuquerque S. S., Corso S., van Gemert G. J., Sauerwein R. W., Rodriguez A., Giordano S., Mota M. M. (2005) HGF/MET signalling protects plasmodium-infected host cells from apoptosis. Cell. Microbiol. 7, 603–609 [DOI] [PubMed] [Google Scholar]
- 13. Carrolo M., Giordano S., Cabrita-Santos L., Corso S., Vigario A. M., Silva S., Leiriao P., Carapau D., Armas-Portela R., Comoglio P. M., Rodriguez A., Mota M. M. (2003) Hepatocyte growth factor and its receptor are required for malaria infection. Nat. Med. 9, 1363–1369 [DOI] [PubMed] [Google Scholar]
- 14. Kaushansky A., Kappe S. H. I. (2011) The crucial role of hepatocyte growth factor receptor during liver-stage infection is not conserved among plasmodium species. Nat. Med. 17, 1180–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huber M. A., Kraut N., Beug H. (2005) Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr. Opin. Cell Biol. 17, 548–558 [DOI] [PubMed] [Google Scholar]
- 16. Horth P., Miller C. A., Preckel T., Wenz C. (2006) Efficient fractionation and improved protein identification by peptide OFFGEL electrophoresis. Mol. Cell. Proteomics 5, 1968–1974 [DOI] [PubMed] [Google Scholar]
- 17. Yin H., Killeen K., Brennen R., Sobek D., Werlich M., van de Goor T. (2005) Microfluidic chip for peptide analysis with an integrated HPLC column, sample enrichment column, and nanoelectrospray tip. Anal. Chem. 77, 527–533 [DOI] [PubMed] [Google Scholar]
- 18. Washburn M. P., Wolters D., Yates J. R., 3rd. (2001) Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242–247 [DOI] [PubMed] [Google Scholar]
- 19. Mosley A. L., Sardiu M. E., Pattenden S. G., Workman J. L., Florens L., Washburn M. P. (2011) Highly reproducible label free quantitative proteomic analysis of RNA polymerase complexes. Mol. Cell. Proteomics 10, M110.000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wojta J., Nakamura T., Fabry A., Hufnagl P., Beckmann R., McGrath K., Binder B. R. (1994) Hepatocyte growth factor stimulates expression of plasminogen activator inhibitor type 1 and tissue factor in HepG2 cells. Blood 84, 151–157 [PubMed] [Google Scholar]
- 21. Norrgran J., Williams T. L., Woolfitt A. R., Solano M. I., Pirkle J. L., Barr J. R. (2009) Optimization of digestion parameters for protein quantification. Anal. Biochem. 393, 48–55 [DOI] [PubMed] [Google Scholar]
- 22. Wisniewski J. R., Zougman A., Nagaraj N., Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 [DOI] [PubMed] [Google Scholar]
- 23. Vizcaíno J. A., Côté R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Perez-Riverol Y., Reisinger F., Ríos D., Wang R., Hermjakob H. The Proteomics Identifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013. January;41(Database issue):D1063–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tao D., Zhang L., Shan Y., Liang Z., Zhang Y. (2011) Recent advances in micro-scale and nano-scale high-performance liquid-phase chromatography for proteome research. Anal. Bioanal Chem. 399, 229–241 [DOI] [PubMed] [Google Scholar]
- 25. Guo W. H., Yuan L. H., Xiao Z. H., Liu D., Zhang J. X. (2011) Overexpression of SUMO-1 in hepatocellular carcinoma: A latent target for diagnosis and therapy of hepatoma. J. Cancer Res. Clin. Oncol. 137, 533–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Amente S., Lavadera M. L., Palo G. D., Majello B. (2012) SUMO-activating SAE1 transcription is positively regulated by myc. Am. J. Cancer Res. 2, 330–334 [PMC free article] [PubMed] [Google Scholar]
- 27. Wagner H. E., Thomas P., Wolf B. C., Rapoza A., Steele G., Jr. (1990) Inhibition of sialic acid incorporation prevents hepatic metastases. Arch. Surg. 125, 351–354 [DOI] [PubMed] [Google Scholar]
- 28. Rousseau B., Menard L., Haurie V., Taras D., Blanc J. F., Moreau-Gaudry F., Metzler P., Hugues M., Boyault S., Lemiere S., Canron X., Costet P., Cole M., Balabaud C., Bioulac-Sage P., Zucman-Rossi J., Rosenbaum J. (2007) Overexpression and role of the ATPase and putative DNA helicase RuvB-like 2 in human hepatocellular carcinoma. Hepatology 46, 1108–1118 [DOI] [PubMed] [Google Scholar]
- 29. Yokobori T., Iinuma H., Shimamura T., Imoto S., Sugimachi K., Ishii H., Iwatsuki M., Ota D., Ohkuma M., Iwaya T., Nishida N., Kogo R., Sudo T., Tanaka F., Shibata K., Toh H., Sato T., Barnard G. F., Fukagawa T., Yamamoto S., Nakanishi H., Sasaki S., Miyano S., Watanabe T., Kuwano H., Mimori K., Pantel K., Mori M. (2013) Plastin3 is a novel marker for circulating tumor cells undergoing the epithelial-mesenchymal transition and is associated with colorectal cancer prognosis. Cancer Res. 73, 2059–2069 [DOI] [PubMed] [Google Scholar]
