Abstract
Unspecific peroxygenase (UPO) represents a new type of heme-thiolate enzyme with self-sufficient mono(per)oxygenase activity and many potential applications in organic synthesis. With a view to taking advantage of these properties, we subjected the Agrocybe aegerita UPO1-encoding gene to directed evolution in Saccharomyces cerevisiae. To promote functional expression, several different signal peptides were fused to the mature protein, and the resulting products were tested. Over 9,000 clones were screened using an ad hoc dual-colorimetric assay that assessed both peroxidative and oxygen transfer activities. After 5 generations of directed evolution combined with hybrid approaches, 9 mutations were introduced that resulted in a 3,250-fold total activity improvement with no alteration in protein stability. A breakdown between secretion and catalytic activity was performed by replacing the native signal peptide of the original parental type with that of the evolved mutant; the evolved leader increased functional expression 27-fold, whereas an 18-fold improvement in the kcat/Km value for oxygen transfer activity was obtained. The evolved UPO1 was active and highly stable in the presence of organic cosolvents. Mutations in the hydrophobic core of the signal peptide contributed to enhance functional expression up to 8 mg/liter, while catalytic efficiencies for peroxidative and oxygen transfer reactions were increased by several mutations in the vicinity of the heme access channel. Overall, the directed-evolution platform described is a valuable point of departure for the development of customized UPOs with improved features and for the study of structure-function relationships.
INTRODUCTION
The unspecific peroxygenase (UPO) (EC 1.11.2.1; also known as aromatic peroxygenase [APO]) is secreted by the edible mushroom Agrocybe aegerita, and it belongs to a new type of peroxide-using enzymes that are of considerable interest due to their wide range of potential biotechnological applications (1). UPO is a compact protein with a thiolate axial ligand of the pivotal Fe3+ that governs the heme domain. Accordingly, UPO is classified as a member of the heme-thiolate peroxidase (HTP) superfamily, along with chloroperoxidase (CPO) from the ascomycete Leptoxyphium fumago, even though CPO is not capable of epoxidizing aromatic rings or hydroxylating alkanes like UPO (2).
The diversity of enzymes in the HTP superfamily is conferred by 2 distinct clusters (“long” and “short” UPOs) that are included in basidiomycetes, ascomycetes, and other fungal sequences. UPO-type genes and proteins have been isolated and characterized in Coprinellus radians, Marasmius rotula, and Coprinopsis cinerea (3–5). Indeed, to date, over 1,000 UPO-like genes have been identified in genetic databases and in basidiomycete genome sequencing, indicating an ancient origin and a widespread distribution of UPO in nature (6, 7). With over 300 identified substrates, UPOs exhibit considerable promiscuity in oxidation reactions, making them potentially attractive industrial biocatalysts. The versatile peroxide-dependent monooxygenase activity of UPO, which is based on a 2-electron oxygenation mechanism (i.e., peroxygenase activity), is of particular significance, as selective oxyfunctionalizations are among the most important reactions in organic synthesis (8). The array of oxygen transfer reactions catalyzed by UPO includes bromide oxidation, sulfoxidation, N-oxidation, aromatic peroxygenation, double-bond epoxidation, hydroxylation of aliphatic compounds, and ether cleavage (2). Fueled by catalytic amounts of H2O2, UPO acts as a self-sufficient monooxygenase through a complex catalytic mechanism that joins the reactive intermediates of heme-peroxidases and P450s (the “peroxide shunt” pathway) (7). Moreover, a UPO oxoiron(IV) protoporphyrin radical cation intermediate (UPO compound I) was recently described and proposed as the main active oxygen species involved in the mono(per)oxygenase activity of UPO (9, 10). Thus, UPO may be considered to be the missing link between P450 monooxygenases and heme-peroxidases (7).
Despite much biotechnological interest in this enzyme, no protein-engineering studies have attempted to adapt its unique features to the requirements of specific biotransformation processes in industrial settings. Moreover, endeavors to gain a better understanding of UPO's complex mechanism of action are hindered by the absence of tools with which to design mutants. Indeed, the successful adaptation of UPO has been hampered by many of the same bottlenecks that have precluded the engineering of CPO for decades, including several obstacles that prevent successful functional expression of HTPs in heterologous hosts (e.g., different codon usage, cumbersome posttranslational modifications, and heme-thiolate prosthetic group attachments [11, 12]).
Escherichia coli and Saccharomyces cerevisiae are the most attractive heterologous hosts in which directed evolution can be performed (13). S. cerevisiae is a particularly versatile vehicle for the functional expression and directed evolution of fungal genes involved in lignin modification (including laccases and peroxidases), and it has been used in the directed evolution of versatile peroxidases (VP) for functional expression and stabilization, whereby medium-redox-potential laccases have been engineered to confer high secretion levels and activity on organic cosolvents (14–16). More recently, this host has been used in the design of high-redox-potential laccases (HRPLs) that are active in human blood and to develop chimeric laccases with combined properties (17–20). The number of protocols developed for the generation of DNA diversity in yeast is steadily increasing, and as such, the in vivo homologous-recombination machinery of the host can be used to enrich mutant libraries (21–23). These strategies have helped to extend the study of S. cerevisiae into the fields of synthetic biology and metabolic engineering, highlighting a wide range of potential applications ranging from biofuel production to novel green processes (24–26).
Here, for the first time, we describe the use of directed evolution to produce a soluble, active, and highly stable form of UPO in S. cerevisiae. Several fusion genes were tested to increase initial secretion levels, which were then further optimized by iterative rounds of random mutagenesis, DNA recombination, and semirational strategies. The enzyme's substrate promiscuity was maintained by simultaneously performing a dual high-throughput screening (HTS) assay to efficiently explore mutant libraries without altering protein stability. The final mutant produced was comprehensively characterized and exhibited markedly improved kinetic properties, secretion, and stability over a range of temperatures, as well as in the presence of high concentrations of cosolvents.
MATERIALS AND METHODS
Laboratory evolution: general aspects.
The original parental n-UPO1 and the α-UPO1, α*-UPO1, n*-UPO1, and n*-3F10 fusion genes were constructed as described in the supplemental material. After each round of directed evolution, the PCR products were loaded onto a preparative agarose gel and then purified using the Zymoclean Gel DNA Recovery kit (Zymo Research). The recovered DNA fragments were cloned under the control of the GAL1 promoter of the pJRoC30 expression shuttle vector, using BamHI and XhoI to linearize the plasmid and remove the parent gene. The linearized vector was loaded onto a low-melting-point preparative agarose gel and purified as described above. The mutational loads, recombination strategies, library sizes, and general conditions for each cycle of evolution are described in Table S1 in the supplemental material. All the primers used in this study are listed in Table S3 in the supplemental material.
First generation.
Four libraries were devised, using n-UPO1 and α-UPO1 as the parental types. For each parent, 2 different mutagenic PCR strategies were used: Taq DNA polymerase (Sigma) in the presence of MnCl2 (1 to 3 mutations/1,000 bp) and the Genemorph II kit (4 to 9 mutations/1,000 bp) (Stratagene; Mutazyme II). The PCR for Taq/MnCl2 was performed in a final volume of 50 μl containing 3% dimethyl sulfoxide (DMSO), 90 nM RMLN, 90 nM RMLC, 0.3 mM deoxynucleoside triphosphates (dNTPs) (0.075 mM each), 0.01 mM MnCl2, 1.5 mM MgCl2, 0.05 U/μl Taq DNA polymerase, and 0.1 ng/μl of the corresponding template. The PCRs for Mutazyme II were carried out in a final volume of 50 μl containing 3% DMSO, 0.37 μM RMLN, 0.37 μM RMLC, 0.8 mM dNTPs (0.2 mM each), 0.05 U/μl Mutazyme II, and 300 ng of the corresponding initial target template (2,800 ng of pJR-n-upo1 and 2,566 ng of pJR-α-upo1). Error-prone PCR was performed on a gradient thermocycler (Mycycler; Bio-Rad) using the following parameters: 95°C for 2 min (1 cycle); 94°C for 45 s, 53°C for 45 s, and 74°C for 3 min (28 cycles); and 74°C for 10 min (1 cycle). The PCR products (200 ng) were mixed with the linearized plasmid (100 ng) and transformed into competent S. cerevisiae cells using the Yeast Transformation kit (Sigma). To promote in vivo ligation, ∼50-bp overhangs homologous to the linear vector were designed. Transformed cells were plated on synthetic complete (SC) dropout plates and incubated for 3 days at 30°C. Colonies containing the whole autonomously replicating vector were selected and subjected to the dual HTS assay and additional rescreening, as described in the supplemental material.
