Abstract
Reactivation of intracellular protein aggregates after a severe stress is mandatory for cell survival. In bacteria, this activity depends on the collaboration between the DnaK system and ClpB, which in vivo occurs in a highly crowded environment. The reactivation reaction includes two steps: extraction of unfolded monomers from the aggregate and their subsequent refolding into the native conformation. Both steps might be compromised by excluded volume conditions that would favor aggregation of unstable protein folding intermediates. Here, we have investigated whether ClpB and the DnaK system are able to compensate this unproductive effect and efficiently reactivate aggregates of three different substrate proteins under crowding conditions. To this aim, we have compared the association equilibrium, biochemical properties, stability, and chaperone activity of the disaggregase ClpB in the absence and presence of an inert macromolecular crowding agent. Our data show that crowding i), increases three to four orders of magnitude the association constant of the functional hexamer; ii), shifts the conformational equilibrium of the protein monomer toward a compact state; iii), stimulates its ATPase activity; and iv), favors association of the chaperone with substrate proteins and with aggregate-bound DnaK. These effects strongly enhance protein aggregate reactivation by the DnaK-ClpB network, highlighting the importance of volume exclusion in complex processes in which several proteins have to work in a sequential manner.
Introduction
A functional and healthy proteome requires the successful folding of newly synthesized proteins and the maintenance of their functionally active conformations, as many proteins are structurally dynamic and could be expressed at concentrations at which they are poorly soluble (1,2). Protein solubility might be further compromised by the high macromolecular concentration estimated in the intracellular medium (3). In stark contrast to the total concentration of macromolecules in typical in vitro experiments, which is usually kept around or below 1 g/l (4), in the cytoplasm of Escherichia coli macromolecules reach concentrations of 300–400 g/l and occupy a significant fraction (up to 40%) of the cellular volume (5,6). Thus, molecular crowding will favor compact over expanded states and association reactions (7), and will increase the effective concentration of dilute macromolecules, affecting the kinetic and thermodynamic properties of reactions (4). It follows from these considerations that to determine the physiological role of a particular reaction in vitro, it is important to consider the possible influence of molecular crowders on the reaction (8).
Crowding will strongly affect aggregation-prone conformations that populate a (re)folding pathway (9), promoting aggregation of partially (un)folded proteins because aggregates exclude less volume to other macromolecules than isolated subunits. A strategy that the cell has developed during evolution to avoid aggregation of partially (un)folded polypeptide chains is to employ molecular chaperones (10), which essentially reduce aggregation because they either bind them and thus reduce their concentration or allow their (re)folding (11). Chaperones and other components involved in degradation of misfolded proteins form the network necessary to maintain a healthy proteome balance, known as proteostasis (12). Under severe stress conditions or at the onset of age-related degenerative disorders, the chaperone network protective capacity can be overwhelmed, resulting in protein aggregation of partially (un)folded conformations. Cell survival requires recycling of these toxic aggregates and reactivation of at least part of the lost protein material, which involves the concerted action of different types of chaperones. Although a huge effort has been made to elucidate the mechanism of individual chaperones, how they cooperate in a network to ensure proteome integrity is still far from being understood.
The disaggregase activity in bacteria requires the cooperation of representatives of the Hsp70, i.e., DnaK, and Hsp100, i.e., ClpB, chaperone families. The DnaK system is composed of the chaperone DnaK, an Hsp40 component, DnaJ, which carries substrate proteins and accelerates the chaperone ATPase activity, and a nucleotide exchange factor, GrpE. The ATPase activity of DnaK regulates its sequential interaction with substrate proteins and cochaperones during the functional cycle (13). The DnaK system is by itself able to reactivate small protein aggregates where the unfolded protein adopts a conformation similar to the native state. However, when the aggregate is formed by extensively unfolded substrate protein enriched in intermolecular β-structure, reactivation becomes strictly ClpB-dependent. The disaggregase ClpB is an AAA+ ATPase that belongs to the Clp protein family (14), and adopts a ring-like structure built by six identical subunits. Each monomer contains four domains: an N-terminal domain connected through a conserved linker to the first nucleotide binding domain (NBD1) in which the middle (M) domain is inserted, and a second NBD (NBD2). All NBDs bind and hydrolyze ATP, an essential process to drive the conformational cycle of the chaperone. CryoEM studies have suggested that in the ADP, closed-state, which shows low affinity for substrate proteins, the N-terminal domain docks onto the rest of the protein oligomeric barrel, and in the ATP, open-state, which displays a higher affinity for substrates, it projects toward the outside of the oligomer (15,16). In the presence of ATP, DnaK and ClpB form a weak, Kd around 5–25 μM (17,18), complex at the aggregate surface, where they pull unfolded monomers out of the aggregate for their subsequent refolding (19).
In this work, we aim to find out whether excluded volume conditions regulate chaperone-assisted protein aggregate reactivation. The significance of macromolecular crowding in chaperone-mediated protein folding is beginning to be studied and it might be a critical parameter affecting the efficiency of intracellular protein folding. Early studies showed that the GroEL system was not only active but even more efficient under excluded volume conditions (20), most likely due to an increase in the association between GroEL and GroES, which caps the internal cavity in which partly unfolded polypeptides are protected from aggregation while they attain their native conformation (21). This efficient capping avoided substrate leakage from the cavity, thus improving the refolding yield. Reactivation of protein aggregates is a complex process that involves binding of chaperones to protein substrates, interaction between different chaperones, and extraction of unfolded protein monomers from the aggregate for their refolding. Crowding could also affect the structural rearrangement that the chaperones, well-recognized molecular gymnasts (22), undergo during their functional cycle, i.e., to their conformational equilibrium. In the case of oligomeric proteins, such as ClpB, excluded volume conditions might also alter its association equilibrium, which in turn controls its ATPase and chaperone activities (23). Therefore, the effect of crowding on ClpB oligomerization, conformation, and association with the DnaK system should be characterized under crowding conditions.
We have found that crowding shifts the association equilibrium of ClpB toward the active, hexameric conformation, increasing the association constant for hexamer formation by three to four orders of magnitude. Our data also indicate that excluded volume conditions favor a compact conformation of the chaperone and accelerates its ATPase activity. Finally, crowding strongly enhances the functional association between the ClpB and the DnaK system, which results in a significant increase (up to 40 times) of the reactivation ability of the bichaperone network, indicating that protein aggregate reactivation in vivo might be more efficient than in vitro experiments would suggest.
Materials and Methods
Reagents, protein production, purification, and labeling
Ficoll 70, α-casein, G6PDH from Leuconostoc mesenteroides, and α-glucosidase from Saccharomyces cerevisiae were purchased from Sigma, sucrose from Serva, and luciferase from Photinus pyralis (American Firefly) from Roche. Wild-type (WT) ClpB and ClpB mutants (ClpB trap (E276/E678A) and ΔN-ClpB) were expressed in a ΔclpB::kan strain derived from MC4100 and purified as previously described (24). DnaK, DnaJ, and GrpE were obtained as reported (25,26). Protein concentration was determined by the colorimetric Bradford assay (Bio-Rad), and expressed as monomers for ClpB, DnaK, and DnaJ or dimers for GrpE. ClpB labeling with Alexa Fluor 647, succinimidyl ester (Life technologies) was carried out as previously described (27), in 50 mM Tris-HCl (pH 7.5), 50 mM KCl, and 5 mM MgCl2, conditions that favor protein oligomerization. The degree of labeling was estimated to be around 1 mol of fluorophore per protein subunit. Control experiments demonstrated that labeling had no effect on the biochemical properties of the chaperone.
