Abstract
Pathogens are recognized by hosts by use of various receptors, including the Toll-like receptor (TLR) and Nod-like receptor (NLR) families. Ligands for these varied receptors, including bacterial products, are identified by the immune system, resulting in development of innate immune responses. Only a couple of components from type III secretion (T3S) systems are known to be recognized by TLR or NLR family members. Known T3S components that are detected by pattern recognition receptors (PRRs) are (i) flagellin, detected by TLR5 and NLRC4 (Ipaf); and (ii) T3S rod proteins (PrgJ and homologs) and needle proteins (PrgI and homologs), detected by NAIP and the NLRC4 inflammasome. In this report, we characterize the induction of proinflammatory responses through TLRs by the Yersinia pestis T3S needle protein, YscF, the Salmonella enterica needle proteins PrgI and SsaG, and the Shigella needle protein, MxiH. More specifically, we determine that the proinflammatory responses occur through TLR2 and -4. These data support the hypothesis that T3S needles have an unrecognized role in bacterial pathogenesis by modulating immune responses.
INTRODUCTION
Type III secretion (T3S) systems are conserved bacterial nanomachines that are involved in the pathogenesis of many important human, animal, and plant diseases (1). T3S systems are also important in several known symbiotic relationships, demonstrating a duality of T3S functions ranging from beneficial to detrimental manipulation of eukaryotic biology by Gram-negative bacteria (1, 2). The hallmark of T3S is the vectorial injection of toxins into eukaryotic cells to alter host signaling pathways. However, little is known of other roles that these exposed structural components may have in disease.
T3S systems are widely distributed in Gram-negative pathogens and are structurally conserved across species (3). An important component of T3S systems is a needle-like appendage that extends from the bacterial surface. The needle is a hollow structure formed by a homopolymer of the needle protein(s), e.g., PrgI and SsaG (encoded in Salmonella pathogenicity island 1 [SPI-1] and SPI-2, respectively) in Salmonella enterica serovar Typhimurium, MxiH in Shigella flexneri, and YscF in Yersinia pestis. X-ray crystallography and nuclear magnetic resonance (NMR) analysis have been utilized to examine the structures of various needle proteins; MxiH from Shigella (4), BsaL from Burkholderia pseudomallei (5), and PrgI from Salmonella Typhimurium (6) have solved structures.
Because the needle acts as an important conduit of bacterial effectors to the host cell, current theory hypothesizes the importance of the needle as a regulatory element as well. In vivo, contact with the host cell membrane is required to initiate translocation of effectors (7). However, in vitro secretion can be triggered by artificial means, e.g., depletion of calcium ions with Yersinia and Pseudomonas T3S systems or addition of Congo red to the Shigella T3S system (8). One hypothesis of regulation via the needle is akin to how flagella turn off secretion of the hook. This hypothesis involves a protofilament that stretches the length of the needle, with the N-terminal end able to sense host cell contact while the C terminus relays the signal to the base of the T3S system in the bacterial cytosol (9). Another hypothesis suggests that the signal is relayed structurally via conformational changes of the needle from the tip to the base (9). Supporting the conformational change hypothesis, several mutants of needle proteins have been isolated that alter regulatory control of secretion (10–13); however, an exact mechanism has not been confirmed in these mutants.
As described above, contemporary research directed toward T3S systems largely centers on the structure, regulation, and role of the translocated toxins in bacterial diseases (3). Currently, only a few studies are being conducted on the roles of T3S structural components within the innate immune response. Logically, one would expect that T3S structural proteins could have important interactions with host cells. Many pathogen-associated molecular patterns (PAMPs) are already known to interact with host pattern recognition receptors (PRRs), such as Nod-like receptors (NLRs) or Toll-like receptors (TLRs) (14). Specifically recognized patterns within T3S systems include flagellin, the major component of the bacterial flagellar shaft, known to be an important molecule that interacts with TLR5 and Nlrc4 to induce cytokine expression (15). Homologs of PrgJ, a needle rod protein, interact with Nlrc4 to induce cytokines (16), and the NLRP12 inflammasome has also been shown to recognize Yersinia pestis (17). Finally, some needle proteins have been shown to activate caspase 1 via NAIP1 and Nlrc4 when delivered intracellularly, demonstrating that needle proteins can be PAMPs (18–20). The lack of evidence in the literature showing needle proteins injected naturally into host cells led us to hypothesize that the needle's location on the outside of bacteria appears to be a prime location for needle proteins to interact with PRRs, such as TLRs, outside the host cell.
The present study reveals that needle proteins induce signaling by NF-κB and/or AP-1 following interaction with TLR2 or TLR4. This activation is MyD88 dependent and results in increased cytokine expression. Crude needle protein preparations from Yersinia pestis were also able to increase NF-κB and/or AP-1 activation.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and plasmids.
Bacterial strains and plasmids used in this study are listed in Table 1. All strains were stored at −80°C in 25% glycerol (vol/vol). Escherichia coli strains were grown at 37°C in LB broth or on tryptose blood agar base (TBA) plates, with antibiotics added as needed. Antibiotics were used at the following concentrations: kanamycin, 50 μg/ml; and carbenicillin, 50 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant properties | Source or reference |
|---|---|---|
| Strains | ||
| Yersinia pestis | ||
| KIM8 | pCD1 (Lcr+) pMT1 Pla− | S. Straley |
| KIM8.P61 | pCD1 ΔyscF (Lcr+) pMT1 Pla− | 12 |
| E. coli | ||
| Novablue | recA1 endA1 hsdR17 (rK− mK+) supE44 thi-1 gyrA96 relA1 lac (F′ proA+B+) lacIqZΔM15::Tn10 | Novagen |
| BL21(DE3) Star | F− ompT hsdSB (rB− mB−) gal dcm (DE3) | Invitrogen |
| TOP10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80dlacZΔM15 ΔlacX74 nupG recA1 araD139 Δ(ara-leu)7697 galE15 galK16 rpsL (Strr) endA1 λ− | Invitrogen |
| Plasmids | ||
| pET15b MxiH | W. Picking | |
| pET200 PrgI | This study | |
| pET200 SsaG | This study | |
| pJM119 | pET24b-YscF Kmr | 21 |
| pET200 LcrG | Lab stock |
Some plasmids used to overexpress needle proteins in this study were constructed in pET200 TOPO, using Champion TOPO expression kits (Invitrogen, Carlsbad, CA). Primers for gene amplification were made by Eurofins MWG Operon, Inc. (Huntsville, AL). Primers used for cloning were as follows: previously published YscF primers (21), PrgI forward (5′-CAC CAT GGC AAC ACC TTG GTC-3′), PrgI reverse (5′-TTA ACG GAA GTT CTG AAT AAT GGC AG-3′), SsaG forward (5′-CAC CAT GGA TAT TGC ACA ATT AGT GGA TAG CTC TCC-3′), and SsaG reverse (5′-TCA GAT TTT AGC AAT GAT TCC ACT AAG CAT ATC C-3′). Template DNA for amplification was generated using a DNeasy kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. PCR was performed using Pfu Ultra polymerase (Agilent Technologies, Santa Clara, CA). Amplified DNA was then placed in pET200 by using a Champion TOPO expression kit according to the manufacturer's instructions. All of the plasmid constructs were verified by sequencing by Eurofins MWG Operon, Inc. (Huntsville, AL). The MxiH-encoding plasmid was a kind gift from William Picking, Oklahoma State University. Plasmids for protein expression were purified from E. coli TOP10 by use of a Qiaprep miniprep kit (Qiagen). Purified plasmid DNA was then transformed into the expression host, BL21(DE3) Star (Invitrogen, Carlsbad, CA).
