Abstract
Given the ease of whole genome sequencing with next-generation sequencers, structural and functional gene annotation is now purely based on automated prediction. However, errors in gene structure are frequent, the correct determination of start codons being one of the main concerns. Here, we combine protein N termini derivatization using (N-Succinimidyloxycarbonylmethyl)tris(2,4,6-trimethoxyphenyl)phosphonium bromide (TMPP Ac-OSu) as a labeling reagent with the COmbined FRActional DIagonal Chromatography (COFRADIC) sorting method to enrich labeled N-terminal peptides for mass spectrometry detection. Protein digestion was performed in parallel with three proteases to obtain a reliable automatic validation of protein N termini. The analysis of these N-terminal enriched fractions by high-resolution tandem mass spectrometry allowed the annotation refinement of 534 proteins of the model marine bacterium Roseobacter denitrificans OCh114. This study is especially efficient regarding mass spectrometry analytical time. From the 534 validated N termini, 480 confirmed existing gene annotations, 41 highlighted erroneous start codon annotations, five revealed totally new mis-annotated genes; the mass spectrometry data also suggested the existence of multiple start sites for eight different genes, a result that challenges the current view of protein translation initiation. Finally, we identified several proteins for which classical genome homology-driven annotation was inconsistent, questioning the validity of automatic annotation pipelines and emphasizing the need for complementary proteomic data. All data have been deposited to the ProteomeXchange with identifier PXD000337.
Recent developments in mass spectrometry and bioinformatics have established proteomics as a common and powerful technique for identifying and quantifying proteins at a very broad scale, but also for characterizing their post-translational modifications and interaction networks (1, 2). In addition to the avalanche of proteomic data currently being reported, many genome sequences are established using next-generation sequencing, fostering proteomic investigations of new cellular models. Proteogenomics is a relatively recent field in which high-throughput proteomic data is used to verify coding regions within model genomes to refine the annotation of their sequences (2–8). Because genome annotation is now fully automated, the need for accurate annotation for model organisms with experimental data is crucial. Many projects related to genome re-annotation of microorganisms with the help of proteomics have been recently reported, such as for Mycoplasma pneumoniae (9), Rhodopseudomonas palustris (10), Shewanella oneidensis (11), Thermococcus gammatolerans (12), Deinococcus deserti (13), Salmonella thyphimurium (14), Mycobacterium tuberculosis (15, 16), Shigella flexneri (17), Ruegeria pomeroyi (18), and Candida glabrata (19), as well as for higher organisms such as Anopheles gambiae (20) and Arabidopsis thaliana (4, 5).
The most frequently reported problem in automatic annotation systems is the correct identification of the translational start codon (21–23). The error rate depends on the primary annotation system, but also on the organism, as reported for Halobacterium salinarum and Natromonas pharaonis (24), Deinococcus deserti (21), and Ruegeria pomeroyi (18), where the error rate is estimated above 10%. Identification of a correct translational start site is essential for the genetic and biochemical analysis of a protein because errors can seriously impact subsequent biological studies. If the N terminus is not correctly identified, the protein will be considered in either a truncated or extended form, leading to errors in bioinformatic analyses (e.g. during the prediction of its molecular weight, isoelectric point, cellular localization) and major difficulties during its experimental characterization. For example, a truncated protein may be heterologously produced as an unfolded polypeptide recalcitrant to structure determination (25). Moreover, N-terminal modifications, which are poorly documented in annotation databases, may occur (26, 27).
Unfortunately, the poor polypeptide sequence coverage obtained for the numerous low abundance proteins in current shotgun MS/MS proteomic studies implies that the overall detection of N-terminal peptides obtained in proteogenomic studies is relatively low. Different methods for establishing the most extensive list of protein N termini, grouped under the so-called “N-terminomics” theme, have been proposed to selectively enrich or improve the detection of these peptides (2, 28, 29). Large N-terminome studies have recently been reported based on resin-assisted enrichment of N-terminal peptides (30) or terminal amine isotopic labeling of substrates (TAILS) coupled to depletion of internal peptides with a water-soluble aldehyde-functionalized polymer (31–35). Among the numerous N-terminal-oriented methods (2), specific labeling of the N terminus of intact proteins with N-tris(2,4,6-trimethoxyphenyl)phosphonium acetyl succinamide (TMPP-Ac-OSu)1 has proven reliable (21, 36–39). TMPP-derivatized N-terminal peptides have interesting properties for further LC-MS/MS mass spectrometry: (1) an increase in hydrophobicity because of the trimethoxyphenyl moiety added to the peptides, increasing their retention times in reverse phase chromatography, (2) improvement of their ionization because of the introduction of a positively charged group, and (3) a much simpler fragmentation pattern in tandem mass spectrometry. Other reported approaches rely on acetylation, followed by trypsin digestion, and then biotinylation of free amino groups (40); guanidination of lysine lateral chains followed by N-biotinylation of the N termini and trypsin digestion (41); or reductive amination of all free amino groups with formaldehyde preceeding trypsin digestion (42). Recently, we applied the TMPP method to the proteome of the Deinococcus deserti bacterium isolated from upper sand layers of the Sahara desert (13). This method enabled the detection of N-terminal peptides allowing the confirmation of 278 translation initiation codons, the correction of 73 translation starts, and the identification of non-canonical translation initiation codons (21). However, most TMPP-labeled N-terminal peptides are hidden among the more abundant internal peptides generated after proteolysis of a complex proteome, precluding their detection. This results in disproportionately fewer N-terminal validations, that is, 5 and 8% of total polypeptides coded in the theoretical proteomes of Mycobacterium smegmatis (37) and Deinococcus deserti (21) with a total of 342 and 278 validations, respectively.
An interesting chromatographic method to fractionate peptide mixtures for gel-free high-throughput proteome analysis has been developed over the last years and applied to various topics (43, 44). This technique, known as COmbined FRActional DIagonal Chromatography (COFRADIC), uses a double chromatographic separation with a chemical reaction in between to change the physico-chemical properties of the extraneous peptides to be resolved from the peptides of interest. Its previous applications include the separation of methionine-containing peptides (43), N-terminal peptide enrichment (45, 46), sulfur amino acid-containing peptides (47), and phosphorylated peptides (48). COFRADIC was identified as the best method for identification of N-terminal peptides of two archaea, resulting in the identification of 240 polypeptides (9% of the theoretical proteome) for Halobacterium salinarum and 220 (8%) for Natronomonas pharaonis (24).
Taking advantage of both the specificity of TMPP labeling, the resolving power of COFRADIC for enrichment, and the increase in information through the use of multiple proteases, we performed the proteogenomic analysis of a marine bacterium from the Roseobacter clade, namely Roseobacter denitrificans OCh114. This novel approach allowed us to validate and correct 534 unique proteins (13% of the theoretical proteome) with TMPP-labeled N-terminal signatures obtained using high-resolution tandem mass spectrometry. We corrected 41 annotations and detected five new open reading frames in the R. denitrificans genome. We further identified eight distinct proteins showing direct evidence for multiple start sites.
EXPERIMENTAL PROCEDURES
Preparation of Protein Extracts
R. denitrificans cells were grown in Erlenmeyer flasks containing 400 ml of Marine Broth (MB, Difco, Detroit, MI), incubated at 30 °C and agitated at 180 rpm. Cells were harvested by centrifugation at 6000 x g and 4 °C. Cell pellets were washed twice with ice cold 20 mm Tris/HCl buffered at pH 8.0. A quantity of 400 mg of cells (wet weight) was resuspended in 2 ml of cold buffer P consisting of 100 mm NaH2PO4/Na2HPO4 buffered at pH 8.2 (at 20 °C) and containing a protease inhibitor mixture (Complete, Roche Applied Science, six tablets in 250 ml). Cells were disrupted using a sonicator Bioblock 75042 with a total applied energy of 60 J. The sample was centrifuged for 30 min at 16,000 x g and 4 °C to remove cellular debris. The resulting supernatant was further desalted by chromatography on two 5 ml HiTrap Desalting columns mounted in series (GE Healthcare) and operated with an Äkta Purifier 100 FPLC system (GE Healthcare) with buffer P. The sample was applied at a flow rate of 1 ml/min to columns previously equilibrated with buffer P.
