Abstract
The CAR (constitutive androstane receptor; NR1I3) is a critical xenobiotic sensor that regulates xenobiotic metabolism, drug clearance, energy and lipid homoeostasis, cell proliferation and development. Although constitutively active, in hepatocytes CAR is normally held quiescent through a tethering mechanism in the cytosol, anchored to a protein complex that includes several components, including heat-shock protein 90. Release and subsequent nuclear translocation of CAR is triggered through either direct binding to ligand activators such as CITCO {6-(4-chlorophenyl)imidazo[2,1-b][1,3]thiazole-5-carbaldehyde O-(3,4-dichlorobenzyl)oxime} or through indirect chemical activation, such as with PB (phenobarbital). In the present study, we demonstrate that proteasomal inhibition markedly disrupts CAR function, repressing CAR nuclear trafficking, disrupting CAR’s interaction with nuclear co-activators and inhibiting induction of CAR target gene responses in human primary hepatocytes following treatment with either PB or CITCO. Paradoxically, these effects occur following accumulation of ubiquitinated hCAR (human CAR). Furthermore, a non-proteolytic function was indicated by its interaction with a SUG1 (suppressor for Gal1), a subunit of the 26S proteasome. Taken together, these data demonstrate that the proteasome complex functions at multiple levels to regulate the functional biology of hCAR activity.
Keywords: constitutive androstane receptor, hepatocyte, human, lactacystin, MG-132, NR1I3, proteasome
INTRODUCTION
Functioning as a xenobiotic sensor, the CAR (constitutive androstane receptor; NR1I3) is predominantly expressed in the liver, and following xenobiotic activation it modulates the expression of numerous hepatic genes that encode enzymes catalysing phase I oxidation, phase II conjugation and phase III drug transporters [1–3]. Furthermore, CAR also regulates pathways impinging on the metabolism and secretion of endogenous molecules such as cholesterol and bilirubin, energy homoeostasis, cell proliferation/apoptosis and promotion of rodent hepatocarcinogenesis [4–6].
In hepatocytes in vivo or in primary cultures, hCAR [human CAR; also known as hCAR1 (wild-type)] is tethered in a cytoplasmic complex and, following exposure to specific chemical activators, CAR is transported to the nucleus, where it heterodimerizes with the RXRα (retinoid X receptor α), interacts with DNA-targeting motifs and recruits co-regulators to modulate the transcription of associated genes [7,8]. Xenobiotic CAR activators are classified either as direct-acting, bound within the ligand-binding pocket of the receptor, or indirect-acting compounds such as PB (phenobarbital), that activate CAR through mechanisms that include receptor dephosphorylation [8,9]. However, in transformed cell lines, CAR exhibits spontaneous nuclear accumulation and therefore constitutive activation, independent of xenobiotic stimuli. Therefore these latter models present challenges for deciphering mechanisms underlying CAR regulation [10,11]. In humans, two prominent splice variants of CAR have been characterized, hCAR2 and hCAR3, which possess four or five amino acid insertions respectively, within the LBD (ligand-binding/heterodimerization domain) of hCAR1, the reference form of the receptor. Interestingly, and unlike hCAR1, the splice variants require ligands for their activation and may display altered ligand-binding preferences [12].
Currently, the mechanisms that control CAR activation and nuclear translocation are poorly understood. With regard to its cytoplasmic sequestration, CAR forms a complex with HSP90 (heat-shock protein 90), CCRP (cytoplasmic CAR retention protein), PPP1R16A (membrane-associated subunit of protein phosphatase 1; also known as protein phosphatase 1, regulatory subunit 16A) and likely other unknown proteins [13,14]. Exposure to direct or indirect CAR activators results in dephosphorylation of CAR, triggering its dissociation from the cytoplasmic complex and enabling the receptor’s nuclear translocation [11,15]. Furthermore, previous studies indicate that the activation and nuclear translocation of CAR is directly or indirectly regulated by multiple cellular signalling molecules. For example, PKC (protein kinase C) and ERK1/2 (extracellular-signal-regulated kinases 1/2) may directly regulate hCAR phosphorylation status at Thr38 or mCAR (mouse CAR) at Thr48, to repress CAR activation and translocation [9,15]. Furthermore, although PP2A (protein phosphatase 2A) modulates mCAR Ser202 dephosphorylation and nuclear translocation; it appears to have no effect on hCAR [16–18]. Several reports have identified hCAR signalling through calcium/calmodulin-dependent kinase or AMP-activating protein kinase in PB-mediated CYP2B (cytochrome P450, family 2, subfamily B) induction. However, inhibition of these two kinases does not affect PB-mediated CAR nuclear translocation, suggesting that they do not directly modulate CAR [19–21]. These results point to the involvement of yet other distinct cellular signalling pathways in the regulation of CAR activation. In these respects, the involvement of the epidermal growth factor receptor has been advanced recently as a PB signalling regulator [22].
Accumulating evidence suggests that the UPS (ubiquitin-proteasome system) functions in part to regulate NR (nuclear receptor)-mediated transcription. The UPS exists as both a cytosolic and nuclear multi-protein complex and is responsible for 80–90 % of the total cellular protein degradation. Many reports have demonstrated that the UPS is capable of targeting a large number of key transcriptional factors, including p53 and c-Jun, as well as nuclear receptors and their co-regulators. In addition to regulating their turnover, the UPS also influences other aspects of NR function. For example, proteasomal activity appears necessary for the exchange of NR co-regulatory proteins, allowing NR-mediated transcription to proceed efficiently [23,24]. Inhibition of proteasome activity is also reported to modulate the cellular localization of select NRs [25,26]. On the basis of these observations, we hypothesized that the proteasome may similarly modulate CAR activation and nuclear translocation. In the present study, we demonstrate that proteasomal inhibition directly affects CAR-mediated regulation at several biological levels, and identify a novel role for the proteasomal complex as a critical mediator of CAR activation.
EXPERIMENTAL
Chemicals and biological reagents
DMSO, PK-11195 [1-(2-chlorophenyl)-N-methyl-N-(1-methylpropyl)-3-isoquinolinecarboxamide], Rif (rifampicin), Lac (lactacystin), Andro (5α-androstan-3α-ol) and DEHP (di-2-ethylhexyl phthalate) were purchased from Sigma-Aldrich. CITCO {6-(4-chlorophenyl)imidazo[2,1-b][1,3]thiazole-5-carbaldehyde O-(3,4-dichlorobenzyl)oxime} and MG-132 were obtained from Biomol Research Laboratories. PB was obtained from the Division of Drug Services at the University of Washington Medical Center (Seattle, WA, U.S.A.). TCPOBOP {1,4-bis[2-(3,5-dichloropyridyloxy)]benzene} was obtained from the Environmental Chemistry Laboratory at the University of Washington (Seattle, WA, U.S.A.). COS-1 and HepG2 cell lines were purchased from A.T.C.C. (Manassas, VA, U.S.A.). DNA primers were purchased from Integrated DNA Technologies.
Cell culture conditions
COS-1 and HepG2 cells were cultured in DMEM (Dulbecco’s modified Eagle’s medium), pH 7.0–7.4, containing 10% FBS (HyClone), 2mM l-glutamine, 10 mM Hepes, 0.15 % sodium bicarbonate, 50 units/ml penicillin G and 50 µg/ml streptomycin (Gibco/Invitrogen). All cell transfections were performed in the same medium, but normal FBS was replaced with 10% dextran/charcoal-treated FBS (HyClone).
Plasmid DNAs
The pTracer vectors, CMV2-hCAR1, CMV2-hCAR2, CMV2-hCAR3, CMV2-mCAR and CMV2-hPXR (human pregnane X receptor), as well as p3 × FLAG-hCAR1, pcDNA3.1-RXRα, CYP2B6-PBREM (phenobarbital-responsive enhancer module)/XREM (xenobiotic-responsive enhancer module) or CYP3A4-XREM reporter and pRL-TK were used as described previously [27,28]. The pcDNA3.1-RXRα LBD, VP16 vectors expressing hCAR1-LBD, pM vector expressing GAL4-SRC1 (steroid receptor co-activator)-RID (receptor-interaction domain) and GAL4-GRIP1 (glutamate-receptor-interacting protein 1)-RID used in mammalian two-hybrid experiments were also reported previously [29]. The p3 × FLAG-ubiquitin plasmid, which expresses the 76-amino-acid ubiquitin protein tagged with three copies of the FLAG epitope (Sigma-Aldrich), was provided by Dr Adriano Marchese (Department of Molecular Pharmacology and Therapeutics, Loyola University, Stritch School of Medicine, Chicago, IL, U.S.A.) and Dr Jeffrey Benovic (Department of Biochemistry & Molecular Biology, Thomas Jefferson University, Philadelphia, PA, U.S.A.). The adenoviral-YFP-fused hCAR vector was provided by Dr Hongbing Wang (Department of Pharmaceutical Sciences, School of Pharmacy, University of Maryland, Baltimore, MD, U.S.A.). The pCMV6-SUG1 (suppressor for Gal1) was purchased from Origene.
HPH (human primary hepatocyte) culture and treatments
HPHs seeded on collagen-coated plates were obtained through the Liver Tissue Procurement and Distribution System, University of Pittsburgh, funded by the NIH (contract number N01-DK-7-0004/HHSN267200700004C). Studies with HPHs were approved by the Institutional Review Board of the Pennsylvania State University (IRB #31438). Once received, the cells were recovered for 48 h in fresh William’s E medium, pH 7.0–7.4, containing 1 % penicillin/streptomycin, 1 % Hepes, 20 µM glutamine, 25 nM dexamethasone, 10 nM insulin, 1 % linoleic acid/BSA, 5 ng/ml selenious acid and 5 µg/ml transferrin, as described previously [30]. Primary hepatocytes (12-well plates) were harvested in 600 µl of TRIzol® (Invitrogen) after 24 h treatment with 1 mM PB or 2 µM CITCO in the presence and absence of 10 µM MG-132 (or with concentrations otherwise indicated) and total RNA was isolated following the manufacturer’s protocol. RNA concentration was determined using a Nanodrop 2000 spectrophotometer (Thermo Scientific). In separate experiments, after 48 h of treatment, primary hepatocytes (six-well plates) were harvested in Nonidet P40 lysis buffer (20 mM Tris/HCl, pH7.5, 100 mM NaCl, 0.5% Nonidet P40 and 1 × protease inhibitor; Calbiochem, catalogue number 539131) followed by short-term sonication to permit isolation of total cellular protein fractions. Protein concentrations were measured using the Pierce 660 assay configured for the Nanodrop 2000 platform. Cell viability was assessed using the Cell Titer-Glo® luminescent cell viability assay (Promega) according to the manufacturer’s instructions (Supplementary Figure S1 at http://www.biochemj.org/bj/458/bj4580095add.htm). For these assays, HPHs in 24-well plates were treated for 24 h with vehicle control (0.1% DMSO), 1 mM PB or 2 µM CITCO in the presence and absence of 10 µM MG-132.
Real-time PCR analysis
cDNA was synthesized from 1 µg of total hepatocyte RNA using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems/Life Technologies) following the manufacturers’ instructions. Real-time PCR assays were performed in 96-well optical plates on an Applied Biosystems 7300 system with 2 × PerfeCTa® SYBR® Green SuperMix (Quanta Bioscience) as described previously [28]. Primers for CYP2B6, CYP3A4 and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA are as follows [18]: CYP2B6, 5′-AGACGCCTTCAATCC-TGACC-3′ (forward) and 5′-CCTTCACCAAGACAAATCC-GC-3′ (reverse); CYP3A4,5′-GTGGGGCTTTTATGATGGTCA-3′ (forward) and 5′-GCCTCAGATTTCTCACCAACACA-3′ (reverse); and GAPDH, 5′-CCCATCACCATCTTCCAGGAG-3′ (forward) and 5′-GTTGTCATGGATGACCTTGGC-3′ (reverse). Fluorescence data were collected at the same CT (cycle threshold) level for each sample within the logarithmic phase of the amplification and analysed using the ΔΔCT method as described previously [30]. The mRNA expression of CYP2B6 and CYP3A4 was normalized against that of GAPDH. All results were presented as fold-change over control samples and conducted in accordance with MIQE recommendations.
