Summary
Background
Studies show that high phosphotidylinositol 3,4,5 tris phosphate (PIP3) promotes cytoskeletal rearrangements and alters cell motility and chemotaxis, possibly through activation of PKBs. However, chemotaxis can still occur in the absence of PIP3 and the identities of the PIP3 independent pathways remain unknown.
Results
Here, we outline a PIP3-independent pathway linking temporal and spatial activation of PKBs by Tor complex 2 (TorC2) to the chemotactic response. Within seconds of stimulating Dictyostelium cells with chemoattractant, two PKB homologs, PKBA and PKBR1, mediate transient phosphorylation of at least eight proteins, including Talin, PI4P 5-kinase, two RasGefs, and a RhoGap. Surprisingly, all of the substrates are phosphorylated with normal kinetics in cells lacking PI 3-kinase activity. Cells deficient in TorC2 or PKB activity show reduced phosphorylation of the endogenous substrates and are impaired in chemotaxis. The PKBs are activated through phosphorylation of their hydrophobic motifs via TorC2 and subsequent phosphorylation of their activation loops. These chemoattractant-inducible events restricted to the cell’s leading edge even in the absence of PIP3. Activation of TorC2 depends on heterotrimeric G-protein function and intermediate G-proteins, including Ras GTPases.
Conclusions
The data lead to a model where cytosolic TorC2, encountering locally activated small G-protein(s) at the leading of the cell, becomes activated and phosphorylates PKBs. These in turn phosphorylate a series of signaling and cytoskeletal proteins, thereby regulating directed migration.
Keywords: TorC2, PKB, chemotaxis
Introduction
Chemotaxis, the directed movement of cells along chemical gradients, is a fundamental cellular response critical to numerous biological processes, including embryogenesis, inflammation, and wound healing. An understanding of chemotaxis could lead to treatments for inflammatory diseases as well as methods to control cancer metastasis. The basic issue in chemotaxis is how shallow chemoattractant gradients differentially bias pseudopod activity at the front and rear of the cell to direct movement. Previous studies have shown that chemoattractant receptors are uniformly distributed along the membrane but that activation of signaling pathways can be confined at the cell’s leading edge. Current research is focused on the mechanisms that spatially restrict signaling and on the events that link local activation to regulation of the cytoskeleton.
Dictyostelium discoideum is the most extensively characterized model system for chemotaxis and its study has provided important insights into the process in highly motile cells such as leukocytes. Chemoattractant signaling triggers a series of stereotypical reactions that regulate cell motility, polarity, and directional sensing. One well-studied reaction is accumulation of phosphatidylinositol-3,4,5-trisphosphate (PIP3) [1]. In D. discoideum, PIP3 accumulation occurs at the front of chemotaxing cells because PI3K is recruited to the membrane facing the chemoattractant source and PTEN dissociates from this region [2, 3]. In pten− cells, excess PIP3 diffuses back from the leading edge, the cells extend pseudopods around much of their perimeter, and lose polarity. In neutrophils, chemoattractants similarly promote PIP3 accumulation at the leading edge and forced elevation of PIP3 is sufficient to cause the cells to polarize [T. Inoue, personal communication, 4, 5]. While these observations demonstrate the involvement of PIP3 signaling in chemotaxis, studies suggest there are additional pathways. For instance, D. discoideum cells treated with the PI3K inhibitor LY294002 as well as pi3k1-5− cells, in which all of the type-I phosphoinositide 3-kinases are disrupted, are still able to carry out chemotaxis [6, 7]. Furthermore, neutrophils from PI3Kγ knockout mice are able to carry out chemotaxis under certain conditions [8, 9]. Recently, it was revealed that a pathway containing a phopholipase A2 (PLA2) functions in parallel with the PIP3 pathway in D. discoideum [10, 11]. Thus, it is likely that in most cells multiple pathways participate in a network that mediates chemotaxis.
A critical regulator of chemotaxis, found through genetic analysis in D. discoideum is Pianissimo (PiaA), now known to be a component of the “Tor complex 2” (TorC2) [12, 13]. PiaA and other subunits, Rip3 and Lst8, are the orthologs of mammalian Rictor, mSin1, and mLst8, respectively, and form a complex with a unique Tor kinase [13]. TorC2 may also regulate cytoskeletal-based events in other cells. In cultured cells knockdown of a subunit of TorC2 altered cell morphology indicating that it may regulate the actin meshwork [14, 15]. Knockout mice lacking TorC2 subunits die in embryogenesis although fibroblasts from these embryos did not show cytoskeletal defects [16, 17]. In addition, TorC2 controls cell cycle dependent actin polarization in budding yeast [18]. One biochemical role of TorC2 is phosphorylation of the hydrophobic motifs (HMs) of PKBs and PKCs [15, 19]. In knockout mice of mSin1, Rictor, or Lst8 [20], the hydrophobic motif of AKT1 was not phosphorylated [16, 17, 21]. Surprisingly, in these studies HM phosphorylation was not required for phosphorylation of all PKB substrates in vivo.
D. discoideum has two PKB homologs, PKBA and PKBR1 [22, 23]. Whereas PKBA has a PIP3-specific PH domain at its N-terminus like the mammalian PKBs, PKBR1 is myristoylated, persistently localized on the membrane, and relatively independent of PIP3. Otherwise, the two enzymes are highly homologous to mammalian PKBs, including within the activation loops (ALs) and the HMs. Cells lacking PKBA are reported to have a weak chemotaxis defect in early development [22]. While pkbR1− cells arrest development after aggregation, they are reported to have little defect in chemotaxis [13, 23]. However, in light of the observations of PIP3 independent chemotaxis, we examined the role of TorC2 and PKBR1 in more detail. These studies led us to a PIP3-independent pathway by which receptor/G-protein stimulation leads to activation of TorC2 and phosphorylation of the HMs in the PKBs. Our findings show that activation of TorC2 involves a RasGEF and an intermediate G-protein and occurs selectively at the leading edge of the cell. Importantly, we find PKB substrates include Talin B, several RasGEFs, a RhoGAP, and PI4P 5-kinase providing possible links to the cytoskeleton.
