Abstract
Solid-state NMR spectroscopy is increasingly used in the high-resolution structural studies of membrane-associated proteins and peptides. Most such studies necessitate isotopically labeled (13C, 15N and 2H) proteins/peptides, which is a limiting factor for some of the exciting membrane-bound proteins and aggregating peptides. In this study, we report the use of a proton-based slow magic angle spinning (MAS) solid-state NMR experiment that exploits the unaveraged 1H-1H dipolar couplings from a membrane-bound protein. We have shown that the difference in the buildup rates of cross peak intensities against the mixing time - obtained from 2D 1H-1H radio frequency-driven recoupling (RFDR) and nuclear Overhauser effect spectroscopy (NOESY) experiments on a 16.7-kDa micelle-associated full-length rabbit cytochrome-b5 (cytb5) - can provide insights into protein dynamics and could be useful to measure 1H-1H dipolar couplings. The experimental buildup curves compare well with theoretical simulations and are used to extract relaxation parameters. Our results show that due to fast exchange of amide protons with water in the soluble heme-containing domain of cyb5, coherent 1H-1H dipolar interactions are averaged out for these protons while alpha and side chain protons show residual dipolar couplings that can be obtained from 1H-1H RFDR experiments. The appearance of resonances with distinct chemical shift values in 1H-1H RFDR spectra enabled the identification of residues (mostly from the transmembrane region) of cytb5 that interact with micelles.
Keywords: Membrane protein, cytochrome-b5, RFDR, NOESY
Introduction
Atomic-level characterization of structure and dynamics using solid-state nuclear magnetic resonance (NMR) spectroscopy can provide piercing insights into macromolecular systems, even though it is a challenging task due to low spectral resolution and sensitivity. Significant advances in the development of pulse sequences, ultrahigh field magnets, and a very fast (in the range of 30–40 kHz) to ultra-fast (≥ 60 kHz) magic angle spinning (MAS) techniques with a low radio-frequency decoupling power requirements have opened up new avenues to investigate a variety of biomolecules such as membrane proteins.1–8 Particularly, the recoupling techniques9,10 have facilitated an efficient measurement of internuclear distances from both small (uniformly labeled) and large (selectively labeled) biomolecules. Most of these recoupling-based structural studies utilize the homonuclear dipolar couplings among low-abundant nuclei (13C, 15N) due to the larger spread of their chemical shift frequency. On the other hand, homogeneously broadened 1H resonances resulting from strong 1H-1H dipolar interactions have not been suitable to fully utilize the high sensitivity and abundance of protons. However, the availability of ultrafast MAS probes and the use of heavily deuterated samples have made the complete suppression of homonuclear 1H-1H dipolar couplings feasible; this has paved the way for the development of proton-detection based multi-dimensional experiments in the solid state.5,11–22 Recent studies have also demonstrated the applicability of MAS experiments on soft solids like micelles and fluid lamellar phase bilayers which, unlike rigid solids, exhibit weak 1H-1H dipolar couplings due to their high molecular mobility.23 In this regard, one particular approach is to employ Radio Frequency-Driven Recoupling (RFDR)24,25 and Nuclear Overhauser Effect SpectroscopY (NOESY)26 experiments under MAS to obtain 2D 1H-1H isotropic chemical shift correlation spectra via coherent 1H-1H dipolar coupling and NOE, respectively. This approach has been previously applied to investigate the motional characteristics of resin and membrane-bound peptides.27,28 A recent study on a membrane-bound antimicrobial peptide (MSI-78, also commercially known as pexiganan) also highlights the importance of this approach for the measurement of 1H-1H residual dipolar couplings.29 In this study, we extend this objective on a relatively large micelle-associated unlabeled cytochrome-b5 (cytb5) to gain better insights into its structure and dynamics.
