ABSTRACT
The brown planthopper (BPH), Nilaparvata lugens (Hemiptera:Delphacidae), is one of the most destructive insect pests of rice crops in Asia. Nudivirus-like sequences were identified during the whole-genome sequencing of BPH. PCR examination showed that the virus sequences were present in all of the 22 BPH populations collected from East, Southeast, and South Asia. Thirty-two of the 33 nudivirus core genes were identified, including 20 homologues of baculovirus core genes. In addition, several gene clusters that were arranged collinearly with those of other nudiviruses were found in the partial virus genome. In a phylogenetic tree constructed using the supermatrix method, the original virus was grouped with other nudiviruses and was closely related to polydnavirus. Taken together, these data indicated that the virus sequences belong to a new member of the family Nudiviridae. More specifically, the virus sequences were integrated into the chromosome of its insect host during coevolution. This study is the first report of a large double-stranded circular DNA virus genome in a sap-sucking hemipteran insect.
IMPORTANCE This is the first report of a large double-stranded DNA virus integrated genome in the planthopper, a plant sap-sucking hemipteran insect. It is an exciting addition to the evolutionary story of bracoviruses (polydnaviruses), nudiviruses, and baculoviruses. The results on the virus sequences integrated in the chromosomes of its insect host also represent a story of successful coevolution of an invertebrate virus and a plant sap-sucking insect.
INTRODUCTION
Brown planthoppers (BPH) (Nilaparvata lugens) are insect herbivores that feed mainly on rice. They damage rice plants by sucking sap from the vascular bundle and by transmitting the rice ragged stunt virus (RRSV) and rice grassy stunt virus (RGSV) (1). In addition, three commensal viruses have been characterized in BPH, i.e., Nilaparvata lugens reovirus (NLRV) (2), himetobi P virus (HiPV), and Nilaparvata lugens commensal X virus (NLCXV) (3). Recently, an iflavirus found in the honeydews of BPH was reported as Nilaparvata lugens honeydew virus 1 (NLHV-1) (4). These viruses infect BPH without visible symptoms, raising the question of how the insect host copes with various foreign microbes.
In our study on the whole genome sequence of BPH, sequences that likely belonged to a previously unknown virus were identified (5). Homology analysis indicated that the sequences came from an uncharacterized virus that was related to the family Nudiviridae. Nudiviruses (Latin nudi = naked) are a highly diverse group of invertebrate viruses that have rod-shaped nucleocapsids and large, double-stranded DNA (dsDNA) genomes. They were once described as “nonoccluded baculoviruses” but were later excluded from the family Baculoviridae (6). These baculovirus-like particles have been reported in a wide range of host species of insects and other arthropods (7, 8); however, although related to baculoviruses, they form a distinct lineage.
To date, only a few nudiviruses have been well studied, such as the cricket (Gryllus bimaculatus) virus GbNV (9), the palm rhinoceros beetle (Oryctes rhinoceros) virus OrNV (10), Heliothis zea nudivirus 1 (HzNV-1) (11), and the gonad-specific HzNV-2 (12, 13). To classify these viruses, a new family, Nudiviridae, was created (http://talk.ictvonline.org/files/proposals/taxonomy_proposals_invertebrate1/m/inv04/4770.aspx) with two new genera: Alphanudivirus (OrNV and GbNV) and Betanudivirus (HzNV). Based on phylogenetic inference, the Penaeus monodon nucleopolyhedrovirus was also reassigned as P. monodon nudivirus (PmNV) (14). More recently, a viral metagenomic study revealed the presence of a Drosophila innubila nudivirus (DiNV) (15). Thus, the nudivirus group currently comprises six different viruses. Furthermore, a nudivirus appears to have been integrated into the genome of parasitoid wasps, the Braconidae, and encodes a variety of proteins in female wasps, including the structural proteins of their symbiotic polydnaviruses (16).
It was surprising that nudivirus-like sequences were detected in the sap-sucking BPH, but when we tried to isolate and purify this virus, we failed. A study of horizontal transmission also failed because negative colonies were absent. PCR detection showed that all of the 22 BPH populations collected from Asian countries carried the viral sequence. Examinations of eggs, nymphs of different instars, and adults all produced positive results. Combining these results with the assembled BPH genome sequencing data, we hypothesized that the virus sequences were chromosomally integrated during evolution. In this study, we investigated mainly the gene composition and organization of the sequences derived from the new virus and its relationship to nudiviruses, baculoviruses, and other invertebrate DNA viruses. Following the suggested nomenclature and taking into account the endogenous nature of the sequences, the original virus is referred to as Nilaparvata lugens endogenous nudivirus (NlENV) in this paper.
MATERIALS AND METHODS
Insects.
The BPH were reared on rice seedlings (Xiushui 134) under constant conditions of 27 ± 0.5°C and 14 h of daily illumination. The Hangzhou population was originally collected from a rice field located in the Huajiachi Campus of Zhejiang University, Hangzhou, China. BPH from other locations were collected and preserved in 80% ethanol.
PCR examination of nudivirus genes.
Genomic DNA was extracted from BPH using the Genomic DNA Easy Minikit (Life Science, China) according to the manufacturer's instructions. The primer pair dnahelF (5′-CTATGAAAGAACACGCAATACCA-3′) and dnahelR (5′-ATTTCCCGAATAGCTAGAGTCT-3′) was used to amplify a 544-bp sequence within the dnahel gene and thus to verify the presence of nudivirus. To confirm the gene organization, PCR amplification was performed using the primer sets listed in Table S1 in the supplemental material.
Sequence analysis.
The predicted amino acid sequences of OrNV, GbNV, and HzNV-1 proteins were downloaded from GenBank and searched against the assembled genome sequence of BPH using a TBLASTN algorithm with a cutoff E value of ≤10−3. Annotation of scaffolds containing NlENV sequences are available upon request. The results were subjected to homology searches against the NCBI's nonredundant protein databases using a BLASTX program to avoid any false predictions. In addition, we extracted all the raw reads which associated with the identified scaffolds and contigs and reassembled de novo the sequences (see Data Set S1 in the supplemental material, scaffolds containing NlENV sequences). Open reading frames (ORFs) were found and translated into amino acid sequences by the DNAStar EditSeq program (Lasergene, WI, USA). Sequence comparisons of all predicted proteins to those in public databases were carried out using the BLASTP and PSI-BLAST programs (http://www.ncbi.nlm.nih.gov/).
