Abstract
Tissue-targeted expression is of major interest for studying the contribution of cellular subpopulations to neurodegenerative diseases. However, in vivo methods to investigate this issue are limited. Here, we report an analysis of the cell specificity of expression of fluorescent reporter genes driven by six neuronal promoters, with the ubiquitous phosphoglycerate kinase 1 (PGK) promoter used as a reference. Quantitative analysis of AcGFPnuc expression in the striatum and hippocampus of rodents showed that all lentiviral vectors (LV) exhibited a neuronal tropism; however, there was substantial diversity of transcriptional activity and cell-type specificity of expression. The promoters with the highest activity were those of the 67 kDa glutamic acid decarboxylase (GAD67), homeobox Dlx5/6, glutamate receptor 1 (GluR1), and preprotachykinin 1 (Tac1) genes. Neuron-specific enolase (NSE) and dopaminergic receptor 1 (Drd1a) promoters showed weak activity, but the integration of an amplification system into the LV overcame this limitation. In the striatum, the expression profiles of Tac1 and Drd1a were not limited to the striatonigral pathway, whereas in the hippocampus, Drd1a and Dlx5/6 showed the expected restricted pattern of expression. Regulation of the Dlx5/6 promoter was observed in a disease condition, whereas Tac1 activity was unaffected. These vectors provide safe tools that are more selective than others available, for the administration of therapeutic molecules in the central nervous system (CNS). Nevertheless, additional characterization of regulatory elements in neuronal promoters is still required.
Delzor and colleagues analyze the cell-type specificity of lentiviral vector expression of fluorescent reporter genes driven by six neuronal promoters. While all vectors exhibited neuronal tropism, substantial diversity of transcriptional activity and cell-type specificity of expression were observed.
Introduction
Lentiviral vectors (LV) have been successfully used for preclinical and clinical gene delivery in the central nervous system (CNS) (Nanou and Azzouz, 2009). A strong neuronal tropism was observed for vesicular stomatitis virus G protein (VSV-G) pseudotyped lentiviral vectors expressing transgenes under the control of the ubiquitous cytomegalovirus (CMV) or phosphoglycerate kinase 1 (PGK) promoters (Naldini et al., 1996; Blömer et al., 1997; Déglon et al., 2000). However, to explore pathophysiological conditions and, in particular, the selective vulnerability of neurons and contribution of various cell types in neurodegenerative diseases (Saxena and Caroni, 2011), restricting transgene expression to specific subpopulations of neurons would be useful. Cell-type specific transgene expression would help dissect the intrinsic components of the disease process, including cell-autonomous vs. non–cell-autonomous mechanisms (Gu et al., 2007; Lobsiger and Cleveland, 2007; Hsiao and Chern, 2010). In addition, the ability to control transgene expression levels (low, medium, and high expression levels) would be particularly valuable to ensure physiological experimental conditions: Potential deleterious effects associated with massive overexpression of the gene of interest could thereby be avoided, and biological benefits could be ensured by sufficiently strong transgene expression.
Most core promoters available were identified and characterized on a gene-by-gene basis. Human genome sequencing has provided very substantial information about protein-coding sequences, but regulatory regions that temporally and spatially regulate gene expression are still poorly understood. The situation is, however, progressively changing with transcriptomic analysis, the Allen Brain Atlas maps of genes expressed in the adult mouse brain (Jones et al., 2009), and GENSAT with bacterial artificial chromosome (BAC) mice expressing GFP under different promoters (Gong et al., 2003). In parallel to these studies, large initiatives, such as the ENCODE and Pleiade projects, are providing information about the functional elements of the human genome and developing minipromoters for the CNS (Birney et al., 2007; Portales-Casamar et al., 2010).
Despite these efforts, only a limited number of promoters with regulatory domains spanning a restricted area of the genome and suitable in size for cloning in LV have been characterized. Most tissue-specific promoters have weaker transcriptional activities than viral (CMV) promoters. This limitation might be overcome, at least in part, by using transcriptional amplification systems (Zhang et al., 2002), integrated enhancers (Liu et al., 2004), or post-transcriptional regulatory elements (Donello et al., 1998; Zufferey et al., 1999), which in all cases maintain the tissue-specific patterns of gene expression. For example, the presence of the woodchuck post-regulatory element (WPRE) in an expression cassette resulted in two- to fivefold increases in transgene expression, depending both on the viral vector and the transcriptional activity of the promoter (Déglon et al., 2000; Xu et al., 2001; Fitzsimons et al., 2002); hybrid regulatory elements containing the CMV enhancer and tissue-specific or housekeeping (chicken β-actin) promoters also enhance transgene expression (Liu et al., 2004; Hioki et al., 2007; Gruh et al., 2008). Inducible systems, such as the tetracycline-regulated system, have been successfully used to create an amplification loop and enhance transcriptional activities of weak promoters (Nettelbeck et al., 1998; Régulier et al., 2002; Liu et al., 2008).
The cell-specific expression in the brain of a transgene carried by an LV is the combined result of host cell-surface factors interacting with the VSV-G envelope and the activity of the promoter. HIV-1 sequences surrounding the promoter in LV also influence the transcriptional activity and tropism of the vector. Neither the receptor of the VSV-G protein nor specific host factors have yet been identified. However, the transduction efficiency of neuronal cells by LV is high (Naldini et al., 1996; Kordower et al., 1999; Déglon et al., 2000). The contribution of the viral backbone is illustrated by the very well-characterized rat neuron-specific enolase (NSE) promoter: If the 1.8 kb fragment carrying this promoter is inserted into an adenoviral vector, both neuronal and glial expression is observed (Kugler et al., 2003), whereas the same promoter integrated into an adeno-associated viral vector (AAV2) results in >99% neuronal transduction (Xu et al., 2001).
We report the integration of six pan-neuronal and cell-type specific neuronal promoters into constitutive and tetracycline-regulated LV containing the WPRE element and compared with the ubiquitous PGK promoter. We assessed the tropism and transcriptional activity of these constructs in the striatum and hippocampus of rodents—two brain regions that are of particular relevance to neurodegenerative disorders.
Material and Methods
Plasmids
Various promoters were inserted into lentiviral backbones to study their transcriptional activity. We used self-inactivated lentiviral vectors containing the WPRE element (SIN-W) (Déglon et al., 2000) and encoding the nuclear-localized green fluorescent protein (pAcGFP1-Nuc; Clontech, Saint-Germain-en-Laye, France) (SIN-W-AcGFPnuc). We used tetracycline-regulated expression as an amplification loop to enhance promoters with weak transcriptional activity. Hence, we used the Tet-off system with the tTA/S2 transactivator (SIN-W-tTA/S2) (Régulier et al., 2002). We used an LV vector expressing the nuclear-localized red fluorescent protein (pDsRed2-Nuc; Clontech, Saint-Germain-en-Laye, France) to assess the specificity of promoters for neuronal subpopulations in transgenic mice expressing GFP (SIN-W-DsRednuc).
The following were used as internal promoters: the ubiquitous murine phosphoglycerate kinase 1 promoter (PGK; from −430 to +74 relative to the transcriptional start site (TSS); GenBank M18735.1 nt: 423 to 931) (Adra et al., 1987; Déglon et al., 2000) and the pan-neuronal 1.8 kb rat neuron specific enolase promoter (pAAV2ss-NSE-eGFP-WPRE-bGH; kindly provided by Prof. MJ During; GenBank AB038993.1, nt: 1029 to 2715 and AH002215.1, nt: 1039 to 1155) (Sakimura et al., 1987; Forss-Petter et al., 1990; Xu et al., 2001).
The following promoters were used for tissue-specific expression: the 369 bp murine homeobox Dlx5/6 (pmI56i; courtesy of Dr. Ekker; GenBank AY168010.1 nt: 8476 to 9012) (Zerucha et al., 2000) fused to the minimal cytomegalovirus promoter (CMVmin; Genbank DQ000970.1, nt: 706 to 834); the 3.7 kb fragment of the gene for the murine glutamic acid decarboxylase of 67 kDa (pXGAD4eGFP; courtesy of Dr. Teschemacher; GenBank AB006974.1, nt: 6442 to 10166) (Szabo et al., 1996; Katarova et al., 1998); a chimeric promoter (570 bp) composed of the murine Drd1a dopamine receptor promoter (pCAT-ActAR1-MU-MA; kindly provided by Dr. Mouradian; GenBank AF171079.1; nt: 21 to 201 and 434 to 790) preceded by the human activator sequence ActAR1 (GenBank M85247.1; nt: 1403 to 1437) (Minowa et al., 1993; Lee et al., 1999; Kim et al., 2003); the rat glutamate receptor 1 promoter (pGluR1-1395/+8; kindly provided by Dr. Dingledine; GenBank AF302117.1; nt: 3641 to 5042) (Borges and Dingledine, 2001); and 868 bp of the rat preprotachykinin 1 promoter (PPT, Tac1, or substance P; GenBank M34159.1; nt: 1 to 868), which was amplified from rat genomic DNA with the following primers: 5’-CACCACCTGCAGAGCTCCAAA GGTAAGC-3’ and 5’-TCTAGTCCCTGCTCCTGCTTCG-3’ (GenBank M34159.1 nt: 1 to 22—with a CACCAC flanking sequence for oriented cloning—and 847 to 868 for forward and reverse primer, respectively) (Carter and Krause, 1990; Mendelson et al., 1995).