N-terminal Chemical Labeling of R. denitrificans Protein Extracts
TMPP reagent (TMPP-AcOSu) from Fluka was dissolved at 200 mm in 20% acetonitrile (ACN)/80% H2O (v/v) immediately before use. To each protein extract (500 μg), 15.6 μl of 200 mm TMPP-AcOSu was added (ratio of 250 TMPP molecules per protein). The mixture was incubated at room temperature for 60 min under gentle agitation. To stop the reaction, 417 μl of 1.5 m Tris/HCl buffered at pH 8.5 (ratio of 200 Tris molecules per TMPP) were added. The mixture was incubated at room temperature for 30 min and then desalted on two 5 ml HiTrap Desalting columns in series in 100 mm sodium phosphate buffered at pH 7.2 and containing 150 mm NaCl at a flow rate of 1 ml/min. After desalting, proteins were reduced with 0.05% tributylphosphine (Aldrich, Milwaukee, WI) at room temperature for 90 min and then alkylated with 5 mm iodoacetamide (Sigma) in the dark for 30 min at 37 °C. The mixture was again desalted as above.
Acetylation of Intact Proteins
Sulfo-NHS acetate (Thermo) was hydrated at a concentration of 1 m in 100 mm sodium phosphate buffered at pH 7.2 and containing 150 mm NaCl immediately before use, as it readily hydrolyzes and becomes unreactive. Sulfo-NHS acetate solution was added to the TMPP-labeled protein extract to a final concentration of 10 mm and incubated 90 min at 30 °C. The mixture was desalted on two 5 ml HiTrap Desalting columns operated in series at 1 ml/min in 50 mm NH4HCO3 buffered at pH 7.8.
Proteolysis
The proteins were digested either with sequencing grade trypsin, chymotrypsin, or endoproteinase Glu-C (Roche Applied Science) reconstituted in 0.01% TFA, 1 mm HCl, or milliQ water, respectively, at a final concentration of 0.1 μg/ml. The enzyme solution (10 μg) was added to the proteins (2% w/w) for overnight proteolysis at 37 °C for trypsin and 25 °C for both chymotrypsin and endoproteinase Glu-C.
COFRADIC
Samples from each enzyme were dried separately using a SPD121 SpeedVac (Savant) and redissolved in 110 μl of solvent A consisting in 0.1% trifluoroacetic acid (TFA) in water/acetonitrile (98:2 v/v). The peptide mix (100 μl) was resolved on a reverse phase 300SB-C18 Zorbax HPLC column (4.6 mm x 150 mm, 5 μm) from Agilent Technologies operated at room temperature with an Agilent 1100 Series HPLC system equipped with G1315B diode array detector, G1322A degasser, G1311A quaternary pump, G1329A autosampler, and G1364C Analyst FC fraction collector. A linear gradient from 0 to 100% solvent B (30:70 v/v 0.1% TFA in water/acetonitrile) was applied over 100 min with a flow rate of 0.4 ml/min. Peptides eluting between 40 and 76 min were collected in 9 fractions of 1.6 ml each. These fractions were dried in a vacuum centrifuge. Each fraction was then redissolved in 70 μl of 50 mm sodium borate (Na2B4O7/H3BO3), pH 9.2. The 2,4,6-trinitrobenzensulfonic acid (TNBS) reagent (Fluka, Ronkonkoma, NY) was diluted to 1 mm in a solution of 50 mm sodium borate (Na2B4O7/H3BO3), pH 9.2. TNBS (4 nmol) was added to each fraction and incubated for 90 min at 37 °C under gentle agitation. The samples were dried before the second HPLC run. Each fraction was redissolved in 75 μl of 0.5% TFA from which 70 μl were loaded onto the reverse phase column. The solvent gradient was performed as in the primary run, and N-terminal peptides were collected in the same 4 min intervals. The nine resulting fractions were dried, redissolved in 11 μl of 0.1% TFA, and 10 μl were analyzed using nanoLC-MS/MS.
NanoLC-MS/MS Analysis
NanoLC-MS/MS experiments were performed on an LTQ-Orbitrap XL hybrid mass spectrometer (ThermoFisher) coupled to an UltiMate 3000 LC system (Dionex-LC Packings). The parameters for recording MS/MS spectra were as described previously (21). Peptide mixtures were loaded and desalted on line in a reverse phase precolumn (Acclaim PepMap 100 C18, 5 μm bead size, 100-Å pore size, 5 mm x 300 μm) from LC Packings at a flow rate of 0.3 μl/min. The previously optimized gradient (21) was applied.
Whole Genome Database Mining
Peak lists from the LC-MS/MS raw data were generated with the MASCOT DAEMON version 2.3.2 software (Matrix Science, Boston, MA) using the extract_msn.exe data import filter Xcalibut FT package (version 2.0.7) proposed by ThermoFisher, generated as previously described (49). To compare and optimize the different steps of the protocol, MS/MS spectra were searched against a coding domain sequence (CDS) database comprising the 4129 polypeptides sequences annotated in the genome of R. denitrificans (50), totaling 1,288,451 amino acids with an average of 312 amino acids per polypeptide (NCBI download, 2010/02/17). The 22 most common keratin contaminants and the proteases used in this study were also included in the database. Searches for peptides were performed with Mascot using the following parameters: a mass tolerance of 5 ppm on the precursor ion and 0.5 Da on the MS/MS, static modification of carboxamidomethylated Cys (+57.0215), and variable modifications of oxidized Met (+15.9949), N-terminal acetylation (+42.0106), and TMPP+-Ac (C29H35O10P, +572.1811) or TNBS (C6H2O6N3, +210.9865) labels on the N termini or Lys lateral chains. The variable modification of TMPP+-Ac-Met (C34H44N1O11P1S1, +703.2216 amu as calculated after taking into account the additional positive charge introduced by TMPP) on the N termini was also included; this modification, in combination with a semi-specific digestion requirement, allows the identification of translation initiation at non-AUG codons, such as when GTG codons, which normally code for valine residues, actually encode the initial methionine. Mascot will treat the resulting peptide as if the C terminus is specifically cleaved, but not the N terminus, which begins with a methione, contrary to the theoretically encoded valine. Mis-cleavages were set at 3 for trypsin or chymotrypsin, and 2 for endoproteinase Glu-C. All peptides with a score above its peptide identity threshold set at p < 0.01 with the CDS database and rank 1 were filtered by the IRMa 1.28.0 software (51). At the proteogenomic stage of genome refinement of R. denitrificans OCh114, the MS/MS datasets were searched against an amino acid sequence database corresponding to a six-frame stop-to-stop translation of the genome sequence and that was restricted to open reading frames (ORFs) with at least 40 amino acids. This ORF database comprised 61,652 polypeptide sequences, totaling 6,201,552 amino acids with an average of 101 amino acids per polypeptide. The sequence of the proteases and the 22 most common keratin contaminants were also included in this database. The maximum numbers of mis-cleavages were set as above. Fixed and variable modification searches were performed with the same parameters as described for the CDS database searches.
Criteria for Extracting N-terminal Peptides
Each nanoLC-MS/MS experiment was analyzed first with specific protease cleavages and then in semi-specific mode to identify TMPP-derivatized peptides. All peptide matches with the ORF database were filtered with the IRMA 1.28.0 software with cutoff values at p < 0.01. Redundant assignments to the same from the dual assignment strategy or multiple records by the mass spectrometer were reduced to a single event. Peptides having an acetylation or TMPP N-terminal label were extracted, and only non-redundant N-terminal most peptides corresponding to a possible start (ATG, GTG, and TTG codons) were conserved. Further validation was applied as follows: peptide score, coding length to total ORF length ratio greater than 0.45, similarities detected with other known proteins, and manual annotation of MS/MS spectra.
In Silico Analysis of Protein Sequences
Prediction of secreted proteins was performed by analyzing proteins with evidence of multiple translational starts or incorrect annotations using the prediction tools SignalP 4.0, SecretomeP 2.0, and LipoP 1.0 as described previously (49). PSI-BLAST (http://blast.ncbi.nlm.nih.gov/blast) searches were performed using the NCBI non-redundant protein sequence database and default parameters.
Proteomic Data Repository
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (http://www.ebi.ac.uk/pride/) with the dataset identifier PXD000337.
