Abstract
Peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) inhibits skin tumorigenesis through mechanisms that may be dependent on HRAS signaling. The present study examined the hypothesis that PPARβ/δ promotes HRAS-induced senescence resulting in suppression of tumorigenesis. PPARβ/δ expression increased p-ERK and decreased p-AKT activity. Increased p-ERK activity results from the dampened HRAS-induced negative feedback response mediated in part through transcriptional upregulation of RAS guanyl-releasing protein 1 (RASGRP1) by PPARβ/δ. Decreased p-AKT activity results from repression of integrin-linked kinase (ILK) and phosphoinositide dependent protein kinase-1 (PDPK1) expression. Decreased p-AKT activity in turn promotes cellular senescence through upregulation of p53 and p27 expression. Both over-expression of RASGRP1 and shRNA-mediated knockdown of ILK partially restore cellular senescence in Pparβ/δ-null cells. Higher PPARβ/δ expression is also correlated with increased senescence observed in human benign neurofibromas and colon adenoma lesions in vivo. These results demonstrate that PPARβ/δ promotes senescence to inhibit tumorigenesis and provide new mechanistic insights into HRAS-induced cellular senescence.
Keywords: peroxisome proliferator-activated receptor-β/δ, HRAS-induced senescence, mechanisms of senescence, inhibition of tumorigenesis
INTRODUCTION
PPARβ/δ is a nuclear receptor that has many normal biological functions. Acting primarily as a transcription factor, PPARβ/δ promotes terminal differentiation 1, 2, inhibits inflammatory signaling 3 and increases skeletal muscle fatty acid catabolism (reviewed in 4). The role of PPARβ/δ in some cancers remains controversial, but there is strong experimental evidence that PPARβ/δ attenuates non-melanoma skin cancer (reviewed in 2). Pparβ/δ-null mice exhibit exacerbated skin tumorigenesis and ligand activation of PPARβ/δ inhibits chemically-induced skin tumorigenesis; this is likely mediated by PPARβ/δ-dependent induction of terminal differentiation and inhibition of cell proliferation and mitosis 5–9. Mutations in the Harvey sarcoma ras virus gene (Hras) is found in over 90% of 7,12-dimethylbenz[a]anthracene (DMBA) initiated skin tumors 10 and increased activity associated with mutant HRAS is one mechanism that can cause cancer 11. An activated mutant HRAS (Hras-V12) triggers cellular senescence (irreversible cell cycle arrest), concomitant with increased expression of p16 and p53 tumor suppressors 12. Oncogene-induced senescence has also been observed for a number of mutations in other genes including KRAS, BRAF, PTEN and NF1 13. Thus, it is widely believed that oncogene-induced senescence serves as a self-defense mechanism to suppress tumor development by preventing the progression of benign lesions to malignancies in the absence of additional cooperating mutations 13. The present study examined the mechanisms by which PPARβ/δ attenuates HRAS-dependent skin tumorigenesis, with an emphasis on cellular senescence.
RESULTS
PPARβ/δ promotes HRAS-induced senescence and suppresses malignant conversion
A complete carcinogen bioassay using DMBA was performed because mutant Hras skin tumors produced through this method have differential sensitivity to malignant conversion 14. A higher percentage of malignant squamous cell carcinomas (SCC) and a lower percentage of benign papillomas in Pparβ/δ-null mice compared to wild-type controls were observed (Figure 1a). The distribution of tumor type was not influenced by ligand activation of PPARβ/δ with GW0742 (Figure 1a). This suggests that PPARβ/δ may suppress malignant conversion of skin tumors. The lack of effect by an exogenous ligand could be due to the presence of high affinity endogenous ligand(s). To further characterize the role of PPARβ/δ in malignant transformation of skin tumors with an Hras mutation, an in vitro malignant conversion assay was performed. This assay uses primary keratinocytes infected with an oncogenic HRAS retrovirus 15 that confers a malignant phenotype characterized by resistance to calcium-induced differentiation and cell cycle arrest 16. Expression of proteins downstream of HRAS activation in HRAS-expressing keratinocytes, including p-ERK and p-AKT, is comparable to that of two skin cancer cell lines with mutant Hras (Supplementary Figure S1a). HRAS-expressing Pparβ/δ-null cells developed calcium-resistant foci (Figure 1b) that are bromodeoxyuridine (BrdU) positive while HRAS-expressing wild-type cells do not (Figure 1c). Ligand activation of PPARβ/δ had no influence on foci in either genotype (Figure 1b). Since oncogenic HRAS could trigger cellular senescence to prevent malignant transformation in primary cells 12, 17 and wild-type HRAS-expressing keratinocytes cultured in high calcium medium exhibited morphology reminiscent of cellular senescence (data not shown), the expression of senescence-associated β-galactosidase (β-gal) and BrdU labeling indices were examined. A higher percentage of BrdU labeling and lower percentage of β-gal-positive cells was noted in HRAS-expressing Pparβ/δ-null compared to wild-type cells (Figure 1d, e). Surprisingly, ligand activation of PPARβ/δ decreased both the percentage of β-gal- and BrdU-positive cells in HRAS-expressing wild-type but not in Pparβ/δ-null cells (Figure 1d, e). 75% of papillomas from wild-type mice and 33% of papillomas from Pparβ/δ-null mice stained positive for β-gal, and no SCC from either genotype were positive for β-gal (Figure 1f). This is consistent with previous studies showing that benign skin papillomas undergo senescence while malignant tumors do not 18, 19.
Figure 1.
PPARβ/δ suppresses malignant conversion by promoting HRAS-induced senescence. (a) Distribution of skin tumor types from wild-type (+/+) or Pparβ/δ-null (−/−) mice topically treated with or without GW0742 was quantified. SCC: squamous cell carcinoma. SCP: squamous cell papilloma. KA: keratoacanthoma. BBCT: benign basal cell tumor. Using a two proportion z-test, significant differences (P ≤ 0.05) were found in the percentage of squamous cell papillomas between wild-type and Pparβ/δ-null controls, the percentage of squamous cell papillomas between GW0742-treated wild-type and Pparβ/δ-null, and the percentage of squamous cell carcinomas between GW0742-treated wild-type and Pparβ/δ-null. No significant differences in the percentage of squamous cell papillomas or squamous cell carcinomas were found between control and GW0742 groups in either genotype. The percentage of squamous cell carcinomas between wild-type and Pparβ/δ-null controls was not statistically significant. However, a higher percentage of keratoacanthomas was found in control Pparβ/δ-null mice compared to wild-type. Keratoacanthomas can convert to squamous cell carcinoma in mice with prolonged promotion 72, signaling typically observed in Pparβ/δ-null mice 5. Thus, it is important to note that there are significantly more combined squamous cell carcinomas and keratoacanthomas in control and/or GW0742-treated Pparβ/δ-null mice compared to similarly treated wild-type mice. These data suggest that loss of PPARβ/δ promotes the conversion of keratoacanthomas to squamous cell carcinomas. (b, c) HRAS-expressing (+/+) or (−/−) keratinocytes were cultured in high Ca2+ medium for 4 weeks and Ca2+ resistant foci were stained with rhodamine. (b) The number and size of foci were quantified and (c) proliferating (+/+) or (−/−) cells (BrdU positive) in the foci are shown. DIC = representative photomicrograph using differential interference contrast microscopy. (d, e) HRAS-expressing (+/+) or (−/−) keratinocytes treated with or without GW0742 were either labeled with BrdU or stained for β-gal. The percentage of BrdU and β-gal positive cells was quantified. (f) β-gal staining of skin tumor samples from (+/+) or (−/−) mice isolated from one stage bioassay. Values represent the mean ± S.E.M.. *significantly different from (+/+) control (P ≤ 0.05). #significantly differerent from (−/−) control (P ≤ 0.05).