For tetracycline-regulated expression, an LV encoding the AcGFPnuc reporter gene under the control of the tetracycline-response element (TRE) with the CMVmin promoter was used (SIN-W-TRE-AcGFPnuc). A NheI-BamHI AcGFPnuc fragment was excised from pAcGFP1-Nuc (Clontech, Saint-Germain-en-Laye, France), end-filled with T4 DNA polymerase (Invitrogen, Cergy-Pontoise, France) and inserted, as a blunt-ended fragment, into the reporterless SIN-W-TRE plasmid to generate the construct SIN-W-TRE-AcGFPnuc. The second vector encodes the tTA/S2 transactivator under the control of the different promoters (SIN-W-promoter-tTA/S2).
To study the effect of overexpression of mutant huntingtin (Htt) on transcriptional activity, LV encoding the first 171 amino acids of Htt with either 18Q or 82Q (SIN-W-PGK-Htt171-18Q/82Q) were used (de Almeida et al., 2002).
Lentiviral vector production
Lentiviral vectors were produced in 293T cells, by a four-plasmid system, as previously described (Hottinger et al., 2000). The HIV-1 vectors were pseudotyped with the VSV-G protein. Viruses were concentrated by ultracentrifugation and resuspended in phosphate-buffered saline (PBS) with 1% bovine serum albumin (BSA). The viral particle content of each batch was determined by p24 antigen enzyme-linked immunosorbent assay (RETROtek; Gentaur, Paris, France). The stocks were stored at −80°C until use.
Animals
Male Wistar rats (weight, 200 g; Iffa Credo/Charles River, Wilmington, MA), GENSAT transgenic Drd1a-eGFP mice (weight, 20 g; Tg(Drd1a-eGFP)X60Gsat/Mmmh strain; MMRRC, Columbia, MO) and GENSAT Drd2-eGFP mice (weight, 20 g;Tg(Drd2-eGFP)S118Gsat/Mmnc; MMRRC, Chapel Hill, NC) (Gong et al., 2003) were used. The animals were housed in a temperature-controlled room and maintained on a 12 hr day/night cycle. Food and water were available ad libitum. The experiments were carried out in accordance with the European Community directive (86/609/EEC) for the care and use of laboratory animals.
Stereotaxic injections
Concentrated viral stocks were thawed on ice and resuspended by vortexing and repeated pipetting. Then, they were diluted in PBS/1%BSA to a final concentration of 67 ng p24/μl for single transduction in rats and mice, and to 33 ng p24/μl for co-transduction in rats (ratio 1:1). These low doses of vectors were chosen to limit transduction in the CNS and therefore avoid saturation in transgene expression and reveal the different transcriptional activity of the various promoters. As a result, 15,000–20,000 cells were infected in striatum as compared with approximately 100,000–300,000 infected cells with an optimal injection of lentiviral vector (Naldini et al., 1996; de Almeida et al., 2002). Rats were anesthetized with isoflurane (CSP, Cournon d'Auvergne, France) and then with 75 mg/kg ketamine, 10 mg/kg xylazine (Coveto, Montaigu, France) delivered by intraperitoneal (i.p.) injection. Mice were anesthetized by i.p. injection with a mixture of 150 mg/kg ketamine and 10 mg/kg xylazine (Coveto, Montaigu, France). Suspensions of lentiviral vectors were injected into the brain through a 34-gauge blunt-tip needle linked to a Hamilton syringe by a polyethylene catheter.
Stereotaxic coordinates for injection into the rat striatum were, from bregma: anteroposterior+0.5 mm; lateral+/–3 mm and ventral −4.5 mm from the dura, with tooth bar set at −3.3 mm (Paxinos and Watson, 2004). Rats received a total volume of 3 μl of the vector preparation, administered at a rate of 0.25 μl/min. For the hippocampus, the coordinates used were as follows, from bregma: anteroposterior −4.1 mm; lateral+/–3 mm and ventral −3.2 mm from the skull surface, with tooth bar set at −3.3 mm.
Stereotaxic coordinates for injection into mouse striatum were, from bregma: anteroposterior+1 mm; lateral+/–2 mm and ventral −2.5 mm from the dura, with tooth bar set at 0 mm (Franklin and Paxinos, 1996). Mice received a total volume of 3 μl of the vector preparation, administered at a rate of 0.2 μl/min.
At the end of injections, needles were left in place for 5 min and then slowly removed. The skin was sutured and animals were allowed to recover.
Histological processing
Tissue preparation
Three weeks post-lentiviral injection, the animals were given an overdose of sodium pentobarbital and were transcardially perfused with a 4% paraformaldehyde (PFA; Sigma-Aldrich, Saint-Quentin Fallavier, France) solution. The brains were removed and post-fixed in 4% PFA for about 12 hr, and then cryoprotected first in 15% sucrose / 0.1 M PBS overnight and then in 30% sucrose / 0.1 M PBS for 24 hr for mice or 48 hr for rats. A sledge microtome with a freezing stage at −30°C (SM2400; Leica, Nanterre, France) was used to cut brain coronal sections 30 μm thick. Slices throughout the entire striatum or the hippocampus were collected and stored in tubes as free-floating sections in PBS supplemented with 0.12 μM sodium azide. Tubes were stored at 4°C until immunohistochemical processing.
Primary antibodies
The following primary antibodies were used: mouse monoclonal anti-neuronal nuclei antibody (NeuN, dilution 1/200; MAB377, Millipore, Molsheim, France); mouse monoclonal antibody recognizing the β subunit of the S100 protein (S100β, dilution 1/500; S2532, Sigma-Aldrich, Saint-Quentin Fallavier, France); rabbit polyclonal anti-GFP antibody (A6455, dilution 1/250; Life Technologies, Saint Aubin, France), the epitope of which is contained in AcGFPnuc; and rabbit polyclonal anti-ubiquitin antibody (Ubi dilution 1/500; Z0458, DakoCytomation, Trappes, France).
Immunohistochemical procedure
Striatal sections from rats injected with SIN-W-promoter-AcGFPnuc or co-injected with SIN-W-promoter-tTA/S2 and SIN-W-TRE-AcGFPnuc were mounted directly in an aqueous medium (FluorSave; Life Technologies). Striatal sections from Drd1a- and Drd2-eGFP mice injected with SIN-W-promoter-DsRednuc were directly mounted in FluorSave.
To overcome the weak transcriptional activity of the NSE promoter, striatal sections from rats injected with SIN-W-NSE-AcGFPnuc vector were processed for immunofluorescence with anti-GFP and the following protocol. Fixed slices were washed with PBS and incubated in a blocking solution of PBS supplemented with 3% NGS, 0.2% gelatin, and 0.3% Triton X-100 for 1 hr at room temperature. They were then incubated overnight at 4°C with the anti-GFP antibody in PBS supplemented with 3% NGS, 0.2% gelatin, and 0.1% Triton X-100. The next day, slices were washed three times in PBS, the fluorescent secondary antibody diluted 1/500 in PBS (AlexaFluor 488 anti-rabbit; Life Technologies) was added, and the samples incubated for 1 hr at room temperature. Finally, slices were washed three times in PBS and mounted in FluorSave (Life Technologies).
For the study of LV tropism, striatal and hippocampal sections from rats were labeled with NeuN and S100β antibodies. The fluorescent secondary antibodies used were AlexaFluor 350 (blue) or AlexaFluor 594 (red) anti-mouse diluted 1/500 (Life Technologies). The protocol was as described above.
To study the effect of Htt171-82Q on the transcriptional activity of Dlx5/6 and Tac1 promoters, ubiquitin was labeled with AlexaFluor 488 (green) or 594 (red) anti-rabbit antibodies diluted at 1/500 (Life Technologies) by the protocol described above.
Quantitative analyses
Co-localization with neuronal or astrocytic markers
Sections labeled for NeuN or S100β were analyzed by epifluorescence microscopy with a Leica DM6000B microscope (Leica, Nanterre, France) equipped with an automated motorized stage and image acquisition software (MorphoStrider software; Explora Nova, La Rochelle, France). The numbers of AcGFPnuc-NeuN-positive cells and AcGFPnuc-S100β-positive cells were determined on images acquired with a 40x objective (six animals, three sections per animal, six images per section) by ImageJ software (National Institutes of Health, Bethesda, MD). Similarly, ImageJ software was used to identify co-localization of DsRednuc and Drd1a- or Drd2-eGFP on images acquired with a 40x objective.
Mean fluorescence intensity per cell
In the present study, we use GFP as a quantitative reporter of gene expression and therefore transcriptional activity. Previous studies have demonstrated that GFP fluorescence is directly proportional to the number of virally delivered gene copies and that the fluorescence intensity is directly correlated to the messenger RNA (mRNA) levels in infected cells (Soboleski et al., 2005). In a recent publication, Xu et al. (2012) demonstrate that GFP mRNA abundance measured by quantitative reverse transcription-PCR is correlated with GFP expression monitored using fluorescence microscopy.
We therefore acquired photomicrographs of AcGFPnuc-positive striatal rat sections with a 10x objective. Mosaic pictures covering the entire striatum were generated and used to measure the fluorescence intensity (Morphostrider software, Explora Nova). The acquisition parameters of the Leica DM6000B microscope (excitation neutral attenuator filters and obturator) were maintained equivalent for all acquisitions while camera parameters (time of exposure from 100 to 800 ms) were adjusted for each group (e.g., each promoter) to maximize fluorescence but without saturation (avoided using display of pixel fluorescence intensity histogram). The section closest to the injection site was used to measure the mean fluorescence intensity (MFI) per cell.
