Abstract
C. elegans has proven to be a useful model organism for investigating molecular and cellular aspects of numerous human diseases. More recently, investigators have explored the use of this organism as a tool for drug discovery. Although earlier drug screens were labor-intensive and low in throughput, recent advances in high-throughput liquid workflows, imaging platforms and data analysis software have made C. elegans a viable option for automated high-throughput drug screens. This review will outline the evolution of C. elegans-based drug screening, discuss the inherent challenges of using C. elegans, and highlight recent technological advances that have paved the way for future drug screens.
Keywords: Whole organism model, high-throughput screening, small molecules, pharmaceuticals, drug target identification, phenotypic analysis, human diseases models
Chemical compounds
Chemical compounds studied in this article Fluphenazine (PubChem CID:3372); Nemadipine-A (PubChem CID:2856102); Clozapine (PubChem CID:2818)
Drug discovery using C. elegans
C. elegans is a microscopic soil nematode that has built a solid reputation as a powerful genetic model organism. Since its first introduction to biology in the early 1960’s, it has played a pivotal role in elucidating genetic pathways controlling important cellular processes such as development [1], cell death [2, 3], ageing [4–6] and RNA-mediated interference of gene expression (RNAi) [7] among others. C. elegans homologs have been identified for 60–80% of human genes [8–12] and counterparts for many human disease-causing genes are present in C. elegans [12, 13]. For these reasons, C. elegans has been extensively used to model complex human diseases including Alzheimer’s disease [14–17], Parkinson’s disease [18] diabetes [19], Duchenne muscular dystrophy [20, 21] and cancer [22, 23]. In the past decade, C. elegans has emerged as a tool for drug discovery. The same properties that make them versatile tools for genetic investigations, for example, small size (~ 1 mm in length), short generation time (~ 3 days) and ability to produce ~300 offspring in ~3 days, genetic amenability, and conservation of cellular processes across species, make C. elegans an excellent candidate for whole organism-based high-throughput screening (HTS). Major advantages of using C. elegans in HTS include: 1) the ability to model complex human diseases that can not be easily reproduced in vitro or in unicellular models, 2) the ability to simultaneously evaluate drug efficacy and absorption, distribution, metabolism, excretion or toxicity (ADMET) characteristics at the initial stages of the drug discovery pipeline, 3) a large repertoire of scorable phenotypes, 4) the multi-cellular and multi-organ system complexity existing in a whole organism improves the chances of identifying drugs that will ultimately be more efficacious in more complex multicellular organisms such as humans, and 5) the availability of time-proven genetic tools and genomic resources (e.g., RNAi-feeding library) simplifies drug target identification. Since several prior reviews have extensively described various C. elegans disease models and discussed how they can be elegantly used in drug discovery [13, 24–28], this review will focus on the current status of drug discovery, challenges associated with C. elegans-based HTS, and how these challenges are currently being addressed.
Evolution of C. elegans-based drug discovery
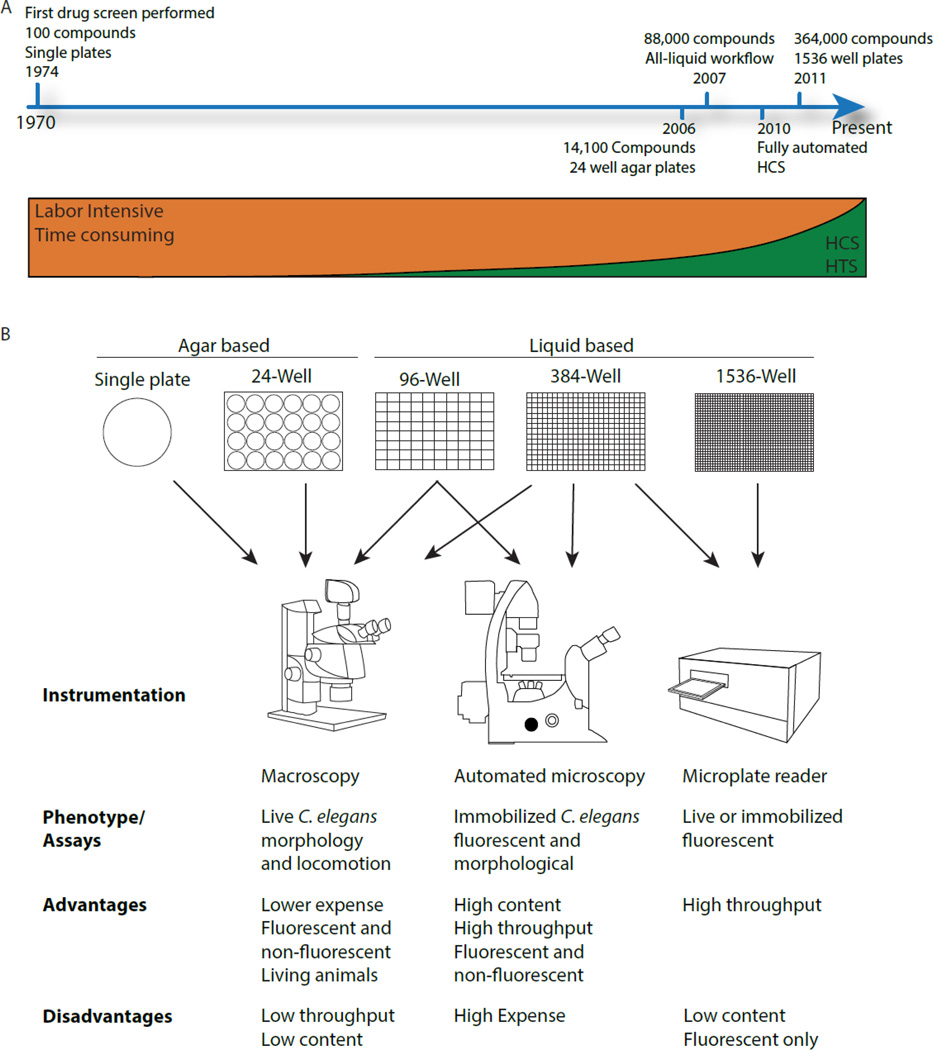
Despite an illustrious history as a model genetic organism, C. elegans was overlooked as a tool for HTS until about a decade ago. This late emergence was due, in part, to culture conditions that were not easily amenable to HTS protocols. C. elegans lives in the soil and feeds on microbes. To emulate their natural environment, C. elegans is typically cultured in the laboratory on agar plates seeded with a lawn of E. coli. The earliest drug testing protocol incorporated compounds into the agar during plate preparation [1]. This method consumed large amounts of compounds, was labor-intensive and was not amenable to HTS. Despite these disadvantages, this approach remained essentially unchanged until recently. Below we highlight major milestones in C. elegans-based drug discovery (Fig. 1).