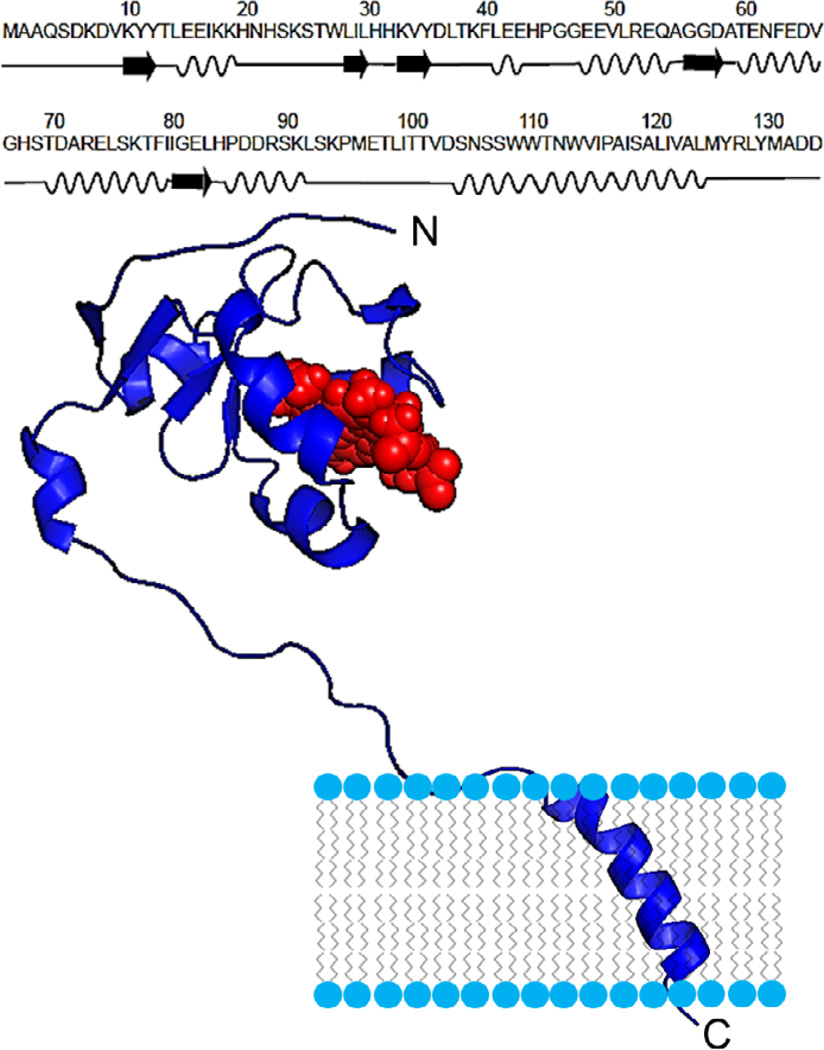
Cytb5 is a 16.7-kDa electron transfer protein consisting of 134 amino acid residues. It is composed of three distinct regions - namely a structured water-soluble paramagnetic ferric low-spin heme-containing domain, a transmembrane helical domain, and an unstructured ~14-residue long linker region.30 The NMR structure along with the amino acid sequence of the full-length membrane-bound rabbit cytb5 (PDB # 2M33) is shown in Figure 1. The structure contains five α-helices (α1, L14-H20; α2, K39-E43; α3, E49-Q54; α4, T60-V66 and α5, T70-F79), five β-strands (β1, K10-Y12; β2, W27-L30; β3, K33-D36 and β4, G56-D58) and one 310 helix (P86-R89). The flexible linker domain (S90-D104) of cytb5 lacks a defined secondary structure and connects the helical transmembrane and heme domains. Cytb5, in combination with its redox partners such as cytochrome-P450 (cytP450) and cytP450 reductase (CPR), is essential for enzyme kinetics to metabolize 70% of the drugs presently available in the market.31 In addition, cytb5 is vital to carryout important biochemical reactions such as the synthesis of testosterone and lipids which are important to preserve the cellular integrity.31 Previous solid-state NMR studies determined the topology of the transmembrane domains of cytb532 and cytP45033, reported the drastic difference between the timescales of motions for the soluble and transmembrane domains,34,35 and also characterized the dynamics of side chains.32 Further, solid-state NMR studies revealed the interaction between the transmembrane domains of cytb5 and cytP450 by using magnetically-aligned bicelles.36 Recently, we reported the first high resolution structure of a highly dynamic membrane-bound full-length cytb5-cytP4502B4 electron transfer complex (~72 kDa) employing experimentally derived NMR constraints from solution and solid-state NMR techniques, and mutagenesis data.30,32 Additionally, amino acid residues in the interacting interface of the cytb5-cytP4502B4 complex and an electron transfer pathway were also identified.30 These findings were well corroborated with our results based on backbone amide-15N chemical shift anisotropy (CSA) tensors derived from free and P450-bound cytb5 using solution NMR and quantum chemical studies.37–39
Figure 1.
Amino acid sequence and NMR structure (PDB # 2M33) of the full-length membrane-bound rabbit cytochrome-b5.30,40
In the present study, we restrict our focus on 2D 1H-1H correlation spectra obtained from the RFDR and NOESY experiments and analyze the intensity difference for the cross-peaks observed from these two MAS experiments. Experimental details and discussion of the NMR structure of micelle-associated cytb5 (PDB # 2M33) are described elsewhere30,40 and will not be discussed further. We believe that the RFDR experiment could be more sensitive compared to the routinely employed solution NMR NOESY experiment as the cross-peak intensities are higher due to additional dipolar recoupling from nearby spins. More importantly, this method can be potentially used to determine the membrane interacting residues, which otherwise may not be easily possible by solution NMR experiments due to their fast spin-spin relaxation.
