Abstract
We previously reported that self-complementary adeno-associated virus (scAAV) type 2 genomes of up to 3.3 kb can be successfully encapsidated into AAV2 serotype capsids. Here we report that such oversized AAV2 genomes fail to undergo packaging in other AAV serotype capsids, such as AAV1, AAV3, AAV6, and AAV8, as determined by Southern blot analyses of the vector genomes, although hybridization signals on quantitative DNA slot-blots could still be obtained. Recently, it has been reported that quantitative real-time PCR assays may result in substantial differences in determining titers of scAAV vectors depending on the distance between the primer sets and the terminal hairpin structure in the scAAV genomes. We also observed that the vector titers determined by the standard DNA slot-blot assays were highly dependent on the specific probe being used, with probes hybridizing to the ends of viral genomes being significantly overrepresented compared with the probes hybridizing close to the middle of the viral genomes. These differences among various probes were not observed using Southern blot assays. This overestimation of titer is a systemic error during scAAV genome quantification, regardless of viral genome sequences and capsid serotypes. Furthermore, different serotypes capsid and modification of capsid sequence may affect the ability of packaging intact, full-length AAV genomes. Although the discrepancy is modest with wild-type serotype capsid and short viral genomes, the measured titer could be as much as fivefold different with capsid mutant vectors and large genomes. Thus, based on our data, we suggest that Southern blot analyses should be performed routinely to more accurately determine the titers of recombinant AAV vectors. At the very least, the use of probes/primers hybridizing close to the mutant inverted terminal repeat in scAAV genomes is recommended to avoid possible overestimation of vector titers.
Wang and colleagues report that 3.3 kb self-complementary (sc) AAV2 genomes fail to undergo packaging into AAV serotype capsids other than AAV2. Serotypes tested include AAV1, AAV3, AAV6, and AAV8. The authors also address the issue of qPCR titering assays leading to overestimation of scAAV vector titers and suggest Southern blot analyses as a more accurate and reliable titering method.
Introduction
Adeno-associated viral (AAV) vectors have begun to show remarkable efficacy in the potential gene therapy of human diseases such as Leber's congenital amaurosis and hemophilia B (Mingozzi and High, 2011). To date, 12 distinct human and nonhuman AAV serotypes (AAV1–12) have been described, and recombinant AAV (rAAV) vectors have been developed (Agbandje-McKenna and Kleinschmidt, 2011). In addition, new technologies have also been developed to generate capsid gene libraries through error-prone PCR (Maheshri et al., 2006) and DNA shuffling (Li et al., 2008). Recently, we substituted surface-exposed tyrosine residues with phenylalanine residues on AAV2 capsids to achieve dramatically improved transgene expression (Zhong et al., 2008). All of these and other strategies involve modification of AAV capsid sequence. However, the effect of modified capsids on packaging intact, full-length AAV genomes has not been studied in detail. A wide variety of methods have been developed to determine AAV vector genome quantitation (Samulski et al., 1989; Halbert et al., 1997; Clark et al., 1999; Sommer et al., 2003), in which dot/slot-blot hybridization and quantitative real-time PCR (qPCR) are the most frequently used (Gray et al., 2011). The main drawback of these two assays is that they do not provide information about viral genome integrity. It has been reported that qPCR assays may result in substantial differences in determining the titers of self-complementary AAV (scAAV) vectors, depending on the localization of the primer sets in the scAAV genomes (Fagone et al., 2012). The discrepancy could be nearly 10-fold (Allay et al., 2011). The same group subsequently developed alternative qPCR protocols that circumvent this issue and enable more reliable determination of scAAV vector titers. However, the effects of hybridization probes on dot/slot-blot assays, another frequently used method for viral titration, have not been studied.
It is now well accepted that the ubiquitin–proteasome pathway plays an essential role in AAV intracellular trafficking (Duan et al., 2000; Douar et al., 2001; Yan et al., 2002; Cheng et al., 2012). In more recent studies by us and by others (Zhong et al., 2007; Kauss et al., 2010; Markusic et al., 2010; Qiao et al., 2010; Petrs-Silva et al., 2011; Cheng et al., 2012), next generation rAAV vectors have been developed containing point mutations in surface-exposed tyrosine residues to avoid phosphorylation of these residues and subsequent ubiquitination and proteasome-mediated degradation of the viral capsid. In addition to tyrosine residues, surface-exposed serine residues can also undergo phosphorylation (Aslanidi et al., 2012), and thus trigger ubiquitination and subsequent proteasome-mediated degradation. Thus, we hypothesized that mutation on surface-exposed serine residues may also increase the transduction efficiency of AAV3 vectors.
In the present studies, we document that: (i) Overestimation of vector titers by DNA slot-blot hybridization using probes specific to the ends of viral genomes leads to systemic errors during scAAV genome quantitation, regardless of viral genome sequences and capsid serotypes; (ii) A significant extent of this hybridization is due to the existence of large amounts of fragmented AAV genomes in the packaged virions; (iii) Different serotype capsids and modification of capsid sequences affect the ability of packaging intact, full-length scAAV genomes; and (iv) The use of methods based on agarose gel electrophoresis and Southern blot assays are the most accurate and reliable ways to determine the titers of scAAV vectors.
Material and Methods
Cell lines and cultures
Human cervical cancer (HeLa) and hepatocellular carcinoma (Huh7) cell lines were purchased from American Type Culture Collection (Manassas, VA), and maintained in complete Dulbecco's modified Eagle's medium (DMEM, Mediatech Inc., Manassas, VA) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Sigma-Aldrich, St. Louis, MO), 1% penicillin and streptomycin (Lonza, Walkersville, MD). A newly established human hepatoblastoma (Hep293TT) cell line (Chen et al., 2009) was generously provided by Dr. Gail E. Tomlinson, University of Texas Health Science Center at San Antonio, and was maintained in complete RPMI medium 1640 (Invitrogen, Camarillo, CA) supplemented with 15% heat-inactivated FBS and 1% penicillin and streptomycin. Cells were grown as adherent cultures in a humidified atmosphere at 37°C in 5% CO2 and were subcultured after treatment with trypsin-versene mixture (Lonza, Walkersville, MD) for 2–5 min at room temperature, washed, and resuspended in complete medium. A human breast cancer cell line, T47D, stably transfected with a hHGFR expression plasmid (T47D+hHGFR), were maintained in complete DMEM (Mediatech Inc.) with 600 μg/ml of G418, supplemented with 10% heat-inactivated FBS (Sigma-Aldrich) and 1% penicillin and streptomycin (Lonza).
