Motility analysis of collectively migrating epithelial sheets reveals the interplay between cellular density and leader cell positioning throughout a collective group, as well as the functional role of leader cell constraint in directing a migrating group of cells.
Abstract
During wound healing and cancer metastasis, cells are frequently observed to migrate in collective groups. This mode of migration relies on the cooperative guidance of leader and follower cells throughout the collective group. The upstream determinants and molecular mechanisms behind such cellular guidance remain poorly understood. We use live-cell imaging to track the behavior of epithelial sheets of keratinocytes in response to transforming growth factor β (TGFβ), which stimulates collective migration primarily through extracellular regulated kinase 1/2 (Erk1/2) activation. TGFβ-treated sheets display a spatial pattern of Erk1/2 activation in which the highest levels of Erk1/2 activity are concentrated toward the leading edge of a sheet. We show that Erk1/2 activity is modulated by cellular density and that this functional relationship drives the formation of patterns of Erk1/2 activity throughout sheets. In addition, we determine that a spatially constrained pattern of Erk1/2 activity results in collective migration that is primarily wound directed. Conversely, global elevation of Erk1/2 throughout sheets leads to stochastically directed collective migration throughout sheets. Our study highlights how the spatial patterning of leader cells (cells with elevated Erk1/2 activity) can influence the guidance of a collective group of cells during wound healing.
INTRODUCTION
Cellular migration in response to environmental cues is an important process that mediates embryonic development, organogenesis, immune response, metastasis, wound healing, and tissue maintenance (Lauffenburger and Horwitz, 1996; Martin, 1997; Locascio and Nieto, 2001; Chambers et al., 2002; Keller, 2005; Wang et al., 2005; Friedl and Gilmour, 2009). Individual cell migration is primarily driven by a dynamic, polarized, and coordinated process of actin polymerization and depolymerization in concert with focal adhesion assembly/disassembly and actinomyosin contractions (Mogilner and Oster, 1996; Servant et al., 2000; Webb et al., 2002; Horwitz and Webb, 2003; Pollard and Borisy, 2003; Van Haastert and Devreotes, 2004; Arrieumerlou and Meyer, 2005). However, many processes in development and cancer progression depend not only on the motility of individual cells, but also on the collective guidance of groups of cells (Bronner-Fraser, 1993; Lois et al., 1996; Zhang et al., 2003; Martin and Parkhurst, 2004; Murase and Horwitz, 2004; Xia and Karin, 2004; Rorth, 2009). Collective cell migration occurs when physically linked cells migrate in a common direction (Rorth, 2009). In developmental processes, this type of cell movement has been observed in sheets of cells, sprouting and branching groups of cells, and streams of cells (Rorth, 2009). In cancer metastasis, the invasive behavior of tumors frequently relies on the collective migration of globular groups, strands, or sheets of cells (Friedl and Gilmour, 2009; Mayor and Carmona-Fontaine, 2010). Unlike the autonomous movement of an individual cell, the collective migration of a group of cells requires constant intercellular communication for the coordination of movement.
During collective migration in sheets of cells, cells at the leading edge of the sheet, with free space around them to spread, adopt “leader” or “pioneer” behaviors (Omelchenko et al., 2003; Poujade et al., 2007; Vitorino and Meyer, 2008). These cells express “guidance receptors” such as receptor tyrosine kinases (epidermal growth factor receptor, platelet-derived growth factor receptor, vascular endothelial growth factor) and display elevated signaling of at least one of these pathways (Gabay et al., 1997; Jekely et al., 2005; Mine et al., 2005). Leader cells are particularly sensitive to growth factor stimulation, which is in agreement with a model that signaling mainly regulates leader cells in order to control directed migration (Khalil and Friedl, 2010). Studies of sheets of Madin–Darby kidney cells show that cells in close proximity to a wound display elevated extracellular regulated kinase 1 and 2 (Erk1/2) activity, suggesting that these cells represent a spatially distinct population of leader cells (Matsubayashi et al., 2004; Nikolic et al., 2006). The mechanism by which these cells achieve activated Erk1/2 in a spatially confined manner is poorly understood but requires local generation of reactive oxygen species (ROS; Matsubayashi et al., 2004; Nikolic et al., 2006). In addition, it is unclear exactly how these leader cells can influence the motility of neighboring follower cells toward the rear of a sheet. One model postulates that follower cells migrate in accordance with diffusive chemical cues such as cAMP, Ca2+, or ROS, where a propagating wave of Erk1/2 activity leads to a propagating wave of cellular motility throughout a sheet (Matsubayashi et al., 2004; Nikolic et al., 2006). Although this model explains how leader cells may activate motility in follower cells, it does not fully explain how each cell in the collective group successfully orients motility toward the wound during sheet migration. Another model for sheet migration proposes that the mechanical forces generated by leader cells coordinates the migratory direction of follower cells (Vitorino and Meyer, 2008; Vitorino et al., 2011). Together these two proposed mechanisms can explain how cells coordinate both cellular direction and cellular speed during wound healing.
Transforming growth factor β (TGFβ) is one of the key growth factors implicated in wound healing in vivo (Martin, 1997; Martin and Parkhurst, 2004); upon injury to skin, TGFβ secretion coincides with the early stages of tissue repair (Werner and Grose, 2003). In addition, TGFβ mediates the activation of cell migration during embryonic eyelid and dorsal closure, both of which are developmental processes that involve the collective migration of epithelial sheets of cells (Zhang et al., 2003; Martin and Parkhurst, 2004; Xia and Karin, 2004). TGFβ signals through both Smad and non-Smad pathways to elicit appropriate cellular responses (Massague, 1998). Smads are transcription factors that are largely responsible for transcriptional regulation of a majority of TGFβ-responsive genes. Activation of non-Smad signaling pathways, including Erk1/2, p38, c-Jun N-terminal kinase, RhoA, and phosphoinositide 3’-kinase, often depends on the specific cellular context (Mulder, 2000; Zhang et al., 2003; Zhang, 2009 ; Moustakas and Heldin, 2005; Chapnick et al., 2011; Mu et al., 2012). Although both Smad and non-Smad pathways have been implicated in modulating epithelial sheet migration in various systems, the underlying mechanisms by which epithelial sheets respond to global TGFβ stimulation to produce directed movement and coordinate individual cell directions are largely undefined.
