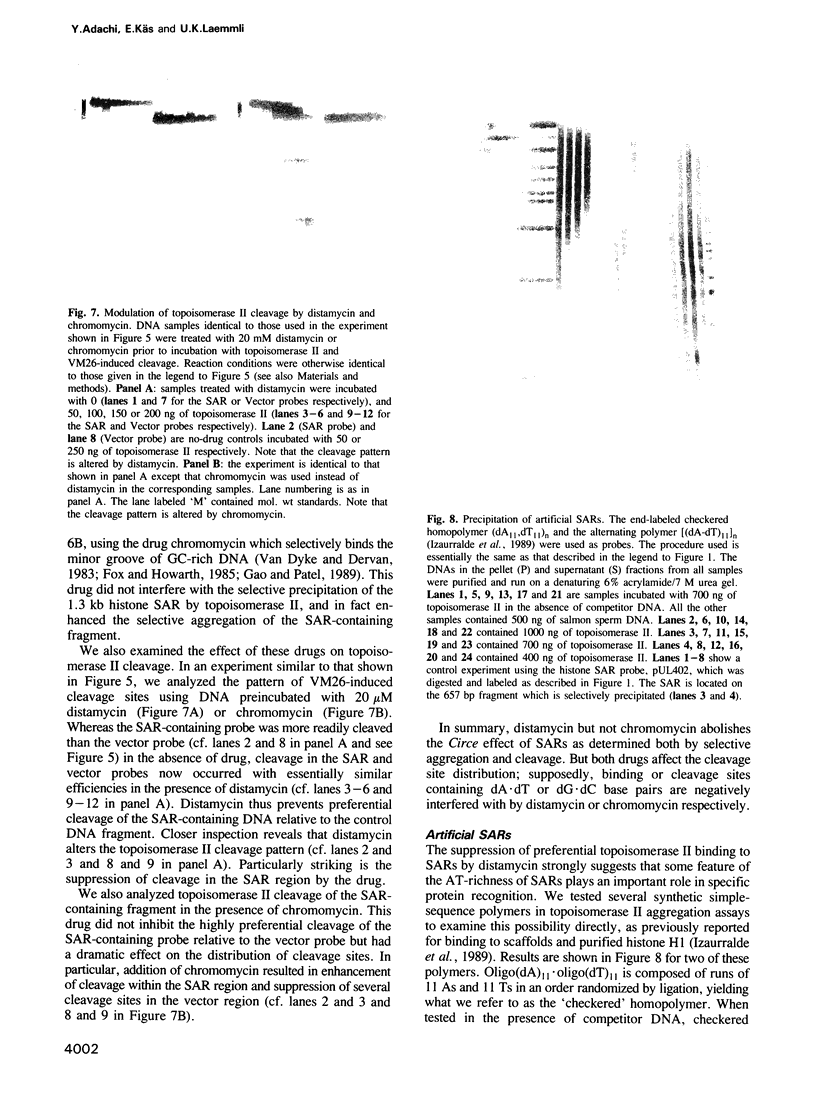
Abstract
DNA elements termed scaffold-associated regions (SARs) are AT-rich stretches of several hundred base pairs which are known to bind specifically to nuclear or metaphase scaffolds and are proposed to specify the base of chromatin loops. SARs contain sequences homologous to the DNA topoisomerase II cleavage consensus and this enzyme is known to be the major structural component of the mitotic chromosome scaffold. We find that purified topoisomerase II preferentially binds and aggregates SAR-containing DNA. This interaction is highly cooperative and, with increasing concentrations of topoisomerase II, the protein titrates quantitatively first SAR-containing DNA and then non-SAR DNA. About one topoisomerase II dimer is bound per 200 bp of DNA. SARs exhibit a Circe effect; they promote in cis topoisomerase II-mediated double-strand cleavage in SAR-containing DNA fragments. The AT-rich SARs contain several oligo(dA).oligo(dT) tracts which determine their protein-binding specificity. Distamycin, which is known to interact highly selectively with runs of A.T base pairs, abolishes the specific interaction of SARs with topoisomerase II, and the homopolymer oligo(dA).oligo(dT) is, above a critical length of 240 bp, a highly specific artificial SAR. These results support the notion of an involvement of SARs and topoisomerase II in chromosome structure.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amati B. B., Gasser S. M. Chromosomal ARS and CEN elements bind specifically to the yeast nuclear scaffold. Cell. 1988 Sep 23;54(7):967–978. doi: 10.1016/0092-8674(88)90111-0. [DOI] [PubMed] [Google Scholar]
- Boy de la Tour E., Laemmli U. K. The metaphase scaffold is helically folded: sister chromatids have predominantly opposite helical handedness. Cell. 1988 Dec 23;55(6):937–944. doi: 10.1016/0092-8674(88)90239-5. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brill S. J., Sternglanz R. Transcription-dependent DNA supercoiling in yeast DNA topoisomerase mutants. Cell. 1988 Jul 29;54(3):403–411. doi: 10.1016/0092-8674(88)90203-6. [DOI] [PubMed] [Google Scholar]
- Chen G. L., Yang L., Rowe T. C., Halligan B. D., Tewey K. M., Liu L. F. Nonintercalative antitumor drugs interfere with the breakage-reunion reaction of mammalian DNA topoisomerase II. J Biol Chem. 1984 Nov 10;259(21):13560–13566. [PubMed] [Google Scholar]
- Cockerill P. N., Garrard W. T. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986 Jan 31;44(2):273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- Cockerill P. N., Garrard W. T. Chromosomal loop anchorage sites appear to be evolutionarily conserved. FEBS Lett. 1986 Aug 11;204(1):5–7. doi: 10.1016/0014-5793(86)81377-1. [DOI] [PubMed] [Google Scholar]
- Earnshaw W. C., Heck M. M. Localization of topoisomerase II in mitotic chromosomes. J Cell Biol. 1985 May;100(5):1716–1725. doi: 10.1083/jcb.100.5.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox K. R., Howarth N. R. Investigations into the sequence-selective binding of mithramycin and related ligands to DNA. Nucleic Acids Res. 1985 Dec 20;13(24):8695–8714. doi: 10.1093/nar/13.24.8695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox K. R., Waring M. J. DNA structural variations produced by actinomycin and distamycin as revealed by DNAase I footprinting. Nucleic Acids Res. 1984 Dec 21;12(24):9271–9285. doi: 10.1093/nar/12.24.9271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X. L., Patel D. J. Solution structure of the chromomycin-DNA complex. Biochemistry. 1989 Jan 24;28(2):751–762. doi: 10.1021/bi00428a051. [DOI] [PubMed] [Google Scholar]