Cell culture.
THP-1 cells were acquired from the ATCC (Manassas, VA) and grown in RPMI medium (Mediatech, Manassas, VA) supplemented with 10% (vol/vol) fetal bovine serum (Atlanta Biologicals, Flowery Branch, GA), 2 mM l-glutamine (Mediatech), 2 mM sodium pyruvate (Mediatech), 0.05 μM β-mercaptoethanol (Fisher BioReagents, Fairlawn, NJ), and 50 μg/ml Pen-Strep (Mediatech) at 37°C with 10% CO2. THP1-XBlue cells were acquired from Invivogen (San Diego, CA) and grown in RPMI medium supplemented with 10% fetal bovine serum, 100 μg/ml Normocin (Invivogen), and 50 μg/ml Pen-Strep at 37°C with 5% CO2. HEK293 cells expressing human TLR2 (HEK-Blue hTLR2 cells), TLR4 (HEK-Blue hTLR4 cells), or TLR5 (HEK-Blue hTLR5 cells) were acquired from Invivogen and grown in Dulbecco's modified Eagle's medium (DMEM) (Mediatech) supplemented with 4.5 g/liter glucose, 10% fetal bovine serum, 50 μg/ml Pen-Strep, 100 μg/ml Normocin, and 2 mM l-glutamine at 37°C and 5% CO2. THP1-XBlue and HEK-Blue TLR cells contain the secreted embryonic alkaline phosphatase (SEAP) reporter gene under the control of NF-κB and AP-1 (THP1-XBlue).
His-tagged protein isolation.
Escherichia coli BL21(DE3) carrying plasmids for a given protein was grown overnight in noninducing medium (50× M, 1 M MgSO4, 40% glucose, 5% aspartic acid) (22) supplemented with the correct antibiotic. Bacteria were then inoculated into autoinducing medium (50× M, 1 M MgSO4, 50× 5052, NZ-amine S, yeast extract, distilled water) (22) with antibiotic and grown to an A620 of 0.6 to 0.8. Cells were harvested by centrifugation at 4,000 × g for 10 min at 4°C and resuspended on ice in wash buffer (50 mM NaH2PO4, 300 mM NaCl, 10% [wt/vol] glycerol). The resulting cellular suspension was then French pressed twice at 20,000 lb/in2 to lyse cells. The lysate was clarified by centrifugation at 10,000 × g for 20 min. The clarified supernatant was collected and diluted with 1,000 ml of wash buffer before application to a preequilibrated Talon metal affinity resin (Clontech, Mountain View, CA) column. The lysate was applied to the columns twice before washing with new wash buffer. Bound protein was eluted in buffer containing 50 mM sodium phosphate, 200 mM NaCl, 150 mM imidazole, and 20% glycerol (wt/vol). Purified protein was concentrated with Amicon Ultra centrifugal filters (Millipore, Billerica, MA) and dialyzed against phosphate-buffered saline (PBS) plus 10% glycerol (wt/vol) in Slide-A-Lyzer dialysis cassettes (Thermo Fisher Scientific, Rockford, IL). Protein concentrations were determined using a Bradford protein assay kit (Thermo Fisher Scientific, Chicago, IL), and proteins were stored at −20°C for future use. Purified proteins were visualized by Coomassie blue staining of 4 to 20% SDS-PAGE gradient gels (GelCode Blue stain [Thermo Scientific, Rockford, IL]) (see Fig. S1 in the supplemental material). Monomers of purified needle proteins appeared as a dominant species (except for SsaG), but various multimers and larger bands of the proteins also appeared on the gels, consistent with previous results (e.g., purification of His6-YscF [see Fig. 1 in reference 21]). The multimers and larger bands on the gels reacted with an anti-His5 antibody (EMD Millipore, Darmstadt, Germany), demonstrating that they were composed of the recombinant proteins.
FIG 1.
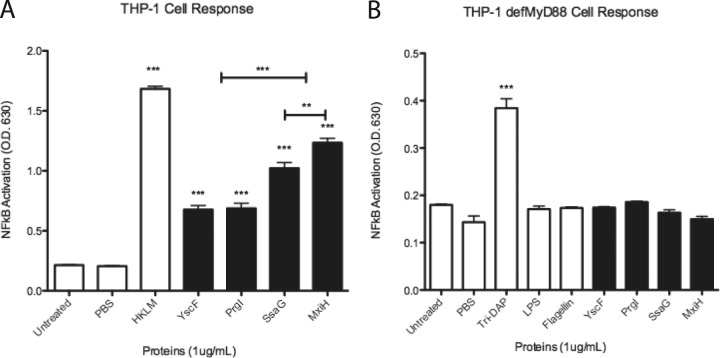
Activation of NF-κB/AP-1 in THP1-XBlue cells by needle proteins is MyD88 dependent. (A) THP1-XBlue and (B) THP1-XBlue defMyd88 cells were seeded in wells and treated with PBS or 1 μg/ml of the following: HKLM, LPS, flagellin, l-Ala-γ-d-Glu-meso-diaminopimelic acid (tri-DAP), or needle proteins dissolved in PBS. SEAP activity was measured as a representation of NF-κB/AP-1 activation. Error bars represent SEM (n = 3). Data shown are representative of at least three experiments. ***, P < 0.001; **, P is between 0.01 and 0.001. O.D.630, optical density at 630 nm.
Stimulation of cell lines by needle proteins.
THP-1 and THP1-XBlue cells were seeded at 2 × 106 cells/ml, and HEK-Blue cells at 2.5 × 105 cells/ml. Cells were suspended in infection medium (RPMI or DMEM alone) as described by the manufacturer. Proteins were added to a final concentration of 1 μg/ml or as indicated. As designated, 20 μg/ml of antibodies (control polyclonal antibody [PAb] and human TLR2 [hTLR2] PAb; Invivogen), anti-YscF, or the TLR4 inhibitor CL1-095 (1 μg/ml; Invivogen) was added to cell cultures prior to addition of needle proteins. Cells were stimulated at 37°C and 5% CO2 for 5 h or 24 h, as indicated.
Enzyme digestion and lipoprotein treatment of needle proteins or purified needles and polymyxin B treatment of THP-1 cells.
Needle proteins and flagellin (standard flagellin from Salmonella Typhimurium; Invivogen) were incubated with 40 μg/ml proteinase K (Thermo-Fisher) at 37°C for 16 h. Proteinase K was then inactivated with 1.6 mg/ml phenylmethylsulfonyl fluoride (PMSF; Sigma-Aldrich, St. Louis, MO). Needle proteins and Pam3CSK4 (Invivogen) were incubated with 50 μg/ml of bacterial lipoprotein lipase (LPL) (lipoprotein lipase from Pseudomonas sp.; Sigma-Aldrich) in 1× PBS at 37°C for 12 h. THP-1 cells were stimulated with 20 μg/ml of polymyxin B (Invivogen) and needle proteins or lipopolysaccharide (LPS) as a control for 24 h. PBS treated with proteinase K, lipoprotein lipase, or polymyxin B was used as a negative control.
