Abstract
Exposures to environmental toxicants and toxins cause epigenetic changes that likely play a role in the development of diseases associated with exposure. The mechanism behind these exposure-induced epigenetic changes is currently unknown. One commonality between most environmental exposures is that they cause DNA damage either directly or through causing an increase in reactive oxygen species, which can damage DNA. Like transcription, DNA damage repair must occur in the context of chromatin requiring both histone modifications and ATP-dependent chromatin remodeling. These chromatin changes aid in DNA damage accessibility and signaling. Several proteins and complexes involved in epigenetic silencing during both development and cancer have been found to be localized to sites of DNA damage. The chromatin-based response to DNA damage is considered a transient event, with chromatin being restored to normal as DNA damage repair is completed. However, in individuals chronically exposed to environmental toxicants or with chronic inflammatory disease, repeated DNA damage-induced chromatin rearrangement may ultimately lead to permanent epigenetic alterations. Understanding the mechanism behind exposure-induced epigenetic changes will allow us to develop strategies to prevent or reverse these changes. This review focuses on epigenetic changes and DNA damage induced by environmental exposures, the chromatin changes that occur around sites of DNA damage, and how these transient chromatin changes may lead to heritable epigenetic alterations at sites of chronic exposure.
Keywords: toxicants, reactive oxygen species, histone modifications, DNA methylation
Background
Epigenetics is the study of heritable gene expression changes that are not due to a change in the DNA sequence. These changes can involve changes in DNA methylation, histone modifications, and/or microRNA expression (reviewed in [Baylin and Jones 2011; Klaunig et al. 2011]). DNA methylation involves the addition of a methyl group to the cytosine of a CpG dinucleotide pair. CpGs in intergenic and repetitive regions of the genome tend to be highly DNA methylated. Environmental exposures and disease development have been associated with loss of DNA methylation from these regions, which may cause reactivation of transposable elements and re-expression of adjacent genes [Kulis and Esteller 2010; Hou et al. 2012]. In contrast to repetitive DNA, about 60% of gene promoters contain dense regions of CpGs, called CpG islands. DNA methylation of a CpG island causes transcriptional repression of the associated gene and aberrant DNA methylation of these islands has been highly studied in carcinogenesis [Baylin and Jones 2011].
In addition to DNA being modified to alter gene expression, histones can also be modified. DNA in the nucleus of cells is wrapped around nucleosomes that are made up of histones. Each histone has a histone tail that protrudes out of the nucleosome and can be post-translationally modified at various residues in many different ways. These modifications are called histone marks. Histone marks are either active marks allowing for open chromatin and expression of the associated genes or repressive marks resulting in compact chromatin and reduced expression of the associated genes [Baylin and Jones 2011]. Environmental exposures have been associated with global and locus-specific changes in histone mark levels [Cortessis et al. 2012; Hou et al. 2012].
The third epigenetic change involves microRNAs (miRNAs), which are a family of small non-coding RNAs that modulate expression of other genes in a sequence specific manner by binding to 3′-untranslated regions of target mRNAs (reviewed in [Di Leva et al. 2013]). miRNAs are short single-stranded RNAs of approximately 20–24 nucleotides in length that are transcribed from DNA but not translated into proteins. miRNAs negatively regulate expression of target genes at the post-transcriptional level by binding to 3′-untranslated regions of target mRNAs. The expression of miRNAs has been shown to change with exposures to toxicants and at sites of inflammation [Cortessis et al. 2012; Hou et al. 2012; Tili et al. 2013].
Exposure to environmental toxicants and toxins plays a role in the etiology of a diverse array of diseases and can lead to epigenetic changes [Cortessis et al. 2012; Hou et al. 2012]. Since by definition epigenetic changes heritably alter gene expression levels, it is thought that such exposure-induced epigenetic alterations may play a direct role in disease formation and/or progression. However, the mechanisms by which exposures cause epigenetic changes are unclear. Exposures to toxicants and toxins can cause DNA damage either directly by the parent compound, via the production of reactive intermediates, or through the generation of reactive oxygen species (ROS) [Poirier 2004; Klaunig et al. 2011]. DNA damage plays a known role in disease etiology through inducing mutation that leads to gain of function or loss of function for key genes. The epigenetic changes discussed above also occur in a transient manner during chromatin-based processes including DNA damage repair, transcription, and DNA replication. However, these transient changes are not considered true epigenetic changes unless they are stable and heritable, being passed on to daughter cells during cell division. For the purpose of this review there is no delineation between epigenetic changes that are permanent versus those that are heritable through a finite number of cell divisions, but ultimately are reversible as both types of epigenetics changes could conceivably play roles in disease development. In this review, I will first discuss the types of DNA damage and epigenetic changes associated with specific environmental exposures and the chromatin changes that are associated with DNA damage repair. I will then discuss the possibility that at least some epigenetic changes are initiated by normally transient chromatin-based changes associated with DNA damage repair.
Environmental exposure-induced DNA damage
A common toxicity inflicted upon cells following environmental exposures is DNA damage. DNA damage can occur directly by exposure to ionizing or ultraviolet (UV) radiation, which ionize components of DNA and are absorbed directly by DNA bases, respectively. Chemicals or their reactive intermediates can react with DNA to modify DNA bases, form DNA adducts, and/or form DNA interstrand crosslinks [Poirier 2004]. Exposures can also act through a mechanism by which elevated levels of ROS cause oxidative DNA damage. ROS may be generated from the agent itself or from many sources in the cell including xenobiotic metabolism, damaged mitochondria, or an immunologic response to the exposure [Klaunig et al. 2011]. ROS are a set of reactive compounds including superoxide, the hydroxyl radical, and hydrogen peroxide (H2O2). Increases in cellular ROS, whether through physiological modification or through chemical exposure contribute to the process of carcinogenesis [Klaunig et al. 2011]. Furthermore, environmental exposures can indirectly cause genotoxcitiy through interference with the mitotic spindle, causing an unbalanced nucleotide pool, interference with DNA damage repair processes, or allowing for bypass of cell cycle checkpoints [Kirsch-Volders et al. 2003]. The above mechanisms of DNA damage can result in single or double strand DNA breaks (DSBs) and base modifications [Klaunig et al. 2011]. Below I briefly discuss both the epigenetic changes associated with various environmental exposures, namely heavy metals, particulate matter (PM), polycyclic aromatic hydrocarbons (PAH), other chemicals, and radiation, and the DNA damage caused by these exposures.
Epigenetic changes resulting from exposure to heavy metals include global changes in DNA methylation after exposure to arsenic, cadmium, and lead; arsenic, chromium, and nickel-associated hyper and hypomethylation of key gene promoters; arsenic, chromium, and nickel-associated changes in histone marks; and arsenic and cadium-induced changes in miRNA expression [Cheng et al. 2012; Chervona and Costa 2012; Chervona et al. 2012]. These heavy metals, as well as beryllium, chromium, cobalt, and iron, are demonstrated carcinogens [Bal and Kasprzak 2002]. There are many molecular mechanisms suggested for the carcinogenicity of these metals. However, several of them, including cobalt, chromium, copper, iron, and nickel, induce reactive oxygen species and/or high valence metal ions that can act as catalytic centers for redox reactions that directly oxidize DNA [Bal and Kasprzak 2002]. Moreover, millions of people consume water contaminated by inorganic arsenic, an environmental carcinogen, and the ROS produced by arsenic exposure is a key factor in arsenic carcinogenicity [De Vizcaya-Ruiz et al. 2009; Chervona and Costa 2012]. Further supporting the importance of ROS in arsenic exposure, several studies have demonstrated that the use of antioxidants significantly reduces the toxic and genotoxic effects of arsenic [Mandal et al. 2007; Mittal and Flora 2007].
Another kind of environmental exposure widely implicated in human cardiovascular and lung disease/carcinogenesis is PM, which can be found in air pollution. Exposure to PM with a diameter of < 10 μm has been associated with decreased DNA methylation levels of repetitive elements and global histone mark and miRNA expression changes in blood cells [Hou et al. 2012; Motta et al. 2013]. In vivo and in vitro studies suggest that PM exposure causes ROS production [Li et al. 2002; Li et al. 2003; Xia et al. 2004; Hou et al. 2010]. At least in part, this ROS generation is by mechanisms that involve the mitochondria-regulated death pathway and the generation of iron-derived free radicals [Upadhyay et al. 2003; Xia et al. 2004].
PAHs are combustion-related environmental pollutants that are found at high levels in charcoal broiled foods, cigarette smoke, and diesel exhaust and are known DNA damaging agents. The PAH benzo[a]pyrene (BaP) is one of the most studied PAHs with a well understood mechanism of action. BaP can be metabolized to form anti-BP-7,8-dihydrodiol-9,10-epoxide (BPDE), a carcinogen that covalently binds to DNA causing DNA damage that can lead to mutations. Other PAHs operate by similar mechanisms to cause DNA damage and mutation [Luch and Baird 2010]. Additionally, PAH-metabolites can be further converted to PAH o-quinones and diesel exhaust particles contain quinoid PAHs, some of which are highly redox active compounds that participate in redox cycling resulting in the generation of ROS [Park et al. 2006; Chung et al. 2008; Zhang et al. 2012]. In children and workers exposed to PAHs, level of exposure positively correlates with levels of DNA damage in the form of DNA strand breaks and levels of oxidative DNA damage [Ruchirawat et al. 2007; Cavallo et al. 2009; Wang et al. 2010; Kuang et al. 2013]. PAH exposure has also been associated with changes in both global and locus specific DNA methylation levels, genome-wide lysine 9 histone H3 (K9H3) acetylation profiles, and expression of miRNAs [Sadikovic et al. 2008; Pavanello et al. 2009; Halappanavar et al. 2011; Herbstman et al. 2012; Lizarraga et al. 2012; Alegria-Torres et al. 2013].
Another environmental toxicant known to cause epigenetic changes is bisphenol A (BPA), a well-known endocrine-disrupting chemical that has received particular attention because of its widespread use in food containers and its potential for reproductive effects. In a mouse model, in utero exposure to BPA causes DNA methylation changes associated with increased obesity and insulin resistance [Dolinoy et al. 2007]. BPA exposure also induces changes in miRNA expression and global levels of trimethyl K27H3 [Avissar-Whiting et al. 2010; Doherty et al. 2010; Tilghman et al. 2012]. Results for the genotoxicity of BPA are still inconclusive, but recent studies suggest that BPA exposure may induce DNA damage accumulation in cells via oxidative stress [Tiwari et al. 2012; Wu et al. 2013].
Pesticides constitute a wide variety of chemicals including herbicides, insecticides, and fungicides and exposure to them may cause acute and delayed health effects. Exposure to many of these chemicals has been linked to epigenetic changes including DNA methylation, histone modification, and miRNA expression changes (reviewed in [Collotta et al. 2013]). In both pesticide factory and agriculture workers, exposure to pesticides has been demonstrated to result in genomic DNA damage, which was evaluated either by using the comet assay or the frequency of micronuclei, although the exact mechanism by which pesticide exposure induces DNA damage is unclear [Bhalli et al. 2006; Sailaja et al. 2006; Benedetti et al. 2013; Khayat et al. 2013].