Experimental
A micelle-associated form of full-length rabbit cytochrome-b5 was expressed in E.Coli and purified as previously reported.30 2.5 mM unlabeled cytb5 was incorporated into 45 mM perdeuterated dodecylphosphocholine (DPC) detergent micelles in 100 mM potassium phosphate (pH 7.4) containing 5% D2O. 40 µL of this sample was packed into a 4 mm zirconium rotor. All NMR measurements were performed on a Varian VNMRS 600 MHz solid-state NMR spectrometer using a 4 mm double-resonance nanoprobe at a spinning speed of 2.7 kHz at 25 °C. Deuterium field-lock was used throughout the experiment and water suppression was achieved using a 1.5 s saturation pulse with a 5 Hz radio frequency power at the beginning of each scan. The proton carrier frequency was set at water proton peak for all the experiments. For assignment of resonances, a 2D 1H-1H TOCSY (TOtal Correlation Spectroscopy)41 experiment was performed under MAS at a spinning speed of 2.7 kHz. The isotropic mixing in the TOCSY pulse sequence was attained using the DIPSI (Decoupling In the Presence of Scalar Interactions)42 spin-lock scheme applied for a duration of 60.4 ms. A proton radio frequency field strength of 65.8 kHz was used for pulses in TOCSY, NOESY and RFDR experiments. 2D 1H-1H NOESY and RFDR spectra were recorded with an acquisition time of 1.6 s, 48 scans, 400 t1 increments, 27028 t2 complex points while the 2D 1H-1H TOCSY spectrum was recorded with an acquisition time of 200 ms, 124 scans, 512 t1 increments, 3380 t2 complex point and a spectral width of 14.1 ppm in both frequency dimensions. All spectra were processed using NMRpipe,43 by applying phase-shifted sine bell multiplication prior to Fourier transformation with zero filling in both t1 and t2 dimensions. Resonance peak assignment and intensity measurements were carried out using Sparky.44
Results and discussion
One-dimensional 1H NMR spectra of micelle-associated cytb5
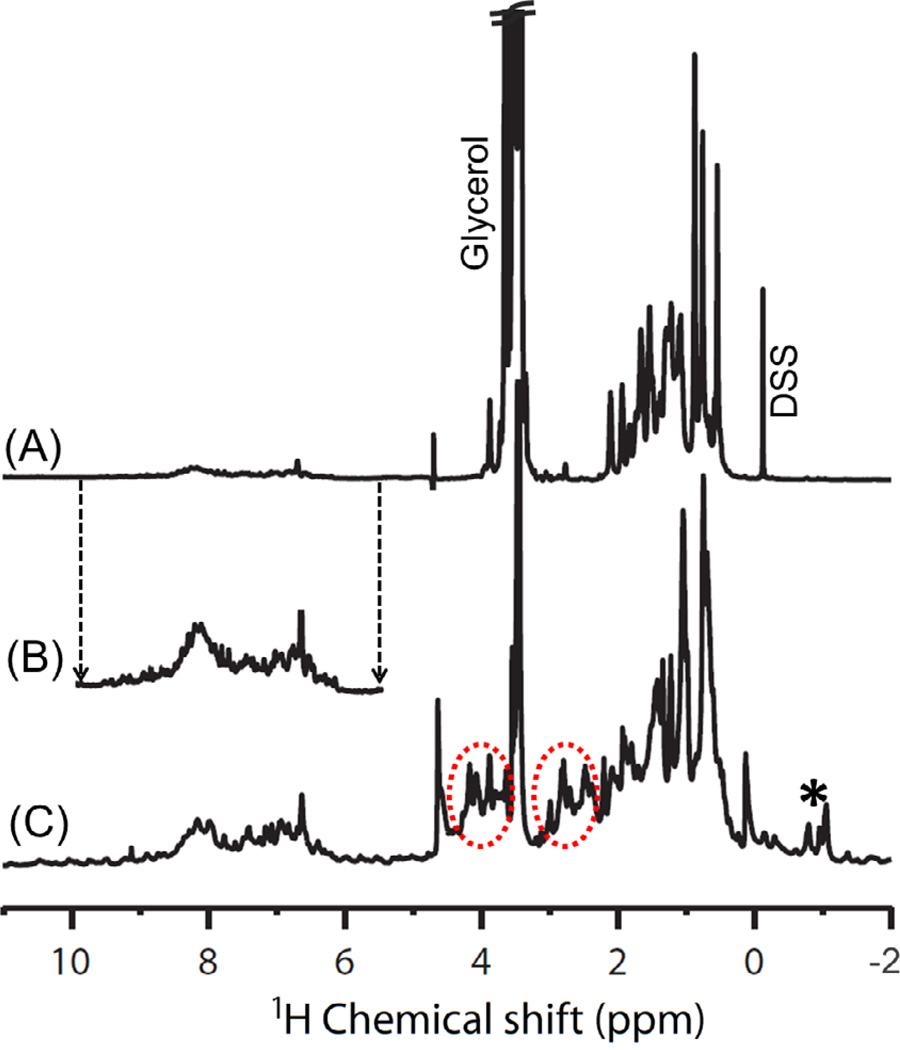
Proteins embedded in a membrane mimetic environment undergo slow tumbling on the NMR timescale and as a result the static NMR spectrum can suffer from significant line broadening caused by the anisotropic interactions, such as the chemical shift anisotropy and the dipolar couplings. In order to enhance the resolution and sensitivity for such macromolecular systems, performing experiments under high magnetic field strengths has always been an obvious choice. However, comparable resolution and sensitivity could also be obtained by performing experiments at a low magnetic field strength under MAS, which narrows the spectral lines by averaging out the orientation dependent anisotropic interactions. To validate this fact, we obtained 1D 1H NMR spectra of full-length cytb5 incorporated in DPC detergent micelles under static and MAS conditions (Figure 2). It is obvious from Figure 2 that the MAS spectrum of cytb5 shows additional resonances in the 2.1 to 4.6 ppm region in comparison to the static spectrum. On the basis of these results from 1D experiments, we carried out 2D 1H-1H RFDR and NOESY experiments under MAS to get additional insights into high-resolution structural characterization of micelle-associated cytb5.
