Abstract
Purpose
Non-Small Cell Lung Cancer (NSCLC) is the leading cause of cancer mortality in the world. Novel diagnostic biomarkers may augment both existing NSCLC screening methods as well as molecular diagnostic tests of surgical specimens to more accurately stratify and stage candidates for adjuvant chemotherapy. Hypermethylation of CpG islands is a common and important alteration in the transition from normal tissue to cancer.
Experimental Design
Following previously validated methods for the discovery of cancer-specific hypermethylation changes we treated 8 NSCLC cell lines with the hypomethylating agent deoxyazacitidine or trichostatin A. We validated the findings using a large publically available database and two independent cohorts of primary samples.
Results
We identified >300 candidate genes. Using The Cancer Genome Atlas (TCGA) and employing extensive filtering to refine our candidate genes for the greatest ability to distinguish tumor from normal, we define a three-gene panel, CDO1, HOXA9, and TAC1, which we subsequently validate in two independent cohorts of primary NSCLC samples. This 3-gene panel is 100% specific, showing no methylation in 75 TCGA normal and 7 primary normal samples and is 83–99% sensitive for NSCLC depending on the cohort.
Conclusion
This degree of sensitivity and specificity may be of high value to diagnose the earliest stages of NSCLC. Addition of this 3-gene panel to other previously validated methylation biomarkers holds great promise in both early diagnosis and molecular staging of NSCLC.
Introduction
Non-Small Cell Lung Cancer (NSCLC) is the leading cause of cancer related mortality worldwide.(1, 2) While improvements in the treatment of advanced stage lung malignancies have been made, including agents targeting specific genetic aberrations, epigenetic therapies, and exploiting the potential of the immune system to assert control over tumor growth, lung cancer remains the main cause of cancer related deaths.(3–5) Cancer-specific molecular changes have utility not only as targets for therapy but also as biomarkers for the determination of risk of recurrence for early stage lung cancer. Such prognostic capability may be due to the biological significance of the alteration or because detection of molecular alterations in lymph nodes may herald a higher stage of disease than is detectable by current pathology standards.(6, 7)
There is also much interest in early detection strategies to improve outcomes in lung cancer, which have culminated in the landmark National Lung Screening Trial (NLST). Although the 20% relative reduction in lung cancer mortality in the NLST low-dose CT screening arm is encouraging, it belies a false positive rate among screening results of 96.4% which has resulted in some pause among clinicians and payers alike for immediate widespread adoption of the technique.(8) Improved techniques or ancillary testing methods to augment both the sensitivity and specificity of screening for lung cancer could augment CT screening.
The most promising non-radiologic ancillary tests involve the detection of cancer-specific events in tissues or fluids carrying tumor cells or tumor DNA, such as lymph node samples, sputum, or plasma. Since cancer specific DNA methylation events are common and occur early in lung cancer progression, recent studies have used nested methylation-specific PCR (MSP) for detection of promoter methylation in sputum.(9, 10) For example, using PAX5α, GATA5, and SULF2, genes derived from studies of genes with known biologic importance in NSCLC demonstrated the ability to predict the outcome of a diagnosis of lung cancer in two high risk cohorts.(11–14) While these studies demonstrate the feasibility of molecular detection of altered, cancer-specific DNA methylation in sputum, there remains a need for improvement in the panel of markers employed. The measure of success expected from a test lies in the frequency of the event (sensitivity) and the absence of the event in normal samples (specificity). In this work, we seek to build upon approaches that define the most highly sensitive and specific markers of cancer, which have often been found to be linked to polycomb-associated sites in embryonic stem cells, towards the deployment of a clinically useful assay.(15–17) We hypothesized that the current genes employed in strategies to assess presence or absence of lung cancer based on sputum and other bodily tissues and fluids may be augmented by a method combing pre-clinical and population-based studies to identify the most highly sensitive and specific methylation events in lung cancer.
Here we report the discovery and characterization of genomic changes in DNA methylation occurring in association with a described biologic program, moving from the study of individual loci to a comprehensive analysis of alterations in NSCLC with the intention of uncovering epigenetic events which may predict a cancer’s natural history or be utilized for the molecular detection of disease. This study provides a method for systematic discovery of epigenetic biomarkers which may be used for improving the screening and diagnosis of this deadly disease.