Western blot analyses
All Western blotting gels, membranes and electrophoresis equipment were purchased from Bio-Rad Laboratories. Total protein (20–30 µg) from treated HPHs were separated on SDS/PAGE (10% gels) and electrophoretically transferred on to PVDF membranes. Subsequently, membranes were blocked for 1 h at room temperature (24 °C) with 5 % non-fat dried skimmed milk powder in 0.1% TBS-T (Tris-buffered saline containing 0.1% Tween 20) before the incubation of primary antibody diluted in blocking buffer. Mouse monoclonal antibodies against human CYP2B6 and CYP3A4 were provided by Dr Frank Gonzalez (Laboratory of Metabolism, National Cancer Institute/National Institute of Health, Bethesda, MD, U.S.A.) and were diluted to 1:1000 for overnight incubation at 4°C. Anti-β-actin antibody (1:1000 dilution; Santa Cruz Biotechnology) was included as an internal control. The membranes were then washed three times with 0.1% TBS-T buffer and incubated with the appropriate secondary HRP (horseradish peroxidase)-conjugated antibody diluted to 1:4000–5000 in 2% non-fat dried skimmed milk powder in 0.1 % TBS-T for 2 h at room temperature. The membranes were developed using the Pierce ECL detection kit (Thermo Scientific).
Cellular localization assay in HPHs
The adenoviral-YFP-tagged hCAR1 vector was provided by Dr Hongbing Wang (School of Pharmacy, University of Maryland). All adenoviral particles were generated and purified by SignaGen Laboratories. The titre was measured at 5.5 × 1011 PFU (plaque-forming units)/ml. HPHs plated on to 12-well collagen-coated dishes were infected with 1–2 µl of adenoviral-eYFP-hCAR1 for 12 h before treatment with either vehicle control (0.1 % DMSO) or test chemicals. After 24 h of treatment, cells were fixed for 30 min in 4 % buffered paraformaldehyde, and then cellular nuclei were stained with 300 nM DAPI for 30 min. Using fluorescence microscopy (Nikon TE-2000s), the subcellular localization of hCAR was visualized and quantitatively characterized as nuclear, cytosolic or mixed (nuclear plus cytosolic) by counting a minimum of 100 positive infected primary hepatocytes per treatment group.
In other experiments, HPHs cultured in six-well plates were infected with 3–4 µl of adenoviral-YFP-hCAR1 for 12 h, subsequent to 24 h treatments with test reagents. Cytoplasmic and nuclear protein fractions of cells were then extracted from two wells each of the culture plates using the NE-PER kit (Thermo Scientific) according to the manufacturer’s instructions. Western blotting was performed on the different protein fractions as detailed above. Rabbit ChIP-grade anti-GFP antibody (1:1000 dilution, Abcam) was used to determine the subcellular localization of YFP-hCAR1. Rabbit anti-TBP (TATA-binding protein) antibody and mouse anti-β-actin antibody (Santa Cruz Biotechnology) were used to determine the relative distribution of respective nuclear and cytoplasmic proteins.
Transactivation and mammalian two-hybrid assays
For transactivation assays, HepG2 or COS-1 cells in 48-well plates were transfected with 100 ng of CYP2B6-PBREM/XREM or CYP3A4 reporter vector, 25 ng of pcDNA3.1-RXRα and 5 ng of pRL-TK in the presence of 25 ng of pTracer CMV2-hCAR1, -hCAR2, -hCAR3, CMV2-mCAR or CMV2-hPXR expression constructs using the FuGENE® 6 reagent (Promega) according to the manufacturer’s instructions. Then, 18 h after transfection, cells were treated for 24 h with a vehicle control (0.1 % DMSO), a positive control (PK-11195, Rif, Andro, DEHP, TCOBOP or CITCO), and corresponding proteasome inhibitors (MG-132 or Lac) with indicated concentrations. For DEHP treatment, the medium was replaced with DMEM containing 10% dextran/charcoal-treated FBS. Cells were harvested in 1 × passive lysis buffer (Promega) and assayed for firefly activities normalized against the activities of co-transfected Renilla using the Dual-Glo Reporter Assay System (Promega) and a Veritas Microplate Luminometer (Promega). In a separate transactivation assay, COS-1 cells were co-transfected with hCAR1 and various amounts of pCMV6-SUG1 plasmid DNA, and then treated for 24 h with vehicle control (0.1 % DMSO), or 10 µM Andro in the presence and absence of 2 µM CITCO. Luciferase activity was determined as described above.
For mammalian two-hybrid assays, COS-1 cells in 48-well plates were transfected with 100 ng of pFR-luciferase reporter vector, 20 ng of VP16-hCAR1-LBD, 20 ng of pM-SRC1-RID or pM-GRIP1-RID, 10 ng of pcDNA3.1-RXRα-LBD plasmid and 10 ng of pRL-TK using FuGENE® 6. At 18 h post-transfection, cells were treated with vehicle control (0.1% DMSO); 2 µM CITCO as an activator control; 0.1, 1 and 10 µM PK-11195 as an inhibitor control; and 0.1, 1 and 10 µM tested MG-132 for 24 h. Luciferase activity was detected as described above. All data are represented as means ± S.D. for three individual transfections.
Co-immunoprecipitation with cytoplasmic HSP90
COS-1 cells in 60-mm dishes were transfected with 2 µg of 3 × FLAG-hCAR1 plasmid using the FuGENE® 6 reagent. Then 36 h later, the cells were treated for 5 h with vehicle control (0.1% DMSO) or 1 mM PB in the presence and absence of 10 µM MG-132. Cells were then washed, scraped with ice-cold PBS, and then collected by centrifugation (1000g, 5 min, 4°C). The cytoplasmic fraction was extracted using the NE-PER kit (Thermo Scientific) according to the manufacturer’s instructions. The concentration of cytoplasmic protein was determined as detailed above. Anti-(HSP90 IgM) antibody (5 µl; Thermo Scientific) or 5 µl of control IgM (Santa Cruz Biotechnology) was added to 300 µg of the cytoplasmic fraction and incubated overnight at 4 °C. After incubation, 20 µl of pre-washed Protein L-Agarose (Santa Cruz Biotechnology) was added, and the mixture was then incubated with rocking for additional 2 h at 4°C. The resin was recovered by centrifugation (7500 g, 1 min, 4 °C) and rinsed five times with ice-cold immunoprecipitation-washing buffer (50 mM Tris/HCl, pH7.5, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.4% Triton X-100, 0.2% Nonidet P40 and protease inhibitor cocktail; Calbiochem, catalogue number 539131). Finally, the protein complexes were extracted from the resin with 5 × SDS loading buffer and then subjected to Western blotting as described above. Primary HRP-conjugated anti-FLAG antibody (1:1000 dilution) was used for detection with the Pierce ECL detection kit. The endogenous HSP90 as an input was detected with anti-HSP90α/β antibody (1:1000 dilution, Santa Cruz Biotechnology).
In vitro ubiquitination assay
COS-1 cells in 60-mm dishes were transfected with CMV2-hCAR1 plus 3 × FLAG-ubiquitin or 3 × FLAG-empty vectors with the FuGENE® 6 reagent. At 36 h post-transfection, the cells were treated for 5h with vehicle control (0.1 % DMSO) or 1 mM PB in the presence and absence of 10 µM MG-132. Cell pellets were resuspended in 500 µl of ice-cold lysis buffer (50 mM Tris/HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 % Triton X-100, 0.5 % Nonidet P40 and protease inhibitor cocktail; Calbiochem, catalogue number 539131) for 15 min and sonicated (ten pulses) using a Branson Sonifier 250 (VWR Scientific). The lysates were centrifuged at 14000 g for 20 min at 4 °C and supernatants were retained. A total of 300 µg of protein was incubated with 20 µl of pre-washed anti-FLAG M2 antibody resin in a volume of 500 µl lysis buffer on a rotator overnight at 4 °C. The tubes were centrifuged at 7500 g to collect the resin and the supernatant was removed. The resin was then washed five times with ice-cold wash buffer (lysis buffer with 0.4% Triton X-100 and 0.2% Nonidet P40). Protein samples were eluted with 20 µl of 2 × sample loading buffer (125 mM Tris/HCl, pH 6.8, with 4 % SDS, 20% glycerol and 0.004% Bromophenol Blue), and heated at 95 °C for 5 min. The resulting samples were analysed by Western blotting using Bio-Rad Laboratories reagents and equipment. The blots were incubated with anti-hCAR antibody (1:1000 dilution, R&D Systems) followed by HRP-conjugated anti-mouse antibody (1:4000 dilution) and detection was performed with the Pierce ECL detection kit. The input of 3 × FLAG-ubiquitin was detected with HRP-conjugated anti-FLAG antibody (Sigma-Aldrich).
Statistical analysis
Experimental data are presented as a means ± S.D. for triplicate determinations unless otherwise noted. Statistical comparisons for hepatocyte induction, transactivation and mammalian two-hybrid studies were made using one- or two-way ANOVA, followed by either Bonferroni tests for comparison of all groups or Dunnett’s tests for comparison of experimental groups to controls. For hCAR localization studies, separate χ2 tests were used to compare each treatment group to the DMSO group, and to compare the PB with PB plus MG-132 groups. The statistical significance was set at P values of <0.05 or <0.01.