Figure 2.
One-dimensional 1H NMR spectra of micelle-associated cytb5 under (A) static condition at 900 MHz and (C) 2.7 kHz magic angle spinning at 600 MHz. (B) Peaks from the amide and aromatic proton chemical shift regions are shown with a magnified intensity scale. Circled regions show additional resonances observed under MAS, while the asterisk represents the spinning sidebands. DSS (4,4-dimethyl-4-silapentane-1-sulfonic acid) (at 0 ppm) was used as a reference for 1H chemical shift.
RFDR and NOESY experiments under MAS
2D RFDR and NOESY pulse sequences are depicted in Figure S1 of the supporting information. These two pulse sequences differ only in the mixing period: the RFDR pulse sequence is composed of a series of rotor-synchronised 180° pulses in the mixing period, whereas the NOESY pulse sequence is devoid of such pulses. The XY16 phase cycling scheme was used for the 180° pulses in the finite-pulse-RFDR to suppress the effects of pulse imperfections like RF field inhomogeneity and offset.45 The rotor-synchronized 180° pulses in the RFDR experiment reintroduces the homonuclear dipolar couplings under MAS. Consequently, the cross-peak intensities in the 2D 1H-1H correlation RFDR spectrum arise from the combined effect of NOE cross-correlation and recoupled 1H-1H dipolar interactions. Hence, the difference in peak intensities observed in the RFDR and NOESY spectra is due to residual (or unaveraged and recoupled by MAS) dipolar couplings that can be used to determine interatomic distances.
The time evolution of the longitudinal proton magnetization is represented by the rate equation, ; where R and L are the kinetic matrices.46–48 The diagonal elements (Rii) of the R matrix represent the auto-relaxation rate while the off-diagonal elements represent the cross-relaxation rate between spins i and j; where aH=5.6965×1010 Å6s−1, rij is the internuclear distance and Jn represents the spectral density function. In the above equation, Lij is the magnetization transfer between spins calculated