Analytical ultracentrifugation
Sedimentation velocity (SV)
SV analysis of WT- and ΔN-ClpB was performed at 10 μM protein in buffer 50 mM Tris-HCl (pH 7.5), 5 mM MgCl2, containing the appropriated KCl concentration (50–500 mM). Experiments were conducted at 40.000 rpm and 18°C in a XL-A analytical ultracentrifuge (Beckman Coulter) with a UV-Vis optics detection system, using an An50Ti rotor and 12-mm double-sector centerpieces. The sedimentation coefficient distributions were calculated by least-squares boundary modeling of sedimentation velocity data using the c(s) method (28) as implemented in the software SEDFIT (National Institutes of Health, Bethesda, MD). The s-values were corrected to standard conditions (water, 20°C, and infinite dilution) using the software SEDNTERP (Biomolecular Interaction Technologies Center, University of New Hampshire), yielding the corresponding standard s-values.
Tracer sedimentation equilibrium
Low-speed tracer sedimentation equilibrium experiments at different angular velocities (4000, 8000, and 9000 rpm) were performed to characterize the effect of Ficoll 70 (acting as a macromolecular crowder) on the self-association properties of WT- and ΔN-ClpB at increasing protein concentration. Experiments were carried out as described in the SV section, both in the absence and presence of 15% (w/v) Ficoll 70, using short columns (80 μl). At 1 and 3 μM ClpB, all the protein was labeled with Alexa 647, and at higher concentrations samples contained a fixed amount of labeled protein (3 μM). This experimental protocol was employed because of the weak experimental signal (absorbance at 650 nm) below 1 μM protein. The dynamic nature of the ClpB assembly ensures subunit exchange between labeled and unlabeled oligomers and formation of hybrid hexamers (29,30). Data analysis was performed as detailed in the Supporting Material.
Fluorescence anisotropy
Measurements were recorded on a Fluorolog spectrofluorimeter (Jobin Yvon) at 25°C. Different concentrations of ClpB trap were added to buffer 50 mM Tris-HCl (pH 7.5), 50 mM KCl, 5 mM MgCl2, and 2 mM dithiothreitol (DTT) containing 0.1 μM fluorescein isothiocyanate (FITC)-α-casein in the presence of 2 mM ATP. This ClpB variant contains mutations in both nucleotide binding domains (E279A/E678A) that allow binding but not hydrolysis of ATP, therefore being able to form stable complexes with protein substrates (31). Experiments were performed in the absence and presence of 15% Ficoll 70. Samples were incubated overnight at 4°C and equilibrated at room temperature 1 h before measuring fluorescence anisotropy. Excitation and emission wavelengths were 492 and 515 nm, and both slits were set at 7 nm. Data were fitted to a quadratic equation modeling a single binding site.
Circular dichroism (CD)
Experiments were carried out in a Jasco J-810 spectropolarimeter equipped with a Peltier Type Control System PFD 425S. Samples contained 3 μM of protein monomer in 20 mM Hepes (pH 7.6), 50 mM KAc- and 5 mM MgAc-, and, when required, 1 mM nucleotide. CD spectra were collected at a scan rate of 50 nm/min using 0.1 or 1 mm pathlength cuvettes. Four spectra were averaged for the native (25°C) and unfolded state (90°C) of each sample. Protein unfolding was followed by measuring the ellipticity at 222 nm at increasing temperatures, using a scan rate of 1 or 0.3°C/min. Tm was the temperature at the midpoint of the unfolding transition, and was estimated from the first derivative of the denaturation profile.
ATPase activity measurements
The ATPase activity of the ClpB variants used in this work was measured spectrophotometrically in 50 mM Tris-HCl (pH 7.5), 5 mM MgCl2, and different concentrations of KCl (50, 150, 300, and 500 mM), Ficoll 70 (0%, 10%, 15%, 20%, and 30%) or sucrose (30% and 40%), which was used as a control to account for the increase in viscosity. Experiments were performed at 25°C in the presence of an ATP-regenerating system. Protein and ATP concentrations were 2 μM and 1 mM, respectively. Substrate-induced stimulation of the chaperone ATPase activity was determined in the presence of 2 μM α-casein (Sigma), a concentration lower than that usually employed (10 μM) in this type of experiment, to avoid substrate aggregation at high Ficoll 70 concentration. The same method was used to estimate the apparent affinity of ClpB for α-casein using 2 μM ClpB and increasing α-casein concentrations.
Reactivation of G6PDH aggregates
G6PDH (10 μM; Sigma) was heat-denatured and aggregated by incubating the protein 30 min at 70°C in 50 mM Tris-HCl (pH 7.5), 150 mM KCl, 20 mM MgCl2, 10 mM DTT. The aggregated substrate was diluted to 0.4 μM in the same buffer containing 50 mM KCl and different concentrations of Ficoll 70 (0%, 5%, 15%, and 30%) or sucrose (30% and 40%). The sample also contained DnaK (1 μM), DnaJ (0.1 μM), GrpE (1.2 μM), and different concentrations of ClpB. Reactivation was started by adding 2 mM ATP to the sample in the presence of an ATP regenerating system (30 mM phosphoenolpyruvate and 20 ng/ml pyruvate kinase) at 30°C. Chaperone-mediated aggregate reactivation was measured as previously reported (32).
Reactivation of α-glucosidase aggregates
α-glucosidase (10 μM; Sigma) was heat-denatured and aggregated by incubating the protein 30 min at 50°C in 50 mM Tris-HCl (pH 7.5), 150 mM KCl, 20 mM MgCl2, 10 mM DTT. Aggregated protein was diluted to 0.4 μM in the same buffer containing 50 mM KCl and increasing concentrations of Ficoll 70 or sucrose and WT ClpB. The concentrations of DnaK, DnaJ, and GrpE were 1 μM, 0.1 μM, and 1.2 μM. Reactivation started by adding 2 mM ATP to the sample that contained the ATP regenerating system at 30°C. Aggregate reactivation was measured by recording the activity of α-glucosidase at different times as reported (33).
Reactivation of luciferase denatured in urea
Luciferase (2.5 μM; Roche) was denatured in 7 M urea, 30 mM Hepes (pH 7.6), 60 mM KCl, 10 mM MgCl2, and 10 mM DTT (45 min at 25°C). The denatured protein was diluted to 25 nM in buffer, 30 mM Hepes (pH 7.6), 50 mM KCl, 10 mM MgCl2, 2 mM DTT, containing the desired Ficoll 70 or sucrose concentration and the ATP-regenerating system. The diluted sample was incubated 10 min to obtain protein aggregates and afterward 1 μM DnaK, 1.2 μM GrpE, 0.1 μM DnaJ, and different concentrations of ClpB were added. Reactivation at distinct Ficoll 70 concentrations was initiated by the addition of 2 mM ATP, and luciferase activity was measured as described (23).
