Abstract
Background
The gastric pathogen Helicobacter pylori relies on nickel-containing urease and hydrogenase enzymes in order to colonize the host. Incorporation of Ni2+ into urease is essential for the function of the enzyme and requires the action of several accessory proteins, including the hydrogenase accessory proteins HypA and HypB and the urease accessory proteins UreE, UreF, UreG and UreH.
Methods
Optical biosensing methods (biolayer interferometry and plasmon surface resonance) were used to screen for interactions between HypA, HypB, UreE and UreG.
Results
Using both methods, affinity constants were found to be 5 nM and 13 nM for HypA–UreE and 8 µM and 14 µM for UreG-UreE. Neither Zn2+ nor Ni2+ had an effect on the kinetics or stability of the HypA–UreE complex. By contrast, addition of Zn2+, but not Ni2+, altered the kinetics and greatly increased the stability of the UreE–UreG complex, likely due in part to Zn2+-mediated oligomerization of UreE. Finally our results unambiguously show that HypA, UreE and UreG cannot form a heterotrimeric protein complex in vitro; instead, HypA and UreG compete with each other for UreE recognition.
General significance
Factors influencing the pathogen's nickel budget are important to understand pathogenesis and for future drug design.
Keywords: Metalloenzyme, Metal homeostasis, Hydrogenase, Urease, Nickel, Zinc
1. Introduction
Helicobacter pylori is a spiral, Gram-negative microaerophilic bacterium that has been shown to be the etiological agent of chronic atrophic gastritis and peptic ulcers [1], which can eventually develop into gastric cancers [2]. Since about 50% of the world's population is thought to be infected by the pathogenic bacterium[3], it is considered a major health threat. Despite H. pylori being a neutralophilic bacterium, it is able to persist for decades in the acidic habitat of the human stomach mucosa. In order to achieve this, the pathogen increases the pH in its surrounding microenvironment by using urease, an enzyme that converts urea into ammonia and carbon dioxide [4]. In addition, the gastric pathogen relies on a H2-uptake [NiFe] hydrogenase to survive [5]. Both the urease and the hydrogenase enzymes require nickel (Ni2+) in order to be catalytically active, and the Ni-maturation process involves a battery of accessory proteins.
Urease accessory proteins include UreE, UreF, UreG and UreH, the latter being homologous to UreD of other bacteria [6]. Based on extensive studies done with Klebsiella aerogenes genes expressed in Escherichia coli, UreD, UreF, UreG and UreE are expected to sequentially bind to the urease catalytic enzyme [7].While all four proteins have been well characterized in K. aerogenes, most studies have focused on UreE and UreG in H. pylori. Hp, UreE is a dimeric Ni2+ and Zn2+-binding protein [8–11] thought to be the final Ni2+-donor (to urease). Hp, UreG is a GTPase enzyme [12, 13] and GTP hydrolysis is needed for the activation of urease, since site-directed mutagenesis of the conserved P-loop region of UreG abolished urease activity [13]. Like Hp, UreE, Hp, UreG can also bind Ni2+ but it has a much higher affinity for Zn2+ [12]. While apo-Hp, UreG has been shown to exist as a monomer in solution, interestingly Zn2+ binding, but not Ni2+ binding, appears to promote UreG dimerization [12].
Besides the four UreE GH accessory proteins described above, a unique feature of the H. pylori urease maturation pathway is that two hydrogenase accessory proteins, HypA and HypB, are also required for full urease activity under nickel-limited conditions [14, 15]. Indeed, H. pylori hypA and hypB mutants are deficient in urease activity, a phenotype that can be partially rescued by addition of Ni2+ into the growth medium[14] or by expression of additional UreE engineered to have increased Ni-binding capacity [9]. Various studies in E. coli have suggested that HypA would serve as a scaffold for assembly of the nickel insertion proteins along with the hydrogenase precursor protein after delivery of the iron center [16, 17]. A similar role is expected for H. pylori HypA; previous studies have shown that HypA is found as a dimeric protein in solution, capable of binding two Ni2+ ions per dimer [18]. Additional work indicated that HypA contains two metal sites, an intrinsic Zn2+, site and a low-affinity Ni2+ binding site [19–21]. Similar to Hp, UreG, HypB is a GTPase [18]with a highly conserved P-loop motif whose inactivation by site-directed mutagenesis leads to abolishment of urease activity [13]. However, in contrast to Hp, UreG, only Ni2+ induces the dimerization of Hp, HypB [22].
In vitro and in vivo interactions between H. pylori accessory proteins specifically needed for urease maturation have been reported using an array of different methods. For instance, interactions between H. pylori UreE and UreG were initially reported by Rain and coworkers, using yeast two-hybrid (Y2H) technology [23]. The interaction was later confirmed by Y2H and coimmunoprecipitation [24], tandem affinity purification (TAP) [25], as well as nuclear magnetic resonance (NMR) and isothermal titration calorimetry (ITC) [8]. Results from the latter study suggested that two monomers of UreG could interact with one dimer of UreE, with a dissociation constant KD of 4 µM; the same study also revealed a role for Zn2+ (but not Ni2+) in stabilizing the UreE–UreG complex [8]. Similarly, interactions between H. pylori hydrogenase accessory proteins HypA and HypB were first suggested by Y2H studies [23]; crosslinking studies using purified HypA and HypB subsequently demonstrated that both proteins could interact, regardless of nickel or GTP provided in the reaction mixture [18]. This in vitro interaction was recently confirmed by Xia and coworkers who also showed that the low affinity complex (KD of 52 µM) was present in vivo [26].
