ABSTRACT
During HIV-1 assembly, Gag polypeptides target to the plasma membrane, where they multimerize to form immature capsids that undergo budding and maturation. Previous mutational analyses identified residues within the Gag matrix (MA) and capsid (CA) domains that are required for immature capsid assembly, and structural studies showed that these residues are clustered on four exposed surfaces in Gag. Exactly when and where the three critical surfaces in CA function during assembly are not known. Here, we analyzed how mutations in these four critical surfaces affect the formation and stability of assembly intermediates in cells expressing the HIV-1 provirus. The resulting temporospatial map reveals that critical MA residues act during membrane targeting, residues in the C-terminal CA subdomain (CA-CTD) dimer interface are needed for the stability of the first membrane-bound assembly intermediate, CA-CTD base residues are necessary for progression past the first membrane-bound intermediate, and residues in the N-terminal CA subdomain (CA-NTD) stabilize the last membrane-bound intermediate. Importantly, we found that all four critical surfaces act while Gag is associated with the cellular facilitators of assembly ABCE1 and DDX6. When correlated with existing structural data, our findings suggest the following model: Gag dimerizes via the CA-CTD dimer interface just before or during membrane targeting, individual CA-CTD hexamers form soon after membrane targeting, and the CA-NTD hexameric lattice forms just prior to capsid release. This model adds an important new dimension to current structural models by proposing the potential order in which key contacts within the immature capsid lattice are made during assembly in cells.
IMPORTANCE While much is known about the structure of the completed HIV-1 immature capsid and domains of its component Gag proteins, less is known about the sequence of events leading to formation of the HIV-1 immature capsid. Here we used biochemical and ultrastructural analyses to generate a temporospatial map showing the precise order in which four critical surfaces in Gag act during immature capsid formation in provirus-expressing cells. Because three of these surfaces make important contacts in the hexameric lattices that are found in the completed immature capsid, these data allow us to propose a model for the sequence of events leading to formation of the hexameric lattices. By providing a dynamic view of when and where critical Gag-Gag contacts form during the assembly process and how those contacts function in the nascent capsid, our study provides novel insights into how an immature capsid is built in infected cells.
INTRODUCTION
The HIV-1 immature capsid (also called the immature lattice) is the spherical protein shell found in the immature virus and is composed of 1,500 to 5,000 copies of the HIV-1 Gag polyprotein (1), which encapsidate the viral genome. Gag, the only viral protein needed for assembly of the HIV-1 immature capsid, undergoes translation, myristoylation, and oligomerization in the cytoplasm of the infected host cell (reviewed in references 2 and 3). Gag oligomers likely associate with HIV-1 genomic RNA (gRNA) in the cytoplasm (4). Subsequently, following a conformational change that exposes the myristate at the N terminus of Gag, Gag oligomers target to the plasma membrane (PM), where they multimerize further to form spherical immature capsids that undergo budding and release (reviewed in references 3 and 5). The final step of particle production involves maturation, in which Gag is cleaved by the HIV-1 protease into its four constituent domains (matrix [MA], capsid [CA], nucleocapsid [NC], and p6) as well as two spacer peptides (sp1 and sp2). Cleavage results in a rearrangement to form the infectious mature viral capsid.
Mutational analyses have identified the Gag residues that are critical for virus replication (reviewed in reference 6). Because Gag or its cleavage products function at multiple stages of the life cycle (e.g., assembly, maturation, and postentry), only a subset of these residues is likely to act during immature capsid assembly. When residues critical for immature capsid assembly are mutated, Gag expression is largely unaffected but virus particle production is significantly reduced; additionally, electron microscopy (EM) reveals structures that are consistent with incomplete assembly. Mutational analyses have shown that residues critical for immature capsid assembly are located in regions of MA, CA, and NC but not in the p6 domain, which is important for HIV-1 budding (reviewed in reference 7). Among the three domains important for assembly, we focused here on the first two because high-resolution three-dimensional structures are available for MA, as well as for the N-terminal and C-terminal subdomains of CA (CA-NTD and CA-CTD, respectively).
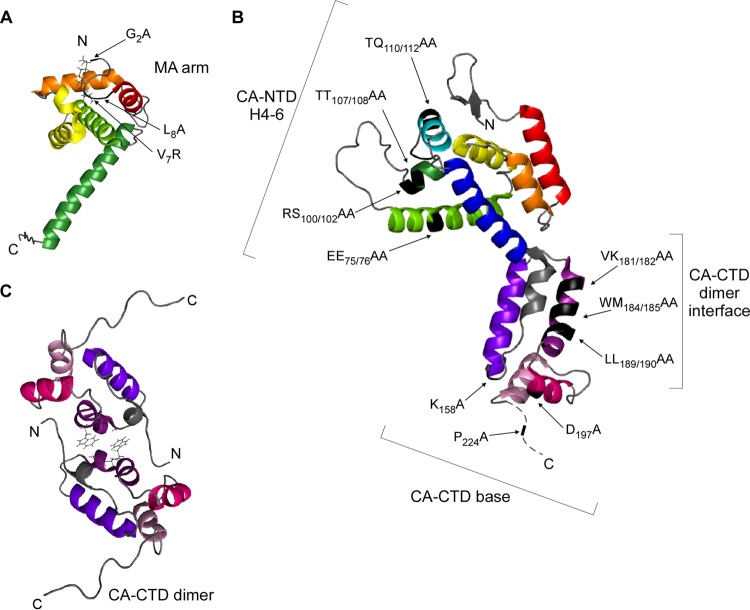
Notably, many of the residues required for proper assembly of HIV-1 immature capsids (8–18) are located on four surfaces that are exposed in three-dimensional structures of MA, CA-NTD, and CA-CTD (Fig. 1A to C; Table 1). The first of these surfaces is located on a flexible arm at the extreme N terminus of MA and governs early events in targeting of Gag to membranes. This surface, termed the MA arm, includes residues required for Gag myristoylation as well as adjacent residues that govern phosphatidylinositol-4,5-bisphosphate-dependent exposure of the sequestered myristate (e.g., see references 8, 10, 12, 15 to 17, and 19 to 22) (Fig. 1A). The other three exposed surfaces involve CA residues that likely play a role in Gag-Gag interactions. One of these CA surfaces is located in CA-NTD, and the other two are in CA-CTD (Fig. 1B and C). In HIV-1 CA-NTD—an arrow-shaped domain composed of an N-terminal β hairpin, seven α helices, and an extended loop (23, 24)—the residues critical for immature capsid assembly are located on a contiguous surface formed by helices 4, 5, and 6 (18) (Fig. 1B), here termed the CA-NTD H4-6 surface. CA-NTD is connected by a flexible linker to CA-CTD, a globular domain consisting of a sheet-turn-helix motif called the major homology region (MHR), whose primary amino acid sequence is conserved among orthoretroviruses, followed by four α helices (helices 8 to 11) (11, 25). Recombinant CA-CTD forms a dimer in solution (11, 26–28), and most high-resolution X-ray structures of wild-type (WT) CA-CTD support a side-by-side model of CA-CTD dimerization with residues in helix 9 forming the dimer interface (11, 25, 27) (Fig. 1C). One set of residues in CA-CTD that is critical for immature capsid assembly maps to the dimer interface in helix 9 (9, 11, 13, 14, 18) (Fig. 1B and C), while a second set of critical residues in CA-CTD maps to an exposed surface at the crystallographic base of CA-CTD (18) and is therefore termed the CA-CTD base surface (Fig. 1B). The CA-CTD base includes residues in the MHR as well as residues within helices 10 and 11 (18). Moreover, because similar phenotypes are observed upon mutation of residues downstream of helix 11 and in the sp1 domain, others have proposed that residues from the CA-CTD–sp1 junction also interact with the CA-CTD base (e.g., see references 18 and 29 to 31). While CA-CTD residues downstream of helix 11 form a disordered tail in CA-CTD crystal structures (11), studies suggest that in full-length Gag this region forms a continuous α helix that spans the CA-CTD–sp1 junction (here referred to as helix 12) and is lost upon Gag cleavage (31–35). Not surprisingly, it is thought that these four exposed surfaces containing residues required for assembly—the MA arm, CA-NTD H4-6, the CA-CTD dimer interface, and the CA-CTD base—play critical roles during immature capsid assembly.
FIG 1.
Critical residues for immature capsid assembly map to four distinct surfaces within HIV-1 Gag. (A) Ribbon diagram of myristoylated MA (PDB accession number 1UPH). The locations of three MA arm mutations analyzed in this study are shown in black (arrows). Helices are colored as follows: helix 1, red; helix 2, orange; helix 3, yellow; helix 4 light green; helix 5, dark green. (B) Ribbon diagram of full-length CA (PDB accession number 3NTE) based on crystal structures of the individual CA-NTD and CA-CTD domains. The locations of the four CA-NTD H4-6 mutations, three CA-CTD dimer mutations, and three CA-CTD base mutations analyzed in this study are shown in black (arrows). Helices are colored as follows: helix 1, red; helix 2, orange; helix 3, yellow; helix 4, light green; helix 5, dark green; helix 6, cyan; helix 7, dark blue; sheet-turn-helix, gray; helix 8, violet; helix 9, purple; helix 10, magenta; helix 11, pink. (C) Ribbon diagram of CA-CTD (PDB accession number 2KOD) shown as a dimer, with side chains for WM184/185 within helix 9 depicted. Helices are colored as described for panel C.
TABLE 1.
Mutants analyzed
| Group/surface of Gag | Construct namea | Location of residues |
|---|---|---|
| Controlsb | WT Gag | NA |
| Gag MACA | Δsp1-NC-sp2-p6 | |
| MA arm | G2A | Extreme N terminus of MA |
| V7R | Extreme N terminus of MA | |
| L8A | Extreme N terminus of MA | |
| CA-NTD H4-6 | EE75/76AA | CA helix 4 |
| RS100/102AA | In or near CA helix 5 | |
| TT107/108AA | Between CA helices 5 and 6 | |
| TQ110/112AA | In or near CA helix 6 | |
| CA-CTD dimer | VK181/182AA | CA-CTD dimer interface (helix 9) |
| WM184/185AA | CA-CTD dimer interface (helix 9) | |
| LL189/190AA | CA-CTD dimer interface (helix 9) | |
| CA-CTD base | K158A | MHR |
| D197A | CA helix 10 | |
| P224A | Preceding CA helix 12 |
Constructs were named according to the matrix or capsid amino acid number(s) that was mutated.
WT Gag and Gag MACA were used as assembly-competent and assembly-incompetent controls, respectively.
Given that the immature capsid is composed of the full-length Gag polyprotein, a three-dimensional structure of full-length Gag would provide important insights into immature capsid assembly that cannot be obtained from structures of individual domains. Unfortunately, such a structure has not been generated because Gag contains the disordered p6 domain as well as flexible linkers between its structured domains (29). However, low-resolution structures of fully assembled immature retroviruses and capsid-like particles (31, 36, 37) have revealed that the immature retroviral capsid is composed of a curved and flexible hexameric protein lattice in which extended Gag polyproteins are arranged radially in a beads-on-a-string configuration, with their N termini tethered to the membrane by the MA domain and the C terminus pointing to the virion's center (reviewed in reference 38). Moreover, a recent high-resolution (8-Å) structure of Mason-Pfizer monkey virus (MPMV) CA-NC assembled in vitro has allowed identification of residues that form inter- and intrahexameric contacts in the immature capsid (29).
While such high-resolution structural studies are leading to a relatively detailed picture of critical Gag contacts present in the completed immature capsid, much less is known about when and where these Gag-Gag interfaces are formed during capsid assembly in cells. The observation, by many groups, that assembly is inhibited by mutations in the four critical surfaces described above implies that these surfaces function at specific points during the assembly process. It is well accepted that the MA arm functions in membrane targeting; in contrast, the function of exposed surfaces in CA likely involves interactions with other exposed protein surfaces, for example, in neighboring Gag proteins (reviewed in reference 3). With respect to timing, membrane flotation and morphological analyses of MA and CA mutants have revealed that some exposed surfaces act while Gag is in the cytoplasm and others act after PM targeting (13, 15, 17, 18, 39); however, the sequential order in which these surfaces act is not known. Our goal here was to generate a temporospatial map that defines the exact order in which critical surfaces function during the process of immature capsid assembly in cells and identifies the specific Gag-containing complexes in which these surfaces act and their subcellular locations.
To generate this temporospatial map, we examined how mutations of 13 residues, located on four critical surfaces in MA and CA, impact progression through a previously described host-catalyzed capsid assembly pathway in cells. During assembly in cells, Gag forms four complexes, designated by their sedimentation values (∼10S, ∼80S, ∼150S, and ∼500S). These complexes are true assembly intermediates, since they appear in a sequential progression in pulse-chase analyses, with newly synthesized Gag first forming the ∼10S complex, followed by the ∼80S intermediate, ∼150S intermediate, and ∼500S intermediate, before culminating in the ∼750S completely assembled HIV-1 immature capsid (40, 41). The order of intermediates in the assembly pathway initially indicated by pulse-chase experiments has been confirmed through the analysis of assembly-defective mutants (40, 42–44). In these studies, cells expressing an assembly-defective Gag mutant accumulate only those intermediates that precede the step at which assembly is blocked. The ∼80S, ∼150S, and ∼500S assembly intermediates (termed the high-molecular-mass intermediates) contain Gag as well as cellular proteins derived from a host precursor complex, at least two of which—the ATP-binding cassette protein E1 (ABCE1) and the DEAD box RNA helicase DDX6—facilitate immature capsid assembly (40, 42, 43, 45, 46). Formation of the ∼80S intermediate, in which Gag first associates with ABCE1 and DDX6, occurs when Gag co-opts the cellular precursor complex that contains these and other host proteins (43). When assembly of the immature capsid is completed, ABCE1, DDX6, and other cellular proteins from the precursor complex dissociate from the immature capsid (40, 43, 46).
In the current study, we first analyzed the subcellular location (cytosol versus membrane) of each of the assembly intermediates, thereby adding a spatial dimension to the temporally ordered immature capsid assembly pathway. We also demonstrated that the high-molecular-mass intermediates contain HIV-1 genomic RNA, as would be expected. Next, by mutating residues within MA and CA that are critical for assembly, we defined the temporospatial order in which these key residues are required in the assembly pathway. Additionally, we used biochemical and ultrastructural approaches to show that each of the critical residues acts while Gag is associated with ABCE1 and DDX6. Finally, we integrated our results with those of previous structural analyses to generate a model showing the temporal and spatial order in which Gag-Gag interfaces may be formed during assembly in cells. Thus, our study bridges the gap between structural data generated from recombinant proteins assembled by mass action in vitro and events of assembly that occur in a facilitated manner in cells, and it provides insights into the assembly process that should spur the development of compounds that inhibit capsid assembly.
MATERIALS AND METHODS
Cells and proviral constructs.