Results
Crowding shifts the association equilibrium of ClpB toward the active hexamer
Before analyzing the effect of crowding on the oligomerization state of ClpB, it should be noted that neither protein labeling with Alexa 647 nor deletion of its N-terminal domain ((34); Fig. 1 A) significantly modified the association properties of the chaperone, as seen by sedimentation velocity experiments (Fig. S1 in the Supporting Material). Excluded volume effects in highly crowded solutions are expected to increase the tendency of ClpB, a relatively large protein, to self-associate, which will increase the concentration of functionally active oligomers, especially at high potassium concentration (150–300 mM). To determine the impact of crowding on ClpB oligomerization at these KCl concentrations, a set of sedimentation equilibrium experiments at increasing chaperone concentrations were carried out in the absence and presence of 15% (w/v) Ficoll 70, and analyzed to obtain the corresponding apparent buoyant molecular weights (Fig. 1). Ficoll 70 was selected as the crowding agent because its interaction with proteins had been previously shown to be mainly repulsive and may be described by steric exclusion (35).
Figure 1.
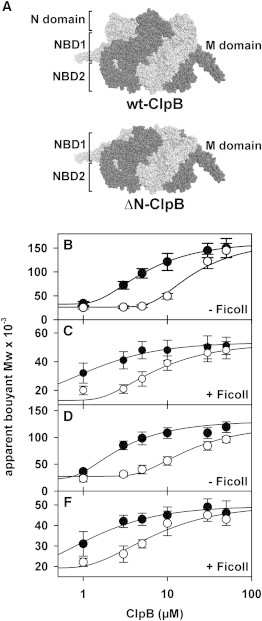
Crowding shifts the association equilibrium of ClpB toward the active hexamer. (A) Atomic model of the E. coli ClpB hexamer (34). Different domains of each identical subunit are located in three tiers as indicated in the structure of WT- (top) and ΔN- (bottom) ClpB. Sedimentation equilibrium measurements were performed in the absence (B and D) and presence (C and E) of 150 g/l Ficoll 70 to obtain the dependency of the buoyant molecular weight of WT ClpB (B and C) or ΔN-ClpB (D and E). The buffer contained 150 (black circles) or 300 mM KCl (white circles). Solid lines are the best fit of the experimental data to the model described in the Experimental section. Measures are represented as the mean ± SE from three independent experiments.
The dependence of M∗i,app upon ClpB concentration was best described by a two-state monomer-hexamer model, both with and without Ficoll 70 (Fig. 1). In the absence of crowder, the association constant for hexamer formation (K6) was almost three orders of magnitude higher at 150 mM than at 300 mM KCl (Fig. 1 B; Table 1), thus confirming the dependence of ClpB self-association on salt concentration (23). The apparent buoyant molar mass values of ClpB in the presence of Ficoll 70 were significantly lower than those obtained in its absence, because of the nonideal behavior of the dilute protein in a highly crowded solution (36). Addition of 150 g/l Ficoll 70 increased the K6 value around three or four orders of magnitude at 300 or 150 mM KCl (Fig. 1 C; Table 1). This corresponds to a decrease in the free energy of hexamer formation of 7 and 9 RT, respectively. Assuming that the hexamer is a cyclic structure (i.e., it has six intermolecular contacts), this free energy change would correspond to an average decrease of 1.1–1.5 RT per intermonomer contact. When the same experiments were performed with a ClpB variant that lacks the N-terminal domain (Fig. 1, D and E; Table 1), the results show that crowding favors ΔN-ClpB association to an extent similar to that described for WT ClpB. The deletion mutant has a stronger tendency to associate into functional oligomers, especially in the absence of crowder (Table 1), indicating that steric repulsions between adjacent N-terminal domains destabilize the protein hexamer. These findings show that crowding shifted the ClpB association equilibrium toward the functional hexamer at salt conditions that otherwise would promote chaperone dissociation in dilute solutions, and that the N-terminal domain of the protein does not significantly alter this behavior.
Table 1.
Effect of crowding on the association equilibrium of ClpB.
| Protein | KCl (mM) | Ficoll (g/l) | log K6 (l/g)5 | ΔG6° (RT)a | ΔΔG6° (RT) |
|---|---|---|---|---|---|
| WT ClpB | 150 | 0 | 2.4 ± 0.4 | −61.2 | 8.8 |
| 150 | 150 | 6.1 ± 1.2 | −70.0 | ||
| 300 | 0 | −0.8 ± 0.3 | −53.8 | 7.2 | |
| 300 | 150 | 2.3 ± 0.6 | −61.0 | ||
| ΔNClpB | 150 | 0 | 4.3 ± 0.5 | −64.5 | 6.5 |
| 150 | 150 | 6.6 ± 1.4 | −70.0 | ||
| 300 | 0 | 0.1 ± 0.1 | −54.8 | 7.1 | |
| 300 | 150 | 3.2 ± 0.8 | −61.9 |
Log K6 and ΔG6o values for WT ClpB and ΔN-ClpB at different concentrations of KCl in the absence or presence (15% w/v) of Ficoll 70. Data shown in Fig. 1 were fitted to the model described in the Experimental section. Measures are represented as the mean ± SE from three independent experiments.
ΔG6° = −RT lnK′6, where K′6 is the equilibrium association constant expressed in molar concentration units: K′6 = K6 (M15/6), being M1 the monomer molar mass of the protein.
Crowding enhances the ATPase activity of ClpB
Crowding could favor a compact conformation of the chaperone, which might alter the conversion rate between the open, ATP- and closed ADP-states, and thus its biochemical properties. To find out if this was the case, the effect of Ficoll 70 on the ATPase activity of ClpB and ΔN-ClpB was characterized. Crowder concentrations above 10% (w/v) enhanced the ATPase activity of ClpB regardless of the ionic strength (Fig. 2 A). Similar nonlinear tuning of the catalytic parameters of Fet3p by crowding has been reported (37). The activation detected at 500 mM KCl indicated that Ficoll 70 reverts to the salt-induced dissociation of the chaperone hexamer, which is known to drastically reduce the protein ATPase activity (23). A greater crowder-dependent activation was observed for ΔN-ClpB (Fig. 2 B), which might be due to the effect that Ficoll 70 could have on the conversion rate between different conformations during the ATPase cycle. If Ficoll 70 favors a compact conformation in which the N-terminal domain docks onto the protein hexamer, the rate of ATP hydrolysis by WT ClpB would be lower due to the energetic penalty that movement of the N-domain during the functional cycle of the protein might cause. The effect of crowding on the ATPase activity of ΔN-ClpB was even greater in the presence of the substrate α-casein, known to stimulate the chaperone ATPase activity. The substrate-induced activation factor of ΔN-ClpB increased from 1.5 to 3.5 with Ficoll 70 concentration (Fig. 2 B; inset), whereas that of WT ClpB slightly decreases above 15% Ficoll 70 and was never higher than 1.5 (Fig. 2 B; inset). As a control to account for the effect of Ficoll 70 on the viscosity of the medium, the ATPase activity of both proteins was characterized in 30% and 40% sucrose. These sucrose concentrations were selected because in terms of bulk viscosity 10% and 20% (w/v) Ficoll 70 are equivalent to 30% and 40% (w/v) sucrose, respectively (38,39). Sucrose addition reduces ClpB activity (Fig. 2 C), an expected consequence of the higher viscosity (40), suggesting that the additional excluded-volume conditions that Ficoll 70 imposes, compensate the otherwise inhibiting effect of viscosity.
Figure 2.