Materials and Methods
Cell Culture and Treatment
All NSCLC cell lines were purchased from the American Type Culture Collection (ATCC). H838, H23, H1993, H1568, H2170 and H520 were cultured in RPMI 1640 medium (Mediatech, Inc.); H1869 was cultured in DMEM/F-12 Medium and SK-MES-1 was cultured in DMEM (Mediatech, Inc.). Cell lines H838, H23, H1993, H1568 were derived from adenocarcinomas and H2170, H520, H1869, SK-MES-1 were derived from squamous cell carcinomas. Cell lines of squamous carcinoma and adenocarcinoma histology are represented equally so that cancer specific, rather than histology specific markers may be elicited by the experimental method. All cell culture media were supplemented with 10% BCS and incubated in humidified air and 5% CO2 at 37°C. For drug treatments, log phase cells were cultured in growth media containing 10% BCS and 1x penicillin/streptomycin with 5 μM decitabine (DAC) (Sigma; stock solution: 1mM in PBS) for 96 hours, replacing fresh media and DAC every 24 hours. Cell treatment with 300 nM Trichostatin A (TSA) (Sigma; stock solution: 1.5 mM dissolved in ethanol) was performed for 18 hours. Control cells underwent mock treatment in parallel with addition of equal volumes of PBS or ethanol without drugs.
Microarray analysis
RNA was harvested from cells in log phase growth using TRIzol (Invitrogen) and the RNeasy kit with DNase digestion (Qiagen) according to the manufacturer’s instructions. RNA was quantified using the NanoDrop ND-100 followed by quality assessment with the 2100 Bioanalyzer (Agilent Technologies). RNA concentrations for each sample was greater than 200ng/μl, with 28S/18S ratios greater than 2.2 and RNA integrity scores of 10 (10 scored as the highest). Sample amplification and labeling procedures were carried out using the Low RNA Input Fluorescent Linear Amplification Kit (Agilent Technologies). The labeled cRNA was purified using the RNeasy mini kit (Qiagen) and quantified. RNA spike-in controls (Agilent Technologies) were added to RNA samples before amplification. Samples (0.75 μg) labeled with Cy3 or Cy5 were mixed with control targets (Agilent Technologies), assembled on Oligo Microarray, hybridized and processed according to the Agilent microarray protocol. Scanning was performed with the Agilent G2505B microarray scanner using settings recommended by Agilent Technologies. Microarray data are available in the ArrayExpress database under accession number E-MTAB-1939.
Data analysis for microarray
Quality checks for all arrays included visual inspection for artifacts and the distribution of signal and background intensity for red and green channels. All arrays passed quality checks and were used. The statistical platform R and packages from Bioconductor were used for all computation.(18, 19) The log ratio of red signal to green signal was calculated after background subtraction and LoEss normalization as implemented in the limma package from Bioconductor.(20) Individual arrays were scaled to have the same inter-quartile range (75th percentile–25th percentile).
Methylation and gene expression analysis
RNA was isolated with TRIzol Reagent (Invitrogen) according to the manufacturer’s instructions. For RT-PCR, 1μg of total RNA was reverse transcribed using SuperScript™ First-Strand Synthesis System for RT-PCR (Invitrogen). For methylation-specific PCR (MSP) analysis, DNA was extracted following a standard phenol-chloroform extraction method. Bisulfite modification of genomic DNA was carried out using the EZ DNA methylation Kit (Zymo Research). Primer sequences specific to unmethylated and methylated promoter sequences were designed using MSPPrimer.(21) MSP was performed as previously described.(22) 10 μl of all PCR products were loaded directly onto 2% agarose gels containing GelStar Nucleic Acid Gel Stain (Cambrex Corp.) and visualized under ultraviolet illumination. Primer sequences and conditions for MSP are available upon request.