RESULTS
Proteosome inhibition decreases CAR-mediated induction of CYP2B6 and CYP3A4 in HPHs
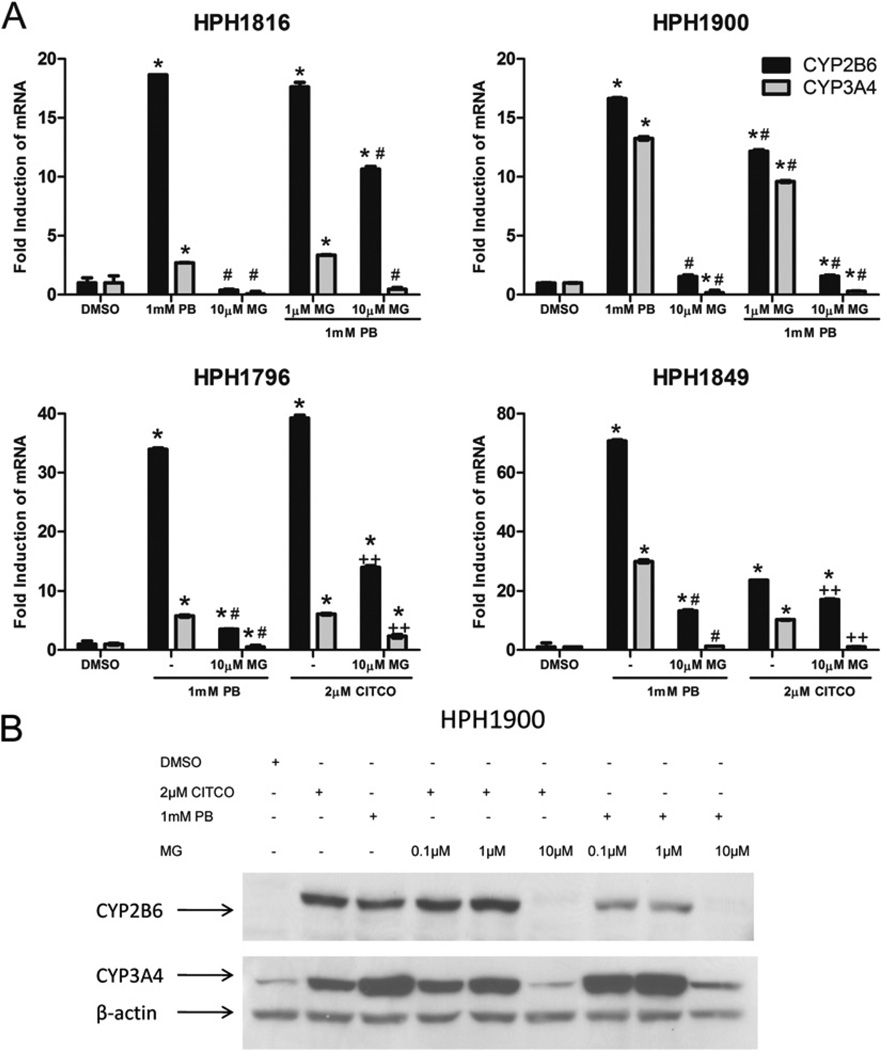
To examine the potential of the reversible proteasome inhibitor MG-132 to modulate the induction of hCAR target genes, HPHs from four different human liver donors were treated with direct or indirect hCAR activators in the presence or absence of MG-132. No obvious changes in cell morphology were observed after any of the treatments (work not shown). Supporting this observation, results from an in vitro cytotoxicity luminescence assay demonstrated that following combined inhibitor/inducer chemical treatments the cells maintained >75 % viability compared with vehicle controls (see Supplementary Figure S1). Assessing mRNA levels, 1 mM PB, a prototypical hCAR indirect activator, exhibited higher induction of CYP2B6 compared with CYP3A4in all four donors (Figure 1A). In HPH1816 and HPH1900, CYP2B6 and CYP3A4 induction by PB was significantly abrogated by MG-132 at 1 and 10 µM, whereas MG-132 alone (10 µM) had no effect on either of the CYPs. Furthermore, treatment of HPH1796 and HPH1849 with a human-specific direct CAR agonist, 2 µM CITCO, markedly induced both CYP2B6 and CYP3A4 RNA expression, however, these responses were similarly inhibited with 10 µM MG-132 co-treatment. MG-132 additions resulted in ~ 85 % reduction in the PB-induced CYP2B6 and CYP3A4 levels in all four of the HPH donor samples.
Figure 1. Inhibition of PB or CITCO-mediated CYP2B6/CYP3A4 induction in HPHs by MG-132.
(A) HPHs from four donors were treated with vehicle control (0.1 % DMSO), PB (1 mM) or CITCO (2 µM) in the presence and absence of proteasome inhibitor [MG-132 (MG); 1 or 10 µM]. Total RNA, extracted after 24 h treatment, was subjected to real-time PCR analysis. The results were normalized against GAPDH and are represented as the means ± S.D. Significantly different from DMSO (*), PB (#) or CITCO (+ +), P < 0.05. (B) In a separate experiment, homogenates were harvested from liver donor HPH1900 treated for 48 h with DMSO, PB (1 mM), CITCO (2 µM) in the presence and absence of MG-132 (0.1, 1 and 10 µM). CYP2B6/CYP3A4 protein levels were assessed with Western immunoblotting.
The HPH cultures were similarly assessed for levels of CYP2B6 and CYP3A4 protein expression. A representative immunoblot from one donor is presented in Figure 1(B). The positive control agents, 2 µM CITCO or 1 mM PB, substantially increased the protein expression levels of these CAR targets relative to vehicle control. However, consistent with the RNA results, co-treatment with MG-132 markedly reduced both PB- and CITCO-induced CYP2B6 and CYP3A4 protein levels in a dose-dependent manner.
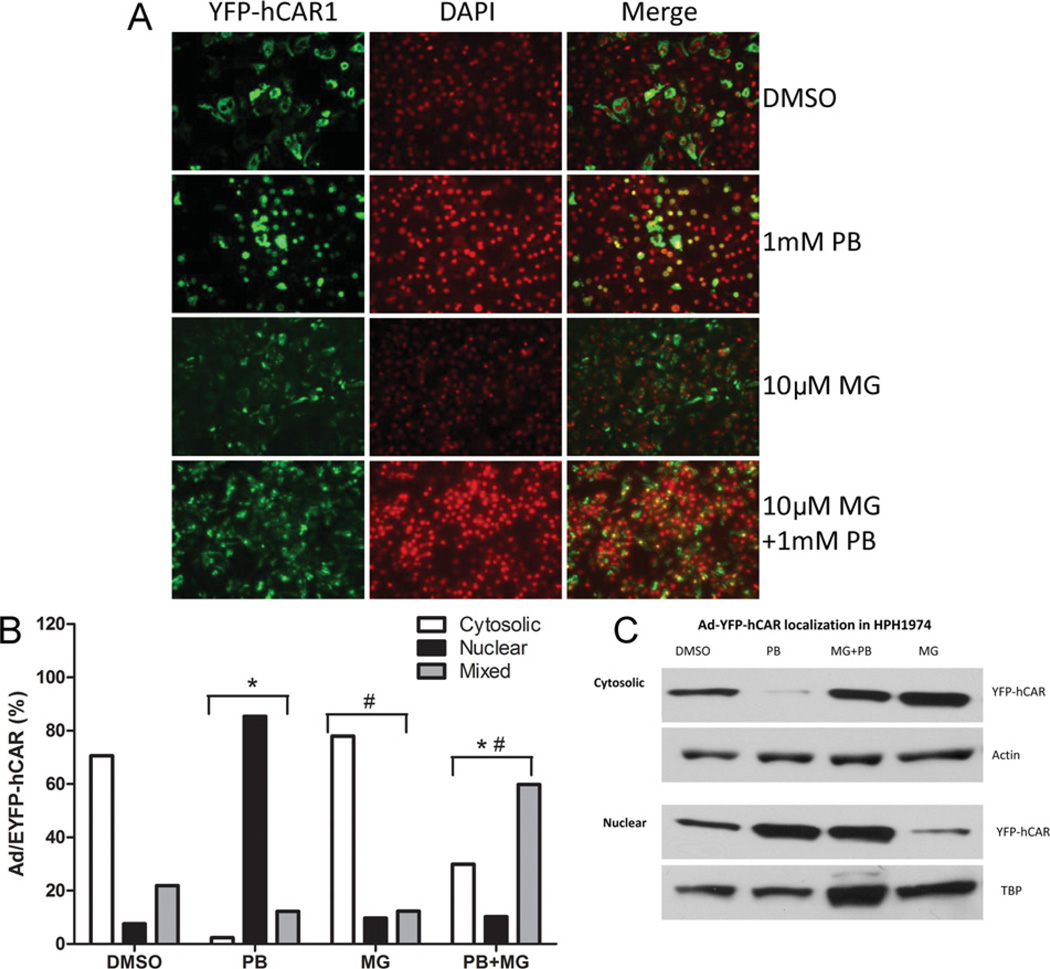
Proteasome inhibition attenuates PB-mediated nuclear translocation of hCAR in HPHs
Whereas transfected hCAR1 spontaneously migrates to the nucleus in transformed cell lines independently of chemical activation, CARs in liver tissues or primary hepatocytes typically translocate from the cytosol to the nuclear compartment following chemical activation. To determine the effect of MG-132 on PB-mediated nuclear translocation of hCAR1, we used an established in vitro translocation assay in HPHs that relies on the transduction of a recombinant adenoviral YFP-tagged hCAR1 vector [18]. Following DMSO treatment, the majority of HPHs (~75%) displayed cytosolic YFP-hCAR1 compartmentalization, whereas ~ 10 % exhibited nuclear localization (Figure 2A, top row). The remaining cells exhibited mixed receptor distribution. Localization of YFP-hCAR1 in HPHs treated only with MG-132 was similar to the control DMSO-treated HPHs (Figure 2A, third row). Following treatment with PB, ~85% of HPH displayed nuclear YFP-hCAR1, whereas ~5% exhibited cytosolic localization (Figure 2A, second row). However, PB-mediated nuclear translocation of YFP-hCAR1 was markedly inhibited in cells co-treated with MG-132, with only ~ 10 % of HPHs now exhibiting complete nuclear localization, ~28% displaying cytosolic localization, and the remaining ~62% showing a mixed distribution (Figure 2A, fourth row). MG-132 treatments similarly inhibited CITCO-mediated hCAR translocation in HPHs (results not shown). The calculated percentages of YFP-hCAR subcellular distribution following the various treatments is presented in Figure 2(B). Furthermore, cytoplasmic and nuclear protein fractions were extracted for immunoblotting analysis (Figure 2C). In agreement with the microscopic observations, PB treatment stimulated nuclear accumulation of YFP-hCAR protein relative to the vehicle control. In the presence of MG-132, the effect of PB on nuclear accumulation of YFP-hCAR was clearly diminished. Of note, in cells treated with MG-132 alone, detectable levels of YFP-hCAR protein in the cytoplasm were increased, whereas nuclear levels decreased.
Figure 2. Attenuation of PB-induced nuclear translocation of adenoviral-YFP-hCAR in HPH by MG-132.
HPHs were infected with adenoviral-YFP-hCAR vectors as described in the Experimental section, and treated with vehicle control (0.1 % DMSO), 10 µM MG-132 (MG), and 1 mM PB in the presence and absence of 10 µM MG-132. (A) After 24 h treatment, hepatocytes were DAPI-stained and subjected to fluorescence microscopy analysis. (B) For each treatment, approximately 100 hCAR-expressing cells were counted and classified on the basis of cytosolic, nuclear or mixed (cytosolic plus nuclear) hCAR cellular localization. The percentages of CAR localization on the basis of the microscopy analysis are indicated. Significantly different from DMSO (*), PB (#), P < 0.05. (C) In a separate experiment, cytoplasmic and nuclear fractions were extracted from the treated HPHs for immunoblotting analysis.
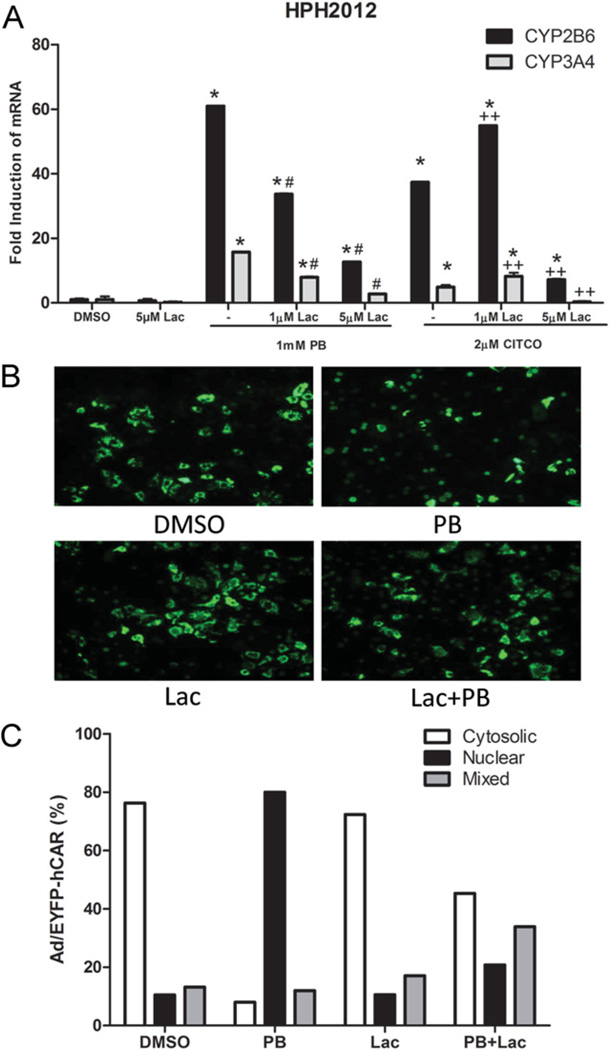
Highly similar results to those with MG-132 were obtained using Lac, an agent that inhibits proteasomal activity though a distinct irreversible mechanism [31]. As shown in Figure 3(A), co-treatment with Lac resulted in dose-dependent inhibition of PB (1 mM) or CITCO (2 µM) mediated CYP2B6 and CYP3A4 target gene induction, confirming the previous observations with MG-132. Treatment of adenoviral-YFP-tagged hCAR infected HPH with PB in the presence of Lac (10 µM) inhibited translocation of hCAR to the nucleus, compared with cells treated with PB alone (Figure 3B). Taken together, these results indicate that inhibiting proteasomal activity decreased both CAR translocation and CAR-mediated transcription responses in human hepatocytes following treatment with receptor activators.