The H9-HIV cell line stably expresses the HIV-1 provirus encoding deletions in env and vif, a frameshift in vpr, and replacement of nef with a puromycin resistance gene and has been described previously (47). H9-HIV cells were maintained under puromycin selection in RPMI (Life Technologies, Carlsbad, CA) with 10% fetal bovine serum (FBS). COS-1 and 293T cells were maintained in Dulbecco modified Eagle medium (Life Technologies) with 10% FBS. All plasmids utilized in this study were generated via site-directed mutagenesis in HIV-1 provirus strain LAI with a deletion in env (HIV-1ΔEnv) (48). Some mutant proviruses also encoded an inactive protease (HIV-1ΔEnv, Pro−), where indicated, as described previously (40). The sequences of the oligonucleotides used to generate each mutation are available upon request.
Transfection, immunoprecipitations, and WB assays.
Cells were transfected with 1 to 5 μg DNA using polyethylenimine (Polysciences, Warrington, PA). Cell lysates were harvested in the presence of freshly prepared protease inhibitor cocktail (Sigma, St. Louis, MO) and RNaseOUT recombinant ribonuclease inhibitor (Invitrogen) either in 1× Triton-X buffer (20 mM HEPES, pH 7.9, 10 mM NaCl, 10 mM EDTA, pH 8.0, 0.35% Triton X-100) followed by immunoprecipitation with antibody to ABCE1 or in 1× NP-40 buffer with EDTA (10 mM Tris acetate, pH 7.4, 50 mM KCl, 100 mM NaCl, 0.625% NP-40, 10 mM EDTA) followed by immunoprecipitation with antibody to DDX6 (Bethyl, Montgomery, TX) using protein G-coupled Dynabeads (Life Technologies), as described previously (43). Immunoprecipitation eluates were analyzed by SDS-PAGE, followed by Western blotting (WB) using a murine monoclonal antibody directed against HIV-1 Gag p24 (HIV-1 p24 hybridoma [183-H12-5C] obtained from Bruce Chesebro through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH) and horseradish peroxidase (HRP)-conjugated anti-mouse IgG1 (Santa Cruz, Dallas, TX). Where indicated, immunoprecipitation eluates were also probed with antibody to ABCE1 (46), followed by HRP-tagged protein A (Thermo Fisher Scientific, Rockford, IL), and detected using Pierce enhanced chemiluminescence substrate (Thermo Fisher Scientific). For detection of Gag in total cell lysates, velocity sedimentation fractions, and membrane flotation fractions, WB assays were performed as described above or by using goat anti-mouse IgG-IRDye 800CW (LI-COR, Lincoln, NE). Quantification of Gag was performed using a LI-COR Odyssey system and LI-COR software or ImageJ software.
Analysis of VLP production.
Cells and supernatants were collected separately at 30 h posttransfection. Cell lysates were harvested in 1× NP-40 buffer plus EDTA with freshly prepared protease inhibitor cocktail (Sigma) and RNaseOUT (Invitrogen). For collection of virus-like particles (VLPs), supernatants were centrifuged at 1,000 rpm (225 × g) for 10 min at 4°C, filtered (pore size, 0.45 μm) to remove remaining cells, and purified through a 30% sucrose cushion in an SW60Ti rotor (60,000 rpm, 30 min, 4°C), as described previously (42).
Velocity sedimentation.
Cells were harvested at 36 to 38 h posttransfection, and lysates were collected in 1× NP-40 buffer. Equivalent amounts of lysate were layered on a step gradient (10%, 15%, 40%, 50%, 60%, 70%, and 80% sucrose in lysis buffer) for each mutant and subjected to velocity sedimentation in a Beckman MLS50 rotor at 45,000 rpm (217,000 × g) for 45 min at 4°C, as described previously (42). Gradients were fractioned from top to bottom.
Membrane flotation.
Transfected cells were collected at 36 h posttransfection, and postnuclear supernatants were harvested in a 1-ml Dounce homogenizer in hypotonic lysis buffer (10 mM Tris acetate, pH 7.4, 50 mM KCl, 100 mM NaCl), followed by centrifugation at 1,000 rpm (225 × g) for 10 min at 4°C to remove nuclei. Postnuclear supernatants were mixed with 87% sucrose to a final concentration of ∼75% sucrose and used as the bottom layer of a 5-ml membrane flotation sucrose gradient, with 65% and 10% sucrose layered on top. Flotation was performed by centrifugation in a Beckman MLS50 rotor at 35,000 rpm (98,400 × g) for 4 h at 4°C. Twelve fractions were collected starting from the top of the gradient, along with an additional 13th fraction that consisted of any denatured protein in the pellet. For experiments involving subsequent sedimentation analyses of selected fractions, the membrane (M) fractions (fractions 3 to 4) or nonmembrane (NM) fractions (fractions 9 to 12) were immediately pooled. NM fractions were passed through a Zeba Desalt spin column (Thermo Scientific) to replace the sucrose with buffer. NP-40 was then added to a final concentration of 0.625% to both the M and NM fractions, and these were incubated with rotation for 30 min at 4°C. Subsequently, the M fraction was diluted to decrease the sucrose percentage in the sample to below 10%, and equivalent amounts (∼3%) of the M and NM fractions were loaded onto separate velocity sedimentation gradients, as described above.
RT-qPCR.
Total H9-HIV cell lysate containing 50 μg of cellular protein was analyzed via velocity sedimentation, after which pooled gradient fractions representing each assembly intermediate were collected and ∼16% of each pooled fraction was subjected to duplicate immunoprecipitations with antibody to ABCE1 or nonimmune rabbit IgG, as described above. Total cell lysate containing 20 μg of cellular protein was also subjected to immunoprecipitation without velocity sedimentation in parallel. One set of immunoprecipitations and inputs was used for WB, as described above. From the other set of immunoprecipitations and inputs, RNA was extracted using the TRIzol reagent (Invitrogen) and precipitated with isopropanol. Purified RNA was resuspended in 15 μl of RNase-free water. To generate cDNAs, reaction mixtures were programmed to contain 5 μl of RNA using random hexamers as primers and reverse transcribed using a SuperScript VILO cDNA synthesis kit (Invitrogen). For every sample, a negative control lacking master mix (reverse transcriptase negative) was analyzed in parallel (and typically contained <16 gRNA copies), and every experiment included a nontemplate control. cDNA obtained from immunoprecipitation samples and inputs was diluted 1:2, and cDNA from total cell lysates was diluted 1:10. HIV-1 genomic RNA was quantified via quantitative PCRs (qPCRs) performed in duplicate, and the reaction mixtures were programmed to contain 2 μl of diluted cDNA and the following oligonucleotides: forward primer 5′-AGAAGGCTGTAGACAAATACTGGG-3′ and reverse primer 5′-TGATGCACACAATAGAGGGTTG-3′. These primers amplified a 108-bp product between bp 162 and 270 within the N terminus of the Gag open reading frame in HIV-1 strain LAI. In parallel, a standard curve was generated using serial 10-fold dilutions of a Gag amplicon that was previously analyzed using picogreen to quantify the number of DNA copies per μl in the immunoprecipitation samples. qPCR products were detected with iQ SYBR green Supermix (Bio-Rad) and analyzed using a Bio-Rad MyiQ reverse transcription (RT)-PCR detection system and iQ5 software (Bio-Rad).
Preparation of virus for immunolabeling experiments.
Vesicular stomatitis virus G glycoprotein (VSV-G)-pseudotyped VLPs were generated by transfecting 293T cells with an HIV-1 provirus carrying the green fluorescent protein (GFP) in place of nef (43), and containing a deletion in env (HIV-1-GFPΔEnv), a second-generation Gag Pol-containing packaging vector (psPAX2), and VSV-G (pMD2.G) at a ratio of 3:2:1. The last two plasmids were obtained from Didier Trono (Swiss Institutes of Technology, Lausanne, Switzerland). At 6 h posttransfection, cells were washed three times with phosphate-buffered saline (PBS) and replaced with complete medium. To harvest virus, supernatants were collected at 48 h and 72 h posttransfection, pooled, centrifuged at 900 × g for 10 min at 4°C, and filtered (pore-size, 0.45 μm) to remove cells. Virus was pelleted by centrifugation at 50,000 × g for 90 min at 20°C, and infectivity was measured by determining the titers of the stocks on TZM-bl cells (from Tranzyme Inc. through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH). Infection of TZM-bl cells was facilitated by the addition 20 μg/ml DEAE-dextran and by spinoculation (2,500 × g, 2 h, 20°C).
Immunogold labeling, electron microscopy, and quantitation.
COS-1 cells were infected with VSV-G-pseudotyped virus at a multiplicity of infection of 40 to 50 using 20 μg/ml DEAE-dextran and spinoculation, as described above. Following incubation with virus for 12 to 15 h, cells were washed 3 times with medium to remove the virus inoculum. Infected cells were harvested at 36 h postinfection in fixative (3% paraformaldehyde, 0.025% glutaraldehyde in 0.1 M phosphate buffer, pH 7.4), pelleted, and subjected to high-pressure freezing using a Leica EMPACT2 system, followed by freeze substitution. Samples were infiltrated overnight with LR White embedding resin (London Resin Company Ltd., Reading, Berkshire, England) in ethanol, which was then changed to straight LR White; embedded in gelatin capsules (Electron Microscopy Sciences [EMS], Hatfield, PA); and cured overnight in a UV light cryochamber at 4°C. Sections (∼50 nm) were placed on grids, treated with 0.05 M glycine for 20 min at room temperature, rinsed in PBS, blocked for 45 min with 1% bovine serum albumin (BSA; EMS), and washed in PBS with 0.1% bovine serum albumin C (BSA-C; EMS). For immunogold labeling, peptide-specific antisera directed against ABCE1 (46) and DDX6 (43) were affinity purified, desalted, and concentrated to 1 to 2 mg/ml. Grids were double labeled with either ABCE1 or DDX6 and Gag. Labeling with anti-ABCE1 (0.7 mg/ml in 0.1% BSA-C with 0.001% Tween 20) or anti-DDX6 (0.3 mg/ml in 0.1% BSA-C with 0.005% Tween 20) was detected using goat anti-rabbit IgG conjugated to 15-nm gold particles (EMS). Grids were subsequently labeled with a murine monoclonal antibody directed against HIV-1 Gag p24 (hybridoma 183-H12-5C; 0.125 mg/ml in 0.1% BSA-C), which was detected using goat anti-mouse IgG conjugated to 6-nm gold particles (EMS). Fixation, staining, imaging with a JEOL-1400 transmission electron microscope, and image acquisition have been described previously (43). Each immunolabeling experiment included 2 sections from each of 5 groups (WT HIV-1, an HIV-1 provirus encoding Gag truncated after CA [Gag MACA], and the EE75/76AA, WM184/185AA, and P224A mutants), and each immunolabeling experiment was performed twice for both double-labeling conditions (ABCE1 and Gag versus DDX6 and Gag).
To quantify the stage of assembly at the PM for the WT and the mutants listed above, data from four independent immuno-EM experiments were analyzed (two involved ABCE1 and Gag colabeling, and two utilized DDX6 and Gag colabeling). The total number of images and Gag clusters analyzed is shown in Table 2. All Gag labeling in these images was categorized as either targeted Gag, an early site of assembling Gag, or a late site of assembling Gag. Targeted Gag was defined as no membrane deformation and no electron-dense staining at the membrane. An early site of assembly was defined as electron-dense staining and/or membrane deformation that formed a bud that was <50% complete. A late site of assembly was defined as membrane deformation that formed a bud that was >50% complete. Each of the four independent experiments was quantified separately, and the average ± standard error of the mean (SEM) of the four experiments was determined. After quantitation was completed, images that together represented the approximate distribution of targeted, early, and late sites quantified in Table 2 were chosen for Fig. 12 and 13.
TABLE 2.
Quantification of assembling Gag at the plasma membrane
| Construct | Description | No. of images analyzed | Total membrane length (mm) | Total no. of Gag clusters analyzeda | % (avg ± SEM)b |
||
|---|---|---|---|---|---|---|---|
| Targeted Gagc | Early assembly sitesd | Late assembly sitese | |||||
| WT Gag | Assembly competent | 306 | 678.3 | 932 | 58.4 ± 5.9 | 15.2 ± 2.7f | 26.4 ± 3.4f |
| Gag MACA | Assembly incompetent | 259 | 599.7 | 1,491 | 99.0 ± 0.4 | 0.8 ± 0.3 | 0.2 ± 0.1 |
| EE75/76AA | CA-NTD H4-6 | 317 | 735.1 | 1,384 | 77.2 ± 3.2 | 8.1 ± 2.5 | 14.7 ± 1.5f |
| WM185/185AA | CA-CTD dimer interface | 305 | 662.9 | 1,094 | 98.1 ± 0.8 | 1.2 ± 0.9 | 0.7 ± 0.1 |
| P224A | CA-CTD base | 331 | 764.6 | 1,621 | 93.8 ± 1.0 | 2.9 ± 1.0 | 3.2 ± 0.9 |
A Gag cluster is defined as three gold particles labeling Gag within a 100-nm diameter and within 100 nm of the PM.
Data were averaged across four independent labeling experiments.
Targeted Gag contains a Gag cluster without obvious curvature of the PM.
Early assembly sites contain a Gag cluster with a noticeably increased electron density and/or a ≤50% curvature of the PM.
Late assembly sites contain a Gag cluster with >50% curvature of the PM.
The percentage of early or late assembly sites differs significantly from that observed for Gag MACA (P ≤ 0.05).
FIG 12.
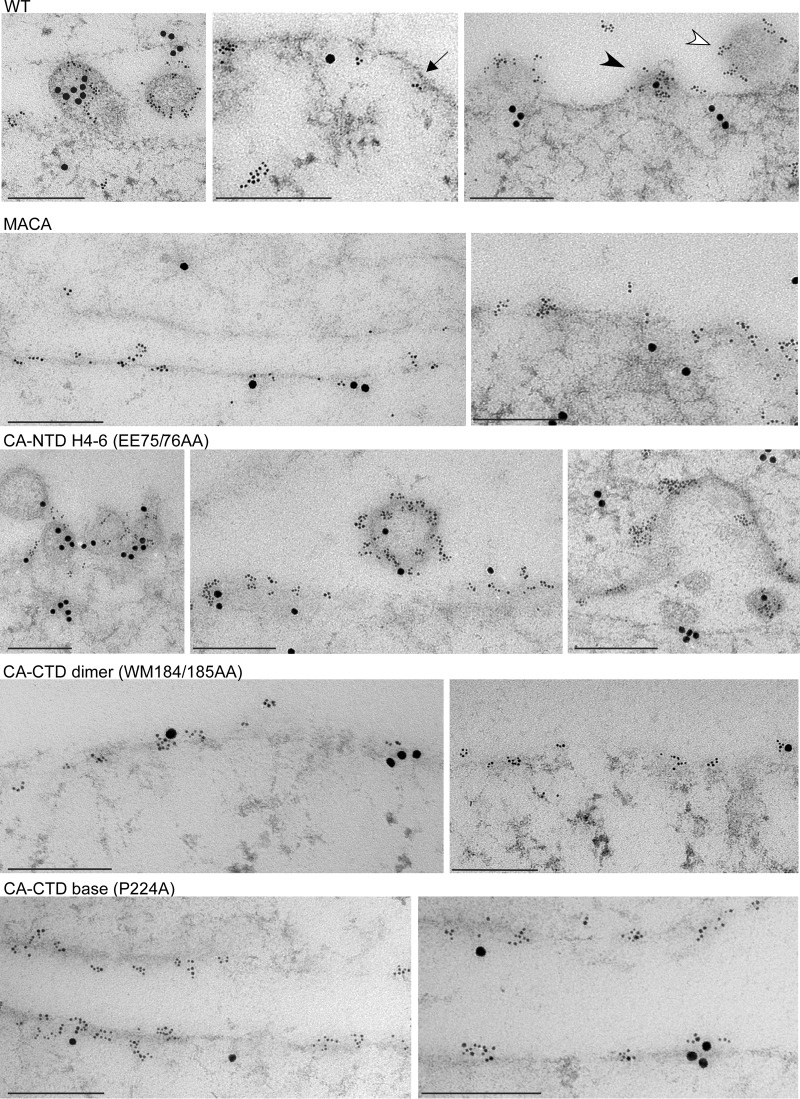
EM analysis of assembly-defective mutants in infected cells using immunolabeling for Gag and ABCE1. Infected COS-1 cells expressing WT Gag or mutations on the indicated critical surfaces were analyzed by immunogold double-label EM, using antibodies to Gag and ABCE1. Small gold particles represent HIV-1 Gag; large gold particles represent ABCE1. Images showing sites of labeling at the PM were chosen to correspond to the approximate distribution of targeted Gag, early assembly sites, and late assembly sites that were observed for each group in the quantitative analysis of EM images (Table 2). One example each of targeted Gag (dark arrow), an early assembly site (dark arrowhead), and a late assembly site (open arrowhead) is shown. Bars, 200 nm.
