Abstract
Processing of mRNA precursors (pre-mRNAs) by polyadenylation is an essential step in gene expression. Polyadenylation consists of two steps, cleavage and poly(A) synthesis, and requires multiple cis elements in the pre-mRNA and a megadalton protein complex bearing the two essential enzymatic activities. While genetic and biochemical studies remain the major approaches in characterizing these factors, structural biology has emerged during the past decade to help understand the molecular assembly and mechanistic details of the process. With structural information about more proteins and higher-order complexes becoming available, we are coming closer to obtaining a structural blueprint of the polyadenylation machinery that explains both how this complex functions and how it is regulated and connected to other cellular processes.
INTRODUCTION
Eukaryotic pre-mRNA 3′-end processing (3′ processing) involves a two-step reaction in which an endonuclease cleaves the pre-mRNA and a poly(A) polymerase (now known as PAPα; referred to here as PAP) synthesizes a polyadenosine tail on the cleaved upstream product. This seemingly simple process involves intricate cis elements on the transcript and a massive and complex machinery consisting of more than 20 polypeptides in yeast cells (1) and as many as 80 in human cells (2). 3′ processing is critical for many cellular events, from upstream coupled transcription and splicing to downstream mRNA export, translation, and decay (3, 4). Defects in 3′ processing can have catastrophic consequences for the cell (1) and have been associated with a variety of human diseases (5). Moreover, 3′ processing can serve as a means of gene expression regulation through alternative polyadenylation (APA) (6, 7), which is widely utilized in modulating mRNA transcript levels in diverse cell types, developmental stages, and diseases (8–10).
Early characterizations of mRNA 3′-end formation focused on genetic and biochemical studies by using a combination of in vivo and in vitro approaches. Subcomplexes, protein binding partners, and individual factors were dissected layer by layer to unravel the complexity of the process (1, 11, 12). In the past decade, structural studies have thrived and played a major role in elucidating the molecular assembly and function of the complex (3, 13). In this minireview, we will summarize the protein factors involved in 3′ processing with an emphasis on structural information and protein interaction networks.
PRE-mRNA cis ELEMENTS
For 3′ processing to occur correctly, pre-mRNAs need specific cis elements to guide the protein factors. In metazoans, the actual site of endonucleolytic cleavage has no apparent consensus, though it is often preceded immediately by a CA dinucleotide (14). Accurate positioning of the 3′ processing complex requires a combination of upstream and downstream sequence elements. First, a highly conserved AAUAAA hexamer, referred to as the polyadenylation signal (PAS), is typically located 10 to 35 nucleotides (nt) upstream of the cleavage site (15, 16), which can display microheterogeneity (17). Second, two downstream elements (DSEs) with lower conservation exist within 30 nt following of the cleavage site (18), featuring GU-rich (19, 20) and U-rich (21, 22) sequences. Third, multiple UGUA motifs are positioned 40 to 100 nt upstream of many cleavage sites (16). A tripartite mechanism has been proposed by which these three core components act cooperatively in directing poly(A) site recognition (13, 23).
Yeast mRNAs have distinct and more diffuse sequences directing polyadenylation (1, 3). The cleavage site usually follows a pyrimidine and a stretch of adenosines (24), the position of which is defined by an A-rich positioning element (PE) located 10 to 30 nt upstream (25) and a UA-rich efficiency element (EE) further upstream (1). Moreover, conserved upstream and downstream U-rich elements have also been identified that appear to synergistically enhance polyadenylation (26, 27).
PROTEIN FACTORS
Pre-mRNA 3′ processing can be reconstituted in vitro with exogenous RNA substrates and cell nuclear extracts (28), and this provided a powerful means to identify active trans-acting components. Five major factors were identified in early biochemical fractionations: cleavage and polyadenylation specificity factor (CPSF), cleavage stimulation factor (CstF), cleavage factor I (CFI), cleavage factor II (CFII), and PAP (29–31). All of these factors, with the exception of PAP, are multisubunit protein complexes. While PAS-directed poly(A) synthesis in the absence of cleavage requires only CPSF and PAP, all of the components are indispensable for efficient cleavage. Some other components of the 3′ processing machinery were discovered subsequently, including nuclear poly(A)-binding protein 1 (PABPN1) (32), RNA polymerase II (RNAP II), especially the C-terminal domain (CTD) of its largest subunit (33, 34), and symplekin (35). A more recent proteomic study revealed a large number of additional proteins associated with the 3′ processing complex (2). Some of these are bona fide components, some serve regulatory roles, and others may assist in coupling 3′ processing to other processes.
Compared to the mammalian 3′ processing complex, the yeast machinery is assembled in a different way. Proteins are found in three main factors: cleavage and polyadenylation factor (CPF), cleavage factor IA (CFIA), and cleavage factor IB (CFIB) (1, 3). Both mammalian and yeast systems contain unique components that do not exist in the other. Despite the evolutionary divergence, evident conservation and similarity prevail. For example, CPF contains all of the homologous proteins from CPSF, and CFIA consists of subunits that share high homology with those in CstF and CFII. In the following sections, we will describe the mammalian pre-mRNA 3′ processing factors and also their respective yeast homologs in more detail. A summary of their genomic symbols and published structures is given in Table 1.
TABLE 1.
Mammalian 3′ processing factors and their genomic symbols, yeast orthologs and published structures
| Mammalian factor | Gene designation | Yeast ortholog | Related structure(s) in PDBa |
|---|---|---|---|
| CPSF-73 | CPSF3 | Ysh1 (Brr5) | 2I7T, 2I7V |
| CPSF-100 | CPSF2 | Ydh1 (Cft2) | 2I7X |
| CPSF-30 | CPSF4 | Yth1 | 2RHK |
| CPSF-160 | CPSF1 | Yhh1 (Cft1) | |
| Fip1 | FIP1L1 | Fip1 | 3C66 |
| WDR33 | WDR33 | Pfs2 | |
| CstF-77 | CSTF3 | Rna14 | 2OND, 2OOE, 2UY1, 4E6H, 4E85, 2L9B, 4EBA |
| CstF-64/τCstF-64 | CSTF2/CSTF2T | Rna15 | 1P1T, 2J8P, 2L9B, 4EBA, 2X1B, 2X1A, 2X1F, 2KM8 |
| CstF-50 | CSTF1 | 2XZ2 | |
| CFI25 | NUDT21 | 2CL3, 2J8Q, 3BAP, 3BHO, 3MDI, 3MDG, 3Q2S, 3Q2T, 3P5T, 3P6Y | |
| CFI68 | CPSF6 | 3Q2S, 3Q2T, 3P5T, 3P6Y | |
| Pcf11 | PCF11 | Pcf11 | 2NPI, 1SZ9, 2BFO, 1SZA |
| Clp1 | CLP1 | Clp1 | 2NPI |
| Symplekin | SYMPK | Pta1 | 3GS3, 3O2T, 3ODR, 3ODS, 3O2S, 3O2Q, 4H3H, 4H3K, 4IMJ, 4IMI |
| RNAP II CTD | RBP1 | RNAP II CTD | 1SZA, 3O2S, 3O2Q, 4H3H, 4H3K, 4IMJ, 4IMIb |
| PAP | PAPOLA | Pap1 | 1FA0, 201P, 2HHP, 2Q66, 1F5A, 1Q78, 1Q79, 3C66 |
| PABPN1 | PABPN1 | Pab1/Nab2 | 3B4D, 3B4M |
PDB, Protein Data Bank.
Only structures of RNAP II CTD in complex with 3′ processing factors are listed here.
CPSF
CPSF defines the specificity of pre-mRNA 3′ processing by recognizing the PAS (30, 31, 36). CPSF also plays important roles in transcription coupling, as it is recruited to the transcription initiation complex and accompanies RNAP II throughout the transcription process (37). Early purifications revealed four major polypeptides in CPSF, i.e., CPSF-160, CPSF-100, CPSF-73, and CPSF-30 (36, 38), but additional subunits were identified later, including Fip1 (39) and WDR33 (2).
A CPSF subunit that has gained considerable attention is CPSF-73, largely because it turns out to be the endonuclease that has been sought for 3 decades (40). The earliest clue to its function came from a sequence analysis showing that CPSF-73 belongs to the metallo-β-lactamase (MβL) superfamily, whose members are mostly hydrolases dependent on metal ions (41). Yeast cells with mutations of the putative residues for zinc binding in Ysh1 (CPSF-73 homolog) are lethal, while zinc chelators added to HeLa cell nuclear extract inhibited or abolished cleavage (42). Despite all of the evidence pointing at CPSF-73, the definitive piece did not arrive until the crystal structure was determined (43).
The N-terminal region of CPSF-73 (residues 1 to 460) contains a canonical MβL domain with a β-CASP domain inserted like a cassette (Fig. 1). Two zinc atoms are octahedrally coordinated with essential motifs in the active site within the MβL domain. However, the active site is buried at the interface between the β-CASP and MβL domains, severely restricting access to the RNA substrate. This likely explains why the bacterially expressed N-terminal domain (NTD) of CPSF-73 displayed minimal nuclease activity in vitro. Unexpectedly, calcium was able to activate nonspecific nuclease activity, through a mechanism that is not understood but was speculated to involve conformational changes triggered by the cation (43).
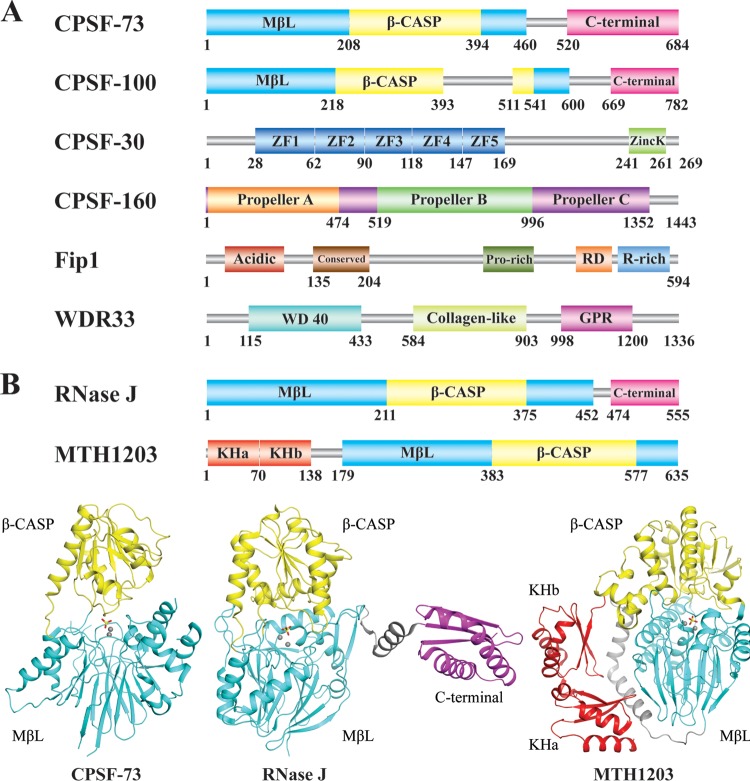
FIG 1.
CPSF. (A) Domain organization of human CPSF subunits. (B) Domain architecture of two CPSF-73 structural homologs, RNase J (Protein Data Bank [PDB] identification [ID] no. 3BK1 [44]) and MTH1203 (PDB ID no. 2YCB [47]), and structural comparison between them and CPSF-73 (PDB ID no. 2I7T [43]).
In recent years, structures of many bacterial and archaeal CPSF-73 homologs belonging to the β-CASP family have been determined, shedding light on the catalytic mechanism of CPSF-73 (44–47) (Fig. 1B). Bacterial RNase J has an overall domain architecture similar to that of CPSF-73 (Fig. 1B). The additional C-terminal region that was missing from the CPSF-73 structure mediates dimerization and is crucial for its nuclease activity (44). Archaeal β-CASP proteins have extra KH motifs at the N terminus that are responsible for RNA recognition (Fig. 1B). They form dimers through their extreme C-terminal region within the MβL domain, distinct from RNase J (45–47). These observations raise the question of whether dimerization through the CTD is also required for CPSF-73 activity. Structural information addressing this is not yet available, nor do we know if CPSF-73 can self-associate, but the fact that full-length CPSF-73 purified from HEK293 cells was not active (48) suggests that CPSF-73 more likely employs a different mechanism (heterodimerization with CPSF-100; see below).