RESULTS AND DISCUSSION
Enrichment of TMPP-labeled N-terminal Peptides
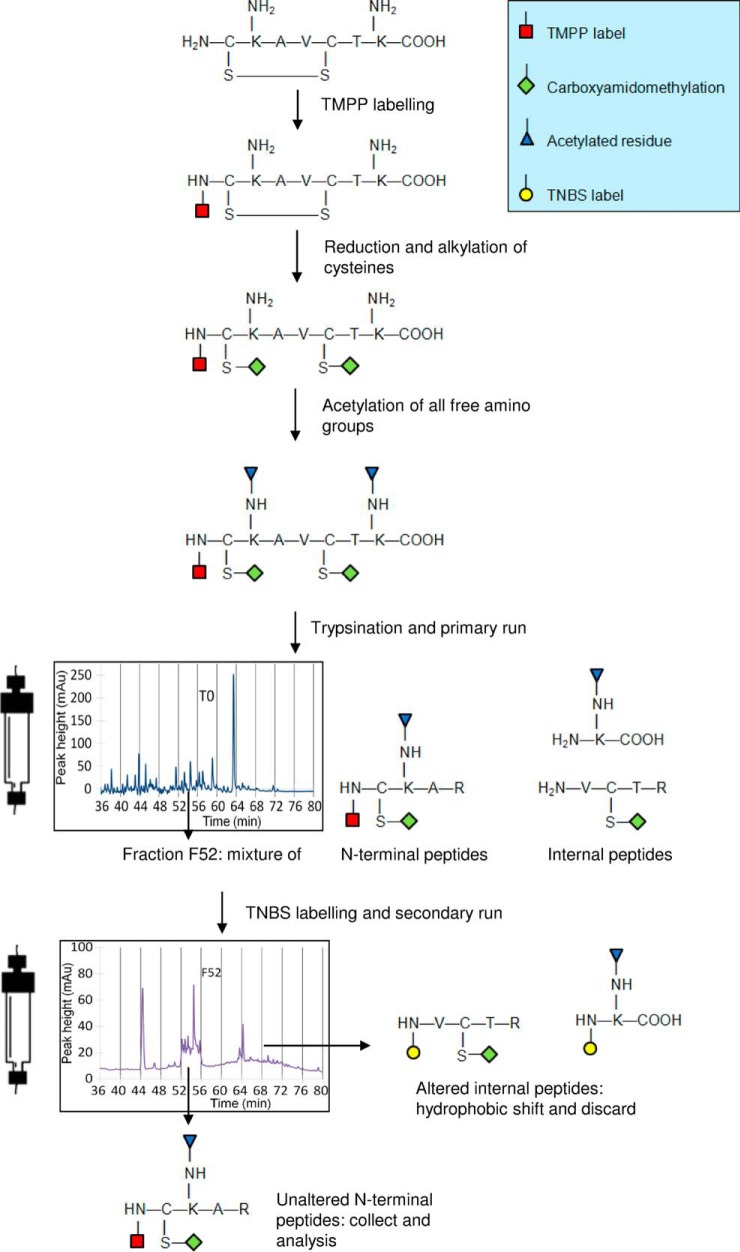
To selectively purify N-terminal peptides, we labeled the whole proteome of R. denitrificans OCh114 with TMPP-Ac-OSu (Fig. 1). After labeling with TMPP, the proteins were proteolyzed (e.g. with trypsin) generating TMPP-labeled N-terminal peptides and unlabeled internal tryptic peptides. The TMPP-labeled peptides, along with co-eluting internal peptides, were collected in an initial chromatographic separation. The co-eluting internal peptides were labeled with a second reagent, which reacts with free N-terminal amines. The newly labeled internal peptides are more hydrophobic and were thus removed in the second chromatographic step. We chose TNBS as the second labeling reagent, and to avoid any labeling of free amino groups from lateral chains with this second reagent that would inhibit enrichment, we introduced a preliminary protection step by acetylating lysine lateral chains with sulfo-NHS-acetate.
Fig. 1.
Strategy applied for N termini enrichment. Proteins are first labeled with TMPP on their N terminus before reduction and alkylation of cysteines. All free amino groups are further protected through an acetylation step. Protein digestion generates a mixture containing both N-terminal TMPP-labeled peptides and non-labeled internal peptides that is resolved by C18 reverse-phase liquid chromatography. Fractions with eluted peptides are isolated and treated separately with TNBS reagent, which reacts with free amino groups of internal peptides. A second chromatographic run carried out in exactly the same conditions is performed to resolve TNBS-altered internal peptides (higher hydrophobicity) from the non-modified TMPP-labeled N-terminal peptides eluted at the same retention time. The chromatogram of fraction F52 after the second chromatography is shown. The N-terminal peptides of interest are further collected and analyzed by tandem mass spectrometry.
To test the efficiency of this acetylation step, we analyzed an acetylated tryptic digest (“Ta” in Table I) and compared the results with those obtained with an equivalent non-acetylated sample (“Tb” in Table I). Both fractions were analyzed in a single nanoLC-MS/MS run, and MS/MS spectra were searched against the CDS database. A complete list of assigned peptides and proteins is given in supplemental Table S1. No acetylated peptides were detected when the sample was not treated with sulfo-NHS-acetate (“Tb” in Table I), whereas the acetylated sample showed 73% protection of lysine residues (“Ta” in Table I). In the acetylated sample, 113 distinct N-terminal peptides matching annotated proteins were found, corresponding to 89 non-redundant polypeptides. Of these, 65 were detected with their N terminus labeled with TMPP; 23 were acetylated at the N terminus, and one was unmodified. Although useful for protecting lysine residues before TNBS labeling, acetylation introduces steric hindrance (42, 52), which inhibits trypsin recognition of the lysine residue as a cleavage site and thus inhibits digestion. Without acetylation, 18% of identified peptides had one mis-cleavage; with acetylation, the percentage of peptides with one mis-cleavage increases to 31. Furthermore, 4% of peptides were identified with three mis-cleavages. In addition to having more mis-cleavages, fewer peptides ending with a lysine residue were found in the acetylated sample. Only 18% of peptides ended with a lysine, compared with 42% in the non-acetylated sample (Supplemental Table S1).
Table I. TMPP assignment results from the CDS database searchesa.
| Number of assigned MS/MS spectra | Number of non-redundant peptides (unique peptide sequences and modifications) | Number of N-terminus peptide signatures | Number of unique proteins with an N-terminus signature | Number of proteins with labeled TMPP peptides | % of acetylated lysines on non-redundant peptides | |
|---|---|---|---|---|---|---|
| Tb | 4183 | 2049 | 128 | 102 | 98 | 0% |
| Ta | 3461 | 1731 | 113 | 89 | 65 | 73% |
| T1 | 10,296 | 3449 | 325 | 230 | 165 | 70% |
| T2 | 3529 | 1559 | 386 | 291 | 212 | 83% |
a Peptide information is indicated in upplementary Table S1.
We resolved the TMPP-labeled and acetylated peptides on a reverse-phase column and collected peptides eluting between 44 and 60 min in four fractions of 1.6 ml each. These fractions were then analyzed using nanoLC-MS/MS. Data from the four runs (“T1” in Table I) resulted in the identification of 3449 different peptides (Table I and supplemental Table S1). Of these, 325 were N-terminal peptides. As expected, increasing the number of spectra acquired in this experiment compared with our previous analysis (“Ta” in Table I) led to a higher number of peptides identified. We then modified the same four fractions with TNBS and subjected them to diagonal reverse-phase chromatography. The four resulting fractions were then analyzed by nanoLC-MS/MS. This dataset (“T2” in Table I) yielded less than half of the total identifications from Data “T1” (1559) but a higher number of N-terminal peptides (386). After the first chromatography step, 9% of identified peptides corresponded to N termini, whereas after the second chromatography step, they increased to 25%, showing a clear enrichment of these specific peptides. Only 25 TNBS-labeled peptides were found in these four fractions (“T2” in Table I) and none corresponded to the N terminus of a protein. As expected, fractions eluting with a delay contained a higher ratio of TNBS-labeled internal peptides (data not shown). A total of 291 unique proteins were detected through their N-terminal peptides, compared with 230 found without the intermediate COFRADIC step. Therefore, this method is efficient at enriching N-terminal peptides.