Figure 1. Historical timeline of C. elegans-based drug discovery.
C. elegans was first used for drug testing in 1974 [1] but not as a tool for HTS until recently. Timeline shows major milestones (A). Common instruments currently used for phenotypic assessment of C. elegans (B).
The first large-scale drug screen using C. elegans was reported in 2006 [29]. Kwok and colleagues applied automated worm transfer using a Complex Object Parametric Analyzer and Sorter (COPAS™ BIOSORT, Union Biometrica) and semi-automated image acquisition to screen 14,100 small molecules. Compounds were assessed for bioactivity by evaluating a variety of phenotypes, including slow growth, lethality, uncoordinated movement and other morphological defects in wild-type animals. Using this approach, 308 bioactive compounds were identified. Although this was the first example of a large-scale drug screen using C. elegans, the screen still relied on agar plates (albeit in the 24-well format), and phenotypes were scored visually.
In 2006, Lehner and colleagues [30] developed an all-liquid workflow to facilitate HTS in C. elegans in a 96-well format. Although this procedure was initially designed to improve the throughput of genome-wide RNAi screens, it was instrumental in overcoming a major hurdle in C. elegans-based drug discovery. The elimination of agar from the workflow allowed the use of automated robotic liquid handlers and the integration of automated imaging platforms. One of the first liquid-based screens was performed by Moy and colleagues using an Enterococcus faecalis infection model in C. elegans to screen for novel antimicrobial compounds [31]. 6,000 synthetic compounds and 1,136 natural product extracts were screened and 16 compounds and 9 extracts that promoted nematode survival were identified. Since then, several other anti-pathogen screens have been reported [32, 33]. In another notable liquid culture screen, Petrascheck and colleagues tested 88,000 compounds for their ability to extend lifespan in adult C. elegans [34, 35]. Worms were cultured in 96- and 384-wells and treated with drugs for 24 days. Following exposure to strong light to stimulate movement, the number of live animals in each well was then scored visually. Using this approach, 48 compounds that extend C. elegans lifespan by 20–60% were identified. While the above studies incorporated some automation in the workflow, they all required pain staking manual inspection of wells and assessment of phenotypes.
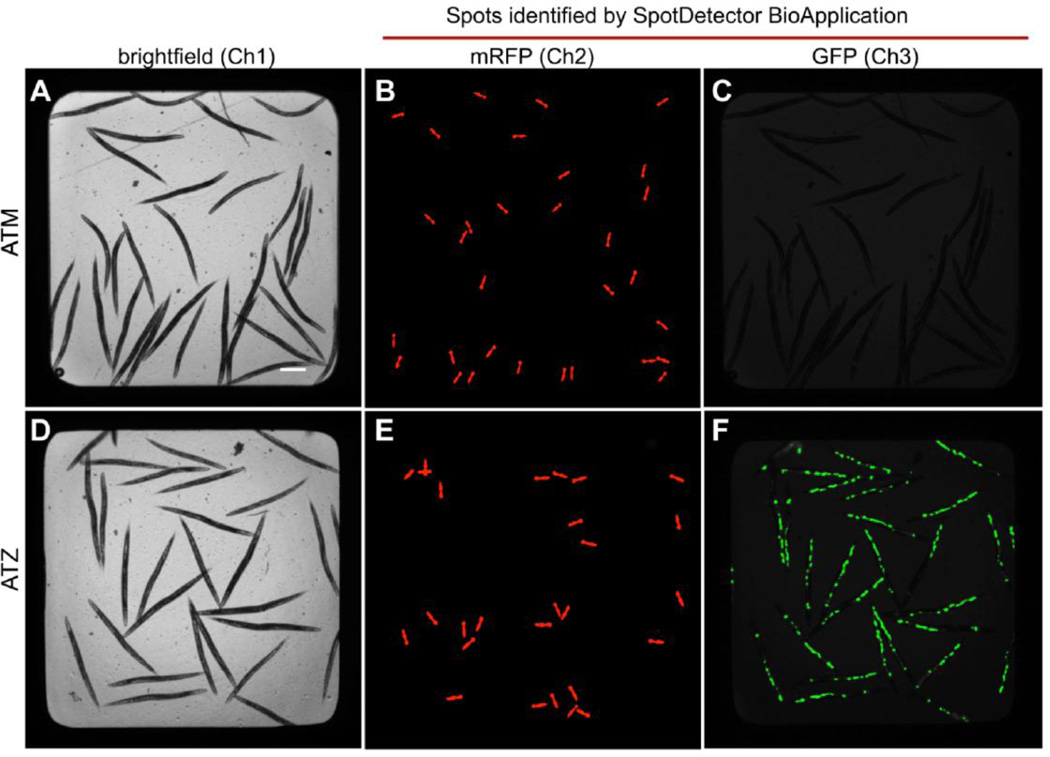
The first automated high-content drug screen integrating automated worm transfer, image acquisition and data analysis was reported by our laboratory in 2010 [36]. Transgenic C. elegans expressing the mutant form of human α1- antitrypsin fused with GFP accumulate misfolded protein within the endoplasmic reticulum similar to that seen in the liver of patients with α1-antitrypsin deficiency (ATD). Thus, to identify drugs that enhance clearance of misfolded proteins, we developed a high-content assay for quantifying protein accumulation within the intestinal cells of C. elegans. Animals were sorted into 384-well plates containing small molecules using the COPAS™ BIOSORT. Images were captured automatically using the high-content screening instrument, Arrayscan VTI (Thermo Scientific Cellomics). Real-time data analysis was performed using the SpotDetector BioApplication to quantify the number, area and intensity of the GFP-positive, protein aggregates (Fig. 2). Using this assay, we conducted a pilot screen of the 1,280 compound LOPAC library and identified 33 hit compounds that significantly reduced the intracellular accumulation of misfolded protein aggregates. One compound, fluphenazine, was subsequently shown to be efficacious in reducing protein aggregates in mammalian cell line- and mouse-models of ATD (Li et al., submitted).