The AcGFPnuc-positive area was delimited and a gray level threshold was applied for automated segmentation and count of infected cells expressing GFP (the lower limit of the gray level was set up at 9). The ImageJ software automatically calculated the mean gray level of the various objects identified. To allow semi-quantitative comparison between the promoters, fluorescence levels for each group (promoter) were normalized to the same time exposure (500 ms). We experimentally checked that fluorescence emission (gray levels) GFP containing brain sections was linearly proportional to exposure time in the range we used (100–800 ms). Graphs representing the distribution of MFI/cell were obtained with Statview software (SAS Institute Inc., Cary, NC).
Quantification of transduced cells
Quantification of transduced cells was analyzed by performing mosaics acquisition over the entire striatum (six animals, all sections with fluorescence separated by 240 μm) with a 10x objective. Acquisition parameters were adjusted for each image to have a maximum of fluorescence without saturation. By performing a threshold analysis based on gray levels, total objects number was automatically counted with the ImageJ software (objects bigger than 20 μm2). This total number of cells was corrected by the Abercrombie factor.
Statistical analysis
Data were analyzed using GraphPad Prism 4 software (GraphPad Software, La Jolla, CA). Two-way or one-way analysis of variance (ANOVA) with a post-hoc Newman-Keuls test analysis were used to assess the significance of differences (set at p<0.05). Data are presented as means±standard error of the mean (SEM).
Results
Characterization of transduced cells
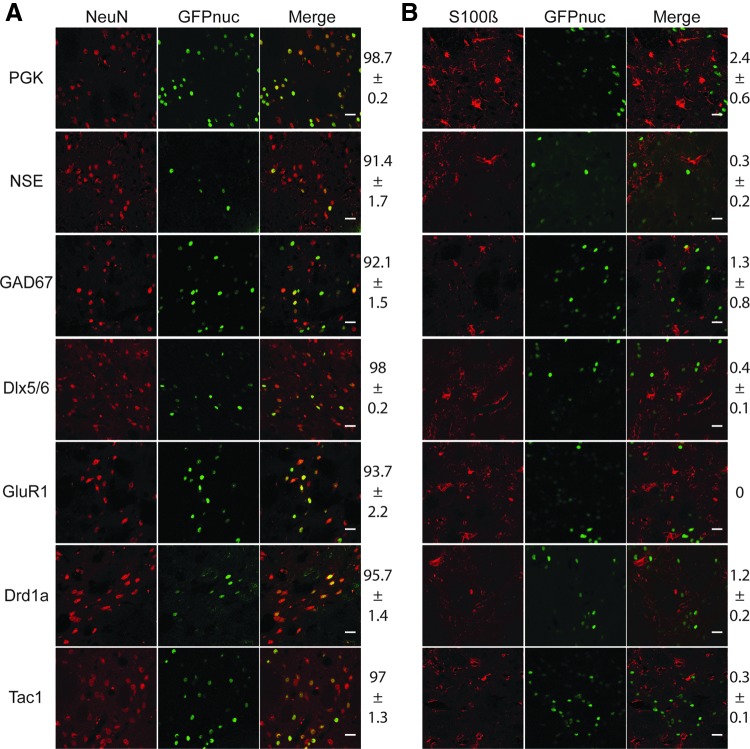
The pan-neuronal promoter NSE and five cell-type specific neuronal promoters (GAD67, Dlx5/6, GluR1, Drd1a, and Tac1) were inserted into SIN-W-AcGFPnuc LV (Supplementary Table S1; Supplementary Material available online at www.liebertonline.com/hum). LV with the housekeeping PGK promoter (SIN-W-PGK-AcGFPnuc) was used as a control (Déglon et al., 2000). The constructs were administered to adult rats by injection, and three weeks later the rats were sacrificed and sections of the striatum were processed for immunofluorescence. Low doses of the vectors were injected to avoid saturation of the system and accurately measure GFP expression. GFP was detected in nuclei around the injection site in all animals; no fluorescence was detected in the untreated contralateral striatum. Antibodies against the neuronal marker NeuN and glial marker S100 β were used to determine the phenotype of the transduced cells (Fig. 1A). Microscopic analysis and cell counts revealed that more than 95% of the transduced cells co-localized with NeuN, whereas only scarce AcGFPnuc-positive cells were double-stained for S100β (Fig. 1A and B). Thus, all the promoters retained neuronal specificity.
FIG. 1.
Transduction efficiency and tropism in adult rats of lentiviral vectors containing the neuronal promoters. (A) Double immunofluorescence staining for the neuronal marker NeuN or (B) the astrocytic marker S100β, and AcGFPnuc after the injection of vesicular stomatitis virus G protein (VSV-G) pseudotyped lentiviral vectors (LVs) into the striatum. Acquisition parameters were optimized for each promoter. Quantification of AcGFPnuc-NeuN- and AcGFPnuc-S100β-positive cells are indicated and confirm the neuronal tropism of all vectors (n=5–6). Scale bar=20 μm.
Striatal transcriptional activity of the neuronal promoters
We assessed the strength of transgene expression driven by these promoters. The GFP protein is a reliable reporter system for the analysis of promoter activity and can be quantified by measuring fluorescence intensity (Soboleski et al., 2005). To evaluate accurately the amount of GFP produced by the neurons and avoid underestimation of GFP due to dendritic and axonal fluorescence, we used a nuclear-localized GFP. In addition, previous studies have shown that GFP fluorescence is directly correlated with the mRNA level (Soboleski et al., 2005; Xu et al., 2012). We chose to compare the promoters in the striatum: the neuronal population in this structure is relatively homogeneous, with more than 95% of all striatal neurons being GABAergic medium-sized spiny neurons (MSNs) (Kawaguchi, 1997). This minimizes potential misinterpretation associated with tissue-specific expression in various subsets of neurons.
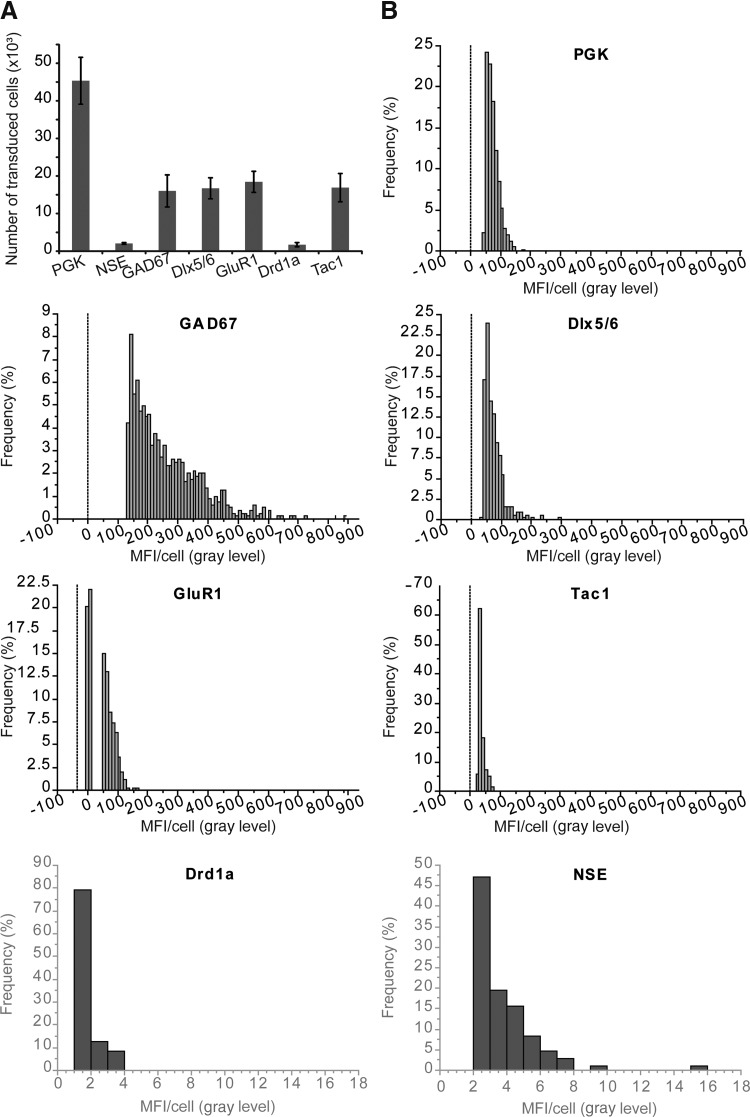
The transcriptional activity of the promoters was measured indirectly by quantifying the mean fluorescence intensity per cell (MFI/cell). This involved a threshold analysis of GFP expression followed by an automatic segmentation of GFP-positive cells. This approach allows the total number of objects determined in each section to be analyzed. For the NSE promoter, the number of infected cells and MFI/cell was determined using GFP-stained sections because of the very weak transcriptional activity of the promoter. The promoter with the highest activity in the striatum was GAD67, followed by PGK, Dlx5/6, GluR1, Tac1, and finally Drd1a and NSE, for which the fluorescence intensity was very low (Fig. 2).
FIG. 2.
