Abstract
Summary: The initial steps in the analysis of next-generation sequencing data can be automated by way of software ‘pipelines’. However, individual components depreciate rapidly because of the evolving technology and analysis methods, often rendering entire versions of production informatics pipelines obsolete. Constructing pipelines from Linux bash commands enables the use of hot swappable modular components as opposed to the more rigid program call wrapping by higher level languages, as implemented in comparable published pipelining systems.
Here we present Next Generation Sequencing ANalysis for Enterprises (NGSANE), a Linux-based, high-performance-computing-enabled framework that minimizes overhead for set up and processing of new projects, yet maintains full flexibility of custom scripting when processing raw sequence data.
Availability and implementation: Ngsane is implemented in bash and publicly available under BSD (3-Clause) licence via GitHub at https://github.com/BauerLab/ngsane.
Contact: Denis.Bauer@csiro.au
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
The initial steps of analyzing next-generation sequencing (NGS) data can be automated in standardized pipelines, for e.g. the many steps in SNP calling and RNA-Seq analysis (Anders et al., 2013). This is critical, as further decreasing sequencing costs and expanding use of replicates to assess biological variability (Auer and Doerge, 2010) will substantially increase future study sizes, therefore making the automated, documented and reproducible processing of large numbers of samples across diverse projects using high-performance computing (HPC) clusters paramount. Yet, because of the constantly evolving technology, software and new application areas, maintaining such production informatics pipelines can be labor intensive.
To address this issue, several software packages have been published in recent years. However, currently available tools are either web-based services, e.g. Galaxy (Goecks et al., 2010), where even API-based access to the web service functionality is not readily amenable to production-scale analysis practices, or heavyweight frameworks written in user-friendly languages, such as snakemake and nestly (Python) (Köster and Rahmann, 2012; McCoy et al., 2013), GATK’s Queue (Scala) – https://github.com/broadgsa/gatk/) or Bpipe (Groovy) (Sadedin et al., 2012), which encapsulate the actual program call in a wrapper script specific syntax, hindering the development of pipeline extensions.
Ngsane is a lightweight, Linux-based, HPC-enabled framework that minimizes overhead for set up and processing of new projects, yet maintains full flexibility of custom scripting for processing raw sequence data. Ngsane allows end users and developers to construct pipelines from call statements that can be tested on the command line directly without syntax alterations or wrapper script involvement providing flexibility in software usage – a substantial advantage when analysis pipelines are constantly revised as new algorithms are developed. We describe Ngsane’s aims below.
Data security and reusability. The framework separates project-specific files from reference data, scripts and software suites that are reusable in other projects (Fig. 1a). Access to confidential data is handled transparently via the underlying Linux permission system. The transaction between projects and framework is facilitated by a project-specific configuration file that defines paths to reference data as well as the analysis tasks to perform. Ngsane supports systems with hierarchical storage management, specifically Data Migration Facility, by ensuring files are online when needed.
Fig. 1.
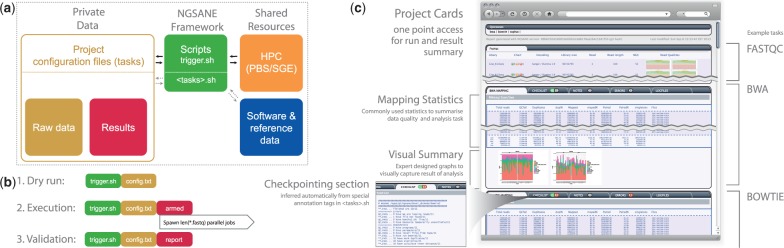
(a) Separation of project data from NGSANE core. (b) Workflow of NGSANE. (c) Example of automatically created project summary
HPC and parallel execution. Ngsane supports Sun Grid Engine and Portable Batch System job scheduling and can be operated in different modes for development and production, thus enabling flexible processing of NGS data. HPC job partitioning and submission is independent from the program calls, therefore enabling new technologies (e.g. Hadoop) to be incorporated.