Figure 2. High-content assay for the quantification of misfolded protein accumulation using C. elegans.
Using a C. elegans model of α1-antitrypsin (AT) deficiency, a high-content assay was developed to quantify accumulation of misfolded AT protein in the intestinal cells of the worm using the ArrayScanVTI. A three-channel assay was used to acquire brightfield (A and D) and fluorescence (B, C, E and F) images of transgenic animals expressing wild-type (ATM) and mutant (ATZ). The SpotDetector BioApplication was then used to quantify: 1) number of animals in each well using the mRFP (red) head marker (B and E) and, 2) the amount of misfolded protein accumulation (GFP-positive spots) in the intestine (C and F). Wild-type AT (ATM) is efficiently secreted and does not accumulate in the intestine (C). In contrast, mutant AT (ATZ) fails to get secreted and instead accumulates as large globules (F). This assay was used to conduct a high-content screen for drugs that alter misfolded protein accumulation (Adapted from Gosai et al., [36]).
Recently, a fluorescence-based, ultra high-throughput screen (uHTS) in a 1536-well format was reported [37]. The investigators used a transgenic C. elegans approach to screen for small molecule inhibitors of SKN-1. SKN-1 is a transcription factor that orchestrates the stress response to oxidants and electrophilic xenobiotics and is responsible for multi-drug resistance in parasitic nematodes. A small molecule inhibitor of SKN-1 could, therefore, be potentially useful for the treatment of parasitic nematode infections that affect almost one third of the world’s population. For the screen, the authors used a fluorescence plate reader to assess the effect of compounds on expression of gst-4::GFP, a well-established target of SKN-1. In total, 364,000 compounds were screened and 125 specific inhibitors of gst-4::GFP expression were identified. This study increased the throughput of C. elegans-based screens by utilizing 1536-well plates; however, a plate reader was used to measure the overall GFP fluorescence in the wells and no images were captured.
Chemical genetics using C. elegans
The identification of drug targets and understanding the mechanism of drug action are critical steps in the drug development process. The availability of powerful forward and reverse genetic tools makes C. elegans an ideal system for investigating chemical genetics. For example, in the aforementioned study, Kwok and colleagues identified a bioactive compound they called nemadipine A [29]. When exposed to worms, nemadipine A induces a variety of defects including those affecting morphology and egg laying. To identify the target of nemadipine A, EMS-mutated animals were treated with nemadipine A and animals no longer responding to the drug (i.e., displaying wild-type phenotype) were isolated. Animals “resistant” to nemadipine A would harbor mutations in the target protein that prevent compound action. Using this strategy, the authors identified a number of suppressor mutations in egl-19, a gene that encodes the only L-type calcium channel α1-subunit in C. elegans. A closer look at nemadipine A structure revealed similarities with a widely prescribed anti-hypertension drug, 1,4-dihydropyridine (DHP), known to antagonize the α1-subunit of L-type calcium channels in humans. This study is an example of how C. elegans genetics can be used to relatively easily identify drug targets.
The ability to specifically knock down gene expression simply by feeding worms with bacteria expressing dsRNA has paved the way for genome-scale chemical genetics investigations. Clozapine is an anti-psychotic drug used to treat treatment-refractory schizophrenia. However, the molecular mechanism of the drug’s therapeutic and toxic action is poorly understood. Worms treated with Clozapine display a robust larval arrest phenotype [38]. To identify suppressors of Clozapine-induced larval arrest, Saur and colleagues [38] performed a genome-wide RNAi screen. Approximately 40 candidate suppressor genes with diverse functions were identified suggesting that Clozapine may act on multiple targets or pathways. Using a candidate gene approach, a α-like nicotinic acetylcholine receptor (nAChR) homolog, ACR-7, was identified as a potential target of Clozapine. Characterization of other suppressor genes and the mammalian homolog of ACR-7 should shed further light on Clozapine’s polypharmocology and side-effects.
Caveats and challenges with C. elegans-based drug discovery
Although the utility of the worm as a versatile genetic tool is undisputed, there are distinct limitations to using C. elegans in drug discovery. C. elegans is an invertebrate that lacks many vital organs and a circulatory system normally found in humans. As such, organ-specific, bone or blood disorders may be difficult to model in C. elegans. According to OrthoDisease, an online database of human disease orthologs, out of the 2,259 human disease genes recently analyzed, only 782 ortholog clusters were identified in C. elegans [39]. The lack of orthologous human disease genes in C. elegans precludes the study of certain diseases, particularly those arising from loss-of-gene-function. From a HTS perspective, this raises the concern that screening campaigns will yield a significant number of false negatives due to the lack of orthologous targets in C. elegans. Conversely, hits targeting C. elegans-specific proteins may not be active in humans leading to a large number of false positives. While this level of uncertainty may be seen as a deterrent to using C. elegans in HTS, the conservation of fundamental biological pathways between the two organisms could be seen as an opportunity to discover novel drugs for diseases for which clear orthologs exist. Indeed, C. elegans is most useful when the molecular mechanism of the disease is well understood. For example, osteogenesis imperfecta (commonly known as brittle bone disease) is caused by mutations in the collagen genes, COL1A1 and COL1A2 [40]. Although C. elegans do not have bones, they do harbor collagen genes, and mutations in these genes lead to dumpy (dpy - short and stubby) phenotypes [41]. These phenotypes could be exploited using forward and reverse chemical genetic approaches to identify drugs that reverse the dpy phenotype. Additionally, even if a disease gene ortholog is not present, relevant information can still be obtained by expressing human transgenes in C. elegans [42, 43]. For example, mutations in α1- antitrypsin (AT) leads to lung and liver diseases via loss-of-function and gain-of-function mechanisms, respectively [44, 45]. While the lung disease associated cannot be effectively modeled in C. elegans due to the absence of lungs and an obvious worm ortholog of AT, the liver disease due to the accumulation of misfolded protein can be studied by expressing the mutant AT transgene in the intestine of the worm. Indeed, using this approach, a high content screen (HCS) for drugs that decrease misfolded protein accumulation has been successfully performed [36].