Spectral Assignment Using the CDS and Six-frame ORF Databases
Using nanoLC-MS/MS, we analyzed all the COFRADIC fractions from the tryptic digest, generating 93,545 MS/MS spectra (supplemental Table S2). After merging the search results and removing the peptides assigned to ambiguous loci, contaminants, and protease autolysis, 61,849 MS/MS spectra assigned to tryptic or semi-tryptic peptides remained. These peptides correspond to 1556 potential N-terminal sequences with the conventional translational start codons ATG, GTG, and TTG (supplemental Table S3). MS/MS data acquired from chymotrypsin and endoproteinase Glu-C digests were processed as for the tryptic digest, allowing 3 and 2 mis-cleavages, respectively. After processing, samples from these two enzymes yielded an additional 24,052 unique MS/MS spectra. In total, 2603 peptides beginning with the possible start codons ATG, GTG, or TTG (supplemental Table S3) were found, corresponding to 840 non-redundant proteins. From the whole peptide pool, the vast majority (1556) arose from trypsin, with 473 and 574 generated by chymotrypsin and endoproteinase Glu-C, respectively. The false discovery rate for peptide assignment, estimated using the same search strategy with a decoy database, was less than 1% with both CDS and six-frame ORF databases. Decoy databases were constructed using the “decoy” function in the Mascot Daemon and consisted of reversed amino acid sequences from the target databases.
Validation of TMPP-labeled N-terminal Peptides
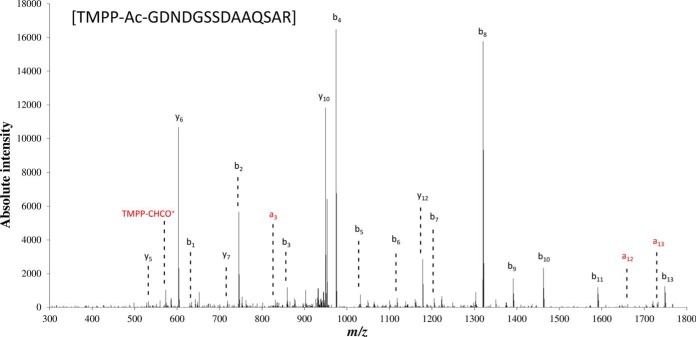
For automatic validation of N-terminal start sites we took advantage of data redundancy arising from differences in terms of mis-cleavages, methionine oxidation, different proteolytic enzymes (i.e. trypsin, chymotrypsin, and endoproteinase Glu-C), and partial removal of the initial methionine. Chymotrypsin specifically hydrolyzes peptide bonds at the C termini of tyrosine, phenylalanine, and tryptophan; leucine, methionine, alanine, aspartic acid, and glutamic acid are cleaved at a lower rate (53). Endoproteinase Glu-C cleaves peptide bonds at the C-terminal of glutamic acid and, at a much lower rate (3000 fold), aspartic acid (54). When the same N-terminal sequence is observed following digestion with two or more enzymes, these multiple pieces of evidence corroborate valid protein N termini. Of the total 2603 N-terminal peptide identifications matching previously annotated proteins, 349 were automatically validated based on redundant identifications. The false discovery rate for N-terminal peptide assignment was estimated to be less than 1% using the same approach and the ORF target-decoy database. Most of the detected proteins (66%) were identified from their N-terminal initiation peptides recorded with only one protease, whereas 25 and 9% of the proteins were detected with a combination of two or all three proteases, respectively. Of those detected with at least two proteases, more proteins are identified with the combination of trypsin and endoproteinase Glu-C (70 proteins), than with trypsin and chymotrypsin (45 proteins) or endoproteinase Glu-C and chymotrypsin (eight proteins). Protease mis-cleavages allowed the corroboration of sequences pointing at the same N-terminal initiation, more so even than parallel proteolysis with different enzymes. Here, trypsin was responsible for most of the mis-cleavages, mainly because of proteolysis inhibition by lysine acetylation. The 491 remainder proteins seen with only one peptide could be further evaluated. We validated 131 protein identifications produced from unique peptides after manual inspection of the corresponding MS/MS spectra. Criteria for inspection included the detection of at least one of the three described TMPP reporter ions, the a1 and b1 ions, and the presence of a ions; only peptides in the top third of the peptide list ranked by ion score, that is, above 23, were considered (supplemental Table S4). As an example, Fig. 2 shows the MS/MS spectrum attributed to the N terminus of YP_684061.1, a hemolysin-like protein. The presence of the TMPP-CHCO+ reporter ion, the b1 ion, the presence of three a ions, and the almost complete b ion series (b1-b11) justifies the assignment of this MS/MS spectrum to the GDNDGSSDAAQSAR peptide.
Fig. 2.
Manual verification of a TMPP-modified peptide. The fragmentation spectrum of the N-terminal TMPP-modified peptide [GDNDGSSDAAQSAR] assigned to the protein YP_684061.1 is shown. The almost complete b ion series and the reporter ion TMPP-CHCO+ give a high confidence in the assignment of this MS/MS spectrum. The a ions series and the reporter ion are indicated in red.
Of the 480 resulting N termini that matched previous annotations, 63% were exclusively detected with a cleaved initial methionine, whereas 9% showed partial removal of this N-terminal amino acid. Methionine removal occurred when alanine, serine, and threonine, and to a lesser extent proline, glycine, valine, and asparagine, are in the second position of the N-terminal peptide sequence, confirming what has been reported previously (55, 56). Incomplete methionine excision was mainly observed with threonine or asparagine in second position of the polypeptide. Their longer lateral chain compared with the other cleavable residues may explain this pattern.
Re-annotation of the R. denitrificans OCh114 Genome
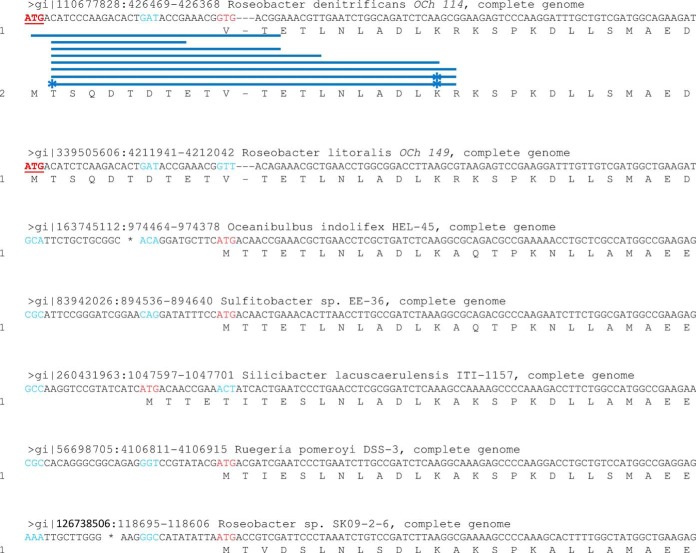
A total of 41 proteins were identified with N termini different from their initial annotation (supplemental Table S4). Of these 41 proteogenomic re-annotations, 18 are longer than previously annotated and 23 are shorter. Fig. 3 shows eight different peptides corresponding to the correct N terminus of YP_680835.1, a Rho transcription termination factor, for which 10 residues were missing in the primary annotation. The true start codon (ATG) is only conserved in the closely related strain R. litoralis OCh149 and not in other Roseobacter strains (Fig. 3). The previously annotated start site (GTG) was likely erroneously determined based on similarity with the ATG start codon of other homologs.
Fig. 3.
Re-annotation of the transcription termination factor Rho from R. denitificans OCh114. Synthesis of the protein YP_680835.1 was predicted to start from a GTG initiation codon based on homologous sequences found in other members of the Roseobacter clade. However, N-terminal peptides demonstrated that the real start site was 30 nucleotides upstream from the initial annotation (ATG codon, underlined and in bold red). This codon was only conserved in the closest-related strain R. litoralis Och149. Previously annotated codons are shown in red and their counterparts in other sequences are in blue. Acetylated residues are indicated by a star.
The gene coding for the fructose-1,6-bisphosphate aldolase (YP_682542.1) was also mis-annotated, as evidenced by four TMPP-labeled peptides originating six amino acids upstream of the previous annotation. Unlike the previous example, the correct start site of this protein is more conserved in closely related homologs than the initial annotation. Ten different peptides indicated a correction of the initiation codon for the 50S ribosomal protein L21 (YP_682897.1) 174 nucleotides downstream from the previously annotated start site, which produces a 58 amino acid truncation. For the transcription elongation factor GreA (YP_683583.1), nine peptides indicated a well-defined start site 468 nucleotides downstream of the current CDS start site (half of the previously annotated sequence). Similarly, TMPP-labeled peptides also indicated a mis-annotation for the DNA-directed RNA polymerase subunit alpha (YP_681761.1) and the polyamine ABC transporter, ATP-binding protein (YP_680591.1). In this later case, the mass spectrometry-certified translation start site is a TTG codon, which is strictly conserved in closely related homologs.