LPS assay.
A ToxinSensor chromogenic LAL endotoxin assay kit (GenScript, Piscataway, NJ) was used to test for endotoxin in protein samples. The kit was used as described by the manufacturer. An amount of the endotoxin standard equivalent to that found contaminating the needle protein samples was then applied to THP1-XBlue cells as described above.
Cytokine analysis.
Cellular supernatants from THP-1 cells stimulated with PBS, heat-killed Listeria monocytogenes (HKLM), LPS, or 1 μg/ml of proteins for 5 to 24 h were collected and stored at −20°C before analysis with Quantikine enzyme-linked immunosorbent assay (ELISA) kits from R&D Systems (Minneapolis, MN). Human tumor necrosis factor alpha (TNF-α), interleukin-6 (IL-6), and IL-8 kits were used as instructed by the manufacturer.
SEAP reporter assays.
Quantification of SEAP from the supernatant was detected using Quanti-Blue reagent (Invivogen) according to the manufacturer's protocol. The color change of the Quanti-Blue reagent was measured by reading the absorbance at 630 nm, using a Synergy HT (BioTek, Winooski, VT) microplate reader, and was analyzed with KC4 v3.3 software (BioTek).
Purification of needles.
Y. pestis KIM8 cultures were grown at 37°C for 8 h in heart infusion broth (HIB). Each 8-h culture was diluted 1:100 in fresh HIB and incubated overnight at 37°C. Samples were then prepared as described by Blaylock et al. (23), with some modifications. Briefly, overnight cultures were harvested by centrifugation (10 min at 10,000 × g) and washed twice in 20 mM Tris-HCl (pH 7.5). Each cell suspension was transferred to a 40-ml Dounce glass-glass tissue grinder (Wheaton, Millville, NJ) to shear needles from the surfaces of cells, using 60 cycles. Unbroken cells and debris were removed (10 min at 10,000 × g), and the supernatant was passed through a 0.45-μm cellulose acetate membrane filter (Whatman) to remove all bacteria and centrifuged at 60,000 × g for 30 min. The pellet was suspended in 50 μl of 5% (wt/vol) sucrose in 20 mM Tris-HCl (pH 7.5), loaded onto a 6-ml step gradient of 70%, 20%, and 10% (wt/vol) sucrose (2 ml each), and centrifuged at 60,000 × g for 19 h. One-milliliter fractions were collected and analyzed by Coomassie blue staining (GelCode Blue stain; Thermo Scientific) and immunoblotting for YscF (9 kDa), which migrates as an 18-kDa dimer.
Data analysis and statistics.
Data were assembled into graphs by using GraphPad Prism, version 5.0d (GraphPad Software). Statistical analysis of SEAP levels and cytokine levels was performed using one-way analysis of variance (calculating the standard error of the mean [SEM]) with Dunnett's multiple-comparison posttest or Bonferroni's multiple-comparison posttest.
RESULTS
THP1-XBlue cells respond to needle proteins.
Evaluation of cellular responses to recombinant needle proteins by THP1-XBlue cells was accomplished by measuring SEAP production. SEAP expression in THP1-XBlue cells is under the control of NF-κB and AP-1. NF-κB and AP-1 are key transcription factors that activate cytokine and chemokine expression critical for innate immune responses. Therefore, increases in SEAP equate to increased NF-κB and/or AP-1 activity. All of the tested needle proteins were used at the same concentration (1 μg/ml of YscF, PrgI, SsaG, or MxiH). The concentration of 1 μg/ml was chosen based on an analysis of SEAP induction by various YscF concentrations. SEAP induction in THP1-XBlue cells by YscF was found to be concentration dependent (see Fig. S2 in the supplemental material). Significant levels of SEAP induction by YscF occurred at 100 ng/ml, and induction levels peaked at 5 μg/ml of YscF.
The addition of needle proteins to cultured THP1-XBlue cells resulted in significantly activated NF-κB/AP-1 (Fig. 1A) compared to that in the untreated controls. Increases in NF-κB/AP-1 activity demonstrated that host cells reacted to needle proteins with the induction of signaling pathways. Additionally, needle proteins from different bacteria triggered TLR responses of various magnitudes. YscF and PrgI increased NF-κB/AP-1 equally, but SsaG and MxiH increased NF-κB/AP-1 significantly more than YscF or PrgI did. The difference in activation seen between SsaG and MxiH was less significant. These results demonstrate that needle proteins from different species of bacteria activated NF-κB/AP-1 to various degrees.
When the needle proteins were incubated with THP1-XBlue cells deficient in MyD88, an adaptor protein required for TLR responses, the activation of NF-κB/AP-1 was abolished (Fig. 1B). The lack of response by cells lacking MyD88 indicates that the response likely occurs through TLR recognition of the needle proteins.
Protein in purified needle protein preparations is responsible for induction of SEAP in THP1-XBlue cells.
Needle proteins and flagellin digested with proteinase K were added to THP1-XBlue cells to confirm that proteins were activating NF-κB/AP-1 (Fig. 2A). Proteinase K is a serine protease that cleaves after hydrophobic amino acids. Proteinase K in PBS was used as a negative control. As expected, flagellin (a TLR5 agonist; positive control) treated with proteinase K abrogated the NF-κB/AP-1 response, and proteinase K alone did not activate NF-κB/AP-1, demonstrating that the proteinase K used did not contain a TLR agonist and that, as expected, proteinase K treatment eliminated the ability of flagellin to activate NF-κB/AP-1 (Fig. 2A). The proteinase K (Fig. 2A)-treated needle proteins also failed to elicit NF-κB/AP-1 activation, confirming that proteins caused the cellular activation of NF-κB/AP-1.
FIG 2.
