Abstract
Development of yellow mustard (Sinapis alba L.) with superior quality traits (low erucic and linolenic acid contents, and low glucosinolate content) can make this species as a potential oilseed crop. We have recently isolated three inbred lines Y1127, Y514 and Y1035 with low (3.8%), medium (12.3%) and high (20.8%) linolenic acid (C18∶3) content, respectively, in this species. Inheritance studies detected two fatty acid desaturase 3 (FAD3) gene loci controlling the variation of C18∶3 content. QTL mapping revealed that the two FAD3 gene loci responsible for 73.0% and 23.4% of the total variation and were located on the linkage groups Sal02 and Sal10, respectively. The FAD3 gene on Sal02 was referred to as SalFAD3.LA1 and that on Sal10 as SalFAD3.LA2. The dominant and recessive alleles were designated as LA1 and la1 for SalFAD3.LA1, and LA2 and la2 for SalFAD3.LA2. Cloning and alignment of the coding and genomic DNA sequences revealed that the SalFAD3.LA1 and SalFAD3.LA2 genes each contained 8 exons and 7 introns. LA1 had a coding DNA sequence (CDS) of 1143 bp encoding a polypeptide of 380 amino acids, whereas la1 was a loss-of-function allele due to an insertion of 584 bp in exon 3. Both LA2 and la2 had a CDS of 1152 bp encoding a polypeptide of 383 amino acids. Allele-specific markers for LA1, la1, LA2 and la2 co-segregated with the C18∶3 content in the F2 populations and will be useful for improving fatty acid composition through marker assisted selection in yellow mustard breeding.
Introduction
Yellow mustard (Sinapis alba L., 2n = 24) is cultivated as an important condiment crop. It has many desirable agronomic traits such as resistance to cabbage aphids [1], flea beetles [2], [3] and blackleg diseases [4]. In addition, it is drought tolerant and resistant to pod shattering. Yellow mustard germplasm with canola quality (low erucic acid and low glucosinolate contents) was developed at Agriculture and Agri-Food Canada-Saskatoon Research Centre (AAFC-SRC) [5], which makes yellow mustard have the potential to become an alternative oilseed crop to canola B. napus, especially in semi-arid areas.
The oil quality of canola B. napus is determined by the proportion of the three major unsaturated fatty acids: oleic acid (C18∶1), linoleic acid (C18∶2) and linolenic acid (C18∶3). Traditional B. napus cultivars contain 9% C18∶3 of the total fatty acids [6]. The high level of linolenic acid in canola oil is undesirable since it shortens the shelf life and causes off-type flavour of the oil due to the three easily oxidized double bonds. A low linolenic acid mutant, containing 3–5% C18∶3, was produced by ethyl methanesulfonate (EMS) treatment of a high C18∶3 B. napus cv. Oro seed [7]. Current low C18∶3 canola cultivars have been developed using this low linolenic gene source.
Linolenic acid content is determined mainly by the embryonic genotype with some influence from temperature, maternal genotype and cytoplasm in B. napus [8]–[10]. QTL mapping identified two major QTLs, accounting for 25.2–28.8% and 52.4–62.7% of the C18∶3 variation, located on the linkage groups A4 and C4, respectively, in B. napus [11], [12]. It was reported that the low C18∶3 variant resulted from mutations of FAD3 genes in B. napus [11]–[13]. The FAD3 gene on A4 harboured a C to T substitution in exon 7, which when translated causes the wild type amino acid arginine to be replaced by cysteine. The FAD3 gene on C4 contained a G to A substitution in the 5′ splice site of intron 6 in the low C18∶3 B. napus line. FAD3 allele-specific markers based on the sequence variation were developed and proved to be useful for identification of different C18∶3 genotypes in canola B. napus [11], [12]. Yellow mustard accessions contain 6.9–12.4% linolenic acid of total fatty acids in the seed [14], [15]. Recently, inbred lines with high (18.5%), medium (13.8%) and low (3.8%) linolenic acid content, respectively, have been obtained through inbreeding of heterozygous open-pollinated plants in yellow mustard [16].
The low linolenic acid variant (3.8%) is a valuable gene source for breeding canola-quality yellow mustard with high stability oil (high oleic and low linolenic acids) as that of canola B. napus. The knowledge about genetic and molecular bases of the variation in C18∶3 content and development of FAD3 allele-specific markers will greatly facilitate the development of low linolenic canola-quality yellow mustard. The objectives of this study were: 1) to determine the inheritance and perform QTL mapping of the C18∶3 content; and 2) to clone the FAD3 genes and further develop allele-specific markers for marker assisted selection.
Materials and Methods
Plant Materials
Linolenic acid contents of the three parental lines Y1127, Y514 and Y1035 are shown in Table 1. Y1127 is an S4 inbred line produced by selfing of the low linolenic S2 line Y158 for two generations and has a low C18∶3 content (average: 3.8%). Y514 is the doubled haploid line SaMD3 [17] and has a medium C18∶3 content (average: 12.3%). Y1035 is an S4 inbred line and has a high C18∶3 content (average: 20.8%).
Table 1. Linolenic acid contents of the parental lines Y1127, Y514, Y1035 and F1 seeds, and the mid-parental value in yellow mustard.
| Genotype | Generation | Linolenic Acid Content* (% of total fatty acids) |
| Y1127 | S4 | 3.8±0.7 |
| Y514 | DH | 12.3±0.7 |
| Y1035 | S4 | 20.8±0.8 |
| Y1127×Y1035 | F1 | 13.7±1.3 |
| Mid parent value | 12.3 | |
| Y1127×Y514 | F1 | 8.9±0.7 |
| Mid parent value | 8.0 | |
| Y514×Y1035 | F1 | 15.3±0.7 |
| Mid parent value | 16.5 |
*: Linolenic acid content is expressed as mean value ± standard deviation.
The F1 seeds of the three crosses Y1127 (low)×Y1035 (high), Y1127 (low)×Y514 (medium) and Y514 (medium)×Y1035 (high) were produced. To produce the BC1 seeds, the F1 plants of the three crosses were crossed as the female with the parental line with a lower C18∶3 content. All plants were raised under the same conditions in the greenhouse at AAFC-SRC.
Regional Linkage Mapping
Regional linkage mapping of the linolenic acid content was performed using intron length polymorphism (ILP) markers and bulked segregant analysis (BSA) [18]. A total of 1478 ILP primer pairs: 380 from Arabidopsis thaliana [19] and 1098 from B. napus [20] were used to screen the three parental lines for polymorphic markers. The high bulk was made by mixing equal amount of DNA from 10 F2 plants with the highest C18∶3 content, while the low bulk was formed from 10 F2 plants with the lowest C18∶3 content for each of the three crosses. The primers detecting polymorphic markers between the two bulks were subsequently used to genotype individual plants of the three F2 populations. Genomic DNA was extracted from young leaves of the parental lines Y1127, Y514 and Y1035, F1 and F2 plants using a modified sodium dodecyl sulfate method [21]. Each PCR (20 µl) contained 1× standard PCR buffer (NEB), 1 U of Taq polymerase (NEB), 0.25 µM forward primer, 0.25 µM reverse primer, 100 µM each dNTP and 50 ng of genomic DNA in a total volume 20 µL. The PCR amplification consisted of an initial denaturation at 94°C for 5 min, 35 cycles consisting of 94°C (45 sec), 55°C (45 sec), 72°C (1 min) terminating with 72°C for 7 min. All PCR products were analyzed by electrophoresis in 2% agarose gels in 1× Tris-acetate-ethylenediaminetetraacetic acid buffer. Gels were visualized by staining in ethidium bromide and photographed on a digital gel documentation system.