C. elegans has traditionally been thought to be a poor candidate for drug testing due to the relatively inefficient drug uptake caused by the impermeability of the cuticle to non-water soluble compounds [46–50] and the selective uptake of drugs by the intestine. To complicate the matter, E. coli (OP50) is typically added to the culture as the primary food source. The use of live bacteria could potentially lower the effective dose delivered to the worms due to compound metabolism or degradation by the bacteria. For these reasons, higher initial compound concentrations (25–100 µM) are typically used in C. elegans-based drug screens. A recent study, however, suggests that drug uptake by C. elegans is comparable to that of mice [51]. The study also found that culture and assay methods - liquid vs agar-based cultures, E. coli preparation and when and how the drugs were added – had significant effects on drug efficacy. The use of heat-killed, rather than live E. coli increased the uptake of drugs as measured by higher internal drug concentrations [51]. The killing of E. coli presumably prevented drug metabolism by the bacteria which in turn increased drug availability [51]. These factors should be weighed carefully in the assay development, in order to optimize drug delivery.
The past decade has seen significant advances in culture conditions and assay development to facilitate the use of C. elegans in HTS. However, an overwhelming obstacle to further development of C. elegans-based HTS has been limitations in automated image acquisition and data analysis. There are several reasons why image acquisition and data analysis are challenging. First, unlike adherent cells in culture that remain static and form relatively uniform monolayers, worms are relatively large, have multiple layers of cells and are highly mobile. As such, worms are inherently more difficult to image. Second, most high-throughput imaging platforms are designed specifically for cell-based assays. Consequently, data analysis applications are optimized for measuring cellular parameters that are not always adaptable to C. elegans applications. Third, imaging platforms are extremely expensive and may be cost-prohibitive to many C. elegans researchers. Below, we summarize recent advances that are paving the way for C. elegans-based HTS.
Recent advances in adapting C. elegans to High Content and High throughput Screening
Automation of animal transfer
The adaptation of C. elegans to an all-liquid workflow has ushered in the way for the development of HTS in 384- and 1536-well plate formats. A wide range of robotic liquid handlers can be used to dispense compounds and reagents to wells with relative ease. However, worms are more difficult to transfer with consistency using pipetting instruments due to their tendency to settle to the bottom and propensity to adhere to pipette tips. Recently, a method for worm transfer using the BioTek MicroFlo volumetric dispenser has been described [37]. For applications in which specific animal size, number and fluorescence intensity are important, a COPAS™ BIOSORT should be used. The BIOSORT can sort through a mixed population of worms and dispense into wells only those that satisfy the target criteria. This specificity, however, comes at a cost. It takes 30–60 mins to sort a 384-well plate at a density of 30 worms/well compared to just 30–60 secs using the MicroFlo. Speed and specificity can be improved if a tightly synchronized population is obtained prior to sorting/dispensing. An alternative method for sorting worms using a Fluorescence Activated Cell Sorter (FACS) has recently been described [52]. The method, called laFACS (for live-animal FACS), allows users to sort >100,000 C. elegans larvae in less than 1 hour. The ability to turn a common FACS into a worm sorter with a few minor modifications may be an attractive option for investigators who are not in the market to purchase a dedicated instrument for worm sorting. The only caveat is that sorting is restricted to L1 larvae due to a nozzle size limitation.
Automated image acquisition and data analysis
A number of sophisticated high-content imaging platforms are available on the market today. While existing platforms are designed primarily with cell-based users in mind, they can nonetheless be useful for imaging whole-organisms with minor adjustments in hardware and software [36]. To acquire images of worms in a 384-well format, we retrofitted the ArrayscanVTI with a 2.5x objective and a 0.63x coupler to enable capture of an entire well in one field (Fig. 2). Similar adjustment could easily be made to other imaging platforms for high-content imaging of worms.
Perhaps the most challenging aspect of C. elegans-based HTS is analysis of captured images for automatic scoring phenotypes. Current high-content imaging instruments are best suited for fluorescence-based imaging and quantifications. As such, simple C. elegans-assays measuring changes in fluorescent protein expression or localization can be relatively easily developed using software available for each platform. However, assays requiring quantitation of more complex phenotypes, such as those involving subtle changes in morphology, behavior or locomotion are far more difficult and may require specialized third-party software. Recently, numerous software programs specifically designed for visualizing and scoring complex worm phenotypes have been developed. CellProfiler WormToolbox is an open-source, image analysis software developed specifically for C. elegans [53]. A major challenge in making measurements on individual worms is that they tend to touch or overlap when imaged in the multi-well formats. WormToolbox can computationally “untangle” a cluster of worms so that data from individual worms can be obtained. In addition to measuring worm size, shape and fluorescence parameters, the machine-learning algorithm in CellProfiler Analyst can be used to detect subtle and complex differences in various measurable phenotypes.
Progressive loss of movement, uncoordinated locomotion, paralysis and other movement defects are common phenotypes in ageing worms and in many other C. elegans disease models [54–57]. However, these are complex phenotypes that lack robust computational methods for quantification. To meet this need, a number of laboratories have worked on developing “worm tracking” systems [58–62]. Multi-Worm Tracker (MWT) is a real-time computer vision system that can simultaneously quantify the movement behaviors of up to 120 animals per plate at video rates including spontaneous movement, turning events and response to touch [63]. A tracking software specifically designed for high-throughput measurements of worm locomotion (swimming or thrashing) in liquid has also been reported [58, 62]. This software can process well images (either previously recorded or directly acquired) and computes a covariance matrix using met lab scripts. It takes 30 seconds to process 30s of movie, and can potentially analyze a 96 well plate in less than 50 minutes [58].