Human Tissue Analysis
Fifty-nine primary lung cancers were obtained from Johns Hopkins Hospital in Baltimore, MD (Cohort A) and 30 from Shinshu University Hospital in Matsumoto, Japan (Cohort B). All tissues were immediately frozen at −80°C after surgical resection. Normal lung cDNA was purchased from DNA Technologies Inc. Six normal lung tissues were obtained from individuals without cancer (five from autopsy and one from lung peripheral to a benign bronchial tumor). Tissue acquisition was conducted under approved guidelines of the institutional review boards from both institutions. Histological examination was based on WHO classification criteria.(23) Clinical staging was done according to Mountain and Dresler’s tumor-node-metastasis classification criteria.(24)
TCGA Analysis Data and Methods
We used the DNA methylation data of 409 lung adenocarcinoma samples with 32 matched normal samples as well as 227 lung squamous cell carcinoma samples with 43 matched normal samples from the Cancer Genome Atlas project (TCGA).(25, 26) DNA methylation was measured on the Illumina HumanMethlation 450K platform.(18, 27)
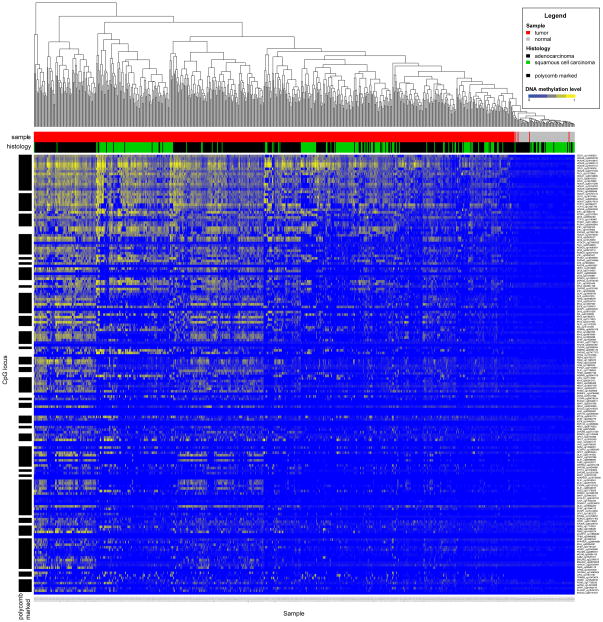
The analysis of DNA methylation data was performed using R/Bioconductor software with the limma package and custom routines for data analysis.(18, 19, 28) We selected only those probes for sites situated within CpG-island promoters of genes unmethylated at their promoter sites in all normal TCGA samples (β value < 0.2). For each probe we estimated a t-statistic and p-value by fitting a linear model of its differential methylation between tumor and normal samples.(29) All probes tested had adjusted p-values less than 1×10−4. Figure 1 shows a heat map of DNA methylation level for each site (in rows) for all tumor and normal samples (in columns). The columns of the heat map were ordered by unsupervised clustering, while rows were ordered top-to-bottom by decreasing value of significance for t-statistic for differential methylation. The sites and corresponding statistics for all probes can be found in Supplemental Table 1.
Figure 1. Cancer Specific DNA Methylation Discriminates NSCLC Tumors from Normal Lung Samples.
Methylation data are derived from 636 NSCLC in the Cancer Genome Atlas representing 227 lung squamous carcinomas with 43 matched normal samples and 409 lung non-squamous carcinomas with 32 matched normal samples. Columns represent tumor or normal tissue samples. Rows represent individual methylation probes from the Infinium methylation array. The ability of each probe to discriminate tumor versus normal and an associated t-statistic was estimated by a linear model for each CpG island promoter probe. Only probes with significant p-values are included in the heatmap. Rows are ordered from top-to-bottom by p-value. All p-values are < 0.0001. Probes with mean Beta-values >0.2 in normal samples were excluded from the analysis. Of the 305 genes exhibiting DAC-specific upregulation, 63 genes represented by 172 methylation probes met the preceding criteria. Columns are ordered by unsupervised hierarchical clustering. A few tumors cluster with normal samples. This is consistent with prior TCGA analyses that demonstrate “normal-like” methylation patterns in a subset of tumors.
Clustering Analysis
DNA methylation clusters were based on the most variable CpG sites from figure 1 and on stage I and II samples. Consensus clustering was applied as implemented in the Bioconductor package ConsensusClusterPlus, with Euclidean distance and partitioning around medoids (pam) was used to derive clusters. (30, 31)
Survival analyses
P-value was computed from the Cox regression (the coxph function of the survival package). (32, 33) Kaplan-Meier curves were made with the help of the survfit function from the same package using TCGA data for stage I and II tumors. The clinical endpoint for analysis was time to death. TCGA samples are not annotated for therapies received, therefore no control for treatment in analysis is possible but may be assumed to represent the standard of care in the United States. Methylation data was obtained by TCGA from fresh frozen tumors examined by Infinium HumanMethylation 450 as previously described.(25) Categorization for groups of comparison for survival outcomes is based on Medoid clustering as described in Clustering Analysis.
Binary DNA Methylation Assessment
We selected the most significant CpG site per gene to define binary DNA methylation. For each gene, a sample was labeled DNA hypermethylated if the individual β-value of the gene was greater than three times the standard deviation of the mean of all combined β-values of normal samples.