The regional linkage map of C18∶3 content was constructed using JoinMap 4.0 [22] with a minimum LOD threshold of 4.0. QTL analysis of C18∶3 content was performed using the interval mapping method of MapQTL 6.0 [23]. A Chi-square test was used for evaluating the genetic model of C18∶3 content in the BC1 and F2 populations, and the ILP markers in the F2 populations.
Cloning of the Coding Region of the FAD3 Gene
Primer pair No 1 (Table S1) was designed based on the conserved coding regions of the FAD3 genes in B. napus and A. thaliana. It was used to clone the coding DNA sequence (CDS) of the FAD3 gene in yellow mustard. Immature seeds at 22 days after pollination were collected from two individual plants from each of the parental lines. Total RNA was extracted from the immature seeds using the RNeasy Plant Mini Kit (Qiagen) as per the manufacturer’s instructions. 750 ng of RNA from each of the parental lines was used to prepare the cDNA using Qiagen’s Omniscript RT Kit as per the manufacturer’s instructions. Each PCR (20 µl) contained 1× PCR standard buffer (NEB), 100 µM of each dNTP, 0.25 µM of each forward and reverse primer, 1 U of Taq polymerase (NEB) and 50 ng of cDNA. Polymerase chain reaction was performed with an initial denaturation at 94°C for 3 min followed by 35 cycles of 45 s at 94°C, 30 s at 55°C and 1 min at 72°C with a final extension cycle of 72°C for 10 min.
Cloning of the 5′ and 3′ Flanking Sequences and the Genomic DNA Sequences of the FAD3 Genes
Primer pairs No 2 and 3 (Table S1) were designed based on the 5′ coding sequences of the cloned SalFAD3.LA1 and SalFAD3.LA2 genes, respectively. They were used to clone the 5′ upstream sequences by PCR walking according to the protocol of Siebert et al. [24]. Primer No 4 (Table S1) was designed based on the 3′ coding sequences of the cloned SalFAD3.LA1 and SalFAD3.LA2 genes, and was used to clone the 3′ flanking sequence by PCR walking. Primer pairs No 5 and 6 (Table S1) were designed based on the 5′ flanking sequence and the 3′ flanking sequence of the cloned SalFAD3.LA1 and SalFAD3.LA2 genes, respectively, and were used to clone the genomic DNA sequences of SalFAD3.LA1 and SalFAD3.LA2 genes. The standard protocol from the Clontech kit (website: www.clontech.com, Protocol PT 3042, Version PR 03300) by Gwyneth Ingram and Karine Coenen was followed to facilitate the PCR walking.
DNA Sequencing
The expected PCR bands were cloned using the pGEM-T Vector System I (Promega) following the provided instructions. The plasmids were extracted using the QiaSpin Kit (Qiagen) following the manufacturer’s instructions and sequenced using the primer pairs No 7–11 (Table S1) at the Plant Biotechnology Institute, National Research Council, Canada.
Phylogenetic Tree
The multiple alignments were performed using ClustalW (http://www.ebi.ac.uk/clustalw/). MEGA software (version 4.0) (http://www.megasoftware.net/index.html)[25] was used to construct a phylogenetic tree with the aligned protein sequences. The neighbor-joining method was used with the pairwise deletion option, poisson correction model, and the 1000 bootstrap replicates test.
Development of the SalFAD3.LA1 and SalFAD3.LA2 Allele-specific Markers
The SalFAD3.LA1 and SalFAD3.LA2 allele-specific markers were generated using primer pair No 12 (Table S1) which was designed based on the conserved flanking sequences of intron 3. The PCR reaction was performed with LongAmp Taq 2× Master Mix (NEB) following the manufacturer’s instructions with a 60°C annealing temperature.
Fatty Acid Analysis