Phylogenetic analysis.
The amino acid sequences of P74 (PIF-0), PIF-1, PIF-2, and PIF-3 were used in this study. Congruence testing and phylogenetic analysis were performed as described in reference 17, with minor alterations. Multiple amino acid alignments were obtained for each gene with the program ClustalX (18), and phylogenetic analyses of the concatenated sequences were performed by using Bayesian inference. ProtTest (19) and Bayesian information criteria were used to select appropriate substitution models and parameters for each gene. MrBayes analyses (20) were run across four Monte Carlo Markov chains for 1 million generations, sampling every 500 generations. The consensus tree was obtained after a burn-in of 500 generations, and the value of average standard deviation (SD) of split frequencies was used as a proof of stationarity if this value was under 0.01.
qRT-PCR analysis.
To quantify virus DNA, quantitative real-time PCR (qRT-PCR) analysis was used. Genomic DNA was extracted, and the concentration of each sample was adjusted to 50 ng/μl. qRT-PCR was performed using the iTaq Universal SYBR green Supermix (Bio-Rad, CA, USA) according to the manufacturer's protocol. A nontemplate control (NTC) sample (nuclease-free water) was included in the experiment to detect contamination and to determine the degree of dimer formation (data not shown).
For gene expression analysis, total RNA was extracted from different tissues of BPH using TRIzol reagent (Invitrogen, USA), and the concentration of each RNA sample was adjusted to 1 μg/μl with nuclease-free water. A total of 0.5 μg RNA was reverse transcribed in a 10-μl reaction mixture using the ReverTra Ace qPCR RT Master Mix with genomic DNA (gDNA) Remover (Toyobo, Japan). qRT-PCR was performed as described above. The constitutively expressed GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene of BPH was used as an internal control in both genomic DNA and cDNA samples. The relative quantitative method (2−ΔΔCT) (21) was used to evaluate the quantitative variation. The sequences of the specific primer sets are listed in Table S1 in the supplemental material.
Nucleotide sequence accession numbers.
BPH nudivirus sequence data were submitted to GenBank under accession numbers KJ566523 to KJ566588.
RESULTS AND DISCUSSION
Nudivirus sequences identified in the BPH genome.
By searching for homology using the protein sequences of GbNV, OrNV, and HzNV-1 as probes, a total of 66 ORFs from the BPH genome were identified as nudivirus-like sequences (Table 1). They distributed in 15 scaffolds or contigs of the assembled BPH genome sequences (Fig. 1) (scaffolds or contigs containing more than one ORFs are shown). The GC content of the predicted ORFs ranged from 31.2 to 43.7%, with an average of 37.4%. The best BLAST hits of these sequences (amino acid identity of 24 to 64%) were known ORFs of other nudiviruses, primarily OrNV. For 10 of the remaining sequences, the best hit was GbNV, and the last one had DiNV as its best hit.
TABLE 1.
Nudivirus-like sequences identified in the BPH genomea
| Scaffold or contig no. (scaffold length, bp) | Position |
Best BLAST hit |
Homologue in other nudivirus (ORF no.) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Start | End | Gene | Species | Amino acid identity (% match identity) | OrNV | GbNV | HzNV-1 | HzNV-2 | |
| sc104 (86,547) | 3895 | 3530 | GrBNV_gp93-like protein | OrNV | 55/180 (30) | 39 | 93 | — | — |
| 3975 | 4418 | GrBNV_gp94-like protein | GbNV | 30/105 (29) | 40 | 94 | — | — | |
| 4382 | 4708 | GrBNV_gp95-like protein | OrNV | 37/96 (39) | 41 | 95 | — | — | |
| 4837 | 5382 | lef-4 | OrNV | 54/147 (36) | 42 | 96 | 98 | 43 | |
| 5854 | 6534 | GrBNV_gp97-like protein | OrNV | 68/225 (30) | 44 | 97 | — | — | |
| 7715 | 6693 | GrBNV_gp23-like protein | OrNV | 124/341 (36) | 45 | 23 | — | — | |
| 9643 | 8093 | GrBNV_gp22-like protein | OrNV | 121/396 (30) | 46 | 22 | — | — | |
| sc5052 (28,801) | 2104 | 1016 | OrNV_gp124-like protein | OrNV | 65/252 (25) | 124 | — | — | — |
| 7274 | 8659 | Flap endonuclease | OrNV | 160/421 (38) | 16 | 65 | 68 | 70 | |
| 8958 | 9908 | 38k | OrNV | 108/276 (39) | 87 | 1 | 10 | 108 | |
| 10731 | 10195 | ac81-like | GbNV | 70/161 (43) | 4 | 14 | 33 | 96 | |
| 11582 | 10758 | GrBNV_gp13-like protein | OrNV | 87/232 (37) | 3 | 13 | — | — | |
| 14327 | 11724 | dnapol | OrNV | 233/812 (29) | 1 | 12 | 131 | 18 | |