In this study, we use two-dimensional epithelial sheets as a model system to understand how TGFβ mediates collective cell migration of human keratinocytes. Using this model system, we aim to answer two outstanding questions about collective migration: how does activated Erk1/2 activity form near the leading edge of a sheet, and does the spatiotemporal regulation of Erk1/2 activity throughout a collective group influence the migration direction of the collective group? We find that TGFβ activation of Erk1/2 activity is cell density dependent, in that high cell density inhibits Erk1/2 activity. TGFβ-treated sheets develop reduced cellular density at the leading edge, which drives elevated Erk1/2 activity at this location. Thus cellular density sensing is likely the primary means by which leader cell identity is achieved near the edge of a wound. In addition, we find that a sheet with spatially confined Erk1/2 activity to the leading edge displays coordination of individual cell migration directions toward the wound, whereas a sheet with global Erk1/2 activation displays significantly reduced ability to migrate toward the wound. These findings highlight the role of cellular density in defining the location of leader cells throughout a collective group, as well as the importance of the spatial confinement of leader cells in governing the overall collective migration direction of a group of cells during wound healing.
RESULTS
TGFβ induces collective migration in epithelial sheets
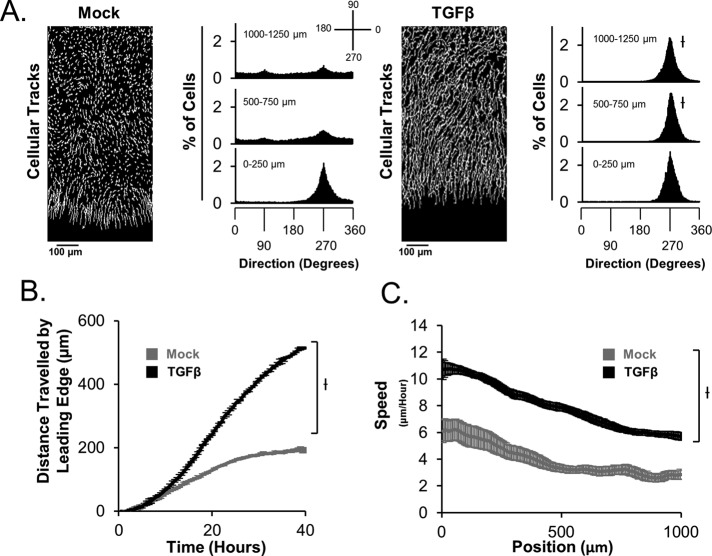
We previously reported a computational tool to analyze cellular migration direction and speed of individual cells in a collective group (Chapnick et al., 2013). This tool, named Pathfinder, was used to analyze the migration behavior of human keratinocytes expressing an exogenous mCherry-tagged histone H2B (HaCaT-H2B cells) in epithelial sheets exposed to mock or TGFβ stimulation. For migration direction analysis, each cell within 1 mm of the wound in an epithelial sheet is assigned a value between 0 and 360°, which represents its migration direction relative to a defined set of axes (Figure 1A). By comparing mock- and TGFβ-stimulated sheets after 24 h, we find that ligand-dependent orientation of cellular motility does not differ significantly from that of mock-treated sheets at the immediate leading edge (0–250 μm; Figure 1A and Supplemental Movie S1). However, for the case of cells distal from the leading edge (500–1250 μm), TGFβ stimulation causes drastically improved cellular orientation toward the wound relative to that of mock stimulation in HaCaT and SCC-13 squamous cell carcinoma cells (Figure 1A and Supplemental Figure S1A, respectively). Thus TGFβ stimulation elicits improved collective migration in these epithelial sheets. In addition, we determined that TGFβ stimulation causes improved wound-healing ability of sheets by enhancing the forward migration ability of the leading edge (Figure 1B). When we measure individual cell speed throughout sheets, we find that not all cells migrate with the same speed in response to TGFβ stimulation, for which the fastest-moving cells are located toward the wound in both HaCaT and SCC-13 cells (Figure 1C and Supplemental Figure S1B, respectively). This procession mode of collective migration suggests that leader cells in the group are likely to be enriched near the wound border.
FIGURE 1:
TGFβ induces collective migration in epithelial sheets. (A) Cellular tracks display the spatial profile of migration throughout sheets between 35 and 40 h for sheets in the presence or absence of TGFβ. Quantification of cellular motility directions at 35 h post–ligand stimulation reveals that TGFβ promotes directional migration toward the wound. Cellular directions are calculated relative to the displayed key, with each cell assigned a value between 0 and 360°. (B) The forward migration of the leading edge of sheets was quantified in the presence or absence of TGFβ as a function of time. (C) The spatial distribution of cellular speed throughout sheets was quantified at 35 h post–ligand stimulation.