FIG 13.
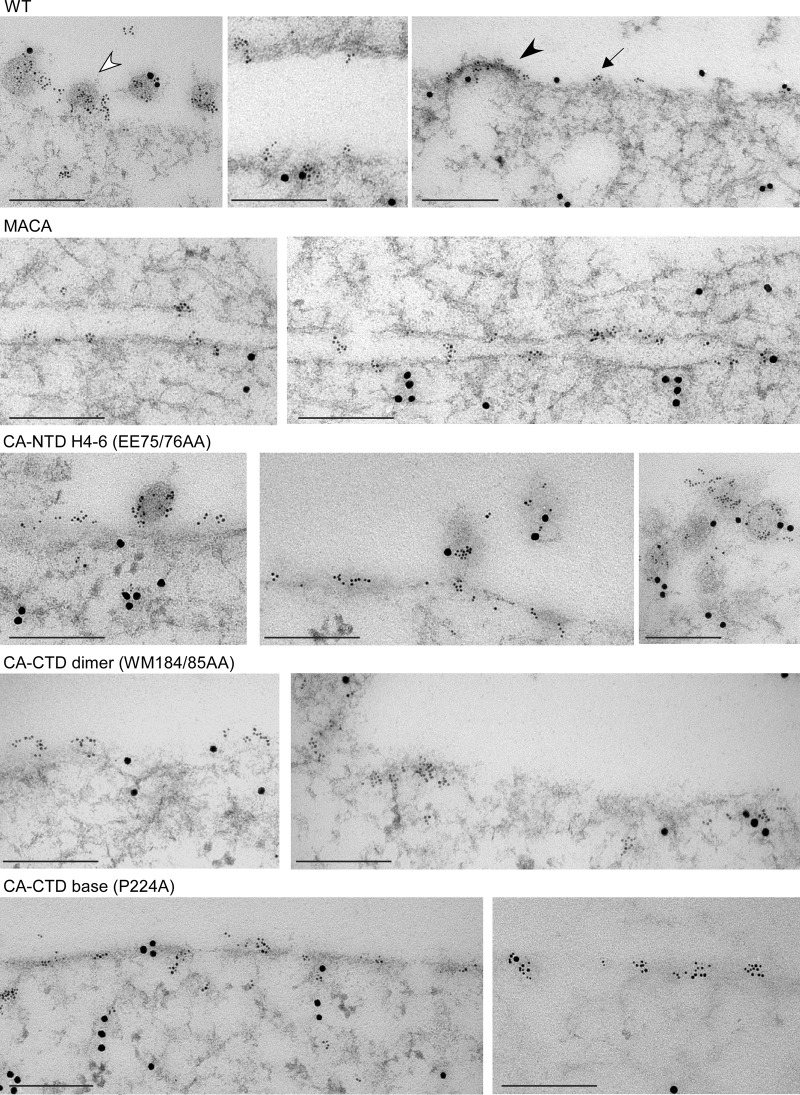
EM analysis of assembly-defective mutants in infected cells using immunolabeling for Gag and DDX6. Infected COS-1 cells expressing WT Gag or mutations on the indicated critical surfaces were analyzed by immunogold double-label EM, using antibodies to Gag and DDX6. Small gold particles represent HIV-1 Gag; large gold particles represent DDX6. Images showing sites of labeling at the PM were chosen to correspond to the approximate distribution of targeted Gag, early assembly sites, and late assembly sites that were observed for each group in the quantitative analysis of EM images (Table 2). One example each of targeted Gag (dark arrow), an early assembly site (dark arrowhead), and a late assembly site (open arrowhead) is shown. Bars, 200 nm.
RESULTS
Confirming the role of exposed surfaces of Gag in VLP release and membrane targeting.
To define the temporospatial order in which four exposed surfaces of Gag act during assembly, we analyzed how 13 mutations (of single residues or two nearby residues) on these surfaces affected the progression of Gag through the HIV-1 assembly pathway (Fig. 1; Table 1). Each of these 13 mutations is known to inhibit virus particle production with minimal effects on Gag expression in cells (8–18, 49); however, with the exception of the G2A mutation within the MA domain (40, 41, 44, 47) and the WM184/185AA mutation within the CA-CTD dimerization interface (42), the effect of these mutations on progression through the ABCE1 capsid assembly pathway had not been examined previously. These 13 mutations can be divided into four groups on the basis of the exposed surface of MA or CA in which they are located (Fig. 1; Table 1). Three of these mutations are located in the N terminus of MA (the MA arm); four mutations are in and around a contiguous surface in the CA-NTD domain formed by helices 4 to 6 (CA-NTD H4-6); three mutations are in helix 9 of CA-CTD, which forms the crystallographic CA dimer interface; and three mutations are dispersed in the CA-CTD sequence but form an exposed surface at the CA-CTD base.
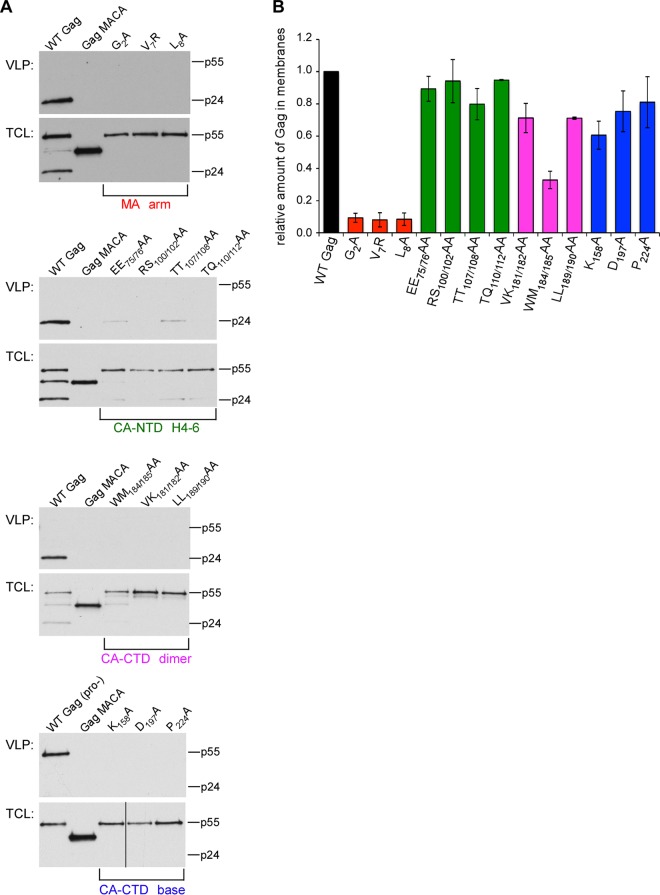
We engineered each Gag mutation into an HIV-1 provirus containing a deletion in env (HIV-1ΔEnv) (40) and verified that the viruses displayed an assembly defect by analyzing the release of virus-like particles (VLPs) following transfection into COS-1 and/or 293T cells. As a negative control, we transfected the same provirus encoding Gag truncated after CA (GagΔsp1-NC-sp2-p6; previously termed Gag p41; here referred to as Gag MACA), which fails to assemble or release because it lacks the entire NC and p6 domains (41, 42, 44, 47, 50–52). While others have analyzed the effects of either assembly-defective MA or CA mutations on VLP production and membrane targeting (8–18), here we compared all of these mutants to each other. As expected, we found that mutations in the MA arm, CA-CTD dimer interface, and CA-CTD base resulted in dramatic reductions in VLP production, even though steady-state Gag levels were unaffected (Fig. 2A). Mutations in the CA-NTD H4-6 group displayed a more modest reduction in particle production compared to that for WT Gag, suggesting that these mutations cause a less severe but still significant defect in assembly, consistent with previous observations (18, 49). Thus, our findings confirm conclusions by other groups that each of these 13 residues/residue pairs plays a major role in virus particle production (8–18, 49). Furthermore, because none of these 13 residues/residue pairs map to regions of Gag important for budding, it is likely that all of these residues function during immature capsid assembly.
FIG 2.
Role of exposed surfaces of Gag in VLP release and membrane targeting. COS-1 or 293T cells were transfected with a provirus encoding the indicated mutation, including a negative-control provirus containing an assembly-defective, truncated Gag (Gag MACA) and a positive-control provirus encoding WT Gag in either a protease-negative (Pro−) or protease-positive context. (A) Total cell lysates (TCL) and VLPs were harvested and analyzed by WB for Gag, with migrations of full-length Gag (p55) and CA (p24) indicated. Data in each panel are representative of those from three independent experiments. (B) In separate experiments, postnuclear supernatants were analyzed by membrane flotation, and fractions were analyzed by WB for Gag. The graph shows the amount of Gag in membrane fractions relative to that in the WT control, which was set to 1.0 in each experiment. Data represent the average ± SEM of three independent experiments.
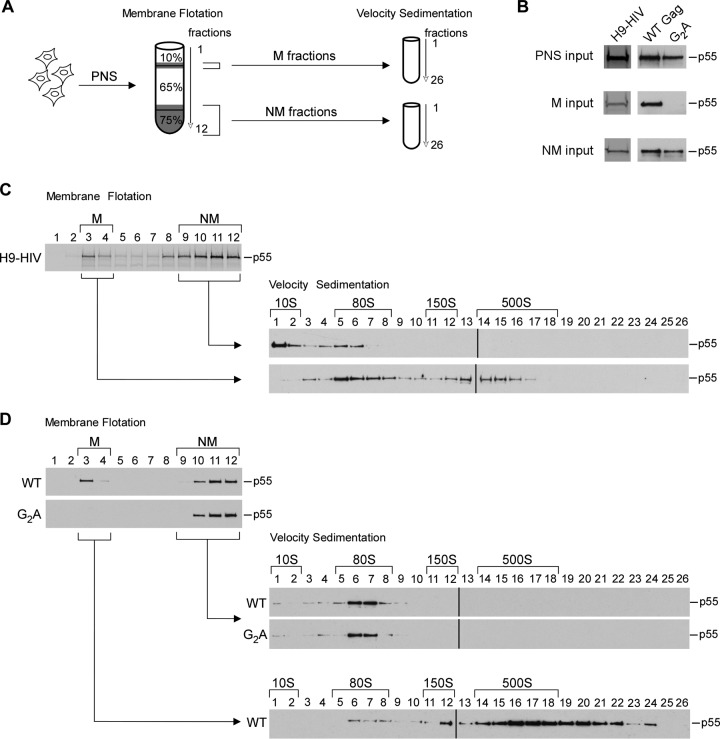
The three mutations located on the MA arm included G2A, which inhibits myristoylation (8, 12), as well as V7R and L8A, which inhibit myristate exposure (10, 15–17). Analysis of cell lysates by membrane flotation revealed that membrane targeting of MA mutants was reduced to less than 10% of that for WT Gag, while mutations within the CA-CTD dimer interface resulted in only modest reductions, and the CA-NTD and CA-CTD base mutations showed little effect on membrane targeting (Fig. 2B). Thus, these data demonstrate that only the MA mutants exhibit a dramatic defect on membrane targeting, which confirms previous findings (13–15, 17, 18).
Since we had previously defined the temporal progression of intermediates during immature capsid assembly (40, 41) but not their subcellular (spatial) localization, we next defined which of the assembly intermediates formed by WT Gag are cytoplasmic and which are localized to membranes. To date, we have found that HIV-1 Gag mutants that fail to target to membranes are arrested as either the ∼10S or ∼80S assembly intermediate, depending on the mutant, leading us to hypothesize that these are the only cytosolic assembly intermediates (42, 45, 47). To test this hypothesis, we performed membrane flotation on lysates of a chronically infected H9 T cell line expressing WT HIV-1ΔEnv (H9-HIV) and analyzed both the membrane and nonmembrane fractions by velocity sedimentation (Fig. 3A). We found that the nonmembrane fraction contained only the ∼10S and ∼80S assembly intermediates, while the membrane fraction contained the ∼80S and ∼500S assembly intermediates (Fig. 3B and C). When integrated with the previously defined temporal order of assembly intermediates (40, 41), these findings support a model in which assembly starts with the formation of the cytoplasmic ∼10S intermediate, which subsequently forms the ∼80S intermediate that takes Gag from the cytosol to membranes, where the ∼500S intermediate is formed. Note that the ∼150S assembly intermediate, which is highly transient and present in small quantities, was not consistently observed in these experiments.
FIG 3.
The ∼80S intermediate is found both in the cytosol and at membranes during immature capsid assembly. (A) Schematic of the experimental setup showing that transfected or infected cells were first subjected to hypotonic lysis. The postnuclear supernatants (PNS) from the lysates were analyzed by membrane flotation. Subsequently, the membrane (M) and nonmembrane (NM) fractions were each pooled and analyzed separately by velocity sedimentation. (B) Aliquots of gradient inputs from panels C and D analyzed by WB for Gag. (C) H9 T cells chronically infected with HIV-1 were treated with 1 μM indinavir for 48 h to inhibit proteolysis and then analyzed as described for panel A. Western blots for Gag are shown for fractions from the membrane flotation gradient and from the subsequent velocity sedimentation analysis of M and NM fractions. (D) COS-1 cells transfected with the indicated constructs were analyzed as described for panel A. Western blots for Gag are shown for fractions from the membrane flotation gradients and from the subsequent velocity sedimentation analyses of the M and NM fractions. Positions of assembly intermediates are indicated by brackets above the velocity sedimentation WB assays. Data are from one experiment that is representative of three independent experiments.