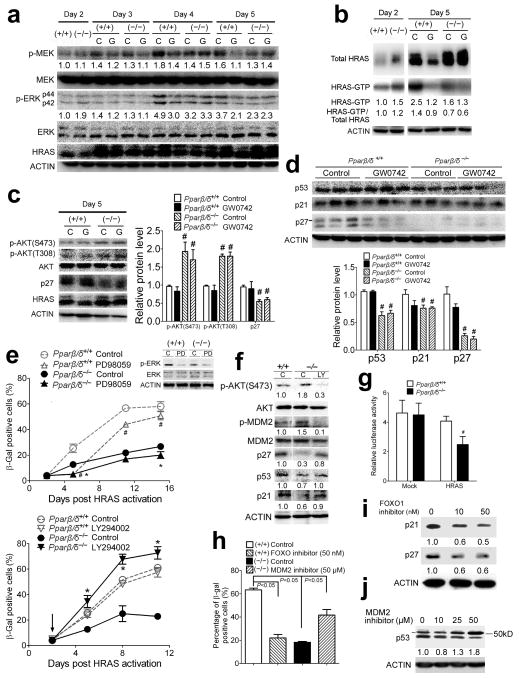
PPARβ/δ promotes senescence by regulating PI3K/AKT and MEK/ERK signaling
Expression of proteins downstream of HRAS signaling was examined to determine if PPARβ/δ differentially regulated these pathways. Four to five days after introduction of oncogenic HRAS, there was higher phosphorylated MEK (p-MEK), p-ERK (Figure 2a) and HRAS-GTP (Figure 2b) in wild-type compared to Pparβ/δ-null keratinocytes. In contrast, a higher level of phosphorylated AKT (p-AKT) was observed in HRAS-expressing Pparβ/δ-null cells as compared to wild-type (Figure 2c). At a later time point (day 11), higher expression of proteins inducing senescence 12, 20, 21 (p53, p21 and p27) was observed in wild-type cells compared to Pparβ/δ-null cells (Figure 2d). While ligand activation of PPARβ/δ with GW0742 caused a decrease in p-MEK, p-ERK, and GTP-bound HRAS in HRAS-expressing wild-type cells (Figure 2a, b), this is likely due to the fact that ligand activated PPARβ/δ selects against cells with higher HRAS expression 9. This might explain why ligand activation of PPARβ/δ had no effect on p53, p21 and p27 in HRAS-expressing wild-type cells.
Figure 2.
PPARβ/δ-dependent regulation of the PI3K/AKT and MEK/ERK pathways. HRAS-expressing wild-type (+/+) or Pparβ/δ-null (−/−) keratinocytes were treated with GW0742 (G) or vehicle control (C). Western blot analysis of: (a) p-MEK, MEK, p-ERK, ERK and HRAS between days 2 and 5, (b) total HRAS and GTP-bound form of HRAS days 2 and 5, (c) p-AKT and p27 on day 5, and (d) senescence markers p53, p27 and p21 on day 11, post HRAS expression, respectively. (e) Quantification of the percentage of β-gal positive HRAS-expressing keratinocytes treated with either a MEK inhibitor (PD98059) or a PI3K inhibitor (LY294002). Arrow indicates the timing of treatment with respective inhibitor. (f) Western blot analysis of HRAS-expressing (+/+) and (−/−) keratinocytes in response to LY294002 8 days post HRAS expression. (g) FOXO transcription factor reporter assay in either mock-infected or HRAS-expressing keratinocytes (5 days post HRAS expression). (h) Quantification of the percentage of β-gal positive HRAS-expressing (+/+) keratinocytes treated with 50 nM FOXO1 inhibitor (AS1842856) and HRAS-expressing (−/−) keratinocytes treated with 50 μM MDM2 inhibitor (trans-4-iodo, 4′-boranyl-chalcone). (i) Western blot analysis of HRAS-expressing (+/+) keratinocytes in response to AS1842856 treatment 9 days post HRAS expression. (j) Western blot analysis of HRAS-expressing (−/−) keratinocytes in response to trans-4-iodo, 4′-boranyl-chalcone treatment 9 days post HRAS expression. Relative expression level of proteins was normalized to that of ACTIN and is shown as the relative fold change as compared to control. Values represent the mean ± S.E.M.. #significantly different than (+/+) control (P ≤ 0.05). *significantly different than (−/−) control (P ≤ 0.05).
The higher levels of p-MEK and p-ERK and lower level of p-AKT in wild-type cells (Figure 2a–c) correlated with higher markers of senescence (Figure 2d) and this was not found in HRAS-expressing Pparβ/δ-null cells (Figure 2a–d). Thus, the hypothesis that PPARβ/δ promotes senescence during the early stages of HRAS activation by inhibiting PI3K/AKT and maintaining MEK/ERK signaling pathways was examined. Inhibition of MEK1 with PD98059 delayed HRAS-induced senescence in both wild-type and Pparβ/δ-null cells (Figure 2e, Supplementary Figure S1b). Over-expression of a constitutively active MEK 22 increased HRAS-induced senescence in both wild-type and Pparβ/δ-null cells, but the effect was more pronounced in Pparβ/δ-null cells (Supplementary Figure S1c, e–f). In contrast, inhibition of PI3K with LY294002 had no effect on HRAS-induced senescence in wild-type cells, but significantly increased HRAS-induced senescence in Pparβ/δ-null cells (Figure 2e). The same effects were also observed with over-expression of PTEN (Supplementary Figure S1d–f). Interestingly, Pparβ/δ-null cells are also more sensitive to inhibition of cell proliferation by treatment with LY294002 and less sensitive to inhibition of cell proliferation by treatment with PD98059 (Supplementary Figure S1g). Further, inhibition of PI3K/AKT by LY294002 caused a modest increased in expression of p53 and p21 and marked increase of p27 in HRAS-expressing Pparβ/δ-null keratinocytes to a level similar to that of HRAS-expressing wild-type keratinocytes (Figure 2f). Combined, these data suggest that PPARβ/δ promotes senescence by regulating PI3K/AKT and the MEK/ERK pathway.
The mechanism by which PPARβ/δ maintains expression of p53, p21 and p27 in HRAS-expressing keratinocytes was investigated. Phosphorylation of MDM2 at S166/S186 by p-AKT can activate its ubiquitin ligase activity toward p53 23 causing reduced expression of p53. Relatively lower expression of p-MDM2 and higher p53 was found in HRAS-expressing wild-type cells compared to Pparβ/δ-null cells (Figure 2f). Inhibition of p-AKT with LY294002 caused a decrease in p-MDM2 and an increase in p53 in Pparβ/δ-null cells (Figure 2f). p27 and p21 are both positively regulated by FOXO, a transcription factor that is inhibited by p-AKT 24. Lower FOXO activity was found in HRAS-expressing Pparβ/δ-null cells compared to controls (Figure 2g). In addition, gene set enrichment analysis from a microarray dataset 9 shows lower FOXO target gene expression in HRAS-expressing Pparβ/δ-null cells compared to control (Supplementary Figure S2a, b). To determine if the higher FOXO activity observed in HRAS-expressing wild-type cells and the higher MDM2 activity observed in Pparβ/δ-null cells underlie the different senescent phenotypes, FOXO and MDM2 activity was blocked using specific inhibitors to FOXO1 25 and MDM2 26 in HRAS-expressing wild-type and Pparβ/δ-null cells, respectively. Inhibition of FOXO1 activity in HRAS-expressing wild-type cells reduced the percentage of β-gal-positive cells (Figure 2h and Supplementary Figure S2c) and the expression of p21 and p27 (Figure 2i). In contrast, inhibition of MDM2 activity in HRAS-expressing Pparβ/δ-null cells increased the percentage of β-gal-positive cells (Figure 2h and Supplementary Figure S2c) and the expression of p53 (Figure 2j). These data collectively suggest that PPARβ/δ represses AKT activity leading to enhanced FOXO activity and decreased MDM2 activity, causing increased expression of proteins that facilitate senescence such as p27, p21 and p53.