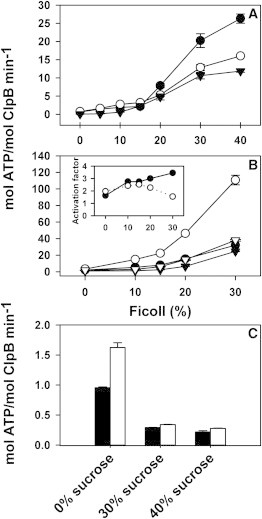
Excluded volume conditions stimulate the ATPase activity of WT ClpB and ΔN-ClpB. (A) ATPase activity of WT ClpB at increasing Ficoll 70 concentrations in buffer containing 50 mM (solid circles), 300 mM (open circles), and 500 mM (inverse triangles) KCl. (B) Effect of Ficoll 70 on the ATPase activity of WT ClpB (triangles) and ΔN-ClpB (circles) in the absence (solid symbols) or presence (open symbols) of 2 μM α-casein. Measurements were carried out in buffer containing 50 mM KCl. Inset: Substrate-induced activation factor for WT ClpB (open circles) and ΔN-ClpB (solid circles). (C) Basal (black bars) and α-casein activated (white bars) ATPase activity of WT ClpB in the presence of different sucrose concentrations. Measures are represented as the mean ± SE (n = 4).
Crowding increases the affinity of active ClpB for substrate proteins
The chaperone activity of ClpB requires its interaction with substrate proteins, and is therefore necessary to estimate the effect that crowding might exert on this association. To this aim, binding of FITC-labeled casein to trap variants of WT and ΔN-ClpB was followed in the presence of ATP. The estimated Kd values were similar for both protein variants in the absence of Ficoll 70 (0.86 ± 0.08 μM), and they did not change significantly in the presence of 15% crowder (0.88 ± 0.06 μM) (Fig. S2). To further characterize this behavior, we followed the activation of the ATPase activity of ClpB at different substrate concentrations. In the absence of Ficoll, the K0.5 values obtained for WT (Fig. 3 A) and ΔN-ClpB (Fig. 3 B) were 3.5 and 1 μM, respectively. The threefold lower value for ΔN-ClpB indicated that under working conditions, deletion of the N-terminal domain increased the affinity of the chaperone for α-casein. In stark contrast with the binding experiments shown previously, 15% Ficoll 70 decreased the K0.5 values 11 and 3 times for WT and ΔN-ClpB, respectively, indicating that crowding facilitates complex formation that leads to chaperone activation.
Figure 3.
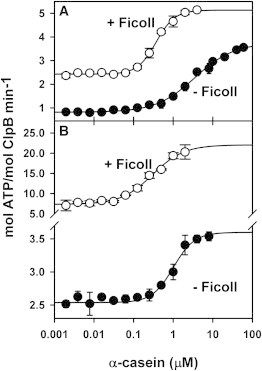
Crowding favors the interaction of substrate proteins with ClpB conformations generated during ATP hydrolysis. ATPase activity of 2 μM WT ClpB (A) or ΔN-ClpB (B) at increasing α-casein concentration. Measurements were performed in the absence (solid symbols) or presence (open symbols) of 15% Ficoll 70. Values of the mean ± SE from at least three independent experiments are shown.
Crowding shifts the conformational equilibrium of ClpB toward a compact state
To prove that Ficoll 70 could influence the conformational transition associated with the ClpB functional cycle, the effect of this crowder on the conformation and stability of the apo and nucleotide-bound states of WT and ΔN-ClpB was studied by CD. The CD spectra of the apo and nucleotide-bound states of WT ClpB (Fig. 4, A, C, and E), and ΔN-ClpB (Fig. 4, B, D, and F), were similar regardless of the presence of Ficoll 70, indicating that neither nucleotides nor crowding significantly modify their secondary structure (Fig. S3). The irreversible thermal denaturation of apo-ClpB showed two events at 56 and 60°C (Fig. 4 A), in agreement with previous DSC data (41), which are shifted to 62 and 76°C upon addition of ADP (Fig. 4 C) or ATP (Fig. 4 E; Table 2). Ficoll displaced to higher temperatures the first thermal event of the WT apo-ClpB, and increased modestly the stability of its ADP-state (Table 2), as it has been shown for other proteins (42,43). The small dependence of the Tm values (ΔT ≈ 0.5–1.2°C) on scan rate suggests that the irreversible process occurs after thermal denaturation. The similarity of the Tm values of apo ClpB in 15% Ficoll 70 and the ADP-state of the protein pointed out that crowding favors an ADP-like conformation of the apo-protein. Two findings suggested the involvement of the N-terminal domain in the crowder-induced structural rearrangement of the apo-protein: i), deletion of the N-terminal domain strongly reduced the Ficoll-induced stabilization of the apo-protein (Fig. 4 B); and ii), the different Tm values of the apo- (59.5°C) and ADP-states (Fig. 4 D; 64.4°C) of the truncated mutant in the presence of Ficoll 70, in contrast to what was observed for WT ClpB. This would be consistent with the current view of protein dynamics, in which the open and closed conformations of ClpB coexist in the absence of ligands, and crowding would displace the conformational equilibrium toward a compact conformation, similar to the ADP state.
Figure 4.
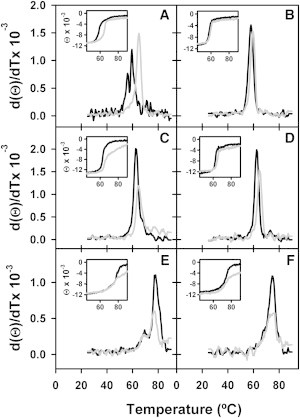
Thermal stability of WT ClpB and ΔN-ClpB under crowding conditions. First derivative of the molar ellipticity at 222 nm as a function of temperature for apo (A and B), ADP-bound (C and D), and ATP-bound (E and F) WT ClpB (A, C, and E) or ΔN-ClpB (B, D, and F) in the absence (black lines) and presence (gray lines) of 15% Ficoll 70. Inset, variation of the molar ellipticity at 222 nm with temperature.
Table 2.
Effect of crowding on the thermal stability of WT ClpB and ΔN-ClpB
| Protein | Ficoll (g/l) |
Tm (°C) |
||
|---|---|---|---|---|
| APO | ADP | ATP-ClpBtrap | ||
| WT ClpB | 0 | 56.3 ± 0.42 | 62.35 ± 0.49 | 77 ± 0.7 |
| 59.25 ± 0.35 | ||||
| 150 | 64.5 ± 0.7 | 64.95 ± 0.63 | 76.75 ± 0.35 | |
| ΔNClpB | 0 | 57.62 ± 0.53 | 62.5 | 75.25 ±0.35 |
| 150 | 59.5 | 64.4 ± 0.14 | 74 ± 0.56 | |
Midpoint denaturation temperature (Tm) values for different states of these proteins obtained from the first derivative of the CD temperature denaturation profile. Date are the mean ± SE (n = 3).
Crowding enhances association of ClpB with the DnaK system and their coordinated aggregate reactivation activity
Reactivation of stable protein aggregates requires the functional association of ClpB and the DnaK system, which could be modulated by crowding. To find out if this was the case, protein aggregate reactivation by this bichaperone network was studied under crowding. Because the activity of the DnaK-ClpB complex is sensitive to the aggregated protein sequence, we have characterized the effect of Ficoll 70 on the reactivation of aggregates of three substrate proteins: dimeric G6PDH (Fig. 5, A–C), tetrameric α-glucosidase (Fig. S4), and monomeric luciferase (Fig. 5, D–F).