Construction of surface-exposed serine residue mutant AAV3 capsid plasmids
Recombinant plasmid pACG2c3 was generously provided by Dr. R. Jude Samulski, University of North Carolina at Chapel Hill. A two-stage procedure, based on QuikChange II site-directed mutagenesis (Stratagene, Santa Clara, CA) was performed as described previously (Zhong et al., 2008; Cheng et al., 2012). Briefly, in stage 1, two PCR extension reactions were performed in separate tubes for each mutant. One tube contained the forward PCR primer and the other contained the reverse primer (Supplementary Table S1). In stage 2, the two reactions were mixed and a standard PCR mutagenesis assay was carried out according to the manufacturer's instructions. PCR primers were designed to introduce changes from serine or threonine to valine residues (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/hgtb). All mutants were sequenced before use.
rAAV vectors
HEK293 cells were transfected using polyethelenimine (PEI, linear, MW 25,000, Polysciences, Inc., Warrington, PA), as described (Reed et al., 2006; Lock et al., 2010). Seventy-two hours post transfection, cells were harvested and vectors were purified by iodixanol (Sigma, St. Louis, MO) gradient centrifugation and ion exchange column chromatography (HiTrap Sp Hp 5 ml, GE Healthcare, Piscataway, NJ). Virus was then concentrated and the buffer was exchanged in three cycles to lactated Ringer's using centrifugal spin concentrators (Apollo, 150-kDa cut-off, 20-ml capacity, Orbital Biosciences, Topsfield, MA).
Quantitative DNA slot blot analyses
The physical particle titers of highly purified of scAAV vector stocks were determined by quantitative DNA slot-blot analyses described previously (Kube and Srivastava, 1997). Briefly, 10 μl of vector stocks was digested with Benzonase (Novagen, Darmstadt, Germany) at 37°C for 1 hr. An equal volume of 100 mM NaOH was added followed by incubation at 65°C for 30 min. A known quantity of plasmid DNA was also denatured in a similar manner for use as a standard reference during quantitation. The denatured DNA was loaded in twofold serial dilutions onto Immobilon-NY+TM membranes (Millipore, Bedford, MA). After cross-linkage, the membranes were then prehybridized for 6 hr at 68°C in 25 ml hybridization solution containing 6×SSC, 100 μg/ml freshly boiled herring sperm DNA, 0.5% sodium dodecyl sulfate (SDS), and 5×Denhardt's reagent. Subsequently, the membranes were hybridized with freshly boiled 32P-labled DNA probe in a total volume of 25 ml of hybridization solution at 68°C for 18–20 hr. Membranes were then washed once in 50 ml of wash solution 1 (2×SSC, 0.1% SDS) at room temperature for 15 min, twice in 50 ml of wash solution 2 (0.1×SSC, 0.1% SDS) at 68°C for 30 min, and then exposed to BIOMAX MRTM X-ray films (Kodak, Rochester, NY) at −70°C.
Southern blot analyses of AAV vector genomes and vector titer determination
Ten microliters vector stocks were digested with Benzonase (Novagen) at 37°C for 1 hr, then for an additional 2 hr with proteinase K (Invitrogen, Grand Island, NY) at 56°C. The reaction mixture was purified by phenol/chloroform, followed by chloroform treatment. DNA was precipitated by ethanol in the presence of 20 μg of glycogen (Invitrogen) and then was dissolved in 100 μl of double-distilled H2O and electrophoresed on 1.2% native agarose gels. DNA was transferred to nylon membranes that were analyzed using the method described by Southern (1975). Briefly, the gel was equilibrated with solution I (0.25 M HCl) for 20 minutes at room temperature in solution II (1 M NaCl, 0.5 M NaOH) for 40 min at room temperature in solution III (1 M NaCl, 0.5 M Tris-HCl) for 40 min. The DNA was transferred to Immobilon-NY+TM membranes (Millipore, Billerica, MA) in 20×SSC. Membranes were prehybridized as before for 6 hr and hybridized overnight with 32P-labled DNA probes at 68°C as already described. Membranes were washed as before and subjected to autoradiography. Twofold serial dilutions of linearized, full-length double-stranded AAV DNA genomes were also electrophoresed as reference standards, and the vector titers were determined using the following equations:
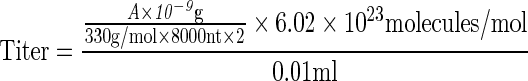 |
(1) |
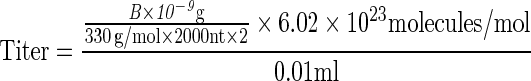 |
(2) |
A=Mr of double-stranded, circular plasmid DNA; B=Mr of XmaI-linearized double-stranded AAV DNA; 330 g/mol represents the average molecular weight of a single deoxyribonucleotide monophosphate; 8000 nt and 2000 nt represent the number of nucleotides in a single-strand of plasmid and XmaI-linearized DNA, respectively; 0.01 ml represents the loading volume for each viral stock; and 6.02×1023 represents the Avogadro constant. The final unit of titer is vector genomes (vg) per milliliter.