Seed fatty acid composition was analyzed according to [26] with the following modification: the gas chromatography of the methyl esters was performed with a HP-INNOWax fused silica capillary column (0.25 mm by 0.5 m and 7.5 µm) (Agilent Technologies) at 250°C using hydrogen as the carrier gas. A minimum of 10 seeds from each of the parental lines and F1 hybrids as well as 160 F2 seeds of each of the three crosses were half-seed analyzed according to [27]. Ninety-six seeds from each of the BC1 populations were analyzed using the single seed method.
Results
Linolenic Acid Content is Controlled by Two Gene Loci in Yellow Mustard
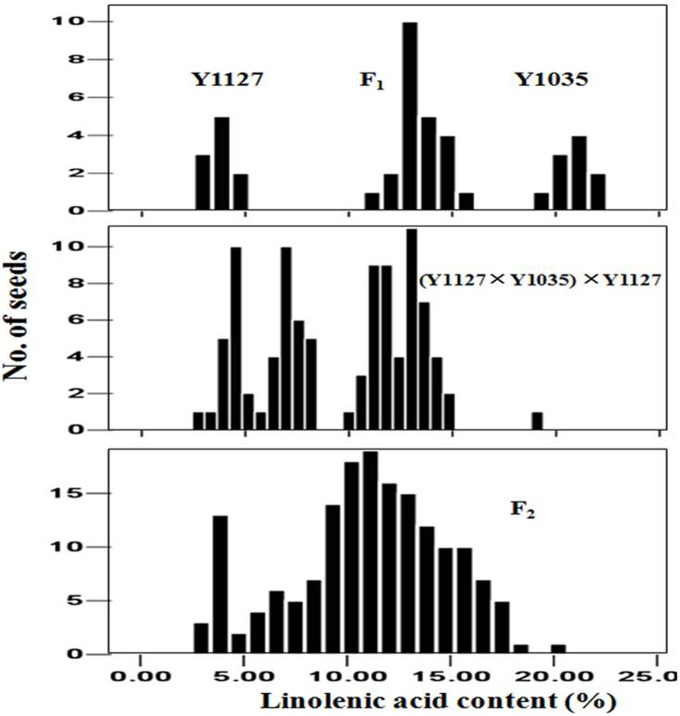
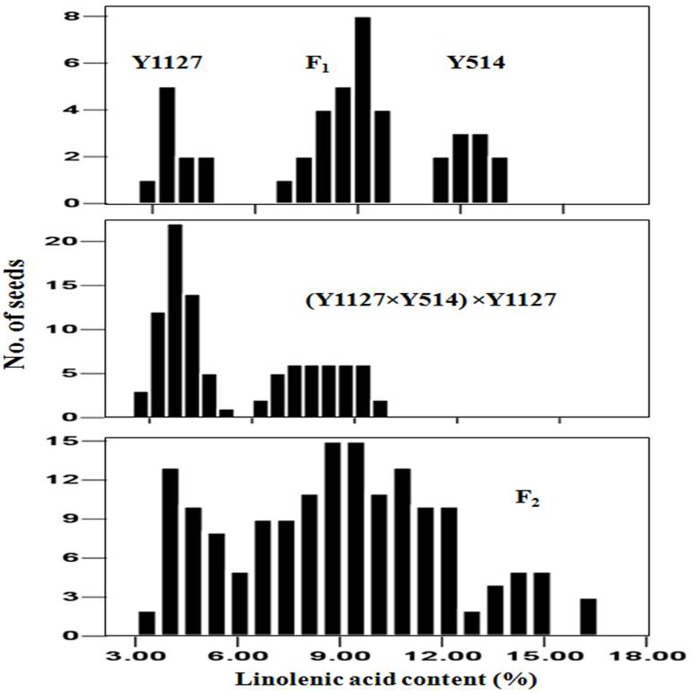
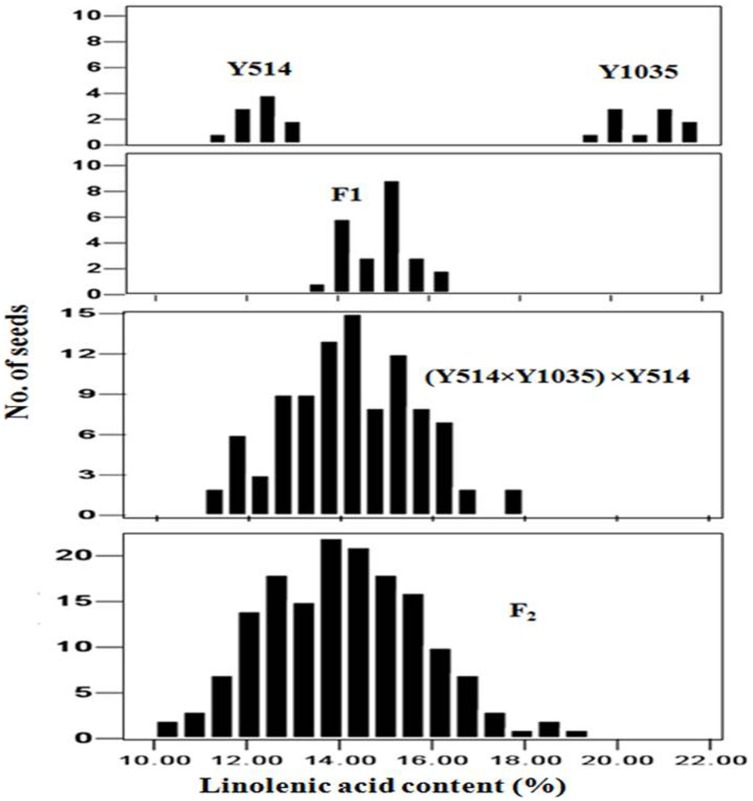
The C18∶3 content of the F1 seeds was significantly higher than the mid-parent value in the crosses of Y1127 (low)×Y1035 (high) (t = 3.84, p<0.01) and Y1127 (low)×Y514 (medium) (t = 5.62, p<0.01) (Table 1, Figure 1 and 2), suggesting a partial dominance of the high/medium over low C18∶3 content. However, in the cross of Y514 (medium)×Y1035 (high) the F1 seeds had significantly lower C18∶3 content (15.3%) than the mid-parent value of 16.5% (t = 6.98, p<0.01) (Table 1, Figure 3), indicating a partial dominance of the medium over high C18∶3 content.
Figure 1. Frequency distributions of linolenic acid contents in individual seeds of Y1127, Y1035, F1, (Y1127×Y1035)×Y1035 and F2 populations.
Figure 2. Frequency distributions of linolenic acid contents in individual seeds of Y1127, Y514, F1, (Y1127×Y514)×Y1127 and F2 populations.
Figure 3. Frequency distributions of linolenic acid contents in individual seeds of Y514, Y11035, F1, (Y514×Y1035)×Y514 and F2 populations.
The BC1 seeds of (Y1127×Y1035)×Y1127 were classified into two groups: seeds with medium to high (5.4–19.1%) C18∶3 content and seeds with low (3.0–4.9%) C18∶3 content (Figure 1), fitting with a segregation ratio of 3∶1 (χ2 = 2.00, p = 0.16). The F2 seeds of Y1127×Y1035 ranged from 2.9% to 20.4% in C18∶3 content (Figure 1) with a segregation ratio of 15∶1 (seeds with 4.5–20.4% versus seeds with 2.9–4.3% C18∶3 content) (χ2 = 3.07, p = 0.08). Therefore, the segregation patterns of C18∶3 content in the BC1 and F2 populations supported a digenic inheritance model in this cross.
The BC1 seeds of (Y1127×Y514)×Y1127 showed a segregation ratio of 1∶1 (seeds with 2.7–5.2% versus seeds with 6.4–9.7% C18∶3 content) (Figure 2) (χ2 = 3.38, p = 0.07), suggesting that the C18∶3 content was controlled by one gene locus in this cross. The F2 seeds of Y1127×Y514 showed a continuous distribution ranging from 3.0% to 16.5% in the C18∶3 content (Figure 2). The BC1 seeds of (Y514×Y1035)×Y514 and the F2 seeds of Y514×Y1035 exhibited a continuous frequency distribution in the C18∶3 content (Figure 3). Therefore, it was not possible to classify the seeds into discrete groups.
Two QTLs Accounting for the Variation of C18∶3 Content are Mapped to Linkage Groups Sal02 and Sal10, Respectively
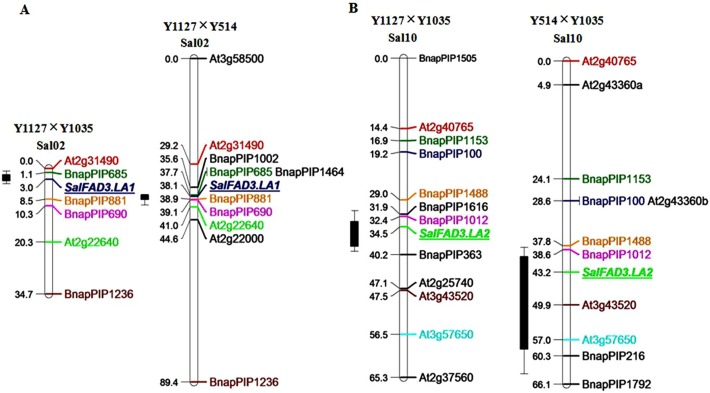
In the F2 population of Y1127 (low)×Y1035 (high), eighteen ILP primer pairs were polymorphic between the high (16.6–20.4%) and low (2.9–4.0%) C18∶3 bulks and generated 18 markers (Table 2). The 18 markers were mapped to two linkage groups, each of which carried one QTL for the C18∶3 content (Figure 4). Based on the common ILP markers, the two linkage groups were revealed to be Sal02 and Sal10 of the constructed S. alba map [28]. One QTL (LOD = 45.43) accounting for 73.0% of the total variation of C18∶3 content was localized between BnapPIP685 and BnapPIP881 in Sal02 (Figure 4). The other QTL (LOD = 9.28) responsible for 23.4% of the total variation was located between BnapPIP1012 and BnapPIP363 in Sal10 (Figure 4). Together, the two QTLs explained 96.4% of the total variation for C18∶3 content in the F2 population.