Results
Functional Identification of Cancer-Specific, Hypermethylated Genes in NSCLC Cell Lines
Based on a previously designed method to unmask epigenetically silenced cancer-specific, DNA-hypermethylated genes, we treated eight NSCLC cell lines with either the DNA-methylation and DNMT inhibitor, DAC, or the HDAC class I/II histone deacetylase inhibitor, TSA. (34, 35) Gene expression changes determined using Affymetrix microarray for DAC or TSA treated cells were compared with mock treated cells. This method enables the identification of genes induced specifically by DAC, an important distinction as DAC has the capacity to induce gene re-expression of loci silenced predominantly by hypermethylation while TSA alone will fail to induce re-expression.(34) The objective of methylation biomarker discovery by DAC-specific re-expression is to generate a list of genes likely to be silenced by methylation of promoter CpG islands. DAC-specific re-expression for a gene is defined as a greater than 2.0 fold re-expression on a microarray with DAC treatment compared to mock treated cells, less than 1.4 fold re-expression with TSA treatment compared to mock treated cells, and no basal expression in mock treated cells as previously described.(34, 35) To find genes which would be expected to have higher frequencies of methylation in lung cancer, we refined this list to require the preceding criteria in at least two of eight cell lines. A total of 305 genes were determined to be up-regulated by DAC using these criteria from eight NSCLC cell lines. (Supplemental Figure 1)
Refining a Diagnostic 3-Gene Panel of Cancer-Specific, Hypermethylated Genes in NSCLC Using the Cancer Genome Atlas Dataset
The comprehensive analysis of 305 genes in primary tumors to determine their utility would represent a challenging task without additional informatics filters to select the most promising candidates. To refine this list of genes, we applied this functionally derived gene list to primary tumors characterized in the TCGA lung cancer project, and then validated the findings in two, independent single-institution cohorts of primary NSCLC tumors (Table 1). We first tested for tumor specificity among the TCGA tumors, comparing DNA methylation between lung tumors and normal lung tissue. Of the 305 DAC up-regulated genes, 63 genes with a total of 172 annotated CpG island promoter probes on the Infinium 450K array had a statistically significant ability to differentiate tumor versus normal in TCGA samples as estimated by a linear regression model. In addition, these genes had extremely low methylation (β-values) in TCGA normal samples, thereby defining a group of DAC-responsive, cancer-specific methylated genes. Data using these probes are represented in a heatmap where rows are ordered from top to bottom by p-values based on the ability of an individual methylation array probe to distinguish tumor vs. normal. Columns are ordered by unsupervised hierarchical clustering. (Figure 1, Supplemental Table 1) Maximum estimated p-value for each probe was 1×10−4. CDO1, HOXA9, and TAC1 were notable for extremely high rates of DNA methylation in tumors and low methylation in normal samples, and were most effective in distinguishing tumor versus normal based on p-value of linear logistic regression model.
Table 1. Clinico-pathological Characteristics of Patient Cohorts.
The Cancer Genome Atlas is a publicly available data base that contains DNA methylation data for hundreds of primary Non-Small Cell Lung Cancer (NSCLC) patients. Cohort A consists of resected NSCLC patients from Johns Hopkins Hospital in Baltimore, MD. Cohort B consists of resected NSCLC patients from Shinshu University Hospital in Matsumoto, Japan.
| Cohort | ||||
|---|---|---|---|---|
|
| ||||
| TCGA (n = 636) | A (n = 59) | B (n = 30) | ||
| Age | Average (years) | 68 | 65.8 | 64.1 |
|
| ||||
| Sex | F (%) | 238 (37.4%) | 27 (45.8%) | 11 (36.7%) |
| M (%) | 306 (48.1%) | 32 (54.2%) | 19 (63.3%) | |
| NA | 92 (14.5%) | 0 | 0 | |
|
| ||||
| Smoking | Ever | 466 (73.3%) | 47 (79.7%) | NA |
| Never | 61 (9.6%) | 4 (6.8%) | NA | |
| NA | 109 (17.1%) | 8 (13.6%) | NA | |
| Histology | Adeno | 409 (64.3%) | 36 (61.0%) | 21 (70%) |
| SCC | 227 (35.7%) | 23 (39.0%) | 9 (30%) | |
|
| ||||
| Stage | Ia | 125 (19.7%) | 16 (27.1%) | 3 (10%) |
| Ib | 159 (25.0%) | 20 (33.9%) | 4 (13.3%) | |
| IIa | 58 (9.1%) | 1 (1.7%) | 3 (10%) | |
| IIb | 84 (13.2%) | 9 (15.3%) | 6 (20%) | |
| IIIa | 78 (12.2%) | 7 (11.9%) | 7 (23.3%) | |
| IIIb | 14 (2.2%) | 3 (5.1%) | 4 (13.3%) | |
| IV | 17 (2.7%) | 3 (5.1%) | 3 (10%) | |
| NA | 101 (15.9%) | 0 | 0 | |
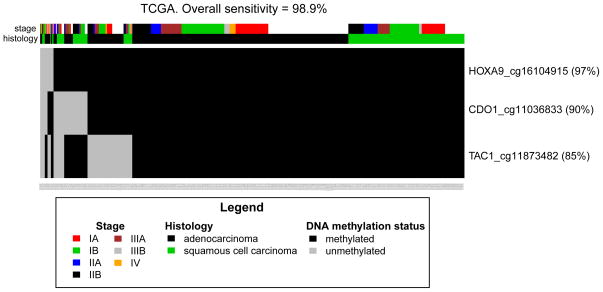