AAV vector transduction assays
Cells were seeded in 96-well plates at a concentration of 10,000 cells per well in complete medium and were transduced with AAV vectors at 2000 vg/cell for 2 hr. After transduction, cells were incubated for additional 72 hr and then the expression of enhanced green fluorescent protein (EGFP) was analyzed by direct fluorescence imaging or by flow cytometry.
Statistical analysis
Results are presented as mean±standard deviation (SD). Differences between groups were identified using a grouped-unpaired two-tailed distribution of Student's t test, and p values <0.05 were considered statistically significant.
Results
Mutations in AAV3 capsids lead to significant differences in vector titers determined by DNA slot-blot and Southern blot assays
We have reported that Huh7 cells, a well-known human hepatocellular carcinoma cell line (Nakabayashi et al., 1982), can be efficiently transduced by AAV3 vectors (Glushakova et al., 2009; Ling et al., 2010, 2011; Cheng et al., 2012), and that the transduction efficiency of these vectors can be further augmented by site-directed mutagenesis of surface-exposed tyrosine residues (Cheng et al., 2012). To further examine whether phosphorylation of surface-exposed serine and/or threonine residues also affects the transduction efficiency of AAV3 vectors, Huh7 cells were either mock-treated or treated with two different serine/threonine kinase inhibitors, c-Jun N-terminal kinase (JNK) inhibitor or p38 mitogen-activated protein kinase (MAPK) inhibitor (SB), followed by infection with scAAV3-CBAp-EGFP vectors, depicted schematically in Fig. 1A, at 2000 vg/cell. Both drug treatments led to a significant increase in AAV3-mediated transgene expression (Supplementary Fig. S1A and S1B), which is consistent with our recent observations with AAV2 vectors (Aslanidi et al., 2012). However, two additional serine/threonine kinase inhibitors, CaMKII and Mek inhibitors had no significant effect (data not shown). To examine whether the treatment with JNK inhibitor II or p38 MAPK inhibitor modulates the Ub/proteasome pathway, cells were also co-treated with MG132, a proteasome inhibitor (Zhong et al., 2007). All treated cells and appropriate controls were infected with scAAV3-CBAp-EGFP vectors. Consistent with our previously published studies (Cheng et al., 2012), treatment with MG132 led to a significant increase in AAV3-mediated transgene expression (Supplementary Fig. S1C). However, co-treatment with either JNK or p38 MAPK inhibitor had no additive effect, which suggests that inhibition of these serine/threonine kinases overlaps with the Ub/proteasome pathway.
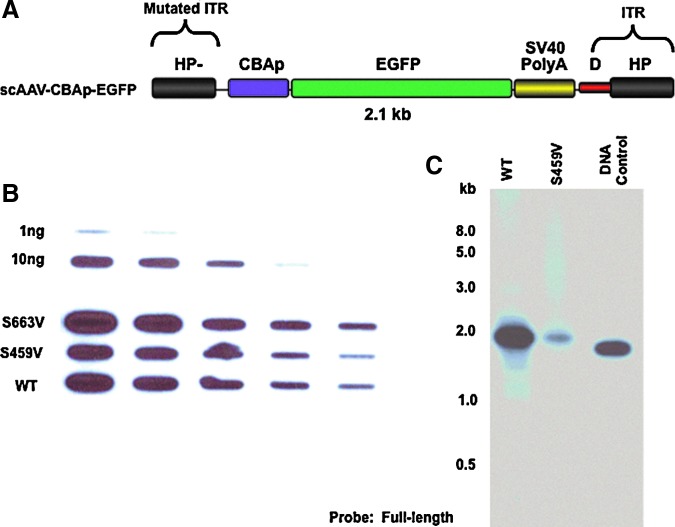
FIG. 1.
Characterization of scAAV3-CBAp-EGFP vectors. (A) Schematic structure of the vector genome. The size of the entire viral genome is 2.1 kb. (B) A representative DNA slot-blot hybridization of the purified vector genomes to determine the vector titers. The indicated amounts of the recombinant plasmids DNA (top two rows) were used as appropriate controls. (C) A representative neutral agarose gel electrophoresis and Southern blot examination of the isolated AAV genomes from the WT and the S549V mutant AAV3 capsids. Both DNA slot-blot and Southern blot membranes were hybridized with 32P-labeled DNA probe specific to the full-length AAV vector genome. Results in each panel are representative of two independent experiments. sc, self-complementary; AAV, adeno-associated viral; EGFP, enhanced green fluorescent protein; WT, wild type. Color images available online at www.liebertpub.com/hgtb
We performed site-directed mutagenesis of two surface-exposed serine (S) residues on AAV3 capsids (S459 and S663), which were substituted with valine (V) residues (Supplementary Table S1). Neither mutations led to any sequence change within the potential AAP gene (Sonntag et al., 2010, 2011). Titers of the recombinant scAAV-CBAp-EGFP vector genomes encapsidated in the wild-type (WT) and the mutant capsids, were determined by quantitative DNA slot-blot assays using a 32P-labeled DNA probe specific to the full-length viral genome, as described previously (Kube and Srivastava, 1997). The results indicated that mutant capsids lead to packaging of the vectors with similar titers (Fig. 1B). Surprisingly, however, analysis of the purified vector DNA by neutral agarose gel electrophoresis and Southern blots using the same DNA probes revealed a substantial difference between the levels of full-length, intact viral genomes in the WT vs. the S459V mutant capsids (Fig. 1C). Thus, it became evident that a specific mutation in AAV3 capsids significantly affects the vector titers depending upon the methodology employed (DNA slot-blots vs. Southern blots).