To verify this model, we also examined the assembly intermediates produced by the targeting-defective G2A Gag, an MA arm mutant (Fig. 3A, B, and D). For these experiments and others involving analysis of assembly intermediates following transfection, we used a cell type, COS-1, in which endocytosis of released virus is minimal, in order to prevent contamination of the intracellular Gag signal with Gag that is released and then internalized. When we analyzed transiently transfected COS-1 cells expressing WT Gag by membrane flotation followed by velocity sedimentation, we observed the same subcellular distribution of assembly intermediates that we had observed with WT Gag from H9-HIV T cells (compare WT in Fig. 3D to H9-HIV in Fig. 3C). In contrast, G2A Gag was entirely localized to the nonmembrane fraction, in the form of the ∼10S and ∼80S intermediates, with essentially no G2A Gag being found in the membrane fraction (Fig. 3D). These findings demonstrate that G2A Gag is arrested as a cytoplasmic ∼80S assembly intermediate, as expected from previous studies (41, 42, 47), and support a model in which the ∼80S intermediate is the complex that takes Gag from the cytosol to membranes.
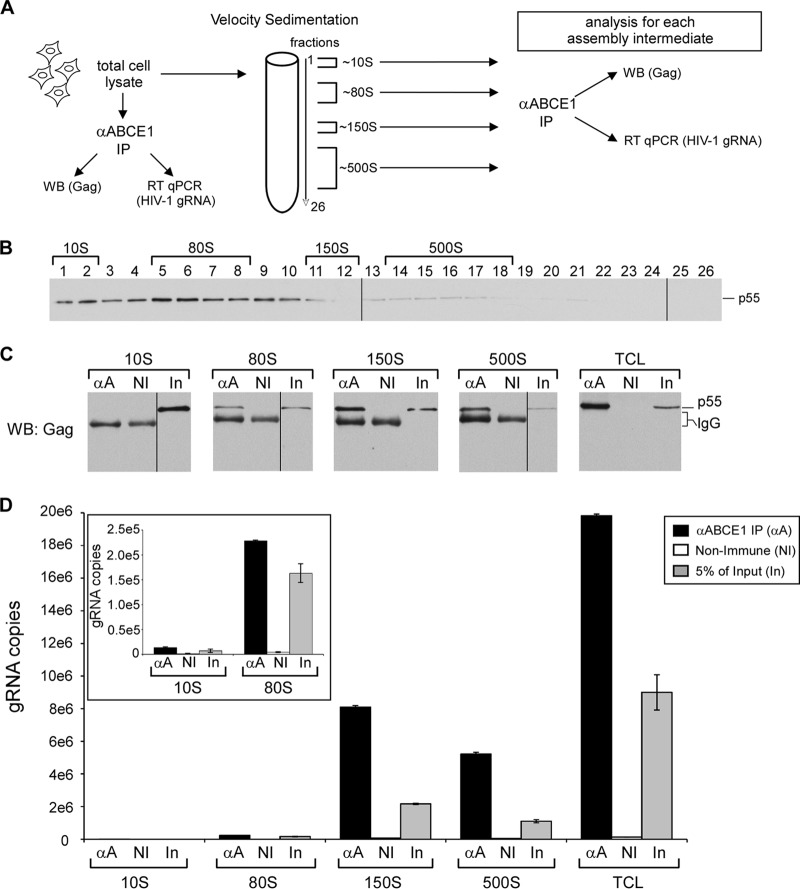
The high-molecular-mass assembly intermediates contain HIV-1 genomic RNA.
Our finding that the earliest assembly intermediates are formed in the cytoplasm while later assembly intermediates are found exclusively at membranes is consistent with what would be predicted for assembling HIV-1 Gag. Based on studies by others (4, 53), one would also predict that HIV-1 gRNA would first associate with Gag in an early assembly intermediate and would also be present in all subsequent assembly intermediates. To test this hypothesis, we separated assembly intermediates from H9-HIV cells by velocity sedimentation, isolated individual assembly intermediates from pooled gradient fractions by immunoprecipitation with antibody to ABCE1 (or a nonimmune control antibody), and subjected immunoprecipitates to RT-qPCR to quantify the number of gRNA copies in each assembly intermediate (Fig. 4A to D). We found that gRNA is associated with ABCE1 in the ∼80S, ∼150S, and ∼500S assembly intermediates (Fig. 4D) but not in the ∼10S fraction. Parallel blots confirmed that Gag is also present in the immunoprecipitated, ABCE1-containing ∼80S, ∼150S, and ∼500S assembly intermediates (Fig. 4C) but is not associated with ABCE1 in the ∼10S fraction, as shown previously (40, 43, 46, 47). Thus, these data demonstrate that the high-molecular-mass assembly intermediates (∼80S or larger) contain HIV-1 gRNA. Together, the subcellular localization and gRNA analyses confirm that the ∼80S, ∼150S, and ∼500S complexes display key characteristics expected of HIV-1 immature capsid assembly intermediates.
FIG 4.
HIV-1 gRNA is present in ∼80S, ∼150S, and ∼500S assembly intermediates in chronically infected T cells. (A) Schematic of experimental setup. (B) Total cell lysates from H9-HIV T cells were examined via velocity sedimentation, and fractions were analyzed for Gag by WB. Brackets identify which fractions contain each assembly intermediate. (C, D) The indicated fractions for each assembly intermediate were pooled, and each was subjected to immunoprecipitation (IP) with antibody to ABCE1 (αA) or a nonimmune control antibody (NI), alongside an immunoprecipitation reaction for total cell lysates (TCL). In, input. Subsequently, immunoprecipitation eluates were analyzed in parallel by either WB for Gag or RT-qPCR for the number of gRNA copies. WB samples were probed together on a single blot, and data shown are from a single exposure. An aliquot of input of lysate (2 to 4% of input) was also analyzed. (Inset) Numbers of gRNA copies in immunoprecipitations from ∼10S and ∼80S assembly intermediates shown using a more sensitive scale. Error bars show standard deviations from duplicate samples in a single experiment. Data are representative of those from three independent experiments.
Identifying residues on exposed Gag surfaces that are required for stability of and/or progression past specific assembly intermediates.
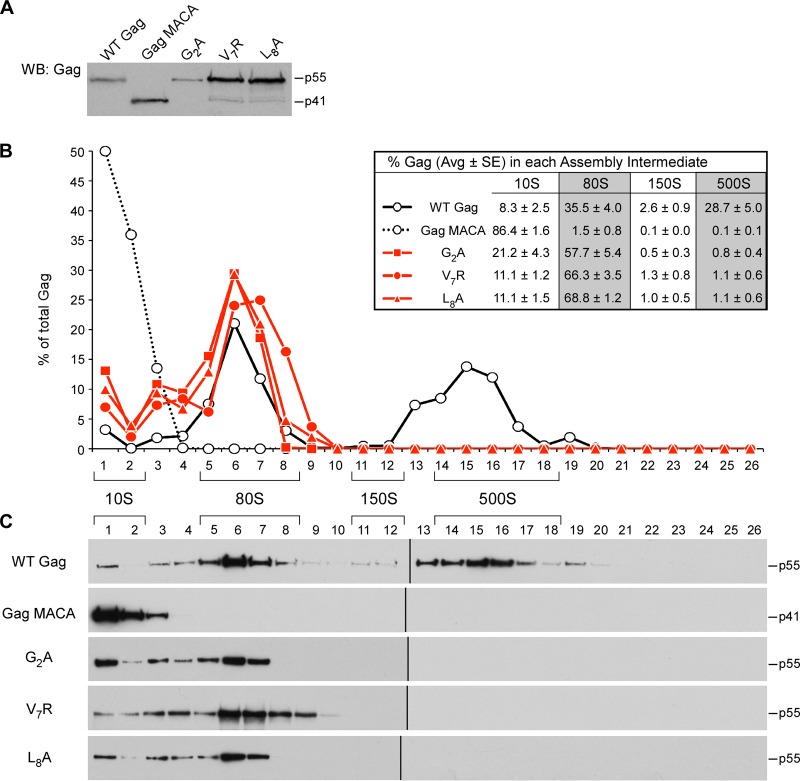
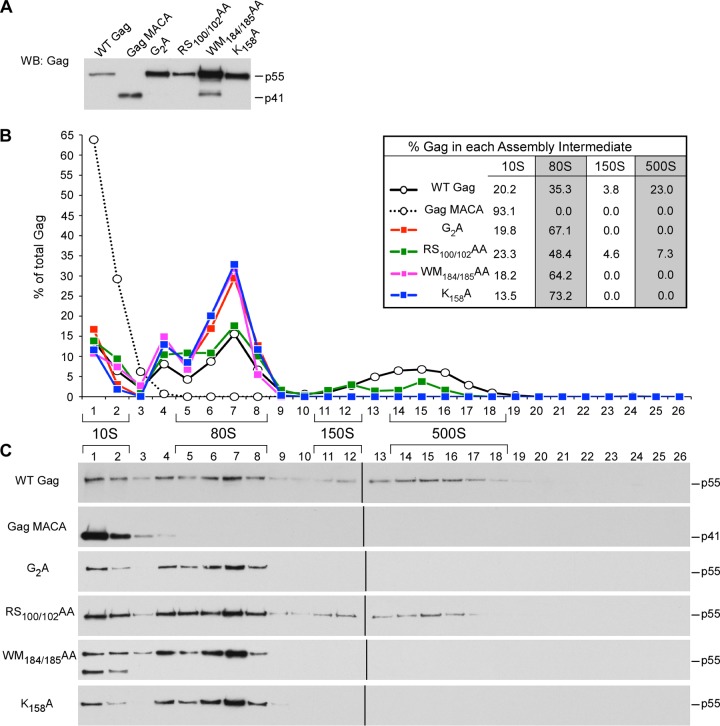
Having defined the subcellular location of each assembly intermediate and shown that the high-molecular-mass assembly intermediates contain HIV-1 gRNA, we next used velocity sedimentation analyses to examine how mutations in each of the four critical surfaces of Gag affects progression through the assembly pathway. These assembly-defective mutants were analyzed in four groups: MA arm mutants, CA-NTD H4-6 mutants, CA-CTD dimer mutants, and CA-CTD base mutants. Velocity sedimentation analysis of COS-1 cells expressing assembly-defective MA arm mutants revealed that V7R and L8A Gag accumulate in the ∼10S and ∼80S assembly intermediates (Fig. 5), with no later intermediates being formed, as was observed with the G2A mutation (Fig. 5). This result contrasts with the result for the WT Gag control, which formed the ∼10S, ∼80S, and ∼500S intermediates at steady state, and the result for the assembly-defective Gag MACA, which formed only the ∼10S intermediate (Fig. 5). Note that while accumulation of Gag in the assembly intermediate that precedes the block was seen for some mutants (e.g., Gag MACA), this was not observed in other cases (Gag G2A). This is most likely because cellular homeostatic degradation mechanisms prevent some arrested assembly intermediates from reaching high levels. Importantly, in all velocity sedimentation experiments in this study, WT and mutant Gag proteins were expressed to similar steady-state levels (e.g., see Fig. 5A); thus, differences in gradient patterns cannot be attributed to differences in intracellular Gag concentration. Together with membrane flotation experiments showing that all three MA mutants remain largely cytoplasmic (Fig. 2B and 3D) (15, 17), these findings indicate that MA residues critical either for myristoylation (G2A) or for myristate exposure (V7R and L8A) are required for progression past the ∼80S cytosolic assembly intermediate.
FIG 5.
Mutations in the N-terminal MA arm arrest immature capsid assembly at the ∼80S intermediate. COS-1 cells were transfected with the indicated WT or mutant provirus. (A) Equivalent aliquots of cell lysates were analyzed by WB for relative Gag expression. (B, C) Equivalent aliquots of cell lysates were also subjected to velocity sedimentation, and gradient fractions were analyzed by WB for Gag. (B) The graph depicts the relative amounts of Gag in each fraction, corresponding to the blots in panel C, and the position of each assembly intermediate is indicated by brackets. Data shown are from one representative experiment, and the table inset shows data averaged from three independent experiments.
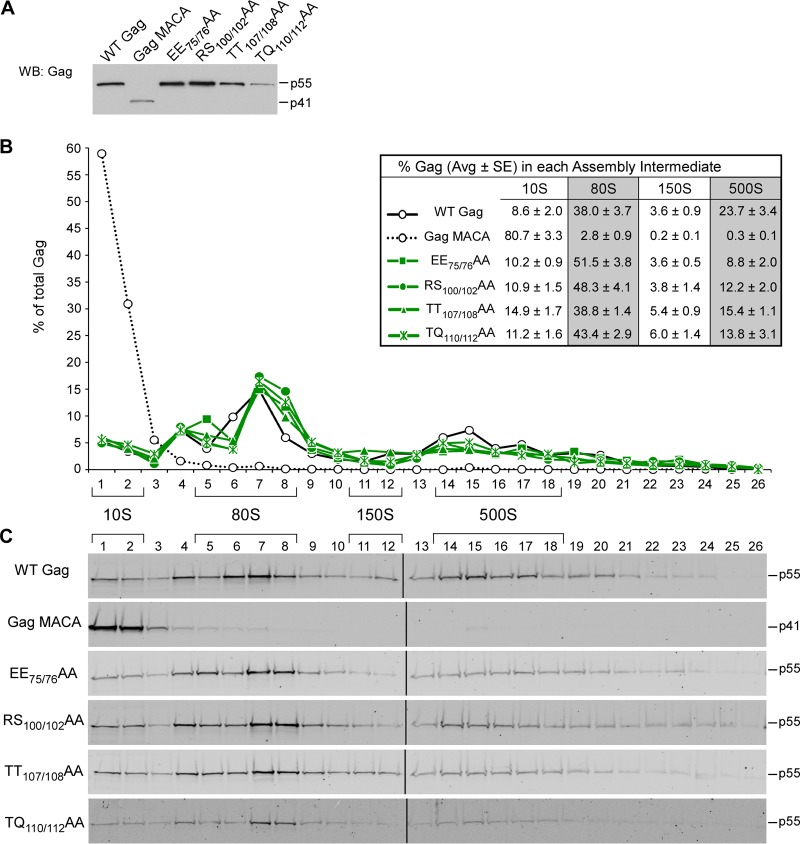
Next, we performed velocity sedimentation analysis on cells expressing Gags containing the CA-NTD H4-6 mutations (EE75/76AA, RS100/102AA, TT107/108AA, and TQ110/112AA), which are located on a continuous exposed surface in the CA-NTD crystal structure (Fig. 1B) (18). This analysis revealed that each of the CA-NTD H4-6 mutations resulted in the formation of the ∼10S, ∼80S, and ∼500S assembly intermediates (Fig. 6). Thus, the pattern of assembly intermediates formed by the CA-NTD H4-6 mutants closely resembled that of WT Gag (Fig. 6B and C), even though the CA-NTD H4-6 mutants are deficient in VLP production (Fig. 2A) (18, 49). Taken together, these data suggest that mutations in this CA-NTD surface result in inefficient progression past the final step of the immature capsid assembly pathway, where assembling Gag exits from the last high-molecular-mass assembly intermediate (the ∼500S intermediate) to form the completely assembled immature capsid.
FIG 6.