Benzene is widely used in chemical industry and is a component of cigarette smoke associated with hematological disorders and cancer formation. Exposure to benzene is associated with changes in DNA methylation, including global hypomethylation of DNA, hypomethylation of repetitive elements, and hypermethylation of specific gene promoters [Hou et al. 2012; Seow et al. 2012; Xing et al. 2013]. Benzene itself is not mutagenic, but benzene is metabolized into toxic metabolites that cause an increase in intracellular production of ROS resulting in oxidative DNA damage [Hiraku and Kawanishi 1996].
Exposure to ionizing radiation (IR) occurs through diagnostic and therapeutic medical devices as well as background radiation, cosmic rays, radioactive waste, radon decay, nuclear tests, and nuclear accidents. Exposure to IR can cause global DNA hypomethylation, DNA methylation changes at specific loci, and changes in expression of the DNA methyltransferases (DNMTs) both acutely and up to 14 days post-irradiation [Kuhmann et al. 2011; Chaudhry and Omaruddin 2012; Antwih et al. 2013] (reviewed in [Kim et al. 2013]). IR exposure also induces phosphorylation of H2AX, a decrease in K20H4 trimethylation, and changes in miRNA expression [Pogribny et al. 2005; Aypar et al. 2011; Metheetrairut and Slack 2013]. Interestingly, both single high dose and chronic low dose whole-body irradiation can significantly alter global and site-specific DNA methylation in mouse tissue [Pogribny et al. 2004; Koturbash et al. 2005; Bernal et al. 2013]. IR is a known carcinogen that damages cellular components, including DNA in the form of single and double strand breaks, base damage, and DNA crosslinks and also increases cellular ROS levels [Klaunig et al. 2011].
UV radiation exposure through sunlight and artificial sources is a primary risk factor for melanoma. Chronic exposure of the skin of mice to UVB radiation or of human keratinocytes in culture to UVA radiation causes chromatin and DNA methylation changes at specific gene promoters [Nandakumar et al. 2011; Chen et al. 2012]. miRNA expression changes have also been demonstrated in mouse skin 24 hours after a single UVB exposure [Xu et al. 2012b]. Exposure of skin to UV radiation induces oxidative stress, inflammation, and DNA damage in the form of DNA photoproducts such as cyclobutane pyrimidine dimers [Xu et al. 2012b].
In addition to directly causing increases in ROS and DNA damage, exposure to environmental toxicants can induce acute and/or chronic inflammation. For example, PM from tobacco smoke and air pollution is associated with chronic obstructive pulmonary disease [Punturieri et al. 2009]. Such inflammation results in increased exposure to ROS. Cells undergo oxidative stress when ROS levels exceed the cell’s ability to balance the oxidative environment. In response to tissue injury caused by exposures, inflammatory cells are activated and directed to the site of injury (reviewed in [Medzhitov 2008]). Chemokines are released that attract specific leukocyte populations, including neutrophils, eosinophils, and macrophages. These cells produce a variety of ROS and release them at sites of inflammation. Inflammatory cells may also use cytokines to stimulate ROS production in neighboring epithelial cells.
Chronic inflammation has been associated with cancer-specific epigenetic changes. Infection with Helicobacter pylori, a carcinogenic bacterium, causes changes in histone modifications, global DNA hypomethylation, and DNA hypermethylation of specific genes in the human gastric mucosa, including those that are DNA methylated and silenced in human gastric cancer cells [Maekita et al. 2006; Ding et al. 2010; Ushijima and Hattori 2012]. H. pylori-mediated inflammation appears to play a key role in these epigenetic changes [Hur et al. 2011]. In the intestine, some Polycomb Repressive Complex 2 (PRC2) target genes, which are a set of genes that have a tendency to DNA methylated and silenced in cancer, are subject to aberrant DNA methylation and decreases in K27H3 trimethylation following chronic inflammation, both in inflamed tissue and tumors [Hahn et al. 2008]. Furthermore, murine infection with a toxigenic bacterium, enterotoxigenic Bacteriodies fragilis, causes acute epigenetic changes in colonic epithelial cells harvested from the areas of highest inflammation. Changes in polycomb group (PcG) protein recruitment have been demonstrated at gene promoters known to become epigenetically silenced in colon cancer [O’Hagan et al. 2011]. miRNA expression is also altered in inflammatory conditions both in immune and epithelial cells and likely plays roles both in controlling chronic inflammation and in promoting cancer development (reviewed in [Chiba et al. 2012]).
Most studies linking environmental exposures and/or inflammation to epigenetic changes have focused on chronic or repetitive exposures. While there is evidence that acute exposure to IR can cause persistent epigenetic changes, chronic low-dose exposures are more potent inducers of lasting epigenetic changes [Kovalchuk et al. 2004; Koturbash et al. 2005]. Furthermore, in mouse a model of colitis, it has been demonstrated that chronic inflammation, as opposed to treatment with a genotoxic agent only, is necessary for the induction of DNA methylation changes also suggesting that most exposure-related epigenetic changes occur through chronic exposure [Katsurano et al. 2012].
Transient chromatin-based changes at sites of DNA damage
DNA in the nucleus is packaged into chromatin, which is a barrier to DNA damage recognition and repair. Compacted chromatin is refractory to full activation of the DNA damage response and disruption of DNA integrity alone can cause a change in chromatin structure that activates DNA damage signaling [Berkovich et al. 2007; Murga et al. 2007; Soria et al. 2012]. In order for repair machinery to access sites of DNA damage chromatin must undergo remodeling. In part this remodeling is done by ATP-dependent nucleosome complexes removing and/or sliding nucleosomes out of the way (reviewed in [Smeenk and van Attikum 2013]). The heterochromatin-associated proteins, HP1 and Kap-1, also participate in the early response to DSBs in euchromatin, although their exact role in DNA repair is still unclear [Luijsterburg et al. 2009; Baldeyron et al. 2011; Soria et al. 2012]. In addition to the movement of nucleosomes, histones are also post-translationally modified at sites of DNA damage to both aid in DNA damage signaling and the recruitment and retention of DNA repair factors [Lukas et al. 2011b]. One of the most well studied histone modifications that occurs in the chromatin surrounding sites of DNA damage is phosphorylation of the histone variant H2AX. phospho-H2AX promotes the retention and accumulation of DNA repair proteins as well as histone modifiers and chromatin remodelers [Lukas et al. 2011b]. Beyond aiding in DNA damage accessibility and signaling, it has been hypothesized that chromatin modifiers are recruited to the sites of DNA damage to locally inhibit transcription to prevent it from interfering with the repair process and/or DNA damage signaling [Price and D’Andrea 2013].
Finally, after DNA damage repair is complete, the chromatin is restored by both repositioning nucleosomes and returning the epigenetic code back to its original state. All of these processes require chromatin-modifying enzymes found at sites of DNA damage. Interestingly, many of these proteins and complexes, particularly those discussed below, are also involved in stable epigenetic changes during development and in cancer bringing into question whether the transient recruitment of these proteins during DNA damage repair can be tied to stable disease-associated epigenetic alterations.
NuRD
The nucleosomes deacetylase and remodeling (NuRD) complex is a co-repressor complex that is recruited to sites of DNA damage induced by UV light and IR [Chou et al. 2010; Larsen et al. 2010; Polo et al. 2010; Smeenk et al. 2010]. A core component of NuRD is the chromodomain-helicase-DNA-binding 4 (CHD4), a member of the SNF2 superfamily of ATPases, which use ATP hydrolysis to move nucleosomes along DNA. NuRD also contains the histone deacetylases, HDAC1 and HDAC2, making the complex capable of forming repressive chromatin. NuRD has been found to be rapidly recruited to sites of DSBs by its interaction with the ubiquitin liagse RNF (RING finger protein) 8. The chromatin-remodeling activity of CHD4 is proposed to decondense the chromatin at the DNA damage site, which stimulates the formation of ubiquitin conjugates by RNF8/RNF168 [Larsen et al. 2010; Luijsterburg et al. 2012; Smeenk et al. 2013]. This ubiquitylation is needed for amplification of the DNA-damage repair signal and recruitment of DNA damage repair proteins including BRCA1 (breast cancer early-onset 1). Loss of BRCA1 recruitment leads to impaired DSB repair and activation of the G2/M checkpoint and increased IR sensitivity. CHD4 also binds BRIT1, a key regulator of homologous recombination, and loss of CHD4 impairs the recruitment of BRIT1 to DNA damage again leading to a reduction in BRCA1 recruitment [Pan et al. 2012]. CHD4 has also been found to be recruited to sites of laser microirradiation in a poly(ADP ribose) polymerase (PARP1) dependent manner [Chou et al. 2010; Polo et al. 2010]. PARP enzymes are activated by DSBs, single-strand breaks, and DNA nicks and, by modifying target proteins with poly(ADP-ribose) chains at DNA damage sites, play a role in the recruitment of DNA repair factors to lesions. CHD4 has been found to directly bind PARylated proteins, including PARP1 itself, at DNA damage sites [Polo et al. 2010].
Polycomb
PcG proteins were originally discovered as chromatin modifiers that control silencing of the homeotic (Hox) genes during embryonic development in fruit flies (reviewed in [Aloia et al. 2013]). In mammals, PcG members are also important in development by regulating expression of key genes in developmental pathways. The PcG proteins make up two main Polycomb Repressive Complexes, PRC1 and PRC2. To initiate silencing, PRC2, which contains the histone methyltransferase EZH2, trimethylates histone H3 at lysine 27. PRC1, which contains E3 ubiquitin ligases and BMI1 (B lymphoma Mo-MLV insertion region 1 homolog), is then recruited to sites of this mark and catalyzes the monoubiquitiylation of K119H2A. While in fruit flies there are DNA motifs for the recruitment for PRC2, there are not similar elements in mammals and it is currently unknown what recruits PRC2 to specific promoters during development. PRC1 and PRC2 also play abnormal roles in cancer. Members of both complexes, especially BMI1 and EZH2, are often over expressed in cancers and are involved with aberrant repression of key tumor suppressor genes [Bracken and Helin 2009].
Recent work has implicated both PRC1 and PRC2 in DNA damage repair mostly at DSBs. Members of PRC1, including BMI1 and Ring1B, have been localized to sites of DSBs [Pan et al. 1074; Chou et al. 2010; Ismail et al. 2010; Chagraoui et al. 2011; Ginjala et al. 2011; Nacerddine et al. 2012]. For an extensive review of this work, see Vissers HA et al [Vissers et al. 2012]. Interestingly, ubiquitylation of H2A on K119 also occurs at sites of DSB in a similar manner as its role in chromatin repression during development [Ginjala et al. 2011]. Recruitment of these proteins and ubK119H2A appear to be early events in the DNA damage repair process as they occur minutes after IR with timing similar to other DNA damage response factors and their presence is sustained for several hours [Chou et al. 2010; Ismail et al. 2010; Ginjala et al. 2011]. Although there are many ubiquitylation events around the sites of DSBs, which cause some discrepancies between the findings of different groups, it is likely that ubK119H2A is dependent on BMI1 as loss of Ring1B and BMI1 interfere with basal and induced monoubiquitylation at sites of DNA damage [Ginjala et al. 2011]. RNF8-RNF168 are responsible for other ubiquitylation events around DSBs, specifically in the generation of K63-linked ubiquitin chains [Shanbhag et al. 2010]. The interaction of these different ubiquitylation events is unclear including which lysine residues of H2A and H2AX are modified by RNF8-RNF168 and how these ubiquitin chains affect monoubiquitylation of K119H2A [Vissers et al. 2012]. Recent work using a small molecule inhibitor of the E3 ubiquitin ligase activity of PRC1, but not RNF8 or RNF168, has suggested that K119H2A monoubiquitylation is required for subsequent RNF8-RNF168-mediated polyubiquitylation at sites of DNA damage [Ismail et al. 2013].