DNA slot-blot assays using probes representing the middle of the viral genome result in more accurate vector titers
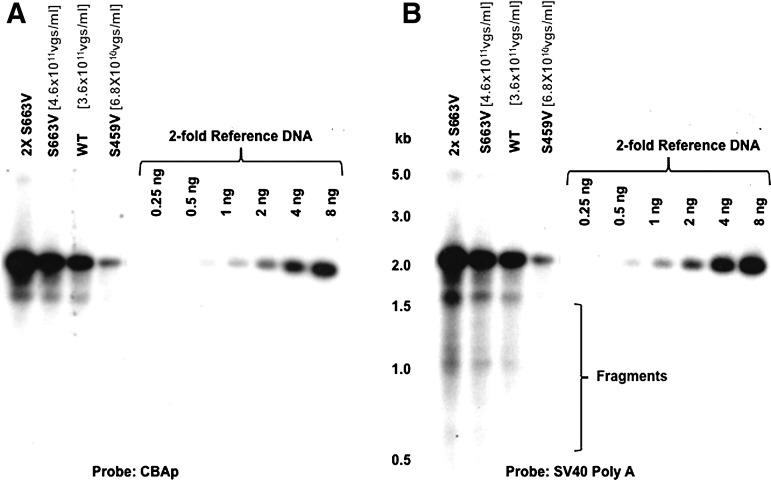
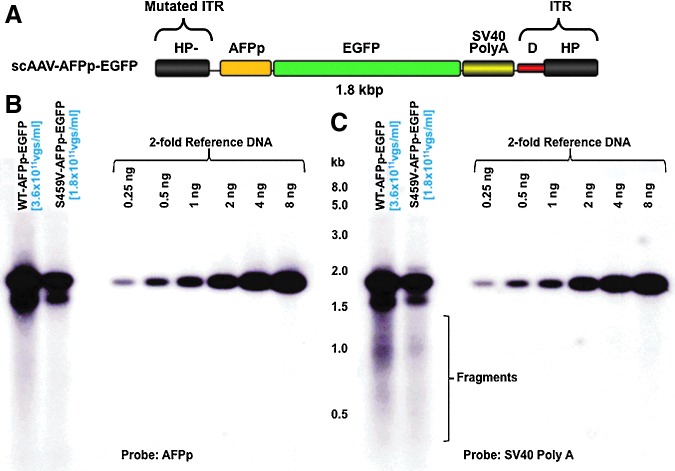
A key drawback of DNA slot-blot assays is that they do not provide information on the viral genome integrity. We hypothesized that the process of viral encapsidation may produce a number of viral particles containing fragments of viral genomes, and that a capsid mutation may affect the frequency of packaging of fragmented viral genomes. It should be noted that in scAAV-CBAp-EGFP genomes, following denaturation, the simian virus 40 (SV40) Poly (A) signal sequences would be present at both 5′- and 3′-ends. On the other hand, DNA sequences involving the CBAp promoter would always be located in the middle of the linearized viral genomes. We performed a series of DNA slot-blot assays using different probes, and the actual measured titers were normalized to that determined for the plasmid DNA reference control by the National Institutes of Health ImageJ software. The following two conclusions could be drawn from these results, which are shown in Table 1. First, in all viral stocks, when analyzed with a probe specific to CBAp, the hybridization signals were reduced. Although the discrepancy was modest with the WT serotypes capsid, the measured titer could be as much as fivefold different with one of the capsid-mutant vectors. The higher titer measurements obtained using probes close to the ends of the genomes suggested that they hybridize to fragmented viral genomes. Second, in DNA slot-blot assays, the ratio between viral stocks using the CBAp probe was much closer to the ratio determined by Southern blots, which suggests that only the results using probes located in the middle of the viral genomes more accurately represent the intact, full-length viral genomes. In addition, qPCR assays using primer-pairs specific for the CBAp region also revealed an approximately sixfold difference between WT and S459V vectors (data not shown), which is close to the results obtained from DNA slot-blots using the CBAp probe. This discrepancy between the two different DNA probes, when used in Southern blot assays, was not observed (Fig. 2). It is evident that the use of the two sets of probes with similar lengths and GC content (Supplementary Table S2), which results in different hybridization signals in DNA slot-blot assays, detected full-length, intact viral genomes in Southern blot assays to nearly the same extent. Correspondingly, it is also noteworthy that under identical conditions, the SV40 Poly (A) probe detected much more fragmented viral genomes (Fig. 2B) compared with the CBAp probe (Fig. 2A). The effect of mutant capsid S459V on rAAV3 viral vector assembly and the detection of fragments by probes close to the ends of the viral genome were further corroborated by encapsidating a different expression cassette, scAFPp-EGFP in WT and S459V capsids, in which the EGFP reporter gene is under the control of a liver cancer cell-specific promoter, the human α-feto-protein (AFP) (Fig. 3A). The use of the AFPp and the SV40 Poly (A) probes also yielded very similar results (Fig. 3B), which further corroborate our hypothesis.
Table 1.
Titers of the Wild-Type and Mutant scAAV3-CBAp-EGFP, scAAV2- and scAAV8-CBAp-Syn Vectors Determined by Different Sets of Probes
 |
FIG. 2.
Neutral agarose gel electrophoresis and Southern blot examination of the isolated vector genomes from WT and serine-mutant AAV3 vectors. Equivalent amounts of DNA samples from the WT and serine-mutant AAV3 vectors (based on DNA slot-blots in Fig. 1) were isolated and analyzed on Southern blots. S663V vector genomes were loaded in twofold dilutions (left two lanes). The membranes were analyzed using 32P-labeled DNA probes specific to (A) CBAp or (B) SV40 Poly (A) sequences. The numbers in parentheses represent vector titers determined by Southern blot assays.
FIG. 3.