CA-NTD H4-6 mutants form all the intermediates in the assembly pathway. COS-1 cells were transfected with the indicated WT or mutant provirus. (A) Equivalent aliquots of cell lysates were analyzed by WB for relative Gag expression. (B, C) Equivalent aliquots of cell lysates were also subjected to velocity sedimentation, and gradient fractions were analyzed by WB for Gag. (B) The graph depicts the relative amounts of Gag in each fraction, corresponding to the blots in panel C, and the position of each assembly intermediate is indicated by brackets. Data shown are from one representative experiment, and the table inset shows data averaged from three independent experiments.
To test the hypothesis that CA-NTD H4-6 mutants progress inefficiently past the ∼500S assembly intermediate relative to WT Gag, we examined the stability of the WT and mutant ∼500S assembly intermediates. We would expect WT assembly intermediates to be relatively stable upon exposure to a higher than physiological salt concentration, while improperly assembled mutant intermediates might disassemble in response to a high salt concentration, allowing inherent defects in assembling capsids to be detected. For this experiment, we analyzed WT and EE75/76AA mutant lysates by membrane flotation in the presence of the standard intracellular NaCl concentration (0.1 M NaCl) or a high salt concentration (0.375 M NaCl) and then analyzed the resulting membrane fractions by velocity sedimentation (Fig. 7A to C). When membrane flotation was performed in the presence of a physiological salt concentration, the membrane fraction of both WT and the EE75/76AA mutant contained intact ∼80S and ∼500S intermediates (Fig. 7C), as expected from our previous velocity sedimentation analyses (Fig. 6). When membrane flotation was performed in the presence of a high salt concentration, the WT ∼500S intermediate retained its integrity, despite the ionic stress (Fig. 7C). In contrast, EE75/76AA Gag in the ∼500S intermediate was lost upon exposure to a high salt concentration, appearing instead in the ∼80S intermediate and in the pellet, which likely contains Gag that had disassembled and formed large aggregates (Fig. 7C). These data demonstrate that residues in the CA-NTD H4-6 surface contribute to the stability of the last assembly intermediate and support the hypothesis that mutations in the CA-NTD H4-6 surface cause inefficient progression past the last step in immature capsid assembly. The instability of the ∼500S intermediate produced by the CA-NTD H4-6 mutants could also explain why the mutant ∼500S intermediates are present in smaller amounts than the WT ∼500S intermediate at steady state (Fig. 6).
FIG 7.
Mutations in CA-NTD H4-6 and the CA-CTD dimer interface result in unstable assembly intermediates. COS-1 cells were transfected with the indicated proviruses, and total cell lysates were subjected to hypotonic lysis. The postnuclear supernatant (PNS) was analyzed by membrane flotation in the presence of a standard intracellular salt concentration (0.1 M NaCl) or a high salt concentration (0.375 M NaCl), and the membrane (M) fraction was subsequently analyzed by velocity sedimentation. (A) Schematic of experimental setup. (B) Equivalent aliquots of input used for membrane flotation gradients (PNS input) and velocity sedimentation gradients (M fraction input) were analyzed by WB for Gag. (C, D) Fractions from the membrane flotation gradients and subsequent velocity sedimentation analysis were analyzed for Gag by WB. Each set of membrane flotation Western blots in panels C and D is from a single exposure, allowing them to be directly compared within each panel. In contrast, the velocity sedimentation Western blots were developed separately for each condition (paired with a control, as shown) to obtain optimal exposures. The Gag present in bottom fractions of the velocity sedimentation gradient (fractions 22 to 25) represents Gag that has likely disassembled and aggregated due to experimental processing. The positions of assembly intermediates are indicated by brackets above the velocity sedimentation Western blots. Data in panels C and D are each from one experiment that is representative of three independent experiments.
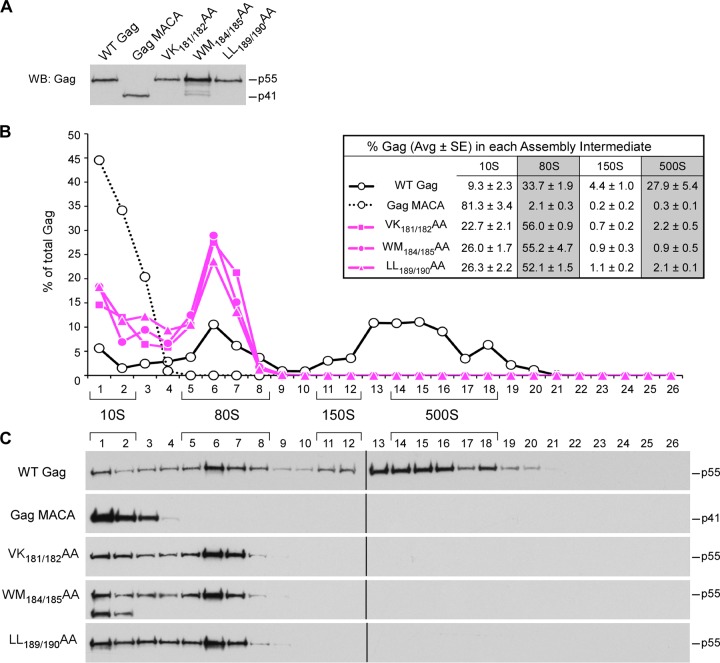
Next, we used velocity sedimentation to analyze the effect of mutations in two critical regions in the CA-CTD subdomain: the CA-CTD dimer interface and the CA-CTD base. A number of studies have shown that mutations of residues in the crystallographic CA-CTD dimer interface, which are exemplified by the WM184/185AA and LL189/190AA mutations, cause large reductions in virus particle production as well as moderately reduced membrane targeting of Gag (9, 11, 13, 14, 18) (Fig. 2B). Previously, we found that the WM184/185AA mutant is arrested as an ∼80S assembly intermediate (42). Here, we also examined two other mutants with mutations in the dimer interface, the previously described LL189/190AA mutant (13) and VK181/182AA mutant, whose mutations were previously studied individually (9). As expected, our membrane flotation experiments confirmed that all three CA-CTD dimer mutants exhibited only modest reductions in the amount of Gag at the membrane (33% to 71% of that for WT Gag), in contrast to the dramatic reductions in the amount observed in mutants with the MA mutations (8% to 9% of that for WT Gag) (Fig. 2B). Velocity sedimentation analyses revealed that all three dimer interface mutants formed only the ∼10S and ∼80S assembly intermediates (Fig. 8). Collectively, our data suggest that CA-CTD dimer mutants form ∼80S intermediates that are capable of membrane targeting but exhibit other defects that prevent further multimerization at the membrane; thus, these mutants fail to form the ∼500S assembly intermediate.
FIG 8.
Mutations in the CA-CTD dimer interface arrest immature capsid assembly at the ∼80S intermediate. COS-1 cells were transfected with the indicated WT or mutant provirus. (A) Equivalent aliquots of cell lysates were analyzed by WB for relative Gag expression. (B, C) Equivalent aliquots of cell lysates were also subjected to velocity sedimentation, and gradient fractions were analyzed by WB for Gag. (B) The graph depicts the relative amounts of Gag in each fraction, corresponding to the blots in panel C, and the position of each assembly intermediate is indicated by brackets. Data shown are from one representative experiment, and the table inset shows data averaged from three independent experiments.
The second critical surface within the CA-CTD subdomain is at the CA-CTD base. K158 (in the MHR), D197 (in helix 10), and P224 (downstream of helix 11) are residues on this surface that are known to be important for assembly (18) (Fig. 2A). Membrane flotation experiments revealed that membrane targeting of the CA-CTD base mutants was only modestly reduced (61% to 81% of that for WT Gag; Fig. 2B), similar to the results for the CA-CTD dimer mutants. Velocity sedimentation analyses revealed that all the CA-CTD base mutants formed the ∼10S and ∼80S assembly intermediates (Fig. 9) but not the ∼500S assembly intermediate, suggesting that they are arrested as membrane-bound ∼80S intermediates. Thus, like the CA-CTD dimer mutants, the CA-CTD base mutants are capable of membrane targeting but incapable of further multimerization and progression to the next step in the assembly pathway.
FIG 9.
Mutations in the CA-CTD base arrest immature capsid assembly at the ∼80S intermediate. COS-1 cells were transfected with the indicated WT or mutant provirus. (A) Equivalent aliquots of cell lysates were analyzed by WB for relative Gag expression. (B, C) Equivalent aliquots of cell lysates were also subjected to velocity sedimentation, and gradient fractions were analyzed by WB for Gag. (B) The graph depicts the relative amounts of Gag in each fraction, corresponding to the blots in panel C, and the position of each assembly intermediate is indicated by brackets. Data shown are from one representative experiment, and the table inset shows data averaged from three independent experiments.
Distinguishing between the action of CA-CTD dimer interface residues and CA-CTD base residues.
The phenotype of arrest as a membrane-bound ∼80S assembly intermediate, observed for both the CA-CTD dimer and CA-CTD base mutants, could be explained either by formation of an unstable membrane-bound ∼80S assembly intermediate or, alternatively, by formation of a stable membrane-bound ∼80S intermediate that is unable to form additional Gag-Gag interactions. To distinguish between these possibilities, we examined the stability of the arrested ∼80S assembly intermediates formed by the CA-CTD mutants (Fig. 7A, B, and D). Lysates of cells expressing the CA-CTD dimer mutation WM184/185AA or the CA-CTD base mutation D197A were analyzed by membrane flotation in the presence of the physiological salt concentration (0.1 M NaCl) versus a high salt concentration (0.375 M NaCl), followed by velocity sedimentation analysis of the membrane fraction. For both mutants, membrane fractions isolated in the presence of salt at the physiological concentration contained only the ∼80S assembly intermediate, as expected from results in Fig. 8 and 9. Interestingly, the high-salt-concentration-treated membrane fraction from cells expressing the CA-CTD dimer WM184/185AA mutant contained a substantial amount of the ∼10S assembly intermediate, as well as an accumulation of Gag near the pellet, which likely represents disassembling Gag that has aggregated (Fig. 7D). Because there was little or no ∼10S Gag observed when WT or mutant membrane fractions were isolated in the presence of salt at the physiological concentration (Fig. 7C and D), the appearance of membrane-associated WM184/185AA Gag in the ∼10S position was likely due to disassembly of an unstable, ∼80S assembly intermediate upon exposure to a high salt concentration. Similar results were obtained for the CA-CTD dimer LL189/190AA mutant (data not shown). In contrast, membrane fractions from cells expressing the CA-CTD base D197A mutant contained only the ∼80S intermediate, with no Gag detected in the ∼10S region, regardless of ionic condition (Fig. 7D). Similar results were obtained for the CA-CTD base K158A mutant (data not shown). Thus, mutations in the CA-CTD dimer interface led to formation of an unstable membrane-bound ∼80S intermediate, while mutations at the CA-CTD base led to formation of a stable membrane-bound ∼80S intermediate that failed to progress to the ∼500S assembly intermediate, most likely because of multimerization defects. Because CA-CTD base mutants were capable of forming a stable ∼80S intermediate, while CA-CTD dimer mutants were not, we concluded that the CA-CTD dimer residues act before the CA-CTD base residues during immature capsid assembly. These findings also suggest that the modest reduction in the amount of membrane-associated Gag observed with the CA-CTD dimer and base mutants (Fig. 2B) could be due to endocytosis and degradation of abnormal assembly intermediates that are arrested at the membrane; alternatively, for the CA-CTD dimer mutants, this reduction could be due to instability of the arrested ∼80S intermediate at the membrane.
Double mutations and analysis using human 293T cells support the proposed order in which exposed surfaces act during assembly.
Thus far, our biochemical studies had shown that each of the four surfaces that we analyzed acts at a different step in the assembly pathway. To confirm the order in which these four Gag surfaces act during assembly, we used analysis of mutants with double mutations (e.g., V7R or L8A paired with EE75/76AA and K158A or P224A paired with EE75/76AA or TQ110/112AA), where we would expect that mutations that act early in the pathway would be dominant over mutations that act late. Indeed, double mutants containing a mutation that causes ∼80S arrest and a mutation causing ∼500S arrest were arrested at the upstream step (∼80S arrest) in all four cases that we examined (data not shown). These results are consistent with the temporal order of assembly intermediates that was established previously (40–42, 44) and confirm our conclusions regarding the order in which exposed surfaces act during assembly.
As mentioned earlier, for analysis of intracellular assembly intermediates following transfection (Fig. 5 to 9), we used COS-1 cells (derived from African green monkey kidney) because they produce and endocytose relatively small amounts of virus. To address whether the same patterns of arrest for assembly-defective Gag mutants would be observed in human cells, we established conditions for studying assembly intermediates in 293T cells, which are derived from human embryonic kidney cells but release and endocytose large amounts of virus (54). We found that unlike in COS-1 cells, where assembly intermediates could be readily identified for up to 36 h posttransfection, in 293T cells the signal from ∼750S completed capsids became progressively larger at longer times after transfection (data not shown), consistent with the time course that others have observed for virus endocytosis in these cells (54). Ultimately, this ∼750S signal became so large that it obscured detection of the ∼500S and ∼150S assembly intermediates (data not shown). However, we were able to circumvent this problem by harvesting 293T cells at early times (15 h) following transfection with small amounts of proviral DNA; under these conditions, little virus was produced or endocytosed, allowing sufficient resolution of assembly intermediates (Fig. 10). Using this approach, we analyzed a subset of representative mutants and confirmed that mutants with mutations on each of the four critical surfaces of Gag displayed the same phenotype in human cells as in nonhuman primate cells (compare Fig. 10 to Fig. 5, 6, 8, and 9).
FIG 10.
Gag mutants display similar assembly phenotypes in COS-1 and 293T cells. 293T cells were transfected with the indicated WT or mutant provirus. (A) Equivalent aliquots of cell lysates were analyzed by WB for Gag expression. (B, C) Equivalent aliquots of cell lysates were also subjected to velocity sedimentation, with gradient fractions analyzed by WB for Gag. (B) The graph depicts the relative amounts of Gag in each fraction, corresponding to the blots in panel C, and the position of each assembly intermediate is indicated by brackets.
Mutations on critical surfaces cause accumulation of assembly-defective Gag at the plasma membrane of infected cells but do not disrupt binding to ABCE1 and DDX6.