Transcriptional activity of the neuronal promoters. (A) Quantification of the number of infected cells for each promoter. To avoid saturation, a low dose of lentiviral vector has been injected, explaining the limited number of transduced cells. (B) Graphs of mean fluorescence intensity per cell (MFI/cell) showing the distribution of the transcriptional activity of the different neuronal promoters in the striatum of adult rats (n=5–6).
Transduction efficiency and transcriptional activity of the neuronal promoters in the hippocampus
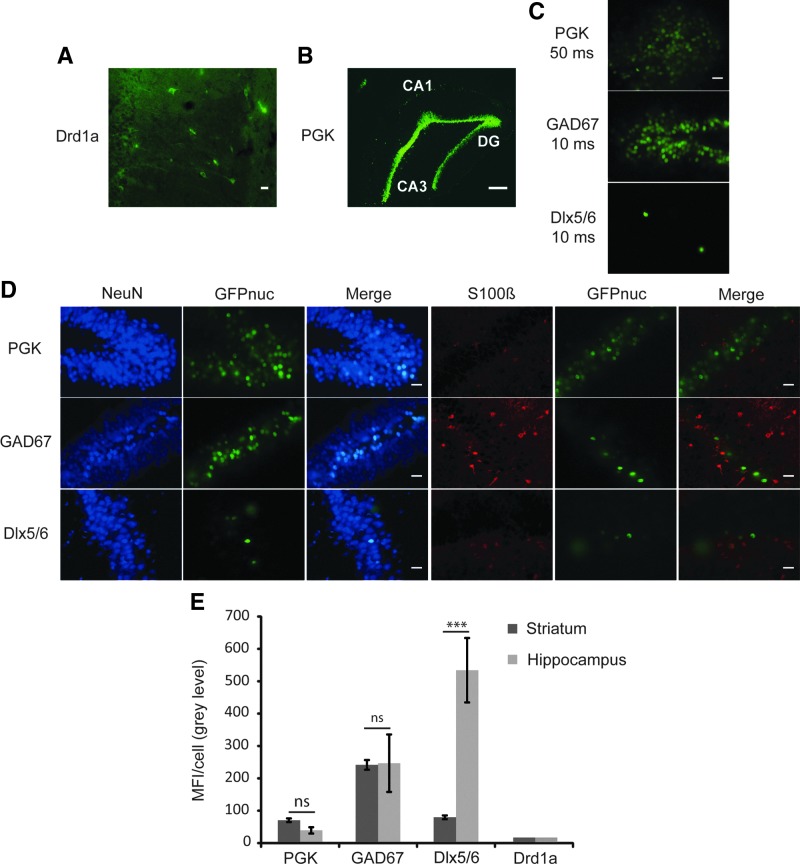
We next investigated whether our LVs restricted transgene expression to neurons in other regions of the brain. We select four promoters—PGK, GAD67, Dlx5/6, and Drd1a—for this study and injected them into the CA1 region of the hippocampus. Expression of the Drd1a-AcGFPnuc vector was poorly detected (Fig. 3A), whereas PGK-, GAD67-, and Dlx5/6-AcGFPnuc were clearly visible in the dentate gyrus, but with different transcriptional activities (Fig. 3B,C,and E). Co-localization of AcGFPnuc with the neuronal marker NeuN shows the neurotropism of the vectors also in the hippocampus (Fig. 3D). Dlx5/6- and Drd1a-AcGFPnuc were expressed in very limited subset of NeuN-positive cells; although, the transcriptional activity of Dlx5/6 in these cells was very high (Fig. 3C–E). This is consistent with published findings that Dlx genes are expressed in GABAergic interneurons of the brain (Cobos et al., 2005; Potter et al., 2009; Wang et al., 2010), and that there is sparse expression of Drd1a in pyramidal cells throughout the CA1, 2, 3 fields and dentate granular cells in the anterior of the dorsal hippocampus (Fremeau et al., 1991). The mean fluorescence intensity per cell for the PGK and GAD67 promoters in the hippocampus were very similar to those in the striatum (Fig. 3E). The transcriptional activity of GAD67 is in agreement with the presence of the enzyme throughout the hippocampus (Jinno et al., 1998).
FIG. 3.
Cell-type specificity and activity of PGK, GAD67, Dlx5/6, and Drd1a promoters in the hippocampus. (A) A weak transgene expression was observed with the SIN-W-Drd1a-AcGFPnuc LV. Scale bar=20 μm. (B) Representative picture of the transduced area, which was localized predominantly in the dentate gyrus. Scale bar=200 μm. (C) Representative images showing difference in fluorescence intensity between promoters. PGK image has been taken with a 50 ms exposition, whereas both GAD67 and Dlx5/6 have been taken with a 10 ms exposition. Scale bar=20 μm. (D) Double-fluorescence immunostaining with either NeuN (blue) or S100β (red), showing co-localization of AcGFPnuc with the neuronal marker. Acquisition parameters were optimized for each promoter. Scale bar=20 μm. (E) Quantitative analysis of MFI/cell in the hippocampus and comparison with that in the striatum (n=5 except for Dlx5/6, n=3).
Tetracycline-inducible LV to create a transcriptional amplification system for weak neuronal promoters
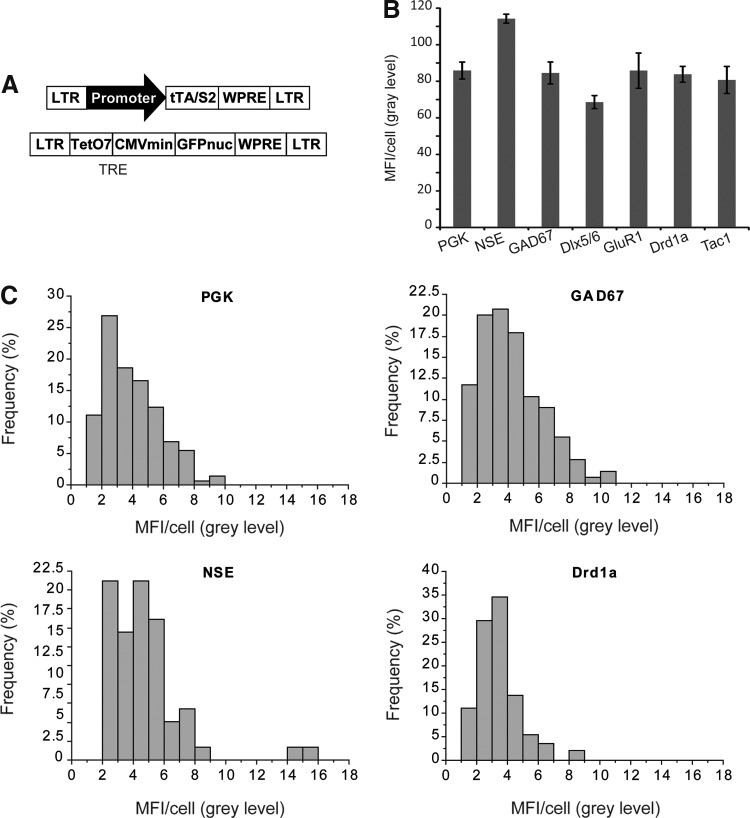
Although the Drd1a promoter may be valuable to obtain transgene expression restricted to a region of interest, its weak transcriptional activity is problematic. One strategy for overcoming this limitation without losing the cell-type specificity is to use inducible expression systems. In tetracycline-regulated LV, the weak neuronal promoter drives the expression of a tetracycline transactivator (tTA/S2), and this molecule then induces strong expression of the AcGFPnuc. We have previously shown that this system is associated with a fivefold stronger transgene expression than obtained with the SIN-W-PGK vector (Régulier et al., 2002). We introduced all the promoters into the SIN-W-tTA/S2 construct (Régulier et al., 2002). Adult rats were injected with a 1:1 ratio of the two vectors of the system (Fig. 4A). A large proportion of infected striatal cells contained a copy of both vectors (Régulier et al., 2002). The mean fluorescence intensity per cell was determined histologically three weeks postinjection. AcGFPnuc expression levels were similar in all groups and also similar to that driven by the PGK promoter. The effect of the inducible system was particularly large for the weakest promoters, NSE and Drd1a, which led to barely detectable AcGFPnuc expression when inserted in the constitutive LV (Fig. 2). AcGFPnuc expression from the GAD67 promoter was no stronger than that from the PGK promoter (Fig. 4B and C); this contrasts with the constitutive expression of AcGFPnuc from the same promoters, where the GAD67 promoter was stronger than the PGK promoter (Fig. 2). This probably reflects saturation of the amplification loop involving the tetracycline-response element: The number of integrated copies of TRE-AcGFPnuc may be limiting such that increasing the expression of tTA/S2 has no effect on AcGFPnuc expression.
FIG. 4.
The tetracycline amplification system improves the transcriptional activity of the weak Drd1a and neuron-specific enolase (NSE) promoters. (A) Scheme of the lentiviral vectors used for regulated expression of the reporter gene. (B) Quantitative analysis of MFI/cell for each promoter (n=6 except for NSE, n=4).
Restricted expression in D1 and D2 receptor-positive medium-sized spiny neurons
Approximately 95% of striatal neurons are GABAergic MSNs (Kawaguchi, 1997). Nevertheless, striatal cells can be classified into two main subgroups according to their neurochemical substance contents and projection sites (Gerfen, 1992). The MSNs projecting to the internal segment of the globus pallidus (GPi) and substantia nigra pars reticulata (SNr), express substance P (or Tac1) and dynorphin, D1 dopamine (D1R, Drd1a) and M4 muscarinic acetylcholine receptors (chrm4), and constitute the direct striatonigral pathway. The MSNs projecting to the entopeduncular nucleus are in the indirect striatopallidal pathway. These neurons are enriched in enkephalin, D2 dopamine (D2R, Drd2), and A2a adenosine receptors (adora2a). In many neurodegenerative diseases, and in particular in Huntington's disease, MSNs of the indirect pathway are affected at earlier stages and to a greater extent than MSNs of the direct pathway (Reiner et al., 1988; Han et al., 2010).