Native agarose gel electrophoresis and Southern blot analyses of the isolated genomes from WT and serine-mutant scAAV3 vectors. (A) Schematic structure of the scAAV-AFPp-EGFP vector genome. The size of the entire viral genome is 1.8 kb. (B) A representative native agarose electrophoresis and Southern blot examination of the isolated AAV genome. The hybridizations were analyzed with 32P-labeled DNA probes specific to (B) the AFPp or (C) the SV40 Poly (A) sequences. Color images available online at www.liebertpub.com/hgtb
Probes close to the ends of the viral genomes consistently detect much more fragmented viral genomes regardless of viral genome sequences and capsid serotypes
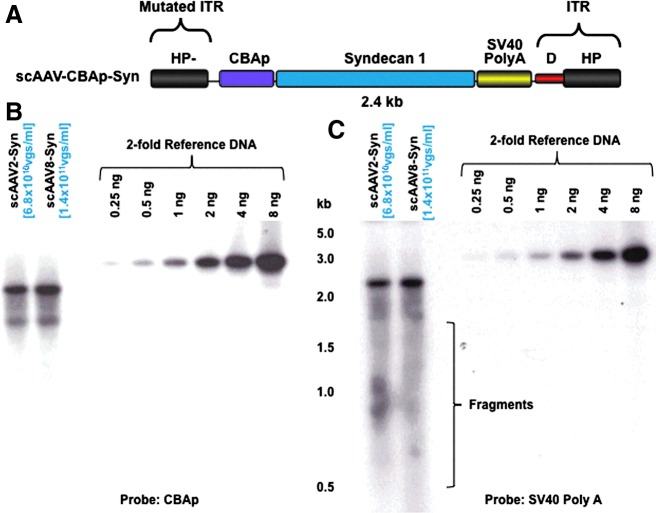
To further corroborate whether the more intense hybridization signal in DNA slot-blot assays resulting from the use of the probe that hybridizes to sequences close to the ends of the viral genomes was a general phenomenon, and not specific to particular scAAV genomes and capsid serotypes, we performed DNA slot-blot and Southern blot assays on additional scAAV genomes encapsidated in AAV2, AAV3, and AAV8 serotype capsids. The genome, scAAV-CBAp-Syn, in which the human Syndecan 1 gene is under the control of the CBAp, depicted schematically in Fig. 4A, is 2.4 kb, which is close to the maximum packaging capacity of AAV vectors (Dong et al., 2010; Lai et al., 2010; Wu et al., 2010), and shares the same CBAp promoter and SV40 Poly (A) sequences with scAAV-CBAp-EGFP vectors. As observed above, in Southern blot assays, the Poly (A) probe consistently detected much more fragmented viral genomes compared with that with the CBAp probe (Fig. 4B and 4C), which may account for the modest discrepancy between the titers of vector stocks determined by different probes in DNA slot-blot assays (Table 1).
FIG. 4.
Characterization of scAAV2- and scAAV8-CBAp-Syn vector genomes by Southern blot assays. (A) Schematic structure of the scAAV-CBAp-Syn vector genome. The size of the entire viral genome is 2.4 kb. (B, C) Representative native agarose gel electrophoresis and Southern blot examination of the isolated genomes from scAAV2 and scAAV8 serotype vectors. The membranes were analyzed using 32P-labeled DNA probes specific to (B) CBAp or (C) SV40 Poly (A) sequences. Results in each panel are representative of two independent experiments. The vector titers determined by Southern blots are indicated in parentheses. Color images available online at www.liebertpub.com/hgtb
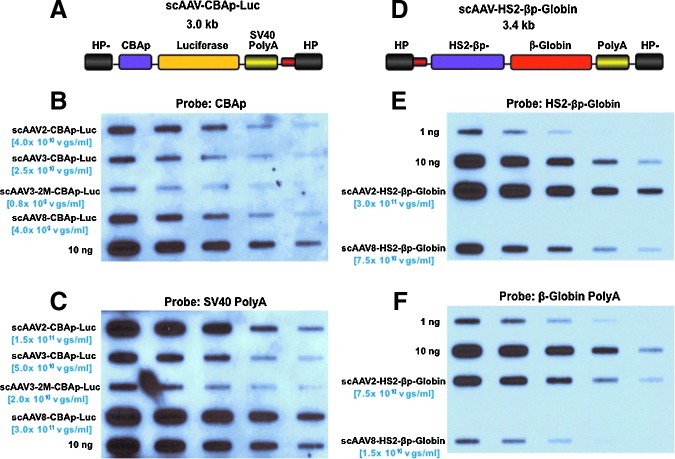
We next carried out experiments with oversized viral genomes, which are generally inefficient in undergoing encapsidation in AAV capsids. scAAV-CBAp-Luc (Fig. 5A) is a rAAV genome containing a 3.0-kb firefly luciferase expression cassette, with the promoter CBAp located in the middle of the linearized viral genome. In contrast, scAAV-HS2-βp-globin (Fig. 5D) contains a 3.4-kb human β-globin expression cassette, with its own Poly (A) sequence located in the middle of viral genome. Both of these genomes exceed the normal ∼2.5-kb packaging capacity of scAAV vectors. Recombinant AAV2, AAV3, and AAV8 serotype vectors were generated and analyzed on DNA slot-blots. As can be seen, regardless of the vectors, probes hybridizing to sequences close to the ends of viral genomes (SV40 Poly (A), Fig. 5C; and HS2-βp, Fig. 5E) resulted in approximately fivefold higher signal compared with probes hybridizing to sequences in the middle in the viral genomes (CBAp, Fig. 5B; and β-Globin Poly (A), Fig. 5F). Again, the fragment lengths and the GC contents of the two probes were very similar (Supplementary Table S2). When these genomes were analyzed on Southern blots, no hybridization signal could be detected for the 3.4-kb scAAV-HS2-βp-globin vectors, as expected (data not shown). However, when the 3.0-kb scAAV-CBAp-Luc vector genomes purified from three different serotypes were analyzed on Southern blots, DNA bands representing intact, full-length vector genomes could be detected only in AAV2 vectors, but not the other two serotypes, AAV3 and AAV8 (Supplementary Fig. S2A and Fig. 2B). These results further corroborate that only AAV2 serotype capsids allow the encapsidation of oversized genomes of scAAV vectors (Wu et al., 2007), and that DNA slot-blot assays alone can lead to overestimation of vector titers, which is largely due to encapsidation of fragmented AAV genomes.