TGFβ-dependent epithelial sheet migration requires MEK/Erk activity
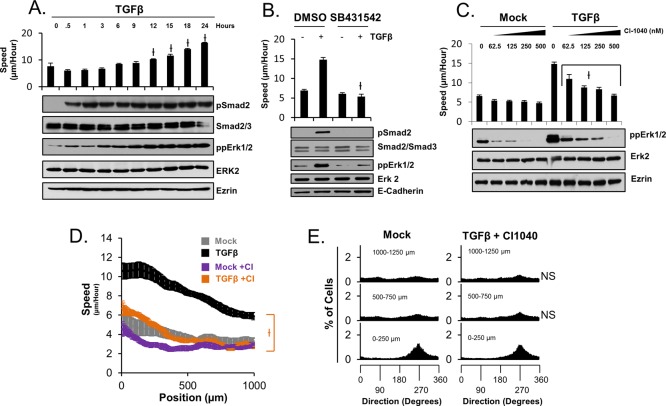
Because Erk1/2 activity was associated with leader cell identity in previous studies, we investigated the potential role of Erk1/2 activity in TGFβ-dependent collective migration with our experimental model system. Using low-density colonies of HaCaT cells, Western blot analysis of phospho-Erk1/2 levels, and live-cell tracking of HaCaT-H2B cells, we determined that TGFβ stimulation results in Erk1/2 activation with similar kinetics to that of TGFβ-dependent stimulation of cellular migration speed (Figure 2A). This ligand-dependent effect on both cellular motility and Erk1/2 activation was confirmed to require TBR1 kinase activity, as chemical inhibition of TBR1 with the compound SB431542 blocks TGFβ-induced Erk1/2 activation (Figure 2B). In addition, inhibition of Erk1/2 activity with a small-molecule inhibitors of MEK1 (CI-1040 and UO126) inhibits both ligand-dependent Erk1/2 activity and migration speed in a dose-dependent manner (Figure 2C and Supplemental Figure S2A, respectively). We additionally determined that TGFβ- and epidermal growth factor (EGF)–induced Erk1/2 activation occurs independently of Smad2 phosphorylation in HaCaT cells (Supplemental Figure S2B), and TGFβ-induced cellular motility throughout sheets and Erk1/2 phosphorylation are unaltered by RNA interference–mediated depletion of either Smad2 or Smad3 in HaCaT cells (Supplemental Figure S2, C and D). Thus TGFβ-dependent regulation of Smad activity appears to be independent of ligand-induced mitogen-activated protein kinase and motility activation. However, TGFβ-dependent effects on spatial patterns of both migration direction and migration speed are inhibited upon MEK1 inhibition (Figure 2, D and E). Together these data show that TGFβ-dependent enhancement of collective migration throughout sheets requires Erk1/2 activity.
FIGURE 2:
Erk1/2 activity is required for TGFβ-induced collective migration. (A) Western blot analysis of low-density colonies (300 cells/mm2) of HaCaT cells and cellular migration speed analysis of HaCaT-H2B cells reveal that the kinetic profile of speed activation correlates with the kinetic profile of Erk1/2, but not Smad2, activation in TGFβ-treated cells. (B) Simultaneous chemical inhibition of TBRI at ligand treatment (t = 0 h) prevents ligand-dependent Erk1/2 activation and stimulation of cellular speed after 24 h. (C) Chemical inhibition of MEK1 at 23 h post–ligand stimulation prevents ligand-dependent Erk1/2 activation and stimulation of cellular speed at 24 h post–ligand stimulation in a dose-dependent manner. (D) MEK1 inhibition at 30 h inhibits TGFβ-dependent cellular speed throughout sheets at 35 h. (E) MEK1 inhibition at 30 h inhibits TGFβ-dependent cellular orientation toward the wound at 35 h.
Spatial patterns of Erk1/2 activity are a consequence of spatial patterns of cellular density
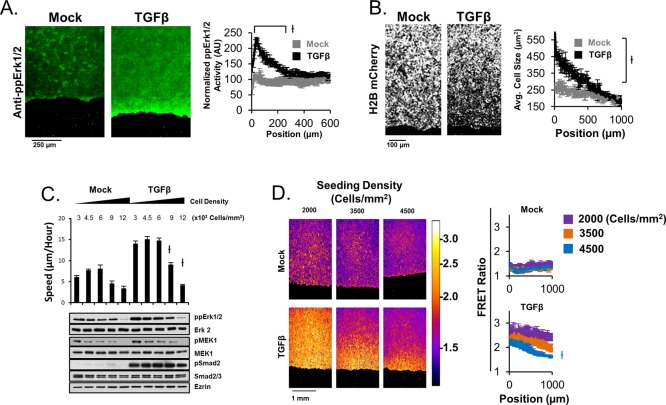
Our spatial measurements of cellular motility speeds throughout TGFβ-stimulated sheets revealed that the fastest-moving cells are located toward the leading edge of sheets (Figure 1C), whether other experiments revealed that cellular migration speed is largely a function of Erk1/2 activity (Figure 2). As a result, we used immunofluorescence experiments to determine whether activated Erk1/2 was spatially constrained in a similar manner to cellular migration speed in ligand-stimulated sheets. Indeed, TGFβ stimulation causes elevated Erk1/2 activity toward the leading edge of sheets (Figure 3A). Similarly, TGFβ stimulation results in decreased cellular density toward the leading edge of epithelial sheets (Figure 3B). Thus we tested the hypothesis that the ligand-dependent pattern in Erk1/2 activity throughout sheets was causally determined by patterns in cellular density. When plated out in increasing density, HaCaT-H2B cells display density-dependent cellular migration speed in both the presence and absence of TGFβ, with high cell density inhibiting migration speed (Figure 3C). This relationship between cellular density and migration speed correlates with Erk1/2 activity in wild-type HaCaT cells (Figure 3C). In addition, using HaCaT cells expressing a cytosolic-targeted Erk sensor (EKAR-NES; Harvey et al., 2008) and live-cell imaging, we found that the spatial pattern of Erk1/2 activity throughout epithelial sheets is modulated by the initial cell density of sheets, by which low-density sheets display global activation of Erk1/2 and high-density sheets display spatially confined Erk1/2 activity toward the leading edge in a TGFβ-dependent manner (Figure 3D). In agreement with our findings in HaCaT cells, our experiments in SCC-13 cells reveal that TGFβ elicits improved spatially constrained Erk1/2 activation, motility, and changes in cell density in sheets (Supplemental Figure S3A), as well as density-dependent regulation of ligand-induced Erk1/2 activation (Supplemental Figure S3B). We investigated whether the known role of TGFβ in epithelial-to-mesenchymal transition (EMT) was possibly responsible for ligand-dependent changes in cellular density that we observe in both HaCaT and SCC-13 cells using immunofluorescence staining of E-cadherin in fixed HaCaT cell sheets in the presence and absence of TGFβ. This experiment shows that TGFβ does cause loss of E-cadherin toward the leading edge of sheets, suggesting that local ligand-induced EMT could be responsible for local elevation of Erk1/2. Although the mechanism by which TGFβ elicits local reduction of cellular density toward the leading edge of sheets is not entirely clear, these experiments provide compelling evidence that density-dependent regulation of Erk1/2 activity in conjunction with TGFβ-dependent localized cell spreading at the leading edge can explain why TGFβ-stimulated sheets display local Erk1/2 activity toward the leading edge.