Results
Role of TorC2 and PKBR1 in chemotaxis
To assess the role of TorC2 and PKBR1 in chemotaxis, we assessed the ability of disruptants to respond to a gradient formed by a cAMP-filled micropipette (Figure 1, movies 1A, 1B, 1C. Wild-type cells responded to the gradient while most pkbR1− cells ignored the gradient and crawled randomly. Compared to wild-type cells, the pkbR1− cells display greatly reduced chemotactic index in spite of a slightly increased speed (Figure 1B and Table 1). Interestingly, detachment of the fronts from the coverslip, a rare phenomenon in wild-type cells, was very prevalent in pkbR1− cells (see Movies 1A and B). Together these results show that pkbR1− cells have a strong defect in chemotaxis which may involve a problem in adhesion. We noted that when pkbR1− cells were cultured for several months, the chemotactic behavior seemed to ameliorate somewhat, possibly due to upregulation of PKBA (data not shown and see below). However, disruptants of PKBA alone showed little chemotactic defect. We should note that our results conflict with previous descriptions of pkbA− and pkbR1− cells [22, 23]. This is possibly due to differences in strains or conditions used in the respective laboratories, nevertheless, these differences have important implications for the mechanism of PIP3-independent chemotaxis as outlined below.
Figure 1. Chemotaxis of WT, pkbR1− and piaA− cells in the micropipette assay.
(A) Wild-type (WT), pkbR1− cells, and piaA− cells were spread on a coverslip and observed at 30 sec intervals for 30 min. Micropipette contains 10 µM cAMP. First and last images are shown. Scale bars represent 50 µm. (B) Tracing of individual cells are shown. (◆) the position of micropipette (C) Distributions of motility speed (black bar) and chemotaxis speed (white bar) are compared from (A).
Table 1.
Quantification of cellular behavior
| strain | Motility speed (µm / min) | Chemotactic index |
|---|---|---|
| AX-3 (wild-type) | 4.9 ± 1.5 (n=51) | 0.65 ± 0.24 |
| pkbR1− | 5.3 ± 2.2 (n=48) | 0.04 ± 0.35 |
| piaA− | 3.7 ± 1.1 (n=52) | 0.31 ± 0.28 |
Motility speed and chemotactic index are calculated by the definition in Experimental Procedures.
To show the chemotaxis defects were specific, we expressed PKBR1 in the pkbR1− cells. Vector expressing pkbR1− cells showed reduced directionality towards the micropipette even after 30 min of exposure to the gradient Figure S1A, left and Movie S1A left). On the other hand, PKBR1 expressing pkbR1− cells showed significantly better chemotaxis, most of the cells faced the micropipette during this assay (Figure S1A right, right and Movie S1A right). Interestingly, the PKBR1 expressing cells took slightly longer than wild-type cells to begin chemotaxis after the micropipette was placed and then they became hyperelongated. These were consistent dominant effects of (over)expression of PKBR1, since the same phenotypes were observed when PKBR1 was expressed in wild-type cells (Figure S1B and Movie S1B left and right).
Since TorC2 is required for the activation of PKBR1, we directly compared the phenotypes of fresh piaA− and pkbR1− cells in the same background. As previously reported, the piaA− cells displayed very impaired chemotaxis with both reduced speed and chemotactic index (Figure 1, Movie 1C, and Table 1). The piaA− cells had a rounded morphology and lacked polarity, which to some extent distinguished them from the polarized pkbR1− cells [12, 13]. This suggests that TorC2 may regulate other factor(s) in addition to PKBR1 to establish polarity.
Chemoattractant-mediated phosphorylation of PKB substrates
To get at the origin of the chemotactic defects, we sought to define the PKB substrates. We used antibodies directed against the phosphorylated consensus motifs of PKB and a variety of other kinases (as controls) to detect proteins modified following chemoattractant stimulation. Cell extracts were prepared for immunoblot analysis at the indicated times after stimulation (Figure 2A). For PKB, multiple bands appeared and declined with different kinetics (Figure 2B). To display the rapid kinetics more clearly, samples were taken at closer intervals. A number of early bands such as pp65/67 and pp90 peaked within 20–60 sec of cAMP addition (Figure 2B right), marginally slower than activation of PKBA or PKBR1 reported previously [22, 23]. The slight delay may stem from our procedure of keeping cells on ice to prevent them from spontaneously responding before stimulation (see Methods). These signals were specific receptor-mediated phosphorylation events. First, the other substrate phosphospecific antibodies showed completely different patterns Figure S2A. In particular, for CDK, many phosphorylated proteins were stained in resting cells and this pattern did not change substantially after stimulation. Second, there were no bands appearing in the absence of stimulation Figure S2B. Third, in subcellular fractionation experiments, we noticed that the signals were greatly reduced in the absence of phosphatase inhibitor cocktails (data not shown).
Figure 2. The TorC2-PKBR1 pathway regulates a major portion of Dictyostelium PKB activity.
(A) Outline of experimental procedure is shown. Differentiated cells were stimulated with 1 µM cAMP and sampled for immunoblot analysis at the indicated time points. All gels were analyzed with α-phosphospecific PKB substrate antibody that recognizes R-X-R-X-X-S/T-X-X in a phosphorylation dependent manner (R: arginine, S: Serine, T: threonine, X: any amino acid). (B) The left panel is a time course to 11 min with molecular weight markers. The right panel is a time course to 120 seconds with major phospho-proteins marked. (C) Comparison of wild-type (WT) and pkbA− cells is shown. (D) Comparison of WT, pkbR1−, and piaA− cells is shown.
The relative contributions of PKBA and PKBR1 to these chemoattractant-induced phosphorylations were strongly correlated with the chemotactic defects (or lack there of) in the pkbR1− and pkbA− cells. As shown in Figure 2C, most proteins were still phosphorylated in the pkbA− cells, indicating that PKBA is not primarily responsible for these modifications. On the other hand, in pkbR1− cells, the signals from pp350, pp280, pp200, pp180, pp110, pp90, and pp65/67 were strongly reduced compared to those in wild-type cells (Figure 2D and S2C). In pkbA− / pkbR1− cells the bands were further reduced (data not shown), suggesting overlap of substrate specificity. Some bands (i.e. pp30) were unaffected in either single or double mutants and we presume these are not PKB substrates. We next determined which bands were recovered when PKBR1 was expressed in pkbR1− cells. Seven signals (pp350, pp280, pp200, pp180, pp110, pp90, and pp65/67) were recovered and an additional band, pp140, also appeared (Figure S2D right). These results identify eight putative substrates of PKBR1 and/or PKBA in vivo and show that chemoattractant is an important regulator of these kinases.