Finally, since hydrogenase and urease pathways share common proteins (e.g. HypA and HypB) in H. pylori, it seems that these proteins may interact at some point with urease specific chaperones, such as UreEFGH. Evidence for such interactions was provided a few years ago, when heterologous HypA–UreE and HypB–UreG complexes were observed by cross-linking of purified proteins [27]; the latter interaction was confirmed by another group via a TAP approach [25]. However cross-linking or TAP approaches permanently capture protein interactions and therefore do not provide any kinetic constants. Therefore the aims of the present study were: (i) to determine whether two complementary optical sensing methods, surface plasmon resonance (SPR) and biolayer interferometry (BLI), can be used to provide association and dissociation constants for heterodimeric complexes formed between hydrogenase and urease accessory proteins and (ii) to determine whether these proteins cooperate to form higher order structures (e.g. heterotrimeric or heterotetrameric structures) or compete with one another. First, we were able to confirm that the hydrogenase maturation protein HypA is able to interact in vitro with the urease maturation protein UreE, a critical “bridge’ between both maturation pathways. Dissociation constant calculations indicated that this complex is three orders of magnitude tighter (nM range) than the KD reported for other complexes (e.g. HypA–HypB or UreE–UreG). In addition, the previously described UreE–UreG complex and its dependence upon Zn2+ but not Ni2+ were further characterized. Finally, our results clearly show that HypA–UreE and UreE–UreG complexes compete with one another, and do not form heterotrimeric complexes, further demonstrating the intricate relationship between hydrogenase and urease maturation pathways.
2. Material and methods
2.1. Expression and purification of HypA, HypB, UreE and UreG
Accessory proteins HypA, HypB, UreE or UreG were expressed in E. coli BL21 (DE3) RIL strain (Novagen, Gibbstown, NJ, USA), from a pET21b derivative plasmid, as previously described [9, 13, 18]. Purification strategies were slightly modified from the ones previously published. All proteins were purified using the ÄKTA FPLC purification system (GE Healthcare, Piscataway, NJ). Briefly, HypA, HypB and UreG proteins were purified from cell-free, membrane-free supernatants, using a combination of anion exchange (5-ml Q sepharose column) followed by size exclusion chromatography (Superdex 75 16/60). The buffer used for purification of HypA, HypB and UreG was 20 mM Tris–HCl, pH 7.5 (with NaCl concentrations ranging from 25 mM to 1 M). The recombinant UreE protein was purified from cell-free, membrane-free supernatants, using a combination of cation exchange (5-ml SP Sepharose) followed by size exclusion chromatography (Superdex 75 16/60). The buffer used was 50 mM Hepes, pH 7.2, 1 mM DTT (with NaCl concentrations ranging from 25 mM to 1 M). After the last step, fractions containing the purified proteins were pooled and concentrated using centricon YM-3 devices (Millipore, Billerica, MA) and dialyzed twice against 1 L of buffer containing 50 mM Hepes, pH 7.0, 25 mM NaCl.
2.2. Protein analysis
Proteins were qualitatively analyzed on denaturing 15% SDS-polyacrylamide gels (Fig. S1). Protein concentration was determined with the BCA protein kit (Thermo Scientific Pierce, Rockford, IL) with bovine serum albumin as standard.
2.3. Inductively-coupled plasma emission spectrometry
ICP-MS was used to determine the metal content of each protein and the dialyzing buffer. All experiments were conducted on an Agilent 7500 CE octopole spectrometer. Twelve different elements (Mg, V, Mn, Fe, Co, Ni, Cu, Zn, Mo, W, Pb and U) were measured in triplicate for each protein and the dialyzing buffer. Finally the buffer-subtracted, average value for each metal was used to determine the molar equivalent per monomer.
2.4. Optical biosensing
All BLI measurements were made on a FortéBio (Menlo Park, CA) Octet QK biosensor using streptavidin sensors. Assays were performed in 96-well microplates at 25 °C. All volumes were 200 µL. Ligand proteins were biotinylated using NHS-LC-LC-biotin (succini-midyl-6-[bio-tinamido]-6-hexanamidohexanoate) (Thermo Scientific) at a 5:1 molar ratio of biotin to protein for 30 min at 25 °C followed by rapid exchange into HBS-P (10 mM HEPES, pH 7.4, 150 mM NaCl, 0.01% Surfactant P-20) by passage over a rapid desalting column. Conditions were chosen according to the manufacturer so that each protein was likely randomly biotinylated at an average of 1–2 positions. After loading biotinylated ligand onto SA sensors, a baseline was established in binding buffer, HBS-P with or without addition of 1 mM NiCl2 or ZnCl2prior to association at varying analyte concentrations. After the association phase, sensors were moved to buffer only to monitor dissociation. When metals were present, a subsequent dissociation phase was performed in buffer with 5 mM EDTA. Nonspecific binding to sensors without ligand was negligible (see supplemental information). Raw sensor data were analyzed using GraphPad Prism.
All SPR measurements were made on a Biacore X100 biosensor (GE Healthcare, Piscataway, NJ) using a biotin CAPture chip. Biotinylated ligand was tethered to the chip via the capture reagent prior to injection of analyte. After dissociation, regeneration was achieved in 6 M guanidium chloride and 250 mM NaOH prior to tethering of ligand for subsequent analyte injections.
3. Results
3.1. Protein analysis
HypA, HypB, UreE and UreG were each independently expressed in E. coli and purified to near homogeneity, as visualized on Coomassie-stained SDS-polyacrylamide gels (Fig. S1). The metal content of each protein was determined by ICP-MS. The Ni2+/monomer molar ratios were 0.02, 0.002, 0.004 and 0.002 for HypA, HypB, UreE and UreG, respectively, indicating that each protein was essentially Ni2+-free. Even though no Zn2+ was added to the growth medium, HypA was almost saturated with Zn2+ with a molar ratio of 0.89±0.09 Zn2+ permonomer, while HypB, UreE and UreG had lower amounts of Zn2+ (0.146±0.005, 0.066±0.003 and 0.050±0.005, respectively). As previously reported, H. pylori HypA possesses an intrinsic Zn2+ binding site involving two conserved CXXC motifs and is able to bind one Zn2+ permonomer [20, 21, 28]. Since previous attempts to express Zn2+-free HypA in E. coli [28] or to remove Zn2+ from purified HypA by chelation [21] were reported to be unsuccessful, Zn2+-bound HypA was used throughout this study.