FIGURE 3:
Cell density–dependent Erk1/2 activation drives spatially constrained Erk1/2 activity throughout sheets. (A) Immunofluorescence staining of active Erk1/2 in fixed sheets of HaCaT cells reveals TGFβ-dependent spatially constrained Erk1/2 activation at the leading edge at 24 h post–ligand stimulation. (B) Cellular density was measured with spatial resolution throughout sheets by quantifying the cellular area as a function of distance from the leading edge (position) after 24 h of ligand stimulation. (C) Titrations of cellular density in the presence or absence of TGFβ at 24 h reveal that high cell density inhibits Erk1/2 activity in a ligand-independent manner. (D) HaCaT EKAR-NES cells were used to measure the effect of cellular density on the spatial profile of Erk1/2 activity at 24 h post–ligand stimulation. The displayed heat map key represents the calculated FRET ratio from the EKAR-NES biosensor.
Sheets with spatially constrained Erk activation display distinct collective migration behavior compared with sheets with global Erk activation
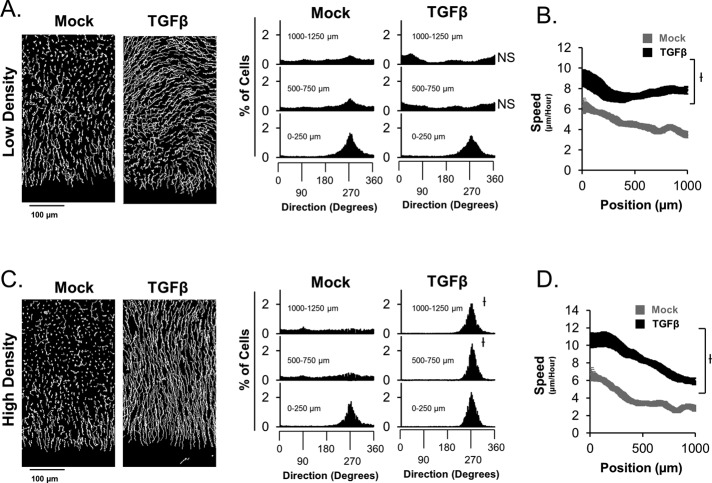
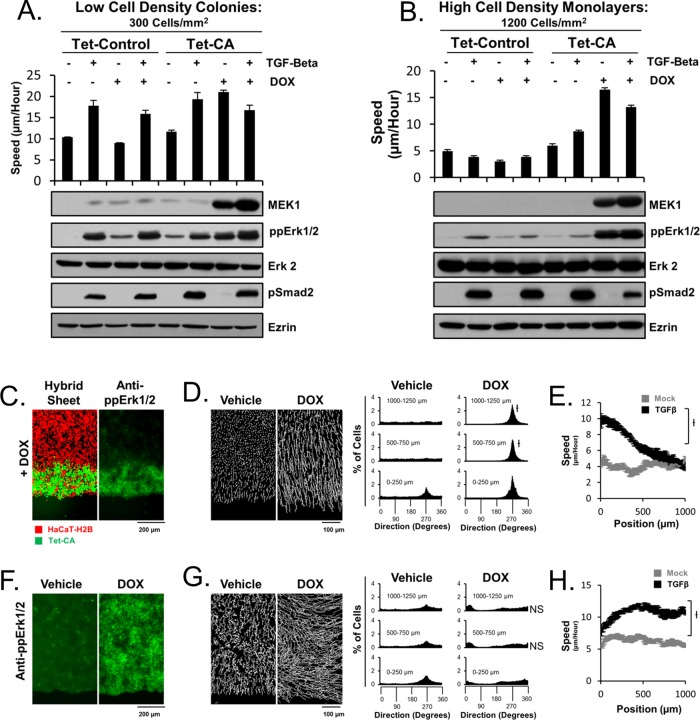
Although spatially constrained Erk1/2 activity has been reported, the importance of such spatial constrain has not been made clear in the literature. As a result, we sought to determine whether an epithelial sheet with spatially constrained Erk1/2 activity migrates differently than that of a sheet in which Erk1/2 activity is globally elevated. To address this problem, we first compared the migration speed and directions of cells throughout low- and high-density sheets in the presence and absence of TGFβ. Low-density sheets display a decreased ability of cells distal from the leading edge to migrate toward the wound in response to ligand stimulation (Figure 4A). However, TGFβ-dependent activation of cellular speeds still occurs throughout low-density sheets, as is the case for high-density sheets (Figure 4, B and D, respectively). Conversely, when the same experiment is conducted in parallel with high-density sheets, all cells throughout a sheet display TGFβ-dependent activation of cellular speed and migration direction toward the wound. Although these experiments suggest that sheets with globally activated Erk1/2 have decreased ability to migrate toward the wound, it is not entirely clear whether this is directly due to cell density–dependent guidance of cells or patterns of Erk1/2 activity throughout the collective group of cells. As a result, we sought to control patterns of Erk1/2 activity while maintaining constant initial cell density throughout a sheet and test directly the role of patterned Erk1/2 activity in the guidance of the collective group of cells. We constructed a stable transgenic HaCaT cell line expressing a constitutively active MEK1 (CA-MEK1) under a doxycycline (DOX)-inducible promoter (Tet-CA cells). Using these cells, live-cell tracking, and Western blot analysis, we found that DOX stimulation leads to efficient induction of CA-MEK1 levels and activation of Erk1/2 and cellular migration speed comparable to that of TGFβ in low- density cells (Figure 5A). When this experiment is repeated under conditions of high cell density, DOX-dependent expression of CA-MEK1 can elicit significantly elevated cellular migration speed and Erk1/2 activity (Figure 5B). Under these conditions, TGFβ fails to stimulate Erk1/2 activity. These Tet-CA cells were used to construct hybrid sheets of HaCaT-H2B cells and Tet-CA cells in which Tet-CA cells were positioned toward the leading edge. These hybrid sheets enable us to activate Erk1/2 specifically toward the leading edge of sheets without the need for exogenous ligand stimulation (Figure 5C). The migration behavior of these sheets was compared with the migration behavior of similar sheets made entirely of Tet-CA cells (Figure 5, D–H, and Supplemental Movie S2). Surprisingly, hybrid sheets