PRC2 members have also been found localized to sites of DNA damage, including after UV light, H2O2, and endonuclease-induced DSBs [Hong et al. 2008; O’Hagan et al. 2008; Chou et al. 2010; O’Hagan et al. 2011; Seiler et al. 2011; Wu et al. 2011; Campbell et al. 2013]. Localization of these proteins in many cases has correlated with an increase in the 3meK27H3 mark, suggesting recruitment of a functional PRC2 complex. However, the exact function of PRC2 at sites of DSBs remains unclear. As mentioned, during development PRC2 recruitment and activity precedes and is necessary for PRC1 recruitment. However, this may not be a requirement at sites of DNA damage. The recruitment of PRC1 is a very early event in the DNA damage response and some groups have demonstrated that PRC1 recruitment is not affected by knockdown of PRC2 constituents [Ismail et al. 2010]. If PRC1 does not require the 3meK27H3 mark for its recruitment to DSBs, then the question remains as to how and why this recruitment is different from what occurs during development.
DNA methyltransferases
DNMTs methylate CpG dinucleotides of DNA and play a normal role in the silencing of developmental genes and repetitive elements. However, in cancer DNMTs are involved in aberrant silencing of 100s to 1000s of gene promoters [Baylin and Jones 2011]. While all of these silencing events are likely not drivers of carcinogenesis, silencing of the promoters of tumor suppressor genes such as MLH1 and p16 play a direct role. DNMT1 is called the maintenance methyltransferase because it prefers methylating the second strand of hemimethylated DNA and is located at DNA synthesis forks for this purpose.
While there is some discrepancy as to which or if all DNMTs are involved in DNA damage repair, it is evident that at least some are. Several groups have demonstrated the presence of DNMT1 at both endonuclease and laser-induced breaks [Mortusewicz et al. 2005; O’Hagan et al. 2008; Ha et al. 2010]. DNMT1 is rapidly and transiently recruited to DSBs through its interaction with proliferating cell nuclear antigen (PCNA) and checkpoint kinase 1 (CHK1) and loss of DNMT1 appears to modulate the rate of repair [Ha et al. 2010]. Because this recruitment appears to be early in the repair process, it is thought that DNMT1 may be functioning in sensing or mobilizing the DNA damage repair response to sites of DNA damage [Jin and Robertson 2013]. DNMT3B, a de novo DNMT, has also both been found at endonuclease-induced DSBs [O’Hagan et al. 2008], although it was not found to be localized to laser-induced breaks [Mortusewicz et al. 2005]. Groups have shown that DNA damage associated with recruitment of DNMTs results in DNA methylation changes that occur immediately after DNA damage or persist after DNA damage repair has been completed [Cuozzo et al. 2007; O’Hagan et al. 2008; O’Hagan et al. 2011]. One possible role of localization of DNMTs at sites of DNA damage may be that DNMT1 is functioning as a scaffolding protein for the recruitment of other epigenetic proteins to sites of DNA damage rather than playing a specific role in methylating DNA. The findings of several groups support this hypothesis. First, the recruitment of DNMT1 to DSBs does not require its catalytic activity, secondly, the DNMTs interact as part of large complex induced by H2O2 treatment, and finally, DNMT1 can play a scaffolding role in silencing genes in cancer cells [Ha et al. 2010; O’Hagan et al. 2011; Clements et al. 2012]. Further work needs to be completed to clarify the role of DNMTs during DNA damage repair.
Histone acetylation and deacetylation
Histone acetylation is a mark that is added and removed by histone acetyl transferases (HATs) and HDACs, respectively. Histone acetylation, unlike some other histone marks, alters the structure and charge of lysine residues and therefore regulates chromatin structure and function by modifying histone-DNA and histone-protein interactions, making histone acetylation directly involved in the regulation of transcription [Gong and Miller 2013]. The HAT, 60 kDa Tat-interactive protein (Tip60) plays key roles in DSB repair by participating in chromatin remodeling at DSBs as part of the NuA4 complex and by activating the ATM kinase [Sun et al. 2010]. The NuA4-Tip60 complex binds to chromatin around sites of DSBs and, p400, a remodeling ATPase that is part of the NuA4-Tip60 complex, exchanges H2A for the histone variant H2AZ in nucleosomes at the DSB [Xu et al. 2010; Xu et al. 2012a]. Tip60 then acetylates histones H2AX and H4, which aids in the turnover of H2AX and modifies the chromatin architecture at break sites [Ikura et al. 2000; Downs et al. 2004; Kusch et al. 2004; Murr et al. 2006; Robert et al. 2006; Ikura et al. 2007; Jha et al. 2008]. Interestingly, recently it has been demonstrated that by modulating histone acetylation TIP60 promotes homologous recombination, while inhibiting nonhomologous end joining [Hsiao and Mizzen 2013; Tang et al. 2013]. The HATs, GCN5, p300, and CBP have also been implicated in DNA damage repair of DSBs and UV lesions [Tini et al. 2002; Kim et al. 2009; Vempati et al. 2010; Guo et al. 2011; Ogiwara et al. 2011].
The most studied HDAC in terms of DNA damage repair is sirtuin1 (SIRT1). In yeast, the SIRT1 homolog relocalizes from telomeres to DSBs and is needed for efficient non-homologous end joining [Tsukamoto et al. 1997; Martin et al. 1999; McAinsh et al. 1999; Mills et al. 1999]. SIRT1 also localizes to sites of DNA damage in mammalian cells and has been implicated in deacetylation of histones at sites of DNA damage as well as deacetylation of DNA repair proteins and other proteins involved in the DNA damage response [Jeong et al. 2007; Yuan and Seto 2007; Li et al. 2008; O’Hagan et al. 2008; Fan and Luo 2010]. Such deacetylation of histones would result in more compacted chromatin, whereas deacetylation of proteins has been shown to change their activity level. SIRT1 also interacts with Tip60 to negatively regulate Tip60-mediated acetylation of H2AX [Yamagata and Kitabayashi 2009]. HDAC1 and HDAC2 have also been implicated in DNA damage repair [Miller et al. 2010; Polo et al. 2010]. HDAC1 and HDAC2 have been shown to associate with many other epigenetic proteins as part of the NuRD complex and with the DNMTs [Zhang et al. 1999; Fuks et al. 2000]. Therefore, it is possible that they are being recruited to sites of DNA damage as part of these complexes. Further studies are required to define how the deacetylation of histones affects the other functions of these complexes.
Both increases and decreases of histone acetylation have been associated with DNA damage repair, likely because DNA damage repair is dynamic process that requires both the opening and closing of chromatin and there are several different histone tail lysines that can be acetylated [Tamburini and Tyler 2005]. It has been difficult to determine the exact relationship between the acetylation/deacetylation dynamics at sites of DNA damage because most studies use chromatin immunoprecipitation (ChIP) assays to look at changes in histone marks. These assays are only capable of resolving the locations of the modified histones based on the size of the sonicated DNA fragments (typically 200 to 1000 bp). Acetylation and deacetylation events may be very specific in terms of the distance to the break site, with acetylation happening near the break and deacetylation occurring a few nucleosomes away, or they may differ in timing, with acetylation occurring right after DNA damage followed by deacetylation when DNA damage repair is completed. Further work needs to be done to understand the exact relationship between histone acetylation and deacetylation at sites of DNA damage.
Non-coding RNAs
The newest type of epigenetic modulators to be studied are non-coding RNAs, with miRNAs being the most studied of this group. miRNAs post-transcriptionally regulate the expression of target genes, have also been found to be involved in the DNA damage response, and, as discussed above, their expression can be altered by environmental exposures (reviewed in [Chowdhury et al. 2012]). The set of miRNAs that change in expression in response to DNA damage depends both on the cell type and on the damaging agent used and can play a role in DNA damage signaling as well as DNA damage repair. Overall their role in the DNA damage response is still poorly understood, but some examples are provided. DNA repair and signaling proteins including ATM, H2AX, BRCA1, and p53 can be directly inhibited by miRNAs [Sharma and Misteli 2013]. Irradiation results in the downregulation of miR-335, a miRNA that targets CtIP, a protein involved in end resection during homologous recombination, and therefore influences the selection of homologous repair for the DNA repair of DSBs [Martin et al. 2013]. miRNA-7 is a negative regulator of SET8 (SET domain containing 8), a H4K20 monomethyltransferase that plays a role in DNA repair [Yu et al. 2013]. Loss of miRNA-7 promotes spontaneous DNA damage and sensitizes cells to induced DNA damage. Recent studies suggest that another class of small RNAs, called DSB-induced small RNAs (diRNAs), are induced around DSB sites and promote DNA damage repair [Francia et al. 2012; Wei et al. 2012]. The exact function of these diRNAs is unclear, however, one theory is that they may help generate either an open or closed chromatin structure at the break site [Ohsawa et al. 2013]. Because there is no evidence that changes in miRNA expression alter the chromatin directly at the site of DNA damage and because little is known at this point about diRNAs, they will have to be a topic for a future review.
Combination of chromatin-based changes at sites of DNA damage
While above the various chromatin modifying proteins and histone mark changes are discussed separately, it is likely that they are acting together or sequentially during the repair of damaged DNA. Several of the proteins are known to interact with each other including the HDACs interacting with NuRD and DNMT1 as discussed [Zhang et al. 1999; Fuks et al. 2000], PcG members interacting with DNMTs [Vire et al. 2006], and SIRT1 interacting with DNMTs and PcG proteins [Kuzmichev et al. 2005; Espada et al. 2007; O’Hagan et al. 2011]. Therefore, these proteins may be recruited to sites of DNA damage as part of larger complexes that are capable of modifying histones and other proteins in several different ways. Multiple histone modifications have been demonstrated to also occur in the chromatin surrounding DNA damage and in some cases multiple modifications are required for the recruitment of proteins involved in the DNA damage repair process. For example, 53BP1, a chromatin-associated factor that promotes DSB repair by non-homologous end joining, needs histones to be modified by both methylation and ubiquitylation for its proper binding to nucleosomes [Fradet-Turcotte et al. 2013].
Inhibition of transcription and DNA damage repair
Both transcription and the DNA damage repair process are intimately involved with chromatin changes. Certain DNA lesions and/or DNA repair processes, including high levels of oxidative stress, result in inhibition of transcription on a global level [Berthiaume et al. 2006]. RNA polymerases (RNAPs) are stalled at bulky DNA lesions reducing levels of transcription and, during transcription coupled DNA damage repair, they become ubiquitylated and undergo proteasome-mediated degradation [Pankotai et al. 2012]. Stalled elongating RNAPII provides a recognition signal for chromatin remodeling and recruitment of proteins involved in DNA damage repair [Fousteri et al. 2006]. Even though ionizing radiation does not reduce transcription on a global level, it has been demonstrated that the induction of DSBs specifically causes a reduction in activity of both RNAPI and RNAPII and, in the case of RNAPII a reduction of transcription of the associated gene [Kruhlak et al. 2007; Shanbhag et al. 2010; Pankotai et al. 2012]. As DNA damage repair is completed both global and local transcription return to normal.