Second generation.
The best mutants obtained from the first generation (1A11 and 3C2) were submitted to error-prone PCR (Taq/MnCl2 and Mutazyme II), as well as in vivo DNA shuffling. The mutagenic rates, the PCR conditions, and the thermal-cycling program employed were the same as those described for the first generation. The mutated PCR products were mixed with the linearized vector (at a 4:1 ratio of PCR products to linearized plasmid) and transformed into competent S. cerevisiae cells in order to promote in vivo DNA shuffling.
Third generation.
The best mutant from the second round of evolution (mutant 12C12) was subjected to 2 different processes.
(i) In vivo assembly of mutant libraries.
A recombined mutant library was built by in vivo assembly of mutant libraries constructed with different mutational spectra (IvAM [27]). Taq/MnCl2 and Mutazyme II libraries were mixed in equimolar amounts and transformed into competent S. cerevisiae cells along with the linearized vector, as described above (at an 8:1 ratio of mutant library to vector).
(ii) Focused domain mutagenesis at the signal peptide.
The 12C12 signal sequence was independently subjected to random mutagenesis by MORPHING (mutagenic organized recombination process by homologous in vivo grouping) (28). Mutagenic PCR was prepared in a final volume of 50 μl containing 3% DMSO, 90 nM RMLN, 90 nM Morphing psn apo1 rev, 0.3 mM dNTPs (0.075 mM each), 0.1 mM MnCl2, 1.5 mM MgCl2, 0.05 U/μl Taq polymerase DNA, and 0.92 ng/μl template. The amplification parameters were 95°C for 2 min (1 cycle); 94°C for 45 s, 50°C for 45 s, and 74°C for 30 s (28 cycles); and 74°C for 10 min (1 cycle). The remaining portion of the whole UPO1 gene was amplified by high-fidelity PCR in a final volume of 50 μl containing 3% DMSO, 0.5 μM Morphing psn apo1 dir, 0.5 μM RMLC, 1 mM dNTPs (0.25 mM each), 0.02 U/μl iProof DNA polymerase, and 0.2 ng/μl template. High-fidelity PCR was carried out on a gradient thermocycler using the following parameters: 98°C for 30 s (1 cycle); 98°C for 10 s, 55°C for 25 s, and 72°C for 45 s (28 cycles); and 72°C for 10 min (1 cycle). The whole gene was in vivo reassembled and recombined by transforming the different PCR products into S. cerevisiae competent cells, a process facilitated by ∼50-bp overhangs flanking each recombination area. The DNA transformation mixture was composed of linearized plasmid (100 ng) mixed with the mutagenized leader (200 ng) and the mature nonmutagenized protein (200 ng).
Fourth generation. (i) Error-prone PCR and in vivo DNA shuffling.
Mutagenic PCRs were performed separately with mutants I13D3, M5D2, and M4D8. The mutated PCR products were mixed with the linearized vector (at a PCR product/linearized plasmid ratio of 6:1) and transformed into competent S. cerevisiae cells to promote in vivo DNA shuffling.
(ii) Site-directed mutagenesis.
The I13D3 mutant from the third generation was used as a template to introduce F[12]Y, A[14]V, and R[15]G mutations (positions in the signal peptide are indicated in brackets) using in vivo overlap extension (IVOE) (29). Two high-fidelity PCRs were performed in a final volume of 50 μl containing (i) 3% DMSO, 0.5 μM RMLN, 0.5 μM PSN*R, 1 mM dNTPs (0.25 mM each), 0.02 U/μl iProof DNA polymerase, and 0.2 ng/μl template or (ii) 3% DMSO, 0.5 μM PSN*F, 0.5 μM RMLC, 1 mM dNTPs (0.25 mM each), 0.02 U/μl iProof DNA polymerase, and 0.2 ng/μl template. The following PCR parameters were used for each reaction: (i) 98°C for 30 s (1 cycle), 98°C for 10 s, 47°C for 25 s, 72°C for 15 s (28 cycles), and 72°C for 10 min (1 cycle); (ii) 98°C for 30 s (1 cycle), 98°C for 10 s, 52°C for 25 s, 72°C for 40 s (28 cycles), and 72°C for 10 min (1 cycle). Both PCR products (200 ng each) were mixed with the linearized vector (100 ng) and transformed into S. cerevisiae for in vivo gene reassembly and cloning. Overlapping areas of ∼50 bp flanking each segment were created to maximize the efficiency of in vivo DNA splicing between fragments.
Fifth generation.
The V57A mutation from 22A10 was introduced into 2A12 by mutational recovery through IVOE. Two high-fidelity PCRs were performed in a final volume of 50 μl containing (i) 3% DMSO, 0.5 μM RMLN, 0.5 μM 2A12*REV, 1 mM dNTPs (0.25 mM each), 0.02 U/μl iProof DNA polymerase, and 0.2 ng/μl template or (ii) 3% DMSO, 0.5 μM 2A12*DIR, 0.5 μM RMLC, 1 mM dNTPs (0.25 mM each), 0.02 U/μl iProof DNA polymerase, and 0.2 ng/μl DNA template. The following PCR parameters were used for each reaction: (i) 98°C for 30 s (1 cycle), 98°C for 10 s, 47°C for 25 s, 72°C for 15 s (28 cycles), and 72°C for 10 min (1 cycle); (ii) 98°C for 30 s (1 cycle), 98°C for 10 s, 52°C for 25 s, 72°C for 35 s (28 cycles), and 72°C for 10 min (1 cycle). Both PCR products (200 ng each) were mixed with the linearized vector (100 ng) and transformed into S. cerevisiae for in vivo gene reassembly and cloning as described above.
High-throughput screening.
Peroxidative and peroxygenase activities were screened with the help of a dual HT assay based on the oxidation of ABTS [2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid)] and the hydroxylation of 5-nitro-1,3-benzodioxole (NBD) (which is in turn spontaneously cleaved to form the chromophore 4-nitrocatechol), as indicated in the supplemental material. Three consecutive rescreenings were carried out to rule out the selection of false positives, which included a thermostability assay for the estimation of T50 (the temperature at which the enzyme retains 50% of its initial activity after 10 min of incubation) values.
Purification and biochemical characterization.
n*-UPO1, the PaDa-I mutant, and wtUPO1 were produced, purified, and biochemically characterized as described in the supplemental material.
RESULTS AND DISCUSSION
Point of departure: construction of fusion genes and design of the HTS assay.
The starting point of this study was the cDNA (upo1 gene) coding for the unspecific peroxygenase from A. aegerita (accession no. FM872457) (30, 31). This gene encodes a protein of 328 amino acids plus a 43-amino-acid signal peptide that directs secretion in A. aegerita. To achieve sufficient expression in the heterologous host in order to begin directed evolution, several constructs were prepared that contained the native signal sequence (n-UPO1), the α-factor preproleader from S. cerevisiae (α-UPO1), and the evolved α-factor preproleader (α*-UPO1). The evolved α-factor preproleader (α*) was previously engineered in association with an HRPL, achieving functional levels of expression after 8 rounds of laboratory evolution (18). The α* construct contained V10D-N23K-A87K mutations that boosted the expression of other HRPL genes (17, 20, 32) and that may also enhance UPO1 secretion. The secretion of each of the fusion genes was determined in 96-well plate microfermentations (mU ABTS/liter): n-UPO1, 149; α-UPO1, 74; α*-UPO1, negligible. While the use of evolved α-factor preproleaders as universal peptides for heterologous expression has been proposed (33), this approach appears to work only when signal sequences are switched between protein templates with a high degree of sequence identity (as in the case of HRPLs), indicating that they cannot be used in other, less related systems.