using the expression where D and r are the diffusion constant and distance between two spins, respectively. The application of rotor-synchronized 180° pulses in the RFDR experiment allows for the magnetization transfer via the recoupled homonuclear dipolar coupling (~1/r3)49,50 in addition to the NOE (~1/r6), and thus resulting in a faster buildup rate along with the increased intensity for the cross peaks measured as a function of the mixing time in comparison to the NOESY experiment. The RFDR and NOESY buildup rates were calculated using a diagonalization scheme by implementing the above rate equation in an in-house Matlab script. The buildup curves for the RFDR and NOESY experiments were fitted using three parameters; relaxation time T1*, buildup rate R and the peak intensity I0. T1* represents the combined contributions from the spin-lattice relaxation, T1, of protons and the imperfection of 180° pulses in RFDR. The buildup curves plotted against the mixing time have both exponential growth and decay fit parameters.
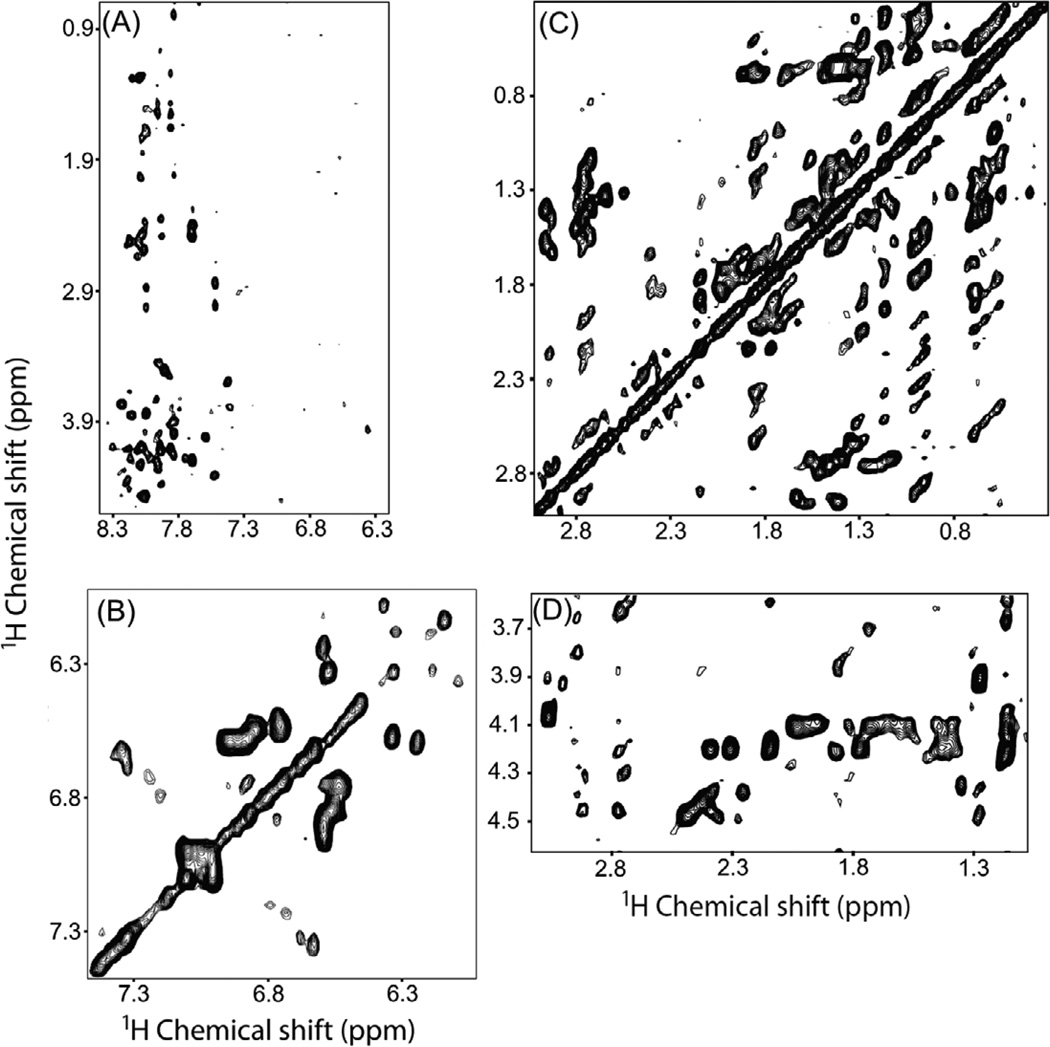
Resonances in 2D 1H-1H RFDR and NOESY spectra for micelle-associated cytb5 recorded under MAS were assigned using a combination of 2D 1H-1H NOESY and 3D HSQC-NOESY spectra with a 100 ms mixing time obtained on a 900 MHz solution NMR spectrometer, and a 2D 1H-1H TOCSY spectrum at 60.4 ms mixing time recorded on a 600 MHz solid-state NMR spectrometer under 2.7 kHz MAS (Figure 3). Isotropic chemical shift values for resonances obtained from 3D NOESY are reported elsewhere.40
Figure 3.
Representative 2D 1H-1H TOCSY spectral regions of cytb5 incorporated in DPC micelles recorded using a mixing time of 60.4 ms under 2.7 kHz MAS showing chemical shift correlations for (A) amide 1H-alpha and side chain protons, (B) aromatic protons, (C) 1Hβ-side chain protons and (D) 1Hα-side chain protons.
Residual 1H-1H dipolar couplings observed in the Hα and side chain regions of cytb5
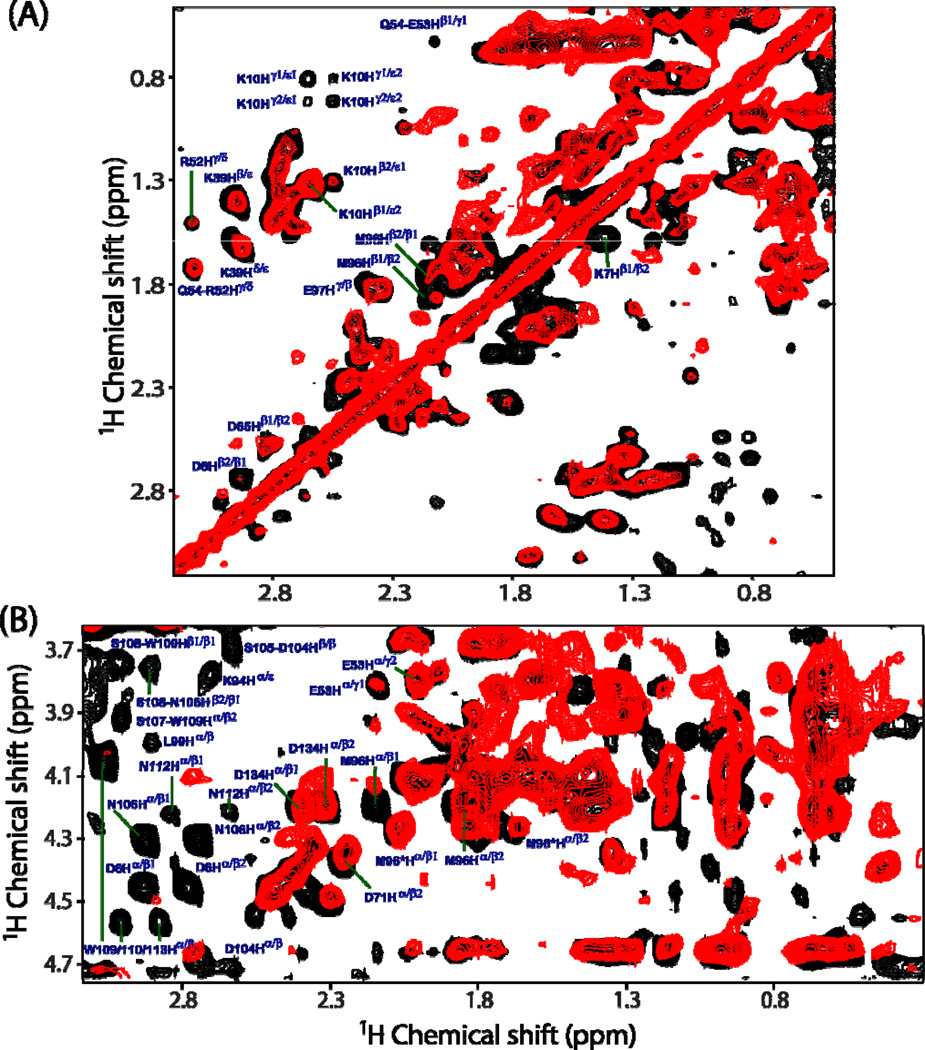
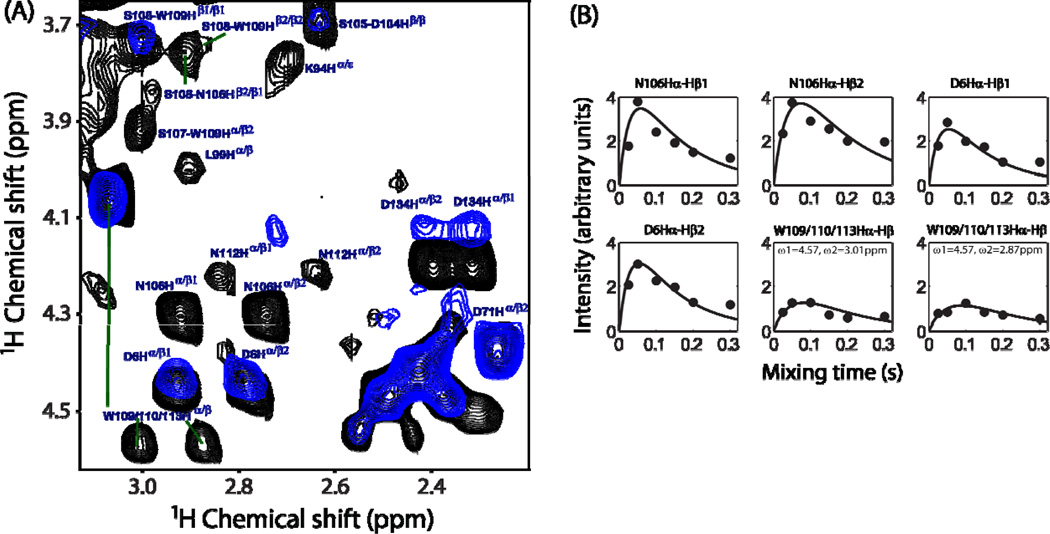