Bright field images are considerably more difficult to analyze than fluorescent images. Although fluorescent reporters or dyes can be used to “label” particular cells, organs or the whole animal, in many cases, such approaches may not be practical or feasible. Recently, White and colleagues (2013) developed an automated computer vision and machine learning system for high-throughput quantification of C. elegans developmental stages called DevStaR (Developmental Stage Recognition) [64]. This software can rapidly segment and count developmental stages in populations of C. elegans and allows phenotypic analysis of bright field images with greater accuracy and speed than otherwise possible via manual analysis. A similar high-throughput software application for computing worm size and shape from bright field images has been recently been reported [65]. WormSizer is an open-source software that is useful for detecting relatively subtle phenotypes and morphological changes that may have been difficult to assess upon visual inspection.
Other technological advances
A novel high-throughput assay for monitoring of nematode motility was recently developed using a real-time cell-monitoring device, xCELLigence [66]. This system monitors electrical resistance using gold micro-electrodes integrated on the bottom of reusable 96-well tissue culture E-plates [66] to accurately and objectively assess nematode motility. Using this assay, the authors assessed the effectiveness of anthelmintic drugs and obtained IC50 values. Although the assay was initially developed for assessing parasite motility, it should be relatively easy to adapt this system for monitoring motility and viability of C. elegans.
An economical alternative to expensive automated microscope-based screening platforms is WormScan [67]. WormScan uses a standard flatbed scanner to monitor C. elegans movement. Two consecutive scans are taken, and movement is measured by the open-source Image J program. WormScan is useful for measuring mortality, movement, fecundity and size. Recently, WormScan was further improved to become a “Lifespan Machine”, which combines multiple flatbed scanners with custom image processing and data validation software to automate the collection of lifespan data. This system allows hourly observation of thousands of nematodes over the course of several weeks [68].
Microfluidic devices have recently emerged as potentially powerful tools for high-throughput analysis of C. elegans [69]. These devices are useful in isolating worms, delivering chemicals with simple microscale manipulations, and observing worms in parallel or in series in a high throughput fashion [29, 70–73]. This technology has opened up new experimental possibilities for studying drug effects on animal behavior, calcium imaging of neuron activity, memory and learning, as well as tissue regeneration after injury. Since microfluidic devices can deliver a continuous supply of food, drugs and even gases to the immobilized animal, brightfield and fluorescent images of live-animals can be acquired for several days. Micropillars can also be inserted to create an artificial soil environment for movement assays [74, 75] and force sensors can be installed to measure forces and mechanical properties of worm locomotion [76, 77]. As microfluidic devices are further refined, they will no doubt open new possibilities for novel and more complex C. elegans-based HTS.
Closing remarks
While C. elegans has been extensively used as a genetic model organism for almost half a century, the use of this model system in high-throughput drug discovery began less than a decade ago. Since then, significant progress has been made in each step of the HTS workflow from automated worm transfer to image acquisition and data analysis. In the next several years it will be important to develop more innovative HTS instrumentation and software to specifically address the limitations of image acquisition and data analysis for assessing complex phenotypes. As more C. elegans-specific technology becomes available, C. elegans will become an even more attractive model system for HTS campaigns.
Numerous proof-of-principle studies have already demonstrated the versatility of C. elegans in compound screening, drug target identification and in deciphering mechanisms of drug action [29, 33, 36–38, 78]. Furthermore, an increasing number of companies are utilizing C. elegans models in various stages of the drug discovery pipeline and a handful of drugs are currently undergoing clinical trials. Although it is too early to say whether C. elegans-based HTS campaigns will lead to the development of effective therapies for human diseases. Given the progress made thus far, we predict that C. elegans will become an important tool in the drug discovery process.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hengartner MO, Horvitz HR. The ins and outs of programmed cell death during C. elegans development. Philos Trans R Soc Lond B Biol Sci. 1994;345:243–246. doi: 10.1098/rstb.1994.0100. [DOI] [PubMed] [Google Scholar]
- 3.Hengartner MO, Horvitz HR. Programmed cell death in Caenorhabditis elegans. Curr Opin Genet Dev. 1994;4:581–586. doi: 10.1016/0959-437x(94)90076-f. [DOI] [PubMed] [Google Scholar]