Several substrates were tested to develop a screening assay for directed UPO evolution. They included benzyl alcohol, veratryl alcohol, NBD, 2,6-dimethoxyphenol (DMP), ABTS, and p-nitro-phenoxy carboxylic acid (pNCA). Given the low levels of UPO1 secretion in microtiter plates, only the ABTS oxidation assay was reliable and stable, with good signal response and a low level of interference in culture broth. This assay was adjusted for the appropriate substrate concentrations (0.3 and 2 mM ABTS and H2O2, respectively) and the optimum pH (4.4). After improving secretion levels (from the second round of evolution onward), a peroxygenase (oxygen transfer) assay using NBD as the substrate was also incorporated into the screening protocol to maintain or even improve mono(per)oxygenase activity. UPO converts NBD into 4-nitrocatechol (yellow) via an initial hydroxylation and the subsequent spontaneous release of formic acid. 4-Nitrocatechol can be deprotonated at basic pH values to produce a strong red color (34). The microfermentation conditions were optimized to minimize interference during screening, evaluating several heme sources (δ-aminolevulinic acid, hemine, and hemoglobin), the concentration of MgSO4 (a source of structural Mg2+), and the effect of ethanol on membrane permeability, as well as a range of temperatures, stirring rates, and periods of incubation (see the supplemental material for details). The heme source chosen for UPO expression (hemoglobin) generated unwanted background activity during screening, and it was removed from the expression medium in the last rounds of evolution, as secretion was sufficiently high in its absence. Moreover, the coefficient of variance of the assays was reduced by up to 12% in the final cycles of evolution due to the stronger activity displayed by the variants. Three consecutive rescreenings were performed to rule out the presence of false positives. To protect UPO stability during the course of the evolution, the T50 was estimated for each mutant studied during the third rescreening.
Directed evolution.
The total activity of UPO1 was improved ∼3,250-fold with respect to the parental type after 5 generations (9,000 clones screened) of directed evolution. Moreover, expression levels of ∼8 mg/liter were achieved with activity values of 6,500 U ABTS/liter and 1,300 U NBD/liter. A combination of several error-prone PCR strategies with in vivo DNA recombination protocols, focused domain mutagenesis on the signal peptide, and mutational recovery was performed. In the first generation, both the n-UPO1 and α-UPO1 fusion genes were used as starting points to enhance expression by subjecting the complete constructs to random mutagenesis and recombination. Accordingly, 4 mutant libraries were designed using different DNA polymerases and mutational loads, 2 for each construct. Mutants selected after several consecutive rescreenings were derived exclusively from n-UPO1 libraries. The 2 best variants from this round were 1A11 (L67F) and 3C2 (I248V-F311L), with 13- and 9-fold improvements in total activity compared to n-UPO1, respectively (Fig. 1; see Table S1 in the supplemental material).
FIG 1.
Route for the directed evolution of UPO1 toward functional expression and improved activity. New mutations are depicted as stars and accumulated mutations as squares. Mutations in the mature PaDa-I mutant and their origins are boxed. The signal peptide is represented in dark red and the mature protein in red. In the parental n-UPO1, the glycosylation sites (Asn11, Asn141, Asn161, Asn182, Asn286, and Asn295) are represented as blue stars, the Phe triad (Phe69, Phe121, and Phe199) involved in the binding of aromatic substrates is marked with green arrows, and the acid-base pair for peroxide cleavage (Glu196 and Arg189) is indicated with blue arrows. TAI represents the improvement in UPO1 activity detected in S. cerevisiae microcultures for each mutant compared with the parental n-UPO1. Thermostability (T50) was estimated from culture supernatants (Fig. 2B). The breakdown of the TAI into specific activity and expression is shown in Fig. 3. The dashed arrows indicate the parental types used for each round of evolution. In the 3rd generation, “a” indicates the offspring obtained by IvAM of 12C12 and “b” indicates the offspring obtained by MORPHING in the signal peptide of 12C12. In the 4th generation, “c” indicates the triple mutant at the signal peptide constructed using I13D3 as a template and “d” the offspring obtained by mutagenic PCR and shuffling of parents I13D3, M5D2, and M4D8. n.d., not determined; n.m., not measurable. (See also Table S1 in the supplemental material.)
The mutations of 1A11 and 3C2 were sufficiently distant (L67F of 1A11 was located at a distance of 181 residues from I248V in the 3C2 mutant) to permit a suitable crossover event to take place in the next cycle of evolution via the yeast in vivo recombination machinery. Accordingly, in the second generation, these 2 variants were subjected to random mutagenesis and in vivo DNA shuffling. As planned, all variants selected from this round combined L67F-I248V-F311L, and they included some extra point mutations in either the mature protein or the signal peptide. The mutant best secreted in this generation (12C12) contained the aforementioned L67F-I248V-F311L mutations, as well as A[21]D in the signal peptide. At this stage of evolution, the NBD assay could be incorporated into the screening protocol to measure oxygen transfer activity, thanks to the high levels of secretion observed for the 12C12 mutant. The use of the dual screening assay based on the NBD/ABTS ratio allowed mutant hits to be selected without jeopardizing activity and variants with improved NBD/ABTS ratios to be identified (see below).
In the third generation, a dual approach was taken. First, mutant libraries with different mutational spectra were assembled in vivo by IvAM (27), using the 12C12 mutant (including its signal peptide) as a template (Fig. 1a). In addition, the signal peptide of 12C12 was subjected to focal mutagenesis by MORPHING (28) in an attempt to enrich the signal peptide in mutations that favor secretion (Fig. 1b). Library landscapes revealed a higher tolerance for mutations in the leader than in the whole UPO1 gene. This is consistent with the observation that mutations in the leader affect only secretion whereas mutations at the whole-gene level may also modify catalytic properties. The most promising mutant from the IvAM library, I13D3, contained the new V75I mutation, and it displayed a 70-fold total activity improvement with respect to n-UPO1. By focusing mutational loads in the signal peptide, 3 beneficial mutations were introduced between positions 12 and 15 in 3 independent mutant winners. Mutations F[12]Y, A[14]V, and R[15]G in the leader were so close to one another that the likelihood of recombination by in vitro or in vivo methods was very low. Thus, for the fourth generation, we constructed a triple mutant by site-directed mutagenesis using the I13D3 mutant as a template (Fig. 1c). The resulting mutant (2A12) contained 4 beneficial mutations, F[12]Y-A[14]V-R[15]G-A[21]D, in the signal sequence, and it showed markedly improved secretion. In addition, a new round of random mutagenesis and recombination was also performed using I13D3, M5D2, and M4D8 as templates. Although M4D8 was not the best variant in generation 3, it was chosen as the parent due to the mutational redundancy observed at position 4, as this substitution also appeared in the 3B5 mutant from generation 2. From this set of experiments, we identified the 22A10 mutant, generated by a recombination event between M5D2 and I13D3 and containing the new mutation V57A (Fig. 1d). Finally, the V57A mutation was introduced into 2A12 by mutational recovery, giving rise to the final mutant, PaDa-I.
Biochemical characterization.
Wild-type UPO1, produced homologously by A. aegerita (wtUPO1), and the PaDa-I mutant secreted in S. cerevisiae were purified to homogeneity (Reinheitszahl value [Rz] [A418/A280], ∼2) and characterized biochemically (Table 1; see Fig. S1 in the supplemental material). The average molecular mass measured by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) was 51,100 Da for PaDa-I (i.e., ∼5,000 Da higher than that of wtUPO1), and the contribution of glycosylation deduced by deglycosylation gels was around 22% for the wild type and 30% for the mutant (see Fig. S1 in the supplemental material). S. cerevisiae tends to hyperglycosylate foreign proteins up to levels of ∼50%, conferring on them increased stability and protection against proteolytic degradation. The glycosylation of wtUPO1 observed was exclusively dependent on 6 predicted N-glycosylation sites (O-glycosylation sites are not described for the enzyme), associated with up to 8 moieties of the high-mannose type (30, 35). None of the amino acid substitutions in PaDa-I introduced new glycosylation motifs, and thus, the higher sugar content in the mutant may be due to an increased Golgi residence time that leads to the addition of more mannose moieties, as described in other directed-evolution studies in yeast (14, 17). The PaDa-I and wtUPO1 proteins had similar spectroscopic characteristics in the Fe3+ resting state, with both enzymes showing a maximum in the Soret region of around 418 nm and 2 Q bands at 570 and 540 nm (Fig. 2A).
TABLE 1.