Binary methylation values as determined by the single best methylation probe from the promoter CpG islands of CDO1, HOXA9, and TAC1, and plotted for all NSCLC stages together as well as for stage I alone. (Figure 2, Supplemental Figure 2, Supplemental Table 1). Sensitivity is not limited by histology or tumor stage in the TCGA dataset. In fact, methylation of at least one of these 3 genes is 98.9% sensitive for tumors stage I–IV and 98.7% sensitive for stage I tumors alone. HOXA9 alone is methylated in 97% of NSCLC TCGA samples. There are limited descriptions of DNA methylation of these genes in human lung cancer in previous studies. While TAC1 promoter methylation has not been described in lung malignancies, highly prevalent HOX cluster gene methylation, including HOXA9, has been reported in cell lines and a small number of squamous stage I tumors (n=4) as well as a pool of mixed stage and mixed histology tumors (n=20).(17, 36) HOXA9 hypermethylation has been described as a potential screening test in combination with SOX1 hypermethylation and DDR1 hypomethylation as assayed by pyrosequencing.(37) CDO1 has been reported as a methylated gene in squamous lung tumors (n=30).(38) CDO1 and TAC1 have been described as high-prevalence cancer-specific methylated genes in breast cancer.(35) However, no previous study has described the sensitivity and specificity for a combination of these genes in a large population of NSCLC tumors and validation cohorts.
Figure 2. DNA Methylation of CDO1, HOXA9, and TAC1 is Highly Sensitive for NSCLC in the Cancer Genome Atlas.
A single Infinium methylation probe with the best discriminative capacity between tumor and normal samples was selected for each of the 3 genes. A sample is considered methylated for a gene if its β-value was larger than three times the standard deviation of the mean of β-values of normal samples. Methylation of at least one gene-promoter among CDO1, HOXA9, and TAC1 by Infinium array identifies 98.9% of NSCLC cases in 636 cases in The Cancer Genome Atlas.
In addition to their diagnostic utility, we examined the potential prognostic significance of this functionally derived list of cancer-specific methylation. As would be expected from a list of genes with an extremely high prevalence of methylation and no described biologic role in lung cancer, none of the 63 genes examined individually was associated with survival outcome in TCGA. (Data not shown) In order to examine whether methylation of these genes taken as a group reflect biological differences in tumors, we clustered all TCGA lung cancer samples using medoid clustering, a method for defining optimal numbers of groups within a data set. When taken together, the 63 cancer-specific hypermethylated genes form three groups, adenocarcinoma-predominant, squamous-predominant, and a mixed group. These clusters demonstrate a marginal association with survival in the TCGA tumors (p=0.04). (Supplemental Figure 3) From our previously published markers of outcome in early stage, resected lung cancer our strongest associations with outcome came from questions pertaining to cancer-specific methylation confirmed in lymph nodes, thus a diagnostic or staging paradigm. As the TCGA contains only samples of primary tumors and no associated lymph nodes, there is no ability to assess concordance of methylation between tumor and lymph node. When examining tumor-only questions from our previous work, we find general agreement with the moderate prognostic capacity of methylation of 4 genes when examined in tumor only, highlighting the need to refine a highly sensitive and specific diagnostic markers for the molecular staging of NSCLC.(6) (Supplemental Figure 4)
Association of Progenitor Cell Polycomb-Associated Genes with Cancer-Specific Methylation Marks
Previous studies have suggested that genes with polycomb marks in chromatin surrounding the transcription start sites are predisposed to aberrant DNA methylation silencing in cancer.(15, 39, 40) In embryonic stem (ES) cells, polycomb association occurs in the context of bivalent chromatin marks containing both active histone 3 lysine 4 trimethylation (H3K4me3) and repressive histone 3 lysine 27 trimethylation (H3K27me3) marks. Of the 63 cancer-specific hypermethylated genes, 45 (71.4%) are considered bivalent genes silenced by polycomb repressive complex in progenitor cell states, a rate much higher than the presence of these marks among all genes (21% using estimated 4413 bivalent genes among an estimated 21,000 total human genes, p <0.0001). (15, 38) CDO1, HOXA9, and TAC1, are all polycomb-associated in ES cells. (Figure 1, Supplementary Table 1)
Validating the Diagnostic Utility of a 3-Gene Panel in Two Cohorts of Primary Tissue
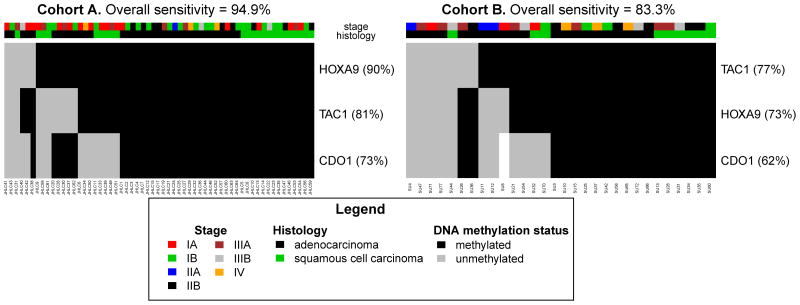