| 15807 | 16496 | vp39 | OrNV | 93/224 (41) | 15 | 64 | 89 | 52 | |
| 17134 | 18216 | rr2 | OrNV | 139/319 (44) | 102 | 63 | 73 | 65 | |
| 19880 | 19569 | GrBNV_gp62-like protein | OrNV | 18/45 (40) | 104 | 62 | — | — | |
| 20693 | 20836 | GrBNV_gp58-like protein | OrNV | 13/29 (44) | 76 | 58 | 143 | 9 | |
| 21128 | 21448 | GrBNV_gp59-like protein | GbNV | 13/24 (54) | 79 | 59 | — | — | |
| 22289 | 21561 | GrBNV_gp60-like protein | OrNV | 45/177 (25) | 80 | 60 | — | — | |
| 23012 | 22365 | GrBNV_gp61-like protein | OrNV | 75/209 (35) | 86 | 61 | — | — | |
| 24854 | 25297 | OrNV_gp098-like protein | OrNV | 60/138 (43) | 98 | — | — | — | |
| 25457 | 27430 | lef-9 | OrNV | 204/525 (39) | 96 | 24 | 75 | 63 | |
| c211476 (9,270) | 1297 | 2535 | dnalig | OrNV | 122/400 (30) | 121 | 38 | 36 | 94 |
| 2660 | 4429 | GrBNV_gp37-like protein | OrNV | 164/499 (32) | 120 | 37 | — | — | |
| 5714 | 4557 | GrBNV_gp36-like protein | OrNV | 91/326 (27) | 119 | 36 | — | — | |
| 6486 | 5797 | GrBNV_gp35-like protein | OrNV | 118/267 (44) | 118 | 35 | — | — | |
| 6654 | 7649 | tk | OrNV | 70/166 (42) | 117 | 34 | 111 | 34 | |
| 7800 | 9236 | GrBNV_gp33-like protein | OrNV | 78/272 (29) | 116 | 33 | — | — | |
| c29512 (5,207) | 602 | 3 | 19kda/pif-4 | GbNV | 73/138 (52) | 33 | 87 | 103 | 39 |
| 819 | 4781 | Helicase | OrNV | 329/1302 (25) | 34 | 88 | 104 | 38 | |
| c302951 (15,503) | 1370 | 57 | GrBNV_gp83-like protein | OrNV | 27/103 (26) | 54 | 83 | — | — |
| 1514 | 2149 | GrBNV_gp84-like protein | OrNV | 72/208 (34) | 53 | 84 | — | — | |
| 2226 | 2522 | lef-5 | OrNV | 29/85 (34) | 52 | 85 | 101 | 40 | |
| 4660 | 2606 | p74 | GbNV | 236/619 (38) | 126 | 45 | 11 | 106 | |
| 4812 | 5840 | GrBNV_gp44-like protein | OrNV | 120/322 (37) | 125 | 44 | 71 | 67 | |
| 7729 | 6584 | pif-2 | OrNV | 180/360 (50) | 17 | 66 | 123 | 26 | |
| 7784 | 9163 | GrBNV_gp67-like protein | OrNV | 38/109 (34) | 18 | 67 | — | — | |
| 9269 | 9574 | OrNVorf19-like protein | DiNV | 30/89 (33) | 19 | — | — | — | |
| 9712 | 11052 | p47 | OrNV | 146/399 (36) | 20 | 69 | 75 | 63 | |
| 13320 | 14105 | GrBNV_gp72-like protein | OrNV | 59/168 (35) | 22 | 72 | — | — | |
| 15421 | 14519 | Guanylate kinase | GbNV | 52/129 (40) | 58 | 74 | 115 | 32 | |
| c357974 (12,543) | 765 | 10 | OrNV_gp028-like protein | OrNV | 28/86 (32) | 28 | — | — | — |
| 1164 | 2435 | GrBNV_gp78-like protein | OrNV | 93/292 (31) | 27 | 78 | — | — | |
| 7503 | 5596 | GrBNV_gp76-like protein | OrNV | 192/551 (34) | 25 | 76 | — | — | |
| 7502 | 8083 | GrBNV_gp75-like protein | OrNV | 64/188 (34) | 24 | 75 | — | — | |
| 11009 | 9570 | GrBNV_gp17-like protein | GbNV | 152/466 (32) | 137 | 17 | 51 | 85 | |
| 11190 | 11600 | OrNV_gp129-like protein | OrNV | 42/115 (36) | 129 | — | — | — | |
| 11659 | 12489 | GrBNV_gp19-like protein | GbNV | 99/214 (46) | 47 | 19 | 30 | 99 | |
| c597385 (3,824) | 1786 | 2 | rr1 | OrNV | 214/516 (41) | 51 | 82 | 95 | 47 |
| 2490 | 1879 | GrBNV_gp81-like protein | OrNV | 31/117 (26) | 29 | 81 | — | — | |
| 2599 | 3783 | vlf-1 | OrNV | 116/327 (35) | 30 | 80 | 121 | 28 | |
| c115904 (4,541) | 370 | 1419 | GrBNV_gp43-like protein | OrNV | 67/243 (27) | 105 | 43 | — | — |
| 1490 | 2971 | pif-1 | GbNV | 209/460 (45) | 60 | 52 | 55 | 82 | |
| 3496 | 3221 | ac68-like | OrNV | 52/81 (64) | 72 | 55 | 74 | 64 | |
| c589636 (9,281) | 3240 | 643 | lef-8 | OrNV | 453/909 (49) | 64 | 49 | 90 | 51 |
| 3440 | 5149 | GrBNV_gp48-like protein | OrNV | 175/627 (27) | 132 | 48 | — | — | |
| 5861 | 7309 | GrBNV_gp06-like protein | OrNV | 87/313 (27) | 114 | 6 | — | — | |
| 7498 | 8472 | ac92-like | OrNV | 72/237 (30) | 113 | 7 | 13 | 104 | |
| 9244 | 8576 | pif-3 | GbNV | 81/206 (39) | 107 | 3 | 88 | 53 | |
| sc88 (919,474) | 93362 | 92088 | odv-e56/pif-5 | OrNV | 133/344 (38) | 115 | 5 | 76 | 62 |
| sc1949 (12,187) | 4581 | 2764 | Helicase 2 | OrNV | 247/743 (33) | 108 | 46 | 60 | 76 |
| 5539 | 4991 | lef-3 | OrNV | 24/98 (24) | 59 | 86 | — | — | |
| sc538 (642,765) | 430879 | 432498 | GrBNV_gp28-like protein | OrNV | 74/306 (24) | 90 | 28 | — | — |
| c525069 (2,856) | 1396 | 512 | GrBNV_gp09-like protein | OrNV | 104/253 (41) | 95 | 9 | — | — |
| c564053 (2,681) | 2206 | 512 | vp91 | OrNV | 133/453 (29) | 106 | 2 | 46 | 89 |
| c588530 (1,006) | 12 | 1004 | Integrase (partial) | OrNV | 110/274 (40) | 75 | 57 | 144 | 8 |
—, absent. Homologues to baculovirus core genes are marked in bold.