Biochemical features of the wild type and the evolved UPO variant
| Biochemical or spectroscopy featureh | Value |
|
|---|---|---|
| wtUPO1e | PaDa-I mutantf | |
| Mass (Da)a | 46,000 | 52,000 |
| Mass (Da)b | NDg | 51,100 |
| Mass (Da)c | 35,942 | 35,914 |
| Glycosylation degree (%) | 22 | 30 |
| Thermal stability (T50) (°C)d | 53 | 55 |
| pI | 4.9–5.7 | 5.5 |
| Optimum pH for ABTS | 4.0 | 4.0 |
| Optimum pH for DMP | 7.0 | 6.0 |
| Optimum pH for NBD | 6.5 | 6.0 |
| Rz (A418/A280) | 2.4 | 1.8 |
| Soret region (nm) | 420 | 418 |
| CT1 (nm) | 572 | 570 |
| CT2 (nm) | 540 | 537 |
Estimated by SDS-PAGE.
Estimated by MALDI-TOF mass spectrometry.
Estimated from the amino acid composition.
Estimated from purified variants.
wtUPO1, UPO1 wild type expressed in A. aegerita.
PaDa-I mutant, the ultimate variant of the whole evolution process in S. cerevisiae (containing the evolved signal peptide [n*] plus the evolved UPO1) (see Fig. S1 in the supplemental material).
ND, not determined.
CT1 and CT2, charge transference bands 1 and 2, respectively.
FIG 2.
Biochemical characterization. (A) Spectroscopic characteristics of wtUPO1 (dashed line) and the PaDa-I mutant (solid line). AU, arbitrary units. (B) Thermostability (T50) of PaDa-I and different parental types. Each point represents the mean and standard deviation of 3 independent experiments. 1G to 5G, 1st generation to 5th generation. (C and D) pH activity profiles for wtUPO1 (triangles) and PaDa-I (squares). Activities were measured in 100 mM citrate/phosphate/borate buffer at different pH values with 2 mM H2O2 and 0.3 mM ABTS (C) or with 1 mM H2O2 and 1 mM NBD (D). UPO1 activity was normalized to the optimum activity value, and each point represents the mean and standard deviation of 3 independent experiments.
The very weak expression of the parental n-UPO1 in S. cerevisiae (∼0.007 mg/liter) hampered its purification to homogeneity. To overcome this impediment, we constructed a fusion gene in which the native signal peptide of n-UPO1 was replaced by that obtained after 5 cycles of directed evolution in yeast (n*, containing F[12]Y A[14]V R[15]G A[21]D mutations). Larger amounts of native UPO1 were produced in S. cerevisiae from the n*-UPO1 fusion gene, which was then purified to homogeneity. This approach allowed us to make an accurate breakdown of the total activity improvement (TAI) in terms of both specific activity and heterologous functional expression (Fig. 3). The product of the n*-UPO1 fusion gene showed biochemical characteristics similar to those of the PaDa-I mutant in terms of molecular mass, the degree of glycosylation, and thermal stability. After large-scale fermentation, the PaDa-I protein showed a 3,250-fold TAI compared to the parental n-UPO1 (6,500 U ABTS/liter for PaDa-I versus 2 U ABTS/liter for n-UPO1), an improvement that was 20-fold greater than that obtained in microtiter plates, where stirring conditions and oxygen availability were limiting. The product of n*-UPO1 showed a 27-fold TAI with respect to n-UPO1, reflecting the potency of the evolved signal peptide in promoting UPO1 secretion. The breakdown of the TAI value revealed a 1,114-fold increase in functional expression and a 3.6-fold increase in peroxidative activity (using ABTS) with respect to the parental type. Bearing in mind that n* enhanced functional expression 27-fold, there was a further ∼41-fold improvement in functional expression conferred by mutations in the mature UPO1. Secretion levels were significantly enhanced, from 0.007 mg/liter to 8 mg/liter, i.e., to levels similar to those obtained in the original fungus A. aegerita (31).
FIG 3.
Breakdown of specific activity and functional expression. A fusion gene containing the evolved signal peptide (n*) attached to the native mature UPO1 was engineered. The n-UPO1, n*-UPO1, and PaDa-I variants were produced on a large scale, and their TAIs were measured. n*-UPO1 and PaDa-I were purified, and their specific activities were calculated. n* enhanced functional expression ∼27-fold, whereas mutations in mature PaDa-I resulted in an ∼120-fold increase in total activity. The 3,250-fold increase in the total activity of PaDa-I was broken down as a 3.6-fold improvement in specific activity and a 1,114-fold improvement in functional expression.
The pH profiles for peroxidative (with ABTS and DMP) and peroxygenase (with NBD) activities revealed similar shapes and optimum pH values for wtUPO1, n*-UPO1, and PaDaI (∼4.0, 5.0, and 6.0 for ABTS, DMP, and NBD, respectively) (Fig. 2C and D and Table 1). Kinetic constants for peroxidative and peroxygenase activities were assayed using several compounds, along with aryl alcohols with different redox potentials (Table 2). The kcat for n*-UPO1 expressed in yeast was ∼5-fold lower than that of wtUPO1, although they showed similar substrate affinities, with the exception of the Km for H2O2 (2-fold lower in n*-UPO1). General differences in protein folding in the heterologous host, particularly those affecting posttranslational modifications (e.g., hyperglycosylation), may affect the activity of the recombinant enzyme. These results are consistent with those reported for lignin-modifying enzymes heterologously expressed in yeast, in which a decrease in the kcat of up to 10-fold was observed (36). Both PaDa-I and wtUPO1 showed very similar catalytic efficiencies for all the substrates tested, indicating marked improvements in the activity of UPO1 after each round of evolution. Indeed, a notable improvement in the kcat value was detected when n*-UPO1 was compared with PaDa-I (with increases of 10-, 9-, 8-, and 7-fold for NBD, ABTS, veratryl alcohol, and benzyl alcohol, respectively), and these increases accounted for an overall increase in catalytic efficiency: the 18-fold improvement in the kcat/Km value for NBD hydroxylation comparing n*-UPO1 and PaDa-I was 2.2 times greater than that of wtUPO1 (i.e., 700 and 320 mM−1 s−1 for PaDa-I and wtUPO1, respectively). Thus, the dual screening assay used during in vitro evolution helped to conserve and even improve both peroxidative and mono(per)oxygenase activities.
TABLE 2.
Kinetic parameters of wild-type, recombinant, and evolved UPO variants
| Substrate | Kinetic constant | Valuea |
||
|---|---|---|---|---|
| wtUPO1 | n*-UPO1 | PaDa-I | ||
| ABTS | Km (mM) | 0.025 ± 0.002 | 0.027 ± 0.005 | 0.048 ± 0.004 |
| kcat (s−1) | 221 ± 6 | 45.0 ± 2.7 | 395 ± 13 | |
| kcat/Km (mM−1 s−1) | 8,800 ± 692 | 1,600 ± 37 | 8,200 ± 598 | |
| NBD | Km (mM) | 0.684 ± 0.207 | 0.782 ± 0.352 | 0.483 ± 0.095 |
| kcat (s−1) | 219 ± 25 | 31.7 ± 6.1 | 338 ± 22 | |
| kcat/Km (mM−1 s−1) | 320 ± 64 | 38.0 ± 11 | 700 ± 99 | |
| Benzyl alcohol | Km (mM) | 1.90 ± 0.11 | 1.10 ± 0.23 | 2.47 ± 0.32 |
| kcat (s−1) | 329 ± 7 | 44.8 ± 3.1 | 307 ± 15 | |
| kcat/Km (mM−1 s−1) | 174 ± 7 | 41.0 ± 6.3 | 124 ± 11 | |
| Veratryl alcohol | Km (mM) | 5.20 ± 0.31 | 5.30 ± 0.82 | 7.9 ± 0.7 |
| kcat (s−1) | 88 ± 2 | 15.2 ± 1.1 | 121 ± 5 | |
| kcat/Km (mM−1 s−1) | 17 ± 0.7 | 2.9 ± 0.25 | 15 ± 0.9 | |
| H2O2 | Km (mM) | 1.37 ± 0.16 | 0.69 ± 0.20 | 0.49 ± 0.06 |
| kcat (s−1) | 290 ± 15 | 40.9 ± 3.8 | 238 ± 8 | |
| kcat/Km (mM−1 s−1) | 211 ± 15 | 59.0 ± 12.3 | 500 ± 42 | |
ABTS kinetic constants for UPO1 were estimated in 100 mM sodium citrate/phosphate buffer, pH 4.4, containing 2 mM H2O2 and those for the rest of the substrates in 100 mM potassium phosphate buffer, pH 7.0, containing 2 mM H2O2 (benzyl and veratryl alcohols) or 1 mM H2O2 (NBD). H2O2 kinetic constants were estimated using benzyl alcohol as a reducing substrate under the corresponding saturated conditions. wtUPO1, UPO1 wild-type expressed in A. aegerita; n*-UPO, native UPO1 fused to the evolved signal peptide for secretion in S. cerevisiae; PaDa-I mutant, the ultimate variant of the whole evolution process in S. cerevisiae (containing the evolved signal peptide plus the evolved UPO1).