To confirm the high prevalence of DNA methylation for these genes in other primary lung tumors, we then validated the sensitivity of these 3 genes in two, independent cohorts of NSCLC tumor samples using MSP. (Table 1, Figure 3) Primers for CDO1, HOXA9, and TAC1 were designed and tested on tumor samples from cohorts in the U.S. and Japan. As was observed for these genes on the Infinium platform within TCGA data, there was no methylation in seven normal lung samples when examined using MSP. In contrast to normal lung, among the American cohort A and Japanese cohort B, respectively, 94.9% and 83.3% of the tumor samples were methylated for at least one of these 3 genes. Since this 3-gene panel has near-zero methylation β-values by Infinium and MSP in normal tissues and is found to have stage-independent hypermethylation in cancer, these genes fulfill critical characteristics for designing a threshold for methylation in clinical assays and for identifying the earliest stages of NSCLC. (Figure 3)
Figure 3. Validation of the Sensitivity of Methylation-Specific PCR for 3 Genes in Two Independent Cohorts.
Methylation of at least one gene-promoter among CDO1, HOXA9, or TAC1 by methylation-specific PCR identifies 94.9% of NSCLC cases in 59-patient United States cohort A and 83.3% of NSCLC cases from the independent 30-patient Japanese cohort B.
Discussion
Using an experimental model to derive a list of candidate cancer-specific, hypermethylated, polycomb-associated genes in lung cancer, we validated a 3-gene test in a large publicly available database and two independent cohorts to describe a highly-sensitive, highly-specific diagnostic test for NSCLC. In the present study, we use a functional approach to identify three genes, CDO1, HOXA9, and, TAC1 in which we describe cancer-specific DNA methylation without regard for the biologic implication of that cancer-specific methylation. When examining diagnostic sensitivity, we find a remarkable concordance between TCGA samples, derived entirely from American hospitals, and our American validation cohort with sensitivities of 98.9% and 94.9% respectively. Diagnostic sensitivity in the Japanese cohort is similar but lower at 83.3%. While some variation may be due to sampling, we can also reasonably hypothesize that this reflects other established differences in the NSCLC populations of American and Japan and highlights the need to tailor a test precisely to target populations. While an 83% sensitivity of detection far exceeds any mutational detection approach currently available, it may be possible to provide an even better 3-gene test if these genes were chosen from among highly methylated genes determined from analysis of lung cancers in Japanese populations.
Additionally, we have explored whether these cancer-specific alterations may have prognostic value. As might be expected, these genes without an established role in the pathogenesis of lung cancer and/or an extremely high prevalence of methylation prove to be of no prognostic value when examined individually. Indeed, in our previously published study of 4 genes, there was limited prognostic value when knowledge of methylation status is known for the tumor only. Additionally, our previous study suggested that the presence of cancer-specific methylation in histologically negative lymph nodes, particularly mediastinal (N2) nodes, was most prognostic of recurrence and lung cancer associated.(6)
An interesting characteristic of the genes elicited by this functional screen for novel cancer-specific biomarkers is a high degree of overlap with polycomb-associated genes. Histone 3 lysine 4 and 27 trimethylation (H3K4me3 and H3K27me3) define a bivalent chromatin state that denotes a low-transcriptional, poised state for a group of genes in progenitor and stem cells highly enriched for developmental processes.(41) These genes, largely active during development of differentiated tissues, are down-regulated by the polycomb repressive complex when a chromatin bivalent state exists and are largely devoid of DNA methylation. These loci are particularly vulnerable to DNA methylation during the process of carcinogenesis.(15) While the mechanism that underlies epigenetic silencing transitioning from the polycomb repressive complex to DNA methylation would suggest little or no alteration in gene expression in some cases, assaying these methylation changes remains useful as highly sensitive and specific hallmarks of tumor tissue and are therefore excellent candidates as diagnostic biomarkers. Additionally, because different stem and progenitor populations show variation in distribution of chromatin-bivalency, the methylation marks at polycomb associated DNA may signal subtle differences in the cell of origin.