Like CPSF-73, CPSF-100 also belongs to the β-CASP family and shares high sequence homology (41, 49) (Fig. 1A). While the domain organization of CPSF-100 resembles CPSF-73 and other β-CASP proteins, equivalent motifs critical for zinc binding are missing, making it incapable of catalysis (41, 43). The exact function of CPSF-100 is not clear, but several studies have shown its importance in pre-mRNA 3′ processing. The yeast CPSF-100 homolog Ydh1 is essential for cell viability, as well as cleavage and poly(A) synthesis (50, 51). Ydh1 can also be UV cross-linked to the pre-mRNA in a sequence-dependent manner, indicating possible direct contact with select RNA substrates (26, 52). Glutathione S-transferase pulldown assays suggest that Ydh1 interacts with the RNAP II CTD and Pcf11, raising the possibility of a role in transcription-coupled pre-mRNA processing (50). In mammals, CPSF-100 and CPSF-73 are tightly associated, likely through their CTDs (53). This heterodimeric structure is reminiscent of the aforementioned other β-CASP protein homodimers, providing a possible mechanism by which CPSF-73 dimerization with CPSF-100 is required for catalysis (54).
CPSF-30 is the smallest CPSF subunit and is essential for both cleavage and poly(A) synthesis (55, 56). CPSF-30 consists of five CCCH zinc finger motifs and a CCHC zinc knuckle motif at the C terminus that is not present in its yeast homolog, Yth1 (55) (Fig. 1A). The structures of these motifs in other proteins have been determined, and they often function in RNA recognition (57, 58), strongly suggesting that CPSF-30 binds the pre-mRNA. Indeed, CPSF-30 can be UV cross-linked to RNA oligomers, with a preference for poly(U) sequences (55, 59). A conserved U-rich element is often located next to the PAS (16), presenting a strong candidate for CPSF-30 recognition. Yth1 indeed binds the pre-mRNA near the cleavage site (56), and RNA recognition is impaired by removal of its zinc finger motifs (60).
The zinc fingers in CPSF-30 are also responsible for making contacts with other proteins. In the case of Yth1, the integrity of zinc finger 4 is crucial for binding to Fip1, as well as Ysh1 (55, 56, 60). Besides factors in the 3′ processing complex, CPSF-30 was found to interact with the NS1A protein from influenza A virus, a mechanism employed by the virus to inhibit host pre-mRNA 3′ processing (61). The crystal structure of NS1A in complex with CPSF-30 zinc fingers 2 and 3 has been solved. The two zinc fingers show high structural similarity to other known RNA-binding zinc finger proteins, in agreement with the proposed RNA recognition function (62). CPSF-30 also binds to the body of RNAP II and is likely responsible, at least in part, for the association of CPSF and RNAP II during transcription (63).
The largest subunit, CPSF-160, is composed of tandem WD40 repeats clustered into three major β-propellers (64) (Fig. 1A). CPSF-160 shares low sequence homology with DDB1, a scaffold protein for cullin binding in E3 ubiquitination whose structure has been well characterized, but has a similar domain architecture (65, 66). In fact, the WD40 domain is one of the most abundant domains in eukaryotic proteomes and also the top protein-interacting domain in human and yeast interactome databases (67). They generally serve as protein scaffolds (67), but some can also recognize nucleic acids (66). This is consistent with the fact that CPSF-160 is involved in both protein-RNA and protein-protein interactions. CPSF-160 can be UV cross-linked to pre-mRNA in a PAS-dependent manner (30, 68). Purified recombinant CPSF-160 protein was shown to bind RNA selectively, but its affinity for AAUAAA was lower than that of intact CPSF (69). CPSF-160 also makes direct contacts with CstF-77 and weakly associates with PAP (69). The yeast CPSF-160 homolog Yhh1 interacts with the RNAP II CTD and binds to RNA through the middle β-propeller (70).
Fip1 was identified more than a decade later than the above-mentioned CPSF subunits, though its yeast homolog had been known for a long time. Fip1 stably associates with all other CPSF components and is required for both cleavage and poly(A) synthesis (39). Human Fip1 is almost twice as large as its yeast counterpart and has a C-terminal extension containing two extra domains (39) (Fig. 1A). The N-terminal regions of the two proteins are similar in domain organization but have relatively low sequence conservation (39).
Yeast Fip1 was originally discovered in a two-hybrid screen for binding partners of yeast PAP, Pap1 (71). In the Fip1-Pap1 complex structure, a fragment of Fip1 (residues 80 to 105) binds to the Pap1 CTD (72), which stabilizes Pap1 but induces minimal structural changes and has little effect on catalytic activity (72). Mutations that disrupt this interaction cause cell death (72). Interestingly, residues at the interface in both Fip1 and Pap1 are not conserved but the interaction between these two proteins has been observed across different species (71–73), suggesting that the tethering but not the atomic details of the interaction is essential for polyadenylation (74).
Besides PAP, human Fip1 also binds to CPSF-30, CPSF-160, and CstF-77; this binding is mediated mainly by the N-terminal region (39). The C-terminal arginine-rich domain can also bind to RNA, particularly U-rich sequences (39). In solution, Fip1 seems to be largely disordered (72), and its extended nature can provide scaffolding interactions with multiple proteins (75).
WDR33 is another CPSF component. It was identified in the proteomic study mentioned above and shown to coelute with CPSF during gel filtration and to be essential for cleavage in vitro (2). WDR33 is a 146-kDa protein that consists of an N-terminal WD40 domain, a middle collagen-like domain, and a C-terminal GPR (glycine-proline-arginine) domain (76) (Fig. 1A). However, the exact function of this protein is not known. The yeast homolog of WDR33, Pfs2, is required for cell survival and polyadenylation (77). Pfs2 is smaller than WDR33 and contains only the WD40 domain (77). Pfs2 binds many protein factors in the 3′ processing complex, including Rna14, Ysh1, Fip1 (77), and Clp1 (78). Additionally, a Schizosaccharomyces pombe Pfs2 mutant was shown to have defects in transcription termination, suggesting a potential role for Pfs2 in transcription-coupled polyadenylation (79).
CstF
CstF was initially purified as a factor that is not required for polyadenylation but significantly stimulates the cleavage reaction (31). Subsequently, this was found to likely reflect contamination of other factors, and CstF is now considered an essential polyadenylation factor. Its activity reflects, in part, cooperative binding of CPSF and CstF to the pre-mRNA (80, 81), in which CstF recognizes the DSEs (18). Like CPSF, CstF also associates with RNAP II during transcription elongation and facilitates transcription-coupled 3′ processing (33). Three proteins constitute the CstF complex: CstF-77, CstF-64, and CstF-50 (80, 82).
A computational analysis identified the N-terminal region of CstF-77 as a HAT (half a TPR) domain (83) (Fig. 2A). The crystal structure of the HAT domain revealed an intrinsic dimeric association (Fig. 2B) (84, 85), which is consistent with earlier biochemical characterizations (35), as well as genetic studies with flies (86), suggesting that CstF assembles with two copies of each subunit (84). Similar characteristics have also been observed in the fungal homolog of CstF-77, Rna14. The HAT domain of Rna14 from Kluyveromyces lactis dimerizes in mostly the same way as that of CstF-77, despite significant structural variations (87). Disruption of this dimerization in yeast can severely impair polyadenylation (88).
FIG 2.
CstF. (A) Domain organization of human CstF subunits. (B) Crystal structure of the murine CstF-77 HAT domain (PDB ID no. 2OOE [84]). (C) NMR structure of the Rna14 C-terminal Rna15-binding domain in complex with the hinge region of Rna15 (PDB ID no. 2L9B [91]). (D) Crystal structure of the dimerization domain of CstF-50 (PDB ID no. 2XZ2 [110]). (E) Crystal structure of the RRM domain of Rna15 with RNA bound at primary and secondary sites (PDB ID no. 2X1A, 2X1F [97]). (F) NMR structure of the RRM domain of Rna15, two RRM domains of Hrp1 and RNA ternary complex (PDB ID no. 2KM8 [99]).
CstF-77 interacts with both CstF-50 and CstF-64 in a way that bridges them since the other two factors make no direct contacts (35, 89). The C-terminal region of CstF-77 contains a proline-rich region that is required for binding to CstF-64 (35, 84, 90). In yeast, Rna14 and Rna15 (CstF-64 homolog, see below) assemble through the same regions (85, 87, 91). With the dimerization of Rna14, they constitute a α2β2 tetramer in the shape of a kinked rod (92). The complex structure of the Rna14 hinge domain and the Rna15 CTD has been obtained alone (91) and together with the Rna14 HAT domain (87) (Fig. 2C). The two domains tether as an interlocked structure through which they stabilize each other (87, 91). This formation requires cooperative folding between the two proteins and probably reflects their tight association in vivo (91). Additionally, there is a long linker connecting the HAT and the CTD of Rna14, making the two domains flexible and possibly functionally independent (87).
CstF-64 was the first protein in the 3′ processing machinery shown, by UV-cross-linking, to bind RNA substrates, even before its identity was known (93). Binding is mediated by an RNA recognition motif (RRM) at the protein's N terminus (94) (Fig. 2A). Further investigation revealed that the RRM can specifically recognize the U-rich DSE (18) and it selects GU-rich sequences in vitro (95). In the nuclear magnetic resonance (NMR) structure of the RRM, a binding pocket was identified at the surface of the central β-sheet to accommodate UU dinucleotides, the presence of which enhances the RNA-RRM interaction. By fine-tuning contacts outside this pocket, the RRM is able to modulate its preference for a wide selection of Gs and Us while still discriminating against other RNA sequences (96). This specific binding variability enables CstF-64 to recognize both U-rich and GU-rich DSEs. In fact, the two DSEs are in close proximity (within 15 nt) (16) and might be bound by two copies of CstF-64 simultaneously, bridged by the CstF-77 HAT domains. This is consistent with the dimeric association of CstF, constructed around the CstF-77 HAT domain dimer.
The yeast homolog of CstF-64, Rna15, not only bears high sequence identity but also shares structural similarity in the RRM region. The Rna15 RRM preferentially binds to a U-rich or GU-rich sequence in vitro (95, 97). The affinity for the RNA is generally low (92) but can be significantly enhanced by Rna14 or Hrp1 (a yeast CFI component that binds the EE, no mammalian homolog) (92, 98). Two RNA-binding sites were identified in the RRM structure (97) (Fig. 2E). The primary site, located at a surface loop, mediates specific interactions with the RNA, largely accounting for the selectivity for GU-rich sequences (97). The second site is less specific and is positioned in a canonical RRM-RNA contact region, as in CstF-64 (96, 97). It has been suggested that Rna15, when complexed with Hrp1, utilizes the secondary site to recognize the A-rich PE (99) (Fig. 2F) and by itself recognizes a U-rich consensus upstream of the cleavage site (26, 27) in a scenario in which CFI contains two copies of Rna15 but only one copy of Hrp1 (88, 100).
The remaining region of CstF-64 is less well studied. The hinge region immediately following the RRM interacts with both CstF-77 and symplekin in a mutually exclusive manner (35, 101). The domain at the very C terminus is highly conserved throughout eukaryotes. It comprises a three-α-helix bundle structure, which is required for polyadenylation and for interaction with Pcf11 (102, 103), as well as with the transcriptional coactivator PC4/Sub1 (103, 104). N terminal to this lies a long proline-glycine-rich region (40%) interrupted by a pentapeptide repeat motif (MEARA/G) that is all α-helical (94, 105). This region does not exist in Rna15, and its function is unknown.
A second isoform of CstF-64, termed τCstF-64, was found expressed mainly in brain and testis (106), and it has been implicated in modulating poly(A) site selection during spermatogenesis (107). Other studies, however, have revealed that τCstF-64 is likely more broadly expressed, and may function redundantly with CstF-64 (2, 108).
CstF-50, which lacks a yeast counterpart, contains an N-terminal dimerization domain and seven WD40 repeats at its C terminus (35, 109) (Fig. 2A). The dimerization domain is crucial for self-association (35) and together with CstF-77 accounts for the hexameric architecture of the CstF complex. The crystal structure of this domain suggests a formation of three tandem α-helices tethered to the other protomer, mediated primarily through a conserved hydrophobic core (110) (Fig. 2D). The CstF-50 WD40 domain functions as a binding platform, and truncation of a single repeat abrogates its interaction with CstF-77 (35). This domain may also serve as a regulatory adaptor that signals transient inhibition of 3′ processing upon DNA damage (111, 112).
CFI
The CFI complex associates early with the transcription elongation complex, along with CPSF and CstF, in facilitating transcription-coupled 3′ processing (23). At the polyadenylation site, CFI associates with the pre-mRNA through UGUA motifs and stabilizes the binding of CPSF (113). Unlike other major 3′ processing factors, CFI exists only in metazoans and does not have a yeast equivalent.
The CFI complex is assembled as a heterotetramer with a dimer of the small subunit, CFI25, and two copies of a combination of large subunits, CFI59, CFI68, or CFI72 (113, 114). CFI59 and CFI68 are encoded by two paralogous genes, while CFI72 is an isoform of CFI68 (115). The three large subunits may be functionally redundant because CFI68 alone is capable of reconstituting CFI activity with CFI25 in vitro (114). Meanwhile, all CFI subunits can be UV cross-linked to pre-mRNAs, suggesting roles in RNA recognition (113). SELEX experiments identified a binding consensus sequence for CFI: UGUAN (N: A > U > G or C) (116), which was later shown to be an important cis element for poly(A) site definition (16, 23).