Many of the aromatic substrates and oxyfunctionalized compounds converted by UPO are further oxidized by the enzyme, generating a pool of products of various complexities. Accordingly, for certain applications, the removal of UPO's peroxidative activity (1-electron oxidation) may be considered a biotechnological priority. Using the HTS assay, we attempted to uncouple the peroxidation and monooxygenase activities. Taking the NBD/ABTS ratio as a discriminatory factor, the 3F10 variant (T120P mutation) identified in the fourth generation showed a dramatic decrease in peroxidative activity (a 4-fold decrease in the TAI for ABTS, from 132- to 31-fold) while conserving its peroxygenase activity (with similar TAIs for NBD and the parental 2A12) (Fig. 1). The signal peptide was switched in the 3F10 variant, as described for the n*-UPO1 construct (see the supplemental material), and the corresponding n*-3F10 construct was produced on a larger scale and compared with both wtUPO1 and n*-UPO1. The NBD/ABTS ratios (expressed as percentages) were 19%, 20%, and 61% for wtUPO1, n*-UPO1, and n*3F10, respectively. During the preliminary characterization of this variant, we detected a dramatic decrease in the thermostability produced by the beneficial but destabilizing T120P mutation (with a 7°C decrease in T50), which precluded its purification and further analysis. Although monooxygenase activity is a clear target for directed UPO evolution, our results indicate that the line between peroxidation and mono(per)oxygenase activity in UPO is very fine and that its catalytic mechanisms are strongly implicated in protein stability (10). This problem may be overcome by including iterative rounds of neutral genetic drift in order to introduce stabilizing mutations into the 3F10 mutant before further evolving its monooxygenase activity (37).
Kinetic thermostability was conserved over the course of evolution, with T50 values in the 57 to 59°C range for all the offspring of the mutants used as parental types in each round of directed evolution (Fig. 1 and 2B). This effect was due to the screening assay used during evolution, in which destabilizing mutations were excluded from the evolutionary pathway.
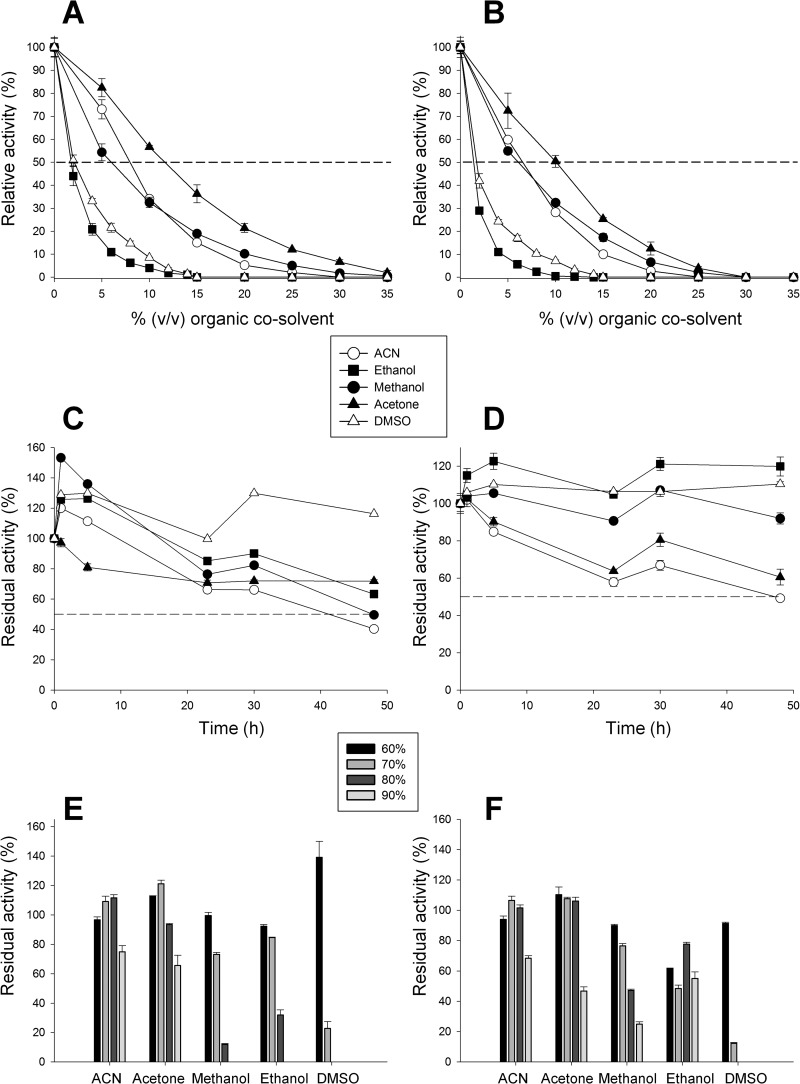
The presence of organic cosolvents is required for many of the transformations mediated by UPO. The activity and stability of wtUPO and the PaDa-I mutant were evaluated in the presence of high concentrations of cosolvents with different polarities (with a logP [octanol-water partition coefficient] ranging from −0.23 to −1.3) and chemical characteristics (Fig. 4A and B). Regardless of the enzyme tested, activity was reduced drastically in the presence of increasing concentrations of cosolvents in the following order: ethanol > DMSO > acetonitrile (ACN) ∼ methanol > acetone. Activities in cosolvents were estimated by measuring the concentration of cosolvent at which the enzyme shows 50% of the corresponding activity in buffer (C50). The strongest activity was observed in acetone (C50, 10 to 12%) and the weakest in ethanol and DMSO (C50, ∼2%). In terms of stability in cosolvents, both enzymes were very stable at concentrations as high as 50% (vol/vol), with a half-life of over 48 h and some hyperactivation (Fig. 4C and D). Under more extreme conditions (concentrations ranging from 60 to 90%), cosolvents exerted detrimental effects in the following order: DMSO > ethanol > methanol > acetone ∼ ACN. Interestingly, PaDa-I was very stable, retaining ∼25% and ∼55% of its activity at concentrations of up to 90% (vol/vol) methanol and ethanol, respectively, when the stability of wtUPO1 was negligible (Fig. 4E and F). The combined effects of hyperglycosylation together with the introduction of stabilizing mutations appears to underlie this resistance.
FIG 4.
Activity and stability in organic cosolvents. (A and B) The relative activities of wtUPO1 (A) and the PaDa-I mutant (B) in organic cosolvents were assessed with 2 mM H2O2 and 0.3 mM ABTS in 100 mM sodium phosphate/citrate buffer (pH 4.4) containing the corresponding concentration of cosolvent. (C and D) The stabilities of wtUPO1 (C) and the PaDa-I mutant (D) after incubation for 48 h in 50% organic cosolvents were assessed by incubating enzyme samples in 100 mM potassium phosphate buffer (pH 7.0) containing 50% (vol/vol) organic cosolvent in screw-cap vials. After 48 h, aliquots were removed and analyzed in an activity assay with 2 mM H2O2 and 0.3 mM ABTS in 100 mM sodium phosphate/citrate buffer (pH 4.4). (E and F) The stabilities of wtUPO1 (E) and the PaDa-I mutant (F) at high concentrations of organic cosolvents were assessed after 5 h of incubation in increasing concentrations of cosolvents and incubating enzyme samples at 20°C in 100 mM potassium phosphate buffer (pH 7.0) containing increasing concentrations (vol/vol) of organic cosolvent (60 to 90%). After 5 h, aliquots were removed and analyzed in the activity assay, as described for panels C and D. Residual activities were expressed as percentages of the original activity at the corresponding concentration of organic cosolvent. The error bars indicate standard deviations.
Mutation analysis.