We tested whether the Dr1da and Tac1 promoter elements in our LV constructs were sufficient to maintain tissue-specific expression (i.e., expression in the direct pathway). We used BAC transgenic mice expressing the GFP reporter gene driven by the regulatory promoter elements of the type 1a dopamine receptor gene (Drd1a, a marker of the direct pathway of the basal ganglia) and the Drd2 promoter (a marker of the indirect pathway). We also assessed the Dlx5/6 promoter for which no data were available.
In BAC-Drd1a-eGFP transgenic mice injected with a Drd1a-DsRednuc LV, both fluorescent proteins were found (and were thus co-localized) in 41.3±3.5% of infected cells. The findings with Drd2 mice were similar, with 48.8±5.6% of Drd2-eGFP neurons showing DsRednuc reporter gene expression (Fig. 5A and B). The results were also similar for the Tac1 promoter with expression of the transgene in 54.3±4.4% of Drd1a-positive neurons and 47.2±1.9% of Drd2-positive neurons (Fig. 5A and B). The Dlx5/6 promoter was associated with equivalent transgene expression in both DR1 and DR2 neurons. These data suggest that the transcriptional regulatory elements of the Drd1a and Tac1 promoters present in the LV are insufficient to restrict transgene expression to D1R receptor-expressing medium-sized spiny neurons.
FIG. 5.
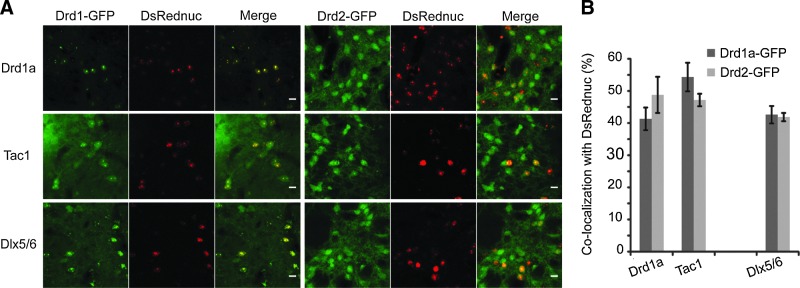
Expression in D1R- or D2R-positive striatal neurons. (A) LVs coding for the nuclear DsRed (DsRed2nuc) under the control of Drd1a, Dlx5/6, or Tac1 promoters were injected into the striatum of transgenic BAC-Drd1a-eGFP or Drd2-eGFP mice. Representative images of co-localization of the DsRednuc, with either the Drd1a-eGFP-positive cells or the Drd2-eGFP-positive cells. (B) Quantitative analysis of the co-localization of DsRed2nuc and GFP expression in BAC-Drd1a-eGFP or BAC-Drd2-eGFP mice (n=5–6, except for Drd1a promoter in BAC-Drd2-eGFP mice, n=3). Scale bar=20 μm.
Effects of pathological conditions on neuronal promoter activities
In the last part of the study, we assessed the effects of environmental conditions on promoter activity. Both physiological and pathological conditions influence promoter activities. For instance, the neuronal activity and, in particular, membrane depolarization that activates the transcription factor CREB (cAMP response element-binding) increases the activity of the CMV promoter, which contains five cAMP-response elements (CREs) (Wheeler and Cooper, 2001). Microarray studies demonstrate that expression profiles are significantly altered by neurodegenerative diseases. In cases of Huntington's disease (HD), the largest abnormalities (differentially expressed mRNAs) were detected in the caudate nucleus (Hodges et al., 2006). To investigate potential modulation of the transcriptional activity in our vectors, we used two promoters, which are differentially affected in HD. Dlx5, but not Tac1, is significantly down-regulated in primary rat striatal cultures infected with an LV expressing a mutant Htt fragment encoding the first 171 amino acids with 82 CAG repeats (Htt171-82Q) (Runne et al., 2008). We therefore expressed Tac1-DsRednuc and Dlx5/6-AcGFPnuc LV in rats also injected with an LV-Htt171-82Q. After three weeks, the misfolded Htt proteins formed typical nuclear inclusions as revealed by labeling with an ubiquitin antibody (Supplementary Fig. S1). At this stage, the GABAergic neurons of the striatum are still dysfunctional, and cell death occurs at 2–3 months (de Almeida et al., 2002). As a control, an analogous fragment encoding the wild-type Htt with 18Q was used (Benchoua et al., 2008). As assessed from the mean fluorescence intensity per cell, the transcriptional activity of the Tac1 promoter was unaffected by the expression of mutant Htt, whereas that of the Dlx5/6 promoter in the presence of the mutant Htt was less than half that in the presence of the wt Htt (Fig. 6).
FIG. 6.
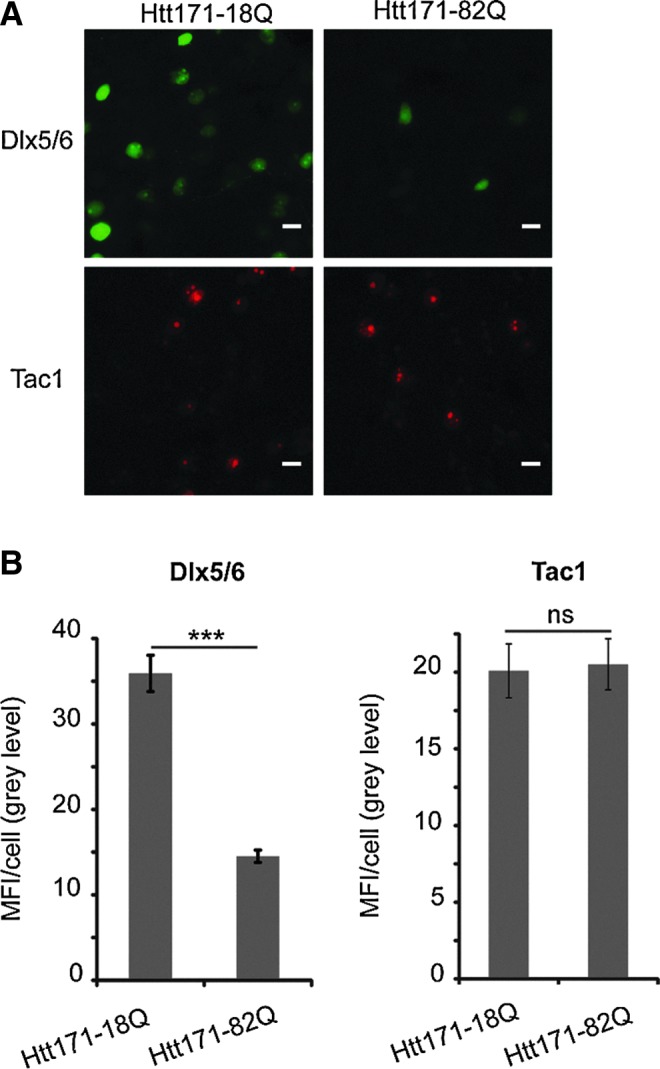
Effect of a disease state on transcriptional activity. (A) LVs encoding a fluorescent reporter gene under the control of Dlx5/6 or Tac1 promoters were co-injected with LVs expressing the first 171 aa of either the wild-type huntingtin (Htt171-18Q) or the mutant huntingtin (Htt171-82Q) into the striatum of adult rats. (B) MFI/cell showing the weaker expression from the Dlx5/6 promoter in animals expressing the mutant than wild-type Htt and indicating that the Tac1 promoter was unaffected (n=5). Scale bar=20 μm.
Discussion
The strength of transgene expression in the brain following LV injection is the combined result of viral vector entry, nuclear import, integration, and transcriptional activity of the promoter driving the expression of the reporter gene. Here, we report that all promoters cloned in VSV-G pseudotyped lentiviral vectors, including the ubiquitous PGK promoter, showed a strong neuronal tropism in the rodent striatum and hippocampus, in agreement with previous studies (Naldini et al., 1996, Déglon et al., 2000).
In this analysis, we assumed that an equal dose of the different constructs produced similar infection patterns and similar numbers of genome copies per cell, and that observed reporter gene expression correlates with the transcriptional activity of the promoters. However, in the case of VSV-G pseudotyped LV, the receptor on target cells is still unknown: Phosphatidylserine has been suggested (Schlegel et al., 1983), but recent studies show that it enhances viral entry but is not the receptor for the VSV-G envelope (Coil and Miller, 2004, 2005). We cannot therefore completely exclude the possibility that the cellular distribution of the VSV-G receptor on the surface of the different neuronal subtypes influences the outcome of LV infection. The relative homogeneity of the neuronal population in the striatum should nevertheless minimize this effect in our experiments. The incorporation of heterologous viral surface glycoproteins has been extensively used, either to acquire new properties, such as retrograde transport (Mazarakis et al., 2001) or alter the tropism in the central nervous system (Desmaris et al., 2001; Watson et al., 2002; Cronin et al., 2005; Cannon et al., 2011). VSV-G, which was the first and still most widely used pseudotyping, confers high vector particle stability, allowing for concentration by ultracentrifugation (Burns et al., 1993; Bartz and Vodicka, 1997). In addition, the great majority of transduced cells showed neuronal morphology (Naldini et al., 1996, Déglon et al., 2000), which dictates the choice of this envelope for the present study.