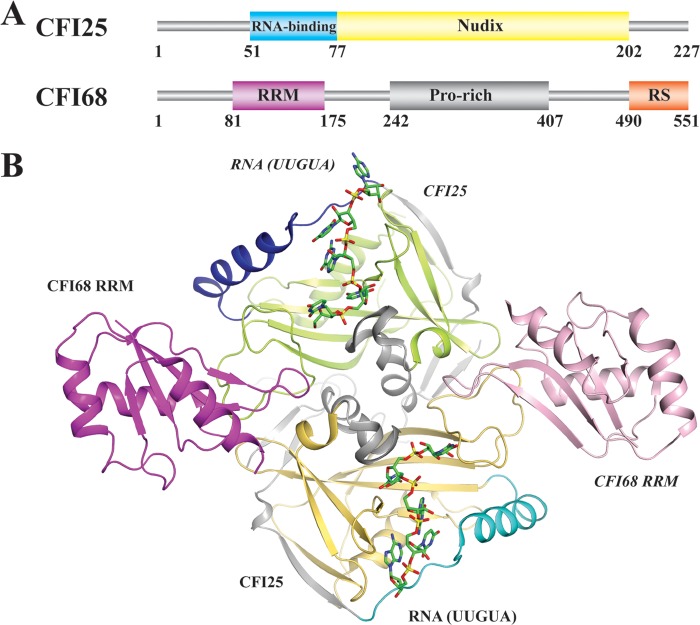
CFI25 encompasses a central Nudix domain (117) (Fig. 3A). The Nudix superfamily is widespread in all kingdoms, and its members function mostly as pyrophosphohydrolases (118). Intriguingly, two signature glutamates that are key for metal coordination and enzymatic catalysis are missing from the Nudix motif of CFI25, which distinguishes CFI25 from most other Nudix proteins. The crystal structure of CFI25 shows a core Nudix domain featuring a canonical α/β/α fold sandwich augmented with N- and C-terminal extensions (119, 120) (Fig. 3B). No metal ions were observed around the Nudix motif, and subsequent biochemical assays failed to detect any enzymatic activity, suggesting that CFI25 is unlikely a hydrolase (119, 120). Instead, CFI25 utilizes its Nudix domain as a platform for binding RNA and other proteins, including CFI68, PAP, and PABPN1 (117). The crystal structure of the CFI25-RNA complex provided insights into the mechanism by which CFI specifically recognizes the UGUA element (121). Unexpectedly, in addition to the Nudix domain, the N-terminal extension also makes direct contact with the RNA (Fig. 3B). Interactions are achieved mainly through hydrogen bonds formed between RNA bases and the protein. Watson-Crick/sugar-edge base interactions within the RNA also contribute to binding specificity (121). Moreover, the dimeric nature of CFI25 enables it to bind two UGUA elements simultaneously.
FIG 3.
CFI. (A) Domain organization of human CFI subunits. (B) Crystal structure of the CFI25-CFI68-RNA complex (PDB ID no. 3Q2T [126]).
The CFI68 subunit is composed of an N-terminal RRM, a middle proline-rich region, and a C-terminal RS domain with alternating arginine and serine residues (114) (Fig. 3A). The domain organization resembles SR proteins involved in pre-mRNA splicing. In fact, CFI68 was shown to copurify with the spliceosome (122, 123) and interacts with splicing factors (117, 124), suggesting its potential role in coordinating pre-mRNA splicing and 3′ processing. The RRM interacts weakly with RNA, but the affinity can be enhanced appreciably by cooperative binding of CFI25 (117). In this case, each CFI68 RRM maintains simultaneous interactions with the CFI25 dimer on the opposite side (125, 126) (Fig. 3B). The presence of the RRM causes little structural change to the CFI25 dimer, nor does it affect RNA binding specificity (125, 126). Two UGUA sequences are bound to CFI25 dimers in an antiparallel fashion. The connecting loop RNA, though not included in the structure, is likely stabilized by the CFI68 RRM (125, 126). On the basis of the structural data, an RNA looping mechanism directed by CFI has been proposed for poly(A) site selection in APA (126, 127), perhaps explaining a correlation between CFI levels and APA observed in several studies (128, 129).
CFII
CFII is perhaps the least well-characterized factor in the mammalian 3′ processing complex, partly because its exact components remain poorly defined. More than 15 proteins were initially copurified in CFII fractions, but only 2, Pcf11 and Clp1, coeluted with CFII activity (130). However, there is no evidence indicating that these two proteins alone are capable of reconstituting CFII activity. Their yeast homologs are essential for 3′-end formation (131, 132), and much of our understanding of CFII comes from studies in yeast. Both Pcf11 and Clp1 are part of the CFIA complex, which also includes Rna14 and Rna15.
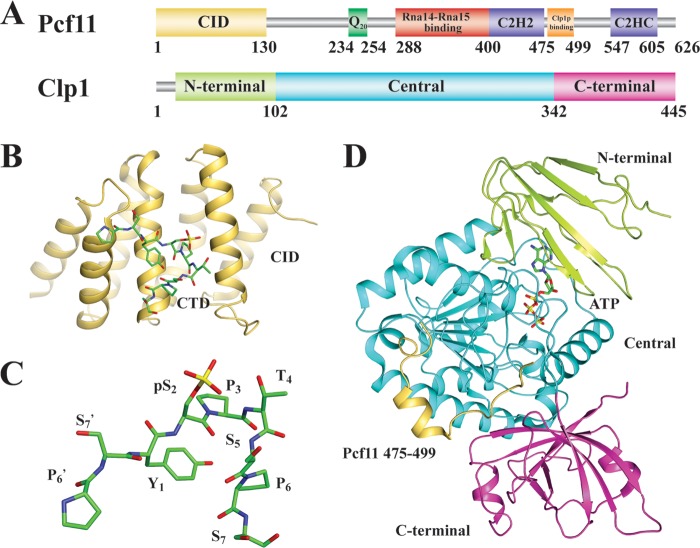
Pcf11 plays a pivotal role in the assembly of CFIA, as it is the only component of the complex that makes direct contact with all other members. The interacting domains have been mapped to the middle and C-terminal regions (102, 103, 131, 133) (Fig. 4A). Two highly conserved zinc fingers were identified flanking the C-terminal Clp1-interating domain, but their function remains unknown. A stretch of 20 consecutive glutamines preceding the middle Rna14-Rna15 binding domain likely serves as a linker to the N-terminal region (133). This region features an RNAP II CTD-interacting domain (CID), which comprises eight α-helices arranged in a right-handed superhelical formation (134, 135) (Fig. 4B). The CID interacts with both unphosphorylated and phosphorylated RNAP II CTDs but has higher affinity for the latter (133, 136, 137). Surprisingly, the CID-CTD interaction is not necessary for 3′ processing, but it is required for proper transcription termination (133, 136). The CID also weakly binds RNA (138) and bridges the RNAP II CTD to the pre-mRNA (139), supporting a role for Pcf11 in coupling transcription to 3′ processing.
FIG 4.
CFII. (A) Domain organization of the yeast homologs of CFII subunits. (B) Crystal structure of the CID of Pcf11 in complex with a RNAP II CTD peptide (PDB ID no. 1SZA [134]). (C) Conformation of the RNAP II CTD peptide bound to the CID of Pcf11. (D) Crystal structure of the Clp1-Pcf11-ATP complex (PDB ID no. 2NPI [141]).
Human Pcf11 is twice as large as its yeast homolog and shares sequence homology only at its N-terminal CID (130). The remainder has not been characterized. Despite differences in its primary sequence, the function of Pcf11 is evolutionarily conserved. Knockdown of Pcf11 in HeLa cells resulted in deficiency in cleavage, as well as transcription termination, and Pcf11 may also be required for degradation of the 3′ product following cleavage (140).
Clp1 consists of a central ATPase domain and two smaller domains at its N and C termini (141) (Fig. 4A). The primary sequence of yeast Clp1 reveals a conserved Walker A/P loop motif (130), which is typically involved in ATP/GTP binding or catalysis (142). Indeed, an ATP molecule was bound to Clp1 in the crystal structure (Fig. 4D), but ATPase activity could not be detected (141). Mutations in the ATP-binding pocket perturb the Clp1-Pcf11 interaction and, in turn, cause defects in 3′ processing and transcription termination, although some of them did not affect ATP binding (78, 143, 144). Given that there is no requirement for ATP in 3′ cleavage, the significance of Clp1 ATP binding is unclear.
In contrast to yeast Clp1, the human homolog is an active 5′-OH polynucleotide kinase (145). The enzymatic activity is required in tRNA splicing and cannot be complemented by yeast Clp1 (145, 146). Although human Clp1 bears high sequence homology with its yeast counterpart, it might have acquired a diverged role during evolution. In 3′ processing, Clp1 interacts with both CPSF and CFI and likely tethers them to CFII (130).
SYMPLEKIN
Symplekin was initially identified not as a polyadenylation factor but as a tight junction plaque protein (147). However, it was subsequently found to associate with the 3′ processing complex, specifically with CstF64, and suggested to function as a scaffold protein (35). Sequence alignment indicated that symplekin bears low sequence similarity to an essential yeast polyadenylation factor, Pta1 (35, 51, 148, 149) (Fig. 5A). Pta1 interacts with various 3′ processing proteins, including Ysh1 (150), Ydh1 (50), Fip1 (148), Pcf11 (50), Clp1 (148), Pap1 (148), and the RNAP II CTD phosphatase Ssu72 (151), consistent with a scaffolding function.
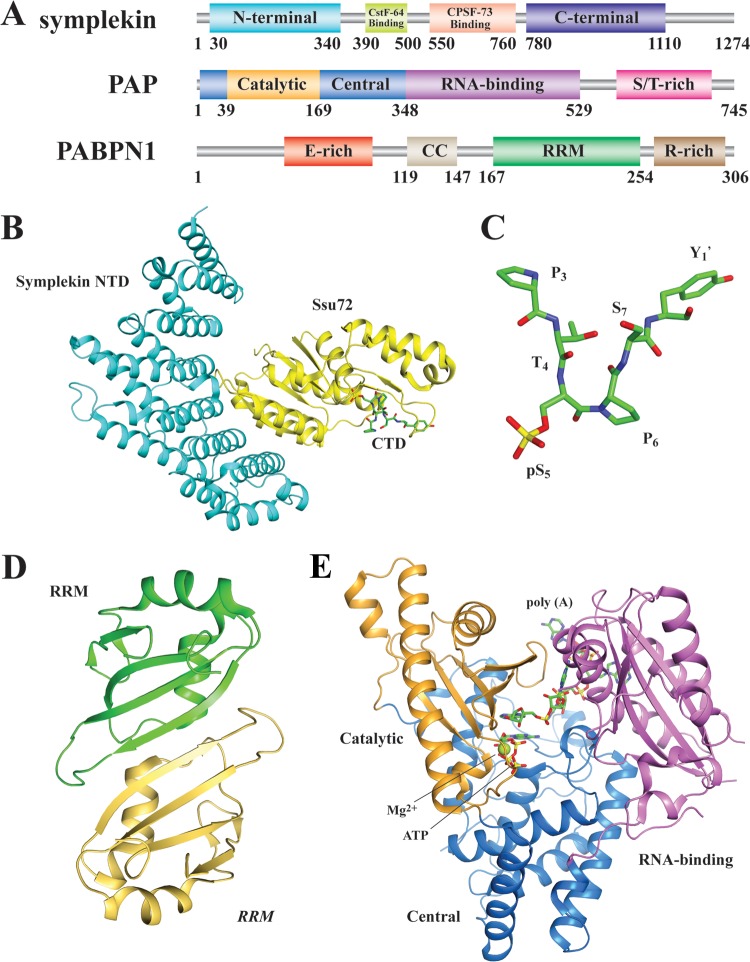
FIG 5.
Other mammalian pre-mRNA 3′ processing factors. (A) Domain organization of human symplekin, PAP, and PABPN1. (B) Crystal structure of the symplekin-Ssu72-RNAP II CTD peptide complex (PDB ID no. 3O2Q [153]). (C) Conformation of the RNAP II CTD peptide in the active site of Ssu72. (D) Crystal structure of the RRM domain dimer of PABPN1 (PDB ID no. 3B4M [192]). (E) Crystal structure of yeast Pap1 in complex with ATP and an oligo(A) sequence (PDB ID no. 2Q66 [176]).
The symplekin NTD contains seven pairs of antiparallel α-helices (152, 153). The overall fold is reminiscent of ARM or HEAT repeats, which are typically involved in protein-protein interactions (154), in agreement with its suggested scaffolding function. Symplekin NTD interacts with Ssu72 and stimulates its phosphatase activity (148, 153) (Fig. 5B). The symplekin-Ssu72 complex also plays important roles in transcription-coupled polyadenylation (153).