The epigenetic proteins and complexes discussed here are all involved in both transcriptional silencing during development and aberrant silencing in cancer [Baylin and Jones 2011]. As discussed above, these proteins are also recruited to sites of DNA damage, which suggests that they may play a role in preventing transcription at sites of DNA damage, likely to prevent transcription from interfering with the DNA damage repair process [Chou et al. 2010; Miller et al. 2010; Lukas et al. 2011b; O’Hagan et al. 2011]. In BMI1 knockout mouse embryonic fibroblasts elongating RNAPII is maintained at DNA damage foci, where as RNAPII is normally lost from these sites, suggesting that BMI1 recruitment is indeed playing a role in reducing transcription at sites of DNA damage [Chagraoui et al. 2011]. Unlike epigenetic silencing during development and disease formation, this DNA damage repair-associated inhibition of transcription is likely a transient event that resumes once DNA damage repair is completed.
In order to prove these hypotheses, models must be developed to examine both transcription and DNA damage repair at known locations in the genome over a given time period. One such system could be one which utilizes the endonuclease assay developed by Shanbhag and Greenberg [Shanbhag and Greenberg 2013].
DNA damage and heritable epigenetic alterations
The involvement of epigenetic silencing proteins in DNA damage repair is most likely a normal part of the DNA damage repair process that causes transient changes in chromatin compaction and transcription, which are restored to normal when DNA damage repair is completed [Soria et al. 2012]. However, if these epigenetic players are not removed correctly during the DNA damage repair process or if in the setting of chronic inflammation or chronic toxicant exposure, there is repetitive DNA damage, these epigenetic changes may result in aberrant permanent silencing of the gene. Aberrant DNA hypermethylation of CpG islands, including promoter CpG islands commonly found DNA hypermethylated in cancer, is observed at sites of chronic inflammation, including chronic biliary tract inflammation, Barrett’s esophagus, H. pylori infection, and inflammatory bowel disease [Niwa and Ushijima 2010]. Global DNA hypomethylation and a decrease in DNA methylation at specific repetitive DNA sequences, similar to what is seen in cancers, have also been observed in chronically inflamed tissue. Oxidative stress has been shown to induce the formation of an epigenetic silencing complex that includes the DNMTs, SIRT1 and PRC2 members [O’Hagan et al. 2011]. This complex is enriched at GC-rich areas of the genome, including CpG islands that become DNA hypermethylated and silenced in cancer. After oxidative damage, these regions gain repressive histone modifications and have reduced transcription of the associated genes, and in the case of low basal expression genes, gain DNA methylation. Similar findings were demonstrated in inflamed tissue using a mouse model of colitis [O’Hagan et al. 2011]. This work led to the hypothesis that in settings of chronic oxidative stress, such changes may persist and become permanent epigenetic changes. Interestingly, it has been hypothesized that transcription protects gene promoters from epigenetic silencing, suggesting that even a transient inhibition of transcription during DNA damage repair may make promoters more susceptible to stable silencing [Thomson et al. 2010]. However, a direct link between inflammation and stable DNA methylation changes has not yet been established.
Recent whole-cell based studies suggest that DSBs themselves or chromatin modifications that are associated with DSBs can at least persist through one cellular generation. DSBs that occur during mitosis persist through mitosis as indicated by foci of DNA repair proteins and are passed on symmetrically to daughter cells as 53BP1 (a DNA damage response protein) nuclear bodies [Giunta et al. 2010; Lukas et al. 2011a]. However, whether successful DNA damage repair of such DSBs occurs and/or the cells survive needs to be demonstrated. Furthermore, exposure of mice to protracted low-dose radiation causes persistent 53BP1 foci in epidermal stem cells that do not colocalize with other DNA repair proteins [Schanz et al. 2012]. The authors suggest that these 53BP1 foci may be permanent chromatin rearrangements caused by the DNA damage repair of radiation-induced DSBs. While chromatin changes need to be more closely examined in this system, this work does suggest that chromatin surrounding a DSB may not always be restored back to normal after DNA damage repair is completed and the altered chromatin can persist for an undetermined amount of time.
While there is still not direct evidence that DNA damage can cause heritable epigenetic changes, several experimental approaches have been utilized to determine if there is a link between DNA damage and stable epigenetic changes. Cuzzo et al. demonstrated that an endonuclease-induced DSB that is repaired by homologous recombination results in sustained DNA methylation and transcriptional silencing of the associated gene [Cuozzo et al. 2007] and the activity of DNMT1 in this process seems to be regulated by GADD45alpha [Lee et al. 2012]. Exposure of cells to the ROS, H2O2, results in the redistribution of SIRT1 away from its normal binding sites in the genome to sites of DNA damage, which results in a derepression of SIRT1-regulated genes that mimics transcriptional changes seen in aging cells [Oberdoerffer et al. 2008]. In another approach, an endonuclease was used to induce a DSB in the CpG island of an exogenously introduced copy of the E-cadherin promoter, which is a promoter that is frequently DNA hypermethylated in epithelial cancers, including the breast cancer cell line used for these studies, MB-MDA-231. The introduced promoter drives expression of the herpes simplex virus gene, thymidine kinase (HSVTK), allowing for negative selection for expression using the drug ganciclovir. Induction of the DSB causes epigenetic silencing of the promoter without causing DNA mutation in a small percentage of cells [O’Hagan et al. 2008]. The epigenetic silencing coincides with a gain in promoter DNA methylation that increases with passage. However, DNA methylation does not appear necessary for the DNA-damage induced epigenetic silencing as knockdown of SIRT1 during the acute phase of DNA damage induction does not affect silencing of the promoter, but does result in a loss of transient enrichment of DNMT3B at the break site and a loss of DNA methylation in the silenced promoters. These studies suggest that at least in experimental systems DNA damage can induce epigenetic changes. Further studies are required to extend these findings to in vivo settings involving environmental exposures and disease developmental to establish firmly if there are direct links between exposure, DNA damage, epigenetic changes, and disease formation.
Conclusions
It is evident that environmental exposures result in stable epigenetic changes. However, the mechanisms of how exposures result in epigenetic alterations are unclear. It is likely that exposures induce epigenetic alterations by more than one mechanism, possibly varying with the specific exposure or tissue being studied. One hypothesis for such a mechanism is driven by known chromatin changes that occur at sites of DNA damage. Many environmental exposures cause an increase in DNA damage in cells either directly or through an increase in ROS. At sites of DNA damage there are many chromatin changes including modifications of histones and repositioning and/or removal of nucleosomes. Additionally, several proteins known to be involved in aberrant epigenetic silencing in cancer, including the NuRD complex, PcG proteins, and the DNMTs, have been localized to sites of DNA damage (Figure 1). These proteins may play a role in inhibiting transcription during the DNA damage repair process. Likely these chromatin changes are transient such that the chromatin is restored back to normal after DNA damage repair is complete. However, at sites of chronic exposure and/or inflammation repeated DNA damage and chromatin rearrangement may occasionally result in aberrant restoration of the chromatin and cause stable epigenetic alterations (Figure 1). If such epigenetic changes result in a survival or proliferative advantage for the cell, they may persist in the cell population. Such a hypothesis is similar in thought to the occasional DNA mutation that escapes proper DNA damage repair and results in activation or repression of an oncogene or tumor suppressor gene, respectively. However, the potential to reverse epigenetic changes after silencing occurs has raised great interest in epigenetic-based therapeutics in the cancer field [Helin and Dhanak 2013]. At this point the published work linking chromatin changes at sites of DNA damage to heritable epigenetic changes is mostly circumstantial and more work needs to be done to demonstrate a direct causative link between the two. Additionally, further work needs to be done to determine other mechanisms by which environmental exposure can induce epigenetic changes. Understanding the mechanisms behind exposure-induced epigenetic changes will hopefully result in the development of biomarkers for exposure in order to better protect the health of our population and possible drug treatments for at-risk populations either undergoing chronic exposure to environmental pollutants or chronic inflammation.
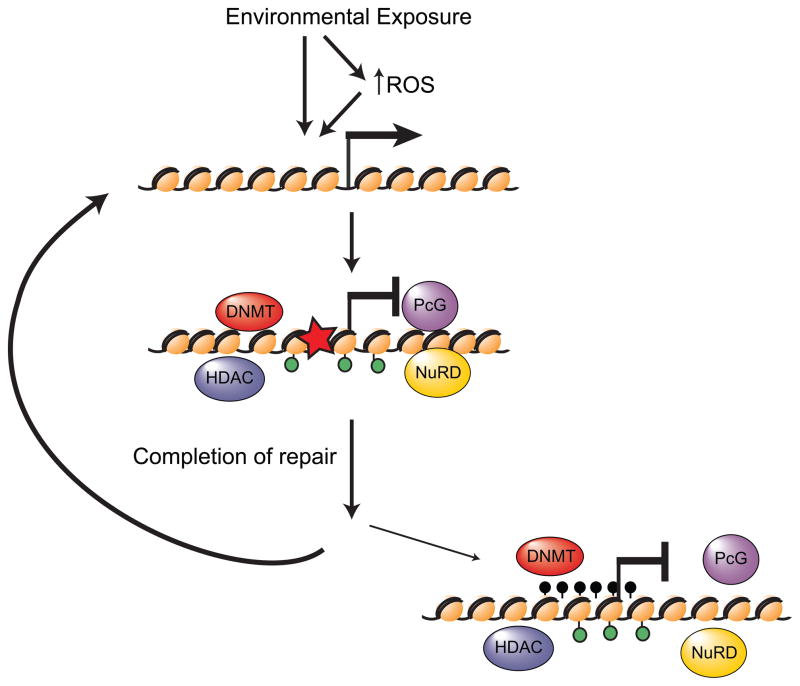
Figure 1. Model for how environmental exposure-induced DNA damage may lead to epigenetic silencing.
At sites of environmental exposure, either the exposure itself or increased levels of ROS caused by exposure cause DNA damage (red star). At the sites of damage histone modifications occur (green circles) and the chromatin is remodeled. Additionally, epigenetic silencing proteins are recruited to the site of damage, possibly to prevent transcription from interfering with the repair process. It is unclear at this time if all of these proteins are being recruited to all sites of damage and whether or not they are interacting with each other. In most instances, after completion of DNA repair the chromatin is restored back to normal, the epigenetic silencing proteins are no longer localized to the area, and transcription resumes. In cases of chronic exposure, DNA damage and repair cycles can happen repeatedly and, in rare instances, the chromatin structure may not be returned to normal and binding of the epigenetic silencing proteins may persist, resulting in stable epigenetic silencing.
Acknowledgments
I thank C. DeStefano Shields and F. Carvalho for critically reading this manuscript. This work was supported by funding from the National Institute of Environmental Health of the National Institutes of Health [R01ES023183].
Footnotes
Statement of Author Contributions.
HOH prepared the manuscript.