FIG. 5.
Characterization of oversized scAAV-CBAp-Luc and scAAV-HS2-βp-globin genomes by DNA slot-blot assays. Schematic structures of (A) the scAAV-CBAp-Luc, and (D) the scAAV-HS2-βp-globin vector genomes. The sizes of the entire viral genome are 3.0 kb and 3.4 kb, respectively. Representative DNA slot-blot analysis of the isolated AAV genomes from scAAV2, scAAV3, and scAAV8 serotype vectors. The membranes were hybridized with the indicated 32P-labeled DNA probes specific to (B, F) the sequences located in the middle of the vector genomes or to (C, E) the sequences located at the ends of the vector genomes. The indicated amounts of the recombinant plasmids for each vector genome were used as appropriate controls. Results in each panel are representative of two independent experiments. The vector titers determined by Southern blots are indicated in parentheses. Color images available online at www.liebertpub.com/hgtb
Discussion
Self-complementary AAV vectors encoding a codon-optimized human factor IX gene under the control of a liver-specific promoter have shown efficacy in preclinical models of hemophilia B gene therapy (Nathwani et al., 2011) and have also yielded exciting results in a recent phase I clinical trial (Mingozzi and High, 2011; Ponder, 2011). During the course of this trial, however, it became apparent that the qPCR method used to determine the vector titers led to an underestimate by a factor of ∼10 due to the presence of covalently closed hairpins at one end of the scAAV vector DNA and rapid self-annealing of the viral genomes (Fagone et al., 2012). Consequently, the use of primers close to the intact inverted terminal repeats (ITRs) during qPCR assays is recommended, when titering regular scAAV genomes (<2.4 kb) encapsidated in WT AAV serotype capsids. It is generally accepted that DNA dot or slot-blot assays overcome the problem of self-annealing. However, in this report, we document that the standard DNA slot-blot assays frequently used to determine the titers of scAAV vectors can also lead to erroneous results and are also highly dependent on the specific probes used, with probes located at the ends of viral genome being significantly overrepresented compared to the probes close to the middle of viral genome, even though these probes (CBAp, Poly (A), full-length) result in no difference in hybridization signals in Southern blot assays (Figs. 1 and 2). Although the discrepancy was modest with WT serotypes capsid and short viral genomes, which is consistent with the observation by Fagone et al. (2012), the measured titer could be as much as fivefold different with capsid-mutant vectors and larger genomes. One of our initial concerns was the different denaturing conditions for vector genomes prior to DNA slot-blot and Southern blot assays. The viral genomes were denatured by treatment with 100 mM NaOH at 65°C for 30 min in DNA slot-blot assays, whereas in Southern blot assays, the genomes were denatured by shaking agarose gels in solution II (1 M NaCl, 0.5 M NaOH) for 40 min. Under both conditions, double-stranded viral genome would be expected to be denatured into single-stranded forms.
Although AAV vectors carrying larger genomes (up to and exceeding 6.0 kb) have been produced and tested in preclinical disease models (Sarkar et al., 2004; Grieger and Samulski, 2005; Monahan et al., 2010), it is unclear to what extent these genomes were in fact encapsidated. Recently, using a single-molecule sequencing method, Kapranov et al. (2012) determined the formation of particles that have smaller genomes, referred to as defective-interfering (DI) particles. Given that they further observed large amounts of DI particles containing only the sequence close to ITRs, we hypothesized that the discrepancy observed in our studies was due to the packaging of fragmented viral genomes. The observation in Southern blot assays that probes specific to the ends of self-complementary viral genomes consistently detect a smear of DNA bands smaller than the unit-length viral genome, which are undetectable using probes specific to the middle of viral genome, corroborates our hypothesis. However, due to the nature of the smear observed in Southern blot assays, it is difficult to document the increased amounts of fragmented viral genomes packaged in mutant capsids compared with the WT capsids. In addition, since we and others have hypothesized that the majority of fragmental DNA is relatively short, close to the size of an ITR, we were unable to detect these fragments in Southern blot assays.
The observation that intact scAAV genomes of up to 3.0 kb could be successfully encapsidated into AAV2 capsids is consistent with our previous studies (Wu et al., 2007). Such oversized scAAV genomes failed to undergo encapsidation in several other serotype capsids, such as AAV1, AAV3, AAV6, and AAV8. This is, perhaps, not surprising since the ITRs in the AAV genomes and the Rep proteins involved in the packaging procedures are of the AAV2 origin, which interact optimally with the AAV2 capsids. Curiously, however, despite the lack of encapsidation of full-length oversized AAV genomes in these serotype capsids, as determined by Southern blot assays, unambiguously similar levels of hybridization signals in all three serotypes vectors, despite prior exhaustive digestion with Benzonase, could be readily detected on DNA slot-blots (Fig. 5). Although we did not examine the exact nature of these signals, it is reasonable to propose that they emanate from encapsidation of fragmented AAV genomes, representing the DI particles (Kapranov et al., 2012).
Although we did not include the conventional single-stranded AAV vectors in our current studies, based on our data, and those of others (Kapranov et al., 2012), we suggest that in standard DNA slot-blot assays, the use of hybridization probes located at the middle of viral genome is the most accurate and reliable way to determine the titers of both single-stranded AAV and scAAV vectors. A universal real-time PCR assay involving the AAV ITR sequences for determining the vector titers has been reported (Aurnhammer et al., 2012), but as stated above, one must exercise caution to rely solely on this method in view of the observation made by Kapranov et al. (2012) that large amounts of DNA sequences close to the ends of viral genomes are encapsidated in AAV vectors. Recently, gel titration assays based on DNA-staining methodology have also been described to provide robust and reliable quantitation (Fagone et al., 2012). However, the method requires large amounts of purified vectors and thus is more suitable for clinical-grade vector preparations. We suggest that the Southern blot hybridization method be used routinely instead. Although it is a little more complex, it requires much lower vector stocks and yields more accurate titers, as demonstrated here.