Symplekin interacts with the hinge region of CstF-64, competitively with CstF-77 (35, 101). A CstF-64 mutant whose interaction with symplekin was abolished maintained its association with CstF-77, and while the mutation did not affect polyadenylation, it impaired histone pre-mRNA 3′ processing (101). (Histone pre-mRNAs are typically cleaved but not polyadenylated. They utilize an overlapping but distinct processing machinery, with symplekin and CPSF-73, for example, functioning in both [155].) It thus appears that CstF-64 associates exclusively with either CstF-77 or symplekin in two separate pre-mRNA processing complexes and perform distinct functions.
Symplekin also interacts with CPSF-73. The binding region was deduced on the basis of the conserved interaction between the CTDs of Pta1 and Ysh1 (148, 150). Symplekin tightly associates with CPSF-73 and CPSF-100 (156, 157), forming a shared stable core complex for both general and histone pre-mRNA 3′ processing (157). As a consequence, it has been speculated that symplekin may regulate the nuclease activity of CPSF-73 through direct interactions or by recruiting additional regulatory factors (40, 157), but further investigation is necessary to test this hypothesis.
RNAP II CTD
The largest subunit of RNAP II contains an extended CTD separated from the globular core structure (158). The CTD consists of consensus repeats Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7, with the number varying from 26 in yeast to 52 in vertebrates (159–163). The CTD is necessary for efficient polyadenylation both in vivo (33) and in vitro (34), but exactly how it promotes mRNA 3′-end formation is still not well understood. A platform role has been proposed since a number of 3′ processing factors have been observed binding to the CTD, such as Ydh1 (50), Yhh1 (70), CstF-77/Rna14 (33, 133), CstF-50 (33), Pcf11 (133, 134, 136, 137), and perhaps Rna15 (133) and Pta1 (164).
The ability of the RNAP II CTD to interact with a variety of 3′ processing factors and thus to link polyadenylation to transcription can be largely explained by its structural diversity, which results from its intrinsic nonuniform and overall disordered architecture (135, 165), various dynamic posttranslational modifications, especially phosphorylation, as well as cis-trans isomerization of prolyl peptide bonds (159–163). Although structural information about the full-length RNAP II CTD is not available, a number of segments ranging from less than one repeat to nearly three repeats have been captured associated with RNAP II CTD-binding proteins. These CTD structures present immensely diverse conformations and modifications (166). Here we will briefly discuss two that are closely related to 3′ processing.
The first is the Pcf11-pSer2 CTD complex (Fig. 4B). In this structure, the bound CTD adopts a β-turn conformation (134) (Fig. 4C). This is likely formed via induced fit through the binding of Pcf11, as NMR experiments suggest that a similar CTD peptide exists as a dynamic unfolded ensemble in solution (135). The phosphate group of pSer2 forms hydrogen bonds within the CTD, in a way that indirectly stabilizes the β-turn structure (134). By iterating the observed CTD repeat, Meinhart and Cramer deduced a compact β-spiral complete CTD model. While Ser2 phosphorylation can be readily accommodated in the model, Ser5 phosphorylation would open up the spiral and induce a more extended structure. With dynamic phosphorylation and dephosphorylation, the CTD conformations would be altered and cycled, so that the spatial and temporal control of mRNA processing factor binding during transcription can be achieved.
The second structure is the Ssu72-pSer5 CTD complex (Fig. 5B). Surprisingly, the CTD captured in the active site of Ssu72 has the peptide bond between pSer5 and Pro6 in the cis configuration (Fig. 5C), in contrast to all earlier known CTD structures, which were exclusively in trans (153, 167). The substrate-binding pocket of Ssu72 has a confined space so that only the CTD with pSer5-Pro6 in a cis configuration can be accommodated. The selectivity of Ssu72 nonetheless severely limits its substrate availability, because less than 20% of the total population of the pSer5-Pro6 peptide bond exists in the cis configuration and natural cis-trans conversion is rather slow (153, 167). Therefore, peptidyl-prolyl isomerases (human Pin1 and yeast Ess1) can promote dephosphorylation of the CTD by accelerating cis-trans conversion, which presents higher-level regulation of the CTD function (153, 167).
PAP
At least four different nuclear PAPs have been identified in metazoans, including canonical PAP, Neo-PAP, Star-PAP, and TPAP (168). The best studied is PAP, which is largely conserved between yeast and humans (169–171). PAP belongs to the DNA polymerase β family (172). The first 500 residues are conserved throughout eukaryotes (173, 174). The crystal structures of bovine PAP and yeast PAP (Pap1) have been determined, revealing a three-globular-domain organization (174, 175) (Fig. 5E). A large open central cleft harboring the active site is encompassed by the three domains. Upon substrate binding, the cleft closes as the NTD and CTD interact (176), suggesting an induced-fit mechanism (177, 178). Vertebrate PAP has a C-terminal extension of ∼20 kDa that does not exist in lower eukaryotes and is not essential for polyadenylation activity (173) (Fig. 5A). This extension, which can vary in sequence because of alternative splicing (179), is enriched with serines and threonines, which are targets for modulating PAP activity and are subject to various posttranslational modifications, including phosphorylation (180), acetylation (181), sumoylation (182), and PARylation (183). PAP has been shown to interact with many protein factors in the 3′ processing complex, such as CPSF-160 (82) and CFI25 (117). In yeast, while the NTD of Pap1 binds to both Pta1 and Yhh1 (184), the CTD interacts with Fip1 (71, 72, 185).
PABPN1
The identity of the mammalian PABPN1 involved in 3′ processing was not unearthed until almost 2 decades after its cytoplasmic counterpart (PABPC) was discovered (32, 186). PABPN1 serves as a stimulatory factor for PAP in PAS-dependent poly(A) synthesis (32). The presence of either CPSF or PABPN1 provides only moderate processivity, but together they promote rapid poly(A) elongation to a length of approximately 200 to 300 nt (187, 188), which matches the size of newly synthesized poly(A) tails in vivo (189). PABPN1 not only ensures the proper length of the poly(A) tail but may also be a significant regulator in APA (190).
PABPN1 has a domain architecture very different from that of PABPC (Fig. 5A). The NTD is acidic and rich in glutamates and may function to prevent undesirable contacts between PAP and PABPN1 (191). The middle region contains a coiled-coil domain that is required for stimulation of PAP (191). Immediately following this is a canonical RRM, which forms a dimer in solution and also in the crystal structure (192) (Fig. 5D). This is compatible with an earlier observation that the CTD (arginine rich) can also self-associate (193). In fact, in the absence of poly(A) and at elevated concentrations, PABPN1 is prone to aggregation into oligomers (194, 195). Each PABPN1 can recognize an ∼10-nt poly(A) sequence (195). Both the RRM and the CTD are required for RNA binding (193). As polyadenylation proceeds, PABPN1 coats the poly(A) tail sequentially, forming a linear filament or a spherical particle up to 21 nm in diameter (194). This structure is thought to restrict CPSF to the PAS while facilitating the physical interaction between CPSF and PAP over the newly synthesized poly(A) sequence until a desired length is reached (196).
In yeast, the apparent PABPN1 sequence homolog is Rbp29, but this protein localizes in the cytoplasm and has a very different function (197). The true functional counterpart of PABPN1 in yeast is still under debate. Two candidates, Pab1 and Nab2, have been proposed, but both of them negatively modulate poly(A) length, which is the opposite to the stimulatory effect of PABPN1 (198).
PERSPECTIVE
Since the start of the second millennium, when the first protein structure of a component of the 3′ processing complex was reported (174, 175), structural studies have flourished and advanced our knowledge of this intricate process at an ever-increasing pace. While a number of structures of protein factors have been determined and analyzed compared to the complicated machinery, what we know is only the tip of the iceberg. Many key proteins have not yet been structurally characterized. More importantly, past studies have focused on individual proteins or even domains rather than looking at the bigger picture. To understand in detail how the polyadenylation complex is assembled and functions as a whole, we need a more complete structural blueprint (Fig. 6). In recent years, several complex structures have been determined, such as CFI (125, 126), symplekin-Ssu72-RNAP II CTD (153, 199), and Rna14-Rna15 (87, 91), which substantially facilitated the molecular mapping of protein interconnections. Nevertheless, structural information about protein complexes in the 3′ processing machinery is still limited and confined within subcomplexes. How different subcomplexes associate and coordinate in the recruitment process and the enzymatic reactions is largely unclear. On the other hand, an electron microscopy study examining the whole 3′ processing complex has provided us a first look at the overall architecture and may provide a powerful future approach (2). Additionally, the coupling of 3′ processing to other nuclear events inevitably raises the complexity but also opens up an interesting and emerging direction in which analyses of bridging factors can be performed from a structural perspective so as to visualize how coupling is achieved, as well as how polyadenylation affects other processes. Finally, the emergence of APA as an important and widespread mechanism of gene control highlights the importance of obtaining a detailed mechanistic understanding of the polyadenylation complex, and structural studies will continue to provide key insights into this large and complex machinery.
FIG 6.
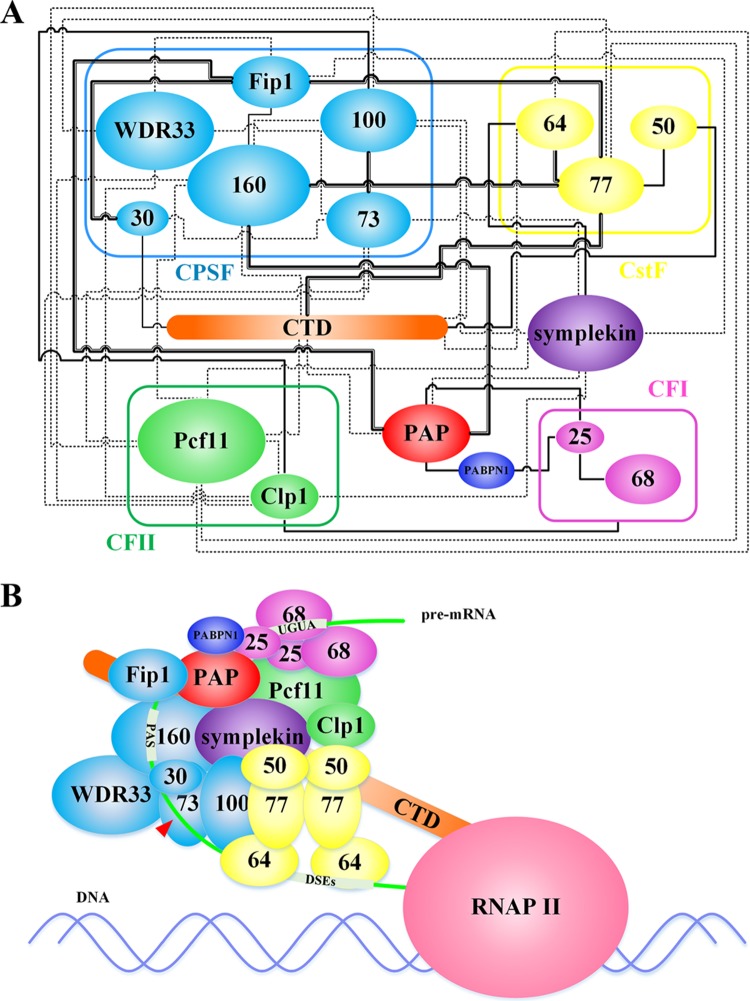
Mammalian pre-mRNA 3′ processing machinery. (A) Wire map of the interaction network of core mammalian pre-mRNA 3′ processing machinery. Thick double lines represent interactions observed in both mammalian and yeast systems. Solid lines represent interactions studied only in mammals, while dashed lines represent interactions studied only in yeast. (B) Model of core mammalian pre-mRNA 3′ processing machinery. cis elements are highlighted in boxes on the pre-mRNA. The red arrowhead indicates the cleavage site.
ACKNOWLEDGMENTS
Kehui Xiang was a joint student in the Tong and Manley laboratories.
The research from our labs described here was supported by grants from the NIH to L.T. (GM077175) and J.L.M. (GM28983).
Biographies

Kehui Xiang received his B.S. from Tsinghua University in China, majoring in mathematics and physics. He was awarded a faculty fellowship to pursue his graduate study at Columbia University in 2007. Comentored by Liang Tong and James L. Manley, he studied protein factors involved in eukaryotic mRNA 3′ processing by using crystallography and biochemistry. His research has been published in several peer-reviewed journals, such as Nature, Nature Communications, and Genes & Development. He received the Peter Sajovic Memorial Prize for outstanding work in biology and his Ph.D. in 2013 at Columbia University. Dr. Xiang is now a postdoctoral associate in David Bartel's lab at the Whitehead Institute, Massachusetts Institute of Technology.