3.2. Initial binding studies
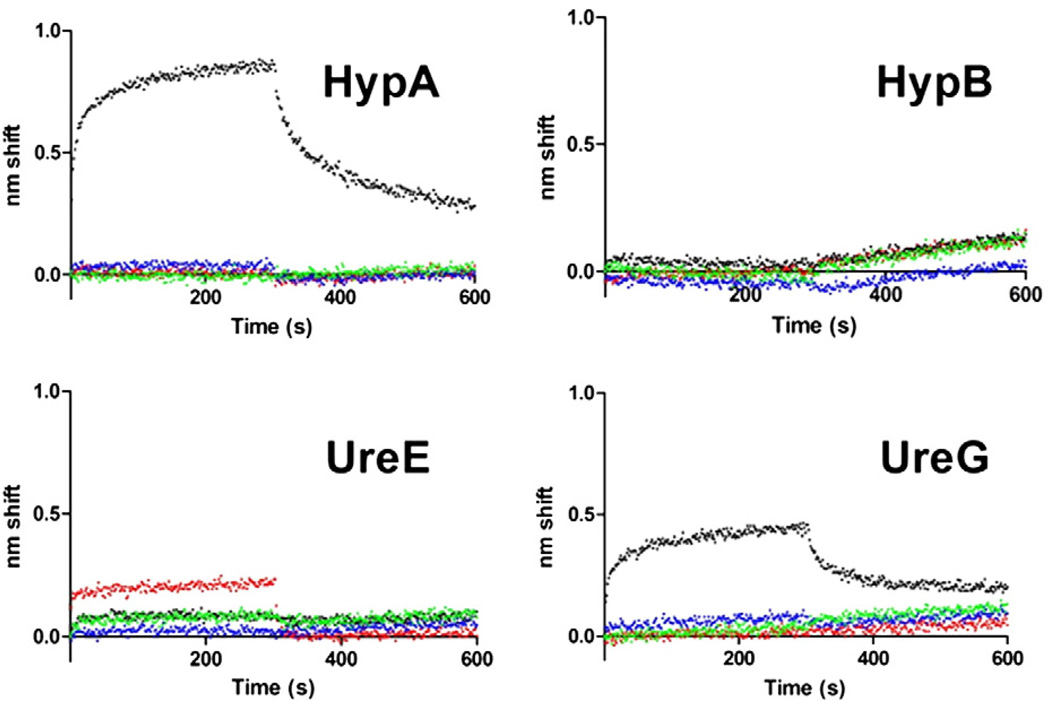
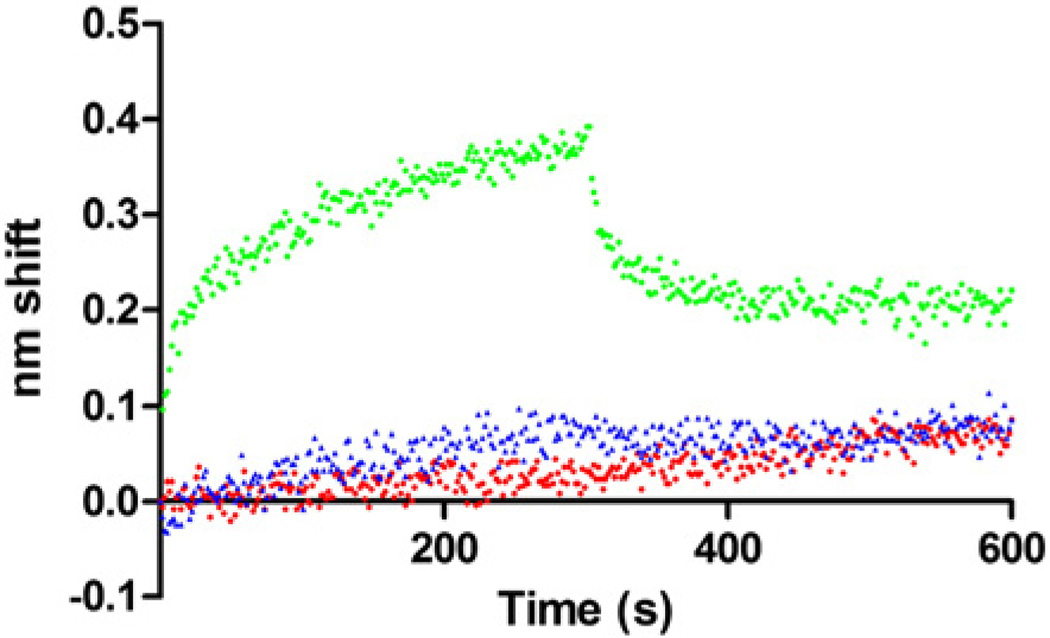
An initial survey of binding among HypA, HypB, UreE and UreG was undertaken (Fig. 1). Each protein was used as biotinylated ligand and exposed to 5 µM of each of the four analyte proteins. Binding was observed between HypA and UreE regardless of which one was tethered to the sensor. Binding between UreE and UreG was observed only when UreG was the ligand. No interaction between HypA and UreG was observed, and there was no binding detected between HypB (either used as ligand or analyte) and any of the three other proteins under the conditions used (Fig. 1).
Fig. 1.
Initial BLI binding survey. For each plot, the indicated ligand was tested for binding against 5 µM HypA (red), HypB (blue), UreE (black) or UreG (green). 0 s corresponds to beginning of association phase. Sensors were moved to buffer only to monitor dissociation at 300 s.
3.3. HypA–UreE interactions
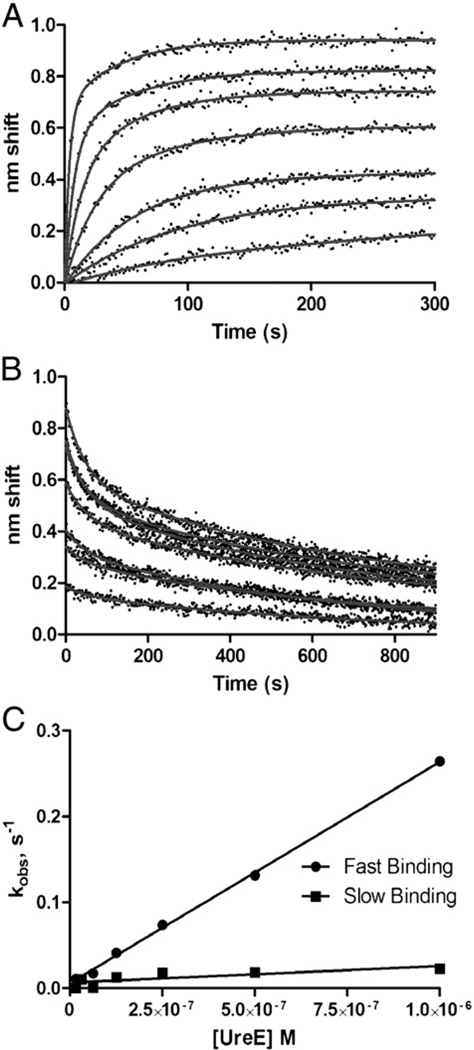
Using BLI and SPR, detailed kinetic analyses were performed to assign association and dissociation rates and affinity constants for the HypA–UreE complex. BLI experiments, using immobilized biotinylated HypA and varying concentrations of UreE analyte, revealed complex binding (Fig. 2). Sensor grams were fit to a two-state parallel model. Most of the amplitudes of association were a result of fast binding though a slow component was also present (Fig. 2A). Two dissociation states were also observed (Fig. 2B) with off rate constants of 0.02 s−1 and 0.0014 s−1. By plotting observed rate constants from the association phases versus UreE analyte concentration, we were able to calculate kon values of 2.6±0.5×105 M−1 s−1 and 1.9±0.7×104 M−1 s−1 (Fig. 2C). Assuming a parallel model, this corresponds to a high affinity KD of 5.4 nM and a low affinity KD of 1.1 µM. Some complexity was likely due to irreversible aggregation on the sensors (see Discussion) and was accounted for in the fits by plateaus in the dissociation phase, the shifts for which range from 0.12 nm (1000 nM UreE) to 0.002 nm (16 nM). Addition of 10-fold molar excess Ni2+ or Zn2+ did not affect HypA–UreE association or dissociation phases (Fig. S2).