For the molecular detection of disease in lymph nodes for staging and for approaches for early detection involving sputum, plasma or fine needle aspirates, molecular alterations present in the vast majority of tumors will be the most sensitive and efficient means of detection. Through the characterization of hypermethylated loci reported here, we have developed a highly-sensitive, highly-specific test for identifying cases of NSCLC which may serve these purposes. A 3-gene methylation assay with sensitivity in tumors approaching 100% may allow for the detection or diagnosis of disease in tissues remote from the primary tumor without specific knowledge of methylation of those genes in the tumor itself. The present study demonstrates the performance of a 3-gene test in primary tumor samples for which inadequate diagnostic methods currently exist. With improvements in detection of DNA methylation in blood and sputum, the sensitivity of detection in additional types of biospecimens including plasma and sputum samples can now be tested.(42)
Supplementary Material
Statement of Translational Relevance.
Lung cancer remains the leading cause of cancer related mortality in the world. The likelihood of mortality related to the disease increases dramatically with the stage of disease. Using a validated experimental method of eliciting frequently methylated genes in cancer which we then examine in hundreds of lung cancer samples in The Cancer Genome Atlas and two, independent cohorts, we describe DNA methylation of one or more of CDO1, HOXA9, and TAC1 as nearly universal in lung cancer in the United States. Such a highly sensitive and specific molecular marker of disease may play a significant role in improving early detection strategies and decreasing NSCLC morbidity and mortality.
Acknowledgments
The authors would like to acknowledge NIH Lung SPORE CA058184, DOD W81XWH-12-1-0323, and MDx-Health for support. The authors would like to acknowledge Leslie Cope. JW would like to acknowledge Mary Lou Wrangle.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Youlden DR, Cramb SM, Baade PD. The International Epidemiology of Lung Cancer: geographical distribution and secular trends. J Thorac Oncol. 2008;3:819–31. doi: 10.1097/JTO.0b013e31818020eb. [DOI] [PubMed] [Google Scholar]
- 3.Vadakara J, Borghaei H. Personalized medicine and treatment approaches in non-small-cell lung carcinoma. Pharmgenomics Pers Med. 5:113–23. doi: 10.2147/PGPM.S24258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Juergens RA, Wrangle J, Vendetti FP, Murphy SC, Zhao M, Coleman B, et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov. 1:598–607. doi: 10.1158/2159-8290.CD-11-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reck M. What future opportunities may immuno-oncology provide for improving the treatment of patients with lung cancer? Ann Oncol. 23(Suppl 8):viii28–34. doi: 10.1093/annonc/mds260. [DOI] [PubMed] [Google Scholar]
- 6.Brock MV, Hooker CM, Ota-Machida E, Han Y, Guo M, Ames S, et al. DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med. 2008;358:1118–28. doi: 10.1056/NEJMoa0706550. [DOI] [PubMed] [Google Scholar]
- 7.Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–24. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 8.Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, Fagerstrom RM, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 365:395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belinsky SA, Nikula KJ, Palmisano WA, Michels R, Saccomanno G, Gabrielson E, et al. Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc Natl Acad Sci U S A. 1998;95:11891–6. doi: 10.1073/pnas.95.20.11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palmisano WA, Divine KK, Saccomanno G, Gilliland FD, Baylin SB, Herman JG, et al. Predicting lung cancer by detecting aberrant promoter methylation in sputum. Cancer Res. 2000;60:5954–8. [PubMed] [Google Scholar]
- 11.Belinsky SA, Klinge DM, Dekker JD, Smith MW, Bocklage TJ, Gilliland FD, et al. Gene promoter methylation in plasma and sputum increases with lung cancer risk. Clin Cancer Res. 2005;11:6505–11. doi: 10.1158/1078-0432.CCR-05-0625. [DOI] [PubMed] [Google Scholar]
- 12.Belinsky SA, Liechty KC, Gentry FD, Wolf HJ, Rogers J, Vu K, et al. Promoter hypermethylation of multiple genes in sputum precedes lung cancer incidence in a high-risk cohort. Cancer Res. 2006;66:3338–44. doi: 10.1158/0008-5472.CAN-05-3408. [DOI] [PubMed] [Google Scholar]
- 13.Machida EO, Brock MV, Hooker CM, Nakayama J, Ishida A, Amano J, et al. Hypermethylation of ASC/TMS1 is a sputum marker for late-stage lung cancer. Cancer Res. 2006;66:6210–8. doi: 10.1158/0008-5472.CAN-05-4447. [DOI] [PubMed] [Google Scholar]