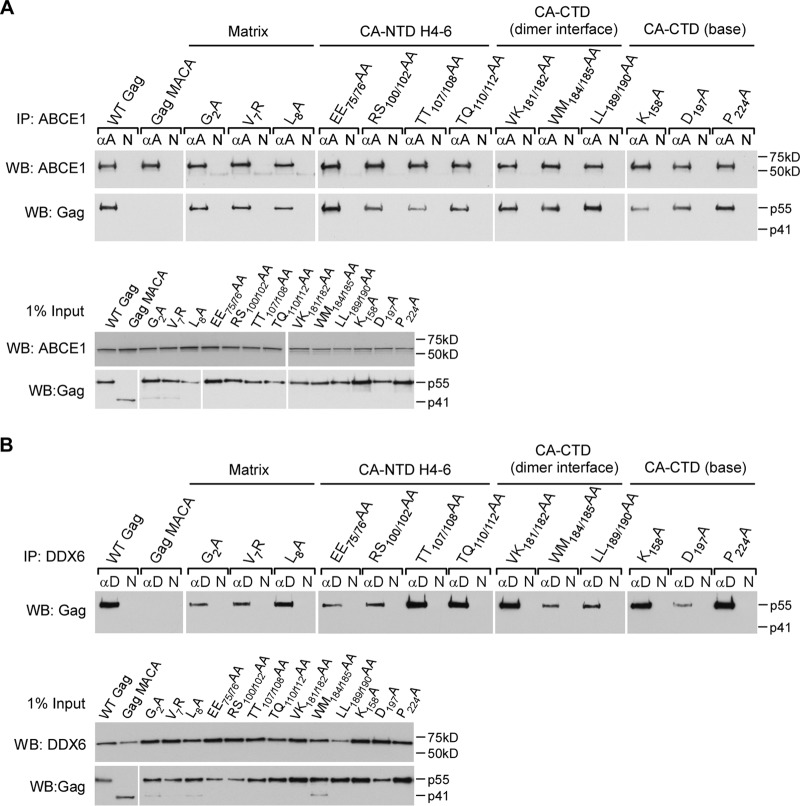
Previously, we had demonstrated that truncations of NC (e.g., Gag MACA) or mutations of basic residues within NC (e.g., Gag KR9A or Gag KR10A) (40–42, 44, 45, 47) arrest assembling Gag within the ∼10S assembly intermediate, which does not contain the cellular facilitators of assembly ABCE1 and DDX6 (40, 43, 45, 46). In contrast, all 13 of the MA and CA mutations that we analyzed here progressed past the ∼10S step, forming either defective ∼80S or defective ∼500S assembly intermediates. Because the WT ∼80S and ∼500S assembly intermediates contain both ABCE1 and DDX6, we asked whether the 13 assembly-defective MA and CA mutants are also associated with ABCE1 and DDX6 in cells. Lysates of cells expressing WT or mutant proviruses were subjected to immunoprecipitation either with antibody to ABCE1 (Fig. 11A) or with antibody to DDX6 (Fig. 11B), followed by WB for Gag. WT Gag and all 13 MA and CA mutants were associated with both ABCE1 and DDX6, unlike Gag MACA (Fig. 11). Thus, mutations within these four critical surfaces of MA and CA that function during assembly do not alter the ability of Gag to associate with previously described cellular facilitators of assembly.
FIG 11.
Mutations on four critical Gag surfaces do not alter the association with cellular facilitators of assembly. COS-1 cells were transfected with the indicated WT or mutant provirus. (A) Lysates were immunoprecipitated with ABCE1 antiserum (αA) or nonimmune IgG (N) and then analyzed by WB for Gag and ABCE1. Equivalent inputs were also analyzed for Gag and ABCE1 by WB. (B) Lysates were immunoprecipitated with DDX6 antibody (αD) or nonimmune IgG (N) and then analyzed by WB for Gag. Equivalent inputs were also analyzed for Gag and DDX6 via WB.
Additionally, we used an ultrastructural approach to verify the results of our biochemical analyses. Given the point at which each mutant was arrested in the assembly pathway, we expected EM analysis to reveal that the CA-NTD H4-6 mutants progress to a late stage in assembly, while the CA-CTD dimer and base mutants would likely be arrested at a relatively early stage at the PM. Moreover, although our biochemical studies revealed differences in the stability of the membrane-bound ∼80S assembly intermediates formed by the CA-CTD dimer and that of the CA-CTD base mutants (Fig. 7D), we would not expect this difference to be apparent upon EM analysis. To test these predictions, we infected COS-1 cells with virus encoding a mutation in either the CA-NTD H4-6 surface, CA-CTD dimer interface, or CA-CTD base surface and examined sections by immunogold double labeling (immuno-EM) using antibodies against Gag and ABCE1 (Table 2; Fig. 12) or Gag and DDX6 (Table 2; Fig. 13). We used sections from four independent experiments to analyze membrane-associated Gag quantitatively, as previously described (43). Labeled Gag at the PM was scored as being in one of three stages of assembly (Table 2): targeted Gag, defined as a cluster of Gag labeling without membrane deformation; an early assembly site, defined as Gag labeling in the presence of significant electron density at the PM or less than 50% of full membrane curvature; or a late assembly site, defined as Gag labeling in the presence of more than 50% of full membrane curvature. We found that 42% of WT Gag clusters were in early and late assembly sites at the PM (15% early, 26% late). In contrast, for the assembly-incompetent MACA Gag mutant (negative control), only 1% of Gag clusters were in the form of early and late assembly sites. For the CA-NTD H4-6 mutant, early and late assembly sites (8% early, 14% late) were readily observed, albeit less frequently than for WT Gag. However, for both the CA-CTD dimer and base mutants, the proportions of Gag clusters in early and late assembly sites were 2% and 6%, respectively, which were not significantly different from the values for the MACA negative control (Table 2). Thus, morphological analysis of the distribution of early and late assembly sites confirmed that the CA-NTD H4-6 mutant is arrested at a very late stage of the assembly pathway, while the CA-CTD dimer and base mutants are arrested soon after PM targeting (as shown quantitatively in Table 2 and with representative images in Fig. 12 and 13). Immuno-EM labeling also revealed extensive colocalization of ABCE1 and DDX6 with Gag in early and late assembly sites at the PM for WT Gag and the CA-NTD H4-6 mutant (Fig. 12 and 13, respectively), as we have shown quantitatively for WT Gag (40, 42, 43).
DISCUSSION
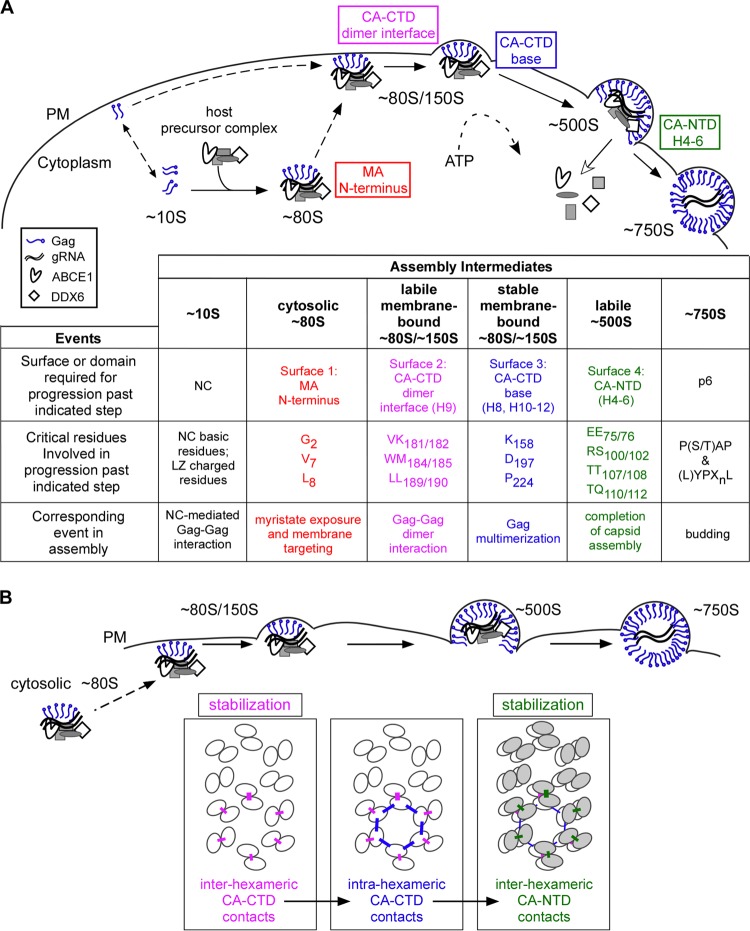
Here we provide, for the first time, a detailed temporospatial map showing when and where critical surfaces of Gag function during HIV-1 immature capsid assembly in cells. Our analysis of the subcellular localization of each WT assembly intermediate provides support for a model in which the ∼80S cytosolic assembly intermediate takes Gag from the cytosol to the membrane, where all subsequent intermediates are located. Additionally, we demonstrated that all of the high-molecular-mass intermediates in the assembly pathway (the ∼80S, ∼150S, and ∼500S complexes) contain HIV-1 gRNA, as would be expected for assembly intermediates. Next we examined four critical surfaces, chosen because they are exposed in crystal structures of MA or the CA subdomains and are known to be important for assembly of the HIV-1 immature capsid in cells. Analysis of three or four mutations in each surface revealed that critical residues on a single surface all function at the same step in the assembly pathway even when these residues are dispersed in the amino acid sequence. Our biochemical studies also showed that each of the four critical surfaces acts at a different step in assembly and allowed us to define the exact order in which these surfaces act (summarized in Fig. 14A): the MA arm is required for membrane targeting of the cytoplasmic assembling Gag oligomer (∼80S intermediate), the CA-CTD dimer interface is critical for stability of the ∼80S intermediate upon initial binding to the membrane, the CA-CTD base is needed for multimerization at the membrane to allow progression past the low oligomer stage, and the CA-NTD H4-6 surface is important for the stability of the nearly completed immature capsid (∼500S intermediate) just prior to budding. These findings were confirmed by immuno-EM analyses of intact cells infected by selected mutants.
FIG 14.
A temporospatial map of HIV-1 capsid assembly in cells. (A) Temporospatial map of HIV-1 assembly highlighting when and where each critical surface of Gag plays a role in the pathway. Assembly begins when newly synthesized Gag first forms the ∼10S assembly intermediate. Progression past the ∼10S intermediate requires Gag oligomerization mediated by either NC or a leucine zipper (LZ) (42). Formation of the cytosolic ∼80S assembly intermediate results when oligomerized ∼10S Gag co-opts a host precursor complex containing the cellular enzymes ABCE1 and DDX6 (43). The ∼80S intermediate targets to the membrane, where it undergoes continued multimerization to form the ∼150S and ∼500S intermediates. ABCE1 and DDX6 dissociate from the immature capsid upon completion of assembly (40, 43, 46), and release of the budding immature capsid requires residues that interact with cellular factors involved in budding, as shown by others (reviewed in reference 3). The chart shows domains/surfaces and critical residues required for progression past each step in the assembly pathway. (B) A model for when three distinct CA-CA interfaces (identified in the high-resolution structure of a completed immature capsid [29]) are formed during assembly. Ovals represent the individual CA subdomains that form the hexameric lattices in the assembling, immature capsid, in which CA-CTD subunits (white ovals) form the lower hexameric lattice and CA-NTD subunits (gray ovals) form the upper hexameric lattice. The three distinct inter- and intrahexameric interfaces in these lattices are shown as colored lines. These three interfaces involve residues that are identical or adjacent to residues analyzed here on the three CA surfaces and are colored accordingly. In this model, the interhexameric CA-CTD contacts (pink), mediated by residues on the CA-CTD dimer interface, are formed upon or just after membrane targeting of the ∼80S intermediate. The intrahexameric CA-CTD contacts (blue), mediated by residues within the CA-CTD base, form at the membrane and are required for multimerization of assembling Gag. The interhexameric CA-NTD contacts (green), mediated by residues within the CA-NTD H4-6 surface, form late during assembly, just prior to completion of the immature capsid. The two interhexameric CA-CA interfaces promote stability of the assembling capsid at two distinct steps in the assembly pathway.
Our data also have implications for our understanding of the structure of the final immature capsid. Structural studies have shown that the immature retroviral capsid consists of two stacked, irregular lattices of CA hexamers connected by inter- and intrahexameric contacts, with the more internal lattice formed by CA-CTD and the lattice outside that formed by CA-NTD (reviewed in reference 38). After we began the studies reported here, an 8-Å-resolution cryo-EM structure of an immature retroviral capsid identified residues responsible for CA-CA interactions within the CA-NTD and CA-CTD hexameric layers of the immature capsid (summarized in Fig. 15; also see Table S2 in reference 29). Interestingly, this new high-resolution structure identified CA-CA contact residues different from those suggested by previous, lower-resolution structures of immature capsids (31, 36). To analyze the structural implications of our findings, we superimposed our temporospatial map of residues critical for HIV-1 immature capsid assembly onto a map of residues found to be important for inter- and intrahexameric CA-CA interfaces in the aforementioned 8-Å-resolution immature retroviral capsid structure (29) and found a striking correspondence (Fig. 15A). Specifically, we observed that all the critical CA residues that we analyzed here overlap or are adjacent to residues identified as making critical CA-CA contacts in the fully assembled immature capsid. Given this correlation and the generally accepted view that the main function of CA during assembly is to form Gag-Gag contacts, it seems reasonable to assume that an important function of the critical CA residues analyzed here is to make CA-CA contacts within the immature capsid structure and that the failure to establish each of these critical CA-CA contacts leads to the arrest in the assembly pathway that we observed. If one accepts these two assumptions, then the temporospatial map in Fig. 14A can be used to generate a CA-CA interaction model (Fig. 14B) that proposes a specific sequence of intermolecular interactions leading to formation of the hexameric CA lattices in the immature capsid. In this sequence of events, the CA-CTD lattice is initiated by dimer contacts between CA-CTD subunits (which will later belong to adjacent hexamers), and this initial contact occurs while Gag is in the ∼80S intermediate, just before or upon membrane targeting. After Gag stably targets to the PM, individual CA-CTD hexamers are completed through formation of the intrahexameric CA-CTD base contacts. Most likely, this process of CA-CTD dimer formation followed by CA-CTD hexamer formation is reiterated as Gag subunits are added during multimerization at the PM. Finally, our data suggest that the outer hexameric lattice is generated very late in the pathway through interhexameric CA-NTD contacts established upon or just before formation of the last assembly intermediate (the ∼500S intermediate). Thus, while the 8-Å-resolution cryo-EM structure identified CA-CA contacts in the completed immature capsid, our study adds a new dimension to our understanding of the immature capsid structure by proposing a model, based on the assumptions described above, for a temporal sequence in which CA-CA contacts are formed during assembly in cells.
FIG 15.