References
- Alegria-Torres JA, Barretta F, Batres-Esquivel LE, Carrizales-Yanez L, Perez-Maldonado IN, Baccarelli A, Bertazzi PA. Epigenetic markers of exposure to polycyclic aromatic hydrocarbons in Mexican brickmakers: a pilot study. Chemosphere. 2013;91:475–480. doi: 10.1016/j.chemosphere.2012.11.077. [DOI] [PubMed] [Google Scholar]
- Aloia L, Di Stefano B, Di Croce L. Polycomb complexes in stem cells and embryonic development. Development. 2013;140:2525–2534. doi: 10.1242/dev.091553. [DOI] [PubMed] [Google Scholar]
- Antwih DA, Gabbara KM, Lancaster WD, Ruden DM, Zielske SP. Radiation-induced epigenetic DNA methylation modification of radiation-response pathways. Epigenetics. 2013:8. doi: 10.4161/epi.25498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avissar-Whiting M, Veiga KR, Uhl KM, Maccani MA, Gagne LA, Moen EL, Marsit CJ. Bisphenol A exposure leads to specific microRNA alterations in placental cells. Reprod Toxicol. 2010;29:401–406. doi: 10.1016/j.reprotox.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aypar U, Morgan WF, Baulch JE. Radiation-induced epigenetic alterations after low and high LET irradiations. Mutat Res. 2011;707:24–33. doi: 10.1016/j.mrfmmm.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Bal W, Kasprzak KS. Induction of oxidative DNA damage by carcinogenic metals. Toxicology Letters. 2002;127:55–62. doi: 10.1016/s0378-4274(01)00483-0. [DOI] [PubMed] [Google Scholar]
- Baldeyron C, Soria G, Roche D, Cook AJ, Almouzni G. HP1alpha recruitment to DNA damage by p150CAF-1 promotes homologous recombination repair. J Cell Biol. 2011;193:81–95. doi: 10.1083/jcb.201101030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti D, Nunes E, Sarmento M, Porto C, Santos CEId, Dias JF, da Silva J. Genetic damage in soybean workers exposed to pesticides: Evaluation with the comet and buccal micronucleus cytome assays. Mutation Research/Genetic Toxicology and Environmental Mutagenesis. 2013;752:28–33. doi: 10.1016/j.mrgentox.2013.01.001. [DOI] [PubMed] [Google Scholar]
- Berkovich E, Monnat RJ, Jr, Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol. 2007;9:683–690. doi: 10.1038/ncb1599. Epub 2007 May 2007. [DOI] [PubMed] [Google Scholar]
- Bernal AJ, Dolinoy DC, Huang D, Skaar DA, Weinhouse C, Jirtle RL. Adaptive radiation-induced epigenetic alterations mitigated by antioxidants. Faseb J. 2013;27:665–671. doi: 10.1096/fj.12-220350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthiaume M, Boufaied N, Moisan A, Gaudreau L. High levels of oxidative stress globally inhibit gene transcription and histone acetylation. DNA Cell Biol. 2006;25:124–134. doi: 10.1089/dna.2006.25.124. [DOI] [PubMed] [Google Scholar]
- Bhalli JA, Khan QM, Nasim A. DNA damage in Pakistani pesticide-manufacturing workers assayed using the Comet assay. Environ Mol Mutagen. 2006;47:587–593. doi: 10.1002/em.20232. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer. 2009;9:773–784. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- Campbell S, Ismail IH, Young LC, Poirier GG, Hendzel MJ. Polycomb repressive complex 2 contributes to DNA double-strand break repair. Cell Cycle. 2013;12:2675–2683. doi: 10.4161/cc.25795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallo D, Ursini CL, Rondinone B, Iavicoli S. Evaluation of a suitable DNA damage biomarker for human biomonitoring of exposed workers. Environ Mol Mutagen. 2009;50:781–790. doi: 10.1002/em.20501. [DOI] [PubMed] [Google Scholar]
- Chagraoui J, Hebert J, Girard S, Sauvageau G. An anticlastogenic function for the Polycomb Group gene Bmi1. Proc Natl Acad Sci U S A. 2011;108:5284–5289. doi: 10.1073/pnas.1014263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry MA, Omaruddin RA. Differential DNA methylation alterations in radiation-sensitive and -resistant cells. DNA Cell Biol. 2012;31:908–916. doi: 10.1089/dna.2011.1509. [DOI] [PubMed] [Google Scholar]
- Chen IP, Henning S, Faust A, Boukamp P, Volkmer B, Greinert R. UVA-induced epigenetic regulation of P16(INK4a) in human epidermal keratinocytes and skin tumor derived cells. Photochem Photobiol Sci. 2012;11:180–190. doi: 10.1039/c1pp05197k. [DOI] [PubMed] [Google Scholar]
- Cheng TF, Choudhuri S, Muldoon-Jacobs K. Epigenetic targets of some toxicologically relevant metals: a review of the literature. J Appl Toxicol. 2012;32:643–653. doi: 10.1002/jat.2717. [DOI] [PubMed] [Google Scholar]
- Chervona Y, Costa M. The control of histone methylation and gene expression by oxidative stress, hypoxia, and metals. Free Radic Biol Med. 2012;53:1041–1047. doi: 10.1016/j.freeradbiomed.2012.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chervona Y, Hall MN, Arita A, Wu F, Sun H, Tseng HC, Ali E, Uddin MN, Liu X, Zoroddu MA, et al. Associations between arsenic exposure and global posttranslational histone modifications among adults in Bangladesh. Cancer Epidemiol Biomarkers Prev. 2012;21:2252–2260. doi: 10.1158/1055-9965.EPI-12-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba T, Marusawa H, Ushijima T. Inflammation-Associated Cancer Development in Digestive Organs: Mechanisms and Roles for Genetic and Epigenetic Modulation. Gastroenterology. 2012;143:550–563. doi: 10.1053/j.gastro.2012.07.009. [DOI] [PubMed] [Google Scholar]
- Chou DM, Adamson B, Dephoure NE, Tan X, Nottke AC, Hurov KE, Gygi SP, Colaiacovo MP, Elledge SJ. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci U S A. 2010;107:18475–18480. doi: 10.1073/pnas.1012946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury D, Choi YE, Brault ME. Charity begins at home: non-coding RNA functions in DNA repair. Nat Rev Mol Cell Biol. 2012;14:181–189. doi: 10.1038/nrm3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung SW, Toriba A, Chung HY, Yu BP, Kameda T, Tang N, Kizu R, Hayakawa K. Activation of 5-lipoxygenase and NF-kappa B in the action of acenaphthenequinone by modulation of oxidative stress. Toxicol Sci. 2008;101:152–158. doi: 10.1093/toxsci/kfm252. [DOI] [PubMed] [Google Scholar]
- Clements EG, Mohammad HP, Leadem BR, Easwaran H, Cai Y, Van Neste L, Baylin SB. DNMT1 modulates gene expression without its catalytic activity partially through its interactions with histone-modifying enzymes. Nucleic Acids Res. 2012;40:4334–4346. doi: 10.1093/nar/gks031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collotta M, Bertazzi PA, Bollati V. Epigenetics and pesticides. Toxicology. 2013;307:35–41. doi: 10.1016/j.tox.2013.01.017. [DOI] [PubMed] [Google Scholar]
- Cortessis VK, Thomas DC, Levine AJ, Breton CV, Mack TM, Siegmund KD, Haile RW, Laird PW. Environmental epigenetics: prospects for studying epigenetic mediation of exposure-response relationships. Hum Genet. 2012;131:1565–1589. doi: 10.1007/s00439-012-1189-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuozzo C, Porcellini A, Angrisano T, Morano A, Lee B, Pardo AD, Messina S, Iuliano R, Fusco A, Santillo MR, et al. DNA Damage, Homology-Directed Repair, and DNA Methylation. PLoS Genet. 2007;3:e110. doi: 10.1371/journal.pgen.0030110. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- De Vizcaya-Ruiz A, Barbier O, Ruiz-Ramos R, Cebrian ME. Biomarkers of oxidative stress and damage in human populations exposed to arsenic. Mutat Res. 2009;674:85–92. doi: 10.1016/j.mrgentox.2008.09.020. [DOI] [PubMed] [Google Scholar]
- Di Leva G, Garofalo M, Croce CM. MicroRNAs in Cancer. Annu Rev Pathol. 2013 doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding SZ, Fischer W, Kaparakis-Liaskos M, Liechti G, Merrell DS, Grant PA, Ferrero RL, Crowe SE, Haas R, Hatakeyama M, et al. Helicobacter pylori-induced histone modification, associated gene expression in gastric epithelial cells, and its implication in pathogenesis. PLoS One. 2010;5:e9875. doi: 10.1371/journal.pone.0009875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty LF, Bromer JG, Zhou Y, Aldad TS, Taylor HS. In utero exposure to diethylstilbestrol (DES) or bisphenol-A (BPA) increases EZH2 expression in the mammary gland: an epigenetic mechanism linking endocrine disruptors to breast cancer. Horm Cancer. 2010;1:146–155. doi: 10.1007/s12672-010-0015-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci U S A. 2007;104:13056–13061. doi: 10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs JA, Allard S, Jobin-Robitaille O, Javaheri A, Auger A, Bouchard N, Kron SJ, Jackson SP, Cote J. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol Cell. 2004;16:979–990. doi: 10.1016/j.molcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Espada J, Ballestar E, Santoro R, Fraga MF, Villar-Garea A, Nemeth A, Lopez-Serra L, Ropero S, Aranda A, Orozco H, et al. Epigenetic disruption of ribosomal RNA genes and nucleolar architecture in DNA methyltransferase 1 (Dnmt1) deficient cells. Nucleic Acids Res. 2007;35:2191–2198. doi: 10.1093/nar/gkm118. Epub 2007 Mar 2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Luo J. SIRT1 regulates UV-induced DNA repair through deacetylating XPA. Mol Cell. 2010;39:247–258. doi: 10.1016/j.molcel.2010.07.006. [DOI] [PubMed] [Google Scholar]
- Fousteri M, Vermeulen W, van Zeeland AA, Mullenders LH. Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol Cell. 2006;23:471–482. doi: 10.1016/j.molcel.2006.06.029. [DOI] [PubMed] [Google Scholar]
- Fradet-Turcotte A, Canny MD, Escribano-Diaz C, Orthwein A, Leung CC, Huang H, Landry MC, Kitevski-LeBlanc J, Noordermeer SM, Sicheri F, et al. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature. 2013;499:50–54. doi: 10.1038/nature12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francia S, Michelini F, Saxena A, Tang D, de Hoon M, Anelli V, Mione M, Carninci P, d’Adda di Fagagna F. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature. 2012;488:231–235. doi: 10.1038/nature11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- Ginjala V, Nacerddine K, Kulkarni A, Oza J, Hill SJ, Yao M, Citterio E, van Lohuizen M, Ganesan S. BMI1 is recruited to DNA breaks and contributes to DNA damage-induced H2A ubiquitination and repair. Mol Cell Biol. 2011;31:1972–1982. doi: 10.1128/MCB.00981-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giunta S, Belotserkovskaya R, Jackson SP. DNA damage signaling in response to double-strand breaks during mitosis. The Journal of Cell Biology. 2010;190:197–207. doi: 10.1083/jcb.200911156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong F, Miller KM. Mammalian DNA repair: HATs and HDACs make their mark through histone acetylation. Mutat Res. 2013 doi: 10.1016/j.mrfmmm.2013.07.002. [DOI] [PubMed] [Google Scholar]