Evidence of Multiple Translational Starts in Eight Proteins
Multiple translational start sites were found for eight proteins, as evidenced by several TMPP-labeled peptides (supplemental Table S4). Alternative start sites both up- and downstream of the original annotated starts were observed. For the ATP-dependent Clp protease proteolytic subunit (YP_683070.1), 16 different TMPP-labeled peptides indicated three start sites: the current CDS annotation starting at position 2,727,465 of the chromosome (confirmed with seven peptides), and two other translational start sites located at the positions 2,727,441 and 2,727,426 confirmed with eight and one peptides, respectively (Fig. 4). These start sites correspond to gene products of 208, 200, and 195 amino acids in length, respectively. PSI-BLAST indicated that all three start codons are strictly conserved in closely related homologs. The N terminus of Clp proteolytic subunit is highly conserved in general, as the 20 most N-terminal amino acids coordinate binding with the ATPase subunit of this multimeric protease (57). The N terminus adopts different conformations to control this coordination and to gate the proteolytic chamber (58), and truncations of the N terminus allow degradation of large substrates in the absense of the ATPase subunit (59) and increase the initial degradation rate while stabilizing the acyl-enzyme intermediate (60). In E. coli, ClpP is first synthesized with a 14-amino acid signal peptide; however, the immature polypeptide has a half-life of ∼2 min, and the mature product is then extremely stable (61). The N termini of ClpP identified here in R. denitrificans Och114 correspond to the mature N terminus of the E. coli homolog, eight amino acids upstream, and five amino acids downstream. The existence of multiple isoforms of this N terminus could be indicative of a prolonged half-life of the immature polypeptide, or it could play a role in controlling the rate of degradation and/or the selectivity of ClpP in this strain.
Fig. 4.
Multiple translation start sites of the ATP-dependent Clp protease proteolytic subunit. Three alternative start sites were recorded by multiple detected N-terminal peptides (represented as blue lines) for protein YP_683070.1. A multiple sequence alignment with closely-related homologs demonstrates the heterogeneity of initiation sites and how this protein should be re-annotated in multiple species as all three codons are strictly conserved (ATG codons underlined and in bold red). The translated polypeptides from the previous annotation and from the new corrected annotation are labeled 1 and 2, respectively.
An alternative translational start was also detected for the ferredoxin-NADP reductase YP_683175.1 (supplemental Fig. S1), corresponding to the original annotation and a six-amino acid truncation. The ferredoxin-NADP oxidoreductase gene petH from the cyanobacterium Synechocystis sp. Strain PCC6803 has three transcriptional start sites, producing two translation products corresponding to a 413- and a 300-amino acid isoforms of this protein, which are differentially expressed in response to nitrogen availability (62). In both R. denitrificans Och114 and Synechocystis, the truncated N-terminal region is outside of the conserved domains. In Synechocystis, this region is responsible for interaction with phycobilisome, and thus binding is inhibited in its absence. Similarly, the six-amino acid N-terminal region of this ferredoxin-NADP reductase in R. denitrificans Och114 may be used to influence its interaction with certain partners.
Similarly, two translational starts were detected for the recombinase A (YP_682935.1), one of which corresponds to the original annotation and the other seven amino acids downstream. The agmatinase (YP_682939.1) was observed with one start site corresponding to the original annotation and another seven amino acids upstream. The dnaK suppressor protein YP_683008.1 similarly had a start site 11 amino acids upstream, in addition to the original annotation. All of the alternative start sites leave the conserved functional domain intact. The hypothetical protein RD1_1231 (YP_681565.1), for which 17 TMPP-labeled peptides were recorded (supplemental Fig. S2 and Table S4), was also observed with two start sites: 12 peptides corresponded to the CDS annotation and five corresponded to a start site nine nucleotides downstream. Alternative in-frame translation starts have been described for several bacterial proteins, such as the outer membrane protease IcsP from Shigella flexneri (63), and the type IV pili component PilA from Geobacter sulfurreducens (64).
Evidence for a Protein with a Non-canonical Translational Start Codon
Two start sites were found for the iojap-like protein (YP_681006.1). Two TMPP-labeled peptides corresponding to the N terminus of the existing annotation were observed; another five peptides corresponded to the unconventional start codon CTG at position 602,762 of the chromosome, implying a 16-amino acid prolongation. The annotated N termini of homologs of this protein from closely-related bacteria are heterogeneous, but the CTG codon is strictly conserved among these genomes (Fig. 5). This non-canonical alternative start codon may therefore be common to all of these organisms. Because a serine residue is found in second position, the initial methionine should be systematically removed, which is in agreement with the observed TMPP-labeled peptides. The human iojap ortholog has been found associated with the large subunit of the mitochondrial ribosome in humans (65), whereas the plant ortholog has been shown to be vital for chloroplast ribosome biogenesis (66), and several bacterial orthologs (RsfA/YbeB) are conserved ribosomal silencing factors (67).
Fig. 5.
Re-annotation of the iojap-like protein initiated with a non-conventional start codon. The sequence alignment of the iojap-like protein (YP_681006.1) is shown with closely-related bacteria. The translated polypeptide from the previous annotation and from the new corrected annotation are labeled 1 and 2, respectively. Previously annotated codons are shown in red and their counterparts in other sequences are in blue. The two alternative start codons are indicated in bold red and underlined. Blue lines represent the N-terminal peptide sequences identified in R. denitrificans OCh114. Initiation heterogeneity is demonstrated and re-annotation of this protein in closely-related species is suggested in red.
Discovery of Five Un-annotated CDS
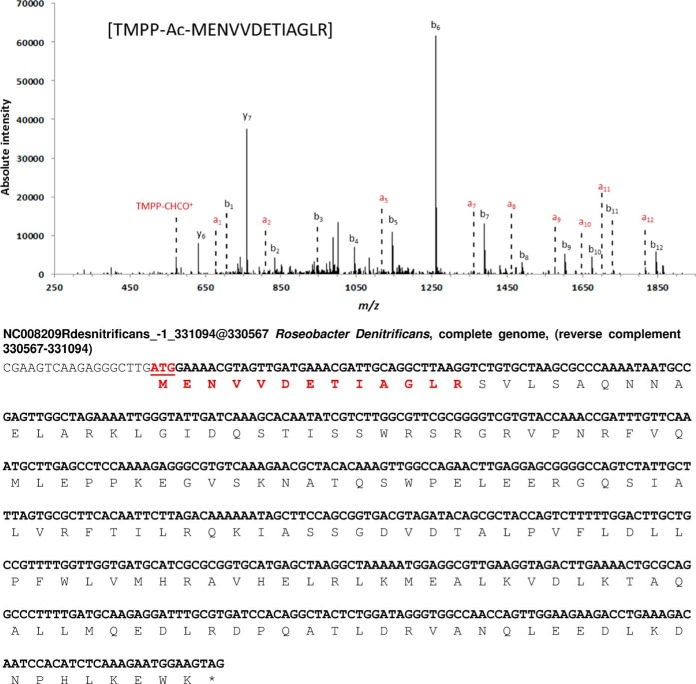
Five previously un-annotated genes in the genome of R. denitrificans OCh114 were detected during this study at loci 331,094–330,567 (frame −1), 338,913–339,212 (frame 3), and 1,376,295–1,377,065 (frame 3) on the chromosome and loci 4139–4615 (frame 2) and 4012–3548 (frame −3) on the pTB4 plasmid (supplemental Table S5). The coordinates of these genes do not overlap with those of previously annotated genes, potential promoters and Shine-Dalgarno sequences exist upstream of each coding sequence, and the corresponding peptides could not be assigned to any other gene. The ORF located at the position 331,094–330,567 on the chromosome was validated with two different peptides obtained during the semi-trypsin and semi-endoproteinase Glu-C analysis; an annotated spectrum from the semi-triptic peptide, as well as the predicted sequence of the entire gene and corresponding product, is shown in Fig. 6. This orphan CDS has no homologs in the NCBI database and thus no function could be assigned. The novel gene at locus 338,913–339,212 on the chromosome was observed with two translation start sites, both of which were detected by two different TMPP-labeled peptides (supplemental Table S5). The closest homolog to this protein was a “hypothetical protein” found in Roseobacter sp. AzwK-3b, and in this case, only the second translation initiation site is given. The novel gene at locus 1,376,295–1,377,065 on the main chromosome was identified by two peptides. Although not previously annotated in the R. denitrificans OCh114 genome, a homolog of this protein is present in the closely related strain R. litoralis OCh149, where it is defined as a glutamate/glutamine/aspartate/asparagine transport ATP-binding protein BztD. Two additional ORFs at positions 4139–4615 and 4012–3548 on the plasmid pTB4 were identified with one N-terminal TMPP-labeled peptide each. Again, low sequence similarities with other proteins were noted for the two polypeptides and no function could be predicted.