Table 2. Polymorphic ILP primers used for regional linkage mapping of C18∶3 content in the three F2 populations of Y1127×Y1035, Y1127×Y514 and Y514×Y1035.
| Primer Name | Locus Name | Y1127×Y1035 | Y1127×Y514 | Y514×Y1035 | |||
| marker type | χ2 Value* | Marker type | χ2 Value | Marker type | χ2 Value | ||
| At2g22640 | At2g22640 | Codominant | χ2 = 2.15 | Codominant | χ2 = 0.03 | - | |
| At2g31490 | At2g31490 | Codominant | χ2 = 0.00 | Codominant | χ2 = 0.21 | - | |
| BnapPIP1236 | BnapPIP1236 | Dominant | χ2 = 0.21 | Dominant | χ2 = 0.61 | - | |
| BnapPIP685 | BnapPIP685 | Dominant | χ2 = 1.57 | Dominant | χ2 = 0.94 | - | |
| BnapPIP690 | BnapPIP690 | Dominant | χ2 = 1.28 | Dominant | χ2 = 0.63 | - | |
| BnapPIP881 | BnapPIP881 | Codominant | χ2 = 2.74 | Codominant | χ2 = 0.41 | - | |
| At2g40765 | At2g40765 | Codominant | χ2 = 1.30 | - | - | Dominant | χ2 = 1.90 |
| At3g43520 | At3g43520 | Codominant | χ2 = 0.46 | - | - | Dominant | χ2 = 0.83 |
| At3g57650 | At3g57650 | Codominant | χ2 = 0.20 | - | - | Codominant | χ2 = 3.27 |
| BnapPIP100 | BnapPIP100 | Dominant | χ2 = 0.10 | - | - | Dominant | χ2 = 1.02 |
| BnapPIP1012 | BnapPIP1012 | Dominant | χ2 = 0.30 | - | - | Dominant | χ2 = 2.70 |
| BnapPIP1153 | BnapPIP1153 | Codominant | χ2 = 1.28 | - | - | Codominant | χ2 = 1.16 |
| BnapPIP1488 | BnapPIP1488 | Dominant | χ2 = 0.05 | - | - | Dominant | χ2 = 1.63 |
| At2g25740 | At2g25740 | Codominant | χ2 = 0.67 | - | - | - | - |
| At2g37560 | At2g37560 | Codominant | χ2 = 1.43 | - | - | - | - |
| BnapPIP1505 | BnapPIP1505 | Codominant | χ2 = 3.27 | - | - | - | - |
| BnapPIP1616 | BnapPIP1616 | Codominant | χ2 = 0.68 | - | - | - | - |
| BnapPIP363 | BnapPIP363 | Dominant | χ2 = 1.54 | - | - | - | - |
| At2g22000 | At2g22000 | - | - | Codominant | χ2 = 1.13 | - | - |
| At3g58500 | At3g58500 | - | - | Codominant | χ2 = 1.70 | - | - |
| BnapPIP1002 | BnapPIP1002 | - | - | Codominant | χ2 = 1.08 | - | - |
| BnapPIP1464 | BnapPIP1464 | - | - | Dominant | χ2 = 1.02 | - | - |
| At2g43360 | At2g43360a | - | - | - | - | Dominant | χ2 = 0.68 |
| At2g43361b | - | - | - | - | Codominant | χ2 = 0.33 | |
| BnapPIP1792 | BnapPIP1792 | - | - | - | - | Codominant | χ2 = 0.53 |
| BnapPIP216 | BnapPIP216 | - | - | - | - | Dominant | χ2 = 0.47 |
*: Codominant markers: Expected Mendelian segregation of 1∶2∶1, χ2 (0.05, 2) = 5.99; Dominant marker: Expected Mendelian segregation of 3∶1, χ2 (0.05, 1) = 3.84.
Figure 4. Mapping QTLs controlling C18∶3 content.
A. The QTL in Sal02 was located between BnapPIP685 and BnapPIP881 in Y1127×Y1035 and Y1127×Y514. B. The QTL in Sal10 was located between BnapPIP1012 and BnapPIP363 in Y1127×Y1035, and between BnapPIP1012 and At3g43520 in Y514×Y1035. 1-LOD and 2-LOD supporting intervals of each C18∶3 QTL were marked by thick and thin bars, respectively. The SalFAD3.LA1 and SalFAD3.LA2 genes co-localized with their C18∶3 QTL peaks in the linkage groups Sal02 and Sal10.
In the F2 population of Y1127 (low)×Y514 (medium), 10 polymorphic ILP primer pairs between the low (3.0–4.0%) and medium (14.5–16.5%) C18∶3 bulks produced 10 markers (Table 2). The 10 markers were all mapped to one linkage group corresponding to Sal02. The QTL (LOD = 46.53) was localized between BnapPIP685 and BnapPIP881 in the linkage group (Figure 4). In the F2 population of Y514 (medium)×Y1035 (high), 11 markers were generated by 10 polymorphic primer pairs between the medium (10.4–11.6%) and high (16.7–19.2%) C18∶3 bulks. The 11 markers were mapped to the linkage group Sal10. The QTL (LOD = 6.09) was located between BnapPIP1012 and At3g43520 in Sal10 (Figure 4). The two FAD3 gene loci controlling the QTLs in Sal02 and Sal10 were referred to as SalFAD3.LA1 and SalFAD3.LA2, respectively. The dominant and recessive alleles of the SalFAD3.LA1 gene were accordingly designated as LA1 and la1, while that of the SalFAD3.LA2 gene as LA2 and la2. Therefore, it could be inferred that the C18∶3 genotypes of Y1127 (low), Y514 (medium) and Y1035 (high) were la1la1la2la2, LA1LA1la2la2 and LA1LA1LA2LA2, respectively.