PaDa-I harbored 9 beneficial mutations (no silent mutations were introduced during evolution), 4 in the signal peptide and 5 in the mature protein. Five of the mutations (3 in the mature protein and 2 in the signal peptide) favored codon usage, which might support secretion (see Table S1 in the supplemental material). The 4 mutations in the signal peptide were located in the hydrophobic core of the leader, and 3 of them (underlined) were nearly consecutive: F[12]Y-A[14]F-R[15]G-A[21]D. Substitutions at positions 12 and 21 enhanced the polarity of this region, while those at positions 14 and 15 had the opposite effect. Overall, these 4 mutations enhanced secretion by up to 27-fold, as seen in the n*-UPO1 fusion gene (Fig. 3). Subtle differences in the adjustment between the signal recognition particle (SRP) and the evolved signal peptide may benefit secretion, bearing in mind that the SRP strongly interacts with the hydrophobic region of the leader. It has been reported that SRP binding to the signal peptide pauses translation at different stages, depending on the nature of the leader (38). In our mutant, this arrest of translation could facilitate proper recognition by the signal peptidase before cleavage and translocation of the nascent UPO polypeptide to the endoplasmic reticulum (39).
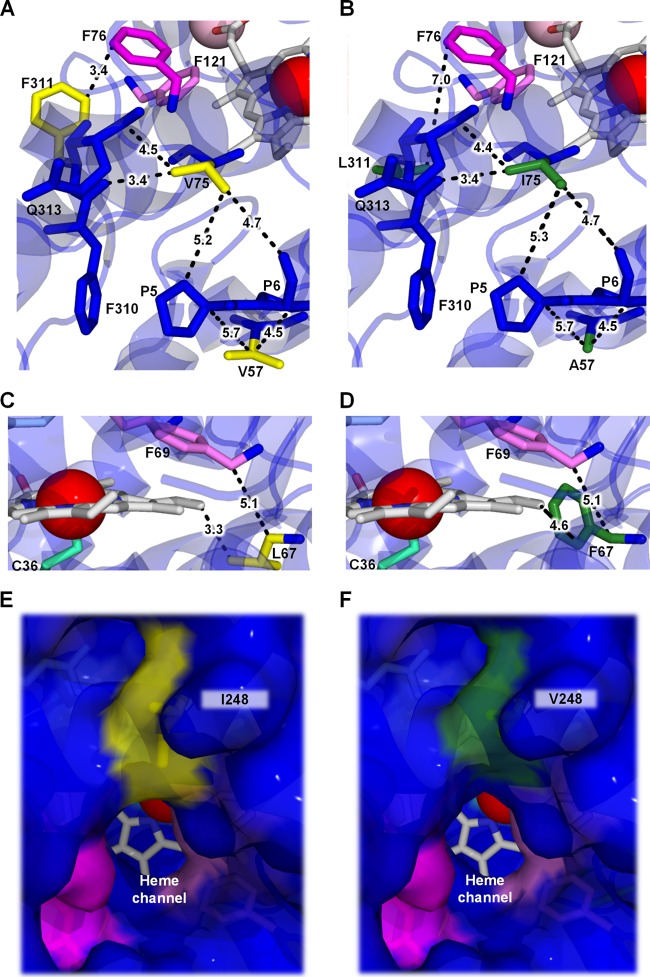
Mutations in the mature UPO1 were mapped onto the recently solved crystal structure of UPO1 (35). UPO1 is mostly formed by helical substructures composed of 1 halide binding site, 1 Mg2+ binding site, and the heme-thiolate domain, with Arg189 and Glu196 forming the acid-base pair for compound I formation. The funnel-shaped access channel to the substrate binding pocket is 8.5 Å in diameter and mainly covered with aromatic residues. This binding pocket is controlled by a Phe triad (Phe69, Phe121, and Phe199), which is essential for the orientation of (aromatic) substrates. All mutations were conservative in terms of polarity and charge, i.e., nonpolar substitutions (V57A, L67F, V75I, I248V, and F311L), leading to few apparent changes in terms of H bond or salt bridge formation/interruption (see Table S2 in the supplemental material). In fact, mutations were located in hydrophobic environments, in some cases far from the catalytic site (see Fig. S2 in the supplemental material). At this point, it is important to note that our aim was to improve total activity while conserving enzyme stability. Thus, we can only speculate as to whether a less rigid directed-evolution approach would have unmasked other, less conservative substitutions at the cost of threatening protein stability (40). The 41-fold enhancement in secretion induced by these mutations may be due to tighter folding during the earliest posttranslational stages, which ultimately favors protein stability and secretion. The V57A mutation lies in a helix at the surface of the protein next to the N terminus (Fig. 5A and B). According to our model, the replacement of Val57 with a less bulky residue may compress this region between adjacent prolines (at positions 5 and 6 of the N terminus) and thereby increase the protein's robustness. The L67F mutation is located in the vicinity of the catalytic pocket, very close to Phe69 of the Phe triad involved in binding aromatic substrates. This mutation may be partially responsible for the kinetic enhancements observed, since after substitution, the aromatic ring of Phe67 is oriented toward the active site (Fig. 5C and D). The V75I mutation is produced by the replacement of a hydrophobic residue with another, slightly larger hydrophobic residue, which may establish new hydrophobic contacts with surrounding residues, thereby favoring protein stability (Fig. 5A and B). Finally, the I248V and F311L mutations are positioned in the surroundings of the heme channel. The I248V mutation is produced by the replacement of Ile248 at the entrance of the channel to the heme cavity with a Val residue, which could favor the access of bulkier substrates (Fig. 5E and F). The F311L mutation is located in front of the heme channel, with Phe76 in between. The replacement of Phe with Leu enlarges the cavity, which may in turn have beneficial effects on kinetics (Fig. 5A and B).
FIG 5.
Mutations in evolved UPO1. A molecular model using as the template the A. aegerita crystal structure (PDB code 2YOR) was prepared to map the mutations. Shown are details of the 5 mutations (green) in the PaDa-I mutant (B, D, and F) compared with the corresponding residues (yellow) in the native UPO1 (A, C, and E). The dashed lines indicate distances (in Å) from the surrounding residues. Phe residues delimiting the active site are highlighted in pink and the Cys36 axial ligand in light blue. The Fe3+ of heme is shown as a red sphere and the structural Mg2+ as a pink sphere. (See also Table S2 and Fig. S2 in the supplemental material.)
Conclusions.
Since its discovery 10 years ago, the potential use of UPO in applications ranging from chemical processes (including some relevant industrial transformations, such as alkane hydroxylation and olefin epoxidation) to the preparation of O- and N-dealkylated human drug metabolites, as well as bioremediation (polycyclic aromatic hydrocarbon [PAH] oxidation) and biosensor development, has been studied exhaustively (41–50). For decades, regio- and enantioselective oxyfunctionalization has been a “forbidden territory” for most biocatalysts, except for P450 monooxygenases. However, unlike the latter, UPO is soluble and requires neither expensive cofactors (NAD[P]H) nor auxiliary flavoproteins. Despite these advantages, the lack of suitable directed-evolution platforms with which to enhance UPO's catalytic properties has limited the exploitation of this versatile biocatalyst. The directed-evolution process presented here describes for the first time an attractive pathway through which ad hoc UPO variants can be tailored for use in several industrial reactions, such as alkane hydroxylation and the transformation of benzene into phenol and naphthalene into naphthol (51, 52).
The evolved UPO1 variant from this study is very active and stable over a wide temperature range, as well as in the presence of a variety of cosolvent types. Easily secreted by yeast, this mutant and any future evolved variants could be translated to other expression systems for overproduction. Preliminary trials in the methylotrophic yeast Pichia pastoris indicate that the evolved enzyme is overproduced 5-fold thanks to the increased density in the host (unpublished data). Future goals for UPO engineering include the conversion of the enzyme into an enantioselective self-sufficient mono(per)oxygenase by quenching its peroxidative activity, the improvement of its activity in the presence of cosolvents, and the enhancement of its oxidative/operational stability in the presence of peroxides. The future combination of directed evolution (including neutral genetic drift) and rational/hybrid design should provide a wealth of information that will help us to better understand the complex mechanism of action as UPO becomes an efficient oxyfunctionalization biocatalyst.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by European Commission Projects (Peroxicats-FP7-KBBE-2010-4-26537 and Indox-FP7-KBBE-2013-7-613549) and the National Project (Evofacel) (BIO2010-19697).
Footnotes
Published ahead of print 28 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00490-14.