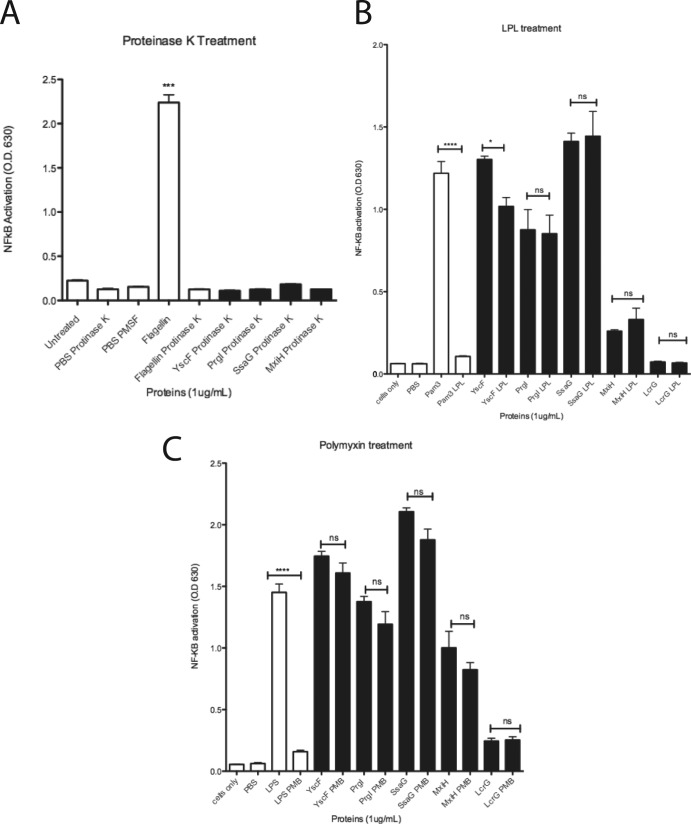
Treatment of needle proteins with proteinase K, lipoprotein lipase, and polymyxin B retains NF-κB/AP-1 activation. Needle proteins (1 μg/ml) (A, B, and C) and LcrG (1 μg/ml) (B and C), PBS, flagellin (1 μg/ml) (A), Pam3 (1 μg/ml) (B), or LPS (1 μg/ml) (C) were incubated with proteinase K (A), lipoprotein lipase (LPL; 50 μg/ml) (B), or polymyxin B (PMB; 20 μg/ml) (C) before addition to THP1-XBlue cells. *, P is between 0.05 and 0.01; ***, P is between 0.001 and 0.0001; ****, P < 0.0001; ns, not significant.
To further demonstrate that the needle proteins, not contaminants, were responsible for induction of cellular responses, the purified needle proteins and His6-LcrG (purified in the same manner as the needle proteins) were incubated with bacterial lipoprotein lipase (LPL) to inactivate bacterial lipoproteins (Fig. 2B) or treated with polymyxin B (40 μg/ml) to neutralize LPS (Fig. 2C). Treatment of YscF with LPL slightly decreased SEAP induction. Treatment of all the other needle proteins with LPL did not affect the ability of needle proteins to induce SEAP expression in THP1-XBlue cells (Fig. 2), showing that the induction seen was not due to contaminating lipoproteins. Treatment with polymyxin B resulted in no significant change in the ability of all tested needle proteins to induce SEAP expression, demonstrating that induction of SEAP was not due to LPS. Treatment of the LPS and Pam3CSK4 positive controls with either polymyxin B or lipoprotein lipase abolished their ability to induce SEAP activity by THP1-XBlue cells (Fig. 2). His6-LcrG failed to induce SEAP expression in THP1-XBlue cells, indicating that the increase in SEAP expression by THP1-XBlue cells in response to the needle proteins does not occur with all proteins purified by our method (Fig. 2A and B).
LPS levels were further analyzed to evaluate potential contamination of the needle protein samples. All samples had approximately 1 endotoxin unit (EU)/ml of LPS (see Fig. S3A in the supplemental material). To determine if 1 EU/ml of LPS had a significant effect on NF-κB/AP-1 activation, the LPS standards from the endotoxin assay kit were used to treat THP1-XBlue cells. The results indicated that 1 EU/ml of LPS did not activate NF-κB/AP-1 (see Fig. S3B). Therefore, the level of LPS in the needle protein samples was not a factor in the activation levels seen in Fig. 1 and 2.
This set of experiments demonstrated the following. (i) The inducing molecule is a protein. (ii) Purified needle proteins contain little LPS. (iii) The induction is not neutralized by polymyxin B, i.e., LPS is not an inducing factor. (iv) The purified needle proteins are not contaminated with bacterial lipoproteins. As a further control, His6-LcrG purified in the same manner as the needle proteins was applied to THP1-XBlue cells and HEK-Blue cells expressing TLR2 or TLR4 (see Fig. S4A to C in the supplemental material). The purified LcrG had no effect on any of these cell types, and SEAP activity was not significantly induced (see Fig. S4 in the supplemental material). Taken together, these results demonstrate that the needle proteins are responsible for inducing SEAP activity by THP1-XBlue cells.
TLR2-expressing HEK-Blue cells respond to needle proteins.
To elucidate the TLRs that needle proteins could signal through, an initial screening for TLR interaction with YscF was conducted by Invivogen (data not shown). HEK-Blue reporter cells transfected with a selected TLR were incubated with YscF (data not shown). The screening indicated that HEK-Blue cells expressing TLR2 or TLR4 responded to YscF (data not shown). The NF-κB/AP-1 reporter in HEK-Blue cells expressing TLR3, -5, -7, -8, or -9 did not respond to YscF (data not shown).
To confirm these results, HEK-Blue cells expressing human TLR2, TLR4, or TLR5 were acquired and used to confirm the screening results. HEK-Blue hTLR2 cells exposed to the various needle proteins showed a pattern similar to that seen with THP1-XBlue cells, where all of the tested needle proteins activated NF-κB/AP-1 (Fig. 3A). A similar result was seen with HEK-Blue cells expressing human TLR4 (Fig. 4A), and HEK-Blue cells expressing hTLR5 did not respond to needle proteins (Fig. 5).
FIG 3.
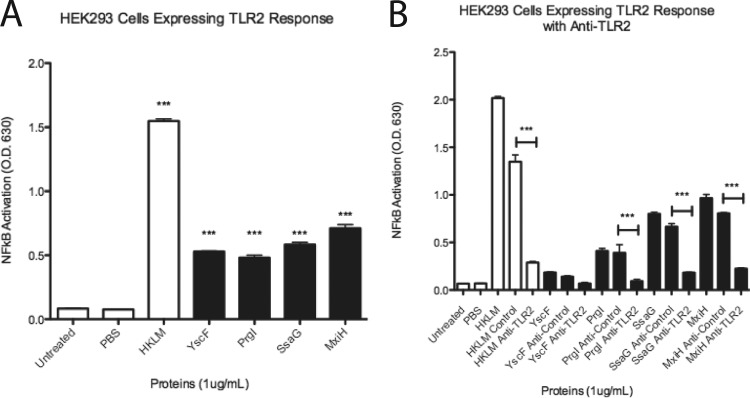
HEK293 cells expressing TLR2 responded to needle proteins, while antibody to TLR2 blocked NF-κB/AP-1 activation. (A) HEK-Blue hTLR2 reporter cells were tested with 1 μg/ml of needle proteins dissolved in PBS. HKLM at 1 μg/ml was used as a control for TLR2 activation. (B) HEK-Blue hTLR2 cells were treated with no antibody, an antibody control, or antibody to TLR2 and then treated with 1 μg/ml of HKLM or needle proteins. SEAP activity was measured as a representation of NF-κB/AP-1 activation. Error bars represent SEM (n = 3). Data shown are representative of at least three experiments. ***, P < 0.001.
FIG 4.