Chemoattractant-induced phosphorylation of Talin, PI4P 5-kinase, two RasGefs, and a RhoGap
We identified five of the putative PKB substrates using an immunoprecipitation strategy followed by SDS gel separation and mass spectrometry (Figure S3). One of the proteins, pp280, was identified as Talin B through recovery of four peptides in mass spectrometry from the purified band (Figure 3 and S3A). Importantly, Talin B (DDB0191526) contains the specific consensus site (RIRGQTP) recognized by the antibody in a phosphorylation dependent manner. Talin B was present at the same levels in wild-type, pkbR1−, piaA− cells (Figure 3B) and was specifically immunoprecipitated with the phospho PKB substrate antibody only in cells expressing PKBR1 and stimulated with cAMP (Figure 3C). At pp180 we found a Ras GEF, GEFN (DDB0167277), containing the PKB motifs (RLRSFTP, RSRAQTP). At pp110 we found another Ras GEF, GEFS (DDB0191324), containing the PKB motifs (RVRHSTP, RIRSPSP) as well as a PI4P 5-kinase domain-containing protein (DDB0234212), containing the PKB motif (RVRLNTP). At pp65/67 we found a RhoGAP domain containing protein, GACQ (DDB0233774), containing the PKB motif (RQRSNTP). For the latter three proteins, we validated chemoattractant-mediated phosphorylation by expressing corresponding GFP-fusion proteins in wild-type cells. In each case a new appropriately sized band appeared when stained with phospho PKB substrate antibody (Figure S3B). This interesting set of proteins suggests mechanisms by which chemoattractant-mediated PKB phosphorylation controls signaling and cytoskeletal events that underlie chemotaxis.
Figure 3. TalB is a substrate of Dictyostelium PKB.
(A) TalB is schematically represented. The FERM, I/LWEQ, and villin head piece (VHP) domain are shown. Four peptides were identified by mass spectrometry analysis. RIRGQT is the site recognized by α-phosphospecific PKB substrate antibodies. TalB also has other plausible PKB phosphorylation sites (KKRKDT and RKKEYS). (B) Whole cell extracts (WCE) from WT, pkbR1−,and piaA− cells were stained with α-Talin B antibody. (C) pkbR1− cells carrying vector (V) or PKBR1-HA plasmid were stimulated with 1 µM cAMP and cell extracts were prepared (WCE). Immunoprecipitation (IP) was performed by α-phosphospecific PKB substrate antibodies and samples were immunoblotted with α-Talin B antibody.
Activation of the PKBs by TorC2 through phosphorylation in the hydrophobic motif
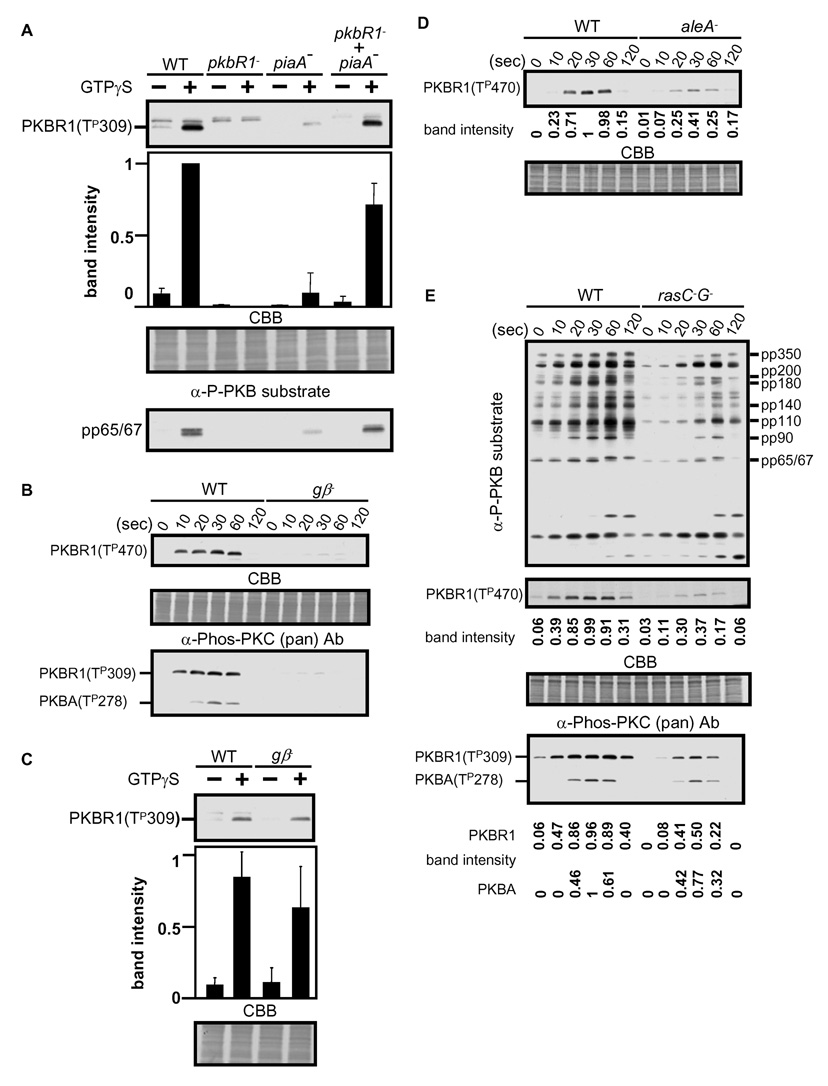
Studies in mammals and Drosophila indicate that PKBs have one phosphorylation site in the AL and another in the HM that are necessary for activity [19]. To confirm the importance of these sites, we performed mutational analysis of each in PKBR1 and examined whether the mutant proteins could complement the developmental defects of pkbR1− cells or support phosphorylation of the endogenous substrates (Figure S4A, S4B, and S4C). Substitution of AL T309 with alanine or phosphomimics, aspartate or glutamate, produced inactive enzymes in both assays, suggesting that T309 is critical for function. Substitution of alanine for HM T470 also yielded an inactive enzyme. However, PKBR1T470D and PKBR1T470E complemented the developmental defect and restored the normal pattern of phosphorylation to the pkbR1− cells. These results strongly suggest that phosphorylation of T470 is important for the in vivo kinase activity of PKBR1.