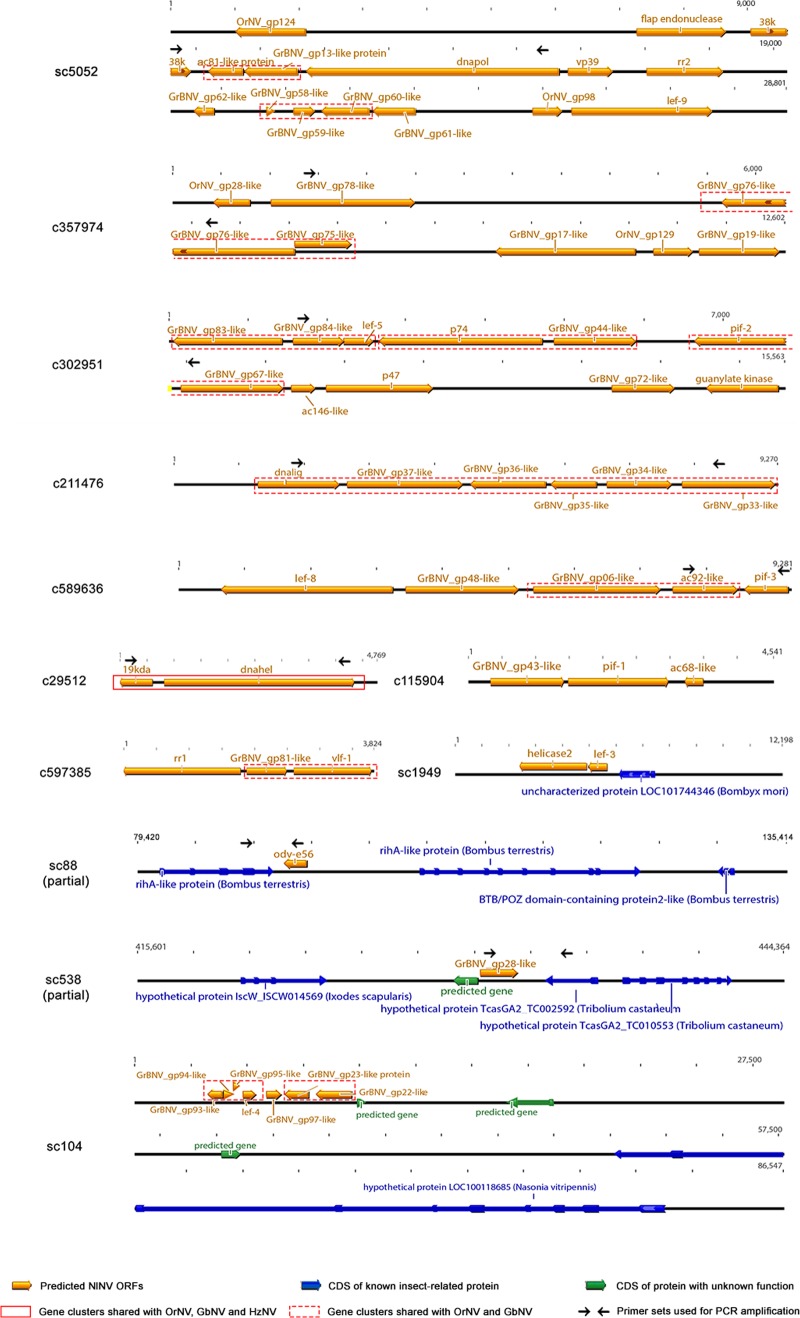
FIG 1.
Organization of NlENV ORFs in BPH scaffolds and contigs. ORFs and their transcriptional directions are indicated as arrows, and predicted BPH cellular genes in the flanking regions are also shown.
The first completely sequenced nudivirus was HzNV-1 (22). Subsequently, the genomes of GbNV (23), OrNV (24, 25), and HzNV-2 (26) have been completely sequenced. Nudiviruses have large genomes. The smallest GbNV genome is 96,944 bp, and those of HzNV-1 and -2 are more than 200 kb. The 66 ORFs of NlENV had a total length of 74,730 bp. Assuming that the coding density of this virus is 90% (similar to that of GbNV and OrNV), its genome size would be approximately 83 kb. However, some of the genes and sequences of nudiviruses are poorly conserved, which means that a substantial proportion of NlENV genes may not have been identified by homology searching.
According to the current data (14, 26, 27), the sequenced nudivirus genomes shared 33 common genes. Of these, 32 were identified in NlENV, including 20 core genes from baculovirus. The only exception was the iap-3 gene. The inhibitor of apoptosis (IAP) protein was first found in baculovirus, and its orthologues are widely distributed in eukaryotes (28). An IAP homologue was also found in our data set, but it is more likely that the sequence belonged to BPH itself because it showed higher similarity to the IAP of Riptortus pedestris (Hemiptera:Alydidae). Besides, the noncore baculovirus genes dnaligase, helicase2, rr1, and rr2 and an OrNV orf19-like homologue were also identified in NlENV. All baculoviruses have 37 core genes (29), which serve key functions in their life cycles. The actual roles of these homologues in nudiviruses are not yet clear, but the conservation of so many baculovirus genes indicates a close relationship between these two groups.
PCR survey of the viral sequence.
The genome sequencing data of BPH were derived from a purified population collected in Hangzhou, China. To investigate whether the viral sequences exist in other geographic populations, PCR examinations were carried out. A total of 22 populations were included in this survey, which were collected from 7 countries in Asia (Fig. 2a). The results suggested that all the BPH populations harbored the nudivirus-like sequence.
FIG 2.