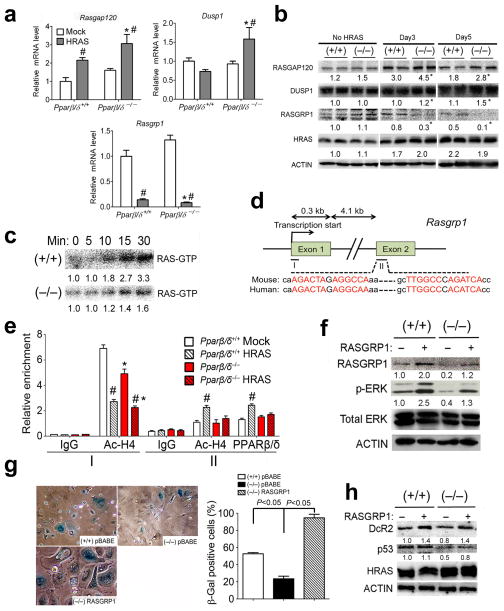
PPARβ/δ up-regulates RASGRP1 to promote HRAS-induced senescence
Microarray analysis from a previously published study 9 identified PPARβ/δ target genes involved in HRAS-induced senescence. Expression of the negative RAS regulator RASGAP120 was increased and the positive RAS regulator RASGRP1 was decreased in response to HRAS activation, respectively (Figure 3a, b). Interestingly, expression of RASGAP120 was significantly higher, whereas expression of RASGRP1 was lower in HRAS-expressing Pparβ/δ-null cells compared to wild-type cells (Figure 3a, b). In response to HRAS activation, expression of mRNA encoding the negative RAS regulator Rasa4 was increased in cells from both genotypes, but relatively higher expression of Rasa4 mRNA was also observed in HRAS-expressing Pparβ/δ-null cells compared to wild-type cells (data not shown). No change in expression of the MAP kinase phosphatase DUSP1, a negative RAS regulator, was found in wild-type cells in response to HRAS activation, however, increased expression of DUSP1 was found in Pparβ/δ-null cells in response to HRAS activation (Figure 3a, b). A similar effect was observed for the mRNA encoding the negative RAS regulator Dusp3 (data not shown). These data are consistent with a previous study showing a negative feedback response to HRAS activation, whereby expression of negative RAS regulators is increased and expression of positive regulators is decreased in an attempt to impede RAS signaling 27. Combined, these results suggest that PPARβ/δ attenuates this HRAS-induced negative feedback response.
Figure 3.
PPARβ/δ up-regulates RASGRP1 to promote HRAS-induced senescence. Wild-type (+/+) or Pparβ/δ-null (−/−) keratinocytes were either mock or HRAS-infected for up to 5 days. (a) qPCR of Rasgap120, Dusp1 and Rasgrp1 mRNA. (b) Western blot analysis of RASGAP120, DUSP1, RASGRP1 and HRAS in mock-infected (No HRAS) or HRAS-expressing keratinocytes at indicated days post HRAS expression. (c) HRAS-GTP loading assays in HRAS-expressing keratinocytes at day 5 post HRAS expression. Expression level of HRAS-GTP at different time points after adding GTPγS was normalized to that found at time zero. (d) Diagram of mouse Ragrp1 promoter and first and second exon regions. Putative PPREs were found in the second exon. PPRE half sites are shown in red letters and sequence comparison between mouse and human is shown below. (e) ChIP analysis of the occupancy of AcH4 on the promoter (I) and occupancy of AcH4 and PPARβ/δ on the PPRE-containing second exon region (II) of the mouse Rasgrp1 gene. (f–h) RASGRP1 was over-expressed in HRAS-expressing (+/+) or (−/−) keratinocytes. (f) Western blot analysis of RASGRP1, p-ERK and ERK at day 5 post HRAS expression. (g) Representative photomicrographs and quantification of percentage of β-gal positive cells and (h) western blot analysis of senescence marker DcR2 and p53 at day 8 post HRAS expression. Relative expression level of proteins was normalized to that of ACTIN and is shown as the relative fold change as compared to control. Values represent the mean ± S.E.M.. *significantly different than (+/+) control (P ≤ 0.05). #significantly different than (+/+) or (−/−) mock control (P ≤ 0.05).
The time course of HRAS-GTP formation was examined to determine if the PPARβ/δ-dependent regulation of the negative and positive RAS regulators modulates HRAS activity. Consistent with relatively higher expression of RASGRP1 in HRAS-expressing wild-type keratinocytes (Figure 3a, b), accumulation of HRAS-GTP was greater in HRAS-expressing wild-type keratinocytes compared to Pparβ/δ-null cells (Figure 3c). This is consistent with a study showing that deficiency in RASGRP1 resulted in decreased level of RAS-GTP in mouse keratinocytes 28. The PPARβ/δ-dependent difference in HRAS-GTP accumulation is unlikely related to differences in expression of RASGAP120 because HRAS with mutations at codons 12 and 61 is resistant to the GTP hydrolase activity associated with RASGAP120 11.
ChIP analysis showed that HRAS caused a decrease of acetylated histone 4 (AcH4) in the Rasgrp1 promoter in both wild-type and Pparβ/δ-null cells (Figure 3e), consistent with the decreased expression of RASGRP1 in response to HRAS activation. Examination of the Rasgrp1 gene revealed two peroxisome proliferator response elements (PPRE) in the second exon (Figure 3d). HRAS expression increased AcH4 and occupancy of PPARβ/δ in the region containing the PPREs in wild-type cells and this effect was absent in Pparβ/δ-null cells (Figure 3e). Increased luciferase activity was also detected in response to HRAS activation using a reporter construct containing the two PPREs in the Rasgrp1 exon (Supplementary Figure 3a). Both PPRE regions in the Rasgrp1 exon were capable of binding with a PPARβ/δ/RXRα heterodimer in gel shift assays (Supplementary Figure S3b, c). This increase in PPARβ/δ occupancy is likely due to an increase of endogenous ligands after HRAS activation since HRAS expression also increased the AcH4 and PPARβ/δ occupancy in a region containing multiple PPREs in the Angtpl4 gene (Supplementary Figure S3d), a well-characterized PPARβ/δ target gene 29.
To determine if higher RASGRP1 expression promotes cellular senescence, RASGRP1 was over-expressed in HRAS-expressing cells. Ectopic expression of RASGRP1 increased p-ERK (Figure 3f) and restored cellular senescence in Pparβ/δ-null cells (Figure 3g), concomitant with increased expression of the senescence markers DcR2 and p53 (Figure 3h). These data suggest that PPARβ/δ positively regulates RASGRP1 to promote HRAS-induced senescence.
PPARβ/δ represses ILK to promote HRAS-induced senescence
ILK and PDPK1 are known to phosphorylate AKT at Ser473 and Thr308, respectively, to fully activate AKT 30, 31. Consistent with past studies 32–34, expression of ILK and PDPK1 was higher in control and HRAS-expressing Pparβ/δ-null cells (Figure 4a, Supplementary Figure S4a). In silico examination of the Ilk gene revealed two potential PPREs in the second intron (Figure 4b). No occupancy of PPARβ/δ on the downstream PPRE was found (data not shown). Higher PPARβ/δ occupancy on the upstream PPRE was associated with increased occupancy of histone deacetylase 1 (HDAC1) and HDAC3 and decreased AcH4 in both mock and HRAS-expressing wild-type cells compared to Pparβ/δ-null counterparts but ligand activation of PPARβ/δ had no effect on these endpoints (Figure 4c). Interestingly, mutating the PPARβ/δ binding half-site in a luciferase reporter construct containing the upstream PPRE (Figure 4b) abolished the repression of ILK by PPARβ/δ (Supplementary Figure S4b). Over-expression of PPARβ/δ in HaCaT cells also caused repression of ILK, PDPK1 and p-AKT, and these changes in expression were associated with decreased AcH4 and increased promoter occupancy of HDAC1 and HDCA3 in the upstream PPRE of the ILK gene (Supplementary Figure S4c–e).
Figure 4.