Since piaA− and pkbR1− cells displayed similar chemotactic defects and TorC2 has been reported to be necessary for activation of AKT in Drosophila and mammalian cells, we next investigated the role of TorC2 in activation of PKBA and PKBR1. In cells lacking PiaA, the phosphorylations of endogenous PKB substrates were greatly reduced (Figure 2D and S2C). In order to show that PKBR1 was a TorC2 substrate, we examined phosphorylation of T470 directly. We noticed that the epitope of the anti phospho-PDK1 docking motif antibody (F/K-X-X-F/Y-S/T-F/Y) matched closely with HM of PKBR1 (FEGFTYVADS) and tested whether this antibody could recognize T470 of PKBR1 in a phosphorylation dependent manner (Figure 4A). In pkbR1− cells expressing PKBR1, the antibody selectively stained the protein following stimulation (Figure S4D). The band was present in PKBR1WT and PKBR1T309A but absent in PKBR1T470A (Figure S4E). The signal in PKBR1T309A was consistently lower than in PKBR1WT, indicating that T309 influences T470 phosphorylation. The staining was disappeared when PKBR1-HA proteins were immunoprecipitated, and treated with λ phosphatase (Figure S4F). We also detected a 75 kDa band during cAMP stimulation of wild-type but not pkbR1− cells, presumably phosphorylation at T470 in endogenous PKBR1 (Figure 4B). Most significantly, T470 phosphorylation was completely absent in piaA− cells (Figure 4B) and it was reduced by over 75% in rip3− cells, which lack another of the TorC2 subunits (Figure S4G). This data strongly suggests that TorC2 is the kinase that phosphorylates the HM of PKBR1 and, since PKBR1 is persistently associated with the membrane, the TorC2 is activated by chemoattractant.
Figure 4. PKBR1 is regulated through phosphorylations in the hydrophobic motif by TorC2 and in the activation loop independently of PIP3.
(A) Schematic representation of PKBR1 shows detection of phosphorylation at T309 and T470 by the α-phospho PKC (pan) antibody and the α-phospho PDK docking motif antibody, respectively. (B) WT, pkbR1−, and piaA− cells were stimulated with 1 µM cAMP and cell extracts taken at the indicated times were probed with α-phospho PDK docking motif antibody. The protein transferred membrane was stained with CBB as a loading control. (C) Cell extracts were prepared as in (B) and were probed with α-phospho PKC (pan) antibody. Shorter and longer exposures are shown. (D) Cell extracts expressing PKBR1(WT), PKBR1(T309A), or PKBR1(T470A) tagged with HA in pkbR1− were prepared at the indicated times after 1 µM cAMP and probed with α-phospho PKC (pan) antibody. α-HA was used to detect expression of each PKBR1 proteins. (E) To evaluate PIP3 dependency of TorC2-PKBR1 pathway, wild-type (WT) and pi3k1-5− cells (in the left panel) and WT cells were treated with or without 30 µM LY294002 (in the right panel) were used. Cell extracts were prepared as in (B). PKBR1 and TorC2 activity were detected with α-phosphospecific PKB substrate antibody (upper panel) and α-phospho PDK docking motif antibody (middle panel) respectively. The α-phospho PKC (pan) antibody was used to show the phosphorylation in the ALs of PKBR1 and PKBA (lower panel).
We also directly observed chemoattractant-induced phosphorylation at T309 of PKBR1 and T278 of PKBA conserved activation loop, using the anti-phospho PKC (pan) antibody that detects phosphorylation of the AL (Figure 4C). The larger band disappeared in pkbR1− cells (Figure 4C) and both bands were absent in pkbA− / pkbR1− cells (data not shown). These phosphorylations peaked at 30 to 60 sec, kinetics nearly identical to those of the HM phosphorylation of PKBR1 by TorC2 (Figure 4B and 4C). Curiously, we noticed that the AL phosphorylation of PKBA was increased in pkbR1− cells compared to wild-type cells, suggesting that it depends on a balance between the amount of PKBR1 and PKBA (Figure 4C, long exposure).
It has been shown that mammalian PKBs can, under some circumstances, be phosphorylated in the AL independently of TorC2 and therefore we asked whether or not this is the case for each PKB in D. discoideum [16, 17, 21]. As shown in Figure 4C, the phosphorylation of PKBR1 completely disappeared in piaA− cells whereas that of PKBA was decreased but still weakly remained at 30 sec (Figure 4C, long exposure). To further confirm the effect of the HM phosphorylation on the AL phosphorylation, we used PKBR1T470A. Consistently, after cAMP stimulation, the anti-phospho PKC (pan) antibody stained neither PKBR1T470A nor PKBR1T309A whereas PKBR1 was stained strongly (Figure 4D). These results suggest that the phosphorylation in the HM is a strict prerequisite for AL phosphorylation in PKBR1 whereas it enhances AL phosphorylation in PKBA .
Activation of PKBR1 by TorC2 is independent of PIP3
The kinetics of T470 phosphorylation and activation of PKBR1 by TorC2 are essentially identical to those for chemoattractant-induced accumulation of PIP3; however, PKBR1 activity measured in vitro is stimulated by cAMP in pi3k1-2− cells [23]. We used pi3k1-5− quintuple knockout cells [7] and LY294002 to ask whether activation of TorC2 and PKBR1 in vivo against endogenous substrates required PIP3. Anti phospho PKB antibody detected nearly the same level of phosphorylation of pp350, pp200, pp180, and pp110 in pi3k1-5− compared to wild-type cells while the phosphorylation of pp90 and pp65/67 were reduced slightly (Figure 4E, left upper). Acute treatment with PI3K inhibitor LY294002 (30 µM) also reduced the signals only slightly (Figure 4E, right upper). These small reductions are likely caused by a failure to activate PKBA activity (see below) and the results suggest that PKBR1 activation is largely independent of PIP3.
Next we assessed whether TorC2 activity is regulated by the PIP3 pathway by monitoring T470 phosphorylation. Again, there were no significant differences between wild-type and pi3k1-5− cells (Figure 4E, left middle). Surprisingly, 30 µM LY294002 reduced T470 phosphorylation of PKBR1 by about 50% (Figure 4E, right middle). Considering the lack of dependence on PI3-kinases, we suggest that this is because LY294002 can directly inhibit TorC2 activity [20]. These results further suggest that the TorC2-PKBR1 pathway is independent of PIP3. Since unlike other PKBs, PKBR1 is persistently present on the membrane while phosphorylation of T470 increases and decreases, it appears that TorC2 is transiently activated by chemoattractant.
We also directly assessed the dependence of the phosphorylation of T309 of PKBR1 and T278 or PKBA on PIP3. For PKBR1, there were no significant differences between wild-type and pi3k1-5− cells (Figure 4E, left lower). In addition, 30 µM LY294002 caused a slight reduction (Figure 4E, right lower), probably because LY294002 inhibits T470 phosphorylation (Figure 4E, right middle) which is required for T309 phosphorylation (Figure 4C and 4D). In both pi3k1-5− and 30 µM LY294002 treated cells, the phosphorylation of PKBA was lost, suggesting that phosphorylation of the AL occurs during its recruitment to the membrane (Figure 4E, left and right lower).