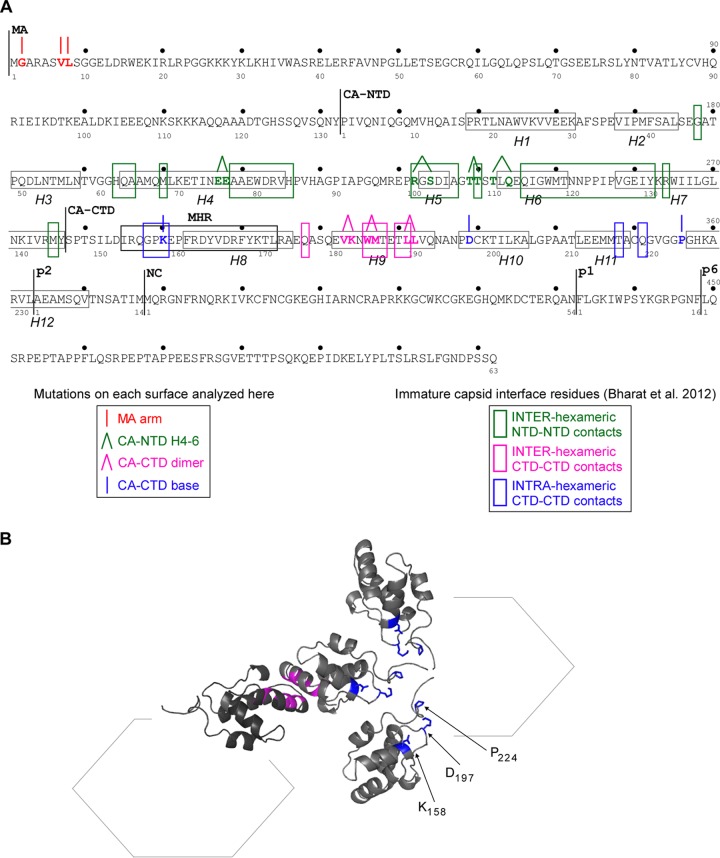
Overlap between Gag residues required for assembly and residues that form CA-CA interfaces in the immature capsid. (A) Amino acid sequence of HIV-1 LAI Gag, with the numbering for individual domains and spacer peptides (MA, CA, sp1, NC, and sp2) shown below and the numbering for the complete polyprotein shown above. The 12 actual or predicted alpha helices within CA (H1 to H12) are enclosed in gray boxes. Residues mutated in this study are indicated in colors that correspond to the surface in which they are located (see the key on the left), with lines above those residues indicating single-residue (|) or double-residue (Λ) mutations. Colored boxes enclose residues that form inter- and intrahexameric interfaces in the fully assembled immature retroviral capsid (see the key on the right), as reported in reference 29 and via personal communication from J. Briggs. Note that because loops connecting alpha helices are flexible, there is little certainty regarding the positions of residues in those regions. While all critical residues in the CA-NTD H4-6 surface and the CA-CTD dimer interface are within 1 residue or identical to residues that form interfaces in the immature capsid (29), the overlap between the CA-CTD base residues analyzed here and residues that form interfaces in the immature capsid (29) was less obvious. Specifically, although one critical residue in the CA-CTD base is identical to an interface residue (K158), two others (D197 and P224) are located further from interface residues in the linear amino acid sequence (29). (B) To analyze whether D197 and P224 are in the right orientation to form intrahexameric interfaces in the three-dimensional structure, we generated a schematic and ribbon diagram showing the spatial relationship of four CA-CTD domains within two adjacent hexamers, oriented as predicted elsewhere (29), using a Protein Data Bank file provided by J. Briggs. Residues critical for the CA-CTD interhexameric interface are colored in pink, and residues at the CA-CTD base studied herein (K158, D197, P224), along with their side chains on three CA-CTD domains, are shown in blue. The ribbon diagram shows the predicted orientations of residues (but not relative distances) and reveals that residues within the CA-CTD base surface are in the correct orientation to form intrahexameric interfaces in the immature capsid.
Overall, the CA-CA interaction model (Fig. 14B) suggests that both the CA-CTD and CA-NTD hexameric lattices form after membrane targeting of the Gag oligomer. More specifically, this model proposes that, following membrane targeting, the interhexameric CA-CTD dimer interaction forms first, the intrahexameric CA-CTD base interactions form next, and the CA-NTD hexameric lattice forms last. Notably, this model is compatible with the possibility that one or more of the exposed surfaces of Gag may transiently interact with a cellular protein(s) present in the assembly intermediates before they form the CA-CA contacts observed in the completed, immature capsid; however, that possibility was not tested here.
Additionally, our stability experiments (Fig. 7), when viewed in the context of our CA-CA interaction model (Fig. 14B), offer insights into the function of CA-CA interactions during assembly. We found that residues important for interhexameric CA-CTD dimer interface contacts act early to confer stability of the Gag oligomer upon membrane targeting, and residues important for interhexameric CA-NTD H4-6 contacts confer stability to the nearly completed immature capsid prior to budding. In contrast, residues important for intrahexameric CA-CTD contacts (the CA-CTD base residues) do not affect stability but instead appear to be required for continued multimerization at the PM. Thus, importantly, our data suggest that additional subunits of Gag are recruited into the targeted Gag oligomer only if that oligomer contains CA-CTD hexamers with properly formed inter- and intrahexameric CA-CTD contacts.
While we do not yet know the exact stoichiometry or oligomerization state of Gag in the assembly intermediates, our data shed some light on this question. Previous live imaging and biochemical studies suggested that the complex that targets Gag to the PM contains gRNA (53), is in the form of a small oligomer (in which the number of Gag proteins is too low to be visualized by live imaging [4, 53]), and undergoes additional multimerization after membrane anchoring (53). Our biochemical studies are consistent with the live imaging observations and suggest that Gag is taken to membranes in an assembly intermediate that is roughly the size of a ribosome (∼80S) and contains HIV-1 gRNA as well as cellular proteins. Our biochemical studies also indicate that the assembling Gag complex increases in size (from ∼80S to ∼500S) after PM targeting, which is consistent with recruitment of additional Gag proteins at the PM. Based on our CA-CA interaction model (Fig. 14B), we speculate that soon after targeting to the PM, assembling Gag progresses from being organized as dimers (held together by the interhexameric CA-CTD dimer interface contacts) to being organized as higher-order multimers (held together by the intrahexameric CA-CTD base contacts). We also speculate that the temporal sequence of contacts for CA-CTD hexamer formation is likely repeated as additional Gag proteins are recruited to the PM, during progression to the ∼500S intermediate. Finally, our data suggest that the hexameric CA-NTD lattice is formed late, after multimerization at the PM is largely completed. In the future, these hypotheses regarding the oligomerization state of Gag in assembly intermediates could be tested using cross-linking of WT and mutant assembly intermediates isolated from cell lysates. This approach is likely to be technically challenging, given the transient nature and very small quantities of assembly intermediates in cells. Note, however, that our hypothesis that Gag is likely dimeric until after it targets to membranes fits well with results obtained by others, in which cytosolic Gag formed complexes no larger than dimers upon cross-linking (4).
Our findings raise a number of questions that will need to be answered in the future. The first relates to our observation that gRNA is associated with all the high-molecular-mass assembly intermediates that contain Gag and ABCE1 (∼80S, ∼150S, and ∼500S intermediates) (Fig. 4). These experiments were performed on intermediates immunoprecipitated using ABCE1 antibody and indicate that gRNA is associated with ABCE1-containing assembly intermediates, as would be expected. Note that similar gRNA immunoprecipitation studies using immunoprecipitation with Gag antibody will be required to determine when Gag first associates with gRNA during assembly and whether gRNA is associated with all Gag-containing complexes or only a subset of Gag-containing complexes. Another set of questions follows from our model that the cytosolic ∼80S assembly intermediate likely takes assembling Gag to the PM. It is clear from pulse-chase analyses that assembling Gag chases out of both the ∼10S and the ∼80S intermediates, indicating that both are intermediates in assembly and not dead-end complexes (40, 41). However, because our pulse-chase analyses did not distinguish between the cytosolic and membrane-associated ∼80S intermediates, we cannot definitively say that the membrane-associated ∼80S intermediate is derived from the ∼80S cytosolic intermediate and not from the ∼10S cytosolic intermediate. However, it seems likely that the ∼80S cytosolic intermediate is the direct precursor of the ∼80S membrane-associated intermediate for two reasons. First, all the mutations that inhibit membrane targeting of Gag cause arrest of Gag as the cytosolic ∼80S intermediate (Fig. 5) and not as the ∼10S intermediate. Second, both the cytosolic and membrane-associated ∼80S intermediates contain ABCE1 and DDX6, as do the subsequent ∼150S and ∼500S intermediates, while the ∼10S intermediate does not. Nevertheless, to acknowledge that the immediate precursor to the ∼80S membrane-associated intermediate remains hypothetical to date, we have used dotted lines in Fig. 14A to show the ∼80S cytosolic intermediate giving rise to the ∼80S membrane-associated intermediate. Future studies aimed at defining all the Gag-containing complexes that contain gRNA may be able to identify the complex responsible for initially anchoring Gag-associated gRNA at the PM. This type of Gag- and gRNA-containing anchoring complex has been proposed to act as the initial nucleation site for further assembly at the PM on the basis of live imaging studies (53).
While previous studies by others had identified which critical surfaces act while Gag is in the cytosol and which act after Gag targets to membranes, the data presented here allow a precise ordering of multiple critical events in assembly for the first time (Fig. 14A). For example, previous studies found that mutations in the three CA surfaces examined here cause defects upon or after membrane targeting (9, 13, 14, 18), but the sequence in which these surfaces act had not been defined. Our study suggests an exact order in which these three critical CA surfaces act (with the CA-CTD dimer interface acting after the MA arm residues but before CA-CTD base residues, which in turn act before the CA-NTD H4-6 residues) and allowed us to make functional distinctions between them by identifying which of these surfaces are important for stabilizing the assembling capsid at the membrane (Fig. 6 to 9).
Our data also shed light on the temporal order in which indirect and direct Gag-Gag interactions are formed early during assembly. Others have shown that early HIV-1 Gag-Gag associations arise in two ways: through indirect interactions mediated by the NC domain of Gag binding to RNA (26, 50–52, 55–65) and through direct CA-CA interactions (9, 11, 13, 18, 26, 29). Previously, we found that NC-mediated interactions are required to form the ∼80S cytosolic assembly intermediate (45). Here we provide data suggesting that MA arm residues are required for membrane targeting of the ∼80S intermediate following formation of indirect NC-mediated interactions, while the CA-CTD dimer interface is required for stability of the membrane-targeted ∼80S assembly intermediate. Together these findings clarify the temporal and spatial order of the indirect versus direct Gag-Gag associations (NC-mediated interactions precede interactions requiring MA arm residues prior to membrane targeting, which in turn precede the interaction at membranes that requires the CA-CTD dimer interface residues; Fig. 14A). Notably, our conclusion that RNA-mediated, indirect Gag-Gag associations are formed prior to the direct CA-CTD dimer interactions makes sense, since the CA-CTD dimer contact is known to be a low-affinity interaction (dissociation constant = 18 μM [11]) and would therefore be facilitated by bringing Gag monomers into proximity with each other through RNA-mediated, indirect interactions.
All the findings reported here are consistent with results reported by other groups, with the exception of our EM analysis of the CA-NTD H4-6 mutant EE75/76AA. Our biochemical analyses revealed that this mutant causes arrest at the ∼500S assembly intermediate but fails to undergo proper release of VLPs, consistent with progression to a late stage of Gag multimerization at the PM. The findings of our quantitative ultrastructural analysis of infected COS-1 cells expressing this mutant were consistent with our biochemical findings and showed abundant early and late assembling forms. In contrast to our results, ultrastructural analysis of the EE75/76AA mutant by other groups (18, 49) revealed large sheets of electron density at the PM with few late budding forms in transfected 293T or HeLa cells. This discrepancy in the ultrastructural phenotype can be explained by the observation that 293T and HeLa cells are highly prone to endocytosis, while COS-1 cells are not (data not shown; supported by results presented in reference 54). Thus, the abnormal, arrested late budding forms that we observed in COS-1 cells may have initially been present in 239T and HeLa cells but were subsequently endocytosed and were therefore not readily visible by EM at late times after transfection. Regardless, the discrepancy in the degree of budding observed for this mutant does not alter our key conclusion regarding the CA-NTD H4-6 mutants: that these mutants achieve a degree of Gag multimerization that allows formation of the ∼500S late assembly intermediate, unlike the other categories of mutants that we examined. In support of this, we found that the EE75/76AA mutant forms the ∼500S intermediate in COS-1 cells as well as in 293T cells at both early (15 h) and late (36 h) times posttransfection (data not shown). Thus, under all conditions that we examined, EE75/76AA forms the ∼500S intermediate, but in some situations, this intermediate may be internalized and therefore not observed at the PM in EM analyses.
Importantly, our data also show that all of the interfaces critical for formation of a stable immature lattice are formed while assembling Gag is in high-molecular-mass assembly intermediates containing host proteins, including the facilitators of assembly ABCE1 and DDX6. Exactly how these cellular facilitators act remains to be determined. In a previous study, we demonstrated that DDX6, an RNA helicase, promotes formation of assembly at or after the membrane-targeting step (43). Clues to how DDX6 may act during assembly come from the observation that purified nucleic acids promote capsid formation in recombinant Gag assembly systems in vitro (55). Because RNA is found only within ribonucleoprotein complexes in cells, we have speculated that DDX6 likely promotes immature capsid assembly by liberating gRNA from ribonucleoprotein complexes and making it available as a scaffold for assembly (43). Moreover, since DDX6 first associates with Gag in the cytosol and remains associated with Gag at the PM (43) (Fig. 12 and 13), such DDX6-dependent RNA remodeling could occur at an early step (i.e., in the cytosolic ∼80S intermediate) or could assist with the formation of one or more of the critical direct CA-CA interactions that occur at the PM.
Finally, our data provide an important link between structural models of assembling Gag and events of immature capsid assembly that occur in cells. While the recent high-resolution structure of an immature retroviral capsid was generated from recombinant Gag domains that were assembled noncatalytically by mass action in vitro (29), our studies were performed in cells that were either transfected with provirus or HIV-1 infected, where immature capsid assembly appears to occur via an enzyme-catalyzed mechanism (40–43, 47). Interestingly, our data fit remarkably well with the structural models generated from recombinant immature capsids, thereby adding a temporospatial dimension to our understanding of CA-CA interactions and immature capsid structure for the first time. This elegant convergence of two fundamentally different approaches argues that the mass action and enzyme-catalyzed models describe the process of assembly from distinct, but complementary, angles. Moreover, this convergence also demonstrates how we can gain valuable insight into immature capsid structure through studying transient assembly intermediates formed in cells.
ACKNOWLEDGMENTS
We thank K. Chutiraka for technical assistance; B. Schneider, S. MacFarlane, S. Knecht, and the Fred Hutchinson Cancer Research Center Electron Microscopy Shared Resources for assistance with transmission EM imaging; the AIDS Research and Reference Reagent Program in the Division of AIDS at NIAID for the HIV-1 Gag p24 hybridoma (183-H12-5C), obtained from B. Chesebro; J. Briggs for the Protein Data Bank file and other structural data used in Fig. 15; and O. Pornillos, M. Tanaka, and K. Nichols for comments on the manuscript.
This work was supported by NIH grant R01AI106397 to J.R.L. and an NIH T32 AI750913 fellowship award to B.A.R.