PPARβ/δ represses ILK to promote HRAS-induced senescence. Wild-type (+/+) or Pparβ/δ-null (−/−) keratinocytes were either mock or HRAS-infected and treated with GW0742 (G) or vehicle control (C). (a) Western blot analysis of ILK and PDPK1 in HRAS-expressing keratinocytes. (b) Diagram of mouse Ilk gene promoter region and regions spanning from the first to the third exons. Putative PPREs (illustrated by pink arrows) were found in the second intron. The PPARβ/δ and RXR binding half sites are shown in red or blue letters, respectively. Sequence comparison of mouse, human and monkey genes are shown below. (c) ChIP analysis of the occupancy of HDAC1, HDAC3, PPARβ/δ and AcH4 on the PPRE1 region of the mouse Ilk gene. (d, e) ILK was knocked down by shRNA in HRAS-expressing (−/−) keratinocytes. (d) Representative photomicrographs and quantification of percentage of β-gal positive cells and (e) western blot analysis of senescence and proliferation markers at day 11 post HRAS expression. Ilk shRNA2 serves as an additional negative control. (f) Representative photomicrographs and quantification of percentage of β-gal positive cells after ILK knockdown in three isolated calcium-resistant HRAS-expressing (−/−) clones. Representative photomicrographs of clone 1 are shown. Relative expression level of proteins was normalized to that of ACTIN and is shown as the relative fold change as compared to control. Values represent the mean ± S.E.M.. #significantly different than (+/+) control (P ≤ 0.05). *significantly different than (−/−) control (P ≤ 0.05).
To determine if higher ILK expression repressed cellular senescence, ILK expression was knocked down by shRNA in Pparβ/δ-null cells. Of the two shRNAs used, only shRNA1 successfully knocked down ILK by 50% (Figure 4e). Decreased ILK expression caused a decrease in p-AKT and restored cellular senescence, concomitant with increased expression of the senescence markers p53, p27 and decreased expression of the cell proliferation marker phospho-retinoblastoma (pRB; S780) in Pparβ/δ-null cells (Figure 4d, e). These effects were not due to an increase in p-ERK following ILK knockdown (Figure 4e). Furthermore, knockdown of ILK partially restored cellular senescence in 3 out of 6 calcium-resistant HRAS-expressing Pparβ/δ-null clones isolated from an assay as shown in Figure 1b (Figure 4f). Results from experiments where RASGRP1 is overexpressed (Figure 3f–h) or ILK is knocked down (Figure 4d–f) collectively suggest that both decreased p-ERK activity and increased p-AKT activity is required to evade HRAS-induced senescence.
PPARβ/δ promotes HRAS-induced senescence in fibroblasts
To confirm that the observed decreased HRAS-induced senescence in Pparβ/δ-null keratinocytes is also observed in other cell types, mouse and human fibroblasts expressing oncogenic HRAS were examined. Decreased HRAS-induced senescence was observed in Pparβ/δ-null dermal fibroblasts as compared to wild-type cells (Fig 5a, b). Decreased mRNA expression of the senescence markers p16 and p21 was observed in HRAS-expressing Pparβ/δ-null dermal fibroblasts as compared to controls (Fig 5c). In addition, Rasgrp1 mRNA was lower and Ilk and Rasgap120 mRNA were higher in HRAS-expressing Pparβ/δ-null dermal fibroblasts (Fig 5c), all of which recapitulates the changes found in primary keratinocytes. The human BJ fibroblast cell line was also examined to confirm the PPARβ/δ-dependent effects observed in mouse cells. Decreased HRAS-induced senescence was observed following knockdown of PPARβ/δ by shRNA in BJ cells expressing mutant HRAS (Figure 5d, e). Further, in BJ cells expressing mutant HRAS following knockdown of PPARβ/δ, RASGRP1 mRNA was decreased, ILK mRNA was increased, and HRAS-induced expression of p16, p21, and DcR2 was reduced (Figure 5f, g). Additionally, HRAS-induced expression of p-ERK and p-AKT was repressed and enhanced, respectively, following knockdown of PPARβ/δ in BJ cells expressing mutant HRAS (Figure 5f, g).
Figure 5.
PPARβ/δ promotes HRAS-induced senescence in fibroblasts. (a–c) Wild-type (+/+) or Pparβ/δ-null (−/−) dermal fibroblasts were infected with HRAS virus for 3 days. (a) Representative photomicrographs of mock and HRAS-expressing dermal fibroblasts and (b) quantification of β-gal-positive cells. (c) qPCR of Hras, p16, p21, Rasgrp1, Rasgap120 and Ilk in dermal fibroblasts. (d–g) Mutant HRAS (G12V) was introduced into BJ cells using a previously described retroviral approach 12. (d) Representative photomicrographs of control (BJ) and PPARβ/δ knockdown cells (BJPPARβ/δ shRNA) and (e) quantification of β-gal-positive cells. (f) qPCR of PPARβ/δ, RASGRP1, ILK, DcR2, p16 and p21 in both cell types. (g) Western blot analysis of p-AKT, p-ERK, senescence markers p16, p21, and HRAS in both cell types. Relative expression level of proteins was normalized to that of LDH and is shown as the relative fold change as compared to control. Values represent the mean ± S.E.M.. *significantly different than control (P ≤ 0.05).
PPARβ/δ attenuates ILK/p-AKT to promote senescence in mouse skin tumors
To test the hypothesis that PPARβ/δ promotes HRAS-induced senescence in vivo, chemically-induced skin tumors were examined. β-gal positive regions of papillomas also stained positive for p27 but were negative for p-AKT (S473) in both wild-type and Pparβ/δ-null mice (Figure 6a). SCC were negative for β-gal but exhibited strong expression of p-AKT (S473) and essentially no p27 expression (Figure 6a). Increased p-AKT staining was found in tumors from Pparβ/δ-null mice compared to wild-type counterparts (Figure 6b, c). These findings are consistent with analysis of HRAS-expressing keratinocytes showing that p-AKT inhibits FOXO and p27 expression (Figure 2f, g). More Ki-67 positive cells were found in tumors from Pparβ/δ-null mice as compared to tumors examined from wild-type mice (Figure 6b). This suggests that PPARβ/δ inhibits the proliferative capacity of skin tumors. Higher expression of ILK, p-AKT (S473, T308) was also found in skin tumors from Pparβ/δ-null mice compared to wild-type mice (Figure 6d). Further, a negative correlation between p-AKT and the senescence marker p16 was also found (Figure 6d). These findings suggest the anti-tumorigenic role of PPARβ/δ in promoting senescence is mediated by repressing ILK/p-AKT signaling.
Figure 6.
PPARβ/δ attenuates ILK/p-AKT to promote senescence in mouse skin tumors. Skin tumors from wild-type (+/+) or Pparβ/δ-null (−/−) mice were obtained from a complete carcinogen bioassay. (a) Immunofluorescence of p-AKT (S473), KERATIN 14 and p27 in squamous cell carcinoma (SCC) and squamous cell papillomas (SCP). Serial sections of tumor samples were also stained for β-gal and H&E. (b) Immunofluorescence of p-AKT (S473) and Ki-67. (c) Cumulative frequency of mean intensity of p-AKT (S473) from at least 1000 cells as shown in b. (d) Western blot analysis and scatter plots of p-AKT and senescence marker p16. Relative expression level of proteins was normalized to that of LDH and are shown as the relative fold change as compared to control. Red circles represent data for tumors from (−/−) mice and black circle represent data for tumors from (+/+) mice. Values represent the mean ± S.E.M.. *significantly different than (+/+) control (P ≤ 0.05).