To reveal whether the phosphorylation of T470 is sufficient for full activation of PKBR1, the phosphomimetic substitution T470E mutant was studied further. As shown in Figure S4A, T470E can complement the biological and biochemical defects in pkbR1− cells. When starved pkbR1− cells carrying PKBR1T470E were stimulated, phosphorylations of endogenous substrate were induced although some bands were slightly reduced compared to PKBR1WT (Figure S4H, left). Both PKBR1wt and PKBR1T470E in pkbR1− cells were phosphorylated at T309 after cAMP addition, although the phosphorylation of PKBR1T470E was weaker and less sustained (Figure S4I). Furthermore, chemoattractant-mediated changes in phosphorylation of PKBR1T470E were observed with the same-kinetics in piaA− cells (Figure S4I). However, this was apparently not sufficient activation to cause full phosphorylation of the endogenous substrates (Figure S4H, right) and PKBR1T470E did not suppress the development defects of piaA− cells (data not shown). These findings have several implications. First, a role of TorC2 in addition to T470 phosphorylation is required for full PKBR1 activation. Second, T309 phosphorylation must be regulated by yet another mechanism besides PIP3 or T470 phosphorylation. D. discoideum contains two PDK1-like enzymes. It is possible that these enzymes act together with TorC2 to activate PKBR1.
Two GTP binding proteins are required for TorC2 activation
To further explore the pathway for activation of TorC2, we devised a cell-free system. Cell lysates were stimulated with GTPγS, incubated briefly, and the phosphorylation of the HM and AL of PKBR1 and phosphorylation of endogenous PKB substrates assessed. We monitored GTPγS-mediated phosphorylation of PKBR1 as well as phosphorylation of endogenous substrate, pp 65/67 (Figure 5A, top and bottom, respectively; data not shown). Consistently, phosphorylation of T309 or pp65/67 was not observed in piaA− cells. However, both could be rapidly reconstituted by mixing lysates of pkbR1− and piaA− cells. This indicates that TorC2 present in the pkbR1− cell lysates can be activated and phosphorylate PKBR1 present in the piaA− cell lysates. This is consistent with previous data showing that wild-type supernatants can reconstitute adenylyl cyclase activation to lysates from piaA− cells prestimulated with cAMP or GTPγS [12, 13].
Figure 5. Two GTP binding proteins are required for TorC2 activity.
(A) Cell lysates from indicated strains were activated with GTPγS and T309 phosphorylation of PKBR1 was evaluated with the α-phospho PKC (pan) antibody. The intensity of each band was quantified from five independent experiments. For PKB activity, cell extracts were probed with α-phosphospecific PKB substrate antibody. (B) Wild-type (WT) and gβ− cells were stimulated with 1 µM cAMP and the phosphorylation level in the HM of PKBR1 (T470) and the AL of PKBR1 (T309) and PKBA (T278) was assessed by the α-phospho PDK docking motif and α-phospho PKC (pan) antibodies. (C) Stimulation of cell lysates from WT and gβ− cells with GTPγS were performed in as (A). (D) The T470 phosphorylation of PKBR1 in WT and aleA− cell was compared. The quantification is the average of three experiments. (E) Phoshorylation of PKB substrates, the HM of PKBR1,and the AL of PKBR1 and PKBA in WT and cAR1 expressing rasC− G− cells were compared. The quantification is the average of three experiments.
The cell-free system together with the assays of phosphorylations mediated by chemoattractant in living cells allowed us to explore the pathway for TorC2 activation more directly. In intact cells chemoattractant-induced phosphorylations of either PKBR1 or PKBA HMs or ALs were strictly dependent on functional heterotrimeric G-proteins since these modifications were completely absent in cells lacking the unique Gβ present in D. discoideum (Figure 5B). However, in lysates from gβ− cells, GTPγS was still able to activate TorC2, as evidenced by phosphorylation of T309 of PKBR1, although the levels were somewhat reduced (Figure 5C). These observations suggest that activation of the heterotrimeric G-protein by chemoattractant is relayed to an intermediate G-protein in the pathway. In vitro, the heterotrimeric G-protein can be bypassed by direct activation of the intermediate G-protein by GTPγs.
One possibility is that the intermediate G-protein(s) are Ras family members since these proteins have been shown to be activated by chemoattractant [24, 25, 26]. The phosphorylation of PKBR1 was reduced by over 60% in cells lacking the RasGEF AleA, suggesting that a Ras-like G-protein could be involved in the pathway (Figure 5D). These results prompted us to test cells lacking RasG or/and RasC. However, in the rasG− cells, phosphorylation events in either in vivo or in vitro assays were basically identical to those wild type cells (data not shown). On the other hand, in rasC− G− cells phosphorylation of PKB substrates, the HM of PKBR1, and the ALs of PKBR1 and PKBA were significantly reduced (Figure 5E). Quantification of the HM of PKBR1 and the ALS of PKBR1 and PKBA were reduced by 70%, 66%, and 38%, respectively. Together, these data suggest that these Ras proteins play a role in TorC2 activation. However, since activity remains in the ras C− G− cells, additional intermediate G-proteins must also be involved.
Local activation of TorC2 and PKB at the leading edge of chemotaxing cells
We wondered whether spatially localized activation of TorC2 and PKBR1 might account for their role in chemotaxis. To investigate TorC2 activation, phosphorylation of the AL of PKBR1 was monitored by staining cells with the anti phos-PKC (pan) antibody. Cells were stimulated uniformly with cAMP and the time course of staining was assessed. Consistent with the immunoblot analysis, there was a transient burst of staining that appeared within 5 sec of stimulation and declined by 60 sec (Figure 6A). This increase was absent in PKBR1T470A cells as expected (see Figure 4C and 4D). The staining with an anti HA antibody was uniform along the cell perimeter and showed no time-dependent changes. These observations suggest that this staining reaction reflects the activation of TorC2 and can be used to monitor the distribution of TorC2 activation in living cells. As shown in 6B, the leading edge of chemotaxing cells were strongly and selectively stained using this approach. These signals were below the level of detection in wild-type cells (data not shown) and were absent in pkbR1− cells expressing PKBR1T470A in which T309 is not phosphorylated (see Figure 4D). Both PKBR1WT and PKBR1T470A are localized uniformly along the membrane as detected by an anti-HA antibody (Figure 6B).
Figure 6. The activity of TorC2 and PKBR1 is spatially regulated in chemotaxing cells.