Another confounding factor in transgene expression is the contribution of cis-acting elements in the HIV-1 sequence, which could interfere with promoter activity. The LTR of HIV-1 contains sequences that alter transgene expression from internal heterologous promoters (Zufferey et al., 1998; Ginn et al., 2003). This promoter interference is cell-type and species-specific and has also been observed with murine leukemia viral vectors (MLV) (Vile et al., 1995). For instance, effects of this type led to loss of tissue specificity of the albumin promoter when integrated into an MLV backbone (Wu et al., 1996). In our study, all promoters were inserted into a self-inactivating version of the vector (deletion of the U3 region), which very substantially reduces promoter interference (Zufferey et al., 1998).
The transcriptional activity differed substantially between the neuronal promoters, with NSE and Drd1a being the weakest. Based on serial analysis of gene expression (SAGE) data in different territories of mouse striatum (Brochier et al., 2008), this result was expected for Drd1a, as very few transcripts were detected (7 Tag counts by sequencing) relative to more abundant mRNAs such as those for proenkephalin 1, Tac1, and DARPP-32 (122, 36, and 27 tag counts, respectively). These findings are also in agreement with the lower fluorescence intensity in BAC-Drd1a mice than that in Drd2 mice (Matamales et al., 2009). The inclusion of the ActAR1 enhancer in our construct was not sufficient to overcome this weak expression. For NSE, there was a discrepancy between our findings and both endogenous expression levels reported in the literature and in SAGE database. NSE mRNA and protein are both among the most abundant in adult brains (Marangos et al., 1979). However, the 1.8 kb promoter has only weak transcriptional activity in our LV and in transgenic mice (Forss-Petter et al., 1990). This low level of transcription corresponds to NSE activity during developmental stages and is probably a consequence of the absence of regulatory elements necessary for the strong expression in differentiated neurons (Forss-Petter et al., 1990). Indeed, our findings are consistent with previous results with AAV and herpes simplex virus (HSV) (Wang et al., 1999; Kugler et al., 2001). The use of an amplification system, however, successfully increased reporter gene expression from the NSE and Drd1a promoters.
To further assess the tissue-specificity of the neuronal promoters, we injected constructs containing four of them into the hippocampus, another structure implicated in neurodegenerative diseases. As expected, the Drd1a promoter drove limited transgene expression in a subset of pyramidal cells. With the PGK and GAD67 promoters, the reporter gene was expressed in hippocampal neurons and the mean fluorescence intensity per cell was similar to that in the striatum. These data for GAD67 are in agreement with a previous study with the same promoter in an AAV (Teschemacher et al., 2005). Finally, in the case of the Dlx5/6 promoter, the reporter gene was expressed at a very high level but in a very small subset of cells, probably GABAergic interneurons (Cobos et al., 2005; Potter et al., 2009; Wang et al., 2010). The homeodomain transcription factors encoded by distal-less homeobox 5 and 6 are transcribed convergently and the mRNAs are detected in virtually all GABAergic neurons of the brain. Several studies implicate these factors in the differentiation of these neurons (Panganiban and Rubenstein, 2002). This promoter has been used to develop a conditional mouse model of HD (Gu et al., 2007). Transgene expression from the Dlx5/6 promoter was detected in striatal MSNs and in a subset of cortical interneurons; however, they did not assess mutant huntingtin expression in the hippocampus.
In the last part of the study, we assessed whether the neuronal promoters segregated neurons expressing dopamine D1 and D2 receptors and tested their response to pathophysiological conditions. To identify the intermingled GABAergic subpopulations, we used Drd1a-eGFP and Drd2-eGFP BAC transgenic mice: Striatonigral and striatopallidal neurons can be specifically labeled in these mice (Gong et al., 2003). In D1R/D2R double transgenic mice (Matamales et al., 2009), it has been shown that 50% of the MSNs express only D1R or D2R and that all striatal MSNs express either Drd1a-eGFP or Drd2-eGFP. Drd1a-eGFP and Drd2-eGFP animals were injected with LV carrying the DsRednuc under the control of Tac1, Drd1a, and Dlx5/6 promoters, and double fluorescent cells were counted: An equal proportion of D1R- and D2R-positive neurons were transduced. This result was expected for the Dlx5/6 promoter, because Dlx5/6 is involved in the differentiation of all GABAergic neurons; however, Tac1 and Drd1a promoters did not display the tissue-specificity of the endogenous gene expression. These observations further illustrate the necessity of a better understanding of the regulatory elements proximal and distal to the transcription start site. Such elements presumably determine both the cell-specificity of expression of these genes and their response to a disease state.
We have previously shown that PGK-driven reporter gene expression is down-regulated in Huntington's disease, such that it may serve as an indirect marker of the pathology in vivo (Zala et al., 2004; Galvan et al., 2012). In animals expressing a mutant huntingtin, a protein altering the function of several transcription factors and regulators, including Sp1, TBP, CREB, and REST (Steffan et al., 2000; Li et al., 2002; Chiang et al., 2007; Ravache et al., 2010), Dlx5/6 transcriptional activity was lower than in animals expressing a wild-type Htt fragment. In contrast, the Tac1 promoter was unaffected by the expression of mutant Htt. Binding sites for some of the transcription factors affected by the mutant Htt are present in Tac1 and Dlx5/6 promoters (for example, those for CREB-binding protein [CBP], SP1, TBP, NFkB, and KLF). The ENCODE and Pleiade projects currently underway should drastically improve our knowledge of regulatory elements. They will provide insight into transcriptional regulation during disease processes and provide useful tools to restrict transgene expression to selected populations of cells. Deciphering the molecular events and signaling pathways in subpopulations of neurons that contribute to behavioral deficits and are differentially involved in pathological conditions might lead to the development of safer and more selective viral vectors for the administration of therapeutic molecules to the CNS.
Supplementary Material
Acknowledgments
This work was partially supported by the CEA and the European Community's Seventh Framework Program FP7/2007-2013 under grant agreement n° HEALTH-F5-2008-222925 (NEUGENE).
Author Disclosure Statement
No competing financial interests exist.
References
- Adra C.N. Boer P.H. Mcburney M.W. Cloning and expression of the mouse pgk-1 gene and the nucleotide sequence of its promoter. Gene. 1987;60:65–74. doi: 10.1016/0378-1119(87)90214-9. [DOI] [PubMed] [Google Scholar]
- Bartz S.R. Vodicka M.A. Production of high-titer human immunodeficiency virus type 1 pseudotyped with vesicular stomatitis virus glycoprotein. Methods. 1997;12:337–342. doi: 10.1006/meth.1997.0487. [DOI] [PubMed] [Google Scholar]
- Benchoua A. Trioulier Y. Diguet E., et al. Dopamine determines the vulnerability of striatal neurons to the N-terminal fragment of mutant huntingtin through the regulation of mitochondrial complex II. Hum Mol Genet. 2008;17:1446–1456. doi: 10.1093/hmg/ddn033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E. Stamatoyannopoulos J.A. Dutta A., et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blömer U. Naldini L. Kafri T., et al. Highly efficient and sustained gene transfer in adult neurons with a lentivirus vector. J. Virol. 1997;71:6641–6649. doi: 10.1128/jvi.71.9.6641-6649.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges K. Dingledine R. Functional organization of the GluR1 glutamate receptor promoter. J Biol Chem. 2001;276:25929–25938. doi: 10.1074/jbc.M009105200. [DOI] [PubMed] [Google Scholar]
- Brochier C. Gaillard M.C. Diguet E., et al. Quantitative gene expression profiling of mouse brain regions reveals differential transcripts conserved in man and affected in disease models. Physiol Genomics. 2008;33:170–179. doi: 10.1152/physiolgenomics.00125.2007. [DOI] [PubMed] [Google Scholar]