REFERENCES
- 1.Hofrichter M, Ullrich R. 2006. Heme-thiolate haloperoxidases: versatile biocatalysts with biotechnological and environmental significance. Appl. Microbiol. Biotechnol. 71:276–288. 10.1007/s00253-006-0417-3 [DOI] [PubMed] [Google Scholar]
- 2.Hofrichter M, Ullrich R, Pecyna MJ, Liers C, Lundell T. 2010. New and classic families of secreted fungal heme peroxidases. Appl. Microbiol. Biotechnol. 87:871–897. 10.1007/s00253-010-2633-0 [DOI] [PubMed] [Google Scholar]
- 3.Anh DH, Ullrich R, Benndorf D, Svatos A, Muck A, Hofrichter M. 2007. The coprophilous mushroom Coprinus radians secretes a haloperoxidase that catalyzes aromatic peroxygenation. Appl. Environ. Microbiol. 73:5477–5485. 10.1128/AEM.00026-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gröbe G, Ullrich R, Pecyna MJ, Kapturska D, Friedrich S, Hofrichter M, Scheibner K. 2011. High-yield production of aromatic peroxygenase by the agaricus fungus Marasmius rotula. AMB Express 1:31. 10.1186/2191-0855-1-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Babot ED, del Rio JC, Kalum L, Martinez AT, Gutierrez A. 2013. Oxyfunctionalization of aliphatic compounds by a recombinant peroxygenase from Coprinopsis cinerea. Biotechnol. Bioeng. 110:2323–2332. 10.1002/bit.24904 [DOI] [PubMed] [Google Scholar]
- 6.Floudas D, Binder M, Riley R, Barry K, Blanchette RA, Henrissat B, Martinez AT, Otillar R, Spatafora JW, Yadav JS, Aerts A, Benoit I, Boyd A, Carlson A, Copeland A, Coutinho PM, de Vries RP, Ferreria P, Findley K, Foster B, Gaskell J, Glotzer D, Gorecki P, Heitman J, Hesse C, Hori C, Igarashi K, Jurgens JA, Kallen N, Kersten P, Kohler A, Kues U, Arun Kumar TK, Kuo A, Labutti K, Larrondo LF, Lindquist E, Ling A, Lombard V, Lucas S, Lundell T, Martin R, McLaughlin DJ, Morgenstern I, Morin E, Murant C, Nagy LG, Nolan M, Ohm RA, Patyshakuliyeva A, Rokas A, Ruiz-Dueñas FJ, Sabat G, Salamov A, Samejima M, Schumtz J, Slot JC, St. John F, Stenlid J, Sun H, Sun S, Syed K, Tsang A, Wiebenga A, Young D, Pisabarro A, Eastwood DC, Martin F, Cullen D, Grigoriev IV, Hibbet DS. 2012. The paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336:1715–1719. 10.1126/science.1221748 [DOI] [PubMed] [Google Scholar]
- 7.Hofrichter M, Kellner H, Pecyna M, Ullrich R. Fungal unspecific peroxygenases: heme-thiolate proteins that combine peroxidase and cytochrome P450 properties. In Hrycay EG. (ed), Monooxygenase, peroxidase and peroxygenase properties and mechanisms of cytochrome P450, in press Springer, Heidelberg, Germany: [DOI] [PubMed] [Google Scholar]
- 8.Torres Pazmino DE, Winkler M, Glieder A, Fraaije MW. 2010. Monooxygenases as biocatalysts: classification, mechanistic aspects and biotechnological applications. J. Biotechnol. 146:9–24. 10.1016/j.jbiotec.2010.01.021 [DOI] [PubMed] [Google Scholar]
- 9.Wang X, Peter S, Kinne M, Hofrichter M, Groves JT. 2012. Detection and kinetic characterization of a highly reactive heme-thiolate peroxygenase compound I. J. Am. Chem. Soc. 134:12897–12900. 10.1021/ja3049223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Peter S, Ullrich R, Hofrichter M, Groves JT. 2013. Driving force for oxygen-atom transfer by heme-thiolate enzymes. Angew. Chem. Int. Ed. Engl. 52:9238–9241. 10.1002/anie.201302137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conesa A, van de Velde F, van Rantwijk F, Sheldon RA, van den Hondel CAMJ, Punt PJ. 2001. Expression of the Caldariomyces fumago chloroperoxidase in Aspergillus niger and characterization of the recombinant enzyme. J. Biol. Chem. 276:17635–17640. 10.1074/jbc.M010571200 [DOI] [PubMed] [Google Scholar]
- 12.Shaw P, Beckwith J. 1960. Chloroperoxidase: a component of the beta-ketoadipate chlorinase system. Federation Proc. 19:47 [Google Scholar]
- 13.Pourmir A, Johannes TW. 2012. Directed evolution: selection of the host microorganism. Comput. Struct. Biotechnol. J. 2:e201209012. 10.5936/csbj.201209012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bulter T, Alcalde M, Sieber V, Meinhold P, Schlachtbauer C, Arnold FH. 2003. Functional expression of a fungal laccase in Saccharomyces cerevisiae by directed evolution. Appl. Environ. Microbiol. 69:987–995. 10.1128/AEM.69.2.987-995.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Ruiz E, Gonzalez-Perez D, Ruiz-Dueñas FJ, Martinez AT, Alcalde M. 2012. Directed evolution of a temperature-, peroxide- and alkaline pH-tolerant versatile peroxidase. Biochem. J. 441:487–498. 10.1042/BJ20111199 [DOI] [PubMed] [Google Scholar]
- 16.Zumarraga M, Bulter T, Shleev S, Polaina J, Martinez-Arias A, Plou FJ, Ballesteros A, Alcalde M. 2007. In vitro evolution of a fungal laccase in high concentrations of organic cosolvents. Chem. Biol. 14:1052–1064. 10.1016/j.chembiol.2007.08.010 [DOI] [PubMed] [Google Scholar]
- 17.Camarero S, Pardo I, Cañas A, Molina P, Record E, Martinez AT, Martinez MJ, Alcalde M. 2012. Engineering platforms for directed evolution of laccase from Pycnoporus cinnabarinus. Appl. Environ. Microbiol. 78:1370–1384. 10.1128/AEM.07530-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mate D, Garcia-Burgos C, Garcia-Ruiz E, Ballesteros AO, Camarero S, Alcalde M. 2010. Laboratory evolution of high-redox potential laccases. Chem. Biol. 17:1030–1041. 10.1016/j.chembiol.2010.07.010 [DOI] [PubMed] [Google Scholar]
- 19.Mate DM, Gonzalez-Perez D, Falk M, Kittl R, Pita M, De Lacey AL, Ludwig R, Shleev S, Alcalde M. 2013. Blood tolerant laccase by directed evolution. Chem. Biol. 20:223–231. 10.1016/j.chembiol.2013.01.001 [DOI] [PubMed] [Google Scholar]
- 20.Pardo I, Vicente AI, Mate DM, Alcalde M, Camarero S. 2012. Development of chimeric laccases by directed evolution. Biotechnol. Bioeng. 109:2978–2986. 10.1002/bit.24588 [DOI] [PubMed] [Google Scholar]
- 21.Gonzalez-Perez D, Garcia-Ruiz E, Alcalde M. 2012. Saccharomyces cerevisiae in directed evolution: an efficient tool to improve enzymes. Bioeng. Bugs 3:172–177. 10.4161/bbug.19544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pirakitikulr N, Ostrov N, Peralta-Yahya P, Cornish VW. 2010. PCRless library mutagenesis via oligonucleotide recombination in yeast. Protein Sci. 19:2336–2346. 10.1002/pro.513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shao ZY, Zhao H, Zhao HM. 2009. DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res. 37:e16. 10.1093/nar/gkn991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Da Silva NA, Srikrishnan S. 2012. Introduction and expression of genes for metabolic engineering applications in Saccharomyces cerevisiae. FEMS Yeast Res. 12:197–214. 10.1111/j.1567-1364.2011.00769.x [DOI] [PubMed] [Google Scholar]
- 25.Hong KK, Nielsen J. 2012. Metabolic engineering of Saccharomyces cerevisiae: a key cell factory platform for future biorefineries. Cell. Mol. Life Sci. 69:2671–2690. 10.1007/s00018-012-0945-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krivoruchko A, Siewers V, Nielsen J. 2011. Opportunities for yeast metabolic engineering: lessons from synthetic biology. Biotechnol. J. 6:262–276. 10.1002/biot.201000308 [DOI] [PubMed] [Google Scholar]