Figure 3. Inhibition of CYP2B6/CYP3A4 induction and nuclear translocation of hCAR in HPHs by Lac.
(A) HPHs were treated with vehicle (0.1 % DMSO), PB (1 mM) or CITCO (2 µM) in the presence and absence of the irreversible proteasome inhibitor Lac. Total RNA, extracted after 24 h treatment, was subjected to real-time PCR analysis. Significantly different from DMSO and Lac (*), PB (#) or CITCO (+ +), P < 0.05. (B and C) In separate experiments, HPHs were infected with adenoviral-YFP-hCAR vectors, and then treated with vehicle control (0.1 % DMSO), 5 µM Lac alone, 1 mM PB or co-treatment with 5 µM Lac. Following 24 h treatment, the localization of hCAR-expressed cells was visualized with fluorescence microscopy.
Effect of proteosome inhibitors on hCAR1 transactivation
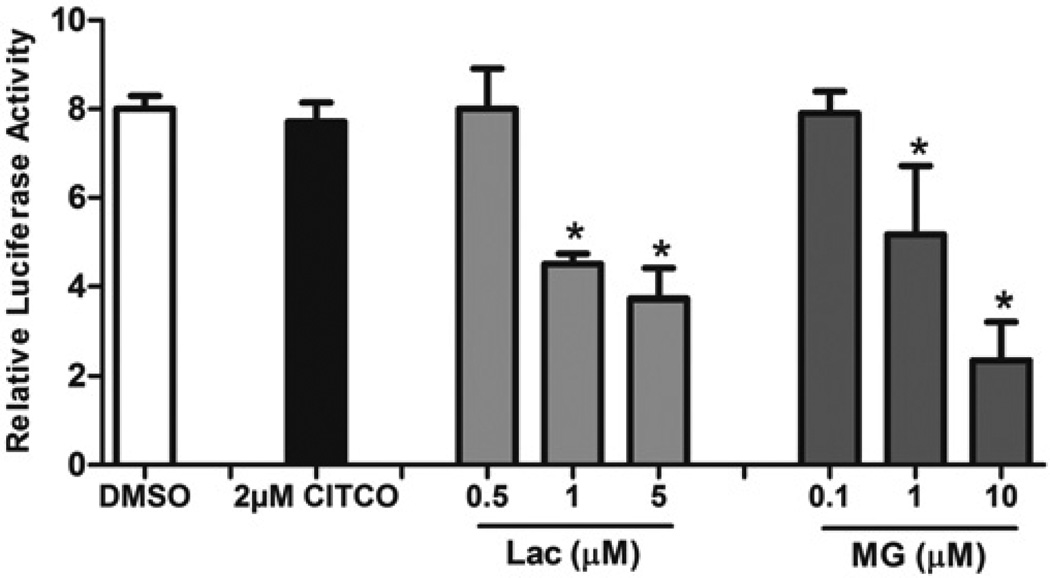
To further these investigations, the effects of MG-132 and Lac on CAR-mediated reporter activity were evaluated in COS-1 cells transfected with hCAR1. The typical high level of hCAR1 constitutive activity was detected in DMSO-treated cells and this activity level was not notably affected with CITCO, an hCAR ligand. However, treatment with either of the proteasomal inhibitors MG-132 or Lac resulted in significant dose-dependent inhibition of hCAR1’s high constitutive activity (Figure 4). Together with the results presented in Figure 1, these effects imply a role for proteasomal regulation of CAR-mediated transcriptional activation.
Figure 4. Proteasome inhibitors MG-132 and Lac suppress hCAR1 constitutive activity.
COS-1 cells were transfected with hCAR1 expression vector along with the 3.1-RXRα expression vector, pRL-CMV normalization vector and CYP2B6-PBREM/XREM reporter. Transfected cells were treated with either vehicle (0.1% DMSO), CITCO (2 µM), MG-132 (MG) (0.1, 1 and 10 µM) or Lac (0.5, 1 and 5 µM). After 24 h treatments, luciferase activities were determined and expressed relative to vehicle control. Data are the means ± S.D. (n = 3). Significantly different from DMSO and CITCO (*), P < 0.05.
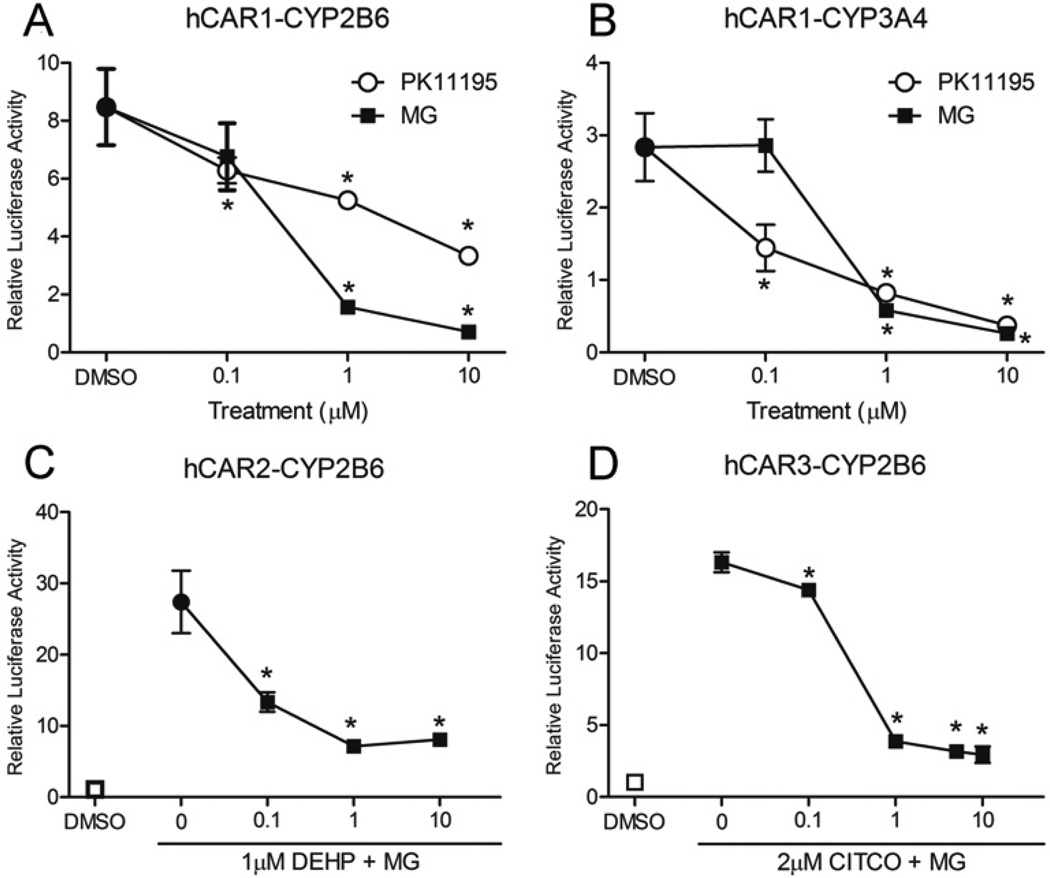
Effect of MG-132 on CAR variants and hPXR in hepatoma cell lines
Given hCAR’s very high constitutive activity, the effects of MG-132 together with an inverse CAR agonist, PK-11195, were evaluated. PK-11195 is a known potent antagonist of hCAR and is commonly used in competition assays to assess potential direct agonists of hCAR1 [32]. These studies were conducted in HepG2 cells transiently transfected with hCAR1 using CYP2B6 and CYP3A4 reporter constructs. Treatment with PK-11195 resulted in dose-dependent inhibition of the high constitutive activities reflected in both CYP2B6 and CYP3A4 reporter assays (Figures 5A and 5B). MG-132 additions similarly suppressed the hCAR1 reporter activities in a dose-dependent manner, consistent with the results obtained in COS-1 cells (Figure 4). Analogous results were also obtained in experiments performed with mCAR (Supplementary Figure S2A at http://www.biochemj.org/bj/458/bj4580095add.htm).
Figure 5. Effect of MG-132 on hCAR1 constitutive activity and ligand-sensitive activation of hCAR splicing variants in HepG2 cells.
HepG2 cells were transfected with hCAR variant expression vector constructs along with 3.1-RXRα expression vector, pRL-CMV normalization vector and either the CYP2B6-PBREM/XREM reporter (A, C and D) or the CYP3A4-XREM reporter (B). (A and B) hCAR1 transfected cells were treated with either vehicle control (0.1 % DMSO), a selective hCAR antagonist (PK-11195) or MG-132 (MG) at the concentrations indicated. (C) hCAR2 transfected cells were treated with vehicle control (0.1 % DMSO), a selective hCAR2 activator (DEHP), or triple doses of MG-132 with a fixed concentration of DEHP (1 µM). (D) hCAR3 transfected cells were treated with vehicle control (0.1 % DMSO), a known hCAR agonist (CITCO), and different doses of MG-132 with a fixed concentration of CITCO (2 µM). Then 24 h after treatments, luciferase activities were determined and expressed relative to vehicle control. Data are the means ± S.D. (n = 3). Significantly different from DMSO (*), P < 0.05.
Next, the effects of MG-132 on the ligand-activated naturally occurring hCAR splice variants hCAR2 and hCAR3 were investigated. In these studies, HepG2 cells were transfected with the CYP2B6 reporter together with hCAR2 or hCAR3 expression vectors, and treated with CAR-selective ligands, either in the absence and presence of MG-132. Consistent with our previous reports [29], CAR2 expressed in HepG2 cells exhibited low basal activity in the absence of ligand, but was markedly activated in the presence of the hCAR2-selective ligand, DEHP 33]. Treatment with MG-132 repressed DEHP-induced hCAR2 activity and exhibited dose dependency (Figure 5C). Similarly, the hCAR direct ligand, CITCO, activated hCAR3 expressed in HepG2 cells, and MG-132 inhibited this activation, also with dose dependency (Figure 5D). Analogous results were obtained with hPXR (Supplementary Figure S2B), a ligand-activated NR that is closely related to hCAR [34].