- Burns J.C. Friedmann T. Driever W., et al. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc.Natl.Acad.Sci.USA. 1993;90:8033–8037. doi: 10.1073/pnas.90.17.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon J.R. Sew T. Montero L., et al. Pseudotype-dependent lentiviral transduction of astrocytes or neurons in the rat substantia nigra. Exp Neurol. 2011;228:41–52. doi: 10.1016/j.expneurol.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter M.S. Krause J.E. Structure, expression, and some regulatory mechanisms of the rat preprotachykinin gene encoding substance P, neurokinin A, neuropeptide K, and neuropeptide gamma. J Neurosci. 1990;10:2203–2214. doi: 10.1523/JNEUROSCI.10-07-02203.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang M.C. Chen H.M. Lee Y.H., et al. Dysregulation of C/EBP{alpha} by mutant Huntingtin causes the urea cycle deficiency in Huntington's disease. Hum Mol Genet. 2007;16:483–498. doi: 10.1093/hmg/ddl481. [DOI] [PubMed] [Google Scholar]
- Cobos I. Calcagnotto M.E. Vilaythong A.J., et al. Mice lacking Dlx1 show subtype-specific loss of interneurons, reduced inhibition and epilepsy. Nat Neurosci. 2005;8:1059–1068. doi: 10.1038/nn1499. [DOI] [PubMed] [Google Scholar]
- Coil D.A. Miller A.D. Phosphatidylserine is not the cell surface receptor for vesicular stomatitis virus. J Virol. 2004;78:10920–10926. doi: 10.1128/JVI.78.20.10920-10926.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coil D.A. Miller A.D. Enhancement of enveloped virus entry by phosphatidylserine. J Virol. 2005;79:11496–11500. doi: 10.1128/JVI.79.17.11496-11500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin J. Zhang X.Y. Reiser J. Altering the tropism of lentiviral vectors through pseudotyping. Curr Gene Ther. 2005;5:387–398. doi: 10.2174/1566523054546224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Almeida L.P. Ross C.A. Zala D., et al. Lentiviral-mediated delivery of mutant huntingtin in the striatum of rats induces a selective neuropathology modulated by polyglutamine repeat size, huntingtin expression levels, and protein length. J Neurosci. 2002;22:3473–3483. doi: 10.1523/JNEUROSCI.22-09-03473.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Déglon N. Tseng J.L. Bensadoun J.C., et al. Self-inactivating lentiviral vectors with enhanced transgene expression as potential gene transfer system in Parkinson's disease. Hum Gene Ther. 2000;11:179–190. doi: 10.1089/10430340050016256. [DOI] [PubMed] [Google Scholar]
- Desmaris N. Bosch A. Salaun C., et al. Production and neurotropism of lentivirus vectors pseudotyped with lyssavirus envelope glycoproteins. Mol. Ther. 2001;4:149–156. doi: 10.1006/mthe.2001.0431. [DOI] [PubMed] [Google Scholar]
- Donello J.E. Loeb J.E. Hope T.J. Woodchuck hepatitis virus contains a tripartite posttranscriptional regulatory element. J.Virol. 1998;72:5085–5092. doi: 10.1128/jvi.72.6.5085-5092.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzsimons H.L. Bland R.J. During M.J. Promoters and regulatory elements that improve adeno-associated virus transgene expression in the brain. Methods. 2002;28:227–236. doi: 10.1016/s1046-2023(02)00227-x. [DOI] [PubMed] [Google Scholar]
- Forss-Petter S. Danielson P.E. Catsicas S., et al. Transgenic mice expressing beta-galactosidase in mature neurons under neuron-specific enolase promoter control. Neuron. 1990;5:187–197. doi: 10.1016/0896-6273(90)90308-3. [DOI] [PubMed] [Google Scholar]
- Franklin B. Paxinos G. The Mouse Brain in Stereotaxic Coordinates. Academic Press; New York: 1996. pp. 1–350. [Google Scholar]
- Fremeau R.T., Jr. Duncan G.E. Fornaretto M.G., et al. Localization of D1 dopamine receptor mRNA in brain supports a role in cognitive, affective, and neuroendocrine aspects of dopaminergic neurotransmission. Proc Natl Acad Sci USA. 1991;88:3772–3776. doi: 10.1073/pnas.88.9.3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvan L. Lepejova N. Gaillard M.C., et al. Capucin does not modify the toxicity of a mutant Huntingtin fragment in vivo. Neurobiol Aging. 2012;33(1845):e5–6. doi: 10.1016/j.neurobiolaging.2012.01.009. [DOI] [PubMed] [Google Scholar]
- Gerfen C.R. The neostriatal mosaic: multiple levels of compartmental organization. TINS. 1992;15:133–138. doi: 10.1016/0166-2236(92)90355-c. [DOI] [PubMed] [Google Scholar]
- Ginn S.L. Fleming J. Rowe P.B., et al. Promoter interference mediated by the U3 region in early-generation HIV-1-derived lentivirus vectors can influence detection of transgene expression in a cell-type and species-specific manner. Hum Gene Ther. 2003;14:1127–1137. doi: 10.1089/104303403322167975. [DOI] [PubMed] [Google Scholar]
- Gong S. Zheng C. Doughty M.L., et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Gruh I. Wunderlich S. Winkler M., et al. Human CMV immediate-early enhancer: a useful tool to enhance cell-type-specific expression from lentiviral vectors. J Gene Med. 2008;10:21–32. doi: 10.1002/jgm.1122. [DOI] [PubMed] [Google Scholar]
- Gu X. Andre V.M. Cepeda C., et al. Pathological cell-cell interactions are necessary for striatal pathogenesis in a conditional mouse model of Huntington's disease. Mol Neurodegener. 2007;2:8. doi: 10.1186/1750-1326-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han I. You Y. Kordower J.H., et al. Differential vulnerability of neurons in Huntington's disease: The role of cell type-specific features. J Neurochem. 2010;113:1073–1091. doi: 10.1111/j.1471-4159.2010.06672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hioki H. Kameda H. Nakamura H., et al. Efficient gene transduction of neurons by lentivirus with enhanced neuron-specific promoters. Gene Ther. 2007;14:872–882. doi: 10.1038/sj.gt.3302924. [DOI] [PubMed] [Google Scholar]
- Hodges A. Strand A.D. Aragaki A.K., et al. Regional and cellular gene expression changes in human Huntington's disease brain. Hum Mol Genet. 2006;15:965–977. doi: 10.1093/hmg/ddl013. [DOI] [PubMed] [Google Scholar]
- Hottinger A.F. Azzouz M. Déglon N., et al. Complete and long-term rescue of lesioned adult motoneurons by lentiviral-mediated expression of glial cell line-derived neurotrophic factor in the facial nucleus. J Neurosci. 2000;20:5587–5593. doi: 10.1523/JNEUROSCI.20-15-05587.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao H.Y. Chern Y. Targeting Glial Cells to Elucidate the Pathogenesis of Huntington's Disease. Mol Neurobiol. 2010;41:248–255. doi: 10.1007/s12035-009-8097-5. [DOI] [PubMed] [Google Scholar]
- Jinno S. Aika Y. Fukuda T. Kosaka T. Quantitative analysis of GABAergic neurons in the mouse hippocampus, with optical disector using confocal laser scanning microscope. Brain Research. 1998;814:55–70. doi: 10.1016/s0006-8993(98)01075-0. [DOI] [PubMed] [Google Scholar]
- Jones A.R. Overly C.C. Sunkin S.M. The Allen Brain Atlas: 5 years and beyond. Nat Rev Neurosci. 2009;10:821–828. doi: 10.1038/nrn2722. [DOI] [PubMed] [Google Scholar]
- Katarova Z. Mugnaini E. Sekerkova G., et al. Regulation of cell–type specific expression of lacZ by the 5′–flanking region of mouse GAD67 gene in the central nervous system of transgenic mice. Eur J Neurosci. 1998;10:989–999. doi: 10.1046/j.1460-9568.1998.00109.x. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y. Neostriatal cell subtypes and their functional roles. Neurosci Res. 1997;27:1–8. doi: 10.1016/s0168-0102(96)01134-0. [DOI] [PubMed] [Google Scholar]
- Kim O.S. Kim H.J. Kwak H.J., et al. Human act and AR1 sequences differentially regulate murine and human D1A dopamine receptor promoters. Molecules and cells. 2003;15:294–300. [PubMed] [Google Scholar]
- Kordower J.H. Bloch J. Ma S., et al. Lentiviral gene transfer to the nonhuman primate brain. Exp Neurol. 1999;160:1–16. doi: 10.1006/exnr.1999.7178. [DOI] [PubMed] [Google Scholar]
- Kugler S. Meyn L. Holzmuller H., et al. Neuron-specific expression of therapeutic proteins: evaluation of different cellular promoters in recombinant adenoviral vectors. Mol Cell Neurosci. 2001;17:78–96. doi: 10.1006/mcne.2000.0929. [DOI] [PubMed] [Google Scholar]
- Kugler S. Kilic E. Bahr M. Human synapsin 1 gene promoter confers highly neuron-specific long-term transgene expression from an adenoviral vector in the adult rat brain depending on the transduced area. Gene Ther. 2003;10:337–347. doi: 10.1038/sj.gt.3301905. [DOI] [PubMed] [Google Scholar]
- Lee S.H. Yajima S. Mouradian M.M. Neural cell line-specific regulatory DNA cassettes harboring the murine D1A dopamine receptor promoter. Neurosci Res. 1999;34:225–234. doi: 10.1016/s0168-0102(99)00055-3. [DOI] [PubMed] [Google Scholar]
- Li S.H. Cheng A.L. Zhou H., et al. Interaction of huntington disease protein with transcriptional activator sp1. Mol Cell Biol. 2002;22:1277–1287. doi: 10.1128/mcb.22.5.1277-1287.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B.H. Wang X. Ma Y.X., et al. CMV enhancer/human PDGF-beta promoter for neuron-specific transgene expression. Gene Ther. 2004;11:52–60. doi: 10.1038/sj.gt.3302126. [DOI] [PubMed] [Google Scholar]