Liang Tong is currently professor and chair of the Department of Biological Sciences at Columbia University in New York City. He received his Ph.D. in 1989 from the University of California at Berkeley and did his postdoctoral research at Purdue University. In 1992, he became a senior scientist (and in 1996 the principal scientist) at Boehringer Ingelheim Pharmaceuticals, Inc., Ridgefield, CT. He is the recipient of the first Boehringer Ingelheim (Worldwide) Research and Development Award. He became an associate professor at Columbia University in 1997 and a professor in 2004. He has published more than 220 papers, and his research focuses on structural and functional studies of proteins involved in mRNA processing and enzymes involved in fatty acid metabolism. He was elected a fellow of the American Association for the Advancement of Science in 2009.

James L. Manley received a B.S. from Columbia University and a Ph.D. from Stony Brook/Cold Spring Harbor Laboratory and did postdoctoral work at the Massachusetts Institute of Technology. He has been in the Department of Biological Sciences at Columbia University since 1980 and was chair from 1995 to 2001 and Julian Clarence Levi professor of life sciences since 1995. His research interests center on understanding the mechanisms and regulation of gene expression in mammalian cells. His work has been supported by many grants, including an NIH MERIT Award. He has authored or coauthored over 300 research articles and reviews on these topics and is an ISI Highly Cited Researcher. Dr. Manley is or has been an editor of three journals and has served on numerous editorial boards and review panels. He is a fellow of the American Academy of Microbiology, the American Academy of Arts and Sciences, and the American Association for the Advancement of Science and a member of the National Academy of Sciences.
Footnotes
Published ahead of print 3 March 2014
REFERENCES
- 1.Zhao J, Hyman L, Moore C. 1999. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63:405–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR, III, Frank J, Manley JL. 2009. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol. Cell 33:365–376. 10.1016/j.molcel.2008.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mandel CR, Bai Y, Tong L. 2008. Protein factors in pre-mRNA 3′-end processing. Cell. Mol. Life Sci. 65:1099–1122. 10.1007/s00018-007-7474-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moore MJ, Proudfoot NJ. 2009. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136:688–700. 10.1016/j.cell.2009.02.001 [DOI] [PubMed] [Google Scholar]
- 5.Danckwardt S, Hentze MW, Kulozik AE. 2008. 3′ end mRNA processing: molecular mechanisms and implications for health and disease. EMBO J. 27:482–498. 10.1038/sj.emboj.7601932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Giammartino DC, Nishida K, Manley JL. 2011. Mechanisms and consequences of alternative polyadenylation. Mol. Cell 43:853–866. 10.1016/j.molcel.2011.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tian B, Manley JL. 2013. Alternative cleavage and polyadenylation: the long and short of it. Trends Biochem. Sci. 38:312–320. 10.1016/j.tibs.2013.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ji Z, Lee JY, Pan Z, Jiang B, Tian B. 2009. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc. Natl. Acad. Sci. U. S. A. 106:7028–7033. 10.1073/pnas.0900028106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mayr C, Bartel DP. 2009. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138:673–684. 10.1016/j.cell.2009.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. 2008. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320:1643–1647. 10.1126/science.1155390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colgan DF, Manley JL. 1997. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 11:2755–2766. 10.1101/gad.11.21.2755 [DOI] [PubMed] [Google Scholar]
- 12.Proudfoot NJ. 2011. Ending the message: poly(A) signals then and now. Genes Dev. 25:1770–1782. 10.1101/gad.17268411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Q, Doublié S. 2011. Structural biology of poly(A) site definition. Wiley Interdiscip. Rev. RNA 2:732–747. 10.1002/wrna.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheets MD, Ogg SC, Wickens MP. 1990. Point mutations in AAUAAA and the poly(A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res. 18:5799–5805. 10.1093/nar/18.19.5799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beaudoing E, Freier S, Wyatt JR, Claverie JM, Gautheret D. 2000. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 10:1001–1010. 10.1101/gr.10.7.1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu J, Lutz CS, Wilusz J, Tian B. 2005. Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. RNA 11:1485–1493. 10.1261/rna.2107305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pauws E, van Kampen AH, van de Graaf SA, de Vijlder JJ, Ris-Stalpers C. 2001. Heterogeneity in polyadenylation cleavage sites in mammalian mRNA sequences: implications for SAGE analysis. Nucleic Acids Res. 29:1690–1694. 10.1093/nar/29.8.1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacDonald CC, Wilusz J, Shenk T. 1994. The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location. Mol. Cell. Biol. 14:6647–6654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hart RP, McDevitt MA, Ali H, Nevins JR. 1985. Definition of essential sequences and functional equivalence of elements downstream of the adenovirus E2A and the early simian virus 40 polyadenylation sites. Mol. Cell. Biol. 5:2975–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McLauchlan J, Gaffney D, Whitton JL, Clements JB. 1985. The consensus sequence YGTGTTYY located downstream from the AATAAA signal is required for efficient formation of mRNA 3′ termini. Nucleic Acids Res. 13:1347–1368. 10.1093/nar/13.4.1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chou ZF, Chen F, Wilusz J. 1994. Sequence and position requirements for uridylate-rich downstream elements of polyadenylation signals. Nucleic Acids Res. 22:2525–2531. 10.1093/nar/22.13.2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gil A, Proudfoot NJ. 1987. Position-dependent sequence elements downstream of AAUAAA are required for efficient rabbit beta-globin mRNA 3′ end formation. Cell 49:399–406. 10.1016/0092-8674(87)90292-3 [DOI] [PubMed] [Google Scholar]
- 23.Venkataraman K, Brown KM, Gilmartin GM. 2005. Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev. 19:1315–1327. 10.1101/gad.1298605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heidmann S, Schindewolf C, Stumpf G, Domdey H. 1994. Flexibility and interchangeability of polyadenylation signals in Saccharomyces cerevisiae. Mol. Cell. Biol. 14:4633–4642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo Z, Sherman F. 1995. 3′-end-forming signals of yeast mRNA. Mol. Cell. Biol. 15:5983–5990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dichtl B, Keller W. 2001. Recognition of polyadenylation sites in yeast pre-mRNAs by cleavage and polyadenylation factor. EMBO J. 20:3197–3209. 10.1093/emboj/20.12.3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graber JH, Cantor CR, Mohr SC, Smith TF. 1999. In silico detection of control signals: mRNA 3′-end-processing sequences in diverse species. Proc. Natl. Acad. Sci. U. S. A. 96:14055–14060. 10.1073/pnas.96.24.14055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore CL, Sharp PA. 1985. Accurate cleavage and polyadenylation of exogenous RNA substrate. Cell 41:845–855. 10.1016/S0092-8674(85)80065-9 [DOI] [PubMed] [Google Scholar]
- 29.Christofori G, Keller W. 1988. 3′ cleavage and polyadenylation of mRNA precursors in vitro requires a poly(A) polymerase, a cleavage factor, and a snRNP. Cell 54:875–889. 10.1016/S0092-8674(88)91263-9 [DOI] [PubMed] [Google Scholar]
- 30.Gilmartin GM, Nevins JR. 1989. An ordered pathway of assembly of components required for polyadenylation site recognition and processing. Genes Dev. 3:2180–2190. 10.1101/gad.3.12b.2180 [DOI] [PubMed] [Google Scholar]
- 31.Takagaki Y, Ryner LC, Manley JL. 1989. Four factors are required for 3′-end cleavage of pre-mRNAs. Genes Dev. 3:1711–1724. 10.1101/gad.3.11.1711 [DOI] [PubMed] [Google Scholar]
- 32.Wahle E. 1991. A novel poly(A)-binding protein acts as a specificity factor in the second phase of messenger RNA polyadenylation. Cell 66:759–768. 10.1016/0092-8674(91)90119-J [DOI] [PubMed] [Google Scholar]
- 33.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL. 1997. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385:357–361. 10.1038/385357a0 [DOI] [PubMed] [Google Scholar]
- 34.Hirose Y, Manley JL. 1998. RNA polymerase II is an essential mRNA polyadenylation factor. Nature 395:93–96. 10.1038/25786 [DOI] [PubMed] [Google Scholar]
- 35.Takagaki Y, Manley JL. 2000. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol. Cell. Biol. 20:1515–1525. 10.1128/MCB.20.5.1515-1525.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bienroth S, Wahle E, Suter-Crazzolara C, Keller W. 1991. Purification of the cleavage and polyadenylation factor involved in the 3′-processing of messenger RNA precursors. J. Biol. Chem. 266:19768–19776 [PubMed] [Google Scholar]
- 37.Dantonel JC, Murthy KG, Manley JL, Tora L. 1997. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature 389:399–402. 10.1038/38763 [DOI] [PubMed] [Google Scholar]
- 38.Murthy KG, Manley JL. 1992. Characterization of the multisubunit cleavage-polyadenylation specificity factor from calf thymus. J. Biol. Chem. 267:14804–14811 [PubMed] [Google Scholar]
- 39.Kaufmann I, Martin G, Friedlein A, Langen H, Keller W. 2004. Human Fip1 is a subunit of CPSF that binds to U-rich RNA elements and stimulates poly(A) polymerase. EMBO J. 23:616–626. 10.1038/sj.emboj.7600070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dominski Z. 2010. The hunt for the 3′ endonuclease. Wiley Interdiscip. Rev. RNA 1:325–340. 10.1002/wrna.33 [DOI] [PubMed] [Google Scholar]
- 41.Callebaut I, Moshous D, Mornon J-P, de Villartay J-P. 2002. Metallo-beta-lactamase fold within nucleic acids processing enzymes: the beta-CASP family. Nucleic Acids Res. 30:3592–3601. 10.1093/nar/gkf470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ryan K, Calvo O, Manley JL. 2004. Evidence that polyadenylation factor CPSF-73 is the mRNA 3′ processing endonuclease. RNA 10:565–573. 10.1261/rna.5214404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, Tong L. 2006. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature 444:953–956. 10.1038/nature05363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li de la Sierra-Gallay I, Zig L, Jamalli A, Putzer H. 2008. Structural insights into the dual activity of RNase J. Nat. Struct. Mol. Biol. 15:206–212. 10.1038/nsmb.1376 [DOI] [PubMed] [Google Scholar]
- 45.Nishida Y, Ishikawa H, Baba S, Nakagawa N, Kuramitsu S, Masui R. 2010. Crystal structure of an archaeal cleavage and polyadenylation specificity factor subunit from Pyrococcus horikoshii. Proteins 78:2395–2398. 10.1002/prot.22748 [DOI] [PubMed] [Google Scholar]
- 46.Mir-Montazeri B, Ammelburg M, Forouzan D, Lupas AN, Hartmann MD. 2011. Crystal structure of a dimeric archaeal cleavage and polyadenylation specificity factor. J. Struct. Biol. 173:191–195. 10.1016/j.jsb.2010.09.013 [DOI] [PubMed] [Google Scholar]
- 47.Silva APG, Chechik M, Byrne RT, Waterman DG, Ng CL, Dodson EJ, Koonin EV, Antson AA, Smits C. 2011. Structure and activity of a novel archaeal β-CASP protein with N-terminal KH domains. Structure 19:622–632. 10.1016/j.str.2011.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kolev NG, Yario TA, Benson E, Steitz JA. 2008. Conserved motifs in both CPSF73 and CPSF100 are required to assemble the active endonuclease for histone mRNA 3′-end maturation. EMBO Rep. 9:1013–1018. 10.1038/embor.2008.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jenny A, Minvielle-Sebastia L, Preker PJ, Keller W. 1996. Sequence similarity between the 73-kilodalton protein of mammalian CPSF and a subunit of yeast polyadenylation factor I. Science 274:1514–1517. 10.1126/science.274.5292.1514 [DOI] [PubMed] [Google Scholar]