(A) Schematic representation of PKBR1 shows phosphorylation of T309 detected by α-phos-PKC (pan) antibody. pkbR1− cells expressing PKBR1 (WT)-HA or PKBR1 (T470A)-HA were fixed at the indicated times after cAMP stimulation and stained with α-phos-PKC (pan) or α-HA antibody. (B) The same cells as (A) were allowed to perform chemotaxis and stained with α-phos-PKC (pan) antibody to detect the phosphorylation of T309 of PKBR1 or α-HA antibody for the localization of PKBR1. (C) Schematic representation of chimera R1-AKT-HA shows phosphorylation at serine (S473) detected by α-phos-AKT (S473) antibody. Cells were treated as in (B) and stained with α-phos-AKT (S473) antibody to detect the phosphorylation of S473 or α-HA antibody for the localization of R1-AKT-HA. Arrow heads show localized staining. Scale bars represent 10 µm.
As a second "biosensor" for the activity of TorC2, we developed a molecular tool, R1-AKT-HA, in which the PH domain of human AKT1 was replaced with the N-terminus of PKBR1 containing the myristoylation site (Figure 6C). Although R1-AKT-HA could not complement the developmental defects of pkbR1− cells (Figure S6A), following cAMP stimulation it was phosphorylated in the HM as assessed by an anti phospho-AKT (S473) antibody in immunoblots (Figure S6B). This phosphorylation disappeared in piaA− cells showing that R1-AKT-HA is phosphorylated by TorC2 (Figure S6B) and that this antibody can be used to explore the spatial pattern of activation of TorC2. While wild-type cells did not show any significant staining with the anti phospho-AKT (S473) antibody, R1-AKT-HA expressing cells were stained selectively at the leading edge (Figure 6C). When stained with anti HA antibody R1-AKT-HA was localized uniformly along the membrane like PKBR1 as expected (Figure 6B). These observations suggest that, although TorC2 is found largely in the cytosol, it is selectively activated by chemoattractant stimulation and at the leading edge of chemotaxing cells.
Discussion
Our observations define a pathway linking chemoattractant receptors through activation of TorC2 and PKBs to regulation of the cytoskeleton at the cell's leading edge. Figure 7 summarizes these findings. Chemoattractant binds to the surface receptor and activates the heterotrimeric G-protein. The activated G-protein signals through a RasGEF to intermediate G-proteins, including RasC. We speculate that activation of the intermediate G-proteins is confined to the front of the cell, leading to spatially restricted activation of TorC2 and PKBR1. Chemoattractant also triggers to local accumulation of PIP3 at the front as previously described. PKBA is recruited to the front through its PIP3–specific PH domain and TorC2 enhances its activation. These events result in chemoattractant-mediated phosphorylation of Talin B and other signaling proteins, providing a possible link to the cytoskeleton.
Figure 7. Schematic diagram of signaling events at the leading edge of a chemotaxing cell.
The chemoattractant, cAMP, signals through its cognate heterotrimeric G-protein and AleA to an intermediate G-protein. We speculate that the intermediate G-protein is locally activated and that TorC2 which is largely in the cytosol transiently encounters it (bent arrows). The activated TorC2 then phosphorylates PKBR1 and PKBA before returning to the cytosol. PKBR1 is a more predominant activity and completely dependent on TorC2 for activation while PKBA is a minor activity and depends on recruitment to PIP3 and TorC2 for activation (heavier or lighter bent arrows). Among PKB substrates is Talin B, which provides a possible link to the cytoskeleton.
Combined genetic and biochemical analyses show there is an intermediate G-protein involved in activation of TorC2. In intact cells, activation of TorC2 by chemoattractant assessed by phosphorylation of HM and AL sites as well as phosphorylation of endogenous substrates, requires heterotrimeric G-protein function. In cells lacking the Ras GEF AleA the activation is attenuated. However, in a cell-free system GTPγS activates TorC2 in the absence of heterotrimeric G-protein function, indicating that a second intermediate G-protein is involved. It has been reported that since RasG interacts with Rip3 in yeast two-hybrid assays, RasG activates TorC2 [13, 25, 27]. Our results indicate that the mechanism is more complex. First, while rip3− cells are significantly impaired, some activation still occurs in the absence of Rip3. Second, our results suggest that RasC, together with AleA, plays a role in the activation since cells lacking RasG show no defect while those lacking RasG and RasC show a partial defect. This suggests that an additional inrtermediate G-protein is also required. Further investigation will be required to determine whether the target(s) of GTPγS stimulation are different Ras family members, a combination of Ras proteins, or as yet undetermined G-proteins and also whether other subunits of TorC2 besides Rip3 participate in the activation.
How might the temporal and spatial regulation of TorC2 activity be mediated? Our studies of the persistently membrane associated PKBR1 force us to consider schemes which do not involve PKB recruitment. We speculate that cytosolic TorC2 encounters membrane associated activators, such as the intermediate G-protein(s) at the leading edge and phosphorylates PKBR1 (and PKBA) before TorC2 returns to the cytosol [28]. It has been shown that RasG and RasC are activated by chemoattractants and that the Ras binding domain of Raf localizes selectively to the front of chemotaxing cells [25]. However, as noted above, cells lacking RasG and RasC still activate PKBR1. Therefore, we seek an additional locally activated intermediate G-protein.
Significantly, one of the PKB substrates we have identified is Talin B and we have traced a pathway from chemoattractant stimulation to Talin B phosphorylation. Furthermore, we speculate that Talin B is selectively phosphorylated at the leading edge of the cell since TorC2 is active there. Cells lacking Talin B display a developmental phenotype similar to cells lacking PKBR1 in that they arrest as late aggregates [29, 30]. Talin controls cell attachment to the substratum by interacting with integrins and D. discoideum cells expresses a series of primitive integrins which regulate cell adhesion and interact with Talin [31]. Interestingly, we find that the effect of loss of PKBR1 activity on chemotaxis seems to include a reduction in cell attachment to the substrate. However, it remains to be determined whether Talin B, and its phosphorylation, regulates cell adhesion or other cytoskeletal events. Furthermore, the other PKB substrates we have identified no doubt also participate in aspects of chemotaxis.
The chemotactic signaling pathways are remarkably resilient. Links from PIP3, localized at the cell's leading edge, to the cytoskeleton have been abundantly documented in D. discoideum and in mammalian cells. Evidence suggests that these connections involve PH-domain containing PKBs. However, an equally strong body of evidence shows that cells can carry out chemotaxis without PIP3. For D. discoideum, these two apparently conflicting observations can be resolved by the scheme shown in Figure 7. We speculate that responses to excessive PIP3 are mediated by PKBA. In the absence of significant changes in PIP3 levels, endogenous PKB substrates still undergo transient phosphorylation due to the chemoattractant-mediated activation of TorC2 and PKBR1 we have described here. If the phosphorylation events are blocked by removal of the PKBs (PKBR1 is predominant) or TorC2, chemotaxis is greatly impaired. Thus, in wild-type cells, the temporal and spatial pattern of the endogeneous phosphorylation events is controlled redundantly by local changes in PIP3 and/or by local activation of TorC2.