- Guo R, Chen J, Mitchell DL, Johnson DG. GCN5 and E2F1 stimulate nucleotide excision repair by promoting H3K9 acetylation at sites of damage. Nucleic Acids Res. 2011;39:1390–1397. doi: 10.1093/nar/gkq983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha K, Lee GE, Palii SS, Brown KD, Takeda Y, Liu K, Bhalla KN, Robertson KD. Rapid and transient recruitment of DNMT1 to DNA double-strand breaks is mediated by its interaction with multiple components of the DNA damage response machinery. Hum Mol Genet. 2010;20:126–140. doi: 10.1093/hmg/ddq451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MA, Hahn T, Lee DH, Esworthy RS, Kim BW, Riggs AD, Chu FF, Pfeifer GP. Methylation of polycomb target genes in intestinal cancer is mediated by inflammation. Cancer Res. 2008;68:10280–10289. doi: 10.1158/0008-5472.CAN-08-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halappanavar S, Wu D, Williams A, Kuo B, Godschalk RW, Van Schooten FJ, Yauk CL. Pulmonary gene and microRNA expression changes in mice exposed to benzo(a)pyrene by oral gavage. Toxicology. 2011;285:133–141. doi: 10.1016/j.tox.2011.04.011. [DOI] [PubMed] [Google Scholar]
- Helin K, Dhanak D. Chromatin proteins and modifications as drug targets. Nature. 2013;502:480–488. doi: 10.1038/nature12751. [DOI] [PubMed] [Google Scholar]
- Herbstman JB, Tang D, Zhu D, Qu L, Sjodin A, Li Z, Camann D, Perera FP. Prenatal exposure to polycyclic aromatic hydrocarbons, benzo[a]pyrene-DNA adducts, and genomic DNA methylation in cord blood. Environ Health Perspect. 2012;120:733–738. doi: 10.1289/ehp.1104056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraku Y, Kawanishi S. Oxidative DNA damage and apoptosis induced by benzene metabolites. Cancer Res. 1996;56:5172–5178. [PubMed] [Google Scholar]
- Hong Z, Jiang J, Lan L, Nakajima S, Kanno S, Koseki H, Yasui A. A polycomb group protein, PHF1, is involved in the response to DNA double-strand breaks in human cell. Nucleic Acids Res. 2008;36:2939–2947. doi: 10.1093/nar/gkn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L, Zhang X, Wang D, Baccarelli A. Environmental chemical exposures and human epigenetics. Int J Epidemiol. 2012;41:79–105. doi: 10.1093/ije/dyr154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L, Zhu ZZ, Zhang X, Nordio F, Bonzini M, Schwartz J, Hoxha M, Dioni L, Marinelli B, Pegoraro V, et al. Airborne particulate matter and mitochondrial damage: a cross-sectional study. Environ Health. 2010;9:48. doi: 10.1186/1476-069X-9-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao KY, Mizzen CA. Histone H4 deacetylation facilitates 53BP1 DNA damage signaling and double-strand break repair. J Mol Cell Biol. 2013;5:157–165. doi: 10.1093/jmcb/mjs066. [DOI] [PubMed] [Google Scholar]
- Hur K, Niwa T, Toyoda T, Tsukamoto T, Tatematsu M, Yang HK, Ushijima T. Insufficient role of cell proliferation in aberrant DNA methylation induction and involvement of specific types of inflammation. Carcinogenesis. 2011;32:35–41. doi: 10.1093/carcin/bgq219. [DOI] [PubMed] [Google Scholar]
- Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- Ikura T, Tashiro S, Kakino A, Shima H, Jacob N, Amunugama R, Yoder K, Izumi S, Kuraoka I, Tanaka K, et al. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol Cell Biol. 2007;27:7028–7040. doi: 10.1128/MCB.00579-07. Epub 2007 Aug 7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail IH, Andrin C, McDonald D, Hendzel MJ. BMI1-mediated histone ubiquitylation promotes DNA double-strand break repair. J Cell Biol. 2010;191:45–60. doi: 10.1083/jcb.201003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail IH, McDonald D, Strickfaden H, Xu Z, Hendzel MJ. A small molecule inhibitor of Polycomb repressive complex 1 inhibits ubiquitin signaling at DNA double-strand breaks. J Biol Chem. 2013;2013:30. doi: 10.1074/jbc.M113.461699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J, Juhn K, Lee H, Kim SH, Min BH, Lee KM, Cho MH, Park GH, Lee KH. SIRT1 promotes DNA repair activity and deacetylation of Ku70. Exp Mol Med. 2007;39:8–13. doi: 10.1038/emm.2007.2. [DOI] [PubMed] [Google Scholar]
- Jha S, Shibata E, Dutta A. Human Rvb1/Tip49 is required for the histone acetyltransferase activity of Tip60/NuA4 and for the downregulation of phosphorylation on H2AX after DNA damage. Mol Cell Biol. 2008;28:2690–2700. doi: 10.1128/MCB.01983-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin B, Robertson KD. DNA methyltransferases, DNA damage repair, and cancer. Adv Exp Med Biol. 2013;754:3–29. doi: 10.1007/978-1-4419-9967-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsurano M, Niwa T, Yasui Y, Shigematsu Y, Yamashita S, Takeshima H, Lee MS, Kim YJ, Tanaka T, Ushijima T. Early-stage formation of an epigenetic field defect in a mouse colitis model, and non-essential roles of T- and B-cells in DNA methylation induction. Oncogene. 2012;31:342–351. doi: 10.1038/onc.2011.241. [DOI] [PubMed] [Google Scholar]
- Khayat CB, Costa EO, Goncalves MW, da Cruz ECDM, da Cruz AS, de Araujo Melo CO, Bastos RP, da Cruz AD, de Melo ESD. Assessment of DNA damage in Brazilian workers occupationally exposed to pesticides: a study from Central Brazil. Environ Sci Pollut Res Int. 2013;20:7334–7340. doi: 10.1007/s11356-013-1747-1. [DOI] [PubMed] [Google Scholar]
- Kim JG, Park MT, Heo K, Yang KM, Yi JM. Epigenetics meets radiation biology as a new approach in cancer treatment. Int J Mol Sci. 2013;14:15059–15073. doi: 10.3390/ijms140715059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MK, Shin JM, Eun HC, Chung JH. The role of p300 histone acetyltransferase in UV-induced histone modifications and MMP-1 gene transcription. PLoS One. 2009;4:e4864. doi: 10.1371/journal.pone.0004864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch-Volders M, Vanhauwaert A, Eichenlaub-Ritter U, Decordier I. Indirect mechanisms of genotoxicity. Toxicology Letters. 2003;140–141:63–74. doi: 10.1016/s0378-4274(02)00498-8. [DOI] [PubMed] [Google Scholar]
- Klaunig JE, Wang Z, Pu X, Zhou S. Oxidative stress and oxidative damage in chemical carcinogenesis. Toxicology and Applied Pharmacology. 2011;254:86–99. doi: 10.1016/j.taap.2009.11.028. [DOI] [PubMed] [Google Scholar]
- Koturbash I, Pogribny I, Kovalchuk O. Stable loss of global DNA methylation in the radiation-target tissue—A possible mechanism contributing to radiation carcinogenesis? Biochemical and Biophysical Research Communications. 2005;337:526–533. doi: 10.1016/j.bbrc.2005.09.084. [DOI] [PubMed] [Google Scholar]
- Kovalchuk O, Burke P, Besplug J, Slovack M, Filkowski J, Pogribny I. Methylation changes in muscle and liver tissues of male and female mice exposed to acute and chronic low-dose X-ray-irradiation. Mutat Res. 2004;548:75–84. doi: 10.1016/j.mrfmmm.2003.12.016. [DOI] [PubMed] [Google Scholar]
- Kruhlak M, Crouch EE, Orlov M, Montano C, Gorski SA, Nussenzweig A, Misteli T, Phair RD, Casellas R. The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature. 2007;447:730–734. doi: 10.1038/nature05842. [DOI] [PubMed] [Google Scholar]
- Kuang D, Zhang W, Deng Q, Zhang X, Huang K, Guan L, Hu D, Wu T, Guo H. Dose-response relationships of polycyclic aromatic hydrocarbons exposure and oxidative damage to DNA and lipid in coke oven workers. Environ Sci Technol. 2013;47:7446–7456. doi: 10.1021/es401639x. [DOI] [PubMed] [Google Scholar]
- Kuhmann C, Weichenhan D, Rehli M, Plass C, Schmezer P, Popanda O. DNA methylation changes in cells regrowing after fractioned ionizing radiation. Radiotherapy and Oncology. 2011;101:116–121. doi: 10.1016/j.radonc.2011.05.048. [DOI] [PubMed] [Google Scholar]
- Kulis M, Esteller M. DNA methylation and cancer. Adv Genet. 2010;70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2. [DOI] [PubMed] [Google Scholar]
- Kusch T, Florens L, Macdonald WH, Swanson SK, Glaser RL, Yates JR, 3rd, Abmayr SM, Washburn MP, Workman JL. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Margueron R, Vaquero A, Preissner TS, Scher M, Kirmizis A, Ouyang X, Brockdorff N, Abate-Shen C, Farnham P, et al. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proc Natl Acad Sci U S A. 2005;102:1859–1864. doi: 10.1073/pnas.0409875102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen DH, Poinsignon C, Gudjonsson T, Dinant C, Payne MR, Hari FJ, Rendtlew Danielsen JM, Menard P, Sand JC, Stucki M, et al. The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. J Cell Biol. 2010;190:731–740. doi: 10.1083/jcb.200912135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Morano A, Porcellini A, Muller MT. GADD45alpha inhibition of DNMT1 dependent DNA methylation during homology directed DNA repair. Nucleic Acids Res. 2012;40:2481–2493. doi: 10.1093/nar/gkr1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Casta A, Wang R, Lozada E, Fan W, Kane S, Ge Q, Gu W, Orren D, Luo J. Regulation of WRN protein cellular localization and enzymatic activities by SIRT1-mediated deacetylation. J Biol Chem. 2008;283:7590–7598. doi: 10.1074/jbc.M709707200. [DOI] [PubMed] [Google Scholar]
- Li N, Kim S, Wang M, Froines J, Sioutas C, Nel A. Use of a stratified oxidative stress model to study the biological effects of ambient concentrated and diesel exhaust particulate matter. Inhal Toxicol. 2002;14:459–486. doi: 10.1080/089583701753678571. [DOI] [PubMed] [Google Scholar]
- Li N, Sioutas C, Cho A, Schmitz D, Misra C, Sempf J, Wang M, Oberley T, Froines J, Nel A. Ultrafine particulate pollutants induce oxidative stress and mitochondrial damage. Environ Health Perspect. 2003;111:455–460. doi: 10.1289/ehp.6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizarraga D, Gaj S, Brauers KJ, Timmermans L, Kleinjans JC, van Delft JH. Benzo[a]pyrene-induced changes in microRNA-mRNA networks. Chem Res Toxicol. 2012;25:838–849. doi: 10.1021/tx2003799. [DOI] [PubMed] [Google Scholar]