- 4.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 5.Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- 6.Larsen PL, Albert PS, Riddle DL. Genes that regulate both development and longevity in Caenorhabditis elegans. Genetics. 1995;139:1567–1583. doi: 10.1093/genetics/139.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 8.Harris TW, Chen N, Cunningham F, Tello-Ruiz M, Antoshechkin I, Bastiani C, Bieri T, Blasiar D, Bradnam K, Chan J, Chen CK, Chen WJ, Davis P, Kenny E, Kishore R, Lawson D, Lee R, Muller HM, Nakamura C, Ozersky P, Petcherski A, Rogers A, Sabo A, Schwarz EM, Van Auken K, Wang Q, Durbin R, Spieth J, Sternberg PW, Stein LD. WormBase: a multi-species resource for nematode biology and genomics. Nucleic Acids Res. 2004;32:D411–D417. doi: 10.1093/nar/gkh066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sonnhammer EL, Durbin R. Analysis of protein domain families in Caenorhabditis elegans. Genomics. 1997;46:200–216. doi: 10.1006/geno.1997.4989. [DOI] [PubMed] [Google Scholar]
- 10.Lai CH, Chou CY, Ch'ang LY, Liu CS, Lin W. Identification of novel human genes evolutionarily conserved in Caenorhabditis elegans by comparative proteomics. Genome Res. 2000;10:703–713. doi: 10.1101/gr.10.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuwabara PE, O'Neil N. The use of functional genomics in C. elegans for studying human development and disease. J Inherit Metab Dis. 2001;24:127–138. doi: 10.1023/a:1010306731764. [DOI] [PubMed] [Google Scholar]
- 12.Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 13.Kaletta T, Hengartner MO. Finding function in novel targets: C. elegans as a model organism. Nat Rev Drug Discov. 2006;5:387–398. doi: 10.1038/nrd2031. [DOI] [PubMed] [Google Scholar]
- 14.Levitan D, Doyle TG, Brousseau D, Lee MK, Thinakaran G, Slunt HH, Sisodia SS, Greenwald I. Assessment of normal and mutant human presenilin function in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1996;93:14940–14944. doi: 10.1073/pnas.93.25.14940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levitan D, Greenwald I. Facilitation of lin-12-mediated signalling by sel-12, a Caenorhabditis elegans S182 Alzheimer's disease gene. Nature. 1995;377:351–354. doi: 10.1038/377351a0. [DOI] [PubMed] [Google Scholar]
- 16.Sundaram M, Greenwald I. Suppressors of a lin-12 hypomorph define genes that interact with both lin-12 and glp-1 in Caenorhabditis elegans. Genetics. 1993;135:765–783. doi: 10.1093/genetics/135.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wittenburg N, Eimer S, Lakowski B, Rohrig S, Rudolph C, Baumeister R. Presenilin is required for proper morphology and function of neurons in C. elegans. Nature. 2000;406:306–309. doi: 10.1038/35018575. [DOI] [PubMed] [Google Scholar]
- 18.Braungart E, Gerlach M, Riederer P, Baumeister R, Hoener MC. Caenorhabditis elegans MPP+ model of Parkinson's disease for high-throughput drug screenings. Neurodegener Dis. 2004;1:175–183. doi: 10.1159/000080983. [DOI] [PubMed] [Google Scholar]
- 19.Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 20.Gieseler K, Grisoni K, Segalat L. Genetic suppression of phenotypes arising from mutations in dystrophin-related genes in Caenorhabditis elegans. Curr Biol. 2000;10:1092–1097. doi: 10.1016/s0960-9822(00)00691-6. [DOI] [PubMed] [Google Scholar]
- 21.Grisoni K, Gieseler K, Mariol MC, Martin E, Carre-Pierrat M, Moulder G, Barstead R, Segalat L. The stn-1 syntrophin gene of C.elegans is functionally related to dystrophin and dystrobrevin. J Mol Biol. 2003;332:1037–1046. doi: 10.1016/j.jmb.2003.08.021. [DOI] [PubMed] [Google Scholar]
- 22.Perrimon N. Signalling pathways initiated by receptor protein tyrosine kinases in Drosophila. Curr Opin Cell Biol. 1994;6:260–266. doi: 10.1016/0955-0674(94)90145-7. [DOI] [PubMed] [Google Scholar]
- 23.Sternberg PW, Han M. Genetics of RAS signaling in C. elegans. Trends Genet. 1998;14:466–472. doi: 10.1016/s0168-9525(98)01592-3. [DOI] [PubMed] [Google Scholar]
- 24.Arya U, Das CK, Subramaniam JR. Caenorhabditis elegans for preclinical drug discovery. Current Science. 2010;99 [Google Scholar]
- 25.Segalat L. Drug discovery: here comes the worm. ACS Chem Biol. 2006;1:277–278. doi: 10.1021/cb600221m. [DOI] [PubMed] [Google Scholar]
- 26.Artal-Sanz M, de Jong L, Tavernarakis N. Caenorhabditis elegans: a versatile platform for drug discovery. Biotechnol J. 2006;1:1405–1418. doi: 10.1002/biot.200600176. [DOI] [PubMed] [Google Scholar]
- 27.Segalat L. Invertebrate animal models of diseases as screening tools in drug discovery. ACS Chem Biol. 2007;2:231–236. doi: 10.1021/cb700009m. [DOI] [PubMed] [Google Scholar]
- 28.Giacomotto J, Segalat L. High-throughput screening and small animal models, where are we? Br J Pharmacol. 2010;160:204–216. doi: 10.1111/j.1476-5381.2010.00725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwok TC, Ricker N, Fraser R, Chan AW, Burns A, Stanley EF, McCourt P, Cutler SR, Roy PJ. A small-molecule screen in C. elegans yields a new calcium channel antagonist. Nature. 2006;441:91–95. doi: 10.1038/nature04657. [DOI] [PubMed] [Google Scholar]
- 30.Lehner B, Tischler J, Fraser AG. RNAi screens in Caenorhabditis elegans in a 96-well liquid format and their application to the systematic identification of genetic interactions. Nat Protoc. 2006;1:1617–1620. doi: 10.1038/nprot.2006.245. [DOI] [PubMed] [Google Scholar]
- 31.Moy TI, Ball AR, Anklesaria Z, Casadei G, Lewis K, Ausubel FM. Identification of novel antimicrobials using a live-animal infection model. Proc Natl Acad Sci U S A. 2006;103:10414–10419. doi: 10.1073/pnas.0604055103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Breger J, Fuchs BB, Aperis G, Moy TI, Ausubel FM, Mylonakis E. Antifungal chemical compounds identified using a C. elegans pathogenicity assay. PLoS Pathog. 2007;3:e18. doi: 10.1371/journal.ppat.0030018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou YM, Shao L, Li JA, Han LZ, Cai WJ, Zhu CB, Chen DJ. An efficient and novel screening model for assessing the bioactivity of extracts against multidrugresistant Pseudomonas aeruginosa using Caenorhabditis elegans. Biosci Biotechnol Biochem. 2011;75:1746–1751. doi: 10.1271/bbb.110290. [DOI] [PubMed] [Google Scholar]