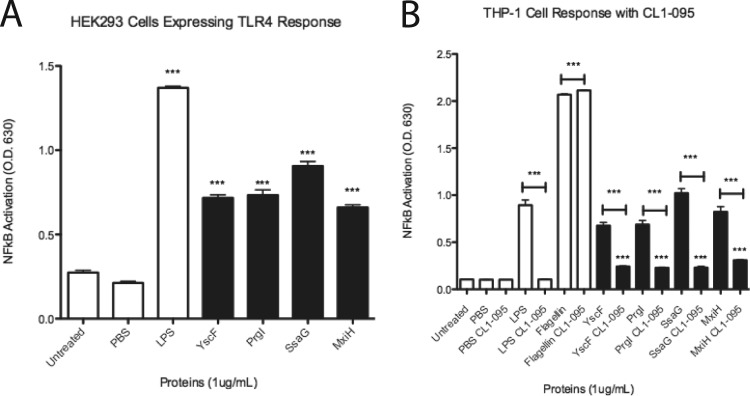
Needle proteins activate NF-κB/AP-1 through TLR4, and NF-κB/AP-1 activation is inhibited by CL1-095, a TLR4 inhibitor. (A) HEK-Blue hTLR4 cells were tested with 1 μg/ml of needle proteins dissolved in PBS. LPS at 1 μg/ml was used as the positive control for TLR4. (B) THP1-XBlue cells were treated with PBS or 1 μg/ml of the following: LPS, flagellin, or needle proteins. Prior to treatment, cells were either left untreated or treated with the TLR4 inhibitor CL1-095. SEAP activity was measured as a representation of NF-κB/AP-1 activation. Error bars represent SEM (n = 3). Data shown are representative of at least three experiments. ***, P < 0.001.
FIG 5.
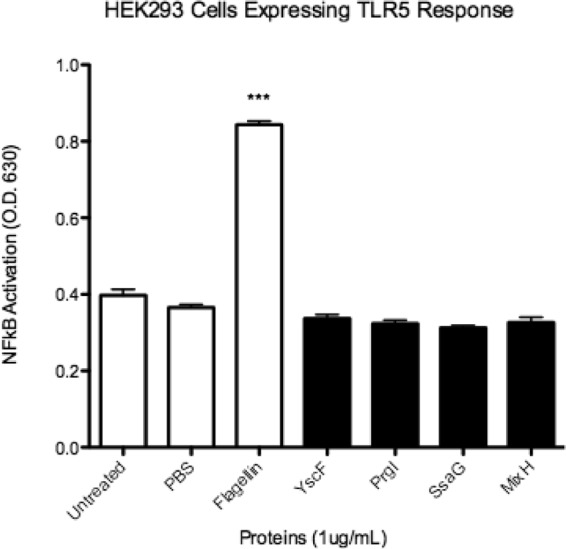
Needle proteins do not activate NF-κB/AP-1 through TLR5. HEK-Blue hTLR5 cells were tested with 1 μg/ml of needle proteins dissolved in PBS. Flagellin (1 μg/ml) was used as a positive control. SEAP activity was measured as a representation of NF-κB/AP-1 activation. Error bars represent SEM (n = 3). Data shown are representative of at least three experiments. ***, P < 0.001.
TLR2 antibodies block activation of HEK 293 cells expressing TLR2 by needle proteins.
Antibodies to TLR2 were utilized to assess interaction of needle proteins with TLR2 in HEK-Blue hTLR2 cells. Control antibodies and TLR2 antibodies were administered at equal concentrations to HEK-Blue hTLR2 cell cultures. Administration of the isotype control antibodies followed by subsequent incubation with needle proteins (YscF, PrgI, MxiH, and SsaG) resulted in no significant decrease of NF-κB/AP-1 activation by the needle proteins (Fig. 3B). After treatment of the cell cultures with TLR2 antibody and subsequent incubation with needle proteins, NF-κB/AP-1 activation was significantly decreased compared to that with control antibody in the case of PrgI, MxiH, and SsaG (Fig. 3B). YscF was not significantly decreased; however, YscF alone had low activation of NF-κB/AP-1 (Fig. 3B) in this experiment. These results confirm that the needle proteins activate NF-κB/AP-1 through TLR2, as neutralization of TLR2 with antibodies abrogated the needle protein-induced response in HEK-Blue hTLR2 cells.
HEK-Blue hTLR4 cells respond to needle proteins.
HEK-Blue hTLR4 cells were also tested, with LPS as a positive control and with the needle proteins YscF, PrgI, SsaG, and MxiH. As expected, LPS increased NF-κB/AP-1 activation of SEAP, as did all the tested needle proteins (Fig. 4A). This result suggests that the needle proteins can interact with TLR4.
TLR4 inhibitor CL1-095 inhibits the response of THP1-Xblue cells to needle proteins.
The TLR4 inhibitor CL1-095 (inhibits the intracellular domain of TLR4) was used to treat THP1-XBlue cells prior to treatment with LPS, flagellin, or needle proteins (YscF, PrgI, MxiH, and SsaG). Cells left untreated with CL1-095 responded to LPS and needle proteins in the same pattern as that seen previously (Fig. 4B). The cells treated with CL1-095 did not respond to LPS, although the response to flagellin was left intact, as expected, and the response to needle proteins was significantly reduced in all cases, although not quite to basal levels, suggesting that activation of THP1-XBlue cells by needle proteins can be reduced by interfering with intracellular signaling induced through TLR4.
HEK-Blue hTLR5 cells do not respond to needle proteins.
Since the HEK-Blue hTLR2 and hTLR4 cells endogenously express TLR5, we also tested HEK-Blue hTLR5 cells with needle proteins. HEK-Blue hTLR5 cells showed no response to any of the needle proteins, indicating that the proteins were specifically targeting TLR2 and TLR4 (Fig. 5). Taken together, the data support the idea that needle proteins are recognized as a PAMP by TLR2 and, potentially, TLR4.
Cytokine expression in response to needle proteins.
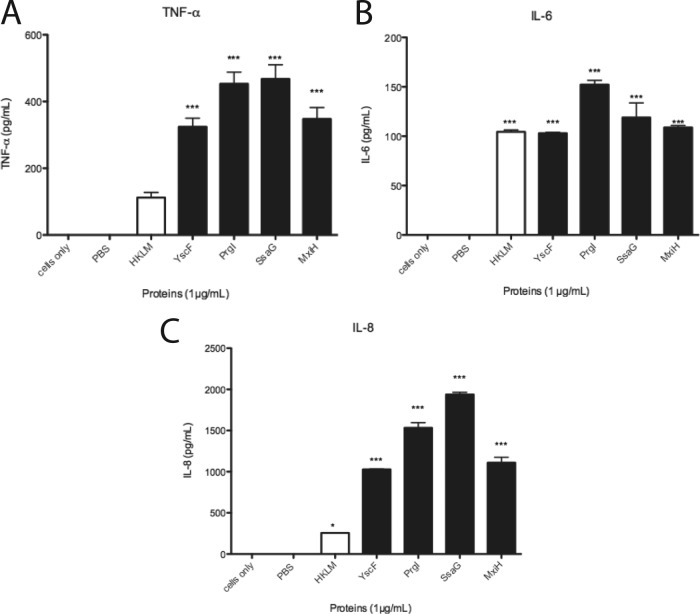
Because the needle protein-induced NF-κB/AP-1 responses in THP1-XBlue cells were detected using a reporter gene, we wanted to examine if those responses translated to cytokine secretion from nontransfected THP-1 cells. Therefore, THP-1 cells were treated with PBS, HKLM, or needle proteins. After 5 h, supernatants were collected and assessed via ELISA for TNF-α, and after 24 h, supernatants were collected and analyzed for IL-6 and IL-8 production. As expected, PBS-treated cells produced no TNF-α, IL-6, or IL-8. HKLM and all needle protein-treated cells led to production of all 3 cytokines (Fig. 6). These results indicated that the NF-κB/AP-1 activation triggered by needle proteins led to increased expression of TNF-α, IL-6, and IL-8 that resulted in secretion of the cytokines into cell culture supernatants.