- Gasser S. M., Laemmli U. K. Cohabitation of scaffold binding regions with upstream/enhancer elements of three developmentally regulated genes of D. melanogaster. Cell. 1986 Aug 15;46(4):521–530. doi: 10.1016/0092-8674(86)90877-9. [DOI] [PubMed] [Google Scholar]
- Gasser S. M., Laroche T., Falquet J., Boy de la Tour E., Laemmli U. K. Metaphase chromosome structure. Involvement of topoisomerase II. J Mol Biol. 1986 Apr 20;188(4):613–629. doi: 10.1016/s0022-2836(86)80010-9. [DOI] [PubMed] [Google Scholar]
- Giaever G. N., Wang J. C. Supercoiling of intracellular DNA can occur in eukaryotic cells. Cell. 1988 Dec 2;55(5):849–856. doi: 10.1016/0092-8674(88)90140-7. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M. Two genes for the major heat-shock protein of Drosophila melanogaster arranged as an inverted repeat. Nucleic Acids Res. 1980 Jan 25;8(2):235–252. doi: 10.1093/nar/8.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiromi Y., Kuroiwa A., Gehring W. J. Control elements of the Drosophila segmentation gene fushi tarazu. Cell. 1985 Dec;43(3 Pt 2):603–613. doi: 10.1016/0092-8674(85)90232-6. [DOI] [PubMed] [Google Scholar]
- Holm C., Goto T., Wang J. C., Botstein D. DNA topoisomerase II is required at the time of mitosis in yeast. Cell. 1985 Jun;41(2):553–563. doi: 10.1016/s0092-8674(85)80028-3. [DOI] [PubMed] [Google Scholar]
- Hsieh T. Knotting of the circular duplex DNA by type II DNA topoisomerase from Drosophila melanogaster. J Biol Chem. 1983 Jul 10;258(13):8413–8420. [PubMed] [Google Scholar]
- Kirchhausen T., Wang J. C., Harrison S. C. DNA gyrase and its complexes with DNA: direct observation by electron microscopy. Cell. 1985 Jul;41(3):933–943. doi: 10.1016/s0092-8674(85)80074-x. [DOI] [PubMed] [Google Scholar]
- Krasnow M. A., Cozzarelli N. R. Catenation of DNA rings by topoisomerases. Mechanism of control by spermidine. J Biol Chem. 1982 Mar 10;257(5):2687–2693. [PubMed] [Google Scholar]
- Käs E., Chasin L. A. Anchorage of the Chinese hamster dihydrofolate reductase gene to the nuclear scaffold occurs in an intragenic region. J Mol Biol. 1987 Dec 20;198(4):677–692. doi: 10.1016/0022-2836(87)90209-9. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Liu L. F., Davis J. L., Calendar R. Novel topologically knotted DNA from bacteriophage P4 capsids: studies with DNA topoisomerases. Nucleic Acids Res. 1981 Aug 25;9(16):3979–3989. doi: 10.1093/nar/9.16.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. F., Wang J. C. DNA-DNA gyrase complex: the wrapping of the DNA duplex outside the enzyme. Cell. 1978 Nov;15(3):979–984. doi: 10.1016/0092-8674(78)90281-7. [DOI] [PubMed] [Google Scholar]
- Liu L. F., Wang J. C. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A. 1987 Oct;84(20):7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loc P. V., Strätling W. H. The matrix attachment regions of the chicken lysozyme gene co-map with the boundaries of the chromatin domain. EMBO J. 1988 Mar;7(3):655–664. doi: 10.1002/j.1460-2075.1988.tb02860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R. F., Holmes N. Use of an 125I-labelled DNA ligand to probe DNA structure. 1983 Mar 31-Apr 6Nature. 302(5907):452–454. doi: 10.1038/302452a0. [DOI] [PubMed] [Google Scholar]