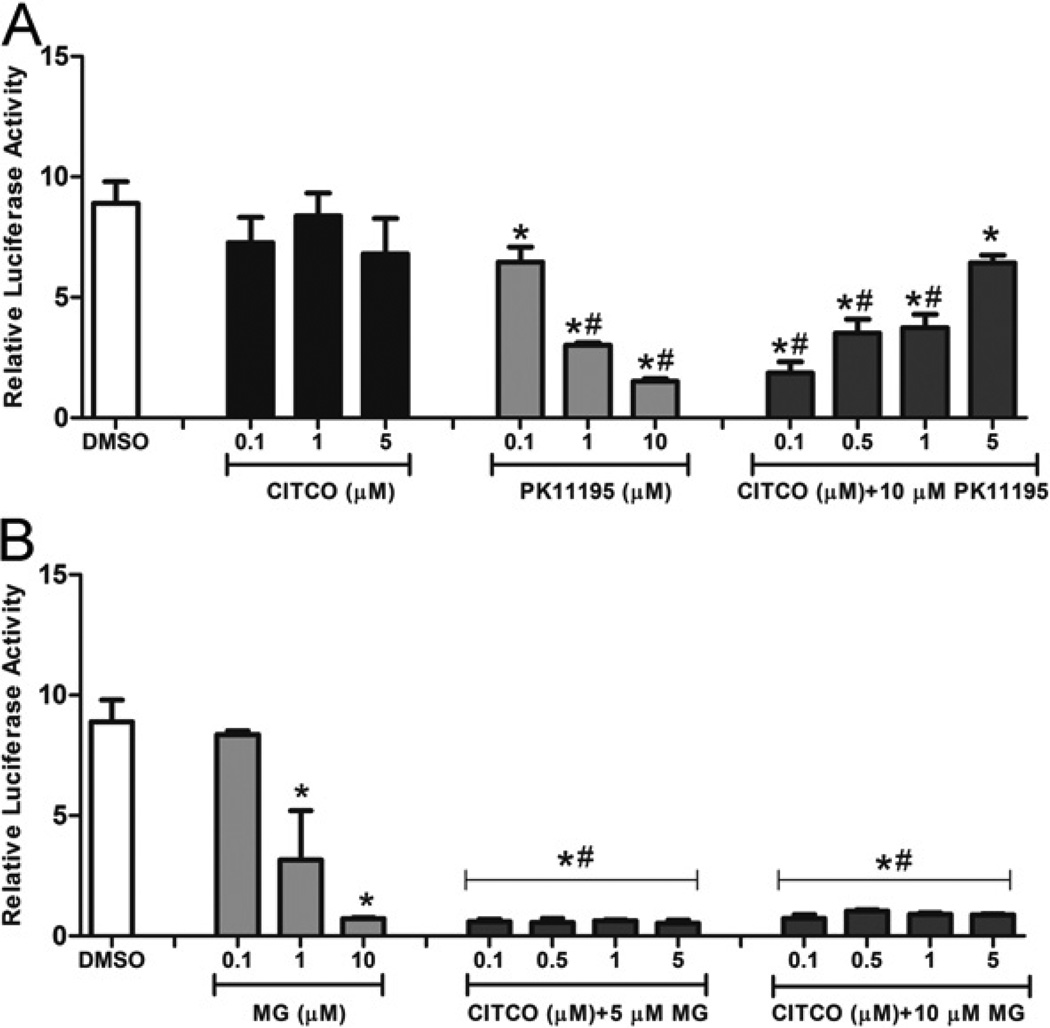
MG-132 is not a direct antagonist of hCAR
To assess mechanistically whether MG-132 inhibits CAR activity through action as a direct inverse agonist, competition assays were conducted in HepG2 cells using the hCARI reporter system (Figure 6A). As expected, PK-11195 antagonized hCAR1 constitutive activity. Using a fixed concentration of PK-11195 (10 µM), the titration of CITCO resulted in a dose-dependent restoration of hCAR constitutive activity (Figure 6A). MG-132 treatment resulted in a dose-dependent deactivation of hCAR1; however, titration of CITCO with MG-132 at either 5 µM or 10 µM concentrations failed to restore hCAR1 activity (Figure 6B). These results indicate that MG-132 is not a direct inverse agonist of hCAR1. Similar results were obtained in reporter competition assays performed with hCAR in COS-1 cells (results not shown).
Figure 6. MG-132 failed to antagonize CITCO-modulated hCAR1 reactivation.
HepG2 cells were transfected with hCAR1 expression vector along with the CYP2B6-PBREM/XREM reporter. (A) Transfected cells were treated with vehicle (0.1 % DMSO), CITCO (0.1, 1 and 5 µM), PK-11195 (0.1, 1 and 10 µM) or increasing doses of CITCO with a fixed PK-11195 dose (10 µM). (B) hCAR1-transfected cells were exposed to vehicle (0.1 % DMS0), MG-132 (0.1, 1 and 10 µM), or increasing doses of CITCO with a fixed MG-132 (MG) dose (either 5 µM or 10 µM). After 24 h treatments, luciferase activities were determined and expressed relative to vehicle control. Data are the means ± S.D. (n = 3). Significantly different from DMSO (*), MG-132 (0.1 and 1 µM, #), P < 0.05.
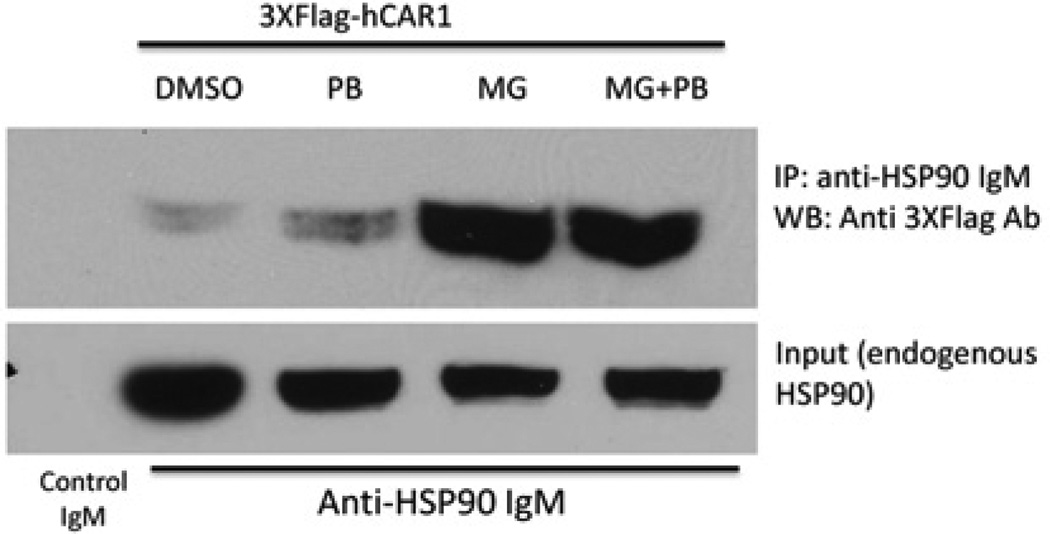
MG-132 enhances the formation of the hCAR/HSP90 protein complex
Given that MG-132 appears to repress chemically mediated hCAR nuclear trafficking in HPHs, the effect of MG-132 on the formation of the hCAR/HSP90 cytoplasmic complex was examined. For these studies, a co-immunoprecipitation assay was used in COS-1 cells to examine the interaction between endogenous HSP90 and transfected 3 × FLAG-hCAR1 following treatment with 10 µM MG-132 or 1 mM PB, or following co-treatments with PB and MG-132. Results of the Western blot analysis conducted following precipitation of the cytosolic fraction with anti-(HSP90 IgM) antibody and detection with anti-3 × FLAG antibody demonstrated marked differences in the interaction between hCAR1 and HSP90 that were modulated by MG-132 treatment. Transfected hCAR spontaneously accumulates into the nucleus of COS-1 cells independently of chemical activation, therefore only a moderate interaction between 3 × FLAG-hCAR and endogenous HSP90 was detected in cells treated with either DMSO or PB (Figure 7, lanes 2 and 3). However, the interaction of hCAR and HSP90 was markedly enhanced by exposure to MG-132 alone or by co-treatment with PB and MG-132 (Figure 7, lanes 4 and 5). These results indicate that proteasomal activity is required for the dissociation of CAR from HSP90.
Figure 7. MG-132 enhanced the interaction between hCAR1 and endogenous HSP90 in cytosolic fractions.
COS-1 cells were transfected with 3 × FLAG-hCAR1 as described in the Experimental section. After 36 h, the cells were treated with vehicle (0.1 % DMSO), PB (1 mM), 10 µM MG-132 (MG) alone, or co-treated with MG-132 plus PB. After 5 h treatment, cytosolic fractions were extracted and immunoprecipitated with anti-(HSP90 IgM) antibody plus Protein L-Agarose. The precipitated protein was subjected to Western blotting with anti-FLAG antibody, followed by anti-HSP90 antibody for endogenous HSP90 detection. The anti-HSP90 antibody precipitated equal amounts of HSP90 from the COS-1 cell cytosolic fractions, whereas control IgM did not precipitate HSP90. IP, immunoprecipitation; WB, Western blot.
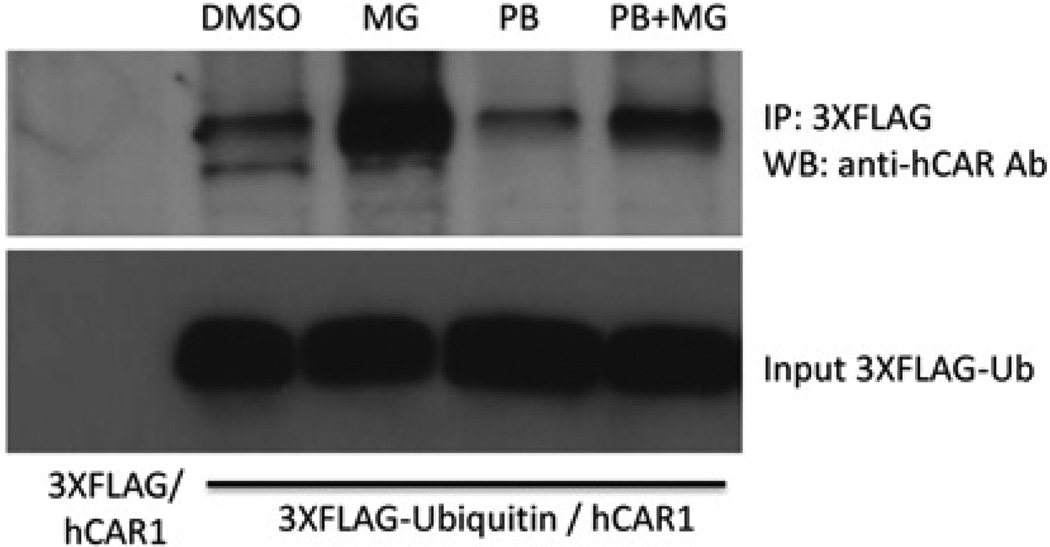
MG-132 increases the ubiquitination of hCAR
Ubiquitination is required for proteasomal regulation of cellular proteins. To determine the effect of MG-132 on levels of ubiquitinated hCAR, an in vitro ubiquitination assay was developed. Cellular extracts from COS-1 cells transfected with hCAR1 and 3 × FLAG-ubiquitin or 3 × FLAG-empty vector were precipitated with anti-(FLAG M2) antibody resin. An hCAR antibody was used to detect ubiquitinated hCAR in an immunoblot of the total precipitated ubiquitinated protein. Using this approach, hCAR protein was not detected in lysates from the 3 × FLAG-empty vector transfection (Figure 8, lane 1); however, ubiquitinated hCAR was clearly detected in lysates from cells transfected with the 3 × FLAG-ubiquitin (Figure 8, lanes 2–5). Treatment with PB did not appear to alter the ubiquitination status of hCAR relative to the DMSO control samples (Figure 8, lane 4). Treatment with the proteasome inhibitor MG-132 alone resulted in increased ubiquitinated hCAR levels (Figure 8, lane 3). It is noteworthy that levels of ubiquitinated hCAR were dramatically increased after co-treatment with both MG-132 and PB relative to PB alone (Figure 8, lane 5).