Fig. 6.
Discovery of a novel gene from Roseobacter denitrificans OCh114. MS/MS spectrum of the TMPP-modified peptide [MENVVDETIAGLR] which indicated the N terminus of the newly-discovered gene encoded on the chromosome in frame −1 between positions 331,094 and 330,567 (nucleotide sequence displayed). The peptide detected by tandem mass spectrometry is highlighted in bold red in the sequence. On the fragmentation spectrum, the reporter ion TMPP-CHCO+ and the a ion series are shown in red. The b ions series is shown in black. The stop codon is indicated with a star symbol.
Concluding Remarks
By combining the TMPP labeling of protein N termini (to increase the ionizability and simplify fragmentation of peptides during mass spectrometry) and the COFRADIC enrichment of such labeled peptides (to remove internal non-informative peptides), we characterized 534 N termini: 480 matching the existing annotation, 41 corrections to the previous annotation, five previously un-annotated proteins, and eight proteins with multiple translational starts. Of the observed proteins, almost 10% were incorrectly annotated in terms of translational start codon. In some cases, classical homology-driven annotation of genomes may lead to incorrect annotation, such as observed for the transcription termination factor Rho, where the corrected start site was not conserved in different Roseobacter strains. This questions the use of ortho-proteogenomics, as first proposed by Gallien and coworkers (37), which uses mass spectrometry-based empirical evidence from one strain to evaluate other related organisms. Indeed, this observation emphasizes the need for empirical evaluation of every sequenced organism for maximum annotation accuracy. Furthermore, multiple translational start sites were observed for several proteins. Although it is widely acknowledged for eukaryotes that a single gene may code for several protein variants (68), the same generality has yet to be demonstrated for prokaryotes. This proteome-wide proteomic analysis of R. denitrificans OCh114 shows that such phenomena exist on a moderate scale in bacteria, an observation that challenges the conventional idea that each gene encodes one unique protein sequence in bacteria (69).
Supplementary Material
Acknowledgments
We thank Attila Csordas and Juan Antonio Vizcaino for their help regarding data repository, as well as our colleague Elizabeth Darrouzet for valuable discussion.
Footnotes
Author contributions: C.B., E.M.H., J.A.C., B.F., and J.A. designed research; C.B., J.A.C., and B.F. performed research; C.B., E.M.H., J.A.C., B.F., and J.A. analyzed data; C.B., E.M.H., J.A.C., and J.A. wrote the paper.
* This work was supported by the Commissariat à l'Energie Atomique et aux Energies Alternatives, and the Région Languedoc-Roussillon (label ≪ Chercheur d'Avenir Confirmé ≫ 2010). CB gratefully acknowledges the Commissariat à l'Energie Atomique et aux Energies Alternatives and the Région Languedoc-Roussillon for their support. JAC-O acknowledges the Ramon Areces foundation and FP7 Marie Curie Actions for financial support.
 This article contains supplemental Figs. S1 and S2 and Tables S1 to S5.
This article contains supplemental Figs. S1 and S2 and Tables S1 to S5.
1 The abbreviations used are:
- TMPP Ac-OSu
- N-tris(2,4,6-trimethoxyphenyl)phosphonium bromide
- COFRADIC
- COmbined FRActional DIagonal Chromatography
- CDS
- coding domain sequence
- ORF
- open reading frame.
REFERENCES
- 1. Armengaud J. (2010) Proteogenomics and systems biology: quest for the ultimate missing parts. Expert Rev. Proteomics 7, 65–77 [DOI] [PubMed] [Google Scholar]
- 2. Armengaud J. (2009) A perfect genome annotation is within reach with the proteomics and genomics alliance. Curr. Opin. Microbiol. 12, 292–300 [DOI] [PubMed] [Google Scholar]
- 3. Armengaud J., Bland C., Christie-Oleza J., Miotello G. (2011) Microbial Proteogenomics, Gaining Ground with the Avalanche of Genome Sequences. J. Bacteriol. Parasitol. S3-001 [Google Scholar]
- 4. Baerenfaller K., Grossmann J., Grobei M. A., Hull R., Hirsch-Hoffmann M., Yalovsky S., Zimmermann P., Grossniklaus U., Gruissem W., Baginsky S. (2008) Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320, 938–941 [DOI] [PubMed] [Google Scholar]
- 5. Castellana N. E., Payne S. H., Shen Z., Stanke M., Bafna V., Briggs S. P. (2008) Discovery and revision of Arabidopsis genes by proteogenomics. Proc. Natl. Acad. Sci. U.S.A. 105, 21034–21038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Castellana N. E., Shen Z., He Y., Walley J. W., Cassidy C. J., Briggs S. P., Bafna V. (2014) An Automated Proteogenomic Method Uses Mass Spectrometry to Reveal Novel Genes in Zea mays. Mol. Cell. Proteomics : MCP 13, 157–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Krug K., Nahnsen S., Macek B. (2011) Mass spectrometry at the interface of proteomics and genomics. Mol. Biosys. 7, 284–291 [DOI] [PubMed] [Google Scholar]
- 8. Renuse S., Chaerkady R., Pandey A. (2011) Proteogenomics. Proteomics 11, 620–630 [DOI] [PubMed] [Google Scholar]
- 9. Jaffe J. D., Berg H. C., Church G. M. (2004) Proteogenomic mapping as a complementary method to perform genome annotation. Proteomics 4, 59–77 [DOI] [PubMed] [Google Scholar]
- 10. Savidor A., Donahoo R. S., Hurtado-Gonzales O., Verberkmoes N. C., Shah M. B., Lamour K. H., McDonald W. H. (2006) Expressed peptide tags: an additional layer of data for genome annotation. J. Proteome Res. 5, 3048–3058 [DOI] [PubMed] [Google Scholar]
- 11. Gupta N., Tanner S., Jaitly N., Adkins J. N., Lipton M., Edwards R., Romine M., Osterman A., Bafna V., Smith R. D., Pevzner P. A. (2007) Whole proteome analysis of post-translational modifications: applications of mass-spectrometry for proteogenomic annotation. Genome Res. 17, 1362–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zivanovic Y., Armengaud J., Lagorce A., Leplat C., Guerin P., Dutertre M., Anthouard V., Forterre P., Wincker P., Confalonieri F. (2009) Genome analysis and genome-wide proteomics of Thermococcus gammatolerans, the most radioresistant organism known amongst the Archaea. Genome Biol. 10, R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. de Groot A., Dulermo R., Ortet P., Blanchard L., Guerin P., Fernandez B., Vacherie B., Dossat C., Jolivet E., Siguier P., Chandler M., Barakat M., Dedieu A., Barbe V., Heulin T., Sommer S., Achouak W., Armengaud J. (2009) Alliance of proteomics and genomics to unravel the specificities of Sahara bacterium Deinococcus deserti. PLoS Genet 5, e1000434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ansong C., Tolic N., Purvine S. O., Porwollik S., Jones M., Yoon H., Payne S. H., Martin J. L., Burnet M. C., Monroe M. E., Venepally P., Smith R. D., Peterson S. N., Heffron F., McClelland M., Adkins J. N. (2011) Experimental annotation of post-translational features and translated coding regions in the pathogen Salmonella Typhimurium. BMC Genomics 12, 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kelkar D. S., Kumar D., Kumar P., Balakrishnan L., Muthusamy B., Yadav A. K., Shrivastava P., Marimuthu A., Anand S., Sundaram H., Kingsbury R., Harsha H. C., Nair B., Prasad T. S., Chauhan D. S., Katoch K., Katoch V. M., Chaerkady R., Ramachandran S., Dash D., Pandey A. (2011) Proteogenomic analysis of Mycobacterium tuberculosis by high resolution mass spectrometry. Mol. Cell. Proteomics 10, M111 011627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. de Souza G. A., Wiker H. G. (2011) A proteomic view of mycobacteria. Proteomics 11, 3118–3127 [DOI] [PubMed] [Google Scholar]