Previous reports have suggested physiological roles for TorC2 and PKBs in regulation of the cytoskeleton in other cells, consistent with our findings of a role in chemotaxis. In budding yeast, TorC2 regulates actin patch polarization via YPKs, yeast PKB homologs [32, 33]. In HeLa cells and 3T3 fibroblasts, reduction of TorC2 activity affects the extent of polymerized actin in cells, leading to different morphological changes [14, 15] and knockouts of TorC2 subunits in mice are lethal [16, 17]. In a variety of cancer cells, alterations in the activity of various PKB isoforms have been reported to enhance or decrease motility, invasion, and metastasis [34]. Furthermore, it has been suggested that PKB, and its kinase PDK1, are required for chemotaxis of endothelial cells during vascular vessel formation [35]. More directly, knockdown of PKB as well as the actin binding protein Girdin, a substrate of PKB, in Vero fibroblasts impaired migration in wound healing assays [36]. Signaling networks similar to the framework outlined in Figure 7 may underlie these various observations. What of the reports of PIP3-independent chemotaxis? The scheme in Figure 7 implies that activation of TorC2 at the cell's leading edge acts redundantly with PIP3. The equivalent of PKBR1 could be a standard, PH-domain containing PKB under unusual conditions when PIP3 levels are extremely low or high and TorC2 would be the dominant regulator [see 37]. Alternatively, TorC2 might act on another kinase such as PKC, which might serve as the equivalent of PKBR1.
Chemotaxis is mediated by a network of signaling events and our results indicate that TorC2-PKB represents an important pathway in addition to the PI3K and PLA2 pathways which have been shown to redundantly regulate chemotaxis [10, 11]. TorC2 activation is independent of both PIP3 and PLA2 (unpublished observations) indicating that there are three parallel pathways. It will be interesting to learn whether simultaneous block of all pathways leads to an even more severe phenotype. It is likely that each of these pathways regulates a specific set of physiological responses that mediate an aspect of chemotaxis. Further understanding may require us to delineate these responses, understand how they act together in chemotaxis, and assign pathways to each response.
Supplementary Material
Acknowledgements
The authors wish to acknowledge members of the Devreotes lab, particularly Jonathan Franca-Koh and Stacey Willard for critical reading. We thank Dr R.R. Kay for pi3k1-5− cells, Dr. M. Iijima for the PKBA disruption plasmid, Dr K. Inouye for α-Talin B antibody, Dr R.H. Insall for aleA− cells, Drs. R. Meile and R.A. Firtel for the PKBR1 disruption plasmid, PKBR1 cDNA, and helpful advice, Dr. G. Weeks for cAR1 expressing rasC− G− cells and proteomics core facility in Johns Hopkins University, School of Medicine, for mass spectrometry analysis. This work was supported by NIH GM 28007 and NIH GM 34933 to P.N.D. and by Uehara memorial foundation to Y.K‥
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Franca-Koh J, Kamimura Y, Devreotes PN. Navigating signaling networks: chemotaxis in Dictyostelium discoideum. Curr. Opin. Genet. Dev. 2006;16:333–338. doi: 10.1016/j.gde.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 2.Iijima M, Devreotes PN. Tumor suppressor PTEN mediates sensing of chemoattractant gradients. Cell. 2002;109:599–610. doi: 10.1016/s0092-8674(02)00745-6. [DOI] [PubMed] [Google Scholar]
- 3.Funamoto S, Meili R, Lee S, Parry L, Firtel RA. Spatial and temporal regulation of 3-phosphoinositides by PI 3-kinase and PTEN mediates chemotaxis. Cell. 2002;109:611–623. doi: 10.1016/s0092-8674(02)00755-9. [DOI] [PubMed] [Google Scholar]
- 4.Wang F, Herzmark P, Weiner OD, Srinivasan S, Servant G, Bourne HR. Lipid products of PI(3)Ks maintain persistent cell polarity and directed motility in neutrophils. Nat. Cell Bio. 2002;4:513–518. doi: 10.1038/ncb810. [DOI] [PubMed] [Google Scholar]
- 5.Wu D, Huang CK, Jiang H. Roles of phospholipid signaling in chemoattractant-induced responses. J. Cell Sci. 2000;113:2935–2940. doi: 10.1242/jcs.113.17.2935. [DOI] [PubMed] [Google Scholar]
- 6.Chen L, Janetopoulos C, Huang YE, Iijima M, Borleis J, Devreotes PN. Two phases of actin polymerization display different dependences on PI(3,4,5)P3 accumulation and have unique roles during chemotaxis. Mol. Biol. Cell. 2003;14(12):5028–5037. doi: 10.1091/mbc.E03-05-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoeller O, Kay RR. Chemotaxis in the absence of PIP3 gradients. Curr. Biol. 2007;17:813–817. doi: 10.1016/j.cub.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Nishio M, Watanabe K, Sasaki J, Taya C, Takasuga S, Iizuka R, Balla T, Yamazaki M, Watanabe H, Itoh R, et al. Control of cell polarity and motility by the PtdIns(3,4,5)P3 phosphatase SHIP1. Nat. Cell Biol. 2007;9:36–44. doi: 10.1038/ncb1515. [DOI] [PubMed] [Google Scholar]
- 9.Ferguson GJ, Milne L, Kulkarni S, Sasaki T, Walker S, Andrews S, Crabbe T, Finan P, Jones G, Jackson S, et al. PI(3)Kγ has an important context-dependent role in neutrophil chemokinesis. Nat. Cell Biol. 2007;9:86–91. doi: 10.1038/ncb1517. [DOI] [PubMed] [Google Scholar]