Fig. 2.
Kinetic analysis of HypA–UreE binding by BLI. A. Association phase. Sensor-immobilized, biotinylated HypA was exposed to (from bottom to top) 16, 32, 63, 125, 250, 500 or 1000 nM analyte UreE for 300 s. B. Dissociation phase. Immediately after association, sensors were moved to buffer only and dissociation was observed for 300 s. Both association and dissociation were fit to parallel two-state models. Off rate constants were determined to be 0.02 s−1 and 1.4×10−3 s−1. All data are reference-subtracted and nonspecific binding was negligible (data not shown). C. Determination of on rate constants. Observed rate constants determined from fits of the association phase plotted against analyte concentration yield slopes (= kon) of 2.6×105, M−1 s−1 for the fast state and 1.9×104 M−1 s−1 for the slow state.
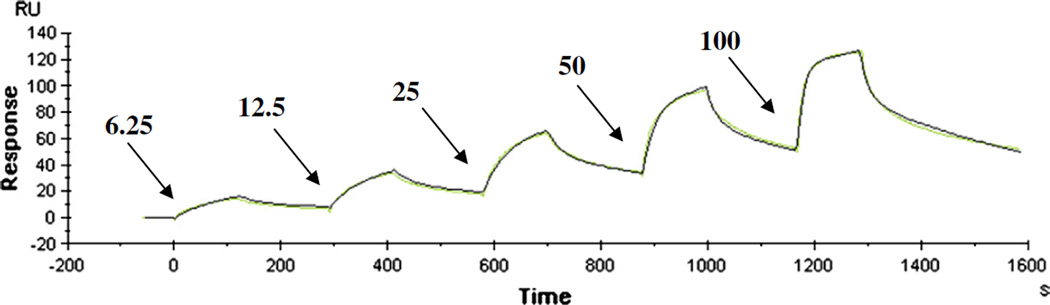
Analysis of HypA–UreE interaction by SPR confirmed complex binding between both proteins. Single-cycle kinetics (Fig. 3) evinced binding that fits a two-state sequential model (A+B↔AB↔AB*) with a fast initial binding followed by slower rearrangement leading to a stable complex (Chi2=2.58). Calculated rate constants were as follows: kon: 2.6×106 M−1 s−1 and koff: 0.15 s−1 for the initial binding; kon: 9×10−3 M−1 s−1 and koff: 0.003 s−1 for the conformational change. The overall KD was 13.5 nM, in good agreement with the high affinity KD determined by BLI analysis. Differences between the constants determined on each platform can be ascribed to various causes such as ligand presentation (see Discussion). However, observed binding on both platforms had a fast initial association and a stable complex described by the low nanomolar KDs.
Fig. 3.
Single-cycle SPR kinetic analysis of HypA–UreE binding. Analyte concentrations of 6.25, 12.5, 25, 50 or 100 nM UreE (as shown by arrows) were injected across a CAP chip with 214 RU tethered biotinylated HypA. Association times were 120 s and were followed by dissociation phases of 180 s prior to the next injection of analyte. Raw data are shown with an overlapping fit to a two-state sequential binding model.
3.4. UreE–UreG interactions
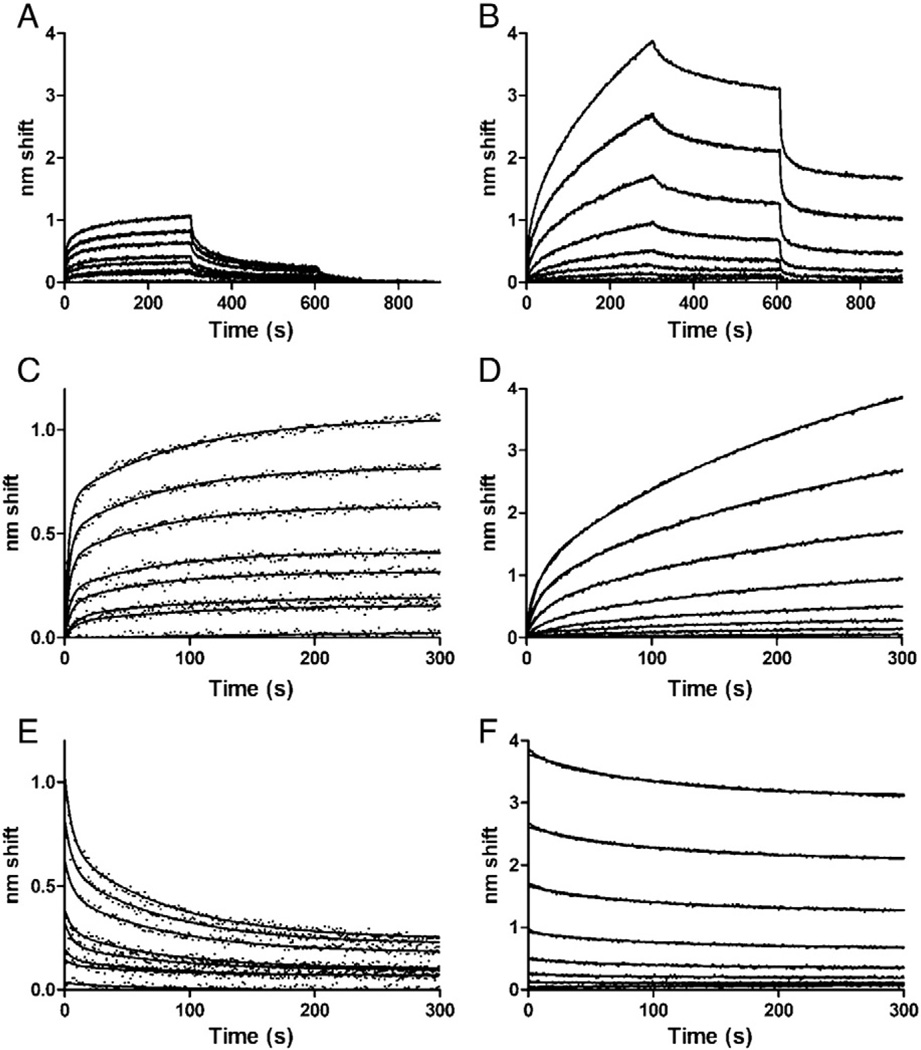
Binding between UreE and UreG was also complex, did not fit a simple model and was dramatically altered by addition of zinc (Fig. 4). First, immobilized UreG was used as ligand and exposed to increasing concentrations of UreE, in the absence (Fig. 4A) or the presence (Fig. 4B) of 1 mM zinc. Three phases were performed; association, dissociation, and then an additional dissociation in the presence of 5 mM EDTA (Fig. 4A and B).