- 30. Wen F., Zhou R., Shen A., Choi A., Uribe D., Shi J. (2012) The tumor suppressive role of eIF3f and its function in translation inhibition and rRNA degradation. PLoS One 7, e34194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shimada H., Nambu-Niibori A., Wilson-Morifuji M., Mizuguchi S., Araki N., Sumiyoshi H., Sato M., Mezaki Y., Senoo H., Ishikawa K., Hatano Y., Okamoto O., Fujiwara S. (2013) Epiplakin modifies the motility of the HeLa cells and accumulates at the outer surfaces of 3-D cell clusters. J. Dermatol. 40, 249–258 [DOI] [PubMed] [Google Scholar]
- 32. Cleynen I., Brants J. R., Peeters K., Deckers R., Debiec-Rychter M., Sciot R., Van de Ven W. J., Petit M. M. (2007) HMGA2 regulates transcription of the Imp2 gene via an intronic regulatory element in cooperation with nuclear factor-kappaB. Mol. Cancer Res. 5, 363–372 [DOI] [PubMed] [Google Scholar]
- 33. Sperry R. B., Bishop N. H., Bramwell J. J., Brodeur M. N., Carter M. J., Fowler B. T., Lewis Z. B., Maxfield S. D., Staley D. M., Vellinga R. M., Hansen M. D. (2010) Zyxin controls migration in epithelial-mesenchymal transition by mediating actin-membrane linkages at cell-cell junctions. J. Cell. Physiol. 222, 612–624 [DOI] [PubMed] [Google Scholar]
- 34. Kim J., Hong S. J., Park J. Y., Park J. H., Yu Y. S., Park S. Y., Lim E. K., Choi K. Y., Lee E. K., Paik S. S., Lee K. G., Wang H. J., Do I. G., Joh J. W., Kim D. S., and Korea Cancer Biomarker Consortium. (2010) Epithelial-mesenchymal transition gene signature to predict clinical outcome of hepatocellular carcinoma. Cancer Sci. 101, 1521–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Greenbaum L., Gozlan Y., Schwartz D., Katcoff D. J., Malik Z. (2002) Nuclear distribution of porphobilinogen deaminase (PBGD) in glioma cells: A regulatory role in cancer transformation? Br. J. Cancer 86, 1006–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ding W., You H., Dang H., LeBlanc F., Galicia V., Lu S. C., Stiles B., Rountree C. B. (2010) Epithelial-to-mesenchymal transition of murine liver tumor cells promotes invasion. Hepatology 52, 945–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bonnomet A., Syne L., Brysse A., Feyereisen E., Thompson E. W., Noel A., Foidart J. M., Birembaut P., Polette M., Gilles C. (2012) A dynamic in vivo model of epithelial-to-mesenchymal transitions in circulating tumor cells and metastases of breast cancer. Oncogene 31, 3741–3753 [DOI] [PubMed] [Google Scholar]
- 38. Matteucci E., Castoldi R., Desiderio M. A. (2001) Hepatocyte growth factor induces pro-apoptotic genes in HepG2 hepatoma but not in B16-F1 melanoma cells. J. Cell. Physiol. 186, 387–396 [DOI] [PubMed] [Google Scholar]
- 39. Radisky D. C., Levy D. D., Littlepage L. E., Liu H., Nelson C. M., Fata J. E., Leake D., Godden E. L., Albertson D. G., Nieto M. A., Werb Z., Bissell M. J. (2005) Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature 436, 123–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ou Z., Shi X., Gilroy R. K., Kirisci L., Romkes M., Lynch C., Wang H., Xu M., Jiang M., Ren S., Gramignoli R., Strom S. C., Huang M., Xie W. (2013) Regulation of the human hydroxysteroid sulfotransferase (SULT2A1) by RORalpha and RORgamma and its potential relevance to human liver diseases. Mol. Endocrinol. 27, 106–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Luo H., Hao X., Ge C., Zhao F., Zhu M., Chen T., Yao M., He X., Li J. (2010) TC21 promotes cell motility and metastasis by regulating the expression of E-cadherin and N-cadherin in hepatocellular carcinoma. Int. J. Oncol. 37, 853–859 [DOI] [PubMed] [Google Scholar]
- 42. Xia F., Lee C. W., Altieri D. C. (2008) Tumor cell dependence on ran-GTP-directed mitosis. Cancer Res. 68, 1826–1833 [DOI] [PubMed] [Google Scholar]
- 43. Radisky D. C., Levy D. D., Littlepage L. E., Liu H., Nelson C. M., Fata J. E., Leake D., Godden E. L., Albertson D. G., Nieto M. A., Werb Z., Bissell M. J. (2005) Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature 436, 123–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Simaga S., Babic D., Osmak M., Ilic-Forko J., Vitale L., Milicic D., Abramic M. (1998) Dipeptidyl peptidase III in malignant and non-malignant gynaecological tissue. Eur. J. Cancer 34, 399–405 [DOI] [PubMed] [Google Scholar]
- 45. Gilkes D. M., Bajpai S., Chaturvedi P., Wirtz D., Semenza G. L. (2013) Hypoxia-inducible factor 1 (HIF-1) promotes extracellular matrix remodeling under hypoxic conditions by inducing P4HA1, P4HA2, and PLOD2 expression in fibroblasts. J. Biol. Chem. 288, 10819–10829 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46. Sillars-Hardebol A. H., Carvalho B., de Wit M., Postma C., Delis-van Diemen P. M., Mongera S., Ylstra B., van de Wiel M. A., Meijer G. A., Fijneman R. J. (2010) Identification of key genes for carcinogenic pathways associated with colorectal adenoma-to-carcinoma progression. Tumour Biol. 31, 89–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.