- 50.Kyburz A, Sadowski M, Dichtl B, Keller W. 2003. The role of the yeast cleavage and polyadenylation factor subunit Ydh1p/Cft2p in pre-mRNA 3′-end formation. Nucleic Acids Res. 31:3936–3945. 10.1093/nar/gkg478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Preker PJ, Ohnacker M, Minvielle-Sebastia L, Keller W. 1997. A multisubunit 3′ end processing factor from yeast containing poly(A) polymerase and homologues of the subunits of mammalian cleavage and polyadenylation specificity factor. EMBO J. 16:4727–4737. 10.1093/emboj/16.15.4727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao J, Kessler MM, Moore CL. 1997. Cleavage factor II of Saccharomyces cerevisiae contains homologues to subunits of the mammalian cleavage/polyadenylation specificity factor and exhibits sequence-specific, ATP-dependent interaction with precursor RNA. J. Biol. Chem. 272:10831–10838. 10.1074/jbc.272.16.10831 [DOI] [PubMed] [Google Scholar]
- 53.Dominski Z, Yang X-C, Purdy M, Wagner EJ, Marzluff WF. 2005. A CPSF-73 homologue is required for cell cycle progression but not cell growth and interacts with a protein having features of CPSF-100. Mol. Cell. Biol. 25:1489–1500. 10.1128/MCB.25.4.1489-1500.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dominski Z. 2007. Nucleases of the metallo-beta-lactamase family and their role in DNA and RNA metabolism. Crit. Rev. Biochem. Mol. Biol. 42:67–93. 10.1080/10409230701279118 [DOI] [PubMed] [Google Scholar]
- 55.Barabino SM, Hübner W, Jenny A, Minvielle-Sebastia L, Keller W. 1997. The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are RNA-binding zinc finger proteins. Genes Dev. 11:1703–1716. 10.1101/gad.11.13.1703 [DOI] [PubMed] [Google Scholar]
- 56.Barabino SM, Ohnacker M, Keller W. 2000. Distinct roles of two Yth1p domains in 3′-end cleavage and polyadenylation of yeast pre-mRNAs. EMBO J. 19:3778–3787. 10.1093/emboj/19.14.3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.D'Souza V, Summers MF. 2004. Structural basis for packaging the dimeric genome of Moloney murine leukaemia virus. Nature 431:586–590. 10.1038/nature02944 [DOI] [PubMed] [Google Scholar]
- 58.Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. 2004. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat. Struct. Mol. Biol. 11:257–264. 10.1038/nsmb738 [DOI] [PubMed] [Google Scholar]
- 59.Jenny A, Hauri HP, Keller W. 1994. Characterization of cleavage and polyadenylation specificity factor and cloning of its 100-kilodalton subunit. Mol. Cell. Biol. 14:8183–8190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tacahashi Y, Helmling S, Moore CL. 2003. Functional dissection of the zinc finger and flanking domains of the Yth1 cleavage/polyadenylation factor. Nucleic Acids Res. 31:1744–1752. 10.1093/nar/gkg265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nemeroff ME, Barabino SM, Li Y, Keller W, Krug RM. 1998. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol. Cell 1:991–1000. 10.1016/S1097-2765(00)80099-4 [DOI] [PubMed] [Google Scholar]
- 62.Das K, Ma L-C, Xiao R, Radvansky B, Aramini J, Zhao L, Marklund J, Kuo R-L, Twu KY, Arnold E, Krug RM, Montelione GT. 2008. Structural basis for suppression of a host antiviral response by influenza A virus. Proc. Natl. Acad. Sci. U. S. A. 105:13093–13098. 10.1073/pnas.0805213105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nag A, Narsinh K, Martinson HG. 2007. The poly(A)-dependent transcriptional pause is mediated by CPSF acting on the body of the polymerase. Nat. Struct. Mol. Biol. 14:662–669. 10.1038/nsmb1253 [DOI] [PubMed] [Google Scholar]
- 64.Neuwald AF, Poleksic A. 2000. PSI-BLAST searches using hidden Markov models of structural repeats: prediction of an unusual sliding DNA clamp and of beta-propellers in UV-damaged DNA-binding protein. Nucleic Acids Res. 28:3570–3580. 10.1093/nar/28.18.3570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N. 2006. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature 443:590–593 (Letter.) 10.1038/nature05175 [DOI] [PubMed] [Google Scholar]
- 66.Scrima A, Konícková R, Czyzewski BK, Kawasaki Y, Jeffrey PD, Groisman R, Nakatani Y, Iwai S, Pavletich NP, Thomä NH. 2008. Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex. Cell 135:1213–1223. 10.1016/j.cell.2008.10.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stirnimann CU, Petsalaki E, Russell RB, Müller CW. 2010. WD40 proteins propel cellular networks. Trends Biochem. Sci. 35:565–574. 10.1016/j.tibs.2010.04.003 [DOI] [PubMed] [Google Scholar]
- 68.Keller W, Bienroth S, Lang KM, Christofori G. 1991. Cleavage and polyadenylation factor CPF specifically interacts with the pre-mRNA 3′ processing signal AAUAAA. EMBO J. 10:4241–4249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Murthy KG, Manley JL. 1995. The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′-end formation. Genes Dev. 9:2672–2683. 10.1101/gad.9.21.2672 [DOI] [PubMed] [Google Scholar]
- 70.Dichtl B, Blank D, Sadowski M, Hübner W, Weiser S, Keller W. 2002. Yhh1p/Cft1p directly links poly(A) site recognition and RNA polymerase II transcription termination. EMBO J. 21:4125–4135. 10.1093/emboj/cdf390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Preker PJ, Lingner J, Minvielle-Sebastia L, Keller W. 1995. The FIP1 gene encodes a component of a yeast pre-mRNA polyadenylation factor that directly interacts with poly(A) polymerase. Cell 81:379–389. 10.1016/0092-8674(95)90391-7 [DOI] [PubMed] [Google Scholar]
- 72.Meinke G, Ezeokonkwo C, Balbo P, Stafford W, Moore C, Bohm A. 2008. Structure of yeast poly(A) polymerase in complex with a peptide from Fip1, an intrinsically disordered protein. Biochemistry 47:6859–6869. 10.1021/bi800204k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Forbes KP, Addepalli B, Hunt AG. 2006. An Arabidopsis Fip1 homolog interacts with RNA and provides conceptual links with a number of other polyadenylation factor subunits. J. Biol. Chem. 281:176–186. 10.1074/jbc.M510964200 [DOI] [PubMed] [Google Scholar]
- 74.Ezeokonkwo C, Zhelkovsky A, Lee R, Bohm A, Moore CL. 2011. A flexible linker region in Fip1 is needed for efficient mRNA polyadenylation. RNA 17:652–664. 10.1261/rna.2273111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gunasekaran K, Tsai C-J, Kumar S, Zanuy D, Nussinov R. 2003. Extended disordered proteins: targeting function with less scaffold. Trends Biochem. Sci. 28:81–85. 10.1016/S0968-0004(03)00003-3 [DOI] [PubMed] [Google Scholar]
- 76.Ito S, Sakai A, Nomura T, Miki Y, Ouchida M, Sasaki J, Shimizu K. 2001. A novel WD40 repeat protein, WDC146, highly expressed during spermatogenesis in a stage-specific manner. Biochem. Biophys. Res. Commun. 280:656–663. 10.1006/bbrc.2000.4163 [DOI] [PubMed] [Google Scholar]
- 77.Ohnacker M, Barabino SM, Preker PJ, Keller W. 2000. The WD-repeat protein pfs2p bridges two essential factors within the yeast pre-mRNA 3′-end-processing complex. EMBO J. 19:37–47. 10.1093/emboj/19.1.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ghazy MA, Gordon JMB, Lee SD, Singh BN, Bohm A, Hampsey M, Moore C. 2012. The interaction of Pcf11 and Clp1 is needed for mRNA 3′-end formation and is modulated by amino acids in the ATP-binding site. Nucleic Acids Res. 40:1214–1225. 10.1093/nar/gkr801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang S-W, Asakawa K, Win TZ, Toda T, Norbury CJ. 2005. Inactivation of the pre-mRNA cleavage and polyadenylation factor Pfs2 in fission yeast causes lethal cell cycle defects. Mol. Cell. Biol. 25:2288–2296. 10.1128/MCB.25.6.2288-2296.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gilmartin GM, Nevins JR. 1991. Molecular analyses of two poly(A) site-processing factors that determine the recognition and efficiency of cleavage of the pre-mRNA. Mol. Cell. Biol. 11:2432–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wilusz J, Shenk T, Takagaki Y, Manley JL. 1990. A multicomponent complex is required for the AAUAAA-dependent cross-linking of a 64-kilodalton protein to polyadenylation substrates. Mol. Cell. Biol. 10:1244–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Takagaki Y, Manley JL, MacDonald CC, Wilusz J, Shenk T. 1990. A multisubunit factor, CstF, is required for polyadenylation of mammalian pre-mRNAs. Genes Dev. 4:2112–2120. 10.1101/gad.4.12a.2112 [DOI] [PubMed] [Google Scholar]
- 83.Preker PJ, Keller W. 1998. The HAT helix, a repetitive motif implicated in RNA processing. Trends Biochem. Sci. 23:15–16. 10.1016/S0968-0004(97)01156-0 [DOI] [PubMed] [Google Scholar]
- 84.Bai Y, Auperin TC, Chou C-Y, Chang G-G, Manley JL, Tong L. 2007. Crystal structure of murine CstF-77: dimeric association and implications for polyadenylation of mRNA precursors. Mol. Cell 25:863–875. 10.1016/j.molcel.2007.01.034 [DOI] [PubMed] [Google Scholar]
- 85.Legrand P, Pinaud N, Minvielle-Sébastia L, Fribourg S. 2007. The structure of the CstF-77 homodimer provides insights into CstF assembly. Nucleic Acids Res. 35:4515–4522. 10.1093/nar/gkm458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Benoit B, Juge F, Iral F, Audibert A, Simonelig M. 2002. Chimeric human CstF-77/Drosophila Suppressor of forked proteins rescue suppressor of forked mutant lethality and mRNA 3′ end processing in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 99:10593–10598. 10.1073/pnas.162191899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Paulson AR, Tong L. 2012. Crystal structure of the Rna14-Rna15 complex. RNA 18:1154–1162. 10.1261/rna.032524.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gordon JMB, Shikov S, Kuehner JN, Liriano M, Lee E, Stafford W, Poulsen MB, Harrison C, Moore C, Bohm A. 2011. Reconstitution of CF IA from overexpressed subunits reveals stoichiometry and provides insights into molecular topology. Biochemistry 50:10203–10214. 10.1021/bi200964p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Takagaki Y, Manley JL. 1994. A polyadenylation factor subunit is the human homologue of the Drosophila suppressor of forked protein. Nature 372:471–474. 10.1038/372471a0 [DOI] [PubMed] [Google Scholar]
- 90.Hockert JA, Yeh H-J, MacDonald CC. 2010. The hinge domain of the cleavage stimulation factor protein CstF-64 is essential for CstF-77 interaction, nuclear localization, and polyadenylation. J. Biol. Chem. 285:695–704. 10.1074/jbc.M109.061705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Moreno-Morcillo M, Minvielle-Sébastia L, Fribourg S, Mackereth CD. 2011. Locked tether formation by cooperative folding of Rna14p monkeytail and Rna15p hinge domains in the yeast CF IA complex. Structure 19:534–545. 10.1016/j.str.2011.02.003 [DOI] [PubMed] [Google Scholar]
- 92.Noble CG, Walker PA, Calder LJ, Taylor IA. 2004. Rna14-Rna15 assembly mediates the RNA-binding capability of Saccharomyces cerevisiae cleavage factor IA. Nucleic Acids Res. 32:3364–3375. 10.1093/nar/gkh664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wilusz J, Shenk T. 1988. A 64 kd nuclear protein binds to RNA segments that include the AAUAAA polyadenylation motif. Cell 52:221–228. 10.1016/0092-8674(88)90510-7 [DOI] [PubMed] [Google Scholar]