Fig. 4.
BLI analysis of UreG-UreE interactions. A. Binding of ligand UreG to analyte UreE in the absence of zinc. Phases are 0–300 s, association; 300–600 s, dissociation; 600–900 s, dissociation in buffer with 5 mM EDTA. Analyte concentrations were (from bottom to top): 0, 0.312, 0.625, 1.25, 2.5, 5, 10 or 20 µM UreE. B. Binding in the presence of zinc. All conditions and concentrations were the same as in A, except that 1 mM ZnCl2 was added to all phases. C. Association phase in the absence of zinc; fits to two-state parallel model. D. Association phase in the presence of zinc; fits to two-state parallel model. E. Dissociation phase in the absence of zinc; fits to a two-state model. F. Dissociation phase in the presence of zinc; fits to a one-state model with plateaus.
In absence of zinc, association (Fig. 4C) and dissociation (Fig. 4E) phases were fit separately to two-state models, but kobs as a function of UreE analyte concentration were not linear, suggesting additional complexity or perhaps a conformational change (data not shown). Simulations of sequential binding could not account for the data either (data not shown). Off-rates in the absence of zinc were 0.16 s−1 and 0.012 s−1. Association rates could not be calculated due to the complexity of the interaction, but steady-state analysis (Fig. S2) yielded an overall affinity of 14 µM, much weaker than that of HypA–UreE binding (nM range). Complementary analysis of UreE–UreG interaction by SPR was performed (Fig. S3, Panel A). SPR steady-state analysis yielded a KD of 7.6 µM, in general agreement with the BLI data (Fig. S3, Panel B).
In comparing the absence (Fig. 4A) and presence (Fig. 4B) of zinc, higher amplitude binding as well as slower dissociation is evident. Association in presence of zinc was fit to a two state model, although rate constants could not be assigned because of complexity (Fig. 4D). Nevertheless, the initial, rapid binding approximates first order and rates are slower than for the same analyte concentrations in the absence of zinc. These results suggest that initial binding is that of a UreE or UreG monomer whose active population is reduced in the presence of zinc, i.e. zinc-induced oligomerization (see Discussion). Nonspecific binding in the presence of zinc remained negligible (Fig. S4).
A one-state model fit initial dissociation (koff: 0.01 s−1) only when a plateau was included in the model (Fig. 4F). However, plateaus represented ~70% or more of binding amplitude, reflecting a practically non dissociable bound state. When sensors were moved to a second dissociation phase in buffer containing EDTA, very rapid dissociation was observed, indicating that a substantial fraction of the tightly bound complex is zinc-dependent (Fig. 4B). The remainder of the binding is likely aggregation of complexes on the sensor. Complexes transferred directly from association phase to dissociation with EDTA showed the same rapid-off dissociation to a non-zero plateau, indicating that the aging occurred during association (Fig. S4).
In addition, since UreG is a GTPase [13], we investigated whether GTP would enhance or prevent the UreE–UreG complex formation. Addition of 5 mM GTP into the mixture did not have any effect on UreE–UreG binding, as monitored by BLI (Fig. S5).
3.5. Competition between HypA and UreG for UreE binding
Competition binding experiments were performed using BLI, with biotinylated UreG as ligand, UreE (5 µM) as analyte and HypA (10 µM) as competitor, in the absence of added nickel or zinc (Fig. 5). When competing HypA was present, UreE–UreG binding was virtually abolished, suggesting that all UreE proteins were now dedicated to the HypA–UreE complex rather than being involved in UreG-UreE binding (Fig. 5). As a negative control, HypA alone (no UreE analyte) did not bind to UreG (Fig. 5). The competition effect was also observed when using HypA as ligand, UreE as analyte and UreG as free competitor, although in this case HypA–UreE binding was only decreased in the presence of UreG, and was not totally abolished (data not shown). Nevertheless our results unambiguously show that HypA and UreG proteins can compete in vitro for UreE binding, instead of forming a trimeric HypA–UreE–UreG complex.
Fig. 5.
BLI analysis of competition binding. Ligand UreG in presence of UreE analyte only (green circle) or HypA analyte only (blue triangle) or UreE analyte in presence of preequilibrated competing HypA (red square). UreE concentration was 5 µM, competing HypA concentration was 10 µM.