- 27.Zumarraga M, Camarero S, Shleev S, Martínez-Arias A, Ballesteros A, Plou FJ, Alcalde M. 2008. Altering the laccase functionality by in vivo assembly of mutant libraries with different mutational spectra. Proteins 71:250–260. 10.1002/prot.21699 [DOI] [PubMed] [Google Scholar]
- 28.Gonzalez-Perez D, Molina-Espeja P, Garcia-Ruiz E, Alcalde M. 2014. Mutagenic organized recombination process by homologous in vivo grouping (MORPHING) for directed enzyme evolution. PLoS One 9:e90919. 10.1371/journal.pone.0090919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alcalde M. 2010. Mutagenesis protocols in Saccharomyces cerevisiae by in vivo overlap extension. Methods Mol. Biol. 634:3–14. 10.1007/978-1-60761-652-8_1 [DOI] [PubMed] [Google Scholar]
- 30.Pecyna MJ, Ullrich R, Bittner B, Clemens A, Scheibner K, Schubert R, Hofrichter M. 2009. Molecular characterization of aromatic peroxygenase from Agrocybe aegerita. Appl. Microbiol. Biotechnol. 84:885–897. 10.1007/s00253-009-2000-1 [DOI] [PubMed] [Google Scholar]
- 31.Ullrich R, Nüske J, Scheibner K, Spantzel J, Hofrichter M. 2004. Novel haloperoxidase from the agaric basidiomycete Agrocybe aegerita oxidizes aryl alcohols and aldehydes. Appl. Environ. Microbiol. 70:4575–4581. 10.1128/AEM.70.8.4575-4581.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mate D, Garcia-Ruiz E, Camarero S, Alcalde M. 2011. Directed evolution of fungal laccases. Curr. Genomics 12:113–122. 10.2174/138920211795564322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rakestraw J, Sazinsky SL, Piatesi A, Antipov E, Wittrup K. 2009. Directed evolution of a secretory leader for the improved expression of heterologous proteins and full-length antibodies in Saccharomyces cerevisiae. Biotechnol. Bioeng. 103:1192–1201. 10.1002/bit.22338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poraj-Kobielska M, Kinne M, Ullrich R, Scheibner K, Hofrichter M. 2012. A spectrophotometric assay for the detection of fungal peroxygenases. Anal. Biochem. 421:327–329. 10.1016/j.ab.2011.10.009 [DOI] [PubMed] [Google Scholar]
- 35.Piontek K, Strittmatter E, Ullrich R, Gröbe G, Pecyna MJ, Kluge M, Scheibner K, Hofrichter M, Plattner DA. 2013. Structural basis of substrate conversion in a new aromatic peroxygenase: P450 functionality with benefits. J. Biol. Chem. 288:34767–34776. 10.1074/jbc.M113.514521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mate DM, Garcia-Ruiz E, Camarero S, Shubin VV, Falk M, Shleev S, Ballesteros AO, Alcalde M. 2013. Switching from blue to yellow: altering the spectral properties of a high redox potential laccase by directed evolution. Biocatal. Biotransform. 31:8–21. 10.3109/10242422.2012.749463 [DOI] [Google Scholar]
- 37.Goldsmith M, Tawfik DS. 2013. Enzyme engineering by targeted libraries. Methods Enzymol. 523:257–283. 10.1016/B978-0-12-394292-0.00012-6 [DOI] [PubMed] [Google Scholar]
- 38.Nothwehr SF, Gordon JI. 1990. Targeting of proteins into the eukaryotic secretory pathway: signal peptide structure/function relationships. Bioessays 12:479–484. 10.1002/bies.950121005 [DOI] [PubMed] [Google Scholar]
- 39.Romanos MA, Scorer CA, Clare JJ. 1992. Foreign gene expression in yeast: a review. Yeast 8:423–488. 10.1002/yea.320080602 [DOI] [PubMed] [Google Scholar]
- 40.Bloom JD, Labthavikul ST, Otey CR, Arnold FH. 2006. Protein stability promotes evolvability. Proc. Natl. Acad. Sci. U. S. A. 103:5869–5874. 10.1073/pnas.0510098103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aranda E, Ullrich R, Hofrichter M. 2010. Conversion of polycyclic aromatic hydrocarbons, methyl naphthalenes and dibenzofuran by two fungal peroxygenases. Biodegradation 21:267–281. 10.1007/s10532-009-9299-2 [DOI] [PubMed] [Google Scholar]
- 42.Barkova K, Kinne M, Ullrich R, Hennig L, Fuchs A, Hofrichter M. 2011. Regioselective hydroxylation of diverse flavonoids by an aromatic peroxygenase. Tetrahedron 67:4874–4878. 10.1016/j.tet.2011.05.008 [DOI] [Google Scholar]
- 43.Gutierrez A, Babot ED, Ullrich R, Hofrichter M, Martinez AT, del Rio JC. 2011. Regioselective oxygenation of fatty acids, fatty alcohols and other aliphatic compounds by a basidiomycete heme-thiolate peroxidase. Arch. Biochem. Biophys. 514:33–43. 10.1016/j.abb.2011.08.001 [DOI] [PubMed] [Google Scholar]
- 44.Kinne M, Zeisig C, Ullrich R, Kayser G, Hammel KE, Hofrichter M. 2010. Stepwise oxygenations of toluene and 4-nitrotoluene by a fungal peroxygenase. Biochem. Biophys. Res. Commun. 397:18–21. 10.1016/j.bbrc.2010.05.036 [DOI] [PubMed] [Google Scholar]
- 45.Kinne M, Poraj-Kobielska M, Aranda E, Ullrich R, Hammel KE, Scheibner K, Hofrichter M. 2009. Regioselective preparation of 5-hydroxypropranolol and 4 ′-hydroxydiclofenac with a fungal peroxygenase. Bioorg. Med. Chem. Lett. 19:3085–3087. 10.1016/j.bmcl.2009.04.015 [DOI] [PubMed] [Google Scholar]
- 46.Kluge M, Ullrich R, Scheibner K, Hofrichter M. 2012. Stereoselective benzylic hydroxylation of alkylbenzenes and epoxidation of styrene derivatives catalyzed by the peroxygenase of Agrocybe aegerita. Green Chem. 14:440–446. 10.1039/c1gc16173c [DOI] [Google Scholar]
- 47.Peng L, Wollenberger U, Kinne M, Hofrichter M, Ullrich R, Scheibner K, Fischer A, Scheller FW. 2010. Peroxygenase based sensor for aromatic compounds. Biosens. Bioelectron. 26:1432–1436. 10.1016/j.bios.2010.07.075 [DOI] [PubMed] [Google Scholar]
- 48.Peter S, Kinne M, Ullrich R, Kayser G, Hofrichter M. 2013. Epoxidation of linear, branched and cyclic alkenes catalyzed by unspecific peroxygenase. Enzyme Microb. Technol. 52:370–376. 10.1016/j.enzmictec.2013.02.013 [DOI] [PubMed] [Google Scholar]
- 49.Poraj-Kobielska M, Atzrodt J, Holla W, Sandvoss M, Groebe G, Scheibner K, Hofrichter M. 2013. Preparation of labeled human drug metabolites and drug-drug interaction-probes with fungal peroxygenases. J. Labelled Comp. Radiopharm. 56:513–519. 10.1002/jlcr.3103 [DOI] [PubMed] [Google Scholar]
- 50.Poraj-Kobielska M, Kinne M, Ullrich R, Scheibner K, Kayser G, Hammel KE, Hofrichter M. 2011. Preparation of human drug metabolites using fungal peroxygenases. Biochem. Pharmacol. 82:789–796. 10.1016/j.bcp.2011.06.020 [DOI] [PubMed] [Google Scholar]
- 51.Karich A, Kluge M, Ullrich R, Hofrichter M. 2013. Benzene oxygenation and oxidation by the peroxygenase of Agrocybe aegerita. AMB Express. 3:5. 10.1186/2191-0855-3-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kluge M, Ullrich R, Dolge C, Scheibner K, Hofrichter M. 2009. Hydroxylation of naphthalene by aromatic peroxygenase from Agrocybe aegerita proceeds via oxygen transfer from H2O2 and intermediary epoxidation. Appl. Microbiol. Biotechnol. 81:1071–1076. 10.1007/s00253-008-1704-y [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.