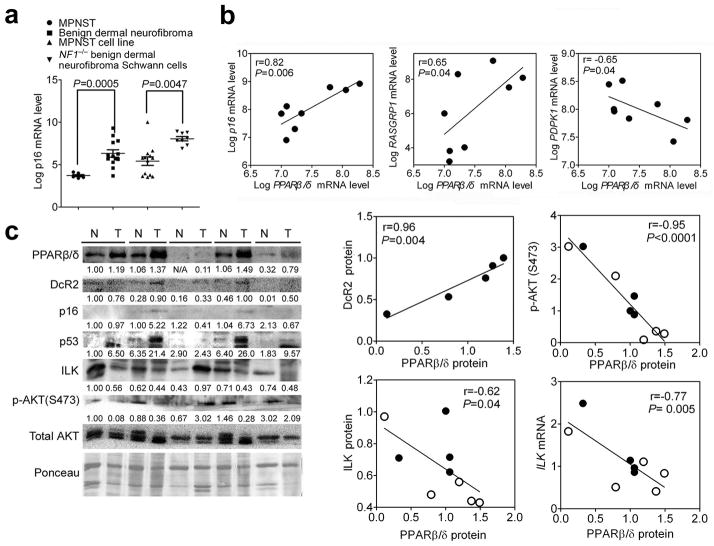
PPARβ/δ promotes cellular senescence in human benign lesions
To determine if the changes observed in mouse models were also found in human tumors, the correlation between PPARβ/δ expression and the senescence marker p16 in human benign dermal neurofibroma lesions was examined. These lesions were chosen because: 1) benign dermal neurofibromas harbor an NF1 loss-of-function mutation, resulting in activation of RAS signaling pathway 27 similar to the activated HRAS model used in the present studies and 2) β-gal positive staining of senescent cells are found in these lesions 27. Examination of a publicly available database 35 revealed that expression of mRNA encoding the senescence marker p16 is significantly higher in benign dermal neurofibromas and NF1-derived primary benign neurofibroma Schwann cells compared to malignant peripheral nerve sheath tumors and cell lines, respectively (Figure 7a). This suggests that p16 mRNA is a good senescence marker in these types of lesions. Neurofibromas are heterogeneous tumors that consist of Schwann cells with initiating homozygous NF1 mutations, but also recruited fibroblasts, peripheral cells, neurons and mast cells that are only heterozygous for NF1 mutations. Since β-gal positive staining was only found in cells with homozygous loss-of-function NF1 mutations 27, eight samples of NF1-derived primary benign neurofibroma Schwann cells with a homozygous NF1 mutation were examined. A positive correlation between PPARβ/δ and p16 mRNA was found in these cells (Figure 7b). In addition, a positive correlation was also found between PPARβ/δ and RASGPR1 (Figure 7b), similar to effects observed in the mouse models. While no correlation between PPARβ/δ and ILK mRNA was found, a negative correlation between PPARβ/δ and PDPK1 mRNA was found in these cells (Figure 7b). Human colon adenomas were also examined because 1) they contain cells exhibiting both strong p16 immunoreactivity and the absence of Ki-67 staining as previously reported 36, 37, and 2) the KRAS oncogene is mutated in approximately 35%–45% of colorectal cancers 38. In the five human colon adenomas that contained a KRAS mutation at codon 13 (data not shown), the average PPARβ/δ protein level was higher compared to untransformed colon (Figure 7c). In addition, PPARβ/δ protein in both normal colon and colon adenomas negatively correlated with both mRNA and protein of ILK and p-AKT (Figure 7c). The expression of PPARβ/δ protein in colon adenomas also positively correlates with expression of senescence markers (Figure 7c). These data suggest that PPARβ/δ promotes cellular senescence in benign human tumors.
Figure 7.
PPARβ/δ promotes cellular senescence in human benign lesions. (a) p16 mRNA expression in different neurofibroma tumors (malignant peripheral nerve sheet tumor (MPNST) or benign dermal neurofibroma) and tumor-derived cell lines (MPNST or NF1−/− benign dermal neurofibroma Schwann cell lines). (b) Scatter plots of the log2 value of PPARβ/δ mRNA compared to p16, RASGRP1 and PDPK1 in NF1-derived primary benign neurofibroma Schwann cells with NF1−/− mutation. (c) Western blot analysis of paired human normal colon and human colon adenomas and scatter plots of PPARβ/δ protein level with protein level of DcR2, p-AKT (S473), ILK and mRNA level of ILK. Relative expression level of proteins was normalized to the intensity of Ponceau staining and is shown as the relative fold change as compared to the normal colon for the first sample in lane 1. The mRNA level of ILK was normalized to that of ACTIN. Close and open circles represent normal tissue and colon adenomas respectively.
DISCUSSION
Results from these studies are the first to show that PPARβ/δ promotes HRAS-induced senescence in keratinocytes and skin tumors. PPARβ/δ promotes senescence by enhancing the RAF/MEK/ERK pathway and inhibiting the PI3K/AKT pathway during HRAS-induced neoplastic transformation. These studies also demonstrated that increased p-ERK and decreased p-AKT activities are both required to facilitate senescence. This conclusion is supported by several studies that showed that higher pERK activity could trigger senescence by upregulating expression of p16 and p21 20, 39, 40. This is consistent with data from the present studies showing that higher PPARβ/δ expression in benign tumors is associated with higher expression of p16. Further, results from the present studies are consistent with other studies showing that high PI3K/AKT activity can prevent HRAS-induced senescence by inhibiting FOXO activity and increasing MDM2 activity that collectively cause reduced expression of p27, p21 and p53 27, 41. Among these senescence markers, the most robust change observed in Pparβ/δ-null keratinocytes was the marked reduction of p27. p27 has been previously shown to promote senescence in multiple tissues and loss of p27 expression led to down-regulation of senescence and progression of cancer 21, 42–44. Collectively, these results suggest that PPARβ/δ inhibits AKT activity causing: 1) increased FOXO activity thereby preventing down-regulation of p27 and p21 caused by activation of HRAS, and 2) decreased p-MDM2 activity thereby preventing down-regulation of p53 mediated by activation of HRAS (Figure 8). Higher PI3K/AKT signaling can also counteract senescence by stimulating cell survival by phosphorylation of many substrates 45. The fact that inhibition of AKT didn’t promote senescence in wild-type HRAS-expressing keratinocytes, whereas inhibition of AKT in Pparβ/δ-null keratinocytes did promote senescence suggests that there may be different threshold levels of p-AKT required for promoting pro-survival versus an anti-senescence function of p-AKT.
Figure 8.
PPARβ/δ promotes HRAS-induced senescence by potentiating p-ERK and repressing p-AKT. HRAS-induced senescence is promoted by RAF/MEK/ERK pathway and inhibited by the PI3K/AKT pathway. The end result is increased expression of proteins that mediate senescence including p16, p21, p27 and p53. PPARβ/δ promotes senescence by inhibiting the PI3K/AKT pathway allowing for increased RAF/MEK/ERK activity. This is mediated by PPARβ/δ-dependent modulation of RASGRP1, PDPK1 and ILK expression.
It was recently shown that mutations in NF1, RAF, and RAS can induce a negative feedback response, whereby expression of negative RAS regulators is increased and expression of positive regulators is decreased to suppress RAS signaling, thereby promoting senescence by subsequent inhibition of PI3K/AKT activities 27. This negative feedback response was also observed in the present studies. Interestingly, PPARβ/δ dampens this negative response without increasing the level of p-AKT. This suggests that PPARβ/δ promotes HRAS-induced cellular senescence by: 1) attenuating the negative feedback response causing higher levels of p-ERK and, 2) preventing an increase of p-AKT level by repressing ILK and PDPK1 expression (Figure 8).
Activation of HRAS repressed expression of RASGRP1 in both wild-type and Pparβ/δ-null keratinocytes. However, increased occupancy of PPARβ/δ and enhanced AcH4 in the exonic PPRE region that was associated with relatively higher expression of RASGRP1 in HRAS-expressing wild-type cells was not found in Pparβ/δ-null counterparts. This suggests that PPARβ/δ helps offset HRAS-induced repression of this positive regulator of RAS signaling causing higher p-ERK levels. Expression of ILK and PDPK1 was higher in Pparβ/δ-null keratinocytes compared to wild-type cells, consistent with past studies 32–34. Additionally, higher expression of PPARβ/δ is associated with lower expression of ILK and PDPK1 in human benign tumors. In clinical studies, higher expression of ILK and PDPK1 are often correlated with human malignancies and poor prognosis 46–48, thus it is not surprising that Pparβ/δ-null mice are more sensitive to DMBA-induced skin tumorigenesis and malignant conversion.
PPARβ/δ represses ILK expression through association with HDACs on chromatin 49, 50. Indeed, binding of PPARβ/δ and occupancy of HDAC1 and HDAC3 on its PPRE is associated with repression of ILK. No functional PPREs within 5 kb downstream or upstream of either the mouse or human PDPK1 gene were found (data not shown). Thus, more distal PPREs for the PDPK1 gene could exist or alternatively, PPARβ/δ could repress PDPK1 expression through a PPRE-independent mechanism.