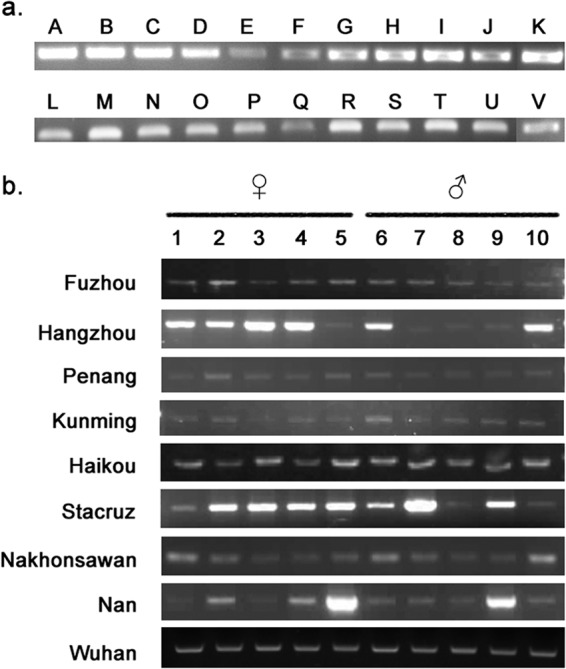
PCR examination of NlENV sequences in different BPH populations. (a) PCR survey of NlENV sequences in 22 BPH populations. Lanes A to V, BPH populations from Iloilo, Davao, and Stacruz (Philippines), Nan (Laos), Penang (Malaysia), Maharashtra (India), Nakhonsawan (Thailand), Hochiminh and Hanoi (Vietnam), and Haikou, Kunming, Yulin, Qingyuan, Wushan, Fuzhou, Wenzhou, Sanmen, Zhuji, Hangzhou, Tongxiang, Wuhan, and Yangzhou (China), respectively. (b) PCR examination performed on 10 individual BPH from nine selected populations.
To determine the percentage of insects carrying the viral sequence, individuals from 9 populations were also subjected to PCR detection. Surprisingly, the experiments revealed a viral sequence carrier rate of 100% (Fig. 2b). Examinations of eggs, nymphs of different instars, and adults all produced positive results (data not shown).
Transcription of NlENV genes.
To confirm that the nudivirus-like genes were expressed in BPH, we searched for putative NlENV mRNAs in the N. lugens transcriptome database (30). A total of 10 transcripts were found, i.e., dnapol, GrBNV gp13-like, ac81, 38k, flap endonuclease, lef-9, 19kda/pif-4, odv-e56/pif-5, helicase, and helicase2. Hence, these 10 ORFs may encode bona fide functional proteins of NlENV.
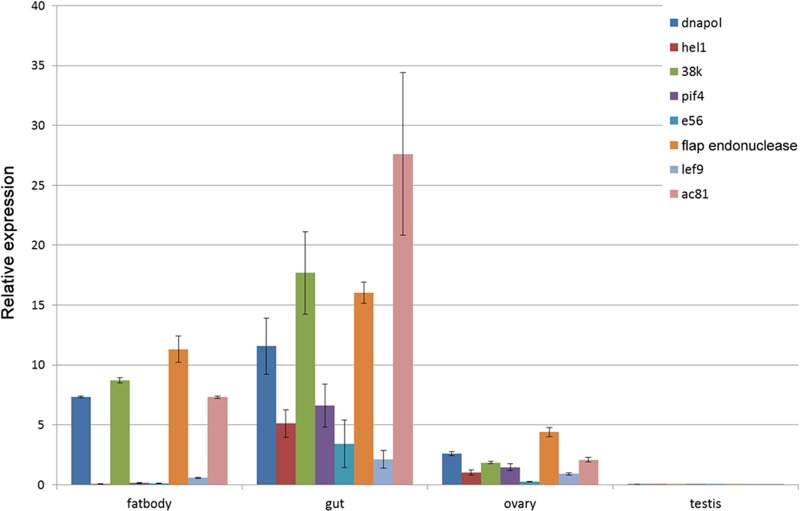
Quantitative real-time PCR was also performed to reveal the gene expression profiles (Fig. 3). All of these transcripts were most abundant in the digestive tract, then in the fat body.
FIG 3.
Transcript levels of putative NlENV genes in BPH. Adults (female for ovary, male for testis, and both for fat body and midgut) at the third day after eclosion were used in the experiment. The GAPDH gene of BPH was used as an internal control to allow for normalization. Shown are the means ± standard deviations of triplicate results.
Arrangement of orthologous genes.
Gene organization varies greatly between nudivirus genomes. However, there are regions of collinearly arranged ORFs in the sequenced nudiviruses (8). These gene clusters also existed in the partial genome of NlENV (Fig. 1). Several assembled scaffolds or contigs contained more than one ORF, which were arranged in the same way as their orthologues in other nudiviruses.
As shown in Fig. 1, the gene arrangement of NlENV was more similar to that of OrNV and GbNV. One of the gene clusters shared by OrNV, GbNV, and NlENV consisted of six genes from orf33 to -38 of GbNV. Another cluster of four genes included orf93 to -96 of GbNV, which corresponded to the orf39 to -42 of OrNV. Three genes (lef-4, tk, and dnalig) in these two clusters were considered nudivirus core genes (14), while others were absent from the HzNV genome. In addition, 9 gene clusters comprising two or three collinearly arranged genes were dispersed in the three genomes.
Only one organizationally conserved region was detected in the five nudiviruses. The gene cluster was composed of dnahel and pif-4, whose relative position and orientation remained unchanged. Notably, the link between these two genes was also conserved in baculoviruses. To date, three core gene clusters whose relative positions are conserved in all of the sequenced baculovirus genomes have been identified (31–33). One of these contained a set of four genes, i.e., dnahel, pif-4, 38k, and lef-5. Homologues of 38k and lef-5 were also present in nudivirus genomes but were separated from the other two (27). The shared gene arrangement again suggested the divergence of nudivirus and baculoviruses from a common ancestor over evolutionary time. As more sequences of nudiviruses are revealed, we will better understand their gene content and conservation.
Phylogeny and evolution.
Of the baculovirus core genes conserved in nudivirus, nine were related to viral structure (p74, pif-1, pif-2, pif-3, 19kda/pif-4, odv-e56/pif-5, vp91, vp39, and 38k) (28). The pif (per os infectivity factor) genes are crucial for successful oral infection of baculovirus occlusion-derived virions (ODVs) and may be associated with host range determination and virulence. Interestingly, homologues of PIF proteins were identified not only in the baculovirus and “nonoccluded baculovirus” but also in several other DNA virus genomes. These included the Musca domestica salivary gland hypertrophy virus (MdSGHV) and Glossina pallidipes salivary gland hypertrophy virus (GpSGHV) (hytrosaviruses) (34, 35) and the Cotesia congregata bracovirus (CcBV) and Chelonus inanitus bracovirus (CiBV) (bracoviruses) (16, 36). Moreover, PIF homologues have been found in the genome of the marine white spot syndrome virus (WSSV) (a nimavirus) by PSI-BLAST searches (25).