- 17. Zhao L., Liu L., Leng W., Wei C., Jin Q. (2011) A proteogenomic analysis of Shigella flexneri using 2D LC-MALDI TOF/TOF. BMC Genomics 12, 528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Christie-Oleza J. A., Miotello G., Armengaud J. (2012) High-throughput proteogenomics of Ruegeria pomeroyi: seeding a better genomic annotation for the whole marine Roseobacter clade. BMC Genomics 13, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Prasad T. S., Harsha H. C., Keerthikumar S., Sekhar N. R., Selvan L. D., Kumar P., Pinto S. M., Muthusamy B., Subbannayya Y., Renuse S., Chaerkady R., Mathur P. P., Ravikumar R., Pandey A. (2012) Proteogenomic analysis of Candida glabrata using high resolution mass spectrometry. J. Proteome Res. 11, 247–260 [DOI] [PubMed] [Google Scholar]
- 20. Chaerkady R., Kelkar D. S., Muthusamy B., Kandasamy K., Dwivedi S. B., Sahasrabuddhe N. A., Kim M. S., Renuse S., Pinto S. M., Sharma R., Pawar H., Sekhar N. R., Mohanty A. K., Getnet D., Yang Y., Zhong J., Dash A. P., MacCallum R. M., Delanghe B., Mlambo G., Kumar A., Keshava Prasad T. S., Okulate M., Kumar N., Pandey A. (2011) A proteogenomic analysis of Anopheles gambiae using high-resolution Fourier transform mass spectrometry. Genome Res. 21, 1872–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Baudet M., Ortet P., Gaillard J. C., Fernandez B., Guerin P., Enjalbal C., Subra G., de Groot A., Barakat M., Dedieu A., Armengaud J. (2010) Proteomics-based refinement of Deinococcus deserti genome annotation reveals an unwonted use of non-canonical translation initiation codons. Mol. Cell. Proteomics : MCP 9, 415–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Poptsova M. S., Gogarten J. P. (2010) Using comparative genome analysis to identify problems in annotated microbial genomes. Microbiology 156, 1909–1917 [DOI] [PubMed] [Google Scholar]
- 23. Venter E., Smith R. D., Payne S. H. (2011) Proteogenomic analysis of bacteria and archaea: a 46 organism case study. PloS One 6, e27587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Aivaliotis M., Gevaert K., Falb M., Tebbe A., Konstantinidis K., Bisle B., Klein C., Martens L., Staes A., Timmerman E., Van Damme J., Siedler F., Pfeiffer F., Vandekerckhove J., Oesterhelt D. (2007) Large-scale identification of N-terminal peptides in the halophilic archaea Halobacterium salinarum and Natronomonas pharaonis. J. Proteome Res. 6, 2195–2204 [DOI] [PubMed] [Google Scholar]
- 25. Sato N., Tajima N. (2012) Statistics of N-terminal alignment as a guide for refining prokaryotic gene annotation. Genomics 99, 138–143 [DOI] [PubMed] [Google Scholar]
- 26. Martinez A., Traverso J. A., Valot B., Ferro M., Espagne C., Ephritikhine G., Zivy M., Giglione C., Meinnel T. (2008) Extent of N-terminal modifications in cytosolic proteins from eukaryotes. Proteomics 8, 2809–2831 [DOI] [PubMed] [Google Scholar]
- 27. Bienvenut W. V., Sumpton D., Martinez A., Lilla S., Espagne C., Meinnel T., Giglione C. (2012) Comparative large scale characterization of plant versus mammal proteins reveals similar and idiosyncratic N-alpha-acetylation features. Mol. Cell. Proteomics : MCP 11, M111.015131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Prudova A., auf dem Keller U., Butler G. S., Overall C. M. (2010) Multiplex N-terminome analysis of MMP-2 and MMP-9 substrate degradomes by iTRAQ-TAILS quantitative proteomics. Mol. Cell. Proteomics : MCP 9, 894–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Van Damme P., Arnesen T., Gevaert K. (2011) Protein alpha-N-acetylation studied by N-terminomics. FEBS J 278, 3822–3834 [DOI] [PubMed] [Google Scholar]
- 30. Kim J. S., Dai Z., Aryal U. K., Moore R. J., Camp D. G., 2nd, Baker S. E., Smith R. D., Qian W. J. (2013) Resin-assisted enrichment of N-terminal peptides for characterizing proteolytic processing. Anal. Chem. 85, 6826–6832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huesgen P. F., Alami M., Lange P. F., Foster L. J., Schroder W. P., Overall C. M., Green B. R. (2013) Proteomic amino-termini profiling reveals targeting information for protein import into complex plastids. PloS One 8, e74483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jefferson T., Auf dem Keller U., Bellac C., Metz V. V., Broder C., Hedrich J., Ohler A., Maier W., Magdolen V., Sterchi E., Bond J. S., Jayakumar A., Traupe H., Chalaris A., Rose-John S., Pietrzik C. U., Postina R., Overall C. M., Becker-Pauly C. (2013) The substrate degradome of meprin metalloproteases reveals an unexpected proteolytic link between meprin beta and ADAM10. Cell. Mol. Life Sci. 70, 309–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kleifeld O., Doucet A., auf dem Keller U., Prudova A., Schilling O., Kainthan R. K., Starr A. E., Foster L. J., Kizhakkedathu J. N., Overall C. M. (2010) Isotopic labeling of terminal amines in complex samples identifies protein N-termini and protease cleavage products. Nat. Biotechnol. 28, 281–288 [DOI] [PubMed] [Google Scholar]
- 34. Kleifeld O., Doucet A., Prudova A., auf dem Keller U., Gioia M., Kizhakkedathu J. N., Overall C. M. (2011) Identifying and quantifying proteolytic events and the natural N terminome by terminal amine isotopic labeling of substrates. Nat. Protoc. 6, 1578–1611 [DOI] [PubMed] [Google Scholar]
- 35. Schlage P., Egli F. E., Nanni P., Wang L. W., Kizhakkedathu J. N., Apte S. S., Auf dem Keller U. (2014) Time-resolved analysis of the matrix metalloproteinase 10 substrate degradome. Mol. Cell. Proteomics. In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen W., Lee P. J., Shion H., Ellor N., Gebler J. C. (2007) Improving de novo sequencing of peptides using a charged tag and C-terminal digestion. Anal. Chem. 79, 1583–1590 [DOI] [PubMed] [Google Scholar]
- 37. Gallien S., Perrodou E., Carapito C., Deshayes C., Reyrat J. M., Van Dorsselaer A., Poch O., Schaeffer C., Lecompte O. (2009) Ortho-proteogenomics: multiple proteomes investigation through orthology and a new MS-based protocol. Genome Res. 19, 128–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. An M., Dai J., Wang Q., Tong Y., Ji J. (2010) Efficient and clean charge derivatization of peptides for analysis by mass spectrometry. Rapid comm. Mass Sp. : RCM 24, 1869–1874 [DOI] [PubMed] [Google Scholar]
- 39. Bertaccini D., Vaca S., Carapito C., Arsene-Ploetze F., Van Dorsselaer A., Schaeffer-Reiss C. (2013) An improved stable isotope N-terminal labeling approach with light/heavy Tmpp to automate proteogenomics data validation: dN-TOP. J. Proteome Res. 12, 3063–3070 [DOI] [PubMed] [Google Scholar]
- 40. McDonald L., Beynon R. J. (2006) Positional proteomics: preparation of amino-terminal peptides as a strategy for proteome simplification and characterization. Nat. Protoc. 1, 1790–1798 [DOI] [PubMed] [Google Scholar]
- 41. Yamaguchi M., Nakayama D., Shima K., Kuyama H., Ando E., Okamura T. A., Ueyama N., Nakazawa T., Norioka S., Nishimura O., Tsunasawa S. (2008) Selective isolation of N-terminal peptides from proteins and their de novo sequencing by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry without regard to unblocking or blocking of N-terminal amino acids. Rapid Comm. Mass Sp.: RCM 22, 3313–3319 [DOI] [PubMed] [Google Scholar]