- 94.Takagaki Y, MacDonald CC, Shenk T, Manley JL. 1992. The human 64-kDa polyadenylylation factor contains a ribonucleoprotein-type RNA binding domain and unusual auxiliary motifs. Proc. Natl. Acad. Sci. U. S. A. 89:1403–1407. 10.1073/pnas.89.4.1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Takagaki Y, Manley JL. 1997. RNA recognition by the human polyadenylation factor CstF. Mol. Cell. Biol. 17:3907–3914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pérez Cañadillas JM, Varani G. 2003. Recognition of GU-rich polyadenylation regulatory elements by human CstF-64 protein. EMBO J. 22:2821–2830. 10.1093/emboj/cdg259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pancevac C, Goldstone DC, Ramos A, Taylor IA. 2010. Structure of the Rna15 RRM-RNA complex reveals the molecular basis of GU specificity in transcriptional 3′-end processing factors. Nucleic Acids Res. 38:3119–3132. 10.1093/nar/gkq002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gross S, Moore CL. 2001. Rna15 interaction with the A-rich yeast polyadenylation signal is an essential step in mRNA 3′-end formation. Mol. Cell. Biol. 21:8045–8055. 10.1128/MCB.21.23.8045-8055.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Leeper TC, Qu X, Lu C, Moore C, Varani G. 2010. Novel protein-protein contacts facilitate mRNA 3′-processing signal recognition by Rna15 and Hrp1. J. Mol. Biol. 401:334–349. 10.1016/j.jmb.2010.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Barnwal RP, Lee SD, Moore C, Varani G. 2012. Structural and biochemical analysis of the assembly and function of the yeast pre-mRNA 3′ end processing complex CF I. Proc. Natl. Acad. Sci. U. S. A. 109:21342–21347. 10.1073/pnas.1214102110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ruepp M-D, Schweingruber C, Kleinschmidt N, Schümperli D. 2011. Interactions of CstF-64, CstF-77, and symplekin: implications on localisation and function. Mol. Biol. Cell 22:91–104. 10.1091/mbc.E10-06-0543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gross S, Moore C. 2001. Five subunits are required for reconstitution of the cleavage and polyadenylation activities of Saccharomyces cerevisiae cleavage factor I. Proc. Natl. Acad. Sci. U. S. A. 98:6080–6085. 10.1073/pnas.101046598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Qu X, Perez-Canadillas J-M, Agrawal S, De Baecke J, Cheng H, Varani G, Moore C. 2007. The C-terminal domains of vertebrate CstF-64 and its yeast orthologue Rna15 form a new structure critical for mRNA 3′-end processing. J. Biol. Chem. 282:2101–2115. 10.1074/jbc.M609981200 [DOI] [PubMed] [Google Scholar]
- 104.Calvo O, Manley JL. 2001. Evolutionarily conserved interaction between CstF-64 and PC4 links transcription, polyadenylation, and termination. Mol. Cell 7:1013–1023. 10.1016/S1097-2765(01)00236-2 [DOI] [PubMed] [Google Scholar]
- 105.Richardson JM, McMahon KW, MacDonald CC, Makhatadze GI. 1999. MEARA sequence repeat of human CstF-64 polyadenylation factor is helical in solution. A spectroscopic and calorimetric study. Biochemistry 38:12869–12875 [DOI] [PubMed] [Google Scholar]
- 106.Wallace AM, Dass B, Ravnik SE, Tonk V, Jenkins NA, Gilbert DJ, Copeland NG, MacDonald CC. 1999. Two distinct forms of the 64,000 Mr protein of the cleavage stimulation factor are expressed in mouse male germ cells. Proc. Natl. Acad. Sci. U. S. A. 96:6763–6768. 10.1073/pnas.96.12.6763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li W, Yeh H-J, Shankarling GS, Ji Z, Tian B, MacDonald CC. 2012. The τCstF-64 polyadenylation protein controls genome expression in testis. PLoS One 7:e48373. 10.1371/journal.pone.0048373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yao C, Choi E-A, Weng L, Xie X, Wan J, Xing Y, Moresco JJ, Tu PG, Yates JR, III, Shi Y. 2013. Overlapping and distinct functions of CstF64 and CstF64τ in mammalian mRNA 3′ processing. RNA 19:1781–1790. 10.1261/rna.042317.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Takagaki Y, Manley JL. 1992. A human polyadenylation factor is a G protein beta-subunit homologue. J. Biol. Chem. 267:23471–23474 [PubMed] [Google Scholar]
- 110.Moreno-Morcillo M, Minvielle-Sébastia L, Mackereth C, Fribourg S. 2011. Hexameric architecture of CstF supported by CstF-50 homodimerization domain structure. RNA 17:412–418. 10.1261/rna.2481011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kleiman FE, Manley JL. 1999. Functional interaction of BRCA1-associated BARD1 with polyadenylation factor CstF-50. Science 285:1576–1579. 10.1126/science.285.5433.1576 [DOI] [PubMed] [Google Scholar]
- 112.Kleiman FE, Manley JL. 2001. The BARD1-CstF-50 interaction links mRNA 3′ end formation to DNA damage and tumor suppression. Cell 104:743–753. 10.1016/S0092-8674(01)00270-7 [DOI] [PubMed] [Google Scholar]
- 113.Rüegsegger U, Beyer K, Keller W. 1996. Purification and characterization of human cleavage factor Im involved in the 3′ end processing of messenger RNA precursors. J. Biol. Chem. 271:6107–6113. 10.1074/jbc.271.11.6107 [DOI] [PubMed] [Google Scholar]
- 114.Rüegsegger U, Blank D, Keller W. 1998. Human pre-mRNA cleavage factor Im is related to spliceosomal SR proteins and can be reconstituted in vitro from recombinant subunits. Mol. Cell 1:243–253. 10.1016/S1097-2765(00)80025-8 [DOI] [PubMed] [Google Scholar]
- 115.Ruepp M-D, Schümperli D, Barabino SML. 2011. mRNA 3′ end processing and more—multiple functions of mammalian cleavage factor I-68. Wiley Interdiscip. Rev. RNA 2:79–91. 10.1002/wrna.35 [DOI] [PubMed] [Google Scholar]
- 116.Brown KM, Gilmartin GM. 2003. A mechanism for the regulation of pre-mRNA 3′ processing by human cleavage factor Im. Mol. Cell 12:1467–1476. 10.1016/S1097-2765(03)00453-2 [DOI] [PubMed] [Google Scholar]
- 117.Dettwiler S, Aringhieri C, Cardinale S, Keller W, Barabino SML. 2004. Distinct sequence motifs within the 68-kDa subunit of cleavage factor Im mediate RNA binding, protein-protein interactions, and subcellular localization. J. Biol. Chem. 279:35788–35797. 10.1074/jbc.M403927200 [DOI] [PubMed] [Google Scholar]
- 118.McLennan AG. 2006. The Nudix hydrolase superfamily. Cell. Mol. Life Sci. 63:123–143. 10.1007/s00018-005-5386-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Coseno M, Martin G, Berger C, Gilmartin G, Keller W, Doublié S. 2008. Crystal structure of the 25 kDa subunit of human cleavage factor Im. Nucleic Acids Res. 36:3474–3483. 10.1093/nar/gkn079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Trésaugues L, Stenmark P, Schüler H, Flodin S, Welin M, Nyman T, Hammarström M, Moche M, Gräslund S, Nordlund P. 2008. The crystal structure of human cleavage and polyadenylation specific factor-5 reveals a dimeric Nudix protein with a conserved catalytic site. Proteins 73:1047–1052. 10.1002/prot.22198 [DOI] [PubMed] [Google Scholar]
- 121.Yang Q, Gilmartin GM, Doublié S. 2010. Structural basis of UGUA recognition by the Nudix protein CFI (m)25 and implications for a regulatory role in mRNA 3′ processing. Proc. Natl. Acad. Sci. U. S. A. 107:10062–10067. 10.1073/pnas.1000848107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Awasthi S, Alwine JC. 2003. Association of polyadenylation cleavage factor I with U1 snRNP. RNA 9:1400–1409. 10.1261/rna.5104603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rappsilber J, Ryder U, Lamond AI, Mann M. 2002. Large-scale proteomic analysis of the human spliceosome. Genome Res. 12:1231–1245. 10.1101/gr.473902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Millevoi S, Loulergue C, Dettwiler S, Karaa SZ, Keller W, Antoniou M, Vagner S. 2006. An interaction between U2AF 65 and CF I(m) links the splicing and 3′ end processing machineries. EMBO J. 25:4854–4864. 10.1038/sj.emboj.7601331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Li H, Tong S, Li X, Shi H, Ying Z, Gao Y, Ge H, Niu L, Teng M. 2011. Structural basis of pre-mRNA recognition by the human cleavage factor Im complex. Cell Res. 21:1039–1051. 10.1038/cr.2011.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yang Q, Coseno M, Gilmartin GM, Doublié S. 2011. Crystal structure of a human cleavage factor CFI(m)25/CFI(m)68/RNA complex provides an insight into poly(A) site recognition and RNA looping. Structure 19:368–377. 10.1016/j.str.2010.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yang Q, Gilmartin GM, Doublié S. 2011. The structure of human cleavage factor I(m) hints at functions beyond UGUA-specific RNA binding: a role in alternative polyadenylation and a potential link to 5′ capping and splicing. RNA Biol. 8:748–753. 10.4161/rna.8.5.16040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kubo T, Wada T, Yamaguchi Y, Shimizu A, Handa H. 2006. Knock-down of 25 kDa subunit of cleavage factor Im in HeLa cells alters alternative polyadenylation within 3′-UTRs. Nucleic Acids Res. 34:6264–6271. 10.1093/nar/gkl794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Martin G, Gruber AR, Keller W, Zavolan M. 2012. Genome-wide analysis of pre-mRNA 3′ end processing reveals a decisive role of human cleavage factor I in the regulation of 3′ UTR length. Cell Rep. 1:753–763. 10.1016/j.celrep.2012.05.003 [DOI] [PubMed] [Google Scholar]
- 130.de Vries H, Rüegsegger U, Hübner W, Friedlein A, Langen H, Keller W. 2000. Human pre-mRNA cleavage factor II(m) contains homologs of yeast proteins and bridges two other cleavage factors. EMBO J. 19:5895–5904. 10.1093/emboj/19.21.5895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Amrani N, Minet M, Wyers F, Dufour ME, Aggerbeck LP, Lacroute F. 1997. PCF11 encodes a third protein component of yeast cleavage and polyadenylation factor I. Mol. Cell. Biol. 17:1102–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Minvielle-Sebastia L, Preker PJ, Wiederkehr T, Strahm Y, Keller W. 1997. The major yeast poly(A)-binding protein is associated with cleavage factor IA and functions in premessenger RNA 3′-end formation. Proc. Natl. Acad. Sci. U. S. A. 94:7897–7902. 10.1073/pnas.94.15.7897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sadowski M, Dichtl B, Hübner W, Keller W. 2003. Independent functions of yeast Pcf11p in pre-mRNA 3′ end processing and in transcription termination. EMBO J. 22:2167–2177. 10.1093/emboj/cdg200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Meinhart A, Cramer P. 2004. Recognition of RNA polymerase II carboxy-terminal domain by 3′-RNA-processing factors. Nature 430:223–226. 10.1038/nature02679 [DOI] [PubMed] [Google Scholar]
- 135.Noble CG, Hollingworth D, Martin SR, Ennis-Adeniran V, Smerdon SJ, Kelly G, Taylor IA, Ramos A. 2005. Key features of the interaction between Pcf11 CID and RNA polymerase II CTD. Nat. Struct. Mol. Biol. 12:144–151. 10.1038/nsmb887 [DOI] [PubMed] [Google Scholar]
- 136.Barillà D, Lee BA, Proudfoot NJ. 2001. Cleavage/polyadenylation factor IA associates with the carboxyl-terminal domain of RNA polymerase II in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 98:445–450. 10.1073/pnas.021545298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Licatalosi DD, Geiger G, Minet M, Schroeder S, Cilli K, McNeil JB, Bentley DL. 2002. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol. Cell 9:1101–1111. 10.1016/S1097-2765(02)00518-X [DOI] [PubMed] [Google Scholar]
- 138.Hollingworth D, Noble CG, Taylor IA, Ramos A. 2006. RNA polymerase II CTD phosphopeptides compete with RNA for the interaction with Pcf11. RNA 12:555–560. 10.1261/rna.2304506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhang Z, Fu J, Gilmour DS. 2005. CTD-dependent dismantling of the RNA polymerase II elongation complex by the pre-mRNA 3′-end processing factor, Pcf11. Genes Dev. 19:1572–1580. 10.1101/gad.1296305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.West S, Proudfoot NJ. 2008. Human Pcf11 enhances degradation of RNA polymerase II-associated nascent RNA and transcriptional termination. Nucleic Acids Res. 36:905–914. 10.1093/nar/gkm1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Noble CG, Beuth B, Taylor IA. 2007. Structure of a nucleotide-bound Clp1-Pcf11 polyadenylation factor. Nucleic Acids Res. 35:87–99. 10.1093/nar/gkl1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Walker JE, Saraste M, Runswick MJ, Gay NJ. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Haddad R, Maurice F, Viphakone N, Voisinet-Hakil F, Fribourg S, Minvielle-Sébastia L. 2012. An essential role for Clp1 in assembly of polyadenylation complex CF IA and Pol II transcription termination. Nucleic Acids Res. 40:1226–1239. 10.1093/nar/gkr800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Holbein S, Scola S, Loll B, Dichtl BS, Hübner W, Meinhart A, Dichtl B. 2011. The P-loop domain of yeast Clp1 mediates interactions between CF IA and CPF factors in pre-mRNA 3′ end formation. PLoS One 6:e29139. 10.1371/journal.pone.0029139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Weitzer S, Martinez J. 2007. The human RNA kinase hClp1 is active on 3′ transfer RNA exons and short interfering RNAs. Nature 447:222–226. 10.1038/nature05777 [DOI] [PubMed] [Google Scholar]