- 34.Petrascheck M, Ye X, Buck LB. An antidepressant that extends lifespan in adult Caenorhabditis elegans. Nature. 2007;450:553–556. doi: 10.1038/nature05991. [DOI] [PubMed] [Google Scholar]
- 35.Petrascheck M, Ye X, Buck LB. A high-throughput screen for chemicals that increase the lifespan of Caenorhabditis elegans. Ann N Y Acad Sci. 2009;1170:698–701. doi: 10.1111/j.1749-6632.2009.04377.x. [DOI] [PubMed] [Google Scholar]
- 36.Gosai SJ, Kwak JH, Luke CJ, Long OS, King DE, Kovatch KJ, Johnston PA, Shun TY, Lazo JS, Perlmutter DH, Silverman GA, Pak SC. Automated highcontent live animal drug screening using C. elegans expressing the aggregation prone serpin alpha1-antitrypsin Z. PLoS One. 2010;5:e15460. doi: 10.1371/journal.pone.0015460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leung CK, Wang Y, Malany S, Deonarine A, Nguyen K, Vasile S, Choe KP. An ultra high-throughput, whole-animal screen for small molecule modulators of a specific genetic pathway in Caenorhabditis elegans. PLoS One. 2013;8:e62166. doi: 10.1371/journal.pone.0062166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saur T, DeMarco SE, Ortiz A, Sliwoski GR, Hao L, Wang X, Cohen BM, Buttner EA. A genome-wide RNAi screen in Caenorhabditis elegans identifies the nicotinic acetylcholine receptor subunit ACR-7 as an antipsychotic drug target. PLoS Genet. 2013;9:e1003313. doi: 10.1371/journal.pgen.1003313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Brien KP, Westerlund I, Sonnhammer EL. OrthoDisease: a database of human disease orthologs. Hum Mutat. 2004;24:112–119. doi: 10.1002/humu.20068. [DOI] [PubMed] [Google Scholar]
- 40.van Dijk FS, Cobben JM, Kariminejad A, Maugeri A, Nikkels PG, van Rijn RR, Pals G. Osteogenesis Imperfecta: A Review with Clinical Examples. Mol Syndromol. 2011;2:1–20. doi: 10.1159/000332228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnstone IL, Shafi Y, Barry JD. Molecular analysis of mutations in the Caenorhabditis elegans collagen gene dpy-7. EMBO J. 1992;11:3857–3863. doi: 10.1002/j.1460-2075.1992.tb05478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McColl G, Roberts BR, Pukala TL, Kenche VB, Roberts CM, Link CD, Ryan TM, Masters CL, Barnham KJ, Bush AI, Cherny RA. Utility of an improved model of amyloid-beta (Abeta(1)(-)(4)(2)) toxicity in Caenorhabditis elegans for drug screening for Alzheimer's disease. Mol Neurodegener. 2012;7:57. doi: 10.1186/1750-1326-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vartiainen S, Aarnio V, Lakso M, Wong G. Increased lifespan in transgenic Caenorhabditis elegans overexpressing human alpha-synuclein. Exp Gerontol. 2006;41:871–876. doi: 10.1016/j.exger.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 44.Perlmutter DH. The role of autophagy in alpha-1-antitrypsin deficiency: a specific cellular response in genetic diseases associated with aggregation-prone proteins. Autophagy. 2006;2:258–263. doi: 10.4161/auto.2882. [DOI] [PubMed] [Google Scholar]
- 45.Silverman GA, Pak SC, Perlmutter DH. Disorders of protein misfolding: alpha-1-antitrypsin deficiency as prototype. J Pediatr. 2013;163:320–326. doi: 10.1016/j.jpeds.2013.03.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Collins JJ, Evason K, Kornfeld K. Pharmacology of delayed aging and extended lifespan of Caenorhabditis elegans. Exp Gerontol. 2006;41:1032–1039. doi: 10.1016/j.exger.2006.06.038. [DOI] [PubMed] [Google Scholar]
- 47.Davies AG, Pierce-Shimomura JT, Kim H, VanHoven MK, Thiele TR, Bonci A, Bargmann CI, McIntire SL. A central role of the BK potassium channel in behavioral responses to ethanol in C. elegans. Cell. 2003;115:655–666. doi: 10.1016/s0092-8674(03)00979-6. [DOI] [PubMed] [Google Scholar]
- 48.Evason K, Huang C, Yamben I, Covey DF, Kornfeld K. Anticonvulsant medications extend worm life-span. Science. 2005;307:258–262. doi: 10.1126/science.1105299. [DOI] [PubMed] [Google Scholar]
- 49.Burns AR, Wallace IM, Wildenhain J, Tyers M, Giaever G, Bader GD, Nislow C, Cutler SR, Roy PJ. A predictive model for drug bioaccumulation and bioactivity in Caenorhabditis elegans. Nat Chem Biol. 2010;6:549–557. doi: 10.1038/nchembio.380. [DOI] [PubMed] [Google Scholar]
- 50.Rand JB, Johnson CD. Genetic pharmacology: interactions between drugs and gene products in Caenorhabditis elegans. Methods Cell Biol. 1995;48:187–204. doi: 10.1016/s0091-679x(08)61388-6. [DOI] [PubMed] [Google Scholar]
- 51.Zheng SQ, Ding AJ, Li GP, Wu GS, Luo HR. Drug absorption efficiency in Caenorhbditis elegans delivered by different methods. PLoS One. 2013;8:e56877. doi: 10.1371/journal.pone.0056877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fernandez AG, Mis EK, Bargmann BO, Birnbaum KD, Piano F. Automated sorting of live C. elegans using laFACS. Nat Methods. 2010;7:417–418. doi: 10.1038/nmeth.f.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wahlby C, Kamentsky L, Liu ZH, Riklin-Raviv T, Conery AL, O'Rourke EJ, Sokolnicki KL, Visvikis O, Ljosa V, Irazoqui JE, Golland P, Ruvkun G, Ausubel FM, Carpenter AE. An image analysis toolbox for high-throughput C. elegans assays. Nat Methods. 2012;9:714–716. doi: 10.1038/nmeth.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brignull HR, Morley JF, Garcia SM, Morimoto RI. Modeling polyglutamine pathogenesis in C. elegans. Methods Enzymol. 2006;412:256–282. doi: 10.1016/S0076-6879(06)12016-9. [DOI] [PubMed] [Google Scholar]
- 55.Darby C, Cosma CL, Thomas JH, Manoil C. Lethal paralysis of Caenorhabditis elegans by Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 1999;96:15202–15207. doi: 10.1073/pnas.96.26.15202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Link CD. Expression of human beta-amyloid peptide in transgenic Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1995;92:9368–9372. doi: 10.1073/pnas.92.20.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Link CD. C. elegans models of age-associated neurodegenerative diseases: lessons from transgenic worm models of Alzheimer's disease. Exp Gerontol. 2006;41:1007–1013. doi: 10.1016/j.exger.2006.06.059. [DOI] [PubMed] [Google Scholar]