FIG 6.
Activation of THP-1 cells by needle proteins results in cytokine secretion. THP-1 cells were treated with PBS, 1 μg/ml of HKLM, or needle proteins (1 μg/ml). After 5 h (TNF-α) and 24 h (IL-6 and IL-8), supernatants were collected and tested by ELISA for production of TNF-α (A), IL-6 (B), and IL-8 (C). Error bars represent SEM (n = 3). Data shown are representative of at least three experiments. ***, P < 0.001; *, P value of 0.01 to 0.05.
THP1-XBlue cells respond to sheared Ysc needles.
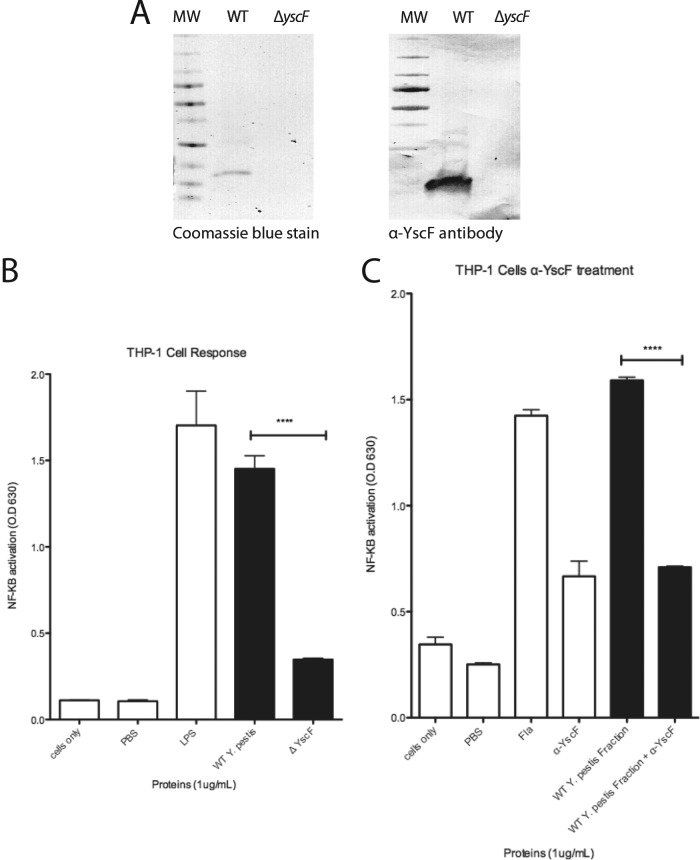
Our results demonstrated that recombinant needle proteins purified from E. coli activated NF-κB/AP-1, leaving the question of whether native needle proteins could activate NF-κB/AP-1. Accordingly, purified needles from Y. pestis were tested for the ability to activate NF-κB/AP-1. Needles from Yersinia pestis were sheared from the bacterial surface to obtain YscF. The presence of purified YscF was confirmed by Coomassie blue staining and immunoblotting with YscF primary antibody (Fig. 7A). Addition of purified YscF to THP1-XBlue cells resulted in SEAP expression, while addition of a mock purified fraction from ΔyscF Y. pestis had no effect, demonstrating that native YscF could also induce NF-κB/AP-1 (Fig. 7B). Treatment of needle fractions with lipoprotein lipase and THP1-XBlue cell treatment with polymyxin B indicated no contamination of lipoproteins or LPS (see Fig. S5 in the supplemental material). Activation of NF-κB/AP-1 in THP1-XBlue cells was seen in response to the YscF-enriched fraction (Fig. 7B), suggesting not only that recombinant needle proteins activate host cells but also that native YscF induces NF-κB/AP-1. This was further confirmed by obtaining a fraction from a strain of Y. pestis lacking yscF that did not induce NF-κB/AP-1. To confirm that YscF was activating NF-κB/AP-1, the THP1-XBlue cells were treated with anti-YscF alone or anti-YscF with native YscF (Fig. 7C). Antibody to YscF significantly inhibited activation of NF-κB/AP-1. This result demonstrates that neutralizing YscF can abrogate the effect of YscF to activate NF-κB/AP-1.
FIG 7.
THP-1 X-Blue cells respond to YscF from Y. pestis. (A) Coomassie blue-stained gel and immunoblot of YscF purified from Y. pestis, detected with rabbit anti-YscF primary antibody. (B) THP1-XBlue cells were seeded in wells and treated with PBS, 1 μg/ml of LPS, purified YscF needle fractions (1 μg/ml) from Y. pestis, or a mock purified preparation from a Y. pestis ΔyscF strain. (C) THP1-XBlue cells were treated with flagellin (1 μg/ml), anti-YscF alone, YscF (1 μg/ml), or YscF incubated with anti-YscF. SEAP activity was measured as a representation of NF-κB/AP-1 activation. Error bars represent SEM (n = 3). Data shown are representative of at least three experiments. ****, P value of 0.001 to 0.01.
DISCUSSION
Characterization of the ability of the needle proteins YscF, PrgI, SsaG, and MxiH to induce innate immune responses revealed the following. (i) NF-κB/AP-1 is activated by needle proteins. (ii) Activation of NF-κB/AP-1 by needle proteins is dependent upon MyD88. (iii) Activation occurs through TLR2 or TLR4. (iv) Native YscF purified from Y. pestis is also able to activate NF-κB/AP-1. (v) The observed variation between needle proteins of different bacteria may indicate that each needle protein uniquely activates eukaryotic cells. Induction of innate immune responses by His6-YscF was concentration dependent and saturable, demonstrating specificity: significant induction began at ∼100 ng/ml of His6-YscF and peaked at 5 μg/ml of His6-YscF. The concentration of YscF during an infection is unknown. However, concentrations of other virulence factors during Y. pestis infection have been measured. Physiologically, the needle tip protein LcrV and the Y. pestis capsule protein F1 are found in mouse serum at concentrations of 1.6 to 26.4 ng/ml and 6.9 to 754 ng/ml, respectively (24).