The SalFAD3.LA1 and SalFAD3.LA2 Genes are Cloned and Exhibit Differences in the Exon and Intron
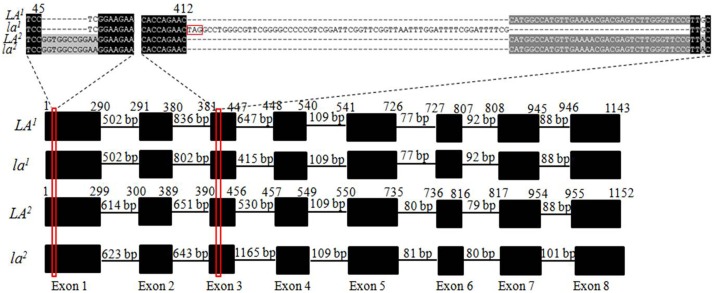
The coding regions of the dominant alleles LA1 and LA2 were cloned from Y1035, while those of the recessive alleles, la1 and la2 from Y1127 using primer pair No 1 (Table S1). LA1 had a coding DNA sequence (CDS) of 1143 bp encoding a polypeptide of 380 amino acids. la1 had a CDS of 1171 bp. Sequence alignment with LA1 indicated that la1 harboured an indel involving a 64 bp insertion and a 36 bp deletion at position 412 (Figure 5 and Figure S1). A stop codon at the beginning of the 64 bp insertion might have resulted in the termination of protein translation after the 137th amino acid residue. Therefore, la1 is a loss-of-function allele. The 5′ flanking sequences from the translation start site were cloned for LA1and la1 using the primer pair No 2 (Table S1). The 5′ fragment of LA1 was 1250 bp, while that of la1 was 621 bp. A 435 bp 3′ flanking sequence from the translation stop codon was cloned for LA1and la1 using the primer pair No 4 (Table S1). The two alleles didn’t exhibit any differences in the cloned 3′ flanking sequences. The genomic DNA sequences of the LA1 and la1 were amplified using the primer pair No 5 (Table S1) which was designed based on the 5′ flanking sequence and the conserved 3′ flanking sequence specific to the candidate SalFAD3.LA1 gene. Comparison of the coding and genomic DNA sequences indicated that the candidate SalFAD3.LA1 gene contained 8 exons and 7 introns (Figure 5). Alignment of the genomic DNA sequences of LA1 and la1 revealed that la1 had an insertion of 584 bp in the third exon. This insertion contained a new intron splicing site GT (Figure S4), which resulted in a 64 bp insertion and a 36 bp deletion (nucleotide 412–447) at position 412 in the CDS (Figure 5). The inserted fragment contained a 5 bp direct repeat (5′-AGAAC-3′) at each end, which is a typical LTR retroelement insertion site (Figure S4). In addition to differences in the CDS, LA1 and la1 exhibited variation in the length of the introns (Figure 5).
Figure 5. Structure of the SalFAD3.LA1 and SalFAD3.LA2 alleles LA1, la1, LA2 and la2 in yellow mustard.
The black boxes represented the exons and the lines between the black boxes indicated the introns. The numbers on the top of black box of LA1 (LA2) indicated the beginning and ending of each exon of LA1 and la1 (LA2 and la2). The number above each black line indicated the intron length. The nucleotide sequences of 9 bp deletion at position 45 of exon 1 of the alleles LA1 and la1, and the 64 bp insertion and the 36 bp deletion at position 412 of exon 3 of la1 were displayed on the top. Allele specific markers were developed based on the variation in intron 3 of the SalFAD3 alleles LA1, la1, LA2 and la2.
Both LA2 and la2 had a CDS of 1152 bp encoding a polypeptide of 383 amino acids (Figure S1 and S5). Six point mutations at positions 567, 579, 666, 699, 777 and 1059 were observed in the CDS of la2 when compared with that of LA2, but did not lead to any amino acid changes. The 5′ flanking sequences from the translation start site were cloned for LA2 and la2 using the primer pair No 3 (Table S1). The 5′ flanking fragments of the two alleles were 444 nucleotides in length and were similar in sequence. A 435 bp 3′ flanking sequence from the translation stop codon was cloned for LA2 and la2 using the primer pair No 4 (Table S1). The two alleles didn’t show any differences in the cloned 3′ flanking sequences. The genomic DNA sequences of LA2 and la2 were cloned using primer pair No 6 (Table S1) which was designed based on the 5′ flanking sequence and the conserved 3′ flanking sequence specific to the candidate SalFAD3.LA2 gene (Figure S3). Comparison of the coding and genomic DNA sequences indicated that the candidate SalFAD3.LA2 gene also contained 8 exons and 7 introns (Figure 5). Variation in the length of the introns was observed between LA2 and la2 (Figure 5). For instance, the third intron of LA2 was 530 bp, while that of la2 was 1165 bp.
Sequence alignment of LA1 and LA2 indicated that LA1 harboured a 9 bp deletion at position 46 (Figure 5 and Figure S1), which resulted in the loss of the three amino acids glycine-arginine-lysine at position 16. In addition, 77 point mutations were observed between LA1 and LA2 (Figure S1), of which 19 mutations led to amino acid changes (Figure S5). The candidate SalFAD3.LA1 and SalFAD3.LA2 genes exhibited differences in the cloned 5′ flanking sequences (Figure S2), but had the same 3′ flanking sequences. Variation in the length of the introns was observed among the four alleles LA1, la1, LA2 and la2 (Figure 5).
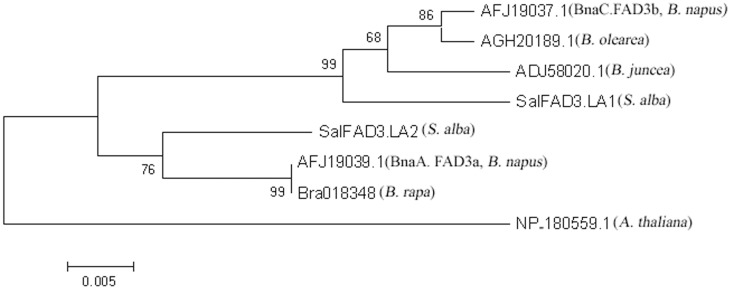
Phylogenetic analysis based on the polypeptide sequences encoded by LA1 and LA2 implied that SalFAD3.LA1 and SalFAD3. LA2 genes in yellow mustard were clustered with FAD3 genes in Brassica species (Figure 6). The SalFAD3.LA1 gene was grouped together with the FAD3 genes of B. oleracea (Genbank accession No.AGH20189), the C genome in B. napus (BnaC.FAD3b, Genbank accession No.AFJ19037.1) and B. juncea (Genbank accession No.ADJ58020.1), whereas the SalFAD3. LA2 gene was in the same cluster with the FAD3 genes of B. rapa (BraA.FAD3a, BRAD accession No. Bra018348) and the A genome in B. napus (BnaA.FAD3a, Genbank accession No.AFJ19039.1).
Figure 6. Phylogentic relationship based on the polypeptide sequences among the LA1 allele of the SalFAD3.LA1 gene, the LA2 allele of the SalFAD3.LA2 gene, and the FAD3 genes of Brassica species and Arabidopsis.