display near-identical migration behavior to that of TGFβ-stimulated HaCaT-H2B sheets in both the spatial pattern of migration speed and the spatial pattern of migration direction (Figures 1A and 5, D and E). Thus TGFβ-dependent collective migration may be entirely determined by spatial Erk1/2 patterns, despite the complex pleiotropic nature of the TGFβ pathway. Of interest, when the same experiment is conducted on sheets of Tet-CA cells, these sheets display a strong tendency not to migrate toward the wound, as is the case for low-density sheets (Figures 5G and 4A, respectively). Of importance, these sheets still display DOX-dependent activation of cellular speed (Figure 5H). Taken together, these data show that spatial patterns of Erk1/2 activity can affect the guidance of collective migration in epithelial sheets.
FIGURE 4:
Cell density throughout sheets influences ligand-dependent collective migration behavior. (A) Analysis of migration directions throughout low-density sheets (2000 cells/mm2) reveals that ligand-dependent orientation of cellular motility is not directed toward the wound for cells distal to the leading edge. (B) These low-density sheets display a significant increase in cell migration speed in response to ligand. The same experiments were repeated for high–cell density sheets (3500 cells/mm2) in C and D, respectively. All experiments depict data after 35 h of ligand stimulation.
FIGURE 5:
Exogenous expression of a constitutively active MEK1 can restore elevated Erk1/2 activity at high cell density. Low-density colonies (A) were compared with high-density colonies (B) using Western blot analysis (HaCaT cells) and cellular migration speed analysis (HaCaT H2B cells). Introduction of a DOX-inducible, constitutively active MEK1 (CA-MEK1) yields DOX-dependent activation of Erk1/2 and cellular speed to similar levels as those of TGFβ in low-density colonies. However, at high cell density, TGFβ treatment no longer stimulates Erk1/2 or cellular speed, both of which can be overcome by DOX-dependent induction of CA-MEK1. An empty vector was used to construct a control cell line (Tet-Control). (C) Hybrid sheets constructed of HaCaT-H2B cells and Tet-CA cells allow for ligand-free specific activation of Erk1/2 toward the leading edge of sheets. (D) Directional analysis of collective migration in hybrid sheets reveals wound-directed migration in a DOX-dependent manner. (E) Hybrid sheets display a similar spatial profile of cellular speed as TGFβ-stimulated sheets. (F) DOX globally elevates Erk1/2 activity in sheets constructed entirely of Tet-CA cells. (G, H) Tet-CA sheets were analyzed as in D and E, respectively. All experiments depict data after 35 h of DOX stimulation.
DISCUSSION
Density-dependent Erk1/2 activation determines leader cell positioning
Although local Erk1/2 activity has been described at the leading edge of sheets of cells (Matsubayashi et al., 2004; Nikolic et al., 2006) and elevated Erk1/2 activity has been identified as a marker of leader cell identity (Khalil and Friedl, 2010), it is unclear exactly how this local biochemical activity is positioned at the leading edge. Our experiments clearly show that the Erk1/2 activity of a cell is not only elevated by TGFβ stimulation but is also inhibited by high cell density (Figure 3C). Migrating sheets of cells experience low resistance to migration at the leading edge, where cells are able to freely diffuse toward the wound, resulting in lower cell density at the wound border (Figure 3B). This asymmetric density throughout a sheet is causally linked to the pattern of Erk1/2 activity throughout the sheet, as modulation of cellular density can affect such a pattern (Figure 3, A and D). Although the molecular mechanism behind density-dependent regulation of Erk1/2 is not known, this regulation aids in the positioning of leader cells (elevated Erk1/2 activity) toward the leading edge of sheets.