- 42. Shen P. T., Hsu J. L., Chen S. H. (2007) Dimethyl isotope-coded affinity selection for the analysis of free and blocked N-termini of proteins using LC-MS/MS. Anal. Chem. 79, 9520–9530 [DOI] [PubMed] [Google Scholar]
- 43. Gevaert K., Van Damme J., Goethals M., Thomas G. R., Hoorelbeke B., Demol H., Martens L., Puype M., Staes A., Vandekerckhove J. (2002) Chromatographic isolation of methionine-containing peptides for gel-free proteome analysis: identification of more than 800 Escherichia coli proteins. Mol. Cell. Proteomics 1, 896–903 [DOI] [PubMed] [Google Scholar]
- 44. Staes A., Van Damme P., Helsens K., Demol H., Vandekerckhove J., Gevaert K. (2008) Improved recovery of proteome-informative, protein N-terminal peptides by combined fractional diagonal chromatography (COFRADIC). Proteomics 8, 1362–1370 [DOI] [PubMed] [Google Scholar]
- 45. Gevaert K., Goethals M., Martens L., Van Damme J., Staes A., Thomas G. R., Vandekerckhove J. (2003) Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nat. Biotechnol. 21, 566–569 [DOI] [PubMed] [Google Scholar]
- 46. Staes A., Impens F., Van Damme P., Ruttens B., Goethals M., Demol H., Timmerman E., Vandekerckhove J., Gevaert K. (2011) Selecting protein N-terminal peptides by combined fractional diagonal chromatography. Nat. Protoc. 6, 1130–1141 [DOI] [PubMed] [Google Scholar]
- 47. Gevaert K., Ghesquiere B., Staes A., Martens L., Van Damme J., Thomas G. R., Vandekerckhove J. (2004) Reversible labeling of cysteine-containing peptides allows their specific chromatographic isolation for non-gel proteome studies. Proteomics 4, 897–908 [DOI] [PubMed] [Google Scholar]
- 48. Gevaert K., Staes A., Van Damme J., De Groot S., Hugelier K., Demol H., Martens L., Goethals M., Vandekerckhove J. (2005) Global phosphoproteome analysis on human HepG2 hepatocytes using reversed-phase diagonal LC. Proteomics 5, 3589–3599 [DOI] [PubMed] [Google Scholar]
- 49. Christie-Oleza J. A., Pina-Villalonga J. M., Bosch R., Nogales B., Armengaud J. (2012) Comparative proteogenomics of twelve Roseobacter exoproteomes reveals different adaptive strategies among these marine bacteria. Mol. Cell. Proteomics 11, M111.013110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Swingley W. D., Sadekar S., Mastrian S. D., Matthies H. J., Hao J., Ramos H., Acharya C. R., Conrad A. L., Taylor H. L., Dejesa L. C., Shah M. K., O'Huallachain M E., Lince M. T., Blankenship R. E., Beatty J. T., Touchman J. W. (2007) The complete genome sequence of Roseobacter denitrificans reveals a mixotrophic rather than photosynthetic metabolism. J. Bacteriol. 189, 683–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dupierris V., Masselon C., Court M., Kieffer-Jaquinod S., Bruley C. (2009) A toolbox for validation of mass spectrometry peptides identification and generation of database: IRMa. Bioinformatics 25, 1980–1981 [DOI] [PubMed] [Google Scholar]
- 52. Gevaert K., Van Damme P., Martens L., Vandekerckhove J. (2005) Diagonal reverse-phase chromatography applications in peptide-centric proteomics: ahead of catalogue-omics? Anal. Biochem. 345, 18–29 [DOI] [PubMed] [Google Scholar]
- 53. Schellenberger V., Braune K., Hofmann H. J., Jakubke H. D. (1991) The specificity of chymotrypsin. A statistical analysis of hydrolysis data. Eur. J. Biochem. 199, 623–636 [DOI] [PubMed] [Google Scholar]
- 54. Breddam K., Meldal M. (1992) Substrate preferences of glutamic-acid-specific endopeptidases assessed by synthetic peptide substrates based on intramolecular fluorescence quenching. Eur. J. Biochem. 206, 103–107 [DOI] [PubMed] [Google Scholar]
- 55. Hirel P. H., Schmitter M. J., Dessen P., Fayat G., Blanquet S. (1989) Extent of N-terminal methionine excision from Escherichia coli proteins is governed by the side-chain length of the penultimate amino acid. Proc. Natl. Acad. Sci. U.S.A. 86, 8247–8251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bonissone S., Gupta N., Romine M., Bradshaw R. A., Pevzner P. A. (2013) N-terminal protein processing: a comparative proteogenomic analysis. Mol. Cell. Proteomics 12, 14–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bewley M. C., Graziano V., Griffin K., Flanagan J. M. (2006) The asymmetry in the mature amino-terminus of ClpP facilitates a local symmetry match in ClpAP and ClpXP complexes. J. Struct. Biol. 153, 113–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Alexopoulos J. A., Guarné A., Ortega J. (2012) ClpP: A structurally dynamic protease regulated by AAA+ proteins. J. Struct. Biol. 179, 202–210 [DOI] [PubMed] [Google Scholar]
- 59. Bewley M. C., Graziano V., Griffin K., Flanagan J. M. (2009) Turned on for degradation: ATPase-independent degradation by ClpP. J. Struct. Biol. 165, 118–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jennings L. D., Bohon J., Chance M. R., Licht S. (2008) The ClpP N-terminus coordinates substrate access with protease active site reactivity. Biochemistry 47, 11031–11040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Maurizi M. R., Clark W. P., Katayama Y., Rudikoff S., Pumphrey J., Bowers B., Gottesman S. (1990) Sequence and structure of Clp P, the proteolytic component of the ATP-dependent Clp protease of Escherichia coli. J. Biol. Chem. 265, 12536–12545 [PubMed] [Google Scholar]
- 62. Omairi-Nasser A., de Gracia A. G., Ajlani G. (2011) A larger transcript is required for the synthesis of the smaller isoform of ferredoxin:NADP oxidoreductase. Mol. Microbiol. 81, 1178–1189 [DOI] [PubMed] [Google Scholar]
- 63. Hensley C. T., Kamneva O. K., Levy K. M., Labahn S. K., Africa L. A., Wing H. J. (2011) Two promoters and two translation start sites control the expression of the Shigella flexneri outer membrane protease IcsP. Arch. Microbiol. 193, 263–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Richter L. V., Sandler S. J., Weis R. M. (2012) Two isoforms of Geobacter sulfurreducens PilA have distinct roles in pilus biogenesis, cytochrome localization, extracellular electron transfer, and biofilm formation. J. Bacteriol. 194, 2551–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wanschers B. F., Szklarczyk R., Pajak A., van den Brand M. A., Gloerich J., Rodenburg R. J., Lightowlers R. N., Nijtmans L. G., Huynen M. A. (2012) C7orf30 specifically associates with the large subunit of the mitochondrial ribosome and is involved in translation. Nucleic Acids Res. 40, 4040–4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Walbot V., Coe E. H. (1979) Nuclear gene iojap conditions a programmed change to ribosome-less plastids in Zea mays. Proc. Natl. Acad. Sci. U.S.A. 76, 2760–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hauser R., Pech M., Kijek J., Yamamoto H., Titz B., Naeve F., Tovchigrechko A., Yamamoto K., Szaflarski W., Takeuchi N., Stellberger T., Diefenbacher M. E., Nierhaus K. H., Uetz P. (2012) RsfA (YbeB) proteins are conserved ribosomal silencing factors. PLoS Genet. 8, e1002815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Menschaert G., Van Criekinge W., Notelaers T., Koch A., Crappe J., Gevaert K., Van Damme P. (2013) Deep proteome coverage based on ribosome profiling aids MS-based protein and peptide discovery and provides evidence of alternative translation products and near-cognate translation initiation events. Mol. Cell. Proteomics 12, 1780–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Benelli D., Londei P. (2009) Begin at the beginning: evolution of translational initiation. Res..Microbiol. 160, 493–501 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.