Figure 5.
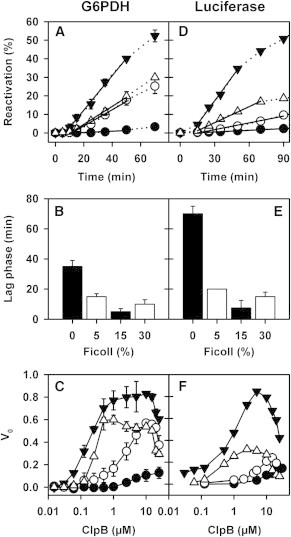
Crowding strongly enhances reactivation of protein aggregates by the DnaK-ClpB bichaperone network. Reactivation of aggregates of G6PDH (A–C) and luciferase (D–F) in the presence of 0% (solid circles), 5% (open circles), 15% (solid triangles), and 30% (open triangles) Ficoll 70. (A and D) Reactivation kinetics at 5 μM WT ClpB monomer. (B and E) Lag phase of the reactivation reaction at 5 μM WT ClpB monomer and increasing crowder concentrations. (C and F) Initial reactivation rates (% refolding × min−1) measured at increasing ClpB and crowder concentrations. Same symbols as in (A). Values of the mean ± SE from four independent experiments are shown.
The DnaK system is composed of three different proteins whose association might also be modulated by crowding. Therefore, to analyze the effect of crowding on the association of ClpB with the DnaK system, the concentration of the components of this system and of substrate proteins are kept constant, and reactivation followed at increasing ClpB amounts for each crowder concentration. This experimental strategy does not require a priori knowledge of the effect of Ficoll 70 on the association of the components of the DnaK system among them or with substrate proteins, and therefore any experimental difference should mainly come from modifications in the association equilibrium of ClpB and/or its interaction with free or aggregate-bound DnaK, which is mandatory to reactivate these aggregates. Addition of Ficoll 70 induced the following changes in the reactivation reaction of the three types of aggregates: i), increased both the maximum rate and reactivation yield at 5% and 15% crowder; ii), decreased one order of magnitude the ClpB concentration that causes half-maximum activation (Table 3); iii), significantly reduced (3–4 times) the lag phase of the reactivation reaction; and iv), at 30% partially inhibited the reactivation reaction, which nevertheless remained more effective than without crowding. The enhancement of the reactivation ability at lower ClpB concentrations in the presence of Ficoll 70 is expected as crowding increases the concentration of functional ClpB hexamers and facilitates their association with aggregate-bound DnaK. The inhibition observed at high crowder and ClpB concentrations might be due to a stabilization of the ClpB hexamer, so that its slower dynamics would also decrease its reactivation efficiency (23) (see below), or of the DnaK-ClpB complex, designed to be transient with an estimated Kd of around 5–20 μM (17,18), which could become detrimental for aggregate reactivation. An increase in ClpB concentration above 2 μM (30% Ficoll 70) or 10–20 μM (5% and 15% Ficoll 70) resulted in an inhibition of the reactivation efficiency, suggesting that stabilization of the interaction between ClpB and aggregate-bound and/or free DnaK could compromise the activity of the bichaperone complex. If this were the case, it would be essential for the cell to control the concentration of the different chaperones to optimize the reactivation efficiency of its functional networks. It is important to note that at 7 μM monomer and 50 mM KCl, ClpB is a hexamer regardless of the presence of Ficoll 70 (23) and therefore the differences in reactivation rate observed at different crowder concentrations do not arise from Ficoll-induced modifications in the oligomerization state of ClpB. They thus reflect primarily changes in the association constant between the two chaperones that build the functional complex. As previously mentioned for the ATPase activity of ClpB, the aggregate reactivation ability of the bichaperone network was abolished at 30% sucrose (not shown), demonstrating that excluded volume conditions are required to observe the aforementioned effects.
Table 3.
Crowding modulates protein aggregate reactivation by the DnaK-ClpB chaperone network
| Ficoll 70 (%) | K0.5 (μM) |
||
|---|---|---|---|
| G6PDH | α-glucosidase | Luciferase | |
| 0 | 3.9 ± 0.5 | 3.4 ± 0.19 | 12.3 ± 2.19 |
| 5 | 2.6 ± 0.4 | 1.6 ± 0.16 | 4.5 ± 0.61 |
| 15 | 0.2 ± 0.006 | 0.1 ± 0.009 | 0.9 ± 0.19 |
| 30 | 0.3 ± 0.01 | 0.2 ± 0.002 | 0.5 ± 0.042 |
The dynamic nature of the ClpB hexamer is maintained under crowding conditions
ClpB is a highly dynamic assembly that easily exchanges subunits at 150 mM KCl (44). The biological relevance of ClpB dynamics remains controversial, and it has been proposed that it might be important to avoid stalling of the chaperone by very stable protein aggregates (44), and that hexamer dissociation is not essential for its chaperone activity (45). The predicted effect of crowding on the association equilibrium of ClpB and the sedimentation equilibrium data shown in Fig. 1 could result in a crowder-induced stabilization of the hexamer that might hamper subunit exchange. To explore this possibility, the aggregate reactivation efficiency was analyzed in the absence and presence of two Ficoll 70 concentrations (15% Fig. S5 and 30% Fig. 6). Subunit exchange was monitored by following the insertion of inactive, trap monomers into the WT hexamer, as it has been shown that incorporation of a single subunit of ClpBtrap within a WT hexamer completely inhibits its chaperone activity (29,30). Therefore, the assay follows the timecourse of the reactivation process by WT ClpB and the effect of adding equimolar amounts of ClpBtrap. Data show that addition of inactive ClpB hexamers arrested reactivation in the absence and presence of both Ficoll 70 concentrations, indicating that crowding did not hamper hexamer disassembly that allows subunit exchange. They also suggest that the aforementioned differences in reactivation are mainly due to crowder-induced modulation of DnaK-ClpB complex stability.
Figure 6.
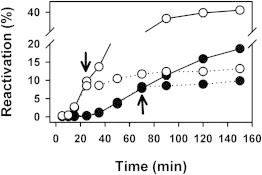
Excluded volume conditions do not hamper ClpB subunit exchange. Folding of G6PDH aggregates at 50 mM KCl and 0% (solid circles) or 30% Ficoll 70 (open circles). Experiments were carried out with 2.5 μM WT ClpB (solid line), and after recovery of a significant reactivation 2.5 μM ClpBtrap (dashed line) was added to the reactivation mixture at the times indicated with black arrows.
Discussion
To our knowledge, this work is the first attempt to estimate the effect of crowding on chaperone-mediated reactivation of protein aggregates, a complex reaction that involves the sequential interaction of different proteins. Crowding effects arise from a balance between steric repulsions that favor compact states, and from nonspecific interactions that can add to the former when they are repulsive or oppose when they are attractive (46,47). The use of synthetic polymers to mimic the intracellular crowded environment has the limitation that natural crowders are proteins and DNA that can establish weak nonspecific interactions with proteins. Despite this potential caveat, studies using synthetic polymeric crowders are necessary to consider the impact that excluded volume conditions might have on complex processes in which several proteins are needed in consecutive steps of a reaction, such as protein aggregate reactivation.