Treatment of HRAS-expressing cells with an exogenous PPARβ/δ ligand did not cause changes in all endpoints including expression of proteins required for senescence. However, genetic disruption of PPARβ/δ resulted in a phenotype indicating that PPARβ/δ promotes HRAS-induced senescence. This phenotype was also observed in mouse dermal fibroblasts and a human fibroblast cell line (BJ) following knockdown of PPARβ/δ. Moreover, higher expression of PPARβ/δ in benign tumors with higher RAS activity is also associated with higher expression of RASGRP1 and p16 in the absence of exogenous ligand. This suggests that high affinity endogenous ligand(s) is/are present in the HRAS-expressing cells that may prevent detection of effects induced by an exogenous ligand. This conclusion is also supported by ChIP and reporter assays showing enhanced PPARβ/δ-dependent activities following activation of HRAS. Further studies are needed to identify these endogenous ligands.
The role of PPARβ/δ in human tumorigenesis remains controversial, in particular for colon cancer, because of conflicting literature (reviewed in 2, 51–54). Some of this controversy arises from discrepancies in the literature suggesting that expression of PPARβ/δ is either higher or lower in tumors as compared to untransformed tissue (reviewed in 2, 53, 54). Whereas some studies have suggested that expression of PPARβ/δ mRNA is higher in human colon tumors compared to control tissue, more recent comprehensive analysis indicates that expression of both PPARβ/δ mRNA and protein is lower in human and rodent colon adenocarcinomas, in particular late stage cancer, as compared to non-transformed colon tissue 55–62. Interestingly, expression of PPARβ/δ protein was higher in benign adenomas in the present studies as compared to normal tissue and this correlated with higher levels of senescence markers in these lesions. There is strong evidence showing that expression of PPARβ/δ is lower in late stage tumors as compared to control tissue 55–62, and a recent study showed that colorectal cancer patients with relatively low expression of PPARβ/δ in the primary tumor were ~4X as likely to die from this disease as compared to patients with relatively higher expression of PPARβ/δ in their primary tumors 63. This suggests that PPARβ/δ may inhibit colon tumorigenesis by promoting cellular senescence resulting in benign lesions and that down-regulation of PPARβ/δ is required to evade senescence and lead to malignant conversion. As colon tumors can have different mutations (KRAS, P53, APC, etc.), it will be of interest to examine whether PPARβ/δ exerts pro-senescence effects only in the presence of specific mutations like KRAS or promotes senescence independently of mutation type(s).
MATERIALS AND METHODS
Plasmids
pBABE-puro-Pten was obtained by cloning mouse Pten into pBABE-puro and pBABE-puro-Mek-DD 64 and pWZL-hygro-H-RASV12 were purchased (Addgene, Cambridge, MA). pBABE-puro-Rasgrp1 was obtained by cloning the mouse Rasgrp1 open reading frame from a shuttle clone vector (GC-Mm04880; GeneCopoeia, Rockville, MD). The plasmid containing a mouse shRNA against Ilk, two human shRNAs against PPARβ/δ or a non-target control shRNA were purchased from Mission shRNA (Sigma-Aldrich, St. Louis, MO). The shRNA catalogue number are: mouse Ilk shRNA1 (TRCN0000022515), mouse Ilk shRNA2 (TRCN0000022518), human PPARβ/δ shRNA (TRCN0000001661), human PPARβ/δ shRNA (TRCN0000010647), and non-target control shRNA (SHC002).
Cell culture
Primary keratinocytes from newborn wild-type and Pparβ/δ-null mice were prepared and cultured as previously described 65. The HaCat cell line was cultured in DMEM medium as previously described 66. To activate PPARβ/δ, GW0742 (1 μM) was used as this is within the range that specifically activates PPARβ/δ in keratinocytes 9, 67. LY294002 (10 μM) was used to inhibit PI3K activity and PD98059 (10 μM) was used to inhibit MEK activity because these concentrations are within the range that specifically inhibits these enzymes 27.
Complete carcinogenesis bioassay
Wild-type or Pparβ/δ-null, female mice (6~8 weeks of age) were initiated with 50 μg of DMBA dissolved in 200 μL acetone (5–7 mice/group). Mice were treated topically with 50 μg of DMBA weekly for 30 weeks. Mice were also topically treated with acetone or 5 μM GW0742 twice a week during this 30 week treatment period. After 30 weeks, mice were euthanized and tumor samples were either fixed or snap frozen in liquid nitrogen for further analysis. Fixed tumor samples were embedded in paraffin, sectioned and stained with hematoxylin and eosin (H&E) and scored for benign or malignant pathology by examination with a light microscope.
Senescence-associated β-galactosidase (β-gal) assay
Cytochemical detection of senescence-associated β-gal activity was determined in cells and tumors as described 68.
Flow cytometry analysis
Cells were stained with bromodeoxyuridine (BrdU) and propidium iodide (PI) and analyzed for cell cycle progression as previously described 69.
RNA isolation and quantitative real-time PCR (qPCR)
Total RNA was isolated using TRIZOL reagent (Invitrogen, Carlsbad, CA) and reverse transcription and qPCR was performed as previously described 70. The relative level of mRNA was normalized to glyceraldehyde 3-phosphate dehydrogenase (Gapdh) or 18s mRNA.
Quantitative western blot analysis
Cell lysates were obtained and western blot analysis using radioactive detection methods was performed as previously described 66. The primary antibodies used were: anti-pRB (Ser780), anti-p-MEK1/2 (Ser217/221), anti-MEK1/2, anti-p-ERK1/2 (Thr202/Tyr204), anti-ERK1/2, anti-p-AKT (Ser473), anti-p-AKT (Thr308), anti-AKT, anti-p27, anti-PTEN, anti-p-MDM2 (Ser166) (Cell Signaling, Beverly, MA), anti-HRAS, anti-RB, anti-p16, anti-p53, anti-p21, anti-DcR2, anti-RASGAP120, anti-RASGRP1, anti-DUSP1, anti-MDM2 (Santa Cruz Biotechnology, Santa Cruz, CA), anti-lactic dehydrogenase (Jackson Immunoresearch, West Grove, PA), anti-β-ACTIN (Rockland, Gilbertsville, PA), anti-ERK1/2 (New England Biolabs, Ipswich, MA), anti-PPARβ/δ (Abcam, San Francisco, CA), anti-PDPK1 (BD Biosciences Pharmingen, San Diego, CA) and anti-ILK (Upstate, Lake Placid, NY),
Chromatin immunoprecipitation (ChIP)
ChIP was performed as previously described 70 using each of the following antibodies: anti mouse PPARβ/δ 71, anti-human PPARβ/δ, anti-HDAC1, anti-HDAC3 (Santa Cruz Biotechnology, Santa Cruz, CA) or acetylated histone 4 (Millipore, Temecula, CA). Rabbit IgG was used as a negative control. qPCR was performed to determine the relative enrichment of specific proteins on different regions of DNA. Relative enrichment of proteins was normalized to the occupancy of each protein on the Ubiquitin C gene (for mouse genes) or β-Actin gene (for human genes).
Statistical analysis
All data analysis was performed using GraphPad Prism v 5.0 (GraphPad Software, La Jolla, CA). Values represent means ± S.E.M., as indicated. Statistical significance was assessed using one-tailed Student t-test or linear regression analysis.
Supplementary Material
Acknowledgments
We gratefully acknowledge Drs. Andrew Billin and Timothy Willson for providing the GW0742, the Center for Quantitative Cell Analysis and the Genomic Core Facility at the Huck Institutes of Life Sciences of The Pennsylvania State University for their technical support with flow cytometry and data analysis. Supported by the National Institutes of Health (CA124533, CA141029, CA140369, AA018863) J.M.P., (CA122109, CA117957) A.B.G., and the National Cancer Institute Intramural Research Program (ZIABC005561, ZIABC005562, ZIABC005708) F.J.G..
Footnotes
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
CONFLICT OF INTEREST
Dr. Peters, Dr. Glick and Dr. Gonzalez work has been funded by the NIH. Dr. Zhu, Dr. Bility, Dr. Khozoie, Dr. Kang, Ms. Ferry, and Mr. Blazanin declare no potential conflict of interest.