Obviously, our study is not an exhaustive characterization of needle proteins but represents only a few bacteria that use T3S systems from different T3S families (3). Interestingly, the T3S systems used in this study utilize T3S in three different manners: anti-inflammatory (YscF), proinflammatory (PrgI and MxiH), and intravacuolar (SsaG). YscF comes from a T3S system from pathogens that have anti-inflammatory infection objectives (25). YscF elicited lower levels of NF-κB/AP-1 activation from THP-1 cells and induced modestly lower levels of TNF-α, IL-6, and IL-8 than those with PrgI or SsaG. The varied responses induced by needle proteins are consistent with the previously published observation that some needle proteins (EscF and Vibrio parahaemolyticus YscF) did not induce caspase-1 through Nlrc4, unlike PrgI and MxiH (18). Shigella and Salmonella are known to cause largely proinflammatory responses to the host in order to cause disease (26). The needle protein MxiH proved to be in accordance with this overarching goal of infection, as it induced increased NF-κB/AP-1 but induced lower levels of TNF-α, IL-6, and IL-8 than those with PrgI or SsaG. PrgI acted similarly to YscF in NF-κB/AP-1 activation but had increased induction of cytokines relative to that with YscF. It is known that other factors (e.g., flagellin, LPS, and SopE) play a significant role in the proinflammatory response of Salmonella (27). The Salmonella SPI-2 needle protein SsaG had the highest levels of activation among the four tested needle proteins. Interestingly, SsaG is not exposed to TLRs expressed on the outside of the host cell, since SPI-2 is expressed only once Salmonella is enclosed in the Salmonella-containing vacuole (27) inside the host cell.
TLR4 is already known to play a distinctive role in Yersinia pestis pathogenesis in that Yersinia pestis produces a unique tetra-acylated LPS (28). The tetra-acylated LPS antagonizes binding of hexa-acylated LPS to TLR4, resulting in deficient activation of the immune system (29). This antagonistic role could prevent other TLR4 ligands from binding as well. TLR4's role in recognition of needle proteins was less expected than that of the more promiscuous TLR2. However, there are several documented cases of TLR4 interaction with pathogen-associated substrates other than LPS, including respiratory syncytial virus (RSV) fusion protein (30), chlamydial Hsp60 (31), pneumolysin (32), Francisella tularensis DnaK (33), Ebola virus glycoprotein (34), and cell wall components from Pseudallescheria boydii (35). The interaction of these PAMPs and TLR4 are well known, and future research will hopefully elucidate how needle proteins interact with TLR4, including whether MD-2 or CD-14 is required for this interaction.
Activation of NF-κB/AP-1 by native YscF could indicate that the key element in activating TLRs is exposed on the outside of the needle. Recent studies of needle structures from Salmonella (36) and Shigella (37) indicate that the nonconserved N terminus of the needle protein is located on the outside of the intact needle structure. The differences in the N termini of different needle proteins could result in the differences seen in NF-κB/AP-1 activation and cytokine production between species and, ultimately, alter the innate host response to the bacteria. Currently, we do not know the specifics of which amino acid sequences play a role in interactions of needle proteins and TLRs; however, this topic is under investigation. Understanding the interaction of needle proteins and TLRs has the potential to increase our knowledge of host responses to T3S-utilizing pathogens and to extend our knowledge of how T3S apparatuses may interact with other host processes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Bill and Wendy Picking (Oklahoma State University, Stillwater, OK) for advice and for providing the MxiH plasmid and Laura Grabanski, Nathaniel D. Lambert, and Thomas Henderson for their assistance with some of the studies.
Funding was provided by a University of North Dakota School of Medicine and Health Sciences faculty seed grant and by Novadigm, Inc.
Footnotes
Published ahead of print 18 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01705-14.
REFERENCES
- 1. Galán JE, Wolf-Watz H. 2006. Protein delivery into eukaryotic cells by type III secretion machines. Nature 444:567–573. 10.1038/nature05272 [DOI] [PubMed] [Google Scholar]
- 2. Preston GM. 2007. Metropolitan microbes: type III secretion in multihost symbionts. Cell Host Microbe 2:291–294. 10.1016/j.chom.2007.10.004 [DOI] [PubMed] [Google Scholar]
- 3. Cornelis GR. 2006. The type III secretion injectisome. Nat. Rev. Microbiol. 4:811–825. 10.1038/nrmicro1526 [DOI] [PubMed] [Google Scholar]
- 4. Deane JE, Cordes FS, Roversi P, Johnson S, Kenjale R, Picking WD, Picking WL, Lea SM, Blocker A. 2006. Expression, purification, crystallization and preliminary crystallographic analysis of MxiH, a subunit of the Shigella flexneri type III secretion system needle. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 62:302–305. 10.1107/S1744309106006555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang L, Wang Y, Picking WL, Picking WD, De Guzman RN. 2006. Solution structure of monomeric BsaL, the type III secretion needle protein of Burkholderia pseudomallei. J. Mol. Biol. 359:322–330. 10.1016/j.jmb.2006.03.028 [DOI] [PubMed] [Google Scholar]
- 6. Wang Y, Ouellette AN, Egan CW, Rathinavelan T, Im W, De Guzman RN. 2007. Differences in the electrostatic surfaces of the type III secretion needle proteins PrgI, BsaL, and MxiH. J. Mol. Biol. 371:1304–1314. 10.1016/j.jmb.2007.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pettersson J, Nordfelth R, Dubinina E, Bergman T, Gustafsson M, Magnusson KE, Wolf-Watz H. 1996. Modulation of virulence factor expression by pathogen target cell contact. Science 273:1231–1233. 10.1126/science.273.5279.1231 [DOI] [PubMed] [Google Scholar]
- 8. Enninga J, Rosenshine I. 2009. Imaging the assembly, structure and activity of type III secretion systems. Cell. Microbiol. 11:1462–1470. 10.1111/j.1462-5822.2009.01360.x [DOI] [PubMed] [Google Scholar]
- 9. Blocker AJ, Deane JE, Veenendaal AK, Roversi P, Hodgkinson JL, Johnson S, Lea SM. 2008. What's the point of the type III secretion system needle? Proc. Natl. Acad. Sci. U. S. A. 105:6507–6513. 10.1073/pnas.0708344105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Harmon DE, Murphy JL, Davis AJ, Mecsas J. 2013. A mutant with aberrant extracellular LcrV-YscF interactions fails to form pores and translocate Yops, but retains the ability to trigger Yop secretion in response to host cell contact. J. Bacteriol. 195:2244–2254. 10.1128/JB.02011-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kenjale R, Wilson J, Zenk SF, Saurya S, Picking WL, Picking WD, Blocker A. 2005. The needle component of the type III secreton of Shigella regulates the activity of the secretion apparatus. J. Biol. Chem. 280:42929–42937. 10.1074/jbc.M508377200 [DOI] [PubMed] [Google Scholar]