Supplementary Material
Acknowledgments
We thank Drs. R. Jude Samulski, James M. Wilson, and Xiao Xiao, for their kind gifts of recombinant AAV2, AAV8, and pdsCBAp-EGFP plasmids, respectively. This research was supported in part by the National Natural Science Foundation of China (30730114), and Public Health Service grants P01 DK-058327 (Project 1), and R01 HL-097088 from the National Institutes of Health, and a Special Project Award from the Bayer Hemophilia Foundation (to AS).
Author Disclosure Statement
None of the authors has any competing financial interest or anything to disclose.
References
- Agbandje-McKenna M. Kleinschmidt J. AAV capsid structure and cell interactions. Methods Mol. Biol. 2011;807:47–92. doi: 10.1007/978-1-61779-370-7_3. [DOI] [PubMed] [Google Scholar]
- Allay J.A. Sleep S. Long S., et al. Good manufacturing practice production of self-complementary serotype 8 adeno-associated viral vector for a hemophilia B clinical trial. Hum. Gene Ther. 2011;22:595–604. doi: 10.1089/hum.2010.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslanidi G.V. Rivers A.E. Ortiz L., et al. High-efficiency transduction of human monocyte-derived dendritic cells by capsid-modified recombinant AAV2 vectors. Vaccine. 2012;30:3908–3917. doi: 10.1016/j.vaccine.2012.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurnhammer C. Haase M. Muether N., et al. Universal real-time PCR for the detection of and quantification of adeno-associated virus serotype 2-derived inverted terminal repeat sequences. Hum. Gene Ther. 2012;23:18–28. doi: 10.1089/hgtb.2011.034. [DOI] [PubMed] [Google Scholar]
- Chen T.T. Rakheja D. Hung J.Y., et al. Establishment and characterization of a cancer cell line derived from an aggressive childhood liver tumor. Pediatr. Blood Cancer. 2009;53:1040–1047. doi: 10.1002/pbc.22187. [DOI] [PubMed] [Google Scholar]
- Cheng B. Ling C. Dai Y., et al. Development of optimized AAV3 serotype vectors: mechanism of high-efficiency transduction of human liver cancer cells. Gene Ther. 2012;19:375–384. doi: 10.1038/gt.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K.R. Liu X. McGrath J.P. Johnson P.R. Highly purified recombinant adeno-associated virus vectors are biologically active and free of detectable helper and wild-type viruses. Hum. Gene Ther. 1999;10:1031–1039. doi: 10.1089/10430349950018427. [DOI] [PubMed] [Google Scholar]
- Dong B. Nakai H. Xiao W. Characterization of genome integrity for oversized recombinant AAV vector. Mol. Ther. 2010;18:87–92. doi: 10.1038/mt.2009.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douar A.M. Poulard K. Stockholm D. Danos O. Intracellular trafficking of adeno-associated virus vectors: routing to the late endosomal compartment and proteasome degradation. J. Virol. 2001;75:1824–1833. doi: 10.1128/JVI.75.4.1824-1833.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan D. Yue Y. Yan Z., et al. Endosomal processing limits gene transfer to polarized airway epithelia by adeno-associated virus. J. Clin. Invest. 2000;105:1573–1587. doi: 10.1172/JCI8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagone P. Wright J.F. Nathwani A.C., et al. Systemic errors in quantitative polymerase chain reaction titration of self-complementary adeno-associated viral vectors and improved alternative methods. Hum. Gene Ther. Methods. 2012;23:1–7. doi: 10.1089/hgtb.2011.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glushakova L.G. Lisankie M.J. Eruslanov E.B., et al. AAV3-mediated transfer and expression of the pyruvate dehydrogenase E1 alpha subunit gene causes metabolic remodeling and apoptosis of human liver cancer cells. Mol. Genet. Metab. 2009;98:289–299. doi: 10.1016/j.ymgme.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray S.J. Choi V.W. Asokan A., et al. Production of recombinant adeno-associated viral vectors and use in in vitro and in vivo administration. Curr. Protoc. Neurosci. 2011 doi: 10.1002/0471142301.ns0417s57. Chapter 4, Unit 4 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieger J.C. Samulski R.J. Packaging capacity of adeno-associated virus serotypes: impact of larger genomes on infectivity and postentry steps. J. Virol. 2005;79:9933–9944. doi: 10.1128/JVI.79.15.9933-9944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbert C.L. Standaert T.A. Aitken M.L., et al. Transduction by adeno-associated virus vectors in the rabbit airway: efficiency, persistence, and readministration. J. Virol. 1997;71:5932–5941. doi: 10.1128/jvi.71.8.5932-5941.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P. Chen L. Dederich D., et al. Native molecular state of adeno-associated viral vectors revealed by single-molecule sequencing. Hum. Gene Ther. 2012;23:46–55. doi: 10.1089/hum.2011.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauss M.A. Smith L.J. Zhong L., et al. Enhanced long-term transduction and multilineage engraftment of human hematopoietic stem cells transduced with tyrosine-modified recombinant adeno-associated virus serotype 2. Hum. Gene Ther. 2010;21:1129–1136. doi: 10.1089/hum.2010.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kube D.M. Srivastava A. Quantitative DNA slot blot analysis: inhibition of DNA binding to membranes by magnesium ions. Nucleic Acids Res. 1997;25:3375–3376. doi: 10.1093/nar/25.16.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y. Yue Y. Duan D. Evidence for the failure of adeno-associated virus serotype 5 to package a viral genome >or=8.2 kb. Mol. Ther. 2010;18:75–79. doi: 10.1038/mt.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. Asokan A. Wu Z., et al. Engineering and selection of shuffled AAV genomes: a new strategy for producing targeted biological nanoparticles. Mol. Ther. 2008;16:1252–1260. doi: 10.1038/mt.2008.100. [DOI] [PubMed] [Google Scholar]