References
- 1.Burdick AD, Kim DJ, Peraza MA, Gonzalez FJ, Peters JM. The role of peroxisome proliferator-activated receptor-β/δ in epithelial cell growth and differentiation. Cell Signal. 2006;18:9–20. doi: 10.1016/j.cellsig.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 2.Peters JM, Shah YM, Gonzales FJ. The role of peroxisome proliferator-activated receptors in carcinogenesis and chemoprevention. Nat Rev Cancer. 2012;12:181–195. doi: 10.1038/nrc3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kilgore KS, Billin AN. PPARβ/δ ligands as modulators of the inflammatory response. Curr Opin Investig Drugs. 2008;9:463–469. [PubMed] [Google Scholar]
- 4.Grimaldi PA. Metabolic and nonmetabolic regulatory functions of peroxisome proliferator-activated receptor β. Curr Opin Lipidol. 2010;21:186–191. doi: 10.1097/mol.0b013e32833884a4. [DOI] [PubMed] [Google Scholar]
- 5.Bility MT, et al. Ligand activation of peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) inhibits chemically-induced skin tumorigenesis. Carcinogenesis. 2008;29:2406–2414. doi: 10.1093/carcin/bgn219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bility MT, Zhu B, Kang BH, Gonzalez FJ, Peters JM. Ligand activation of peroxisome proliferator-activated receptor-β/δ and inhibition of cyclooxygenase-2 enhances inhibition of skin tumorigenesis. Toxicol Sci. 2010;113:27–36. doi: 10.1093/toxsci/kfp212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim DJ, et al. Peroxisome proliferator-activated receptor β (δ)-dependent regulation of ubiquitin C expression contributes to attenuation of skin carcinogenesis. J Biol Chem. 2004;279:23719–23727. doi: 10.1074/jbc.M312063200. [DOI] [PubMed] [Google Scholar]
- 8.Zhu B, et al. Chemoprevention of chemically induced skin tumorigenesis by ligand activation of peroxisome proliferator-activated receptor-β/δ and inhibition of cyclooxygenase 2. Mol Cancer Ther. 2011;9:3267–3277. doi: 10.1158/1535-7163.MCT-10-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu B, et al. Peroxisome proliferator-activated receptor β/δ cross talks with E2F and attenuates mitosis in HRAS-expressing cells. Mol Cell Biol. 2012;32:2065–2082. doi: 10.1128/MCB.00092-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quintanilla M, Brown K, Ramsden M, Balmain A. Carcinogen-specific mutation and amplification of Ha-ras during mouse skin carcinogenesis. Nature. 1986;322:78–80. doi: 10.1038/322078a0. [DOI] [PubMed] [Google Scholar]
- 11.Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- 12.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 13.Courtois-Cox S, Jones SL, Cichowski K. Many roads lead to oncogene-induced senescence. Oncogene. 2008;27:2801–2809. doi: 10.1038/sj.onc.1210950. [DOI] [PubMed] [Google Scholar]
- 14.Brown K, Buchmann A, Balmain A. Carcinogen-induced mutations in the mouse c-Ha-ras gene provide evidence of multiple pathways for tumor progression. Proc Natl Acad Sci U S A. 1990;87:538–542. doi: 10.1073/pnas.87.2.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roop DR, et al. An activated Harvey ras oncogene produces benign tumours on mouse epidermal tissue. Nature. 1986;323:822–824. doi: 10.1038/323822a0. [DOI] [PubMed] [Google Scholar]
- 16.Morgan D, et al. Development of an in vitro model to study carcinogen-induced neoplastic progression of initiated mouse epidermal cells. Cancer Res. 1992;52:3145–3156. [PubMed] [Google Scholar]
- 17.Vijayachandra K, Lee J, Glick AB. Smad3 regulates senescence and malignant conversion in a mouse multistage skin carcinogenesis model. Cancer Res. 2003;63:3447–3452. [PubMed] [Google Scholar]
- 18.Collado M, et al. Tumour biology: senescence in premalignant tumours. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- 19.Sun P, et al. PRAK is essential for ras-induced senescence and tumor suppression. Cell. 2007;128:295–308. doi: 10.1016/j.cell.2006.11.050. [DOI] [PubMed] [Google Scholar]
- 20.de Keizer PL, et al. Activation of forkhead box O transcription factors by oncogenic BRAF promotes p21cip1-dependent senescence. Cancer Res. 2010;70:8526–8536. doi: 10.1158/0008-5472.CAN-10-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin HK, et al. Skp2 targeting suppresses tumorigenesis by Arf-p53-independent cellular senescence. Nature. 2010;464:374–379. doi: 10.1038/nature08815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schramek H, Feifel E, Healy E, Pollack V. Constitutively active mutant of the mitogen-activated protein kinase kinase MEK1 induces epithelial dedifferentiation and growth inhibition in madin-darby canine kidney-C7 cells. J Biol Chem. 1997;272:11426–11433. doi: 10.1074/jbc.272.17.11426. [DOI] [PubMed] [Google Scholar]
- 23.Ogawara Y, et al. Akt enhances Mdm2-mediated ubiquitination and degradation of p53. J Biol Chem. 2002;277:21843–21850. doi: 10.1074/jbc.M109745200. [DOI] [PubMed] [Google Scholar]
- 24.Greer EL, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24:7410–7425. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- 25.Nagashima T, et al. Discovery of novel forkhead box O1 inhibitors for treating type 2 diabetes: improvement of fasting glycemia in diabetic db/db mice. Mol Pharmacol. 2010;78:961–970. doi: 10.1124/mol.110.065714. [DOI] [PubMed] [Google Scholar]
- 26.Busuttil V, et al. NF-kappaB inhibits T-cell activation-induced, p73-dependent cell death by induction of MDM2. Proc Natl Acad Sci U S A. 2010;107:18061–18066. doi: 10.1073/pnas.1006163107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Courtois-Cox S, et al. A negative feedback signaling network underlies oncogene-induced senescence. Cancer Cell. 2006;10:459–472. doi: 10.1016/j.ccr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma A, Luke CT, Dower NA, Stone JC, Lorenzo PS. RasGRP1 is essential for ras activation by the tumor promoter 12-O-tetradecanoylphorbol-13-acetate in epidermal keratinocytes. J Biol Chem. 2010;285:15724–15730. doi: 10.1074/jbc.M109.100016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mandard S, et al. The direct peroxisome proliferator-activated receptor target fasting-induced adipose factor (FIAF/PGAR/ANGPTL4) is present in blood plasma as a truncated protein that is increased by fenofibrate treatment. J Biol Chem. 2004;279:34411–34420. doi: 10.1074/jbc.M403058200. [DOI] [PubMed] [Google Scholar]
- 30.Alessi DR, et al. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Curr Biol. 1997;7:261–269. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- 31.Persad S, et al. Regulation of protein kinase B/Akt-serine 473 phosphorylation by integrin-linked kinase: critical roles for kinase activity and amino acids arginine 211 and serine 343. J Biol Chem. 2001;276:27462–27469. doi: 10.1074/jbc.M102940200. [DOI] [PubMed] [Google Scholar]
- 32.Burdick AD, et al. Ligand activation of peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) inhibits cell growth of human N/TERT-1 keratinocytes. Cell Signal. 2007;19:1163–1171. doi: 10.1016/j.cellsig.2006.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marin HE, et al. Ligand activation of peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) inhibits colon carcinogenesis. Cancer Res. 2006;66:4394–4401. doi: 10.1158/0008-5472.CAN-05-4277. [DOI] [PubMed] [Google Scholar]
- 34.Yang L, et al. Knockdown of peroxisome proliferator-activated receptor-β induces less differentiation and enhances cell-fibronectin adhesion of colon cancer cells. Oncogene. 2010;29:516–526. doi: 10.1038/onc.2009.370. [DOI] [PubMed] [Google Scholar]
- 35.Miller SJ, et al. Integrative genomic analyses of neurofibromatosis tumours identify SOX9 as a biomarker and survival gene. EMBO molecular medicine. 2009;1:236–248. doi: 10.1002/emmm.200900027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuilman T, et al. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008;133:1019–1031. doi: 10.1016/j.cell.2008.03.039. [DOI] [PubMed] [Google Scholar]
- 37.Dai CY, et al. p16(INK4a) expression begins early in human colon neoplasia and correlates inversely with markers of cell proliferation. Gastroenterology. 2000;119:929–942. doi: 10.1053/gast.2000.17952. [DOI] [PubMed] [Google Scholar]