Co-segregation of the SalFAD3.LA1 and SalFAD3.LA2 Allele-specific Markers with C18∶3 Contents in the F2 Populations
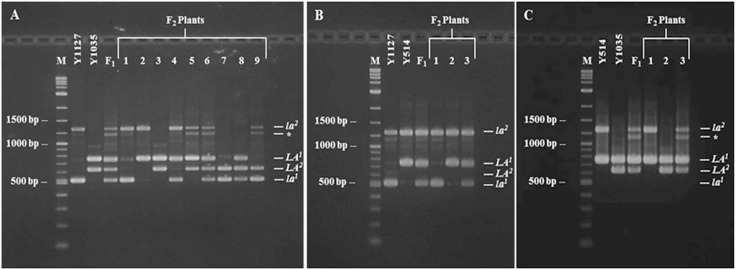
Primer pair 12 (Table S1) produced co-dominant markers of 742 bp, 510 bp, 626 bp and 1273 bp specific for LA1, la1, LA2 and la2, respectively, which co-segregated with the C18∶3 content in all of the F2 populations. In the cross of Y1127 (low)×Y1035 (high), all of the nine possible genotypes were identified using the markers specific for LA1, la1, LA2 and la2 (Figure 7A; Table 3). The homozygous F2 plants (LA1LA1LA2LA2) had a significantly higher C18∶3 content (average: 17.1%) than the heterozygous F2 plants of LA1la1LA2LA2 (average: 13.1%) (t = 6.12, p<0.01) and of LA1LA1LA2la2 (average: 15.5%) (t = 2.23, p = 0.04) (Table 3). The homozygous F2 plants of LA1LA1la2la2 had a higher average C18∶3 content (14.0%) than those of la1la1LA2LA2 (average: 9.1%) (t = 5.81, p<0.01). In the cross of Y1127 (low)×Y514 (medium), the three genotypes for C18∶3 content were differentiated with the markers specific for LA1 and la1 (Figure 7B; Table 3). The average C18∶3 content of the homozygous F2 plants (LA1LA1la2la2) was 12.7%, which was significantly higher than the heterozygous F2 plants (LA1la1la2la2, average: 9.2%) (t = 5.02, p<0.01) (Table 3). In the cross of Y514 (medium)×Y1035 (high), the markers specific for LA2 and la2 distinguished the three C18∶3 genotypes (Figure 7C; Table 3). The homozygous F2 plants (LA1LA1LA2LA2) had an average C18∶3 content of 15.8%, which was higher than the heterozygous F2 plants (LA1LA1LA2la2, average: 13.8%) (t = 2.23, p = 0.04) (Table 3). The SalFAD3.LA1 and SalFAD3.LA2 genes co-localized with the QTL peaks on Sal02 and Sal10, respectively (Figure 4). A new band was observed in the F1 and F2 plants with the heterozygote genotype of LA2la2 (Figs. 7A and 7C).
Figure 7. Identification of different C18∶3 genotypes based on the SalFAD3.LA1 and SalFAD3.LA2 allele-specific markers in the F2 populations of the three crosses Y1127×Y1035 (A), Y1127×Y514 (B) and Y514×Y1035 (C).
M: DNA ladder. Y1127: Low C18∶3 line (la1la1la2la2). Y1035: high C18∶3 line (LA1LA1LA2LA2). A. F1 (Y1127×Y1035): LA1la1LA2la2. Lane 1: la1la1la2la2; Lane 2: LA1LA1la2la2; Lane 3: LA1LA1LA2LA2; Lane 4: LA1la1la2la2; Lane 5: LA1LA1LA2la2; Lane 6: LA1la1LA2la2; Lane 7: la1la1LA2LA2; Lane 8: LA1la1LA2LA2; Lane 9: la1la1LA2la2. B. F1 (Y1127×Y1035): LA1la1la2la2; Lane 1: la1la1la2la2; Lane 2: LA1LA1la2la2; Lane 3: LA1la1la2la2. C. F1 (Y514×Y1035): LA1LA1LA2la2; Lane 1: LA1LA1la2la2; Lane 2: LA1LA1LA2LA2; Lane 3: LA1LA1LA2la2. * indicated new band. The different C18∶3 genotypes were identified using primer No 12 (SalFAD3 F and SalFAD3 R) (Table S1) designed based on the variation in intron 3 of the SalFAD3 alleles LA1, la1, LA2 and la2.
Table 3. Co-segregation of the SalFAD3.LA1 and SalFAD3.LA2 allele-specific markers with C18∶3 contents in the F2 populations of Y1127×Y1035, Y1127×Y514 and Y514×Y1035.
| F2 populations | Allele-specific Markers | Genotype | No. of Plants | C18∶3 Content (% of total fatty acids) | ||||
| LA1 | la1 | LA2 | la2 | Mean | Range | |||
| Y1127×Y1035 | + | − | + | − | LA1LA1LA2LA2 | 9 | 17.1 | 15.4–20.4 |
| + | − | + | + | LA1LA1LA2la2 | 22 | 15.5 | 11.1–18.7 | |
| + | − | − | + | LA1LA1 la2la2 | 5 | 14.0 | 10.0–16.0 | |
| + | + | + | − | LA1la1LA2LA2 | 14 | 13.1 | 10.4–15.5 | |
| + | + | + | + | LA1la1LA2la2 | 44 | 11.1 | 8.5–13.4 | |
| + | + | − | + | LA1la1la2la2 | 20 | 10.2 | 8.3–12.5 | |
| − | + | + | − | la1la1LA2LA2 | 12 | 9.1 | 7.7–10.6 | |
| − | + | + | + | la1la1LA2la2 | 19 | 5.7 | 3.1–7.5 | |
| − | + | − | + | la1la1la2la2 | 12 | 3.9 | 2.9–4.5 | |
| Y1127×Y514 | + | − | − | + | LA1LA1 la2la2 | 36 | 12.7 | 8.6–16.5 |
| + | + | − | + | LA1la1la2la2 | 82 | 9.2 | 5.6–14.8 | |
| − | + | − | + | la1la1la2la2 | 37 | 4.6 | 3.0–5.9 | |
| Y514×Y1035 | + | − | + | − | LA1LA1LA2LA2 | 37 | 15.8 | 13.7–19.2 |
| + | − | + | + | LA1LA1LA2la2 | 90 | 13.8 | 10.1–16.0 | |
| + | − | − | + | LA1LA1 la2la2 | 30 | 12.9 | 10.4–15.0 | |
Discussion
The present paper reported on the inheritance and QTL mapping of C18∶3 content as well as molecular characterization of the FAD3 genes in yellow mustard. Linolenic acid content was controlled by the nuclear genotype of the embryo in yellow mustard as reported in B. napus [8]. Two nuclear gene loci were detected and functioned independently and additively to determine the total C18∶3 content in the seeds. However, maternal effects on the C18∶3 content couldn’t be ruled out since appropriate progeny tests were not performed in the present study. QTL analysis further revealed that the two gene loci SalFAD3.LA1 and SalFAD3.LA2 had a different magnitude of effect and together explained 96.4% of the total variation for C18∶3 content. The residual 3.6% variation of C18∶3 content beyond the two QTLs could be resulted from maternal and environmental effects. It has been reported that temperature, maternal genotype and cytoplasm have effects on C18∶3 content in B. napus [8]–[10]. The duplication of the FAD3 gene provides additional evidence that yellow mustard is a secondary polyploid species as revealed by molecular studies [29], [30]. The two linkage groups Sal02 and Sall0 containing the SalFAD3.LA1 and SalFAD3.LA2 genes, did not share any common ILP markers, suggesting the occurrence of extensive genomic changes during the speciation of yellow mustard.
Molecular cloning and sequencing indicated that the SalFAD3.LA1 and SalFAD3.LA2 genes contained 8 exons and 7 introns in yellow mustard, which is in agreement with that in B. napus [12] and A. thaliana (Locus: AT2G29980, TAIR) [31]. However, the molecular mechanism underlying the naturally occurring C18∶3 variant in yellow mustard was different from that of the EMS-induced C18∶3 variant in B. napus and B. oleracea. The FAD3 gene with reduced C18∶3 content resulted from SNP mutations in B. napus [11], [12] and B. oleracea [32]. However, the recessive allele la1 of the SalFAD3.LA1 gene was a loss-of-function mutant due to an insertion of 584 bp in exon 3. The inserted fragment contained a typical LTR retroelement insertion site (5′-AGAAC-3′) at each end, suggesting that the inserted fragment might be a remnant of a transposable element which had undergone a deletion following the insertion event. The recessive allele la2 of the SalFAD3.LA2 gene was functional and had a CDS encoding the same polypeptide sequence when compared with the dominant allele LA2. However, la2 was different in intron sequence. It remains to be investigated why LA2 and la2 controlled a different C18∶3 content. The SalFAD3.LA1 and SalFAD3.LA2 allele-specific markers proved to be useful for identification of different C18∶3 genotypes in the present study.