- Ling C. Lu Y. Kalsi J.K., et al. Human hepatocyte growth factor receptor is a cellular coreceptor for adeno-associated virus serotype 3. Hum. Gene Ther. 2010;21:1741–1747. doi: 10.1089/hum.2010.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling C. Lu Y. Cheng B., et al. High-efficiency transduction of liver cancer cells by recombinant adeno-associated virus serotype 3 vectors. J. Vis. Exp. 2011:pii–2538. doi: 10.3791/2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock M. Alvira M. Vandenberghe L.H., et al. Rapid, simple, and versatile manufacturing of recombinant adeno-associated viral vectors at scale. Hum. Gene Ther. 2010;21:1259–1271. doi: 10.1089/hum.2010.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshri N. Koerber J.T. Kaspar B.K. Schaffer D.V. Directed evolution of adeno-associated virus yields enhanced gene delivery vectors. Nat. Biotechnol. 2006;24:198–204. doi: 10.1038/nbt1182. [DOI] [PubMed] [Google Scholar]
- Markusic D.M. Herzog R.W. Aslanidi G.V., et al. High-efficiency transduction and correction of murine hemophilia b using AAV2 vectors devoid of multiple surface-exposed tyrosines. Mol. Ther. 2010;18:2048–2056. doi: 10.1038/mt.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingozzi F. High K.A. Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat. Rev. Genet. 2011;12:341–355. doi: 10.1038/nrg2988. [DOI] [PubMed] [Google Scholar]
- Monahan P.E. Lothrop C.D. Sun J., et al. Proteasome inhibitors enhance gene delivery by AAV virus vectors expressing large genomes in hemophilia mouse and dog models: a strategy for broad clinical application. Mol. Ther. 2010;18:1907–1916. doi: 10.1038/mt.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi H. Taketa K. Miyano K., et al. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 1982;42:3858–3863. [PubMed] [Google Scholar]
- Nathwani A.C. Rosales C. McIntosh J., et al. Long-term safety and efficacy following systemic administration of a self-complementary AAV vector encoding human fix pseudotyped with serotype 5 and 8 capsid proteins. Mol. Ther. 2011;19:876–885. doi: 10.1038/mt.2010.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrs-Silva H. Dinculescu A. Li Q., et al. Novel properties of tyrosine-mutant AAV2 vectors in the mouse retina. Mol. Ther. 2011;19:293–301. doi: 10.1038/mt.2010.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponder K.P. Hemophilia gene therapy: a Holy Grail found. Mol. Ther. 2011;19:427–428. doi: 10.1038/mt.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao C. Zhang W. Yuan Z., et al. Adeno-associated virus serotype 6 capsid tyrosine-to-phenylalanine mutations improve gene transfer to skeletal muscle. Hum. Gene Ther. 2010;21:1343–1348. doi: 10.1089/hum.2010.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed S.E. Staley E.M. Mayginnes J.P., et al. Transfection of mammalian cells using linear polyethylenimine is a simple and effective means of producing recombinant adeno-associated virus vectors. J. Virol. Methods. 2006;138:85–98. doi: 10.1016/j.jviromet.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Samulski R.J. Chang L.S. Shenk T. Helper-free stocks of recombinant adeno-associated viruses: normal integration does not require viral gene expression. J. Virol. 1989;63:3822–3828. doi: 10.1128/jvi.63.9.3822-3828.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar R. Tetreault R. Gao G., et al. Total correction of hemophilia a mice with canine FVIII using an AAV8 serotype. Blood. 2004;103:1253–1260. doi: 10.1182/blood-2003-08-2954. [DOI] [PubMed] [Google Scholar]
- Sommer J.M. Smith P.H. Parthasarathy S., et al. Quantification of adeno-associated virus particles and empty capsids by optical density measurement. Mol. Ther. 2003;7:122–128. doi: 10.1016/s1525-0016(02)00019-9. [DOI] [PubMed] [Google Scholar]
- Sonntag F. Schmidt K. Kleinschmidt J.A. A viral assembly factor promotes AAV2 capsid formation in the nucleolus. Proc. Natl. Acad. Sci. U. S. A. 2010;107:10220–10225. doi: 10.1073/pnas.1001673107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonntag F. Kother K. Schmidt K., et al. The assembly-activating protein promotes capsid assembly of different adeno-associated virus serotypes. J. Virol. 2011;85:12686–12697. doi: 10.1128/JVI.05359-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E.M. Detection of specific sequences, among DNA fragments seperated by gel electrophoresis. J. Mol. Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Wu J. Zhao W. Zhong L., et al. Self-complementary recombinant adeno-associated viral vectors: packaging capacity and the role of rep proteins in vector purity. Hum. Gene Ther. 2007;18:171–182. doi: 10.1089/hum.2006.088. [DOI] [PubMed] [Google Scholar]
- Wu Z. Yang H. Colosi P. Effect of genome size on AAV vector packaging. Mol. Ther. 2010;18:80–86. doi: 10.1038/mt.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z. Zak R. Luxton G.W., et al. Ubiquitination of both adeno-associated virus type 2 and 5 capsid proteins affects the transduction efficiency of recombinant vectors. J. Virol. 2002;76:2043–2053. doi: 10.1128/jvi.76.5.2043-2053.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L. Zhao W. Wu J., et al. A dual role of EGFR protein tyrosine kinase signaling in ubiquitination of AAV2 capsids and viral second-strand DNA synthesis. Mol. Ther. 2007;15:1323–1330. doi: 10.1038/sj.mt.6300170. [DOI] [PubMed] [Google Scholar]
- Zhong L. Li B. Mah C.S., et al. Next generation of adeno-associated virus 2 vectors: point mutations in tyrosines lead to high-efficiency transduction at lower doses. Proc. Natl. Acad. Sci. U. S. A. 2008;105:7827–7832. doi: 10.1073/pnas.0802866105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.