- 58.Buckingham SD, Sattelle DB. Fast, automated measurement of nematode swimming (thrashing) without morphometry. BMC Neurosci. 2009;10:84. doi: 10.1186/1471-2202-10-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ramot D, Johnson BE, Berry TL, Jr, Carnell L, Goodman MB. The Parallel Worm Tracker: a platform for measuring average speed and drug-induced paralysis in nematodes. PLoS One. 2008;3:e2208. doi: 10.1371/journal.pone.0002208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stirman JN, Crane MM, Husson SJ, Wabnig S, Schultheis C, Gottschalk A, Lu H. Real-time multimodal optical control of neurons and muscles in freely behaving Caenorhabditis elegans. Nat Methods. 2011;8:153–158. doi: 10.1038/nmeth.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsibidis GD, Tavernarakis N. Nemo: a computational tool for analyzing nematode locomotion. BMC Neurosci. 2007;8:86. doi: 10.1186/1471-2202-8-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zheng M, Gorelenkova O, Yang J, Feng Z. A liquid phase based C. elegans behavioral analysis system identifies motor activity loss in a nematode Parkinson's disease model. J Neurosci Methods. 2012;204:234–237. doi: 10.1016/j.jneumeth.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 63.Swierczek NA, Giles AC, Rankin CH, Kerr RA. High-throughput behavioral analysis in C. elegans. Nat Methods. 2011;8:592–598. doi: 10.1038/nmeth.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.White A, Lees B, Kao HL, Cipriani G, Paaby A, Sontag E, Erickson K, Geiger D, Gunsalus K, Piano F. DevStaR: High-throughput quantification of<i>C. elegans</i> developmental stages. IEEE Trans Med Imaging, 2013 doi: 10.1109/TMI.2013.2265092. [DOI] [PubMed] [Google Scholar]
- 65.Moore BT, Jordan JM, Baugh LR. WormSizer: high-throughput analysis of nematode size and shape. PLoS One. 2013;8:e57142. doi: 10.1371/journal.pone.0057142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smout MJ, Kotze AC, McCarthy JS, Loukas A. A novel high throughput assay for anthelmintic drug screening and resistance diagnosis by real-time monitoring of parasite motility. PLoS Negl Trop Dis. 2010;4:e885. doi: 10.1371/journal.pntd.0000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mathew MD, Mathew ND, Ebert PR. WormScan: a technique for highthroughput phenotypic analysis of Caenorhabditis elegans. PLoS One. 2012;7:e33483. doi: 10.1371/journal.pone.0033483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stroustrup N, Ulmschneider BE, Nash ZM, Lopez-Moyado IF, Apfeld J, Fontana W. The Caenorhabditis elegans Lifespan Machine. Nat Methods. 2013;10:665–670. doi: 10.1038/nmeth.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ben-Yakar A, Chronis N, Lu H. Microfluidics for the analysis of behavior, nerve regeneration, and neural cell biology in C. elegans. Curr Opin Neurobiol. 2009;19:561–567. doi: 10.1016/j.conb.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Clausell-Tormos J, Lieber D, Baret JC, El-Harrak A, Miller OJ, Frenz L, Blouwolff J, Humphry KJ, Koster S, Duan H, Holtze C, Weitz DA, Griffiths AD, Merten CA. Droplet-based microfluidic platforms for the encapsulation and screening of Mammalian cells and multicellular organisms. Chem Biol. 2008;15:427–437. doi: 10.1016/j.chembiol.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 71.Luo L, Gabel CV, Ha HI, Zhang Y, Samuel AD. Olfactory behavior of swimming C. elegans analyzed by measuring motile responses to temporal variations of odorants. J Neurophysiol. 2008;99:2617–2625. doi: 10.1152/jn.00053.2008. [DOI] [PubMed] [Google Scholar]
- 72.Kuwahara T, Koyama A, Koyama S, Yoshina S, Ren CH, Kato T, Mitani S, Iwatsubo T. A systematic RNAi screen reveals involvement of endocytic pathway in neuronal dysfunction in alpha-synuclein transgenic C. elegans. Hum Mol Genet. 2008;17:2997–3009. doi: 10.1093/hmg/ddn198. [DOI] [PubMed] [Google Scholar]
- 73.Shi W, Wen H, Lu Y, Shi Y, Lin B, Qin J. Droplet microfluidics for characterizing the neurotoxin-induced responses in individual Caenorhabditis elegans. Lab Chip. 2010;10:2855–2863. doi: 10.1039/c0lc00256a. [DOI] [PubMed] [Google Scholar]
- 74.Lockery SR, Lawton KJ, Doll JC, Faumont S, Coulthard SM, Thiele TR, Chronis N, McCormick KE, Goodman MB, Pruitt BL. Artificial dirt: microfluidic substrates for nematode neurobiology and behavior. J Neurophysiol. 2008;99:3136–3143. doi: 10.1152/jn.91327.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Park S, Hwang H, Nam SW, Martinez F, Austin RH, Ryu WS. Enhanced Caenorhabditis elegans locomotion in a structured microfluidic environment. PLoS One. 2008;3:e2550. doi: 10.1371/journal.pone.0002550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Doll JC, Harjee N, Klejwa N, Kwon R, Coulthard SM, Petzold B, Goodman MB, Pruitt BL. SU-8 force sensing pillar arrays for biological measurements. Lab Chip. 2009;9:1449–1454. doi: 10.1039/b818622g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Park SJ, Goodman MB, Pruitt BL. Analysis of nematode mechanics by piezoresistive displacement clamp. Proc Natl Acad Sci U S A. 2007;104:17376–17381. doi: 10.1073/pnas.0702138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gaud A, Simon JM, Witzel T, Carre-Pierrat M, Wermuth CG, Segalat L. Prednisone reduces muscle degeneration in dystrophin-deficient Caenorhabditis elegans. Neuromuscul Disord. 2004;14:365–370. doi: 10.1016/j.nmd.2004.02.011. [DOI] [PubMed] [Google Scholar]