Leader cell positioning effectively influences the migration direction of a collective group of cells
Previous studies using sheets of cells concluded that lack of resistance at the wound border plays a role in the guidance of leader cells toward the wound (Vitorino et al., 2011). Our data consistently show that all the cells at the leading edge (0–250 μm) display exceptional orientation toward the wound independent of Erk1/2 levels (Figures 1A and 2E). As a result, we conclude that orientation of these cells is largely independent of Erk1/2 activity and is primarily directed by the low-resistance paths toward the wound. However, the Erk1/2 activity of leader cells does regulate cellular speed, driving the forward migration of a TGFβ-stimulated sheet (Figure 2D). The elevated Erk1/2 activity in leader cells is also required for orienting follower cells in the rear of the sheet toward the wound (Figure 3E), provided that the follower cells do not themselves display elevated Erk1/2 activity (Figures 4 and 5). We interpret these data to suggest that a spatially constrained pattern of Erk1/2 activity throughout a sheet is largely wound directed. However, when the entire sheet consists of leader cells (elevated Erk1/2 activity), the direction of collective migration is largely not wound directed (Figures 4A and 5G). Although it is not entirely clear what directional cues are governing non–wound-directed collective migration, our experiments provide evidence for a functional relationship between leader cell patterning and collective migration guidance during wound healing.
Physiological significance of density-directed collective cell migration
Our novel findings are that spatial positioning of leader cells throughout a collective group of cell is driven by spatial patterns in cell density and that spatial segregation of leader cells affects the direction of collective migration throughout the collective group. These findings may provide insight into not only how epithelial wounds heal, but also how epithelial barriers maintain homogeneous cell density in vivo. In our TGFβ-stimulated epithelial sheets, the direction of collective cell migration aligns such that cells migrate from high to low cell density (Figures 1A and 3B). This type of density-directed cell migration may occur in tissues in order to prevent accumulation of heterogeneous density. In areas where cell proliferation is high, cell density will inevitably become elevated unless a mechanism is in place to relieve accumulated cell density. Similarly, during cancer progression, tumor density may drive tumor expansion through local patterning of Erk1/2 activity and subsequent density-directed collective migration. Thus it is possible that a commonly used mechanism for tissue maintenance is highjacked during tumor progression to aid in the process of metastasis.
MATERIALS AND METHODS
Cell culture and experimental platforms
HaCaT cells were cultured in DMEM supplemented with 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin under 5% CO2 at 37°C. For sheet migration studies, 250,000 cells were seeded into each well of a 96-well plate (MGB096-1-2-HG-L; Matrical Bioscience, Spokane, WA). After 24 h, half of the cells (on one side of the well) were manually removed using a 200-μl pipette tip. Fluorescence imaging of live cells was conducted with phenol-free DMEM (31053028; Life Technologies, Grand Island, NY) that was prebleached for 24 h under a 15-W fluorescent light. All experiments were conducted with a TGFβ concentration of 100 pM (R&D Systems, Minneapolis, MN), EGF concentration of 100 nM (R&D Systems), SB431542 concentration of 10 μM (R&D Systems), CI-1040 concentration of 500 nM (unless explicitly stated otherwise; Tocris Bioscience, Minneapolis, MN), and DOX concentration of 0.5 μg/ml (Sigma-Aldrich, St. Louis, MO). In hybrid sheet migration assays, 250,000 Tet-Ca cells were seeded into each well. After 3 h, half of cells were removed, similar to the procedure for sheets. Then 250,000 HaCaT H2B mCherry cells or EKAR cells were added to each well. After 3 h, all cells were trypsinized for 2 min at 20°C, and a 200-μl pipette tip was used to remove desired cells, leaving behind a sheet with a leading edge of mixed Tet-CA and HaCaT-H2B cells.
Construction of stable cell lines
Transgenic N-terminally tagged H2B mCherry cell lines were generated using murine leukemia virus (MuLV)–mediated gene transfer by which 293T cells were simultaneously transfected with MuLV packaging vector, pHCMV-VSV-g, and pRex-H2B-mCherry. Viral supernatant was used to infect HaCaT cells, supplemented with 10 μg/ml Polybrene (Sigma-Aldrich). Tet-CA cells were generated in HaCaT H2B GFP cells using lentiviral-mediated gene transfer by which 293T cells were simultaneously transfected with pMDLg, pHCMV-VSV-g, pREV and CSIV-TRE-RfA-CMV-KT, or CSIV-TRE-CA-MEK1-CMV-KT plasmids (gifts of Hiroyuki Miyoshi, RIKEN BioResource Center, Tsukuba, Japan). Constitutively active MKK1 is the R4F mutation (a gift of Natalie Ahn, University of Colorado, Boulder, CO) and was cloned into CSIV-TRE-RfA-CMV-KT using Gateway cloning (Invitrogen, Grand Island, NY). Stable transgenic EKAR cells were manufactured using transient transfection of pBSR-cyto-EKAR and pBP into HaCaT or SCC-13 cells and selected using 8 μg/ml blasticidin S (Life Technologies) for 2 wk.
Imaging, cellular tracking, and data analysis
Data acquisition and analysis of live-cell microscopy videos were conducted as previously described (Chapnick et al., 2013). Spatial quantification of sheets of EKAR cells were conducted using Matlab to construct videos of background-corrected fluorescence resonance energy transfer (FRET) ratios, and ImageJ (National Institutes of Health, Bethesda, MD) was used to perform line scans throughout sheets in such videos. The leading edge of sheets were aligned manually in Excel, which was also used to construct final graphs of FRET ratio versus position.
Two-dimensional cell area was quantified by manual segmentation of cells in ImageJ, and conversion of area from pixels squared to micrometers squared used the conversion factor 1 pixel2 = 0.416 μm2 at 20× magnification and binning of 2 × 2 pixels.