Superimposed 2D 1H-1H RFDR and NOESY spectra in Figure 4, showing the chemical shift correlations for 1Hβ-side chain protons (Figure 4A) and 1Hα-side chain protons (Figure 4B), were obtained from an unlabeled cytb5 incorporated in perdeuterated DPC micelles. A comparison is made at two different mixing times (50 ms for RFDR and 200 ms for NOESY) because the maximum intensity for most of the cross-peaks could only be obtained at these values. Substantial differences in the cross-peak intensities are observed in 2D 1H-1H RFDR and NOESY spectra. The variation in the cross-peak intensity could arise from many factors: such as the spin-spin coupling (both through space (or dipolar) and scalar through-bond (or J) couplings) among neighboring protons, electrostatic interactions and chemical shift exchange effects. In Figure 4, it can be seen that the cross-peaks in the RFDR spectrum corresponding to K7Hβ1−Hβ2, K10Hβ−Hε and K10Hγ−Hε correlations have much higher intensities as compared to the corresponding peaks in the NOESY spectrum. This observation indicates that side chains of residues K7 and K10 are associated with strong electrostatic interactions possibly due to the formation of a salt bridge between lysine and aspartate residues (D6/D8) located in the N-terminal region of cytb5 that could prevent the complete averaging of 1H-1H dipolar couplings. Surprisingly, few cross-peaks arising from the side chains of the linker region residues (M96 and E97) of cytb5 show higher peak intensities and hence these residues are associated with 1H-1H dipolar couplings. This could be attributed to the restricted isotropic motion of these residues due to their interaction with surrounding DPC molecules. This finding is vital as our earlier solution NMR investigation suggested the linker region of cytb5 to be highly flexible due to solvent exposure and lacks any definite secondary structure based on the absence of both inter- and intra-residue NOEs for most residues in the linker region.40
Figure 4.