A novel aspect of this study is the experimental characterization and analysis of the effect of crowding on ClpB oligomerization, which extends the previous characterization of ClpB self-association in diluted solutions (23). Enhancement of ClpB oligomer formation in the presence of Ficoll 70 is expected on grounds of excluded volume theory (47). This work provides a quantitative analysis, by means of nonideal tracer sedimentation equilibrium, of ClpB self-association under crowding conditions, an experimental challenge because of the difficulty to study a complex reaction under dilute and crowded solutions. Our data show that Ficoll 70 substantially shifts the overall ClpB isotherm toward the active hexamer. The equilibrium association constant for the active, hexameric conformation (K6) increases between three and four orders of magnitude at 15% Ficoll 70, a similar behavior to that found for the oligomerization equilibrium of the heptameric cochaperonin cpn10 (43). The stabilizing effect of crowding on the active ClpB hexamer suggests that it could be an additional factor that regulates the intracellular activity of this chaperone, as it could widen the range of salt conditions where ClpB hexamer could function in vivo. Interestingly, hexamer stabilization does not avoid subunit exchange, indicating that the dynamic nature of the protein assembly is maintained under crowding conditions even in the presence of nucleotides, natural ClpB ligands that further stabilize the oligomer (23). This could be explained assuming that the difference in free energy of hexamer assembly with and without crowding, which amounts to 14% of the total free energy, is compensated by structural rearrangements of the oligomer during its functional cycle that promote dissociation.
Regarding the effect of crowding on the conformation and activity of ClpB, our data show that crowding shifts the conformational equilibrium of apoClpB toward a compact conformation and stimulates its ATPase activity. This structural transition is not associated with a significant modification of the protein secondary structure, as seen by far-UV CD, and involves the N-terminal domain, which most likely contacts the rest of the hexamer under excluded volume conditions. The change in the relative position of this domain, which is connected to the rest of the protein by a conserved linker, has been observed in cryoEM studies of Hsp104 (48). The biochemical consequence of this structural rearrangement is an activation of the chaperone ATPase activity, which is essential during the disaggregation process. This activation could be related to the crowder-induced faster conformational transition between the open and closed states, and to the ability of ATP binding to induce an open conformation under excluded volume conditions. Nucleotide exchange will shift the conformational equilibrium toward the open, ATP-state, because ATP binding could provide the energy to drive an otherwise unfavorable structural transition, as proposed for DnaK (49). As sucrose significantly reduces the ATPase and chaperone activities of the protein, the stimulating effect of Ficoll comes from excluded volume conditions. A similar activation of phosphoglycerate kinase by Ficoll 70 has been recently reported, and was related to the approximation of the protein catalytic domains under crowding conditions (40). The same argument can be used to explain the higher substrate-induced stimulation of the ATPase activity of ΔN-ClpB at high crowder concentration. If Ficoll 70 favors a compact conformation, the N-terminal domain would complicate the structural transition to the open, ATP-state that shows higher affinity for substrates. Thus, the protein variant that lacks the mobile N-domain could be activated more efficiently than WT ClpB at high crowder concentrations. An intriguing finding of this work is that the expected crowder-induced enhancement of the affinity of ClpB for substrates is only observed under hydrolyzing conditions, i.e., for the active protein in the presence of ATP, and not for inactive trap variants under the same experimental conditions. This apparent discrepancy might be explained if crowding would induce a rigid conformation in the inactive variants that could compensate the expected increase in affinity for the substrate (50). The situation would be different for the active proteins that will be cycling between distinct conformations. Under these conditions, the unfolded substrate translocates through the central channel (51) and the complex could effectively exclude less volume than the isolated components.
Another aspect of this study that shall be discussed is the impact that crowding might have on the functional association of the DnaK system and ClpB. The K0.5 values obtained from aggregate reactivation experiments decrease up to one order of magnitude upon Ficoll addition, pointing out that crowding significantly increases the affinity of ClpB for the DnaK system. This interpretation assumes, as it has been previously demonstrated, that aggregate-bound DnaK recruits ClpB to the aggregate surface (17). The three- to fourfold reduction in the lag phase, i.e., the time required to extract unfolded monomers and refold them into their active conformation, of the reactivation process further supports this hypothesis and indicates that crowding favors the productive interaction of the DnaK system and ClpB at the aggregate surface. The initial aggregate rearrangement most likely needs binding of a productive amount of active chaperones to the aggregate surface, which could be the limiting step of the reactivation process. The similar crowding-induced effects on the reactivation of three different substrate proteins suggests that this complex process seems to be governed by chaperone binding to the aggregate more than by the polypeptide sequence that is being reactivated. The reactivation efficiency will thus depend on i), the amount of aggregate-bound chaperones, assuming that extraction of an unfolded monomer would require the force generated by a minimum number of chaperones, and ii), the ATP hydrolysis rate of both ATPases, DnaK, and ClpB, that would provide the energy necessary for the extraction, which under crowding conditions will be an unfavorable process. Crowding will favor formation of chaperone functional complexes, and at the same time will activate ClpB so that it could exert more work to pull unfolded substrate molecules out of the aggregate during the reactivation reaction. The outcome will be, as experimentally observed, a shortening of the lag phase and an increase in the reactivation rate.
Refolding of the extracted, unfolded monomers and their subsequent oligomerization, if the active protein conformation is a dimer -G6PDH- or tetramer -α-glucosidase-, could also sense excluded volume conditions. below 30% Ficoll, increasing crowder concentrations could favor compaction of the substrate unfolded state to a native-like conformation provided that competing processes, i.e., aggregation of folding intermediates, are conveniently prevented by chaperones. Partial inhibition of reactivation at high crowder concentrations, i.e., 30% Ficoll 70, could be due to any of the following reasons or to a combination of them: i), diffusion-rate limiting of the conformational rearrangement required to fold into the native state due to an increase in viscosity (40); ii), promotion of intermolecular interactions between aggregation-prone folding intermediates; and iii), acceleration of refolding to compact nonnative conformations that could be similar in overall structure to the native substrate but lack enzymatic activity, as it has been found for carbonic anhydrase (52). Interestingly, the observed crowder-induced enhancement of the reactivation rate and yield occurs in the absence of a protein cage that would provide confinement conditions to avoid intermolecular interactions that lead to irreversible aggregation. Under similar conditions, it has been reported that stabilization of the GroEL-GroES complex reduces inefficient nonnative protein release from the Anfinsen’s cage that compromises protein refolding (20). Our data show that in a crowded and nonconfined environment, chaperones are also able to efficiently extract unfolded protein molecules from aggregates and refold them into their native states avoiding aggregation.
Conclusion
The results of this study indicate that crowding strongly modulates protein aggregate reactivation by the DnaK-ClpB bichaperone network. Crowding enhances reactivation by i), shifting the association equilibrium of ClpB toward the functional hexamer; ii), accelerating its functional cycle; iii), enhancing the affinity of ClpB for aggregate-bound DnaK, which results in a higher amount of functional chaperone complexes at the aggregate surface; and iv), increasing the affinity of (a) transient conformations generated during ATP hydrolysis for substrate proteins. They also provide insights into the importance of crowding to optimize complex reactions in which different proteins must interact sequentially, as protein aggregate reactivation.