- 146.Ramirez A, Shuman S, Schwer B. 2008. Human RNA 5′-kinase (hClp1) can function as a tRNA splicing enzyme in vivo. RNA 14:1737–1745. 10.1261/rna.1142908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Keon BH, Schäfer S, Kuhn C, Grund C, Franke WW. 1996. Symplekin, a novel type of tight junction plaque protein. J. Cell Biol. 134:1003–1018. 10.1083/jcb.134.4.1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ghazy MA, He X, Singh BN, Hampsey M, Moore C. 2009. The essential N terminus of the Pta1 scaffold protein is required for snoRNA transcription termination and Ssu72 function but is dispensable for pre-mRNA 3′-end processing. Mol. Cell. Biol. 29:2296–2307. 10.1128/MCB.01514-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhao J, Kessler M, Helmling S, O'Connor JP, Moore C. 1999. Pta1, a component of yeast CF II, is required for both cleavage and poly(A) addition of mRNA precursor. Mol. Cell. Biol. 19:7733–7740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhelkovsky A, Tacahashi Y, Nasser T, He X, Sterzer U, Jensen TH, Domdey H, Moore C. 2006. The role of the Brr5/Ysh1 C-terminal domain and its homolog Syc1 in mRNA 3′-end processing in Saccharomyces cerevisiae. RNA 12:435–445. 10.1261/rna.2267606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.He X, Khan AU, Cheng H, Pappas DL, Jr, Hampsey M, Moore CL. 2003. Functional interactions between the transcription and mRNA 3′ end processing machineries mediated by Ssu72 and Sub1. Genes Dev. 17:1030–1042. 10.1101/gad.1075203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kennedy SA, Frazier ML, Steiniger M, Mast AM, Marzluff WF, Redinbo MR. 2009. Crystal structure of the HEAT domain from the pre-mRNA processing factor symplekin. J. Mol. Biol. 392:115–128. 10.1016/j.jmb.2009.06.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Xiang K, Nagaike T, Xiang S, Kilic T, Beh MM, Manley JL, Tong L. 2010. Crystal structure of the human symplekin-Ssu72-CTD phosphopeptide complex. Nature 467:729–733. 10.1038/nature09391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Andrade MA, Petosa C, O'Donoghue SI, Müller CW, Bork P. 2001. Comparison of ARM and HEAT protein repeats. J. Mol. Biol. 309:1–18. 10.1006/jmbi.2001.4624 [DOI] [PubMed] [Google Scholar]
- 155.Marzluff WF, Wagner EJ, Duronio RJ. 2008. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat. Rev. Genet. 9:843–854. 10.1038/nrg2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hofmann I, Schnölzer M, Kaufmann I, Franke WW. 2002. Symplekin, a constitutive protein of karyo- and cytoplasmic particles involved in mRNA biogenesis in Xenopus laevis oocytes. Mol. Biol. Cell 13:1665–1676. 10.1091/mbc.01-12-0567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sullivan KD, Steiniger M, Marzluff WF. 2009. A core complex of CPSF73, CPSF100, and symplekin may form two different cleavage factors for processing of poly(A) and histone mRNAs. Mol. Cell 34:322–332. 10.1016/j.molcel.2009.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Cramer P, Bushnell DA, Kornberg RD. 2001. Structural basis of transcription: RNA polymerase II at 2.8 Angstrom resolution. Science 292:1863–1876. 10.1126/science.1059493 [DOI] [PubMed] [Google Scholar]
- 159.Bartkowiak B., Mackellar A. L., Greenleaf A. L. 2011. Updating the CTD story: from tail to epic. Genet. Res. Int. 2011:623718. 10.4061/2011/623718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hsin J-P, Manley JL. 2012. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 26:2119–2137. 10.1101/gad.200303.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Egloff S, Dienstbier M, Murphy S. 2012. Updating the RNA polymerase CTD code: adding gene-specific layers. Trends Genet. Trends Genet. 28:333–341. 10.1016/j.tig.2012.03.007 [DOI] [PubMed] [Google Scholar]
- 162.Zhang D. W., Rodríguez-Molina J. B., Tietjen J. R., Nemec C. M., Ansari A. Z. 2012. Emerging views on the CTD code. Genet. Res. Int. 2012:347214. 10.1155/2012/347214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Heidemann M, Hintermair C, Voß K, Eick D. 2013. Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription. Biochim. Biophys. Acta 1829:55–62. 10.1016/j.bbagrm.2012.08.013 [DOI] [PubMed] [Google Scholar]
- 164.Rodriguez CR, Cho EJ, Keogh MC, Moore CL, Greenleaf AL, Buratowski S. 2000. Kin28, the TFIIH-associated carboxy-terminal domain kinase, facilitates the recruitment of mRNA processing machinery to RNA polymerase II. Mol. Cell. Biol. 20:104–112. 10.1128/MCB.20.1.104-112.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Bienkiewicz EA, Moon Woody A, Woody RW. 2000. Conformation of the RNA polymerase II C-terminal domain: circular dichroism of long and short fragments. J. Mol. Biol. 297:119–133. 10.1006/jmbi.2000.3545 [DOI] [PubMed] [Google Scholar]
- 166.Jasnovidova O, Stefl R. 2013. The CTD code of RNA polymerase II: a structural view. Wiley Interdiscip. Rev. RNA 4:1–16. 10.1002/wrna.1138 [DOI] [PubMed] [Google Scholar]
- 167.Werner-Allen JW, Lee C-J, Liu P, Nicely NI, Wang S, Greenleaf AL, Zhou P. 2011. cis-proline-mediated Ser(P)5 dephosphorylation by the RNA polymerase II C-terminal domain phosphatase Ssu72. J. Biol. Chem. 286:5717–5726. 10.1074/jbc.M110.197129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Chan S, Choi E-A, Shi Y. 2011. Pre-mRNA 3′-end processing complex assembly and function. Wiley Interdiscip. Rev. RNA 2:321–335. 10.1002/wrna.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lingner J, Kellermann J, Keller W. 1991. Cloning and expression of the essential gene for poly(A) polymerase from S. cerevisiae. Nature 354:496–498. 10.1038/354496a0 [DOI] [PubMed] [Google Scholar]
- 170.Raabe T, Bollum FJ, Manley JL. 1991. Primary structure and expression of bovine poly(A) polymerase. Nature 353:229–234. 10.1038/353229a0 [DOI] [PubMed] [Google Scholar]
- 171.Wahle E. 1991. Purification and characterization of a mammalian polyadenylate polymerase involved in the 3′ end processing of messenger RNA precursors. J. Biol. Chem. 266:3131–3139 [PubMed] [Google Scholar]
- 172.Edmonds M. 1990. Polyadenylate polymerases. Methods Enzymol. 181:161–170. 10.1016/0076-6879(90)81118-E [DOI] [PubMed] [Google Scholar]
- 173.Martin G, Keller W. 1996. Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and catalytic domain, homologous to the family X polymerases, and to other nucleotidyltransferases. EMBO J. 15:2593–2603 [PMC free article] [PubMed] [Google Scholar]
- 174.Martin G, Keller W, Doublié S. 2000. Crystal structure of mammalian poly(A) polymerase in complex with an analog of ATP. EMBO J. 19:4193–4203. 10.1093/emboj/19.16.4193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Bard J, Zhelkovsky AM, Helmling S, Earnest TN, Moore CL, Bohm A. 2000. Structure of yeast poly(A) polymerase alone and in complex with 3′-dATP. Science 289:1346–1349. 10.1126/science.289.5483.1346 [DOI] [PubMed] [Google Scholar]
- 176.Balbo PB, Bohm A. 2007. Mechanism of poly(A) polymerase: structure of the enzyme-MgATP-RNA ternary complex and kinetic analysis. Structure 15:1117–1131. 10.1016/j.str.2007.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Balbo PB, Toth J, Bohm A. 2007. X-ray crystallographic and steady state fluorescence characterization of the protein dynamics of yeast polyadenylate polymerase. J. Mol. Biol. 366:1401–1415. 10.1016/j.jmb.2006.12.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Martin G, Möglich A, Keller W, Doublié S. 2004. Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) polymerase. J. Mol. Biol. 341:911–925. 10.1016/j.jmb.2004.06.047 [DOI] [PubMed] [Google Scholar]
- 179.Zhao W, Manley JL. 1996. Complex alternative RNA processing generates an unexpected diversity of poly(A) polymerase isoforms. Mol. Cell. Biol. 16:2378–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Colgan DF, Murthy KG, Prives C, Manley JL. 1996. Cell-cycle related regulation of poly(A) polymerase by phosphorylation. Nature 384:282–285. 10.1038/384282a0 [DOI] [PubMed] [Google Scholar]
- 181.Shimazu T, Horinouchi S, Yoshida M. 2007. Multiple histone deacetylases and the CREB-binding protein regulate pre-mRNA 3′-end processing. J. Biol. Chem. 282:4470–4478. 10.1074/jbc.M609745200 [DOI] [PubMed] [Google Scholar]
- 182.Vethantham V, Rao N, Manley JL. 2008. Sumoylation regulates multiple aspects of mammalian poly(A) polymerase function. Genes Dev. 22:499–511. 10.1101/gad.1628208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Di Giammartino DC, Shi Y, Manley JL. 2013. PARP1 represses PAP and inhibits polyadenylation during heat shock. Mol. Cell 49:7–17. 10.1016/j.molcel.2012.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ezeokonkwo C, Ghazy MA, Zhelkovsky A, Yeh P-C, Moore C. 2012. Novel interactions at the essential N-terminus of poly(A) polymerase that could regulate poly(A) addition in Saccharomyces cerevisiae. FEBS Lett. 586:1173–1178. 10.1016/j.febslet.2012.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Helmling S, Zhelkovsky A, Moore CL. 2001. Fip1 regulates the activity of poly(A) polymerase through multiple interactions. Mol. Cell. Biol. 21:2026–2037. 10.1128/MCB.21.6.2026-2037.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Blobel G. 1973. A protein of molecular weight 78,000 bound to the polyadenylate region of eukaryotic messenger RNAs. Proc. Natl. Acad. Sci. U. S. A. 70:924–928. 10.1073/pnas.70.3.924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Bienroth S, Keller W, Wahle E. 1993. Assembly of a processive messenger RNA polyadenylation complex. EMBO J. 12:585–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Wahle E. 1995. Poly(A) tail length control is caused by termination of processive synthesis. J. Biol. Chem. 270:2800–2808 [DOI] [PubMed] [Google Scholar]
- 189.Brawerman G. 1981. The role of the poly(A) sequence in mammalian messenger RNA. CRC Crit. Rev. Biochem. 10:1–38 [DOI] [PubMed] [Google Scholar]
- 190.Jenal M, Elkon R, Loayza-Puch F, van Haaften G, Kühn U, Menzies FM, Oude Vrielink JAF, Bos AJ, Drost J, Rooijers K, Rubinsztein DC, Agami R. 2012. The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell 149:538–553. 10.1016/j.cell.2012.03.022 [DOI] [PubMed] [Google Scholar]
- 191.Kerwitz Y, Kühn U, Lilie H, Knoth A, Scheuermann T, Friedrich H, Schwarz E, Wahle E. 2003. Stimulation of poly(A) polymerase through a direct interaction with the nuclear poly(A) binding protein allosterically regulated by RNA. EMBO J. 22:3705–3714. 10.1093/emboj/cdg347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ge H, Zhou D, Tong S, Gao Y, Teng M, Niu L. 2008. Crystal structure and possible dimerization of the single RRM of human PABPN1. Proteins 71:1539–1545. 10.1002/prot.21973 [DOI] [PubMed] [Google Scholar]
- 193.Kühn U, Nemeth A, Meyer S, Wahle E. 2003. The RNA binding domains of the nuclear poly(A)-binding protein. J. Biol. Chem. 278:16916–16925. 10.1074/jbc.M209886200 [DOI] [PubMed] [Google Scholar]
- 194.Keller RW, Kühn U, Aragón M, Bornikova L, Wahle E, Bear DG. 2000. The nuclear poly(A) binding protein, PABP2, forms an oligomeric particle covering the length of the poly(A) tail. J. Mol. Biol. 297:569–583. 10.1006/jmbi.2000.3572 [DOI] [PubMed] [Google Scholar]
- 195.Nemeth A, Krause S, Blank D, Jenny A, Jenö P, Lustig A, Wahle E. 1995. Isolation of genomic and cDNA clones encoding bovine poly(A) binding protein II. Nucleic Acids Res. 23:4034–4041. 10.1093/nar/23.20.4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Kühn U, Gündel M, Knoth A, Kerwitz Y, Rüdel S, Wahle E. 2009. Poly(A) tail length is controlled by the nuclear poly(A)-binding protein regulating the interaction between poly(A) polymerase and the cleavage and polyadenylation specificity factor. J. Biol. Chem. 284:22803–22814. 10.1074/jbc.M109.018226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Winstall E, Sadowski M, Kuhn U, Wahle E, Sachs AB. 2000. The Saccharomyces cerevisiae RNA-binding protein Rbp29 functions in cytoplasmic mRNA metabolism. J. Biol. Chem. 275:21817–21826. 10.1074/jbc.M002412200 [DOI] [PubMed] [Google Scholar]
- 198.Kühn U, Wahle E. 2004. Structure and function of poly(A) binding proteins. Biochim. Biophys. Acta 1678:67–84. 10.1016/j.bbaexp.2004.03.008 [DOI] [PubMed] [Google Scholar]
- 199.Luo Y, Yogesha SD, Cannon JR, Yan W, Ellington AD, Brodbelt JS, Zhang Y. 2013. Novel modifications on C-terminal domain of RNA polymerase II can fine-tune the phosphatase activity of Ssu72. ACS Chem. Biol. 8:2042–2052. 10.1021/cb400229c [DOI] [PMC free article] [PubMed] [Google Scholar]