- Luch A, Baird WM. 14.06 - Carcinogenic Polycyclic Aromatic Hydrocarbons. In: Charlene AM, editor. Comprehensive Toxicology. 2. Oxford: Elsevier; 2010. pp. 85–123. [Google Scholar]
- Luijsterburg MS, Acs K, Ackermann L, Wiegant WW, Bekker-Jensen S, Larsen DH, Khanna KK, van Attikum H, Mailand N, Dantuma NP. A new non-catalytic role for ubiquitin ligase RNF8 in unfolding higher-order chromatin structure. Embo J. 2012;31:2511–2527. doi: 10.1038/emboj.2012.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luijsterburg MS, Dinant C, Lans H, Stap J, Wiernasz E, Lagerwerf S, Warmerdam DO, Lindh M, Brink MC, Dobrucki JW, et al. Heterochromatin protein 1 is recruited to various types of DNA damage. J Cell Biol. 2009;185:577–586. doi: 10.1083/jcb.200810035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas C, Savic V, Bekker-Jensen S, Doil C, Neumann B, Solvhoj Pedersen R, Grofte M, Chan KL, Hickson ID, Bartek J, et al. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol. 2011a;13:243–253. doi: 10.1038/ncb2201. [DOI] [PubMed] [Google Scholar]
- Lukas J, Lukas C, Bartek J. More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nat Cell Biol. 2011b;13:1161–1169. doi: 10.1038/ncb2344. [DOI] [PubMed] [Google Scholar]
- Maekita T, Nakazawa K, Mihara M, Nakajima T, Yanaoka K, Iguchi M, Arii K, Kaneda A, Tsukamoto T, Tatematsu M, et al. High levels of aberrant DNA methylation in Helicobacter pylori-infected gastric mucosae and its possible association with gastric cancer risk. Clin Cancer Res. 2006;12:989–995. doi: 10.1158/1078-0432.CCR-05-2096. [DOI] [PubMed] [Google Scholar]
- Mandal AK, Das S, Basu MK, Chakrabarti RN, Das N. Hepatoprotective activity of liposomal flavonoid against arsenite-induced liver fibrosis. J Pharmacol Exp Ther. 2007;320:994–1001. doi: 10.1124/jpet.106.114215. [DOI] [PubMed] [Google Scholar]
- Martin NT, Nakamura K, Davies R, Nahas SA, Brown C, Tunuguntla R, Gatti RA, Hu H. ATM-dependent MiR-335 targets CtIP and modulates the DNA damage response. PLoS Genet. 2013;9:e1003505. doi: 10.1371/journal.pgen.1003505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SG, Laroche T, Suka N, Grunstein M, Gasser SM. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell. 1999;97:621–633. doi: 10.1016/s0092-8674(00)80773-4. [DOI] [PubMed] [Google Scholar]
- McAinsh AD, Scott-Drew S, Murray JAH, Jackson SP. DNA damage triggers disruption of telomeric silencing and Mec1p-dependent relocation of Sir3p. Current Biology. 1999;9:963–966. doi: 10.1016/s0960-9822(99)80424-2. [DOI] [PubMed] [Google Scholar]
- Medzhitov R. Origin and physiological roles of inflammation. Nature. 2008;454:428–435. doi: 10.1038/nature07201. [DOI] [PubMed] [Google Scholar]
- Metheetrairut C, Slack FJ. MicroRNAs in the ionizing radiation response and in radiotherapy. Current Opinion in Genetics & Development. 2013;23:12–19. doi: 10.1016/j.gde.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S, Jackson SP. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol. 2010;17:1144–1151. doi: 10.1038/nsmb.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills KD, Sinclair DA, Guarente L. MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell. 1999;97:609–620. doi: 10.1016/s0092-8674(00)80772-2. [DOI] [PubMed] [Google Scholar]
- Mittal M, Flora SJ. Vitamin E supplementation protects oxidative stress during arsenic and fluoride antagonism in male mice. Drug Chem Toxicol. 2007;30:263–281. doi: 10.1080/01480540701380075. [DOI] [PubMed] [Google Scholar]
- Mortusewicz O, Schermelleh L, Walter J, Cardoso MC, Leonhardt H. Recruitment of DNA methyltransferase I to DNA repair sites. Proc Natl Acad Sci U S A. 2005;102:8905–8909. doi: 10.1073/pnas.0501034102. Epub 2005 Jun 8913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta V, Angelici L, Nordio F, Bollati V, Fossati S, Frascati F, Tinaglia V, Bertazzi PA, Battaglia C, Baccarelli AA. Integrative Analysis of miRNA and inflammatory gene expression after acute particulate matter exposure. Toxicol Sci. 2013;132:307–316. doi: 10.1093/toxsci/kft013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murga M, Jaco I, Fan Y, Soria R, Martinez-Pastor B, Cuadrado M, Yang SM, Blasco MA, Skoultchi AI, Fernandez-Capetillo O. Global chromatin compaction limits the strength of the DNA damage response. J Cell Biol. 2007;178:1101–1108. doi: 10.1083/jcb.200704140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, Herceg Z. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol. 2006;8:91–99. doi: 10.1038/ncb1343. Epub 2005 Dec 2011. [DOI] [PubMed] [Google Scholar]
- Nacerddine K, Beaudry JB, Ginjala V, Westerman B, Mattiroli F, Song JY, van der Poel H, Ponz OB, Pritchard C, Cornelissen-Steijger P, et al. Akt-mediated phosphorylation of Bmi1 modulates its oncogenic potential, E3 ligase activity, and DNA damage repair activity in mouse prostate cancer. J Clin Invest. 2012;122:1920–1932. doi: 10.1172/JCI57477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar V, Vaid M, Tollefsbol TO, Katiyar SK. Aberrant DNA hypermethylation patterns lead to transcriptional silencing of tumor suppressor genes in UVB-exposed skin and UVB-induced skin tumors of mice. Carcinogenesis. 2011;32:597–604. doi: 10.1093/carcin/bgq282. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Niwa T, Ushijima T. Induction of epigenetic alterations by chronic inflammation and its significance on carcinogenesis. Adv Genet. 2010;71:41–56. doi: 10.1016/B978-0-12-380864-6.00002-X. [DOI] [PubMed] [Google Scholar]
- O’Hagan HM, Mohammad HP, Baylin SB. Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genet. 2008;4:e1000155. doi: 10.1371/journal.pgen.1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hagan HM, Wang W, Sen S, Destefano Shields C, Lee SS, Zhang YW, Clements EG, Cai Y, Van Neste L, Easwaran H, et al. Oxidative Damage Targets Complexes Containing DNA Methyltransferases, SIRT1, and Polycomb Members to Promoter CpG Islands. Cancer Cell. 2011;20:606–619. doi: 10.1016/j.ccr.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, Hartlerode A, Stegmuller J, Hafner A, Loerch P, et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiwara H, Ui A, Otsuka A, Satoh H, Yokomi I, Nakajima S, Yasui A, Yokota J, Kohno T. Histone acetylation by CBP and p300 at double-strand break sites facilitates SWI/SNF chromatin remodeling and the recruitment of non-homologous end joining factors. Oncogene. 2011;30:2135–2146. doi: 10.1038/onc.2010.592. [DOI] [PubMed] [Google Scholar]
- Ohsawa R, Seol JH, Tyler JK. At the intersection of non-coding transcription, DNA repair, chromatin structure, and cellular senescence. Front Genet. 2013;4:136. doi: 10.3389/fgene.2013.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan MR, Hsieh HJ, Dai H, Hung WC, Li K, Peng G, Lin SY. Chromodomain helicase DNA-binding protein 4 (CHD4) regulates homologous recombination DNA repair, and its deficiency sensitizes cells to poly(ADP-ribose) polymerase (PARP) inhibitor treatment. J Biol Chem. 2012;287:6764–6772. doi: 10.1074/jbc.M111.287037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan MR, Peng G, Hung WC, Lin SY. Monoubiquitination of H2AX protein regulates DNA damage response signaling. J Biol Chem. 1074;286:28599–28607. doi: 10.1074/jbc.M111.256297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankotai T, Bonhomme C, Chen D, Soutoglou E. DNAPKcs-dependent arrest of RNA polymerase II transcription in the presence of DNA breaks. Nat Struct Mol Biol. 2012;19:276–282. doi: 10.1038/nsmb.2224. [DOI] [PubMed] [Google Scholar]
- Park JH, Troxel AB, Harvey RG, Penning TM. Polycyclic aromatic hydrocarbon (PAH) o-quinones produced by the aldo-keto-reductases (AKRs) generate abasic sites, oxidized pyrimidines, and 8-oxo-dGuo via reactive oxygen species. Chem Res Toxicol. 2006;19:719–728. doi: 10.1021/tx0600245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavanello S, Bollati V, Pesatori AC, Kapka L, Bolognesi C, Bertazzi PA, Baccarelli A. Global and gene-specific promoter methylation changes are related to anti-B[a]PDE-DNA adduct levels and influence micronuclei levels in polycyclic aromatic hydrocarbon-exposed individuals. Int J Cancer. 2009;125:1692–1697. doi: 10.1002/ijc.24492. [DOI] [PubMed] [Google Scholar]
- Pogribny I, Koturbash I, Tryndyak V, Hudson D, Stevenson SML, Sedelnikova O, Bonner W, Kovalchuk O. Fractionated Low-Dose Radiation Exposure Leads to Accumulation of DNA Damage and Profound Alterations in DNA and Histone Methylation in the Murine Thymus. Molecular Cancer Research. 2005;3:553–561. doi: 10.1158/1541-7786.MCR-05-0074. [DOI] [PubMed] [Google Scholar]
- Pogribny I, Raiche J, Slovack M, Kovalchuk O. Dose-dependence, sex- and tissue-specificity, and persistence of radiation-induced genomic DNA methylation changes. Biochem Biophys Res Commun. 2004;320:1253–1261. doi: 10.1016/j.bbrc.2004.06.081. [DOI] [PubMed] [Google Scholar]
- Poirier MC. Chemical-induced DNA damage and human cancer risk. Nat Rev Cancer. 2004;4:630–637. doi: 10.1038/nrc1410. [DOI] [PubMed] [Google Scholar]
- Polo SE, Kaidi A, Baskcomb L, Galanty Y, Jackson SP. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. Embo J. 2010;29:3130–3139. doi: 10.1038/emboj.2010.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price BD, D’Andrea AD. Chromatin remodeling at DNA double-strand breaks. Cell. 2013;152:1344–1354. doi: 10.1016/j.cell.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punturieri A, Szabo E, Croxton TL, Shapiro SD, Dubinett SM. Lung cancer and chronic obstructive pulmonary disease: needs and opportunities for integrated research. J Natl Cancer Inst. 2009;101:554–559. doi: 10.1093/jnci/djp023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert F, Hardy S, Nagy Z, Baldeyron C, Murr R, Dery U, Masson JY, Papadopoulo D, Herceg Z, Tora L. The transcriptional histone acetyltransferase cofactor TRRAP associates with the MRN repair complex and plays a role in DNA double-strand break repair. Mol Cell Biol. 2006;26:402–412. doi: 10.1128/MCB.26.2.402-412.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchirawat M, Settachan D, Navasumrit P, Tuntawiroon J, Autrup H. Assessment of potential cancer risk in children exposed to urban air pollution in Bangkok, Thailand. Toxicology Letters. 2007;168:200–209. doi: 10.1016/j.toxlet.2006.09.013. [DOI] [PubMed] [Google Scholar]