Representative regions of 2D 1H-1H RFDR (black) and NOESY (red) spectra of cytb5 incorporated in perdeuterated DPC micelles showing the chemical shift correlations for 1Hβ-side chain protons (panel A) and 1Hα-side chain protons (panel B). RFDR and NOESY spectra were recorded at a mixing time of 50 and 200 ms, respectively, under 2.7 kHz MAS condition. Many additional cross-peaks are observed in the RFDR spectrum, which mostly belong to the side chains of the residues located in the transmembrane region of cytb5.
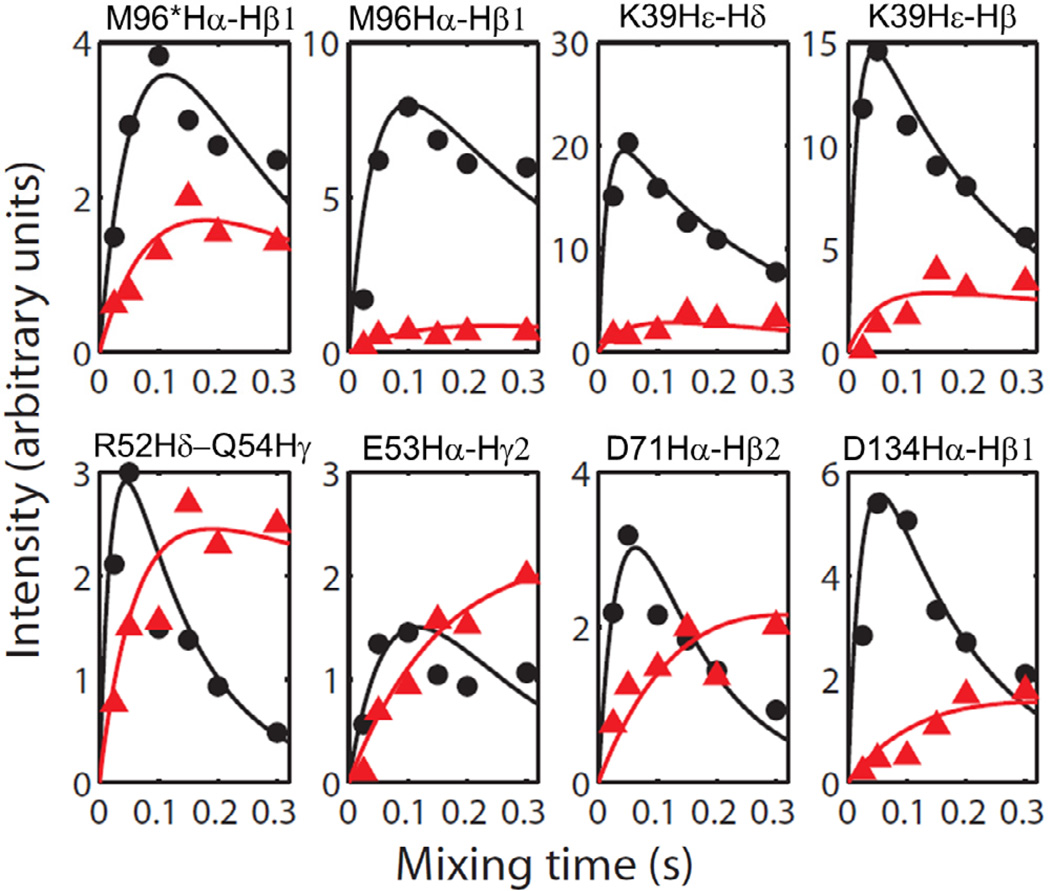
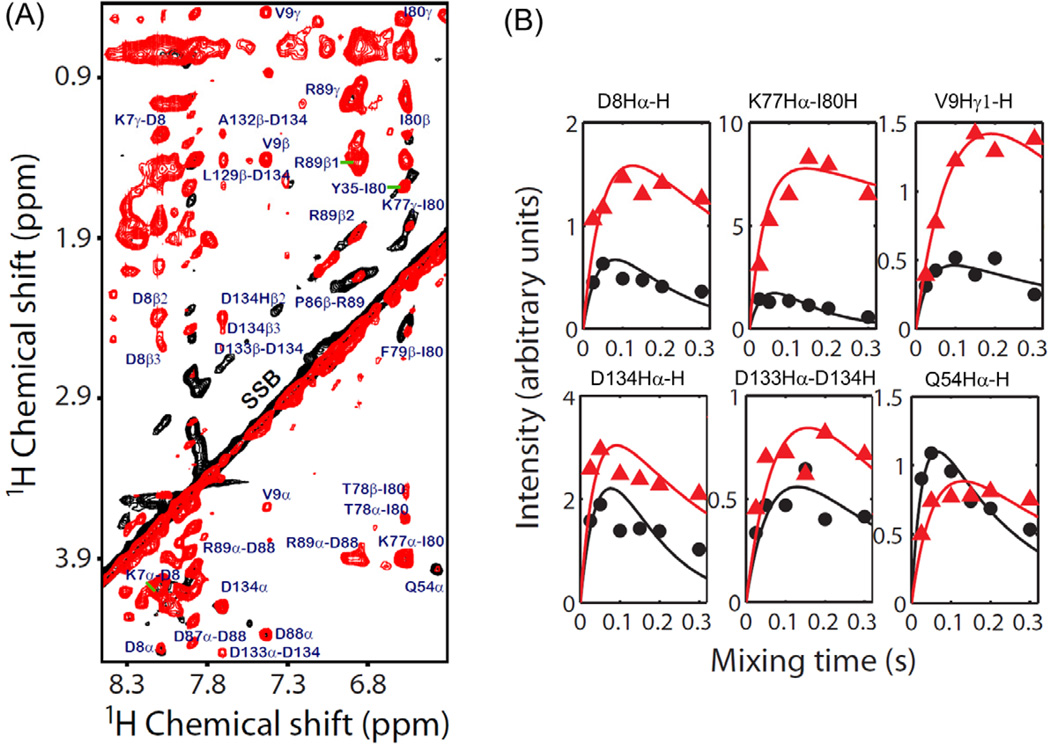
In order to provide a detailed discussion on the observed cross-peak intensity differences and to quantify residual dipolar couplings to extract 1H-1H distances, we have plotted experimentally measured cross-peak intensities against the mixing time (the so called buildup curves) for various residues of cytb5 (Figure 5). The experimental intensities were simulated using the rate equation discussed above and the best fitting parameters are reported in Table 1. The difference in cross-peak intensities measured from the RFDR and NOESY spectra can be used to extract 1H-1H dipolar coupling constraints for the structural studies of the protein. It can be seen from Figure 5 that the simulated buildup curves for different residues of cytb5 are in close agreement with the experimental data. Figure 5 also shows that the cross-peak intensity for Hα and side chain regions reaches a maximum within a short mixing time of ~50–100 ms in the RFDR experiment, which is in contrast to the NOESY experiment where the maximum intensity is obtained at longer mixing times. As discussed above, the transfer of magnetization is mediated through coherent 1H-1H dipolar interactions recovered from the RFDR experiment in addition to the magnetization transfer via an incoherent