- 38.Karapetis CS, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 39.Satyanarayana A, et al. Mitogen stimulation cooperates with telomere shortening to activate DNA damage responses and senescence signaling. Mol Cell Biol. 2004;24:5459–5474. doi: 10.1128/MCB.24.12.5459-5474.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu J, Woods D, McMahon M, Bishop JM. Senescence of human fibroblasts induced by oncogenic Raf. Genes Dev. 1998;12:2997–3007. doi: 10.1101/gad.12.19.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kennedy AL, et al. Activation of the PIK3CA/AKT pathway suppresses senescence induced by an activated RAS oncogene to promote tumorigenesis. Mol Cell. 2011;42:36–49. doi: 10.1016/j.molcel.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alexander K, Hinds PW. Requirement for p27(KIP1) in retinoblastoma protein-mediated senescence. Mol Cell Biol. 2001;21:3616–3631. doi: 10.1128/MCB.21.11.3616-3631.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cao X, et al. WW domain-containing E3 ubiquitin protein ligase 1 (WWP1) delays cellular senescence by promoting p27(Kip1) degradation in human diploid fibroblasts. J Biol Chem. 2011;286:33447–33456. doi: 10.1074/jbc.M111.225565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Majumder PK, et al. A prostatic intraepithelial neoplasia-dependent p27 Kip1 checkpoint induces senescence and inhibits cell proliferation and cancer progression. Cancer Cell. 2008;14:146–155. doi: 10.1016/j.ccr.2008.06.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dai DL, et al. Increased expression of integrin-linked kinase is correlated with melanoma progression and poor patient survival. Clin Cancer Res. 2003;9:4409–4414. [PubMed] [Google Scholar]
- 47.Maurer M, et al. 3-Phosphoinositide-dependent kinase 1 potentiates upstream lesions on the phosphatidylinositol 3-kinase pathway in breast carcinoma. Cancer Res. 2009;69:6299–6306. doi: 10.1158/0008-5472.CAN-09-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu J, Shi R, Zhang D, Wang E, Qiu X. Expression of integrin-linked kinase in lung squamous cell carcinoma and adenocarcinoma: correlation with E-cadherin expression, tumor microvessel density and clinical outcome. Virchows Archiv : an international journal of pathology. 2011;458:99–107. doi: 10.1007/s00428-010-1016-3. [DOI] [PubMed] [Google Scholar]
- 49.Adhikary T, et al. Genomewide analyses define different modes of transcriptional regulation by peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) PLoS ONE. 2011;6:e16344. doi: 10.1371/journal.pone.0016344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi Y, Hon M, Evans RM. The peroxisome proliferator-activated receptor δ, an integrator of transcriptional repression and nuclear receptor signaling. Proc Natl Acad Sci U S A. 2002;99:2613–2618. doi: 10.1073/pnas.052707099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peters JM, Foreman JE, Gonzalez FJ. Dissecting the role of peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) in colon, breast and lung carcinogenesis. Cancer Metastasis Rev. 2011;30:619–640. doi: 10.1007/s10555-011-9320-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peters JM, Gonzalez FJ. Sorting out the functional role(s) of peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) in cell proliferation and cancer. Biochim Biophys Acta. 2009;1796:230–241. doi: 10.1016/j.bbcan.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peters JM, Hollingshead HE, Gonzalez FJ. Role of peroxisome-proliferator-activated receptor β/δ (PPARβ/δ) in gastrointestinal tract function and disease. Clin Sci (Lond) 2008;115:107–127. doi: 10.1042/CS20080022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peters JM, Morales JL, Gonzales FJ. Modulation of gastrointestinal inflammation and colorectal tumorigenesis by peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) Drug Discovery Today: Disease Mechanisms. 2011;8:e85–e93. doi: 10.1016/j.ddmec.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Foreman JE, et al. Functional characterization of peroxisome proliferator-activated receptor-β/δ expression in colon cancer. Mol Carcinog. 2011;50:884–900. doi: 10.1002/mc.20757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hong Y, Downey T, Eu KW, Koh PK, Cheah PY. A ‘metastasis-prone’ signature for early-stage mismatch-repair proficient sporadic colorectal cancer patients and its implications for possible therapeutics. Clinical & experimental metastasis. 2010;27:83–90. doi: 10.1007/s10585-010-9305-4. [DOI] [PubMed] [Google Scholar]
- 57.Kaiser S, et al. Transcriptional recapitulation and subversion of embryonic colon development by mouse colon tumor models and human colon cancer. Genome Biol. 2007;8:R131. doi: 10.1186/gb-2007-8-7-r131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Modica S, et al. The intestinal nuclear receptor signature with epithelial localization patterns and expression modulation in tumors. Gastroenterology. 2010;138:636–648. 648 e631–612. doi: 10.1053/j.gastro.2009.09.060. [DOI] [PubMed] [Google Scholar]
- 59.Notterman DA, Alon U, Sierk AJ, Levine AJ. Transcriptional gene expression profiles of colorectal adenoma, adenocarcinoma, and normal tissue examined by oligonucleotide arrays. Cancer Res. 2001;61:3124–3130. [PubMed] [Google Scholar]
- 60.Sabates-Bellver J, et al. Transcriptome profile of human colorectal adenomas. Mol Cancer Res. 2007;5:1263–1275. doi: 10.1158/1541-7786.MCR-07-0267. [DOI] [PubMed] [Google Scholar]
- 61.Skrzypczak M, et al. Modeling oncogenic signaling in colon tumors by multidirectional analyses of microarray data directed for maximization of analytical reliability. PLoS ONE. 2010:5. doi: 10.1371/journal.pone.0013091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Uhlen M, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 63.Yang L, et al. Biological function and prognostic significance of peroxisome proliferator-activated receptor delta in rectal cancer. Clin Cancer Res. 2011;17:3760–3770. doi: 10.1158/1078-0432.CCR-10-2779. [DOI] [PubMed] [Google Scholar]
- 64.Boehm JS, et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell. 2007;129:1065–1079. doi: 10.1016/j.cell.2007.03.052. [DOI] [PubMed] [Google Scholar]
- 65.Dlugosz AA, Glick AB, Tennenbaum T, Weinberg WC, Yuspa SH. Isolation and utilization of epidermal keratinocytes for oncogene research. Methods Enzymol. 1995;254:3–20. doi: 10.1016/0076-6879(95)54003-2. [DOI] [PubMed] [Google Scholar]
- 66.Borland MG, et al. Stable over-expression of PPARβ/δ and PPARγ to examine receptor signaling in human HaCaT keratinocytes. Cell Signal. 2011;23:2039–2050. doi: 10.1016/j.cellsig.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Borland MG, et al. Ligand Activation of Peroxisome Proliferator-Activated Receptor-β/δ (PPARβ/δ) Inhibits Cell Proliferation in Human HaCaT Keratinocytes. Mol Pharmacol. 2008;74:1429–1442. doi: 10.1124/mol.108.050609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Debacq-Chainiaux F, Erusalimsky JD, Campisi J, Toussaint O. Protocols to detect senescence-associated beta-galactosidase (SA-betagal) activity, a biomarker of senescent cells in culture and in vivo. Nature protocols. 2009;4:1798–1806. doi: 10.1038/nprot.2009.191. [DOI] [PubMed] [Google Scholar]
- 69.He P, et al. Effect of ligand activation of peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) in human lung cancer cell lines. Toxicology. 2008;254:112–117. doi: 10.1016/j.tox.2008.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Palkar PS, et al. Cellular and Pharmacological Selectivity of the PPARβ/δ Antagonist GSK3787. Mol Pharmacol. 2010;78:419–430. doi: 10.1124/mol.110.065508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Girroir EE, et al. Quantitative expression patterns of peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) protein in mice. Biochem Biophys Res Commun. 2008;371:456–461. doi: 10.1016/j.bbrc.2008.04.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Knutsen GL, Kovatch RM, Robinson M. Gross and microscopic lesions in the female SENCAR mouse skin and lung in tumor initiation and promotion studies. Environ Health Perspect. 1986;68:91–104. doi: 10.1289/ehp.866891. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.