4. Discussion
Optical biosensing methods such as SPR and BLI are now widely used to measure biomolecular interactions in vitro. While SPR remains the biosensing method of choice, there is growing interest in BLI as an alternate method. Like SPR, BLI measures surface-induced light refractions, however it usually avoids refractive changes unrelated to binding [29, 30]. In this study we used both complementary methods to analyze interactions between four H. pylori accessory proteins previously shown to be needed either for hydrogenase and urease maturation (HypA and HypB) or solely for urease maturation (UreE and UreG). A preliminary BLI survey among the four proteins of interest in this study revealed the formation of two protein complexes, HypA–UreE and UreE–UreG. Binding between HypA and UreE was observed regardless of which protein was ligand or analyte. Differences in amplitude between the ligand-analyte pairs are ascribable to differences in amount of ligand immobilized, analyte masses or active populations. Binding between UreE and UreG was observed only when UreG was the ligand. Both complexes were expected because they had been previously reported by several groups, using various detection methods [8, 23–25, 27]. Based on previously published studies, we were also expecting interactions between HypA and HypB [18, 23, 26] as well as between HypB and UreG [25, 27]. Despite several attempts using various batches of purified HypB protein, neither optical biosensing method (BLI or SPR) revealed any interaction involving HypB. However, failure to observe binding does not allow the conclusion that the proteins do not bind, as there are reasons why otherwise interacting proteins may give a negative result via biosensing, e.g. biotinylation could alter protein- or metal-binding sites.
Analysis of HypA–UreE and UreE–UreG interactions revealed complex binding, with fast and slow states of association and dissociation. In the case of HypA and UreE analyzed by BLI, binding was fit by a parallel model which ignores non-dissociable binding. The irreversibly bound state was not nonspecific binding, as the analyte bound negligibly to sensors not loaded with ligand; rather, this reflected an aging of HypA–UreE complexes on the sensors, as previously reported for other proteins [31, 32]. Two-state binding was also observed in SPR and was thus unlikely to be solely an artifact of the method, though it could reflect heterogeneous populations. With an unknown stoichiometry, it is possible that multiple binding sites exist. The appreciable off rate correlates with results from a previous study indicating that cross-linking was needed to capture the HypA–UreE interaction [27]. Addition of nickel or zinc did not affect the formation or stability of the HypA–UreE complex, suggesting that neither metal is required for such interaction, in agreement with previous results indicating that nickel was not needed for interaction between both proteins [27]. However, given that the recombinant HypA protein used in the present study was zinc-saturated, the lack of a zinc effect on this particular complex must be analyzed with caution.
SPR analysis of HypA–UreE yielded excellent fits to a sequential (conformational change) model. Given the tethering strategy (biotinylated protein bound to a streptavidin-conjugated oligonucleotide in turn bound to its complement on a dextran matrix) it is likely that the ligand had more freedom than when it was attached to the BLI sensors. Therefore we consider the SPR analysis more likely reflective of the biologically relevant complexity. The same interaction may be happening in BLI, but it may be masked by other events such as anomalies of immobilization and the aging of complexes into irreversibly bound states. Regardless of the complexities inherent in the binding, both methods correlate to low nM KD that results from fast initial binding and relatively slow off rates.
UreG-UreE binding was even more complex in that neither parallel nor sequential models explained the data. While parallel fits are shown in Fig. 4A, C, and E, they allow for plateaus in the dissociation phase which represent up to ~25% of total binding. Another likely reason for complexity was heterogeneity of both ligand and analyte. There was some EDTA-induced rapid dissociation in the no-zinc added experiment, reflecting a small population of stably bound complexes containing endogenous zinc or other metal. Indeed, ICP-MS results showed small fractions of the purified UreE and UreG proteins were zinc-bound. Other factors, including ligand presentation anomalies, may also affect binding.
BLI and SPR steady-state analysis of UreE–UreG binding revealed KD of 7.6 µM and 14 µM respectively, in good agreement with previously published results [8] and much weaker than that of HypA–UreE binding (nM range). In addition, BLI experiments showed a clear effect of zinc on stabilizing the UreE–UreG complex. Our results confirmed those of Bellucci et al., who used size-exclusion chromatography, light scattering and ITC experiments to show that zinc was stabilizing the UreE–UreG complex [8]. This stabilizing role was in part attributed to previously shown zinc-induced UreG dimerization [12]. In the BLI UreE–UreG binding experiments described herein, UreG was used as biotinylated ligand and therefore bound to SA sensor. This condition would theoretically inhibit zinc-induced protein dimerization, but this is a possibility worth considering. Oligomerization on the sensor as multiple analytes bind is another distinct possibility, one supported by the observation than zinc does not significantly alter HypA–UreE binding (Fig. S2). The difference in amplitude of the association phases relative to zinc could also be due to other factors, including oligomerization of UreE. Such oligomerization would correlate with the observed reduced rates of initial binding (Fig. 4D; slower diffusion of larger order structures) and the very slow dissociation (Fig. 4F; higher affinity complexes), in stark contrast to the rapid no-zinc dissociation (Fig. 4E). Previous studies have shown that Hp UreE is usually found as a dimer in solution [8, 9]; in addition crystal structures of metal-bound forms of HpUreE revealed that dimers of UreE dimers (“kissing” dimers) can associate around the metal ion (Cu2+, Ni2+ or Zn2+) through coordination by His102 residue [10]. Similar tetrameric forms have been reported in the case of Zn2+- bound UreE from Bacillus pasteurii [33,34] or Cu2+, Ni2+, or Zn2+- bound UreE from K. aerogenes [35]. However such UreE “kissing dimers” would hinder rather than favor UreE–UreG interaction. In addition, zinc-bound UreG could also induce oligomerization of UreE. Finally, EDTA-driven dissociation of zinc-stabilized complexes demonstrates zinc dependence. The rapid loss of much of the binding probably reflects all the complexes capable of dissociation, the remainder being the age don aggregates observed elsewhere [31, 32].
HypA and UreG compete for binding to UreE. Addition of HypA abolished UreE–UreG binding (Fig. 5), while addition of UreG only partially decreased the formation of HypA–UreE complex (data not shown). Several conclusions can be drawn from this study. First, when HypA, UreE and UreG are present, our results suggest that the HypA–UreE complex would be favored over the UreE–UreG complex, in agreement with the KD values calculated in this study: the HypA–UreE complex is much tighter, with KD approximately 3 orders magnitude lower than that of UreE–UreG (KD between 4 and 14 µM; this study and [8]). Second, the mutual exclusion of both protein complexes suggests that binding sites may overlap. Third, there was no evidence for a ternary complex since response was always lower in the presence of competitor; indeed a HypA–UreE–UreG complex would be expected to generate more signal due to larger mass. Therefore this implicitly suggests that both dimeric complexes (HypA–UreE vs UreE–UreG) happen sequentially and this may influence the maturation of one Ni-sink (hydrogenase) over another (urease). Indeed, HypA is also a hydrogenase accessory protein (besides being a urease accessory protein)while UreE and UreG are not.