- Liu B. Wang S. Brenner M., et al. Enhancement of cell-specific transgene expression from a Tet-Off regulatory system using a transcriptional amplification strategy in the rat brain. J Gene Med. 2008;10:583–592. doi: 10.1002/jgm.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobsiger C.S. Cleveland D.W. Glial cells as intrinsic components of non–cell-autonomous neurodegenerative disease. Nat Neurosci. 2007;10:1355–1360. doi: 10.1038/nn1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marangos P.J. Schmechel D. Parma A.M., et al. Measurement of neuron-specific (NSE) and non-neuronal (NNE) isoenzymes of enolase in rat, monkey and human nervous tissue. J Neurochem. 1979;33:319–329. doi: 10.1111/j.1471-4159.1979.tb11735.x. [DOI] [PubMed] [Google Scholar]
- Matamales M. Bertran-Gonzalez J. Salomon L., et al. Striatal medium-sized spiny neurons: identification by nuclear staining and study of neuronal subpopulations in BAC transgenic mice. PLoS ONE. 2009;4:e4770. doi: 10.1371/journal.pone.0004770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazarakis N.D. Azzouz M. Rohll J.B., et al. Rabies virus glycoprotein pseudotyping of lentiviral vectors enables retrograde axonal transport and access to the nervous system after peripheral delivery. Hum. Mol. Genet. 2001;10:2109–2121. doi: 10.1093/hmg/10.19.2109. [DOI] [PubMed] [Google Scholar]
- Mendelson S.C. Morrison C.F. Mcallister J., et al. Repression of preprotachykinin-A promoter activity is mediated by a proximal promoter element. Neuroscience. 1995;65:837–847. doi: 10.1016/0306-4522(94)00554-i. [DOI] [PubMed] [Google Scholar]
- Minowa M.T. Minowa T. Mouradian M.M. Activator region analysis of the human D1A dopamine receptor gene. J Biol Chem. 1993;268:23544–23551. [PubMed] [Google Scholar]
- Naldini L. Blömer U. Gallay P., et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- Nanou A. Azzouz M. Gene therapy for neurodegenerative diseases based on lentiviral vectors. Prog Brain Res. 2009;175:187–200. doi: 10.1016/S0079-6123(09)17513-1. [DOI] [PubMed] [Google Scholar]
- Nettelbeck D.M. Jerome V. Muller R. A strategy for enhancing the transcriptional activity of weak cell type–specific promoters. Gene Therapy. 1998;5:1656–1664. doi: 10.1038/sj.gt.3300778. [DOI] [PubMed] [Google Scholar]
- Panganiban G. Rubenstein J.L. Developmental functions of the Distal-less/Dlx homeobox genes. Development. 2002;129:4371–4386. doi: 10.1242/dev.129.19.4371. [DOI] [PubMed] [Google Scholar]
- Paxinos G. Watson C. The Rat Brain in Stereotaxic Coordinates. Academic press; New York: 2004. pp. 1–474. [Google Scholar]
- Portales-Casamar E. Swanson D.J. Liu L., et al. A regulatory toolbox of MiniPromoters to drive selective expression in the brain. Proc Natl Acad Sci U S A. 2010;107:16589–16594. doi: 10.1073/pnas.1009158107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter G.B. Petryniak M.A. Shevchenko E., et al. Generation of Cre-transgenic mice using Dlx1/Dlx2 enhancers and their characterization in GABAergic interneurons. Mol Cell Neurosci. 2009;40:167–186. doi: 10.1016/j.mcn.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravache M. Weber C. Merienne K. Trottier Y. Transcriptional activation of REST by Sp1 in Huntington's disease models. PLoS ONE. 2010;5:e14311. doi: 10.1371/journal.pone.0014311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Régulier E. Pereira De Almeida L. Sommer B., et al. Dose-dependent neuroprotective effect of CNTF delivered via tetracycline-regulated lentiviral vectors in the quinolinic acid rat model of Huntington's disease. Hum Gene Ther. 2002;13:1981–1990. doi: 10.1089/10430340260355383. [DOI] [PubMed] [Google Scholar]
- Reiner A. Albin R.L. Anderson K.D., et al. Differential loss of striatal projection neurons in Huntington disease. Proc. Natl. Acad. Sci. USA. 1988;85:5733–5737. doi: 10.1073/pnas.85.15.5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runne H. Regulier E. Kuhn A., et al. Dysregulation of gene expression in primary neuron models of Huntington's disease shows that polyglutamine-related effects on the striatal transcriptome may not be dependent on brain circuitry. J Neurosci. 2008;28:9723–9731. doi: 10.1523/JNEUROSCI.3044-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakimura K. Kushiya E. Takahashi Y. Suzuki Y. The structure and expression of neuron-specific enolase gene. Gene. 1987;60:103–113. doi: 10.1016/0378-1119(87)90218-6. [DOI] [PubMed] [Google Scholar]
- Saxena S. Caroni P. Selective neuronal vulnerability in neurodegenerative diseases: from stressor thresholds to degeneration. Neuron. 2011;71:35–48. doi: 10.1016/j.neuron.2011.06.031. [DOI] [PubMed] [Google Scholar]
- Schlegel R. Tralka T.S. Willingham M.C. Pastan I. Inhibition of VSV binding and infectivity by phosphatidylserine: is phosphatidylserine a VSV-binding site? Cell. 1983;32:639–646. doi: 10.1016/0092-8674(83)90483-x. [DOI] [PubMed] [Google Scholar]
- Soboleski M.R. Oaks J. Halford W.P. Green fluorescent protein is a quantitative reporter of gene expression in individual eukaryotic cells. FASEB journal. 2005;19:440–442. doi: 10.1096/fj.04-3180fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan J.S. Kazantsev A. Spasic–Boskovic O., et al. The Huntington's disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc Natl Acad Sci USA. 2000;97:6763–6768. doi: 10.1073/pnas.100110097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo G. Katarova Z. Kortvely E. Greenspan R.J., et al. Structure and the promoter region of the mouse gene encoding the 67-kD form of glutamic acid decarboxylase. DNA and Cell Biology. 1996;15:1081–1091. doi: 10.1089/dna.1996.15.1081. [DOI] [PubMed] [Google Scholar]
- Teschemacher A.G. Paton J.F. Kasparov S. Imaging living central neurones using viral gene transfer. Adv Drug Deliv Rev. 2005;57:79–93. doi: 10.1016/j.addr.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Vile R.G. Diaz R.M. Miller N. Mitchell S. Tuszyanski A., et al. Tissue-specific gene expression from Mo-MLV retroviral vectors with hybrid LTRs containing the murine tyrosinase enhancer/promoter. Virology. 1995;214:307–313. doi: 10.1006/viro.1995.9923. [DOI] [PubMed] [Google Scholar]
- Wang Y. Yu L. Geller A.I. Diverse stabilities of expression in the rat brain from different cellular promoters in a helper virus-free herpes simplex virus type 1 vector system. Hum Gene Ther. 1999;10:1763–1771. doi: 10.1089/10430349950017446. [DOI] [PubMed] [Google Scholar]
- Wang Y. Dye C.A. Sohal V., et al. Dlx5 and Dlx6 regulate the development of parvalbumin-expressing cortical interneurons. J Neurosci. 2010;30:5334–5345. doi: 10.1523/JNEUROSCI.5963-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson D.J. Kobinger G.P. Passini M.A., et al. Targeted transduction patterns in the mouse brain by lentivirus vectors pseudotyped with VSV, Ebola, Mokola, LCMV, or MuLV envelope proteins. Mol Ther. 2002;5:528–537. doi: 10.1006/mthe.2002.0584. [DOI] [PubMed] [Google Scholar]
- Wheeler D.G. Cooper E. Depolarization strongly induces human cytomegalovirus major immediate-early promoter/enhancer activity in neurons. J Biol Chem. 2001;276:31978–31985. doi: 10.1074/jbc.M103667200. [DOI] [PubMed] [Google Scholar]
- Wu X. Holschen J. Kennedy S.C. Ponder K.P. Retroviral vector sequences may interact with some internal promoters and influence expression. Hum Gene Ther. 1996;7:159–171. doi: 10.1089/hum.1996.7.2-159. [DOI] [PubMed] [Google Scholar]
- Xu R. Janson C.G. Mastakov M., et al. Quantitative comparison of expression with adeno-associated virus (AAV–2) brain-specific gene cassettes. Gene Ther. 2001;8:1323–1332. doi: 10.1038/sj.gt.3301529. [DOI] [PubMed] [Google Scholar]
- Xu W. Russ J.L. Eiden M.V. Evaluation of residual promoter activity in gamma–retroviral self–inactivating (SIN) vectors. Mol Ther. 2012;20:84–90. doi: 10.1038/mt.2011.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zala D. Bensadoun J.-C. Pereira ee Almeida L., et al. Long-term lentiviral-mediated expression of ciliary neurotrophic factor in the striatum of Huntington's disease transgenic mice. Exp. Neurol. 2004;185:26–35. doi: 10.1016/j.expneurol.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Zerucha T. Stuhmer T. Hatch G., et al. A highly conserved enhancer in the Dlx5/Dlx6 intergenic region is the site of cross-regulatory interactions between Dlx genes in the embryonic forebrain. J Neurosci. 2000;20:709–721. doi: 10.1523/JNEUROSCI.20-02-00709.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. Adams J.Y. Billick E., et al. Molecular Engineering of a Two-Step Transcription Amplification (TSTA) System for Transgene Delivery in Prostate Cancer. Mol Ther. 2002;5:223–232. doi: 10.1006/mthe.2002.0551. [DOI] [PubMed] [Google Scholar]
- Zufferey R. Dull T. Mandel R.J., et al. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J Virol. 1998;72:9873–9880. doi: 10.1128/jvi.72.12.9873-9880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufferey R. Donello J.E. Trono D. Hope T.J. Woodschuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J Virol. 1999;73:2886–2892. doi: 10.1128/jvi.73.4.2886-2892.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.