- 10.Chen L, Iijima M, Tang M, Landree MA, Huang YE, Xiong Y, Iglesias PA, Devreotes PN. PLA2 and Pi3K/PTEN pathways act in parallel to mediate chemotaxis. Dev. Cell. 2007;12:603–614. doi: 10.1016/j.devcel.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Haastert PJM, Keizer-Gunnink I, Kortholt A. Essential role of PI3-kinase and phospholipase A2 in Dictyostelium discoideum chemotaxis. J. Cell Biol. 2007;177:809–816. doi: 10.1083/jcb.200701134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen MY, Long Y, Devreotes PN. A novel cytosolic regulator, pianissimo, is required for chemoattractant receptor and G protein-mediated activation of the 12 transmembrane domain adenylyl cyclase in Dictyostelium. Genes Dev. 1997;11:3218–3231. doi: 10.1101/gad.11.23.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee S, Comer FI, Sasaki A, McLeod IX, Duong Y, Okumura K, Yates JR, IIIrd, Parent CA, Firtel RA. TOR complex 2 integrates cell movement during chemotaxis and signal relay in Dictyostelium. Mol. Biol. Cell. 2005;16:4572–4583. doi: 10.1091/mbc.E05-04-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacinto E, Loewith R, Schmidt A, Lin S, Rüegg MA, Hall A, Hall MN. Mammalian TOR complex2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 2004;6:1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- 15.Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway of mTOR that regulates the cytoskeleton. Curr. Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 16.Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCα, but not S6K1. Dev. Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 17.Shiota C, Woo JT, Lindner J, Shelton KD, Magnuson MA. Multiallelic disruption of the rictor gene in mice reveals that mTOR complex 2 is essential for fetal growth and viability. Dev. Cell. 2006;11:583–589. doi: 10.1016/j.devcel.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 18.Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall MN. Two TOR complexes, only ones of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 19.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the Rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 20.Bhaskar PT, Hay N. The two TORs and Akt. Dev. Cell. 2007;12:487–502. doi: 10.1016/j.devcel.2007.03.020. [DOI] [PubMed] [Google Scholar]
- 21.Jacinto E, Facchinetti V, Liu D, Soto N, Wei S, Jung S, Huang Q, Qin J, Su B. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127:125–137. doi: 10.1016/j.cell.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 22.Meili R, Ellsworth C, Lee S, Reddy TBK, Ma H, Firtel RA. Chemoattractant-mediated transient activation and membrane localization of Akt/PKB is required for efficient chemotaxis to cAMP in Dictyostelium. EMBO J. 1999;18:2092–2105. doi: 10.1093/emboj/18.8.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meili R, Ellsworth C, Firtel RA. A novel Akt/PKB-related kinase is essential for morphogenesis in Dictyostelium. Curr. Biol. 2000;10:708–717. doi: 10.1016/s0960-9822(00)00536-4. [DOI] [PubMed] [Google Scholar]
- 24.Kae H, Lim CJ, Spiegelman GB, Weeks G. Chemoattractant-induced Ras activation during Dictyostelium aggregation. EMBO Rep. 2004;5:602–606. doi: 10.1038/sj.embor.7400151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sasaki AT, Chun C, Takeda K, Firtel RA. Localized Ras signaling at the leading edge regulates PI3K, cell polarity, and directional cell movement. J. Cell Biol. 2004;167:505–518. doi: 10.1083/jcb.200406177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilkins A, Szafranski K, Fraser DJ, Bakthavatsalam D, Müller R, Fisher PR, Glöckner G, Eichinger L, Noegel AA, Insall RH. The Dictyostelium genome encodes numerous RasGEFs with multiple biological roles. Genome Biol. 2005;6:R68. doi: 10.1186/gb-2005-6-8-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee S, Parent CA, Insall R, Firtel RA. A novel Ras-interacting protein required for chemotaxis and cyclic adenosine monophosphate signal relay in Dictyostelium. Mol. Biol. Cell. 1999;10:2829–2845. doi: 10.1091/mbc.10.9.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pergolizzi B, Peracino B, Silverman J, Ceccarelli A, Noegel A, Devreotes PN, Bozzaro S. Temperature-sensitive inhibition of development in Dictyostelium due to a point mutation in the piaA gene. Dev. Biol. 2002;251:18–26. doi: 10.1006/dbio.2002.0809. [DOI] [PubMed] [Google Scholar]
- 29.Tsujioka M, Machesky LM, Cole SL, Yahta K, Inouye K. A unique homologue with a villin headpiece-like domain is required for multicellular morphology in Dictyostelium. Curr. Biol. 1999;9:389–392. doi: 10.1016/s0960-9822(99)80169-9. [DOI] [PubMed] [Google Scholar]
- 30.Tsujioka M, Yoshida K, Inouye K. Talin B is required for force transmission in morphogenesis of Dictyostelium. EMBO J. 2004;23:2216–2225. doi: 10.1038/sj.emboj.7600238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cornillon S, Gebbie L, Benghezal M, Nair P, Keller S, Wehrle-Haller B, Charette SJ, Brükert F, Letourneur F, Cosson P. An adhesion molecule in free-living Dictyostelium amoebae with integrin β features. EMBO Rep. 2006;7:617–621. doi: 10.1038/sj.embor.7400701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmidt A, Kunz J, Hall MN. TOR2 is required for organization of the actin cytoskeleton in yeast. Proc. Natl. Acad. Sci. USA. 1996;93:13780–13785. doi: 10.1073/pnas.93.24.13780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamada Y, Fujioka Y, Suzuki NN, Inagaki F, Wullschleger S, Loewith R, Hall MN, Ohsumi Y. Tor2 directly phosphorylates the AGC kinase Ypk2 to regulate actin polarization. Mol. Cell. Biol. 2005;25:7239–7248. doi: 10.1128/MCB.25.16.7239-7248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stambolic V, Woodgett JR. Functional distinctions of protein kinase B/Akt isoforms defined by their influence on cell migration. Trends Cell Biol. 2006;16:461–466. doi: 10.1016/j.tcb.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 35.Primo L, di Blasio L, Roca C, Droetto S, Piva R, Schaffhausen B, Bussolino F. Essential role of PDK1 in regulating endothelial cell migration. J. Cell Biol. 2007;176:1035–1047. doi: 10.1083/jcb.200607053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Enomoto A, Murakami H, Asai N, Morone N, Watanabe T, Kawai K, Murakumo Y, Usukura J, Kaibuchi K, Takahashi M. Akt/PKB regulates actin organization and cell motility via Girdin/APE. Dev. Cell. 2005;9:389–402. doi: 10.1016/j.devcel.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 37.Hietakangas V, Cohen SM. Re-evaluating AKT regulatino: role of TOR complex 2 in tissue growth. Genes & Dev. 2007;21:632–637. doi: 10.1101/gad.416307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hagedorn M, Neuhaus EM, Soldati T. Optimized fixation and immunofluorescence staining methods for Dictyostelium cells. In: Eichinger L, Rivero F, editors. Methods in Molecular Biology, vol. 346: Dictyostelium discoideum protocols. Human Press; 2006. pp. 327–338. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.