Hot swapping and adaptability. Individual task blocks (e.g. read mapping) are packaged in bash script modules, which can be executed locally or on subsets to test module code, submission parameters and compute node environment in stages. During production, Ngsane automatically submits separate module calls for each input file or set of files to the HPC queue. This allows different existing modules, parameter settings or software versions to be executed by changes to the project-specific configuration file rather than the software code (hot swapping).
Reproducibility and checkpoint recovery. A full audit trail is generated recording performed tasks, used reference data, timestamps, software version as well as HPC log files, including any errors. Ngsane gracefully recovers from unsuccessfully executed jobs, be it owing to failed commands, missing or incorrect input or under-resourced HPC jobs by cleanly restarting after the most recent successfully executed checkpoint.
Robust execution and full monitoring. In our experience, modular workflows are executed in stages with optional human quality control; NSANE hence focuses on providing robust checkpointing and intuitive report generation (Fig. 1b). However, workflows can be fully automated by using NGSANE’s control over HPC queuing systems and by leveraging the customizable interfaces between modules when submitting multiple dependent stages at once.
Automated project summary creation. Ngsane generates a high-level summary (Project Card, Fig. 1b and c) to enable informed decisions about the experimental success. This interactive HTML report provides an access point for new lab members or collaborators. Furthermore, the Project Card can be used as a gold standard for software development when using a continuous integration server.
Complete customization. Ngsane’s configuration file contains details about the submission system, typical HPC resource allocations and location of third-party software. However, Ngsane’s credo is that every parameter can be overwritten; hence, default parameters can be adjusted in the project-specific configuration file to indicate different software versions, additional resources or an altered output location. Additional parameters, such as a specific HPC queue, or new parameters in a software release, can be provided to each program via a special ‘free form’ variable in the configuration file.
Repeated calls. As stated by McCoy et al. (2013), pipelines often have to be rerun on the full or a subset of the data with possibly altered parameter settings. Ngsane facilitates and documents this by allowing multiple (automatically created) configuration files.
Knowledge transfer. Ngsane provides a unified framework (i.e. folder structure) for processing data from different experimental protocols. This allows co-investigators and reviewers to easily understand and reproduce work from Ngsane’s log and report files.
Ngsane is open source and available via GitHub. Currently implemented workflows include those for adapter trimming, read mapping, peak calling, motif discovery, transcript assembly, variant calling and chromatin conformation analysis. These workflows use publicly available published software, yet allow the end user to add his/her own code and create new workflows as required. Ngsane is also available as Amazon Machine Image and can be deployed to the Amazon Elastic Compute Cloud (EC2) using StarCluster to allow on-demand processing of samples without requiring software installation or HPC maintenance.
Supplementary Material
ACKNOWLEDGEMENT
The authors thank Piotr Szul for help with AMI and StarCluster.
Funding: The National Health and Medical Research Council (1051757, 1010620 and 1063559 to S.J.C.); the Cancer Institute of New South Wales (11/REG/1-10 to Dr Warren Kaplan); the National Breast Cancer Foundation (program grant to S.J.C.); the Commonwealth Scientific and Industrial Research Organizations Transformational Capability Platform (D.C.B.); Science and Industry Endowment Fund (D.C.B. and S.J.C.); Information Management and Technology Services.
Conflict of Interest: none declared.
REFERENCES
- Anders S, et al. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat. Protoc. 2013;8:1765–1786. doi: 10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
- Auer PL, Doerge RW. Statistical design and analysis of RNA sequencing data. Genetics. 2010;185:405–416. doi: 10.1534/genetics.110.114983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, et al. Galaxy: a comprehensive approach for supporting accessible, reproducible and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köster J, Rahmann S. Snakemake – a scalable bioinformatics workflow engine. Bioinformatics. 2012;28:2520–2522. doi: 10.1093/bioinformatics/bts480. [DOI] [PubMed] [Google Scholar]
- McCoy CO, et al. Nestly – a framework for running software with nested parameter choices and aggregating results. Bioinformatics. 2013;29:387–388. doi: 10.1093/bioinformatics/bts696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadedin SP, et al. Bpipe: a tool for running and managing bioinformatics pipelines. Bioinformatics. 2012;28:1525–1526. doi: 10.1093/bioinformatics/bts167. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