Examination of the effects of zinc and other metals on competition may elucidate some details of the maturation pathways and will be pursued in future studies. For example, the bacterium is likely faced with different conditions in which urease versus hydrogenase maturation is advantageous and cytoplasmic metal levels may influence this metal partitioning.
5. Conclusion
These results shed light on the intertwined relationships between both Ni-enzyme maturation pathways (hydrogenase and urease) in H. pylori. The hydrogenase and urease maturation protein HypA can compete in vitro with the urease maturation protein UreG for recognition of the urease accessory protein UreE, rather than making a trimeric complex HypA–UreE–UreG.
Supplementary Material
Acknowledgements
The authors are grateful to Andrew Lancaster and Mike W. Adams (University of Georgia) for the ICP-MS analysis. This work was supported by NIH RO1 DK062852, NIHR15 GM080701 and NSF CHE0922699 and by the Georgia Foundation.
Abbreviations
- BLI
biolayer interferometry
- SPR
Surface Plasmon Resonance
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.bbagen.2012.06.002.
References
- 1.Blaser MJ. The role of Helicobacter pylori in gastritis and its progression to peptic ulcer disease. Aliment. Pharmacol. Ther. 1995;9(Suppl. 1):27–30. doi: 10.1111/j.1365-2036.1995.tb00780.x. [DOI] [PubMed] [Google Scholar]
- 2.Sipponen P, Hyvarinen H, Seppala K, Blaser MJ. Review article: pathogenesis of the transformation from gastritis to malignancy. Aliment. Pharmacol. Ther. 1998;12(Suppl. 1):61–71. doi: 10.1111/j.1365-2036.1998.00005.x. [DOI] [PubMed] [Google Scholar]
- 3.Covacci A, Telford JL, Del Giudice G, Parsonnet J, Rappuoli R. Helicobacter pylori virulence and genetic geography. Science. 1999;284:1328–1333. doi: 10.1126/science.284.5418.1328. [DOI] [PubMed] [Google Scholar]
- 4.Scott DR, Marcus EA, Weeks DL, Sachs G. Mechanisms of acid resistance due to the urease system of Helicobacter pylori . Gastroenterology. 2002;123:187–195. doi: 10.1053/gast.2002.34218. [DOI] [PubMed] [Google Scholar]
- 5.Olson JW, Maier RJ. Molecular hydrogen as an energy source for Helicobacter pylori . Science. 2002;298:1788–1790. doi: 10.1126/science.1077123. [DOI] [PubMed] [Google Scholar]
- 6.Cussac V, Ferrero RL, Labigne A. Expression of Helicobacter pylori urease genes in Escherichia coli grown under nitrogen-limiting conditions. J. Bacteriol. 1992;174:2466–2473. doi: 10.1128/jb.174.8.2466-2473.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carter EL, Flugga N, Boer JL, Mulrooney SB, Hausinger RP. Interplay of metal ions and urease. Metallomics. 2009;1:207–221. doi: 10.1039/b903311d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bellucci M, Zambelli B, Musiani F, Turano P, Ciurli S. Helicobacter pylori UreE, a urease accessory protein: specific Ni(2+)− and Zn(2+)−binding properties and interaction with its cognate UreG. Biochem. J. 2009;422:91–100. doi: 10.1042/BJ20090434. [DOI] [PubMed] [Google Scholar]
- 9.Benoit S, Maier RJ. Dependence of Helicobacter pylori urease activity on the nickel-sequestering ability of the UreE accessory protein. J. Bacteriol. 2003;185:4787–4895. doi: 10.1128/JB.185.16.4787-4795.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi R, Munger C, Asinas A, Benoit SL, Miller E, Matte A, Maier RJ, Cygler M. Crystal structures of apo and metal-bound forms of the UreE protein from Helicobacter pylori: role of multiple metal binding sites. Biochemistry. 2010;49:7080–7088. doi: 10.1021/bi100372h. [DOI] [PubMed] [Google Scholar]
- 11.Banaszak K, Martin-Diaconescu V, Bellucci M, Zambelli B, Rypniewski W, Maroney MJ, Ciurli S. Crystallographic and X-ray absorption spectroscopic characterization of Helicobacter pylori UreE bound to Ni2+ and Zn2+ reveal a role for the disordered C-terminal arm in metal trafficking. Biochem. J. 2011;441:1017–1026. doi: 10.1042/BJ20111659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zambelli B, Turano P, Musiani F, Neyroz P, Ciurli S. Zn2+-linked dimerization of UreG from Helicobacter pylori, a chaperone involved in nickel trafficking and urease activation. Proteins. 2009;74:222–239. doi: 10.1002/prot.22205. [DOI] [PubMed] [Google Scholar]
- 13.Mehta N, Benoit S, Maier RJ. Roles of conserved nucleotide-binding domains in accessory proteins, HypB and UreG, in the maturation of nickel-enzymes required for efficient Helicobacter pylori colonization. Microb. Pathog. 2003;35:229–234. doi: 10.1016/s0882-4010(03)00151-7. [DOI] [PubMed] [Google Scholar]
- 14.Olson JW, Mehta NS, Maier RJ. Requirement of nickel metabolism proteins HypA and HypB for full activity of both hydrogenase and urease in Helicobacter pylori . Mol. Microbiol. 2001;39:176–182. doi: 10.1046/j.1365-2958.2001.02244.x. [DOI] [PubMed] [Google Scholar]
- 15.Benoit SL, Zbell AL, Maier RJ. Nickel enzyme maturation in Helicobacter hepaticus: roles of accessory proteins in hydrogenase and urease activities. Microbiology. 2007;153:3748–3756. doi: 10.1099/mic.0.2007/010520-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forzi L, Sawers RG. Maturation of [NiFe]-hydrogenases in Escherichia coli . Biometals. 2007;20:565–578. doi: 10.1007/s10534-006-9048-5. [DOI] [PubMed] [Google Scholar]
- 17.Kaluarachchi H, Chan Chung KC, Zamble DB. Microbial nickel proteins. Nat. Prod. Rep. 2010;27:681–694. doi: 10.1039/b906688h. [DOI] [PubMed] [Google Scholar]