Immunofluorescence
Cells grown in 96-well plates were fixed with –20°C 100% MeOH and incubated at 4°C overnight (endogenous) or 3.7% paraformaldehyde for 5 min (fluorescent protein–containing samples). Fixed cells were rehydrated in phosphate-buffered saline and blocked with 0.5% bovine serum albumin (BSA) in 50 mM Tris-HCl supplemented with 0.05% Tween-20 for 1 h. Primary antibody was substituted for BSA at a dilution of 1:200 for 3 h at room temperature. Secondary antibodies were used at a dilution of 1:200 for 1 h at room temperature. The antibodies used were ppERK1/2 (M7802; Sigma-Aldrich), E-cadherin (SC-21791; Santa Cruz Biotechnology, Dallas, TX), Alexa 555 goat anti-rabbit (Molecular Probes, Grand Island, NY), and Alexa 488 goat anti-mouse (Molecular Probes).
Western blotting
The following antibodies were used: pSmad2 (3108; Cell Signaling, Danvers, MA), Smad2/3 (SC-133098; Santa Cruz Biotechnology), ezrin (E8897; Sigma-Aldrich), ppERK1/2 (M9692; Sigma-Aldrich), ERK2 (SC-154; Santa Cruz Biotechnology), pMEK (9154; Cell Signaling), MEK (2352; Cell Signaling), and lamin A/C (SC-376248; Santa Cruz Biotechnology).
Statistical analysis
The error displayed for speed measurements is SEM, and the number of observations is at least three, representing experiments done in triplicate. Statistical significance is denoted by the symbol †, which signifies p < 0.01. This p value was calculated from a two-tailed t test in Excel. For directional measurements the p value represents comparisons of the percentage of cells with cellular directions between 180 and 360°.
Supplementary Material
Acknowledgments
We thank Kazuhiro Aoki and Hiroyuki Miyoshi for EKAR and lentiviral expression plasmids. We thank Tobias Meyer, Sabrina Spencer, Feng-Qiao Tsai, Leslie Leinwand, Kristi Anseth, Natalie Ahn, Zhike Zi, Amy Palmer, Tom Cheung, and members of Xuedong Liu's laboratory for discussion. D.A.C. was supported by a predoctoral training grant from the National Institute of General Medical Sciences (T32GM08759). This work was supported by Grants R01GM083172 and R01CA107098 from the National Institutes of Health to X.L. The ImageXpress MicroXL was supported by National Center for Research Resources Grant S10 RR026680 from the National Institutes of Health.
Abbreviations used:
- EMT
epithelial-to-mesenchymal transition
- Erk
extracellular regulated kinase
- TGF
transforming growth factor
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-01-0697) on March 12, 2014.
REFERENCES
- Arrieumerlou C, Meyer T. A local coupling model and compass parameter for eukaryotic chemotaxis. Dev Cell. 2005;8:215–227. doi: 10.1016/j.devcel.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Bronner-Fraser M. Neural crest cell migration in the developing embryo. Trends Cell Biol. 1993;3:392–397. doi: 10.1016/0962-8924(93)90089-j. [DOI] [PubMed] [Google Scholar]
- Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2:563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- Chapnick DA, Jacobsen J, Liu X. The development of a novel high throughput computational tool for studying individual and collective cellular migration. PloS One. 2013;8:e82444. doi: 10.1371/journal.pone.0082444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapnick DA, Warner L, Bernet J, Rao T, Liu X. Partners in crime: the TGFbeta and MAPK pathways in cancer progression. Cell Biosci. 2011;1:42. doi: 10.1186/2045-3701-1-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol. 2009;10:445–457. doi: 10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- Gabay L, Seger R, Shilo BZ. In situ activation pattern of Drosophila EGF receptor pathway during development. Science. 1997;277:1103–1106. doi: 10.1126/science.277.5329.1103. [DOI] [PubMed] [Google Scholar]
- Harvey CD, Ehrhardt AG, Cellurale C, Zhong H, Yasuda R, Davis RJ, Svoboda K. A genetically encoded fluorescent sensor of ERK activity. Proc Natl Acad Sci USA. 2008;105:19264–19269. doi: 10.1073/pnas.0804598105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz R, Webb D. Cell migration. Curr Biol. 2003;13:R756–R759. doi: 10.1016/j.cub.2003.09.014. [DOI] [PubMed] [Google Scholar]
- Jékely G, Sung HH, Luque CM, Rørth P. Regulators of endocytosis maintain localized receptor tyrosine kinase signaling in guided migration. Dev Cell. 2005;9:197–207. doi: 10.1016/j.devcel.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Keller R. Cell migration during gastrulation. Curr Opin Cell Biol. 2005;17:533–541. doi: 10.1016/j.ceb.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Khalil AA, Friedl P. Determinants of leader cells in collective cell migration. Integr Biol (Camb) 2010;2:568–574. doi: 10.1039/c0ib00052c. [DOI] [PubMed] [Google Scholar]
- Lauffenburger DA, Horwitz AF. Cell migration: a physically integrated molecular process. Cell. 1996;84:359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- Locascio A, Nieto MA. Cell movements during vertebrate development: integrated tissue behaviour versus individual cell migration. Curr Opin Genet Dev. 2001;11:464–469. doi: 10.1016/s0959-437x(00)00218-5. [DOI] [PubMed] [Google Scholar]