Acknowledgments
We are grateful to A. Minton for his help with data analysis, S. Rodziewicz-Motowidlo for the atomic model of ClpB from E. coli, Begoña Monterroso for critically reading the manuscript, and Natalia Orozco for technical assistance.
We acknowledge financial support from the Ministerio de Ciencia e Innovación (grants BFU2010-15443 to A.M. and BIO2011-28941-C03 to G.R.), and the Universidad del País Vasco and Gobierno Vasco (grant IT709-13 to A.M.).
Supporting Material
References
- 1.Kim Y.E., Hipp M.S., Hartl F.U. Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 2013;82:323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- 2.Tartaglia G.G., Dobson C.M., Vendruscolo M. Physicochemical determinants of chaperone requirements. J. Mol. Biol. 2010;400:579–588. doi: 10.1016/j.jmb.2010.03.066. [DOI] [PubMed] [Google Scholar]
- 3.Ellis R.J., Minton A.P. Protein aggregation in crowded environments. Biol. Chem. 2006;387:485–497. doi: 10.1515/BC.2006.064. [DOI] [PubMed] [Google Scholar]
- 4.Ralston G.B. Effects of crowding in protein solution. J. Chem. Educ. 1990;67:857–860. [Google Scholar]
- 5.Zimmerman S.B., Trach S.O. Estimation of macromolecule concentrations and excluded volume effects for the cytoplasm of Escherichia coli. J. Mol. Biol. 1991;222:599–620. doi: 10.1016/0022-2836(91)90499-v. [DOI] [PubMed] [Google Scholar]
- 6.Fulton A.B. How crowded is the cytoplasm? Cell. 1982;30:345–347. doi: 10.1016/0092-8674(82)90231-8. [DOI] [PubMed] [Google Scholar]
- 7.Minton A.P. Implications of macromolecular crowding for protein assembly. Curr. Opin. Struct. Biol. 2000;10:34–39. doi: 10.1016/s0959-440x(99)00045-7. [DOI] [PubMed] [Google Scholar]
- 8.Minton A.P. The influence of macromolecular crowding and macromolecular confinement on biochemical reactions in physiological media. J. Biol. Chem. 2001;276:10577–10580. doi: 10.1074/jbc.R100005200. [DOI] [PubMed] [Google Scholar]
- 9.van den Berg B., Ellis R.J., Dobson C.M. Effects of macromolecular crowding on protein folding and aggregation. EMBO J. 1999;18:6927–6933. doi: 10.1093/emboj/18.24.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellis R.J. Molecular chaperones: avoiding the crowd. Curr. Biol. 1997;7:R531–R533. doi: 10.1016/s0960-9822(06)00273-9. [DOI] [PubMed] [Google Scholar]
- 11.Ellis R.J., Hartl F.U. Principles of protein folding in the cellular environment. Curr. Opin. Struct. Biol. 1999;9:102–110. doi: 10.1016/s0959-440x(99)80013-x. [DOI] [PubMed] [Google Scholar]
- 12.Balch W.E., Morimoto R.I., Kelly J.W. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 13.Mayer M.P. Hsp70 chaperone dynamics and molecular mechanism. Trends Biochem. Sci. 2013;38:507–514. doi: 10.1016/j.tibs.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 14.Kress W., Maglica Z., Weber-Ban E. Clp chaperone-proteases: structure and function. Res. Microbiol. 2009;160:618–628. doi: 10.1016/j.resmic.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 15.Lee S., Sielaff B., Tsai F.T. CryoEM structure of Hsp104 and its mechanistic implication for protein disaggregation. Proc. Natl. Acad. Sci. USA. 2010;107:8135–8140. doi: 10.1073/pnas.1003572107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wendler P., Shorter J., Saibil H.R. Motor mechanism for protein threading through Hsp104. Mol. Cell. 2009;34:81–92. doi: 10.1016/j.molcel.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Acebrón S.P., Martín I., Muga A. DnaK-mediated association of ClpB to protein aggregates. A bichaperone network at the aggregate surface. FEBS Lett. 2009;583:2991–2996. doi: 10.1016/j.febslet.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 18.Rosenzweig R., Moradi S., Kay L.E. Unraveling the mechanism of protein disaggregation through a ClpB-DnaK interaction. Science. 2013;339:1080–1083. doi: 10.1126/science.1233066. [DOI] [PubMed] [Google Scholar]
- 19.Schlieker C., Tews I., Mogk A. Solubilization of aggregated proteins by ClpB/DnaK relies on the continuous extraction of unfolded polypeptides. FEBS Lett. 2004;578:351–356. doi: 10.1016/j.febslet.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 20.Martin J., Hartl F.U. The effect of macromolecular crowding on chaperonin-mediated protein folding. Proc. Natl. Acad. Sci. USA. 1997;94:1107–1112. doi: 10.1073/pnas.94.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin J. Requirement for GroEL/GroES-dependent protein folding under nonpermissive conditions of macromolecular crowding. Biochemistry. 2002;41:5050–5055. doi: 10.1021/bi015925l. [DOI] [PubMed] [Google Scholar]
- 22.Kityk R., Kopp J., Mayer M.P. Structure and dynamics of the ATP-bound open conformation of Hsp70 chaperones. Mol. Cell. 2012;48:863–874. doi: 10.1016/j.molcel.2012.09.023. [DOI] [PubMed] [Google Scholar]
- 23.del Castillo U., Alfonso C., Muga A. A quantitative analysis of the effect of nucleotides and the M domain on the association equilibrium of ClpB. Biochemistry. 2011;50:1991–2003. doi: 10.1021/bi101670s. [DOI] [PubMed] [Google Scholar]
- 24.Woo K.M., Kim K.I., Chung C.H. The heat-shock protein ClpB in Escherichia coli is a protein-activated ATPase. J. Biol. Chem. 1992;267:20429–20434. [PubMed] [Google Scholar]
- 25.Moro F., Fernández V., Muga A. Interdomain interaction through helices A and B of DnaK peptide binding domain. FEBS Lett. 2003;533:119–123. doi: 10.1016/s0014-5793(02)03752-3. [DOI] [PubMed] [Google Scholar]
- 26.Zylicz M., Yamamoto T., Georgopoulos C. Purification and properties of the dnaJ replication protein of Escherichia coli. J. Biol. Chem. 1985;260:7591–7598. [PubMed] [Google Scholar]
- 27.Reija B., Monterroso B., Zorrilla S. Development of a homogeneous fluorescence anisotropy assay to monitor and measure FtsZ assembly in solution. Anal. Biochem. 2011;418:89–96. doi: 10.1016/j.ab.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 28.Schuck P., Perugini M.A., Schubert D. Size-distribution analysis of proteins by analytical ultracentrifugation: strategies and application to model systems. Biophys. J. 2002;82:1096–1111. doi: 10.1016/S0006-3495(02)75469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.del Castillo U., Fernández-Higuero J.A., Muga A. Nucleotide utilization requirements that render ClpB active as a chaperone. FEBS Lett. 2010;584:929–934. doi: 10.1016/j.febslet.2010.01.029. [DOI] [PubMed] [Google Scholar]
- 30.Hoskins J.R., Doyle S.M., Wickner S. Coupling ATP utilization to protein remodeling by ClpB, a hexameric AAA+ protein. Proc. Natl. Acad. Sci. USA. 2009;106:22233–22238. doi: 10.1073/pnas.0911937106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weibezahn J., Schlieker C., Mogk A. Characterization of a trap mutant of the AAA+ chaperone ClpB. J. Biol. Chem. 2003;278:32608–32617. doi: 10.1074/jbc.M303653200. [DOI] [PubMed] [Google Scholar]