Footnotes
Published ahead of print 12 March 2014
REFERENCES
- 1.Briggs JA, Simon MN, Gross I, Krausslich HG, Fuller SD, Vogt VM, Johnson MC. 2004. The stoichiometry of Gag protein in HIV-1. Nat. Struct. Mol. Biol. 11:672–675. 10.1038/nsmb785 [DOI] [PubMed] [Google Scholar]
- 2.Balasubramaniam M, Freed EO. 2011. New insights into HIV assembly and trafficking. Physiology (Bethesda) 26:236–251. 10.1152/physiol.00051.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sundquist WI, Krausslich HG. 2012. HIV-1 assembly, budding, and maturation. Cold Spring Harb. Perspect. Med. 2:a015420. 10.1101/cshperspect.a006924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kutluay SB, Bieniasz PD. 2010. Analysis of the initiating events in HIV-1 particle assembly and genome packaging. PLoS Pathog. 6:e1001200. 10.1371/journal.ppat.1001200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chukkapalli V, Ono A. 2011. Molecular determinants that regulate plasma membrane association of HIV-1 Gag. J. Mol. Biol. 410:512–524. 10.1016/j.jmb.2011.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adamson CS, Jones IM. 2004. The molecular basis of HIV capsid assembly—five years of progress. Rev. Med. Virol. 14:107–121. 10.1002/rmv.418 [DOI] [PubMed] [Google Scholar]
- 7.Freed EO. 1998. HIV-1 Gag proteins: diverse functions in the virus life cycle. Virology 251:1–15. 10.1006/viro.1998.9398 [DOI] [PubMed] [Google Scholar]
- 8.Bryant M, Ratner L. 1990. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc. Natl. Acad. Sci. U. S. A. 87:523–527. 10.1073/pnas.87.2.523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu HH, Chang YF, Wang CT. 2006. Mutations in the alpha-helix directly C-terminal to the major homology region of human immunodeficiency virus type 1 capsid protein disrupt Gag multimerization and markedly impair virus particle production. J. Biomed. Sci. 13:645–656. 10.1007/s11373-006-9094-6 [DOI] [PubMed] [Google Scholar]
- 10.Freed EO, Orenstein JM, Buckler-White AJ, Martin MA. 1994. Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. J. Virol. 68:5311–5320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gamble TR, Yoo S, Vajdos FF, von Schwedler UK, Worthylake DK, Wang H, McCutcheon JP, Sundquist WI, Hill CP. 1997. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science 278:849–853. 10.1126/science.278.5339.849 [DOI] [PubMed] [Google Scholar]
- 12.Gottlinger HG, Sodroski JG, Haseltine WA. 1989. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. U. S. A. 86:5781–5785. 10.1073/pnas.86.15.5781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joshi A, Nagashima K, Freed EO. 2006. Mutation of dileucine-like motifs in the human immunodeficiency virus type 1 capsid disrupts virus assembly, Gag-Gag interactions, Gag-membrane binding, and virion maturation. J. Virol. 80:7939–7951. 10.1128/JVI.00355-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ono A, Waheed AA, Joshi A, Freed EO. 2005. Association of human immunodeficiency virus type 1 Gag with membrane does not require highly basic sequences in the nucleocapsid: use of a novel Gag multimerization assay. J. Virol. 79:14131–14140. 10.1128/JVI.79.22.14131-14140.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ono A, Freed EO. 1999. Binding of human immunodeficiency virus type 1 Gag to membrane: role of the matrix amino terminus. J. Virol. 73:4136–4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ono A, Huang M, Freed EO. 1997. Characterization of human immunodeficiency virus type 1 matrix revertants: effects on virus assembly, Gag processing, and Env incorporation into virions. J. Virol. 71:4409–4418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paillart JC, Gottlinger HG. 1999. Opposing effects of human immunodeficiency virus type 1 matrix mutations support a myristyl switch model of Gag membrane targeting. J. Virol. 73:2604–2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.von Schwedler UK, Stray KM, Garrus JE, Sundquist WI. 2003. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J. Virol. 77:5439–5450. 10.1128/JVI.77.9.5439-5450.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ono A, Ablan SD, Lockett SJ, Nagashima K, Freed EO. 2004. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc. Natl. Acad. Sci. U. S. A. 101:14889–14894. 10.1073/pnas.0405596101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saad JS, Loeliger E, Luncsford P, Liriano M, Tai J, Kim A, Miller J, Joshi A, Freed EO, Summers MF. 2007. Point mutations in the HIV-1 matrix protein turn off the myristyl switch. J. Mol. Biol. 366:574–585. 10.1016/j.jmb.2006.11.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spearman P, Horton R, Ratner L, Kuli-Zade I. 1997. Membrane binding of human immunodeficiency virus type 1 matrix protein in vivo supports a conformational myristyl switch mechanism. J. Virol. 71:6582–6592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang C, Loeliger E, Luncsford P, Kinde I, Beckett D, Summers MF. 2004. Entropic switch regulates myristate exposure in the HIV-1 matrix protein. Proc. Natl. Acad. Sci. U. S. A. 101:517–522. 10.1073/pnas.0305665101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gamble TR, Vajdos FF, Yoo S, Worthylake DK, Houseweart M, Sundquist WI, Hill CP. 1996. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell 87:1285–1294. 10.1016/S0092-8674(00)81823-1 [DOI] [PubMed] [Google Scholar]
- 24.Gitti RK, Lee BM, Walker J, Summers MF, Yoo S, Sundquist WI. 1996. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science 273:231–235. 10.1126/science.273.5272.231 [DOI] [PubMed] [Google Scholar]
- 25.Worthylake DK, Wang H, Yoo S, Sundquist WI, Hill CP. 1999. Structures of the HIV-1 capsid protein dimerization domain at 2.6 Å resolution. Acta Crystallogr. D Biol. Crystallogr. 55:85–92. 10.1107/S0907444998007689 [DOI] [PubMed] [Google Scholar]
- 26.Burniston MT, Cimarelli A, Colgan J, Curtis SP, Luban J. 1999. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J. Virol. 73:8527–8540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Byeon IJ, Meng X, Jung J, Zhao G, Yang R, Ahn J, Shi J, Concel J, Aiken C, Zhang P, Gronenborn AM. 2009. Structural convergence between cryo-EM and NMR reveals intersubunit interactions critical for HIV-1 capsid function. Cell 139:780–790. 10.1016/j.cell.2009.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Momany C, Kovari LC, Prongay AJ, Keller W, Gitti RK, Lee BM, Gorbalenya AE, Tong L, McClure J, Ehrlich LS, Summers MF, Carter C, Rossmann MG. 1996. Crystal structure of dimeric HIV-1 capsid protein. Nat. Struct. Biol. 3:763–770. 10.1038/nsb0996-763 [DOI] [PubMed] [Google Scholar]
- 29.Bharat TA, Davey NE, Ulbrich P, Riches JD, de Marco A, Rumlova M, Sachse C, Ruml T, Briggs JA. 2012. Structure of the immature retroviral capsid at 8 Å resolution by cryo-electron microscopy. Nature 487:385–389. 10.1038/nature11169 [DOI] [PubMed] [Google Scholar]
- 30.Waki K, Durell SR, Soheilian F, Nagashima K, Butler SL, Freed EO. 2012. Structural and functional insights into the HIV-1 maturation inhibitor binding pocket. PLoS Pathog. 8:e1002997. 10.1371/journal.ppat.1002997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wright ER, Schooler JB, Ding HJ, Kieffer C, Fillmore C, Sundquist WI, Jensen GJ. 2007. Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J. 26:2218–2226. 10.1038/sj.emboj.7601664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Accola MA, Hoglund S, Gottlinger HG. 1998. A putative alpha-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J. Virol. 72:2072–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo X, Roldan A, Hu J, Wainberg MA, Liang C. 2005. Mutation of the SP1 sequence impairs both multimerization and membrane-binding activities of human immunodeficiency virus type 1 Gag. J. Virol. 79:1803–1812. 10.1128/JVI.79.3.1803-1812.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang C, Hu J, Russell RS, Roldan A, Kleiman L, Wainberg MA. 2002. Characterization of a putative alpha-helix across the capsid-SP1 boundary that is critical for the multimerization of human immunodeficiency virus type 1 Gag. J. Virol. 76:11729–11737. 10.1128/JVI.76.22.11729-11737.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang C, Hu J, Whitney JB, Kleiman L, Wainberg MA. 2003. A structurally disordered region at the C terminus of capsid plays essential roles in multimerization and membrane binding of the Gag protein of human immunodeficiency virus type 1. J. Virol. 77:1772–1783. 10.1128/JVI.77.3.1772-1783.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Briggs JA, Riches JD, Glass B, Bartonova V, Zanetti G, Kräusslich HG. 2009. Structure and assembly of immature HIV. Proc. Natl. Acad. Sci. U. S. A. 106:11090–11095. 10.1073/pnas.0903535106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Marco A, Davey NE, Ulbrich P, Phillips JM, Lux V, Riches JD, Fuzik T, Ruml T, Kräusslich HG, Vogt VM, Briggs JA. 2010. Conserved and variable features of Gag structure and arrangement in immature retrovirus particles. J. Virol. 84:11729–11736. 10.1128/JVI.01423-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ganser-Pornillos BK, Yeager M, Pornillos O. 2012. Assembly and architecture of HIV. Adv. Exp. Med. Biol. 726:441–465. 10.1007/978-1-4614-0980-9_20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spearman P, Wang JJ, Vander Heyden N, Ratner L. 1994. Identification of human immunodeficiency virus type 1 Gag protein domains essential to membrane binding and particle assembly. J. Virol. 68:3232–3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dooher JE, Schneider BL, Reed JC, Lingappa JR. 2007. Host ABCE1 is at plasma membrane HIV assembly sites and its dissociation from Gag is linked to subsequent events of virus production. Traffic 8:195–211. 10.1111/j.1600-0854.2006.00524.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lingappa JR, Hill RL, Wong ML, Hegde RS. 1997. A multistep, ATP-dependent pathway for assembly of human immunodeficiency virus capsids in a cell-free system. J. Cell Biol. 136:567–581. 10.1083/jcb.136.3.567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klein KC, Reed JC, Tanaka M, Nguyen VT, Giri S, Lingappa JR. 2011. HIV Gag-leucine zipper chimeras form ABCE1-containing intermediates and RNase-resistant immature capsids similar to those formed by wild-type HIV-1 Gag. J. Virol. 85:7419–7435. 10.1128/JVI.00288-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reed JC, Molter B, Geary CD, McNevin J, McElrath J, Giri S, Klein KC, Lingappa JR. 2012. HIV-1 Gag co-opts a cellular complex containing DDX6, a helicase that facilitates capsid assembly. J. Cell Biol. 198:439–456. 10.1083/jcb.201111012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh AR, Hill RL, Lingappa JR. 2001. Effect of mutations in Gag on assembly of immature human immunodeficiency virus type 1 capsids in a cell-free system. Virology 279:257–270. 10.1006/viro.2000.0706 [DOI] [PubMed] [Google Scholar]
- 45.Lingappa JR, Dooher JE, Newman MA, Kiser PK, Klein KC. 2006. Basic residues in the nucleocapsid domain of Gag are required for interaction of HIV-1 Gag with ABCE1 (HP68), a cellular protein important for HIV-1 capsid assembly. J. Biol. Chem. 281:3773–3784. 10.1074/jbc.M507255200 [DOI] [PubMed] [Google Scholar]
- 46.Zimmerman C, Klein KC, Kiser PK, Singh ARS, Firestein BL, Riba SC, Lingappa JR. 2002. Identification of a host protein essential for assembly of immature HIV-1 capsids. Nature 415:88–92. 10.1038/415088a [DOI] [PubMed] [Google Scholar]
- 47.Dooher JE, Lingappa JR. 2004. Conservation of a step-wise, energy-sensitive pathway involving HP68 for assembly of primate lentiviral capsids in cells. J. Virol. 78:1645–1656. 10.1128/JVI.78.4.1645-1656.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kimpton J, Emerman M. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J. Virol. 66:2232–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grover JR, Llewellyn GN, Soheilian F, Nagashima K, Veatch SL, Ono A. 2013. Roles played by capsid-dependent induction of membrane curvature and Gag-ESCRT interactions in tetherin recruitment to HIV-1 assembly sites. J. Virol. 87:4650–4664. 10.1128/JVI.03526-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gheysen D, Jacobs E, de Foresta F, Thiriart C, Francotte M, Thines D, De Wilde M. 1989. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell 59:103–112. 10.1016/0092-8674(89)90873-8 [DOI] [PubMed] [Google Scholar]
- 51.Hockley DJ, Nermut MV, Grief C, Jowett JB, Jones IM. 1994. Comparative morphology of Gag protein structures produced by mutants of the gag gene of human immunodeficiency virus type 1. J. Gen. Virol. 75:2985–2997. 10.1099/0022-1317-75-11-2985 [DOI] [PubMed] [Google Scholar]
- 52.Jowett JB, Hockley DJ, Nermut MV, Jones IM. 1992. Distinct signals in human immunodeficiency virus type 1 Pr55 necessary for RNA binding and particle formation. J. Gen. Virol. 73:3079–3086. 10.1099/0022-1317-73-12-3079 [DOI] [PubMed] [Google Scholar]
- 53.Jouvenet N, Simon SM, Bieniasz PD. 2009. Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles. Proc. Natl. Acad. Sci. U. S. A. 106:19114–19119. 10.1073/pnas.0907364106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jouvenet N, Neil SJ, Bess C, Johnson MC, Virgen CA, Simon SM, Bieniasz PD. 2006. Plasma membrane is the site of productive HIV-1 particle assembly. PLoS Biol. 4:e435. 10.1371/journal.pbio.0040435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Campbell S, Rein A. 1999. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J. Virol. 73:2270–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cimarelli A, Sandin S, Hoglund S, Luban J. 2000. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J. Virol. 74:3046–3057. 10.1128/JVI.74.7.3046-3057.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dawson L, Yu XF. 1998. The role of nucleocapsid of HIV-1 in virus assembly. Virology 251:141–157. 10.1006/viro.1998.9374 [DOI] [PubMed] [Google Scholar]
- 58.Derdowski A, Ding L, Spearman P. 2004. A novel fluorescence resonance energy transfer assay demonstrates that the human immunodeficiency virus type 1 Pr55Gag I domain mediates Gag-Gag interactions. J. Virol. 78:1230–1242. 10.1128/JVI.78.3.1230-1242.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Feng YX, Li T, Campbell S, Rein A. 2002. Reversible binding of recombinant human immunodeficiency virus type 1 Gag protein to nucleic acids in virus-like particle assembly in vitro. J. Virol. 76:11757–11762. 10.1128/JVI.76.22.11757-11762.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Khorchid A, Javanbakht H, Wise S, Halwani R, Parniak MA, Wainberg MA, Kleiman L. 2000. Sequences within Pr160gag-pol affecting the selective packaging of primer tRNA(Lys3) into HIV-1. J. Mol. Biol. 299:17–26. 10.1006/jmbi.2000.3709 [DOI] [PubMed] [Google Scholar]
- 61.Ono A, Demirov D, Freed EO. 2000. Relationship between human immunodeficiency virus type 1 Gag multimerization and membrane binding. J. Virol. 74:5142–5150. 10.1128/JVI.74.11.5142-5150.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sandefur S, Smith RM, Varthakavi V, Spearman P. 2000. Mapping and characterization of the N-terminal I domain of human immunodeficiency virus type 1 Pr55(Gag). J. Virol. 74:7238–7249. 10.1128/JVI.74.16.7238-7249.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sandefur S, Varthakavi V, Spearman P. 1998. The I domain is required for efficient plasma membrane binding of human immunodeficiency virus type 1 Pr55Gag. J. Virol. 72:2723–2732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Y, Barklis E. 1997. Effects of nucleocapsid mutations on human immunodeficiency virus assembly and RNA encapsidation. J. Virol. 71:6765–6776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Y, Qian H, Love Z, Barklis E. 1998. Analysis of the assembly function of the human immunodeficiency virus type 1 Gag protein nucleocapsid domain. J. Virol. 72:1782–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]