The phylogenetic analysis based on the polypeptide sequences indicated that the LA1 and LA2 genes in yellow mustard were clustered with the FAD3 genes in Brassica species and A. thaliana. Interestingly, LA1 and LA2 were clustered into different groups. LA1 was grouped together with the FAD3 genes of B. oleracea and the C genome in B. napus, whereas LA2 was in the same cluster with the FAD3 gene of B. rapa and the A genome in B. napus. In our study, the LA1 gene controlled a higher C18∶3 content than the LA2 gene. It was reported that the FAD3 gene of the C genome in B. napus also contributed more to the total C18∶3 content than that of the A genome [11], [12]. This suggested that the molecular divergence of the LA1 and LA2 genes occurred before the speciation of yellow mustard and Brassica species.
In conclusion, our study revealed the existence of two FAD3 gene loci contributing to the genetic variation of linolenic acid content in yellow mustard. The SalFAD3.LA1 gene was located in the linkage group Sal02, while the SalFAD3.LA2 gene in Sal10. We have cloned the SalFAD3.LA1 and SalFAD3.LA2 genes and developed allele-specific markers for the detection of desirable genotypes, which will be valuable for marker assisted breeding in yellow mustard.
Supporting Information
Alignment of the coding DNA sequences of the SalFAD3.LA1 alleles LA1 , la1 and the SalFAD3.LA2 alleles LA2 and la2 in yellow mustard. The nucleotide sequence alignment was carried out using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalW2/).
(PDF)
Alignment of the 5′ upstream sequences of the SalFAD3.LA1 and SalFAD3.LA2 genes in yellow mustard. UP-LA1 and UP-la1 represented the 5′ upstream sequences of the alleles LA1 and la1 of the SalFAD3.LA1 gene. UP-LA2 and UP-la2 indicated the 5′ upstream sequences of the alleles LA2 and la2 of the SalFAD3.LA2 gene. The nucleotide sequence alignment was carried out using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/).
(PDF)
PCR amplification of the SalFAD3.LA1 and SalFAD3.LA2 genes. A. PCR amplification of the genomic DNA sequences of SalFAD3.LA1 gene using the primer pair No 5 (Table S1). Lanes 1–2: 4268 bp fragment of la1 from Y1127; Lanes 3–6: 4534 bp fragment of LA1 from Y1035. B. PCR amplification of the genomic DNA sequences of SalFAD3.LA2 gene using the primer pair No 6 (Table S1). Lanes 1–4: 4688 bp fragment of la2 from Y1127; Lanes 5–6: 4042 bp fragment of LA2 from Y1035.
(PDF)
Nucleotide sequences of intron 3 and its flanking of SalFAD3.LA1 and SalFAD3.LA2 genes in yellow mustard. The sequence of exon 3 was underlined in red while that of exon 4 was lined in blue. The nucleotide sequence of the inserted fragment in exon 3 of la1 was underlined in pink. The new intron splicing site GT in the inserted fragment was indicated in green rectangle box. The nucleotides in blue rectangle box indicated the inserted fragment that remained in the CDS of exon 3 of la1. The nucleotides in red rectangle box indicated the 5 bp direct repeat (5′-AGAAC-3′). The first and the last nucleotides of intron 3 were indicated by arrowhead and arrow, respectively. The intron 3 of LA1, la1 LA2, la2 are 647 bp, 415 bp, 530 bp and 1165 bp in length, respectively.
(PDF)
Amino acid sequences encoded by the SalFAD3.LA1 allele LA1 and the SalFAD3.LA2 allele LA2 of yellow mustard, AFJ19039.1 ( BraA.FAD3a ) and AFJ19037.1 ( BnaC.FAD3b ) of B. napus , AGH20189.1 of B. oleracea , ADJ58020.1 of B. juncea , Bra018348 of B. rapa , NP_180559.1 of A. thaliana . The amino acid sequence alignment was carried out by ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/).
(PDF)
Primers used in this study.
(PDF)
Funding Statement
The condiment yellow mustard breeding and research programs are supported by the Developing Innovative Agri-Products Initiative of the Growing Canadian Agri-Innovations Program (DIAP), Growing Forward II - Agri-Innovation Program (AIP-P027), Mustard 21 Canada Inc. and the Agriculture Development Fund (ADF) of Saskatchewan (grant No 20100032), Canada. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Thompson KF (1963) Resistance to the cabbage aphid (Brevicoryne brassicae) in Brassica plants. Nature 198: 209. [Google Scholar]
- 2. Bodnaryk RP, Lamb RJ (1991) Mechanisms of resistance to the flea beetle, Phyllotreta cruciferae (Goeze), in mustard seedlings, Sinapis alba L. Can J Plant Sci. 71: 13–20. [Google Scholar]
- 3.Elliott RH, Rakow GFW (1999) Resistance of brassica and sinapis species to flea beetles, phyllotreta cruciferae. in: proc. the 10th international rapeseed congress. Camberra, Australia.
- 4.Gugel RK, Séguin-Swartz G (1997) Introgression of blackleg resistance from Sinapis alba into Brassica napus Brassica 97, int soc hortic sci symp brassicas/10th crucifer genetics workshop, 23–27 Sept 1997, Rennes, France:Abst: p 222.