Figure 8. MG-132 increases the ubiquitination of hCAR.
COS-1 cells transfected with hCAR1 expression vector and 3 × FLAG—ubiquitin or 3 × FLAG-empty vector were exposed to chemical treatments for 5 h. Cellular extracts were precipitated with anti-FLAG M2 antibody resin. The resulting total precipitated ubiquitinated protein was subjected to Western blot detection with an anti-hCAR antibody. Lane 1, precipitates from cells transfected with hCAR1 and 3 × FLAG-empty vector; lanes 2–5, precipitates from cells transfected with hCAR1 and 3 × FLAG—ubiquitin after various chemical treatments. The lower panel shows the level of 3 × FLAG—ubiquitin input. Ab, antibody; IP, immunoprecipitation; WB, Western blot; Ub, ubiquitin.
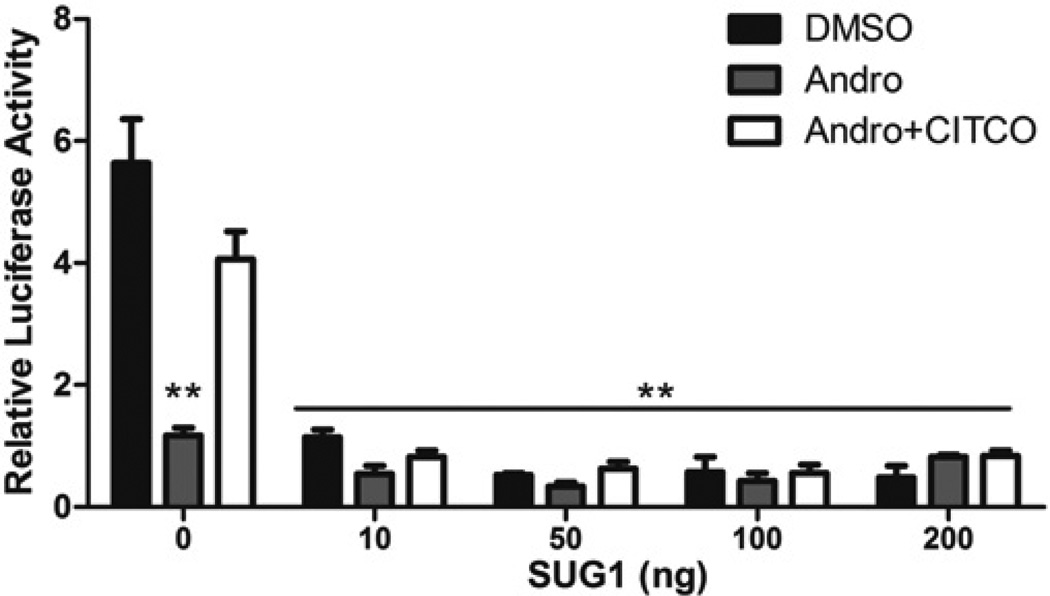
The proteasome subunit SUG1 suppresses the constitutive and ligand-mediated activity of hCAR1
We tested the effect of SUG1, a subunit of 26S proteasome complex, on hCAR1 activity in a transactivation assay. As shown in Figure 9, 10 µM Andro, a CAR inverse agonist, repressed the high basal activity of hCAR1, an inhibition that was competitively restored by exposure to the direct CAR agonist CITCO. SUG1 dramatically decreased the high constitutive activity of hCAR independently of treatments, even at transfection levels as low as 10 ng, indicating mechanistic involvement of this specific component of the 26S proteosome in the regulatory scheme.
Figure 9. The proteasome subunit SUG1 suppresses the constitutive and ligand-mediated activity of hCAR1.
COS-1 cells were co-transfected with hCAR1 expression vector and various amounts of pCMV6-SUG1 expression vector, along with the CYP2B6-PBREM/XREM reporter. Transfected cells were treated for 24 h with vehicle control (0.1 % DMSO) or 10 µM Andro in the presence and absence of 2 µM CITCO. After treatment, luciferase activity was determined. Data are the means ± S.D. (n = 3). Significantly different from hCAR1 activity in control cells (no SUG1) treated with DMSO or Andro plus CITCO (**), P < 0.05.
MG-132 inhibits the recruitment of hCAR with co-activators
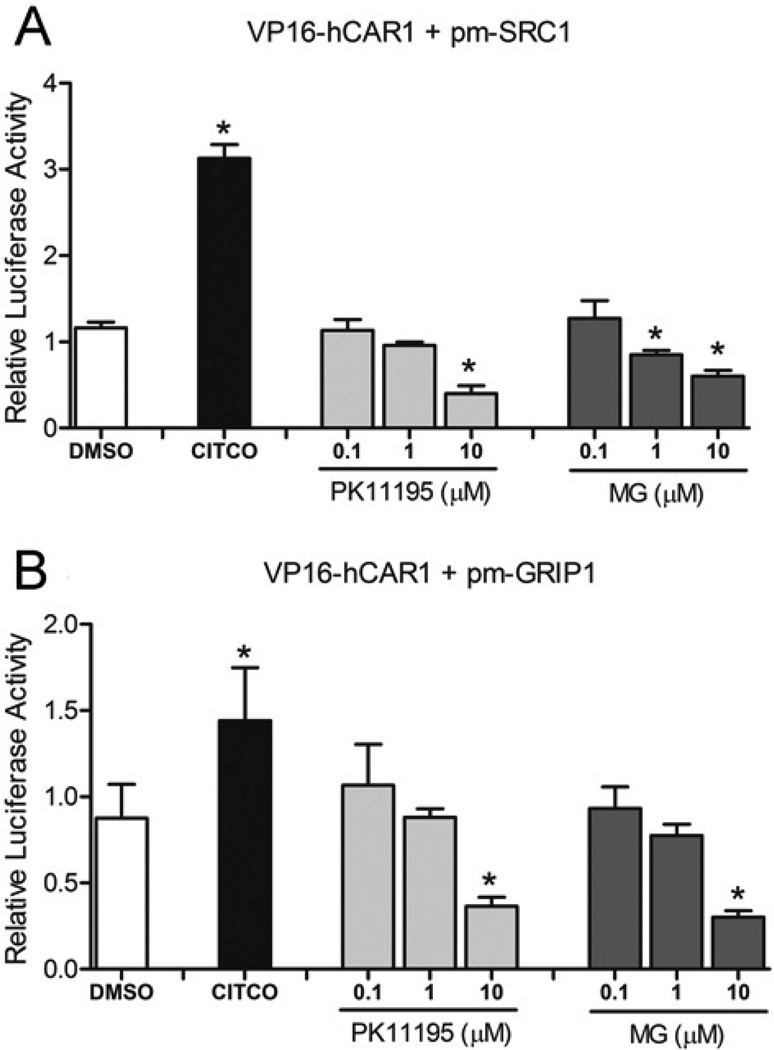
Since the proteasomal complex modulates both cytosolic and nuclear events, a mammalian two-hybrid assay was used to investigate whether MG-132 may also affect the interaction between hCAR and known nuclear co-regulators of CAR, in particular SRC1 and GRIP1. As shown in Figure 10(A), the interaction of hCAR with SRC1 was increased by a single dose of the direct hCAR agonist CITCO (2 µM), when compared with vehicle control (0.1 % DMSO), whereas the specific hCAR antagonist, PK-11195, produced a significant dose-dependent inhibition of the interaction. Treatment with the proteasome inhibitor MG-132 also exhibited dose-dependent inhibition of the interaction of hCAR with SRC1. Furthermore, both PK-11195 and MG-132 similarly inhibited the interaction between hCAR1 and GRIP1 (Figure 10B).
Figure 10. Inhibition of hCAR1 and co-activator (SRC1 or GRIP1) interactions by MG-132.
Mammalian two-hybrid assays were performed in COS-1 cells transfected with indicated expression plasmids encoding VP16-AD/hCAR1 fusion proteins and GAL4-DBD/SRC1 (A) or GRIP1 fusion protein (B), with the reporter plasmid pG5-luciferase. Cells were treated with DMSO (0.1 %), CITCO (2 µM), PK-11195 and MG-132 at the indicated concentrations for 24 h before determination of luciferase activities. Data are the means ± S.D. (n = 3). Significantly different from DMSO (*), P < 0.05.
DISCUSSION
The studies described in the present paper demonstrate a critical role for the 26S proteasome as a modulator of CAR functional activity. The 26S proteasome system is an integral cytosolic and nuclear protease complex responsible for ATP-dependent degradation of most ubiquitinated cellular proteins [35]. The proteasome complex is highly conserved in eukaryotic evolution and consists of two multicatalytic subunit complexes. One is the barrel-like 20S catalytic core that contains multiple peptidase activities for protein hydrolysis. MG-132 and other proteasome inhibitors such as Lac and bortezomid are active site inhibitors that bind within the catalytic core [31]. The second component of the complex is the 19S regulatory particle, composed of a lid- and base-like structure that serves to recognize ubiquitinated proteins, unfold and direct them into the 20S core complex. A wide variety of substrates have been identified for the 26S proteasome, including the transcriptional regulators c-Jun, p53 and NF-κB (nuclear factor κB), and cell cycle regulator proteins such as the cyclins. In addition to the proteolytic roles of the proteasome, important non-proteolytic roles have also emerged.
For example, accumulating evidence indicates that the 26S proteasome participates in the degradation of NR proteins and also functions in NR-mediated gene transcription [24]. Indeed, the results of the present study demonstrate that CAR is a target for ubiquitination and that proteasomal inhibition leads to intracellular accumulation of ubiquitinated CAR. However, despite CAR’s accumulation following proteasomal inhibition, the receptor’s transcriptional activity is markedly down-regulated under these conditions. Another report similarly demonstrated an increase in ubiquitinated PXR (pregnane X receptor) after MG-132 inhibition of the 26S proteasome, resulting in the inhibition of PXR activity [36].
Mechanistically, the interface between CAR and the proteosome occurs at several levels. Exposure of HPHs to MG-132 attenuated CYP2B6 and CYP3A4, target genes for CAR, up-regulation subsequent to treatments with either indirect or direct CAR activators (PB or CITCO respectively). These attenuation responses were noted at both the mRNA and protein levels. Additional mechanistic studies demonstrated that inhibition of the 26S proteasome in HPHs decreased the nuclear translocation of hCAR. During CAR activation, nuclear trafficking is an initial and important step in vivo and in HPH cultures. CAR is tethered in a cytosolic complex that includes HSP90 and other proteins [8]. Following chemical activation, CAR is released from the complex and translocates to the nucleus. The mechanisms underlying the receptor’s release and nuclear transport are unclear, although recruitment of PP2A to the CAR complex followed by dephosphorylation of Thr38 of hCAR is implicated [15].
The results shown in the present study using HPHs and co-immunoprecipitation assays demonstrate that proteasomal inhibition enhances the interaction between hCAR and HSP90, supporting the concept that proteasomal activity is also required for these initial CAR activation steps. Interestingly, others have demonstrated a role for the proteasome in modulating NR cellular trafficking. For example, in mammalian cells and mouse embryo primary fibroblasts, proteasome inhibition results in enhanced nuclear translocation of the aryl hydrocarbon receptor, in the absence of receptor ligand [37,38]. In contrast, proteasomal inhibition by MG-132 decreases the mobility of the GR (glucocorticoid receptor) in the nucleus [26] and inhibits androgen-induced nuclear translocation of the AR (androgen receptor) [25]. The latter results are similar to our findings with hCAR. Although a non-proteolytic role for the proteasome in AR nuclear translocation has been proposed [25], proteolytic activity is required for nuclear translocation of other transcription factors. For example, similar to CAR, NF-κB is sequestered in the cytoplasm and forms a complex with the inhibitory protein IκB (inhibitory κB). Following stimulation, IκB is ubiquitinated and degraded by the proteasome, releasing NFκB to translocate to the nucleus and activate target genes [39]. Further investigations will be required to extract the exact mechanisms of proteasomal regulation of intracellular CAR trafficking.