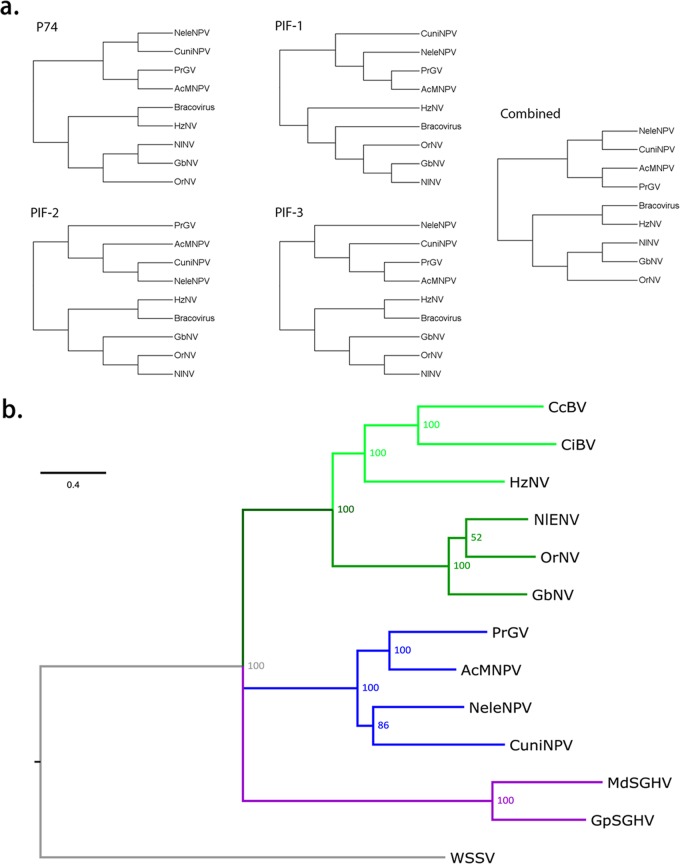
Based on the predicted amino acid sequences of 4 PIF proteins (P74/PIF0, PIF1, PIF2, and PIF3), a phylogenetic tree was constructed using the supermatrix method (Fig. 4). Before concatenating the sequences, congruence analyses on unrooted trees were done to ensure that there was no conflicting signal between the different genes (Fig. 4a; see Table S2 in the supplemental material).
FIG 4.
Nudivirus phylogeny. (a) Tree topologies of 4 single and combined PIF proteins (P74/PIF0, PIF1, PIF2, and PIF3) from nudiviruses and baculoviruses. To avoid missing data, a subset of taxa was selected to construct the unrooted trees. Log-likelihood values were calculated by matching all amino acid alignments with every topology and then subjected to Shimodaira-Hasegawa congruence tests. The results (also see Table S2 in the supplemental material) indicated that all topologies were congruent with the phylogenetic signal of each gene (a value of >0.05) and thus can be combined for phylogenetic analysis. (b) The tree of large arthropod DNA viruses based on the combined amino acid sequences of 4 PIF proteins. Multiple-sequence alignments were performed using ClustalX, and the tree was inferred using a mixed-model Bayesian phylogenetic analysis. The Bayesian inference posterior probabilities are presented at the nodes as percent values. Viruses included in this analysis were Autographa californica nucleopolyhedrovirus (AcMNPV), Pieris rapae granulosis virus (PrGV), Culex nigripalpus NPV (CuniNPV), Neodiprion lecontei NPV (NeleNPV), Cotesia congregata bracovirus (CcBV), Chelonus inanitus bracovirus (CiBV), Musca domestica salivary gland hypertrophy virus (MdSGHV), Glossina pallidipes salivary gland hypertrophy (GpSGHV), white spot syndrome virus (WSSV), Gryllus bimaculatus nudivirus (GbNV), Oryctes rhinoceros nudivirus (OrNV), Heliothis zea nudivirus 1 (HzNV-1), and Nilaparvata lugens endogenous nudivirus (NlENV). WSSV was used to root the tree.
As shown in Fig. 4, the baculoviruses were monophyletic, and bracoviruses were clustered with nudiviruses. NlENV was more closely related to OrNV than it was to GbNV, which was in accordance with the BLAST results. A previous study (17) suggested the paraphyly of the nudivirus clade. Similar to that, here HzNV-1 was grouped with the bracoviruses rather than the branch containing NlENV, OrNV, and GbNV. A phylogenetic tree of the PIF genes is not sufficient to clarify the exact phylogeny of nudiviruses. However, these results raised the possibility that the lineage of nudivirus and its relationship to bracovirus are more complicated than had been estimated.
Including NlENV, the nudivirus group contains seven viruses. The other two nudiviruses, PmNV and DiNV, were not included in analysis because their complete genome data were not available at this time. Shrimp is the host of PmNV, and other viruses infect insects of the Orthoptera, Coleoptera, Lepidoptera, Diptera, and Hemiptera. It has been suggested that the phylogeny of baculoviruses and nudiviruses does not reflect that of their insect hosts and that the host ecology rather than phylogeny was the main driving force of virus evolution (23, 37). Interestingly, the hosts of bracoviruses are wasp species that parasitize lepidopteran larvae, while HzNV infects lepidopteran insects or lepidopterous cell lines. However, N. lugens and O. rhinoceros, the hosts of NlENV and OrNV, respectively, were not closely related either evolutionarily or ecologically. NlENV is the first identified nudivirus sequences from a hemipteran insect. The great diversity and wide host range in arthropods indicate that nudiviruses are very ancient DNA viruses. In a recent study of the phylogeny of large dsDNA viruses, the baculoviruses and nudiviruses were rooted at 310 million years ago (Mya), much older than any other viruses (17).