- 18.Mehta N, Olson JW, Maier RJ. Characterization of Helicobacter pylori nickel metabolism accessory proteins needed for maturation of both urease and hydrogenase. J. Bacteriol. 2003;185:726–734. doi: 10.1128/JB.185.3.726-734.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herbst RW, Perovic I, Martin-Diaconescu V, O'Brien K, Chivers PT, Pochapsky SS, Pochapsky TC, Maroney MJ. Communication between the zinc and nickel sites in dimeric HypA: metal recognition and pH sensing. J. Am. Chem. Soc. 2010;132:10338–10351. doi: 10.1021/ja1005724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia W, Li H, Sze KH, Sun H. Structure of a nickel chaperone, HypA, from Helicobacter pylori reveals two distinct metal binding sites. >J. Am. Chem. Soc. 2009;131:10031–10040. doi: 10.1021/ja900543y. [DOI] [PubMed] [Google Scholar]
- 21.Kennedy DC, Herbst RW, Iwig JS, Chivers PT, Maroney MJ. A dynamic Zn site in Helicobacter pylori HypA: a potential mechanism for metal-specific protein activity. J. Am. Chem. Soc. 2007;129:16–17. doi: 10.1021/ja066958x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sydor AM, Liu J, Zamble DB. Effects of metal on the biochemical properties of Helicobacter pylori HypB, a maturation factor of [NiFe]-hydrogenase and urease. J. Bacteriol. 2011;193:1359–1368. doi: 10.1128/JB.01333-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rain JC, Selig L, De Reuse H, Battaglia V, Reverdy C, Simon S, Lenzen G, Petel F, Wojcik J, Schachter V, Chemama Y, Labigne A, Legrain P. The protein–protein interaction map of Helicobacter pylori . Nature. 2001;409:211–215. doi: 10.1038/35051615. [DOI] [PubMed] [Google Scholar]
- 24.Voland P, Weeks DL, Marcus EA, Prinz C, Sachs G, Scott D. Interactions among the seven Helicobacter pylori proteins encoded by the urease gene cluster. Am. J. Physiol. Gastrointest. Liver Physiol. 2003;284:G96–G106. doi: 10.1152/ajpgi.00160.2002. [DOI] [PubMed] [Google Scholar]
- 25.Stingl K, Schauer K, Ecobichon C, Labigne A, Lenormand P, Rousselle JC, Namane A, de Reuse H. In vivo interactome of Helicobacter pylori urease revealed by tandem affinity purification. Mol. Cell. Proteomics. 2008;7:2429–2441. doi: 10.1074/mcp.M800160-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Xia W, Li H, Yang X, Wong KB, Sun H. Metallo-GTPase HypB from Helicobacter pylori and its interaction with nickel chaperone protein HypA. J. Biol. Chem. 2012;287:6753–6763. doi: 10.1074/jbc.M111.287581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benoit SL, Mehta N, Weinberg MV, Maier C, Maier RJ. Interaction between the Helicobacter pylori accessory proteins HypA and UreE is needed for urease maturation. Microbiology. 2007;153:1474–1482. doi: 10.1099/mic.0.2006/003228-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atanassova A, Zamble DB. Escherichia coli HypA is a zinc metalloprotein with a weak affinity for nickel. J. Bacteriol. 2005;187:4689–4697. doi: 10.1128/JB.187.14.4689-4697.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abdiche Y, Malashock D, Pinkerton A, Pons J. Determining kinetics and affinities of protein interactions using a parallel real-time label-free biosensor, the Octet. Anal. Biochem. 2008;377:209–217. doi: 10.1016/j.ab.2008.03.035. [DOI] [PubMed] [Google Scholar]
- 30.Concepcion J, Witte K, Wartchow C, Choo S, Yao D, Persson H, Wei J, Li P, Heidecker B, Ma W, Varma R, Zhao LS, Perillat D, Carricato G, Recknor M, Du K, Ho H, Ellis T, Gamez J, Howes M, Phi-Wilson J, Lockard S, Zuk R, Tan H. Label-free detection of biomolecular interactions using BioLayer interferometry for kinetic characterization. Comb. Chem. High Throughput Screening. 2009;12:791–800. doi: 10.2174/138620709789104915. [DOI] [PubMed] [Google Scholar]
- 31.McMurry JL, Chrestensen CA, Scott IM, Lee EW, Rahn AM, Johansen AM, Forsberg BJ, Harris KD, Salerno JC. Rate, affinity and calcium dependence of nitric oxide synthase isoform binding to the primary physiological regulator calmodulin. FEBS J. 2011;278:4943–4954. doi: 10.1111/j.1742-4658.2011.08395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morris DP, Roush ED, Thompson JW, Moseley MA, Murphy JW, McMurry JL. Kinetic characterization of Salmonella FliK–FlhB interactions demonstrates complexity of the type III secretion substrate-specificity switch. Biochemistry. 2010;49:6386–6393. doi: 10.1021/bi100487p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ciurli S, Safarov N, Miletti S, Dikiy A, Christensen SK, Kornetzky K, Bryant DA, Vandenberghe I, Devreese B, Samyn B, Remaut H, van Beeumen J. Molecular characterization of Bacillus pasteurii UreE, a metal-binding chaperone for the assembly of the urease active site. J. Biol. Inorg. Chem. 2002;7:623–631. doi: 10.1007/s00775-002-0341-7. [DOI] [PubMed] [Google Scholar]
- 34.Remaut H, Safarov N, Ciurli S, Van Beeumen J. Structural basis for Ni(2+) transport and assembly of the urease active site by the metallochaperone UreE from Bacillus pasteurii . >J. Biol. Chem. 2001;276:49365–49370. doi: 10.1074/jbc.M108304200. [DOI] [PubMed] [Google Scholar]
- 35.Grossoehme NE, Mulrooney SB, Hausinger RP, Wilcox DE. Thermodynamics of Ni2+, Cu2+, and Zn2+ binding to the urease metallochaperone UreE. Biochemistry. 2007;46:10506–10516. doi: 10.1021/bi700171v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.