- 14.Leng S, Do K, Yingling CM, Picchi MA, Wolf HJ, Kennedy TC, et al. Defining a gene promoter methylation signature in sputum for lung cancer risk assessment. Clin Cancer Res. 18:3387–95. doi: 10.1158/1078-0432.CCR-11-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Easwaran H, Johnstone SE, Van Neste L, Ohm J, Mosbruger T, Wang Q, et al. A DNA hypermethylation module for the stem/progenitor cell signature of cancer. Genome Res. 22:837–49. doi: 10.1101/gr.131169.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, et al. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 143:313–24. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rauch T, Wang Z, Zhang X, Zhong X, Wu X, Lau SK, et al. Homeobox gene methylation in lung cancer studied by genome-wide analysis with a microarray-based methylated CpG island recovery assay. Proc Natl Acad Sci U S A. 2007;104:5527–32. doi: 10.1073/pnas.0701059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.team RDc. R: A language and environment for statistical computing. R foundation for statistical computing; Vienna, Austria: 2004. [Google Scholar]
- 19.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–73. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 21.Brandes JC, Carraway H, Herman JG. Optimal primer design using the novel primer design program: MSPprimer provides accurate methylation analysis of the ATM promoter. Oncogene. 2007;26:6229–37. doi: 10.1038/sj.onc.1210433. [DOI] [PubMed] [Google Scholar]
- 22.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–6. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gibbs AR, Thunnissen FB. Histological typing of lung and pleural tumours: third edition. J Clin Pathol. 2001;54:498–9. doi: 10.1136/jcp.54.7.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mountain CF, Dresler CM. Regional lymph node classification for lung cancer staging. Chest. 1997;111:1718–23. doi: 10.1378/chest.111.6.1718. [DOI] [PubMed] [Google Scholar]
- 25.Comprehensive genomic characterization of squamous cell lung cancers. Nature. 489:519–25. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.
- 27.Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, et al. High density DNA methylation array with single CpG site resolution. Genomics. 98:288–95. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 28.Smyth G. Limma: linear models for microarray data. In: Gentleman R, Dudoit VCS, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. NY: Springer; 2005. pp. 397–420. [Google Scholar]
- 29.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Statistical Applications in Genetics and Molecular Biology. 2004 doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 30.Wilkerson M. R package version 1.5.1. 2011. ConsensusClusterPlus: ConsensusClusterPlus. [Google Scholar]
- 31.Reynolds A, Richards G, de la Iglesia B, Rayward-Smith V. Clustering rules: A comparison of partitioning and hierarchical clustering algorithms. Journal of Mathematical Modelling and Algorithms. 1992;5:475–504. [Google Scholar]
- 32.Andersen PaGR. Cox’s regression model for counting processes, a large sample study. Annals of Statistics. 1982:10. [Google Scholar]
- 33.Therneau T, Grambsch P. Modeling Survival Data: Extending the Cox Model. Springer-Verlag; 2000. [Google Scholar]
- 34.Schuebel KE, Chen W, Cope L, Glockner SC, Suzuki H, Yi JM, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007;3:1709–23. doi: 10.1371/journal.pgen.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeschke J, Van Neste L, Glockner SC, Dhir M, Calmon MF, Deregowski V, et al. Biomarkers for detection and prognosis of breast cancer identified by a functional hypermethylome screen. Epigenetics. 7:701–9. doi: 10.4161/epi.20445. [DOI] [PubMed] [Google Scholar]
- 36.Son JW, Jeong KJ, Jean WS, Park SY, Jheon S, Cho HM, et al. Genome-wide combination profiling of DNA copy number and methylation for deciphering biomarkers in non-small cell lung cancer patients. Cancer Lett. 311:29–37. doi: 10.1016/j.canlet.2011.06.021. [DOI] [PubMed] [Google Scholar]
- 37.Nelson HH, Marsit CJ, Christensen BC, Houseman EA, Kontic M, Wiemels JL, et al. Key epigenetic changes associated with lung cancer development: results from dense methylation array profiling. Epigenetics. 7:559–66. doi: 10.4161/epi.20219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwon YJ, Lee SJ, Koh JS, Kim SH, Lee HW, Kang MC, et al. Genome-wide analysis of DNA methylation and the gene expression change in lung cancer. J Thorac Oncol. 7:20–33. doi: 10.1097/JTO.0b013e3182307f62. [DOI] [PubMed] [Google Scholar]
- 39.Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 17:510–22. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pfeifer GP, Rauch TA. DNA methylation patterns in lung carcinomas. Semin Cancer Biol. 2009;19:181–7. doi: 10.1016/j.semcancer.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bailey VJ, Keeley BP, Razavi CR, Griffiths E, Carraway HE, Wang TH. DNA methylation detection using MS-qFRET, a quantum dot-based nanoassay. Methods. 52:237–41. doi: 10.1016/j.ymeth.2010.03.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.