Integrated or free virus?
qRT-PCR indicated that BPH harbored approximately 2 viral genome copies per cell (data not shown). However, electron microscopy of ultrathin sections of different insect tissues did not reveal any rod-shaped virions. As the genome sequencing data suggested, these NlENV sequences were integrated into the chromosomes of their host. The lengths of BPH scaffolds or contigs containing nudivirus sequences ranged from 1,006 bp to 919,474 bp. Some of them contained many host genes, which were located adjacent to the NlENV genes (Fig. 1). In the fosmid library containing large DNA fragments of the BPH genome (up to 40 kbp), these NlENV ORFs were also located in the same fosmids with predicted BPH cellular genes, which reinforced the integration of the sequences. Furthermore, PCR primers were designed to amplify the junctional regions (Fig. 1) between virus and host genes, and the results were in accordance with the scaffold sequences.
The symbiosis between wasps and the nudivirus ancestor was thought to be the origin of bracoviruses (38). Over the course of evolution, the integrated symbiont genome was reduced and fragmented and became a complex example of endogenous viral elements (EVEs) (39). More recently, HzNV-1 was found to integrate its genome into host chromosomes during the infection process (40). There were differences between HzNV-1, a “living” virus, and bracovirus, which was more like a wasp organelle system. Since the NlENV sequence was detected in a wide range of BPH geographic populations, we hypothesized that the integration occurred in the early speciation or had provided some competitive advantages to the host insect. As far as we know, planthoppers carrying NlENV sequences are asymptomatic, and there has not yet been any evidence that the virus has a free stage. More likely, these sequences are ancient viral relics, representing a second group of EVEs derived from nudiviruses. In the case of braconid wasps, nudiviruses were domesticated during coevolution and became gene delivery vectors. However, for the NlENV sequences, their origin and function remain mysteries.
In conclusion, the biological characteristics of NlENV remain largely unknown, and further studies are required to clarify the lineage and evolution of nudiviruses, bracoviruses, baculoviruses, and other large DNA viruses. The accumulated knowledge will also provide insight into the complicated connections between insect hosts and their symbionts.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Basic Research Program of China (2010CB126205), the National Natural Science Foundation of China (31070136), and the Education of Zhejiang Province, project no. Y201225018.
Footnotes
Published ahead of print 26 February 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.03166-13.
REFERENCES
- 1.Hibino H. 1996. Biology and epidemiology of rice viruses. Annu. Rev. Phytopathol. 34:249–274. 10.1146/annurev.phyto.34.1.249 [DOI] [PubMed] [Google Scholar]
- 2.Noda H, Ishikawa K, Hibino H, Omura T. 1991. A reovirus in the brown planthopper, Nilaparvata lugens. J. Gen. Virol. 72:2425–2430. 10.1099/0022-1317-72-10-2425 [DOI] [PubMed] [Google Scholar]
- 3.Nakashima N, Kawahara N, Omura T, Noda H. 2006. Characterization of a novel satellite virus and a strain of Himetobi P virus (Dicistroviridae) from the brown planthopper, Nilaparvata lugens. J. Invertebr. Pathol. 91:53–56. 10.1016/j.jip.2005.10.001 [DOI] [PubMed] [Google Scholar]
- 4.Murakami R, Suetsugu Y, Kobayashi T, Nakashima N. 2013. The genome sequence and transmission of an iflavirus from the brown planthopper, Nilaparvata lugens. Virus Res. 176:179–187. 10.1016/j.virusres.2013.06.005 [DOI] [PubMed] [Google Scholar]
- 5.Bao Y, Qu L, Zhao D, Chen L, Jin H, Xu L, Cheng J, Zhang C. 2013. The genome-and transcriptome-wide analysis of innate immunity in the brown planthopper, Nilaparvata lugens. BMC Genomics 14:160. 10.1186/1471-2164-14-160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mayo MA, Christian PD, Hillmann BI, Brunt AA, Desselberger U. 1995. Unassigned viruses, p 504–507 In Murphy FA, Fauquet CM, Bishop DH, Ghabrial SA, Jarvis AW, Martelli GP, Mayo MA, Summers MD. (ed), Virus taxonomy: classification and nomenclature of viruses. Sixth report of the International Committee on Taxonomy of Viruses. Springer-Verlag, Vienna, Austria [Google Scholar]
- 7.Huger AM, Krieg A. 1991. Baculoviridae. Nonoccluded baculoviruses, p 287–319 In Adams JR, Bonami JR. (ed), Atlas of invertebrate viruses. CRC Press, Boca Raton, FL [Google Scholar]
- 8.Wang Y, Burand JP, Jehle JA. 2007. Nudivirus genomics: diversity and classification. Virol. Sinica 22:128–136. 10.1007/s12250-007-0014-3 [DOI] [Google Scholar]
- 9.Huger AM. 1985. A new virus disease of crickets (Orthoptera:Gryllidae) causing macronucleosis of fatbody. J. Invertebr. Pathol. 45:108–111. 10.1016/0022-2011(85)90055-2 [DOI] [Google Scholar]
- 10.Huger AM. 1966. A virus disease of the Indian rhinoceros beetle, Oryctes rhinoceros(linnaeus), caused by a new type of insect virus, Rhabdionvirus oryctes gen. n., sp. n. J. Invertebr. Pathol. 8:38–51. 10.1016/0022-2011(66)90101-7 [DOI] [PubMed] [Google Scholar]
- 11.Granados RR, Nguyen T, Cato B. 1978. An insect cell line persistently infected with a baculovirus-like particle. Intervirology 10:309–317. 10.1159/000148993 [DOI] [PubMed] [Google Scholar]
- 12.Herzog GA, Phillips JR. 1982. Manifestation of an abnormal reproductive system in a laboratory strain of the bollworm Heliothis zea. J. Georgia Entomol. Soc. 17:506–513 [Google Scholar]
- 13.Raina AK, Adams JR. 1995. Gonad-specific virus of corn-earworm. Nature 374:770. 10.1038/374770a0 [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Jehle JA. 2009. Nudiviruses and other large, double-stranded circular DNA viruses of invertebrates: new insights on an old topic. J. Invertebr. Pathol. 101:187–193. 10.1016/j.jip.2009.03.013 [DOI] [PubMed] [Google Scholar]