- 12. Torruellas J, Jackson MW, Pennock JW, Plano GV. 2005. The Yersinia pestis type III secretion needle plays a role in the regulation of Yop secretion. Mol. Microbiol. 57:1719–1733. 10.1111/j.1365-2958.2005.04790.x [DOI] [PubMed] [Google Scholar]
- 13. Sato H, Frank DW. 2011. Multi-functional characteristics of the Pseudomonas aeruginosa type III needle-tip protein, PcrV; comparison to orthologs in other Gram-negative bacteria. Front. Microbiol. 2:142. 10.3389/fmicb.2011.00142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kawai T, Akira S. 2006. TLR signaling. Cell Death Differ. 13:816–825. 10.1038/sj.cdd.4401850 [DOI] [PubMed] [Google Scholar]
- 15. Miao EA, Andersen-Nissen E, Warren SE, Aderem A. 2007. TLR5 and Ipaf: dual sensors of bacterial flagellin in the innate immune system. Semin. Immunopathol. 29:275–288. 10.1007/s00281-007-0078-z [DOI] [PubMed] [Google Scholar]
- 16. Miao EA, Warren SE. 2010. Innate immune detection of bacterial virulence factors via the NLRC4 inflammasome. J. Clin. Immunol. 30:502–506. 10.1007/s10875-010-9386-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vladimer GI, Weng D, Paquette SW, Vanaja SK, Rathinam VA, Aune MH, Conlon JE, Burbage JJ, Proulx MK, Liu Q, Reed G, Mecsas JC, Iwakura Y, Bertin J, Goguen JD, Fitzgerald KA, Lien E. 2012. The NLRP12 inflammasome recognizes Yersinia pestis. Immunity 37:96–107. 10.1016/j.immuni.2012.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhao Y, Yang J, Shi J, Gong YN, Lu Q, Xu H, Liu L, Shao F. 2011. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature 477:596–600. 10.1038/nature10510 [DOI] [PubMed] [Google Scholar]
- 19. Rayamajhi M, Zak DE, Chavarria-Smith J, Vance RE, Miao EA. 2013. Mouse NAIP1 detects the type III secretion system needle protein. J. Immunol. 191:3986–3989. 10.4049/jimmunol.1301549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yang J, Zhau Y, Shi J, Shao F. 2013. Human NAIP and mouse NAIP1 recognize bacterial type III secretion needle protein for inflammasome activation. Proc. Natl. Acad. Sci. U. S. A. 110:14408–14413. 10.1073/pnas.1306376110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Matson JS, Durick KA, Bradley DS, Nilles ML. 2005. Immunization of mice with YscF provides protection from Yersinia pestis infections. BMC Microbiol. 5:38. 10.1186/1471-2180-5-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Studier FW. 2005. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41:207–234. 10.1016/j.pep.2005.01.016 [DOI] [PubMed] [Google Scholar]
- 23. Blaylock B, Berube BJ, Schneewind O. 2010. YopR impacts type III needle polymerization in Yersinia species. Mol. Microbiol. 75:221–229. 10.1111/j.1365-2958.2009.06988.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Flashner Y, Fisher M, Tidhar A, Mechaly A, Gur D, Halperin G, Zahavy E, Mamroud E, Cohen S. 2010. The search for early markers of plague: evidence for accumulation of soluble Yersinia pestis LcrV in bubonic and pneumonic mouse models of disease. Immunol. Med. Microbiol. 59:197–206. 10.1111/j.1574-695X.2010.00687.x [DOI] [PubMed] [Google Scholar]
- 25. Amedei A, Niccolai E, Marino L, D'Elios MM. 2011. Role of immune response in Yersinia pestis infection. J. Infect. Dev. Ctries. 5:628–639. 10.3855/jidc.1999 [DOI] [PubMed] [Google Scholar]
- 26. Phalipon A, Sansonetti PJ. 2007. Shigella's ways of manipulating the host intestinal innate and adaptive immune system: a tool box for survival? Immunol. Cell Biol. 85:119–129. 10.1038/sj.icb7100025 [DOI] [PubMed] [Google Scholar]
- 27. Broz P, Ohlson MB, Monack DM. 2012. Innate immune response to Salmonella typhimurium, a model enteric pathogen. Gut Microbes 3:62–70. 10.4161/gmic.19141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kawahara K, Tsukano H, Watanabe H, Lindner B, Matsuura M. 2002. Modification of the structure and activity of lipid A in Yersinia pestis lipopolysaccharide by growth temperature. Infect. Immun. 70:4092–4098. 10.1128/IAI.70.8.4092-4098.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Telepnev MV, Klimpel GR, Haithcoat J, Knirel YA, Anisimov AP, Motin VL. 2009. Tetraacylated lipopolysaccharide of Yersinia pestis can inhibit multiple Toll-like receptor-mediated signaling pathways in human dendritic cells. J. Infect. Dis. 200:1694–1702. 10.1086/647986 [DOI] [PubMed] [Google Scholar]
- 30. Rallabhandi P, Phillips RL, Boukhvalova MS, Pletneva LM, Shirey KA, Gioannini TL, Weiss JP, Chow JC, Hawkins LD, Vogel SN, Blanco JC. 2012. Respiratory syncytial virus fusion protein-induced Toll-like receptor 4 (TLR4) signaling is inhibited by the TLR4 antagonists Rhodobacter sphaeroides lipopolysaccharide and eritoran (E5564) and requires direct interaction with MD-2. mBio 3:e00218-12. 10.1128/mBio.00218-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bulut Y, Faure E, Thomas L, Karahashi H, Michelsen KS, Equils O, Morrison SG, Morrison RP, Arditi M. 2002. Chlamydial heat shock protein 60 activates macrophages and endothelial cells through Toll-like receptor 4 and MD2 in a MyD88-dependent pathway. J. Immunol. 168:1435–1440 [DOI] [PubMed] [Google Scholar]
- 32. Malley R, Henneke P, Morse SC, Cieslewicz MJ, Lipsitch M, Thompson CM, Kurt-Jones E, Paton JC, Wessels MR, Golenbock DT. 2003. Recognition of pneumolysin by Toll-like receptor 4 confers resistance to pneumococcal infection. Proc. Natl. Acad. Sci. U. S. A. 100:1966–1971. 10.1073/pnas.0435928100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ashtekar AR, Zhang P, Katz J, Deivanayagam CC, Rallabhandi P, Vogel SN, Michalek SM. 2008. TLR4-mediated activation of dendritic cells by the heat shock protein DnaK from Francisella tularensis. J. Leukoc. Biol. 84:1434–1446. 10.1189/jlb.0308215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Okumura A, Pitha PM, Yoshimura A, Harty RN. 2010. Interaction between Ebola virus glycoprotein and host Toll-like receptor 4 leads to induction of proinflammatory cytokines and SOCS1. J. Virol. 84:27–33. 10.1128/JVI.01462-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Figueiredo RT, Bittencourt VC, Lopes LC, Sassaki G, Barreto-Bergter E. 2012. Toll-like receptors (TLR2 and TLR4) recognize polysaccharides of Pseudallescheria boydii cell wall. Carbohydr. Res. 356:260–264. 10.1016/j.carres.2012.02.028 [DOI] [PubMed] [Google Scholar]
- 36. Demers J-P, Kubori T, Sgourakis NG, Gupta R, Loquet A, Giller K, Riedel D, Laube B, Kolbe M, Baker D, Becker S, Lange A. 2013. The common structural architecture of Shigella flexneri and Salmonella typhimurium type three secretion needles. PLoS Pathog. 9:e1003245. 10.1371/journal.ppat.1003245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Loquet A, Sgourakis NG, Gupta R, Giller K, Riedel D, Goosmann C, Kolbe M, Baker D, Becker S, Lange A. 2012. Atomic model of the type III secretion system needle. Nature 486:276–279. 10.1038/nature11079 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.