- 5. Raney JP, Rakow G, Olson T (1995) Development of low erucic, low glucosinolate Sinapis alba . In proceedings of the 9th international rapeseed congress, Cambridge, United Kindom 2: 416–418. [Google Scholar]
- 6. Scarth R, Tang J (2006) Modification of Brassica oil using conventional and transgenic approaches. Crop Sci 46: 1225–1236. [Google Scholar]
- 7. Röbbelen G, Nitsch A (1975) Genetical and physiological investigations on mutants for polyenoic fatty acids in rapeseed, Brassica napus L. I. selection and description of new mutants. Z Pflanzenzucht 75: 92–105. [Google Scholar]
- 8. Pleines S, Friedt W (1989) Genetic control of linolenic acid concentration in seed oil of rapeseed (Brassica napus L.). Theor Appl Genet 78: 793–797. [DOI] [PubMed] [Google Scholar]
- 9. Rajcan I, Kasha KJ, Kott LS, Beversdorf WD (2002) Evaluation of cytoplasmic effects on agronomic and seed quality traits in two doubled haploid populations of Brassica napus L. Euphytica. 123: 401–409. [Google Scholar]
- 10. Baux A, Hebeisen T, Pellet D (2008) Effects of minimal temperatures on low-linolenic rapeseed oil fatty-acid composition. Eur J Agron 29: 102–107. [Google Scholar]
- 11. Hu X, Sullivan-Gilbert M, Gupta M, Thompson SA (2006) Mapping of the loci controlling oleic and linolenic acid contents and development of fad2 and fad3 allele-specific markers in canola (Brassica napus L.). Theor Appl Genet 113: 497–507. [DOI] [PubMed] [Google Scholar]
- 12. Yang Q, Fan C, Guo Z, Qin J, Wu J, et al. (2012) Identification of FAD2 and FAD3 genes in Brassica napus genome and development of allele-specific markers for high oleic and low linolenic acid contents. Theor Appl Genet 125(4): 715–729. [DOI] [PubMed] [Google Scholar]
- 13. Mikolajczyk K, Dabert M, Karlowski WM, Spasibionek S, Nowakowska J, et al. (2010) Allele-specific SNP markers for the new low linolenic mutant genotype of winter oilseed rape. Plant Breeding 129: 502–507. [Google Scholar]
- 14. Ecker R, Yaniv Z (1993) Genetic control of fatty acid composition in seed oil of Sinapis alba L. Euphytica. 69: 45–49. [Google Scholar]
- 15. Katepa-Mupondwa F, Gugel RK, Raney JP (2006) Genetic diversity for agronomic, morphological and seed quality traits in Sinapis alba L. (yellow mustard). Can J Plant Sci 86: 1015–1025. [Google Scholar]
- 16. Cheng B, Williams DJ, Zhang Y (2012) Genetic variation in morphology, seed quality and self-(in)compatibility among the inbred lines developed from a population variety in outcrossing yellow mustard (Sinapis alba). Plants 1: 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bundrock T (1998) Doubled haploidy in yellow mustard (Sinapis alba L.). Dissertation, University of Saskatchewan.
- 18. Michelmore R, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88: 9828–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Panjabi P, Jagannath A, Bisht N, Padmaja K, Sharma S, et al. (2008) Comparative mapping of Brassica juncea and Arabidopsis thaliana using intron polymorphism (IP) markers: homoeologous relationships, diversification and evolution of the A, B and C brassica genomes. BMC Genomics 9: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yang L, Jin G, Zhao X, Zheng Y, Xu Z, et al. (2007) PIP: a database of potential intron polymorphism markers. Bioinformatics 23: 2174–2177. [DOI] [PubMed] [Google Scholar]
- 21. Somers DJ, Friesen KRD, Rakow G (1998) Identification of molecular markers associated with linoleic acid desaturation in Brassica napus . Theor Appl Genet 96: 897–903. [Google Scholar]
- 22.Van Ooijen JW (2006) JoinMap Version 4.0: Software for the calculation of genetic linkage maps. Plant Research International, Wageningen, The Netherlands.
- 23.Van Ooijen JW (2009) MapQTL 6: Software for the mapping of quantitative trait loci in experimental populations of diploid species. Kyazma BV: Wageningen, Netherlands.
- 24. Siebert PD, Chenchik A, Kellogg DE, Lukyanov KA, Lukyanov SA (1995) An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Research 23: 1087–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kumar S, Tamura K, Nei M (2004) MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5: 150–163. [DOI] [PubMed] [Google Scholar]
- 26. Thies W (1971) Schnelle und einfache Analysen der Fettsäurezusammensetzung in einzelnen Rapskotyledonen I. Gaschromatographische und papierchromatographische Methoden. Z Pflanzenzücht 65: 181–202. [Google Scholar]
- 27. Downey RK, Harvey BL (1963) Methods of breeding for oil quality in rape. Can J Plant Sci 43: 271–275. [Google Scholar]
- 28. Javidfar F, Cheng BF (2013) Construction of a genetic linkage map and QTL analysis of erucic acid content and glucosinolate components in yellow mustard (Sinapis alba L.). BMC Plant Biology 13: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lysak MA, Koch MA, Pecinka A, Schubert I (2005) Chromosome triplication found across the tribe Brassiceae. Genome Res 15: 516–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nelson MN, Lydiate DJ (2006) New evidence from Sinapis alba L. for ancestral triplication in a crucifer genome. Genome 49: 230–238. [DOI] [PubMed] [Google Scholar]
- 31. Nishiuchi T, Nishimura M, Arondel V, Iba K (1994) Genomic nucleotide sequence of a gene encoding a microsomal omega-3 fatty acid desaturase from Arabidopsis thaliana . Plant Physiol 105: 767–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rahman H, Singer S, Weselake R (2013) Development of low-linolenic acid Brassica oleracea lines through seed mutagenesis and molecular characterization of mutants. Theor Appl Genet 126: 1587–1598. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of the coding DNA sequences of the SalFAD3.LA1 alleles LA1 , la1 and the SalFAD3.LA2 alleles LA2 and la2 in yellow mustard. The nucleotide sequence alignment was carried out using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalW2/).
(PDF)
Alignment of the 5′ upstream sequences of the SalFAD3.LA1 and SalFAD3.LA2 genes in yellow mustard. UP-LA1 and UP-la1 represented the 5′ upstream sequences of the alleles LA1 and la1 of the SalFAD3.LA1 gene. UP-LA2 and UP-la2 indicated the 5′ upstream sequences of the alleles LA2 and la2 of the SalFAD3.LA2 gene. The nucleotide sequence alignment was carried out using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/).
(PDF)
PCR amplification of the SalFAD3.LA1 and SalFAD3.LA2 genes. A. PCR amplification of the genomic DNA sequences of SalFAD3.LA1 gene using the primer pair No 5 (Table S1). Lanes 1–2: 4268 bp fragment of la1 from Y1127; Lanes 3–6: 4534 bp fragment of LA1 from Y1035. B. PCR amplification of the genomic DNA sequences of SalFAD3.LA2 gene using the primer pair No 6 (Table S1). Lanes 1–4: 4688 bp fragment of la2 from Y1127; Lanes 5–6: 4042 bp fragment of LA2 from Y1035.
(PDF)
Nucleotide sequences of intron 3 and its flanking of SalFAD3.LA1 and SalFAD3.LA2 genes in yellow mustard. The sequence of exon 3 was underlined in red while that of exon 4 was lined in blue. The nucleotide sequence of the inserted fragment in exon 3 of la1 was underlined in pink. The new intron splicing site GT in the inserted fragment was indicated in green rectangle box. The nucleotides in blue rectangle box indicated the inserted fragment that remained in the CDS of exon 3 of la1. The nucleotides in red rectangle box indicated the 5 bp direct repeat (5′-AGAAC-3′). The first and the last nucleotides of intron 3 were indicated by arrowhead and arrow, respectively. The intron 3 of LA1, la1 LA2, la2 are 647 bp, 415 bp, 530 bp and 1165 bp in length, respectively.
(PDF)
Amino acid sequences encoded by the SalFAD3.LA1 allele LA1 and the SalFAD3.LA2 allele LA2 of yellow mustard, AFJ19039.1 ( BraA.FAD3a ) and AFJ19037.1 ( BnaC.FAD3b ) of B. napus , AGH20189.1 of B. oleracea , ADJ58020.1 of B. juncea , Bra018348 of B. rapa , NP_180559.1 of A. thaliana . The amino acid sequence alignment was carried out by ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/).
(PDF)
Primers used in this study.
(PDF)