- Mirkovitch J., Gasser S. M., Laemmli U. K. Relation of chromosome structure and gene expression. Philos Trans R Soc Lond B Biol Sci. 1987 Dec 15;317(1187):563–574. doi: 10.1098/rstb.1987.0081. [DOI] [PubMed] [Google Scholar]
- Mirkovitch J., Gasser S. M., Laemmli U. K. Scaffold attachment of DNA loops in metaphase chromosomes. J Mol Biol. 1988 Mar 5;200(1):101–109. doi: 10.1016/0022-2836(88)90336-1. [DOI] [PubMed] [Google Scholar]
- Mirkovitch J., Mirault M. E., Laemmli U. K. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell. 1984 Nov;39(1):223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- Newport J. Nuclear reconstitution in vitro: stages of assembly around protein-free DNA. Cell. 1987 Jan 30;48(2):205–217. doi: 10.1016/0092-8674(87)90424-7. [DOI] [PubMed] [Google Scholar]
- Rowe T. C., Wang J. C., Liu L. F. In vivo localization of DNA topoisomerase II cleavage sites on Drosophila heat shock chromatin. Mol Cell Biol. 1986 Apr;6(4):985–992. doi: 10.1128/mcb.6.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander M., Hsieh T. S. Drosophila topoisomerase II double-strand DNA cleavage: analysis of DNA sequence homology at the cleavage site. Nucleic Acids Res. 1985 Feb 25;13(4):1057–1072. doi: 10.1093/nar/13.4.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander M., Hsieh T., Udvardy A., Schedl P. Sequence dependence of Drosophila topoisomerase II in plasmid relaxation and DNA binding. J Mol Biol. 1987 Mar 20;194(2):219–229. doi: 10.1016/0022-2836(87)90370-6. [DOI] [PubMed] [Google Scholar]
- Udvardy A., Schedl P., Sander M., Hsieh T. S. Novel partitioning of DNA cleavage sites for Drosophila topoisomerase II. Cell. 1985 Apr;40(4):933–941. doi: 10.1016/0092-8674(85)90353-8. [DOI] [PubMed] [Google Scholar]
- Udvardy A., Schedl P., Sander M., Hsieh T. S. Topoisomerase II cleavage in chromatin. J Mol Biol. 1986 Sep 20;191(2):231–246. doi: 10.1016/0022-2836(86)90260-3. [DOI] [PubMed] [Google Scholar]
- Uemura T., Ohkura H., Adachi Y., Morino K., Shiozaki K., Yanagida M. DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell. 1987 Sep 11;50(6):917–925. doi: 10.1016/0092-8674(87)90518-6. [DOI] [PubMed] [Google Scholar]
- Uemura T., Tanagida M. Mitotic spindle pulls but fails to separate chromosomes in type II DNA topoisomerase mutants: uncoordinated mitosis. EMBO J. 1986 May;5(5):1003–1010. doi: 10.1002/j.1460-2075.1986.tb04315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke M. W., Dervan P. B. Chromomycin, mithramycin, and olivomycin binding sites on heterogeneous deoxyribonucleic acid. Footprinting with (methidiumpropyl-EDTA)iron(II). Biochemistry. 1983 May 10;22(10):2373–2377. doi: 10.1021/bi00279a011. [DOI] [PubMed] [Google Scholar]
- Van Dyke M. W., Hertzberg R. P., Dervan P. B. Map of distamycin, netropsin, and actinomycin binding sites on heterogeneous DNA: DNA cleavage-inhibition patterns with methidiumpropyl-EDTA.Fe(II). Proc Natl Acad Sci U S A. 1982 Sep;79(18):5470–5474. doi: 10.1073/pnas.79.18.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. C. DNA topoisomerases. Annu Rev Biochem. 1985;54:665–697. doi: 10.1146/annurev.bi.54.070185.003313. [DOI] [PubMed] [Google Scholar]
- Wyckoff E., Natalie D., Nolan J. M., Lee M., Hsieh T. Structure of the Drosophila DNA topoisomerase II gene. Nucleotide sequence and homology among topoisomerases II. J Mol Biol. 1989 Jan 5;205(1):1–13. doi: 10.1016/0022-2836(89)90361-6. [DOI] [PubMed] [Google Scholar]