- Sadikovic B, Andrews J, Carter D, Robinson J, Rodenhiser DI. Genome-wide H3K9 histone acetylation profiles are altered in benzopyrene-treated MCF7 breast cancer cells. J Biol Chem. 2008;283:4051–4060. doi: 10.1074/jbc.M707506200. [DOI] [PubMed] [Google Scholar]
- Sailaja N, Chandrasekhar M, Rekhadevi PV, Mahboob M, Rahman MF, Vuyyuri SB, Danadevi K, Hussain SA, Grover P. Genotoxic evaluation of workers employed in pesticide production. Mutat Res. 2006;609:74–80. doi: 10.1016/j.mrgentox.2006.06.022. [DOI] [PubMed] [Google Scholar]
- Schanz S, Schuler N, Lorat Y, Fan L, Kaestner L, Wennemuth G, Rübe C, Rübe CE. Accumulation of DNA damage in complex normal tissues after protracted low-dose radiation. DNA Repair. 2012;11:823–832. doi: 10.1016/j.dnarep.2012.07.005. [DOI] [PubMed] [Google Scholar]
- Seiler DM, Rouquette J, Schmid VJ, Strickfaden H, Ottmann C, Drexler GA, Mazurek B, Greubel C, Hable V, Dollinger G, et al. Double-strand break-induced transcriptional silencing is associated with loss of tri-methylation at H3K4. Chromosome Res. 2011;19:883–899. doi: 10.1007/s10577-011-9244-1. [DOI] [PubMed] [Google Scholar]
- Seow WJ, Pesatori AC, Dimont E, Farmer PB, Albetti B, Ettinger AS, Bollati V, Bolognesi C, Roggieri P, Panev TI, et al. Urinary benzene biomarkers and DNA methylation in Bulgarian petrochemical workers: study findings and comparison of linear and beta regression models. PLoS One. 2012;7:e50471. doi: 10.1371/journal.pone.0050471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag NM, Greenberg RA. The dynamics of DNA damage repair and transcription. Methods Mol Biol. 2013;1042:227–235. doi: 10.1007/978-1-62703-526-2_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010;141:970–981. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V, Misteli T. Non-coding RNAs in DNA damage and repair. FEBS Lett. 2013;587:1832–1839. doi: 10.1016/j.febslet.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeenk G, van Attikum H. The chromatin response to DNA breaks: leaving a mark on genome integrity. Annu Rev Biochem. 2013;82:55–80. doi: 10.1146/annurev-biochem-061809-174504. [DOI] [PubMed] [Google Scholar]
- Smeenk G, Wiegant WW, Marteijn JA, Luijsterburg MS, Sroczynski N, Costelloe T, Romeijn RJ, Pastink A, Mailand N, Vermeulen W, et al. Poly(ADP-ribosyl)ation links the chromatin remodeler SMARCA5/SNF2H to RNF168-dependent DNA damage signaling. J Cell Sci. 2013;126:889–903. doi: 10.1242/jcs.109413. [DOI] [PubMed] [Google Scholar]
- Smeenk G, Wiegant WW, Vrolijk H, Solari AP, Pastink A, van Attikum H. The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. J Cell Biol. 2010;190:741–749. doi: 10.1083/jcb.201001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria G, Polo SE, Almouzni G. Prime, repair, restore: the active role of chromatin in the DNA damage response. Mol Cell. 2012;46:722–734. doi: 10.1016/j.molcel.2012.06.002. [DOI] [PubMed] [Google Scholar]
- Sun Y, Jiang X, Price BD. Tip60: connecting chromatin to DNA damage signaling. Cell Cycle. 2010;9:930–936. doi: 10.4161/cc.9.5.10931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamburini BA, Tyler JK. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol Cell Biol. 2005;25:4903–4913. doi: 10.1128/MCB.25.12.4903-4913.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Cho NW, Cui G, Manion EM, Shanbhag NM, Botuyan MV, Mer G, Greenberg RA. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat Struct Mol Biol. 2013;20:317–325. doi: 10.1038/nsmb.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JP, Skene PJ, Selfridge J, Clouaire T, Guy J, Webb S, Kerr AR, Deaton A, Andrews R, James KD, et al. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature. 2010;464:1082–1086. doi: 10.1038/nature08924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilghman SL, Bratton MR, Segar HC, Martin EC, Rhodes LV, Li M, McLachlan JA, Wiese TE, Nephew KP, Burow ME. Endocrine disruptor regulation of microRNA expression in breast carcinoma cells. PLoS One. 2012;7:e32754. doi: 10.1371/journal.pone.0032754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tili E, Michaille JJ, Croce CM. MicroRNAs play a central role in molecular dysfunctions linking inflammation with cancer. Immunol Rev. 2013;253:167–184. doi: 10.1111/imr.12050. [DOI] [PubMed] [Google Scholar]
- Tini M, Benecke A, Um SJ, Torchia J, Evans RM, Chambon P. Association of CBP/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Mol Cell. 2002;9:265–277. doi: 10.1016/s1097-2765(02)00453-7. [DOI] [PubMed] [Google Scholar]
- Tiwari D, Kamble J, Chilgunde S, Patil P, Maru G, Kawle D, Bhartiya U, Joseph L, Vanage G. Clastogenic and mutagenic effects of bisphenol A: an endocrine disruptor. Mutat Res. 2012;743:83–90. doi: 10.1016/j.mrgentox.2011.12.023. [DOI] [PubMed] [Google Scholar]
- Tsukamoto Y, Kato J, Ikeda H. Silencing factors participate in DNA repair and recombination in Saccharomyces cerevisiae. Nature. 1997;388:900–903. doi: 10.1038/42288. [DOI] [PubMed] [Google Scholar]
- Upadhyay D, Panduri V, Ghio A, Kamp DW. Particulate matter induces alveolar epithelial cell DNA damage and apoptosis: role of free radicals and the mitochondria. Am J Respir Cell Mol Biol. 2003;29:180–187. doi: 10.1165/rcmb.2002-0269OC. [DOI] [PubMed] [Google Scholar]
- Ushijima T, Hattori N. Molecular pathways: involvement of Helicobacter pylori-triggered inflammation in the formation of an epigenetic field defect, and its usefulness as cancer risk and exposure markers. Clin Cancer Res. 2012;18:923–929. doi: 10.1158/1078-0432.CCR-11-2011. [DOI] [PubMed] [Google Scholar]
- Vempati RK, Jayani RS, Notani D, Sengupta A, Galande S, Haldar D. p300-mediated acetylation of histone H3 lysine 56 functions in DNA damage response in mammals. J Biol Chem. 2010;285:28553–28564. doi: 10.1074/jbc.M110.149393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- Vissers JH, van Lohuizen M, Citterio E. The emerging role of Polycomb repressors in the response to DNA damage. J Cell Sci. 2012;125:3939–3948. doi: 10.1242/jcs.107375. [DOI] [PubMed] [Google Scholar]
- Wang F, He Y, Guo H, Li J, Yang Y, Wu Z, Zheng H, Wu T. Genetic variants of nucleotide excision repair genes are associated with DNA damage in coke oven workers. Cancer Epidemiol Biomarkers Prev. 2010;19:211–218. doi: 10.1158/1055-9965.EPI-09-0270. [DOI] [PubMed] [Google Scholar]
- Wei W, Ba Z, Gao M, Wu Y, Ma Y, Amiard S, White CI, Rendtlew Danielsen JM, Yang YG, Qi Y. A role for small RNAs in DNA double-strand break repair. Cell. 2012;149:101–112. doi: 10.1016/j.cell.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Wu HJ, Liu C, Duan WX, Xu SC, He MD, Chen CH, Wang Y, Zhou Z, Yu ZP, Zhang L, et al. Melatonin ameliorates bisphenol A-induced DNA damage in the germ cells of adult male rats. Mutat Res. 2013;752:57–67. doi: 10.1016/j.mrgentox.2013.01.005. [DOI] [PubMed] [Google Scholar]
- Wu Z, Lee ST, Qiao Y, Li Z, Lee PL, Lee YJ, Jiang X, Tan J, Aau M, Lim CZ, et al. Polycomb protein EZH2 regulates cancer cell fate decision in response to DNA damage. Cell Death Differ. 2011;18:1771–1779. doi: 10.1038/cdd.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia T, Korge P, Weiss JN, Li N, Venkatesen MI, Sioutas C, Nel A. Quinones and aromatic chemical compounds in particulate matter induce mitochondrial dysfunction: implications for ultrafine particle toxicity. Environ Health Perspect. 2004;112:1347–1358. doi: 10.1289/ehp.7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing C, Chen Q, Li G, Zhang L, Zheng M, Zou Z, Hou L, Wang QF, Liu X, Guo X. Microsomal epoxide hydrolase (EPHX1) polymorphisms are associated with aberrant promoter methylation of ERCC3 and hematotoxicity in benzene-exposed workers. Environ Mol Mutagen. 2013;54:397–405. doi: 10.1002/em.21786. [DOI] [PubMed] [Google Scholar]
- Xu Y, Ayrapetov MK, Xu C, Gursoy-Yuzugullu O, Hu Y, Price BD. Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Mol Cell. 2012a;48:723–733. doi: 10.1016/j.molcel.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Sun Y, Jiang X, Ayrapetov MK, Moskwa P, Yang S, Weinstock DM, Price BD. The p400 ATPase regulates nucleosome stability and chromatin ubiquitination during DNA repair. J Cell Biol. 2010;191:31–43. doi: 10.1083/jcb.201001160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Zhou B, Wu D, Yin Z, Luo D. Baicalin modulates microRNA expression in UVB irradiated mouse skin. J Biomed Res. 2012b;26:125–134. doi: 10.1016/S1674-8301(12)60022-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata K, Kitabayashi I. Sirt1 physically interacts with Tip60 and negatively regulates Tip60-mediated acetylation of H2AX. Biochem Biophys Res Commun. 2009;390:1355–1360. doi: 10.1016/j.bbrc.2009.10.156. [DOI] [PubMed] [Google Scholar]
- Yu N, Huangyang P, Yang X, Han X, Yan R, Jia H, Shang Y, Sun L. microRNA-7 suppresses the invasive potential of breast cancer cells and sensitizes cells to DNA damages by targeting histone methyltransferase SET8. J Biol Chem. 2013;288:19633–19642. doi: 10.1074/jbc.M113.475657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Seto E. A functional link between SIRT1 deacetylase and NBS1 in DNA damage response. Cell Cycle. 2007;6:2869–2871. doi: 10.4161/cc.6.23.5026. [DOI] [PubMed] [Google Scholar]
- Zhang L, Jin Y, Huang M, Penning TM. The Role of Human Aldo-Keto Reductases in the Metabolic Activation and Detoxication of Polycyclic Aromatic Hydrocarbons: Interconversion of PAH Catechols and PAH o-Quinones. Front Pharmacol. 2012;3:193. doi: 10.3389/fphar.2012.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]