To further examine the effects of proteasome inhibition on CAR activation, transactivation assays were conducted in the COS-1 and HepG2 cell lines. Following its transfection in cell lines, hCAR1 escapes cytosolic tethering and therefore exhibits high-constitutive transcriptional activation of reporter plasmids in COS-1 cells independently of chemical stimulus. In the cell-based CYP2B6 and CYP3A4 reporter assays conducted in the present study, MG-132 robustly inhibited the high basal activity of hCAR1 in a dose-dependent manner. Although the inhibitory effect of the hCAR1 inverse agonist PK-11195 could be competed with the hCAR agonist CITCO, the inhibitory effect of MG-132 was not affected by agonist exposure, suggesting that MG-132’s effects are not mediated by direct interaction with the ligand-binding pocket of hCAR1. Furthermore, MG-132 does not induce nuclear translocation, whereas the direct inverse agonist PK-11195 does [32], providing further evidence that MG-132 does not interact as a ligand with CAR.
As stated above, since transiently transfected hCAR1 spontaneously accumulates in the nucleus of cell lines [11], it is unlikely that MG-132’s inhibition of transcriptional activity is due to inhibition of translocation, as seen in HPHs. Instead, it appears that proteasome inhibition also directly affects CAR-mediated transcriptional processes. Interestingly, two naturally occurring hCAR splice variants, termed hCAR2 and hCAR3, which together appear to account for up to one-third of the total hCAR transcript pool in human livers, are not constitutively active like hCAR1; rather, they are ligand-activated receptors and exhibit certain distinctive ligand selectivities [12]. Results presented from HepG2-based reporter assays demonstrated that MG-132 dose-dependently down-regulated ligand-induced hCAR2 activation by DEHP, and hCAR3 activation by CITCO, suggesting that proteasome modulation is an important regulatory feature affecting each of these hCARs. In these respects, MG-132 also inhibited TCPOBOP-induced mCAR activity and Rif-induced activity of PXR, a CAR-related NR with overlapping substrate specificity and target gene activation. These results demonstrate that inhibition of proteosome activity suppresses CAR-mediated transcription regardless of ligand activation requirements, and that the effects are not species-specific or limited to CAR.
Indeed, others have reported that inhibition of the proteasome modulates NR-mediated transcription, outlining new regulatory roles for the proteasome. For example, treatment of mammalian cells with MG-132 inhibits ER (oestrogen receptor)-, AR- and PR (progesterone receptor)-mediated gene activation, despite increases in their respective protein levels. In the case of ERs, proteasome inhibition appeared to interfere with the exchange of co-regulatory proteins and the dynamic cycling of the ER transcription complex with DNA-response elements [40,41]. Similarly, proteasome activity is required for proper assembly of PR- and AR-transcription complexes [42,43]. Further studies have shown that recruitment of the proteasome is required for release and subsequent degradation of the NCoR (nuclear receptor co-repressor 1) complex from liganded NR, allowing co-activator proteins to associate [44]. Despite these observations, proteasome inhibition does not always inhibit transcription. For example, MG-132 increases the protein level of GR and also facilitates GR-mediated transactivation [26]. Thus the effects of proteasomal inhibition appear to selectively differ depending on the NR target. For CAR and PXR, proteasomal inhibition decreased transcriptional activity in reporter assays. Mechanistically, the data obtained from mammalian two-hybrid experiments suggests that the inhibition of proteasome activity affects the interaction of CAR with its co-regulatory protein partners.
In early reports, before it was recognized as a component of the 19S proteasome, SUG1 was shown to repress ligand-stimulated activity of the vitamin D3 receptor, ER, thyroid hormone receptor and RXRα in yeast [45]. The hCAR1 transactivation assay data reported in the present paper also demonstrate that SUG1 represses both the constitutive and ligand-activated transcriptional activity of hCAR1. Future studies are required to examine the detailed mechanisms involved in these effects, although we speculate that SUG1 may function to disrupt the assembly of the hCAR1 transcription complex. Indeed, recent studies in mammalian systems demonstrated that SUG1 may directly affect NR-mediated transcription complexes. In these respects, ChIP assays have identified SUG1 complexed with ER, RNA polymerase II and other transcription factors bound to an ER-responsive promoter [41]. The investigators did not detect the 20S PSMA2/α2 subunit, suggesting SUG1 acts independently in transcription initiation. SUG1 also interacts with SRC3 at RAR (retinoic acid receptor) target genes and is thought to aid in the assembly of the transcription complex [46]. The non-proteolytic role of the 19S particle is not limited to NR-mediated transcription. It is also a reported regulator of HIV and MHC II transcription, as well as other systems (reviewed in [47]). Thus, in addition to its role in delivering ubiquitinated proteins to the 20S hydrolytic component, the 19S regulatory particle also appears to serve as a co-regulator, facilitating or inhibiting transcription initiation. However, a recent study suggests that it is unlikely that proteasomal subunits are independently recruited to transcription sites. Instead, it was proposed that the entire 26S proteasome is present and that the subunits function individually, depending on transcriptional status [48].
The results of the present study demonstrate the importance of proteasomal functional interaction with CAR activity at a minimum of two levels. First, the key finding that the proteasome inhibitor MG-132 attenuated PB-modulated nuclear translocation in HPHs suggests that the proteasome system plays a critical role modulating the cellular trafficking of CAR in vivo. Secondly, results of transactivation studies demonstrate that MG-132 inhibits both constitutive hCAR1-mediated, as well as ligand-dependent hCAR2-, hCAR3- and PXR-mediated transcriptional activation, suggesting that proteasome activity is required for gene target activation by these receptors. In these respects, the data presented from mammalian two-hybrid experiments show that MG-132 inhibits the interaction between CAR and the co-activators, SRC1 and GRIP1, further supporting a role for the proteasome at CAR-targeted gene transcription sites. The ability of the proteasome to regulate CAR’s activity at multiple levels may provide a means of fine tuning CAR-mediated activation of target genes in response to different CAR ligands and indirect activators, such as PB. These results and those of ongoing investigations may offer the proteasomal complex as a novel target for the development of therapeutic and research tools for exploiting the functional biology of hCAR.
Supplementary Material
ACKNOWLEDGEMENTS
We gratefully acknowledge the contributions of Dr Stephen Strom at the Department of Pathology, University of Pittsburgh, PA, U.S.A., for provision of primary human hepatocytes.
FUNDING
This research was supported by the USPHS (United States Public Health Service) National Institute of General Medical Sciences [grant number GM066411].
Abbreviations
- Andro
5α-androstan-3α-ol
- AR
androgen receptor
- CAR
constitutive androstane receptor
- CITCO
6-(4-chlorophenyl)imidazo[2,1-b][1,3]thiazole-5-carbaldehyde O-(3,4-dichlorobenzyl)oxime
- CT
cycle threshold
- CYP
cytochrome P450
- DEHP
di-2-ethylhexyl phthalate
- DMEM
Dulbecco’s modified Eagle’s medium
- ER
oestrogen receptor
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GR
glucocorticoid receptor
- GRIP1
glutamate-receptor-interacting protein 1
- hCAR
human CAR
- HPH
human primary hepatocyte
- hPXP
human pregnane X receptor
- HRP
horseradish peroxidase
- HSP90
heat-shock protein 90
- IκB
inhibitory κB
- Lac
lactacystin
- LBD
ligand-binding/heterodimerization domain
- mCAR
mouse CAR
- NF-κB
nuclear factor κB
- NR
nuclear receptor
- PB
phenobarbital
- PBREM
phenobarbital-responsive enhancer module
- PK-11195
1-(2-chlorophenyl)-N-methyl-N-(1-methylpropyl)-3-isoquinolinecarboxamide
- PP2A
protein phosphatase 2A
- PXR
pregnane X receptor
- PR
progesterone receptor
- RID
receptor-interaction domain
- Rif
rifampicin
- RXRα
retinoid X receptor α
- SRC1
steroid receptor co-activator
- SUG1
suppressor for Gal1
- 0.1 % TBS-T
Tris-buffered saline containing 0.1% Tween 20
- TCPOBOP
1,4-bis[2-(3,5-dichloropyridyloxy)]benzene
- UPS
ubiquitin-proteasome system
- XREM
xenobiotic-responsive enhancer module.
Footnotes
AUTHOR CONTRIBUTION
All authors contributed to the design and execution of the experiments. Tao Chen, Elizabeth Laurenzana and Curtis Omiecinski contributed to the writing of the paper.