- 32.Diamant S., Ben-Zvi A.P., Goloubinoff P. Size-dependent disaggregation of stable protein aggregates by the DnaK chaperone machinery. J. Biol. Chem. 2000;275:21107–21113. doi: 10.1074/jbc.M001293200. [DOI] [PubMed] [Google Scholar]
- 33.Motohashi K., Watanabe Y., Yoshida M. Heat-inactivated proteins are rescued by the DnaK.J-GrpE set and ClpB chaperones. Proc. Natl. Acad. Sci. USA. 1999;96:7184–7189. doi: 10.1073/pnas.96.13.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zietkiewicz S., Slusarz M.J., Rodziewicz-Motowidło S. Conformational stability of the full-atom hexameric model of the ClpB chaperone from Escherichia coli. Biopolymers. 2010;93:47–60. doi: 10.1002/bip.21294. [DOI] [PubMed] [Google Scholar]
- 35.Fodeke A.A., Minton A.P. Quantitative characterization of polymer-polymer, protein-protein, and polymer-protein interaction via tracer sedimentation equilibrium. J. Phys. Chem. B. 2010;114:10876–10880. doi: 10.1021/jp104342f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rivas G., Minton A.P. Non-ideal tracer sedimentation equilibrium: a powerful tool for the characterization of macromolecular interactions in crowded solutions. J. Mol. Recognit. 2004;17:362–367. doi: 10.1002/jmr.708. [DOI] [PubMed] [Google Scholar]
- 37.Pozdnyakova I., Wittung-Stafsheda P. Non-linear effects of macromolecular crowding on enzymatic activity of multi copper oxidase. Biochim. Biophys. Acta. 2010;1804:1740–1744. doi: 10.1016/j.bbapap.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 38.Wenner J.R., Bloomfield V.A. Crowding effects on EcoRV kinetics and binding. Biophys. J. 1999;77:3234–3241. doi: 10.1016/S0006-3495(99)77154-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wheast R.C. 63rd ed. CRC Press; Boca Raton, FL: 1982. CRC Handbook of Chemistry and Physics. [Google Scholar]
- 40.Dhar A., Samiotakis A., Cheung M.S. Structure, function, and folding of phosphoglycerate kinase are strongly perturbed by macromolecular crowding. Proc. Natl. Acad. Sci. USA. 2010;107:17586–17591. doi: 10.1073/pnas.1006760107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin I., Underhaug J., Muga A. Screening and evaluation of small organic molecules as ClpB inhibitors and potential antimicrobials. J. Med. Chem. 2013;56:7177–7189. doi: 10.1021/jm400499k. [DOI] [PubMed] [Google Scholar]
- 42.Christiansen A., Wittung-Stafshede P. Quantification of excluded volume effects on the folding landscape of Pseudomonas aeruginosa apoazurin in vitro. Biophys. J. 2013;105:1689–1699. doi: 10.1016/j.bpj.2013.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aguilar X., F Weise C., Wittung-Stafshede P. Macromolecular crowding extended to a heptameric system: the Co-chaperonin protein 10. Biochemistry. 2011;50:3034–3044. doi: 10.1021/bi2002086. [DOI] [PubMed] [Google Scholar]
- 44.Werbeck N.D., Schlee S., Reinstein J. Coupling and dynamics of subunits in the hexameric AAA+ chaperone ClpB. J. Mol. Biol. 2008;378:178–190. doi: 10.1016/j.jmb.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 45.Biter A.B., Lee S., Tsai F.T. Structural basis for intersubunit signaling in a protein disaggregating machine. Proc. Natl. Acad. Sci. USA. 2012;109:12515–12520. doi: 10.1073/pnas.1207040109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sarkar M., Smith A.E., Pielak G.J. Impact of reconstituted cytosol on protein stability. Proc. Natl. Acad. Sci. USA. 2013;110:19342–19347. doi: 10.1073/pnas.1312678110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Minton A.P. Quantitative assessment of the relative contributions of steric repulsion and chemical interactions to macromolecular crowding. Biopolymers. 2013;99:239–244. doi: 10.1002/bip.22163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wendler P., Shorter J., Saibil H.R. Atypical AAA+ subunit packing creates an expanded cavity for disaggregation by the protein-remodeling factor Hsp104. Cell. 2007;131:1366–1377. doi: 10.1016/j.cell.2007.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taneva S.G., Moro F., Muga A. Energetics of nucleotide-induced DnaK conformational states. Biochemistry. 2010;49:1338–1345. doi: 10.1021/bi901847q. [DOI] [PubMed] [Google Scholar]
- 50.Zhou H.X., Rivas G., Minton A.P. Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences. Annu. Rev. Biophys. 2008;37:375–397. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weibezahn J., Tessarz P., Bukau B. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell. 2004;119:653–665. doi: 10.1016/j.cell.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 52.Monterroso B., Minton A.P. Effect of high concentration of inert cosolutes on the refolding of an enzyme: carbonic anhydrase B in sucrose and ficoll 70. J. Biol. Chem. 2007;282:33452–33458. doi: 10.1074/jbc.M705157200. [DOI] [PubMed] [Google Scholar]
- 53.Bocanegra R., Alfonso C., Rodriguez-Huete A., Fuertes M.A., Jimenez M., Rivas G., Mateu M.G. Association equilibrium of the HIV-1 capsid protein in a crowded medium reveals that hexamerization during capsid assembly requires a functional C-domain dimerization interface. Biophys. J. 2013;104:884–893. doi: 10.1016/j.bpj.2012.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cole J.L. Analysis of heterogeneous interactions. Methods Enzymol. 2004;384:212–232. doi: 10.1016/S0076-6879(04)84013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zorrilla S., Jiménez M., Minton A.P. General analysis of sedimentation equilibrium in highly nonideal solutions of associating solutes: application to ribonuclease. Biophys. Chem. 2004;108:89–100. doi: 10.1016/j.bpc.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 56.Rivas G., Minton A.P. Beyond the second virial coefficient: sedimentation equilibrium in highly non-ideal solutions. Methods. 2011;54:167–174. doi: 10.1016/j.ymeth.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saroff H.A. Evaluation of uncertainties for parameters in binding studies: the sum-of-squares profile and Monte Carlo estimation. Anal. Biochem. 1989;176:161–169. doi: 10.1016/0003-2697(89)90287-x. [DOI] [PubMed] [Google Scholar]
- 58.Rivas G., Fernández J.A., Minton A.P. Direct observation of the self-association of dilute proteins in the presence of inert macromolecules at high concentration via tracer sedimentation equilibrium: theory, experiment, and biological significance. Biochemistry. 1999;38:9379–9388. doi: 10.1021/bi990355z. [DOI] [PubMed] [Google Scholar]
- 59.Minton A.P. In: Modern Analytical Ultracentrifugation. Schuster T.H., Lave T.H., editors. Birkhauser; Boston, MA: 1994. pp. 81–93. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