- 15.Unckless RL. 2011. A DNA virus of Drosophila. PLoS One 6:e26564. 10.1371/journal.pone.0026564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bézier A, Annaheim M, Herbinière J, Wetterwald C, Gyapay G, Bernard-Samain S, Wincker P, Roditi I, Heller M, Belghazi M. 2009. Polydnaviruses of braconid wasps derive from an ancestral nudivirus. Science 323:926–930. 10.1126/science.1166788 [DOI] [PubMed] [Google Scholar]
- 17.Thézé J, Bézier A, Periquet G, Drezen J, Herniou EA. 2011. Paleozoic origin of insect large dsDNA viruses. Proc. Natl. Acad. Sci. U. S. A. 108:15931–15935. 10.1073/pnas.1105580108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 19.Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27:1164–1165. 10.1093/bioinformatics/btr088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61:539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔCT) method. Methods 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 22.Cheng C, Liu S, Chow T, Hsiao Y, Wang D, Huang J, Chen H. 2002. Analysis of the complete genome sequence of the Hz-1 virus suggests that it is related to members of the Baculoviridae. J. Virol. 76:9024–9034. 10.1128/JVI.76.18.9024-9034.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Kleespies RG, Huger AM, Jehle JA. 2007. The genome of Gryllus bimaculatus nudivirus indicates an ancient diversification of baculovirus-related nonoccluded nudiviruses of insects. J. Virol. 81:5395–5406. 10.1128/JVI.02781-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Kleespies RG, Ramle MB, Jehle JA. 2008. Sequencing of the large dsDNA genome of Oryctes rhinoceros nudivirus using multiple displacement amplification of nanogram amounts of virus DNA. J. Virol. Methods 152:106–108. 10.1016/j.jviromet.2008.06.003 [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Bininda-Emonds OR, van Oers MM, Vlak JM, Jehle JA. 2011. The genome of Oryctes rhinoceros nudivirus provides novel insight into the evolution of nuclear arthropod-specific large circular double-stranded DNA viruses. Virus Genes 42:444–456. 10.1007/s11262-011-0589-5 [DOI] [PubMed] [Google Scholar]
- 26.Burand J, Kim W, Afonso CL, Tulman ER, Kutish GF, Lu Z, Rock DL. 2012. Analysis of the genome of the sexually transmitted insect virus Helicoverpa zea nudivirus 2. Viruses 4:28–61. 10.3390/v4010028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Bininda-Emonds OR, Jehle JA. 2012. Nudivirus genomics and phylogeny, p 33–52 In Garcia M. (ed), Viral genomes—molecular structure, diversity, gene expression mechanisms and host-virus interactions. InTech, Rijeka, Croatia [Google Scholar]
- 28.Rohrmann GF. 2011. Baculovirus molecular biology, 2nd ed. National Center for Biotechnology Information, Bethesda, MD: [PubMed] [Google Scholar]
- 29.Garavaglia MJ, Miele SAB, Iserte JA, Belaich MN, Ghiringhelli PD. 2012. The ac53, ac78, ac101, and ac103 genes are newly discovered core genes in the family Baculoviridae. J. Virol. 86:12069–12079. 10.1128/JVI.01873-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xue J, Bao Y, Li B, Cheng Y, Peng Z, Liu H, Xu H, Zhu Z, Lou Y, Cheng J. 2010. Transcriptome analysis of the brown planthopper Nilaparvata lugens. PLoS One 5:e14233. 10.1371/journal.pone.0014233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herniou EA, Olszewski JA, Cory JS, O'Reilly DR. 2003. The genome sequence and evolution of baculoviruses. Annu. Rev. Entomol. 48:211–234. 10.1146/annurev.ento.48.091801.112756 [DOI] [PubMed] [Google Scholar]
- 32.McCarthy CB, Theilmann DA. 2008. AcMNPV ac143(odv-e18) is essential for mediating budded virus production and is the 30th baculovirus core gene. Virology 375:277–291. 10.1016/j.virol.2008.01.039 [DOI] [PubMed] [Google Scholar]
- 33.Yuan M, Huang Z, Wei D, Hu Z, Yang K, Pang Y. 2011. Identification of Autographa californica nucleopolyhedrovirus ac93 as a core gene and its requirement for intranuclear microvesicle formation and nuclear egress of nucleocapsids. J. Virol. 85:11664–11674. 10.1128/JVI.05275-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abd-Alla AM, Cousserans F, Parker AG, Jehle JA, Parker NJ, Vlak JM, Robinson AS, Bergoin M. 2008. Genome analysis of a Glossina pallidipes salivary gland hypertrophy virus reveals a novel, large, double-stranded circular DNA virus. J. Virol. 82:4595–4611. 10.1128/JVI.02588-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jehle JA, Abd-Alla AM, Wang Y. 2013. Phylogeny and Evolution of Hytrosaviridae. J. Invertebr. Pathol. 112(Suppl 1):S62–S67. 10.1016/j.jip.2012.07.015 [DOI] [PubMed] [Google Scholar]
- 36.Bézier A, Herbinière J, Lanzrein B, Drezen J. 2009. Polydnavirus hidden face: the genes producing virus particles of parasitic wasps. J. Invertebr Pathol. 101:194–203. 10.1016/j.jip.2009.04.006 [DOI] [PubMed] [Google Scholar]
- 37.Herniou EA, Olszewski JA, O'Reilly DR, Cory JS. 2004. Ancient coevolution of baculoviruses and their insect hosts. J. Virol. 78:3244–3251. 10.1128/JVI.78.7.3244-3251.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Federici BA, Bigot Y. 2003. Origin and evolution of polydnaviruses by symbiogenesis of insect DNA viruses in endoparasitic wasps. J. Insect Physiol. 49:419–432. 10.1016/S0022-1910(03)00059-3 [DOI] [PubMed] [Google Scholar]
- 39.Herniou EA, Huguet E, Thézé J, Bézier A, Periquet G, Drezen J. 2013. When parasitic wasps hijacked viruses: genomic and functional evolution of polydnaviruses. Philos. Trans. R. Soc. B Biol. Sci. 368:20130051. 10.1098/rstb.2013.0051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin C, Lee J, Chen S, Wood HA, Li M, Li C, Chao Y. 1999. Persistent Hz-1 virus infection in insect cells: evidence for insertion of viral DNA into host chromosomes and viral infection in a latent status. J. Virol. 73:128–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.