- Lois C, Garcia-Verdugo JM, Alvarez-Buylla A. Chain migration of neuronal precursors. Science. 1996;271:978–981. doi: 10.1126/science.271.5251.978. [DOI] [PubMed] [Google Scholar]
- Martin P. Wound healing—aiming for perfect skin regeneration. Science. 1997;276:75–81. doi: 10.1126/science.276.5309.75. [DOI] [PubMed] [Google Scholar]
- Martin P, Parkhurst SM. Parallels between tissue repair and embryo morphogenesis. Development. 2004;131:3021–3034. doi: 10.1242/dev.01253. [DOI] [PubMed] [Google Scholar]
- Massague J. TGF-beta signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y, Ebisuya M, Honjoh S, Nishida E. ERK activation propagates in epithelial cell sheets and regulates their migration during wound healing. Curr Biol. 2004;14:731–735. doi: 10.1016/j.cub.2004.03.060. [DOI] [PubMed] [Google Scholar]
- Mayor R, Carmona-Fontaine C. Keeping in touch with contact inhibition of locomotion. Trends Cell Biol. 2010;20:319–328. doi: 10.1016/j.tcb.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mine N, Iwamoto R, Mekada E. HB-EGF promotes epithelial cell migration in eyelid development. Development. 2005;132:4317–4326. doi: 10.1242/dev.02030. [DOI] [PubMed] [Google Scholar]
- Mogilner A, Oster G. Cell motility driven by actin polymerization. Biophys J. 1996;71:3030–3045. doi: 10.1016/S0006-3495(96)79496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustakas A, Heldin C-H. Non-Smad TGF-beta signals. J Cell Sci. 2005;118:3573–3584. doi: 10.1242/jcs.02554. [DOI] [PubMed] [Google Scholar]
- Mu Y, Gudey SK, Landstrom M. Non-Smad signaling pathways. Cell Tissue Res 347, 11–20. 2012. [DOI] [PubMed]
- Mulder KM. Role of Ras and Mapks in TGFbeta signaling. Cytokine Growth Factor Rev. 2000;11:23–35. doi: 10.1016/s1359-6101(99)00026-x. [DOI] [PubMed] [Google Scholar]
- Murase S, Horwitz AF. Directions in cell migration along the rostral migratory stream: the pathway for migration in the brain. Curr Top Dev Biol. 2004;61:135–152. doi: 10.1016/S0070-2153(04)61006-4. [DOI] [PubMed] [Google Scholar]
- Nikolic DL, Boettiger AN, Bar-Sagi D, Carbeck JD, Shvartsman SY. Role of boundary conditions in an experimental model of epithelial wound healing. Am J Physiol Cell Physiol. 2006;291:C68–C75. doi: 10.1152/ajpcell.00411.2005. [DOI] [PubMed] [Google Scholar]
- Omelchenko T, Vasiliev JM, Gelfand IM, Feder HH, Bonder EM. Rho-dependent formation of epithelial “leader” cells during wound healing. Proc Natl Acad Sci USA. 2003;100:10788–10793. doi: 10.1073/pnas.1834401100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- Poujade M, Grasland-Mongrain E, Hertzog A, Jouanneau J, Chavrier P, Ladoux B, Buguin A, Silberzan P. Collective migration of an epithelial monolayer in response to a model wound. Proc Natl Acad Sci USA. 2007;104:15988–15993. doi: 10.1073/pnas.0705062104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorth P. Collective cell migration. Annu Rev Cell Dev Biol. 2009;25:407–429. doi: 10.1146/annurev.cellbio.042308.113231. [DOI] [PubMed] [Google Scholar]
- Servant G, Weiner OD, Herzmark P, Balla T, Sedat JW, Bourne HR. Polarization of chemoattractant receptor signaling during neutrophil chemotaxis. Science. 2000;287:1037–1040. doi: 10.1126/science.287.5455.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Haastert PJ, Devreotes PN. Chemotaxis: signalling the way forward. Nat Rev Mol Cell Biol. 2004;5:626–634. doi: 10.1038/nrm1435. [DOI] [PubMed] [Google Scholar]
- Vitorino P, Hammer M, Kim J, Meyer T. A steering model of endothelial sheet migration recapitulates monolayer integrity and directed collective migration. Mol Cell Biol. 2011;31:342–350. doi: 10.1128/MCB.00800-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitorino P, Meyer T. Modular control of endothelial sheet migration. Genes Dev. 2008;22:3268–3281. doi: 10.1101/gad.1725808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Goswami S, Sahai E, Wyckoff JB, Segall JE, Condeelis JS. Tumor cells caught in the act of invading: their strategy for enhanced cell motility. Trends Cell Biol. 2005;15:138–145. doi: 10.1016/j.tcb.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Webb DJ, Parsons JT, Horwitz AF. Adhesion assembly, disassembly and turnover in migrating cells—over and over and over again. Nat Cell Biol. 2002;4:E97–E100. doi: 10.1038/ncb0402-e97. [DOI] [PubMed] [Google Scholar]
- Werner S, Grose R. Regulation of wound healing by growth factors and cytokines. Physiol Rev. 2003;83:835–870. doi: 10.1152/physrev.2003.83.3.835. [DOI] [PubMed] [Google Scholar]
- Xia Y, Karin M. The control of cell motility and epithelial morphogenesis by Jun kinases. Trends Cell Biol. 2004;14:94–101. doi: 10.1016/j.tcb.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Zhang L, Wang W, Hayashi Y, Jester JV, Birk DE, Gao M, Liu CY, Kao WW, Karin M, Xia Y. A role for MEK kinase 1 in TGF-beta/activin-induced epithelium movement and embryonic eyelid closure. EMBO J. 2003;22:4443–4454. doi: 10.1093/emboj/cdg440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YE. Non-Smad pathways in TGF-beta signaling. Cell Res. 2009;19:128–139. doi: 10.1038/cr.2008.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.