REFERENCES
- 1.Sueyoshi T, Kawamoto T, Zelko I, Honkakoski P, Negishi M. The repressed nuclear receptor CAR responds to phenobarbital in activating the human CYP2B6 gene. J. Biol. Chem. . 1999;274:6043–6046. doi: 10.1074/jbc.274.10.6043. [DOI] [PubMed] [Google Scholar]
- 2.Qatanani M, Moore DD. CAR, the continuously advancing receptor, in drug metabolism and disease. Curr. Drug Metab. 2005;6:329–339. doi: 10.2174/1389200054633899. [DOI] [PubMed] [Google Scholar]
- 3.Stanley LA, Horsburgh BC, Ross J, Scheer N, Wolf CR. PXR and CAR: nuclear receptors which play a pivotal role in drug disposition and chemical toxicity. Drug Metab. Rev. 2006;38:515–597. doi: 10.1080/03602530600786232. [DOI] [PubMed] [Google Scholar]
- 4.Timsit YE, Negishi M. CAR and PXR: the xenobiotic-sensing receptors. Steroids. 2007;72:231–246. doi: 10.1016/j.steroids.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moreau A, Vilarem MJ, Maurel P, Pascussi JM. Xenoreceptors CAR and PXR activation and consequences on lipid metabolism, glucose homeostasis, and inflammatory response. Mol. Pharm. 2008;5:35–41. doi: 10.1021/mp700103m. [DOI] [PubMed] [Google Scholar]
- 6.Gao J, Xie W. PXR and CAR at the crossroad of drug metabolism and energy metabolism. Drug Metab. Dispos. 2010;38:2091–2095. doi: 10.1124/dmd.110.035568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swales K, Negishi M. CAR, driving into the future. Mol. Endocrinol. 2004;18:1589–1598. doi: 10.1210/me.2003-0397. [DOI] [PubMed] [Google Scholar]
- 8.Li H, Wang H. Activation of xenobiotic receptors: driving into the nucleus. Expert Opin. Drug Metab. Toxicol. 2010;6:409–426. doi: 10.1517/17425251003598886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osabe M, Negishi M. Active ERK1/2 protein interacts with the phosphorylated nuclear constitutive active/androstane receptor (CAR; NR1I3), repressing dephosphorylation and sequestering CAR in the cytoplasm. J. Biol. Chem. 2011;286:35763–35769. doi: 10.1074/jbc.M111.284596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen T, Tompkins LM, Li L, Li H, Kim G, Zheng Y, Wang H. A single amino acid controls the functional switch of human constitutive androstane receptor (CAR) 1 to the xenobiotic-sensitive splicing variant CAR3. J. Pharmacol. Exp. Ther. 2010;332:106–115. doi: 10.1124/jpet.109.159210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawamoto T, Sueyoshi T, Zelko I, Moore R, Washburn K, Negishi M. Phenobarbital-responsive nuclear translocation of the receptor CAR in induction of the CYP2B gene. Mol. Cell. Biol. 1999;19:6318–6322. doi: 10.1128/mcb.19.9.6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Auerbach SS, Ramsden R, Stoner MA, Verlinde C, Hassett C, Omiecinski CJ. Alternatively spliced isoforms of the human constitutive androstane receptor. Nucleic Acids Res. 2003;31:3194–3207. doi: 10.1093/nar/gkg419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sueyoshi T, Moore R, Sugatani J, Matsumura Y, Negishi M. PPP1R16A, the membrane subunit of protein phosphatase 1β, signals nuclear translocation of the nuclear receptor constitutive active/androstane receptor. Mol. Pharmacol. 2008;73:1113–1121. doi: 10.1124/mol.107.042960. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi K, Sueyoshi T, Inoue K, Moore R, Negishi M. Cytoplasmic accumulation of the nuclear receptor CAR by a tetratricopeptide repeat protein in HepG2 cells. Mol. Pharmacol. 2003;64:1069–1075. doi: 10.1124/mol.64.5.1069. [DOI] [PubMed] [Google Scholar]
- 15.Mutoh S, Osabe M, Inoue K, Moore R, Pedersen L, Perera L, Rebolloso Y, Sueyoshi T, Negishi M. Dephosphorylation of threonine 38 is required for nuclear translocation and activation of human xenobiotic receptor CAR (NR1I3) J. Biol. Chem. 2009;284:34785–34792. doi: 10.1074/jbc.M109.048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hosseinpour F, Moore R, Negishi M, Sueyoshi T. Serine 202 regulates the nuclear translocation of constitutive active/androstane receptor. Mol. Pharmacol. 2006;69:1095–1102. doi: 10.1124/mol.105.019505. [DOI] [PubMed] [Google Scholar]
- 17.Yoshinari K, Kobayashi K, Moore R, Kawamoto T, Negishi M. Identification of the nuclear receptor CAR:HSP90 complex in mouse liver and recruitment of protein phosphatase 2A in response to phenobarbital. FEBS Lett. 2003;548:17–20. doi: 10.1016/s0014-5793(03)00720-8. [DOI] [PubMed] [Google Scholar]
- 18.Li H, Chen T, Cottrell J, Wang H. Nuclear translocation of adenoviral-enhanced yellow fluorescent protein-tagged-human constitutive androstane receptor (hCAR): a novel tool for screening hCAR activators in human primary hepatocytes. Drug Metab. Dispos. 2009;37:1098–1106. doi: 10.1124/dmd.108.026005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rencurel F, Stenhouse A, Hawley SA, Friedberg T, Hardie DG, Sutherland C, Wolf CR. AMP-activated protein kinase mediates phenobarbital induction of CYP2B gene expression in hepatocytes and a newly derived human hepatoma cell line. J. Biol. Chem. 2005;280:4367–4373. doi: 10.1074/jbc.M412711200. [DOI] [PubMed] [Google Scholar]
- 20.Marc N, Galisteo M, Lagadic-Gossmann D, Fautrel A, Joannard F, Guillouzo A, Corcos L. Regulation of phenobarbital induction of the cytochrome P450 2b9/10 genes in primary mouse hepatocyte culture. Involvement of calcium- and cAMP-dependent pathways. Eur. J. Biochem. 2000;267:963–970. doi: 10.1046/j.1432-1327.2000.01083.x. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto Y, Kawamoto T, Negishi M. The role of the nuclear receptor CAR as a coordinate regulator of hepatic gene expression in defense against chemical toxicity. Arch. Biochem. Biophys. 2003;409:207–211. doi: 10.1016/s0003-9861(02)00456-3. [DOI] [PubMed] [Google Scholar]
- 22.Mutoh S, Sobhany M, Moore R, Perera L, Pedersen L, Sueyoshi T, Negishi M. Phenobarbital indirectly activates the constitutive active androstane receptor (CAR) by inhibition of epidermal growth factor receptor signaling. Sci. Signal. 2013;6:ra31. doi: 10.1126/scisignal.2003705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nawaz Z, O’Malley BW. Urban renewal in the nucleus: is protein turnover by proteasomes absolutely required for nuclear receptor-regulated transcription? Mol. Endocrinol. 2004;18:493–499. doi: 10.1210/me.2003-0388. [DOI] [PubMed] [Google Scholar]
- 24.Keppler BR, Archer TK, Kinyamu HK. Emerging roles of the 26S proteasome in nuclear hormone receptor-regulated transcription. Biochim. Biophys. Acta. 2011;1809:109–118. doi: 10.1016/j.bbagrm.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin HK, Altuwaijri S, Lin WJ, Kan PY, Collins LL, Chang C. Proteasome activity is required for androgen receptor transcriptional activity via regulation of androgen receptor nuclear translocation and interaction with coregulators in prostate cancer cells. J. Biol. Chem. 2002;277:36570–36576. doi: 10.1074/jbc.M204751200. [DOI] [PubMed] [Google Scholar]
- 26.Deroo BJ, Rentsch C, Sampath S, Young J, DeFranco DB, Archer TK. Proteasomal inhibition enhances glucocorticoid receptor transactivation and alters its subnuclear trafficking. Mol. Cell. Biol. 2002;22:4113–4123. doi: 10.1128/MCB.22.12.4113-4123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Auerbach SS, Dekeyser JG, Stoner MA, Omiecinski CJ. CAR2 displays unique ligand binding and RXRalpha heterodimerization characteristics. Drug Metab. Dispos. 2007;35:428–439. doi: 10.1124/dmd.106.012641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Omiecinski CJ, Coslo DM, Chen T, Laurenzana EM, Peffer RC. Multi-species analyses of direct activators of the constitutive androstane receptor. Toxicol. Sci. 2011;123:550–562. doi: 10.1093/toxsci/kfr191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Auerbach SS, Stoner MA, Su S, Omiecinski CJ. Retinoid X receptor-α-dependent transactivation by a naturally occurring structural variant of human constitutive androstane receptor (NR1I3) Mol. Pharmacol. 2005;68:1239–1253. doi: 10.1124/mol.105.013417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goyak KM, Johnson MC, Strom SC, Omiecinski CJ. Expression profiling of interindividual variability following xenobiotic exposures in primary human hepatocyte cultures. Toxicol. Appl. Pharmacol. 2008;231:216–224. doi: 10.1016/j.taap.2008.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meiners S, Ludwig A, Stangl V, Stangl K. Proteasome inhibitors: poisons and remedies. Med. Res. Rev. 2008;28:309–327. doi: 10.1002/med.20111. [DOI] [PubMed] [Google Scholar]
- 32.Li L, Chen T, Stanton JD, Sueyoshi T, Negishi M, Wang H. The peripheral benzodiazepine receptor ligand 1-(2-chlorophenyl-methylpropyl)-3-isoquinoline-carboxamide is a novel antagonist of human constitutive androstane receptor. Mol. Pharmacol. 2008;74:443–453. doi: 10.1124/mol.108.046656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dekeyser JG, Stagliano MC, Auerbach SS, Prabu KS, Jones AD, Omiecinski CJ. Di(2-ethylhexyl) phthalate is a highly potent agonist for the human constitutive androstane receptor splice variant, CAR2. Mol. Pharmacol. 2009;75:1005–1013. doi: 10.1124/mol.108.053702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore JT, Moore LB, Maglich JM, Kliewer SA. Functional and structural comparison of PXR and CAR. Biochim. Biophys. Acta. 2003;1619:235–238. doi: 10.1016/s0304-4165(02)00481-6. [DOI] [PubMed] [Google Scholar]
- 35.Lonard DM, O’Malley BW. Emerging roles of the ubiquitin proteasome system in nuclear hormone receptor signaling. Prog. Mol. Biol. Transl. Sci. 2009;87:117–135. doi: 10.1016/S1877-1173(09)87004-X. [DOI] [PubMed] [Google Scholar]
- 36.Staudinger JL, Xu C, Biswas A, Mani S. Post-translational modification of pregnane X receptor. Pharmacol. Res. 2011;64:4–10. doi: 10.1016/j.phrs.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song Z, Pollenz RS. Ligand-dependent and independent modulation of aryl hydrocarbon receptor localization, degradation and gene regulation. Mol. Pharmacol. 2002;62:806–816. doi: 10.1124/mol.62.4.806. [DOI] [PubMed] [Google Scholar]
- 38.Santiago-Josefat B, Pozo-Guisado E, Mulero-Navarro S, Fernandez-Salguero PM. Proteasome inhibition induces nuclear translocation and transcriptional activation of the dioxin receptor in mouse embryo primary fibroblasts in the absence of xenobiotics. Mol. Cell. Biol. 2001;21:1700–1709. doi: 10.1128/MCB.21.5.1700-1709.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 40.Lonard DM, Nawaz Z, Smith CL, O’Malley BW. The 26S proteasome is required for estrogen receptor-α and coactivator turnover and for efficient estrogen receptor-α transactivation. Mol. Cell. 2000;5:939–948. doi: 10.1016/s1097-2765(00)80259-2. [DOI] [PubMed] [Google Scholar]
- 41.Reid G, Hubner MR, Metivier R, Brand H, Denger S, Manu D, Beaudouin J, Ellenberg J, Gannon F. Cyclic, proteasome-mediated turnover of unliganded and liganded ERα on responsive promoters is an integral feature of estrogen signaling. Mol. Cell. 2003;11:695–707. doi: 10.1016/s1097-2765(03)00090-x. [DOI] [PubMed] [Google Scholar]
- 42.Kang Z, Pirskanen A, Janne OA, Palvimo JJ. Involvement of proteasome in the dynamic assembly of the androgen receptor transcription complex. J. Biol. Chem. 2002;277:48366–48371. doi: 10.1074/jbc.M209074200. [DOI] [PubMed] [Google Scholar]
- 43.Dennis AP, Lonard DM, Nawaz Z, O’Malley BW. Inhibition of the 26S proteasome blocks progesterone receptor-dependent transcription through failed recruitment of RNA polymerase II. J. Steroid Biochem. Mol. Biol. 2005;94:337–346. doi: 10.1016/j.jsbmb.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 44.Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell. 2004;116:511–526. doi: 10.1016/s0092-8674(04)00133-3. [DOI] [PubMed] [Google Scholar]
- 45.vom Baur E, Zechel C, Heery D, Heine MJ, Garnier JM, Vivat V, Le Douarin B, Gronemeyer H, Chambon P, Losson R. Differential ligand-dependent interactions between the AF-2 activating domain of nuclear receptors and the putative transcriptional intermediary factors mSUG1 and TIF1. EMBO J. 1996;15:110–124. [PMC free article] [PubMed] [Google Scholar]
- 46.Ferry C, Gianni M, Lalevee S, Bruck N, Plassat JL, Raska I, Jr, Garattini E, Rochette-Egly C. SUG-1 plays proteolytic and non-proteolytic roles in the control of retinoic acid target genes via its interaction with SRC-3. J. Biol. Chem. 2009;284:8127–8135. doi: 10.1074/jbc.M808815200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhat KP, Greer SF. Proteolytic and non-proteolytic roles of ubiquitin and the ubiquitin proteasome system in transcriptional regulation. Biochim. Biophys. Acta. 2011;1809:150–155. doi: 10.1016/j.bbagrm.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 48.Geng F, Tansey WP. Similar temporal and spatial recruitment of native 19S and 20S proteasome subunits to transcriptionally active chromatin. Proc. Natl. Acad. Sci. U.S.A. 2012;109:6060–6065. doi: 10.1073/pnas.1200854109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.