cross-relaxation from NOE. The T1* of the RFDR cross-peaks are usually much shorter in comparison to that of the NOESY cross-peaks due to the applied rotor-synchronized 180° pulses in the RFDR sequence. The imperfection of 180° pulses in the RFDR causes attenuation of the cross-peak intensity for longer mixing times, due to which a significant difference in the buildup curves obtained from the RFDR and NOESY experiments is observed.
Figure 5.
The simulated buildup curves for various cross-peaks observed in Hα and side chain regions for selected residues of cytb5 obtained from the RFDR (Black) and NOESY (Red) experiments on cytb5 incorporated in perdeuterated DPC micelles. Though the intensity scale was arbitrarily chosen, the intensities of data points in all panels are directly comparable. RFDR and NOESY spectra were collected for six different mixing times, 25, 50 100, 150, 200 and 300 ms.
Table 1.
Relaxation parameters obtained from the simulation of experimentally measured crosspeak intensities from cytb5 incorporated in DPC micelles.
| Residues | RFDR | NOESY | ||||
|---|---|---|---|---|---|---|
| 1/T1* (s−1) |
R (s−1) |
I0 | 1/T1* (s−1) |
R (s−1) |
I0 | |
| M96*Hα-Hβ1 | 4.7 | 5 | 18 | 2.5 | 4 | 7 |
| M96Hα-Hβ1 | 3 | 10 | 25 | 2.5 | 2 | 5 |
| K39Hε-Hδ | 3.7 | 35 | 48 | 2 | 10 | 8 |
| K39Hε-Hβ | 4.3 | 30 | 38 | 1 | 10 | 7 |
| R52Hδ−Q54Hγ | 8 | 20 | 10 | 0.8 | 8 | 6 |
| E53Hα-Hγ2 | 6 | 3 | 12 | 0.2 | 3 | 5 |
| D71Hα-Hβ2 | 8 | 10 | 14 | 1.6 | 2 | 10 |
| D134Hα-Hβ2 | 6 | 18 | 18 | 1 | 3 | 5 |
| W27 | 13.5 | 27 | 8.8 | 4 | 6 | 5.5 |
| Y12 | 15 | 22 | 21 | 7 | 7 | 11 |
| F79Hε-Hδ | 14 | 25 | 42 | 4 | 19 | 21 |
| F40 | 5.5 | 12 | 25 | 5 | 3 | 50 |
| Y11 | 14.8 | 16 | 30 | 4 | 4.5 | 8 |
| D8Hα-H | 6 | 8 | 3 | 2.5 | 8 | 5 |
| K77Hα-I80H | 8 | 10 | 8 | 0.8 | 12 | 18 |
| V9Hγ1-H | 2 | 14 | 1.2 | 2.5 | 3.5 | 6.2 |
| D134Hα-H | 8.9 | 4.7 | 17 | 3 | 12 | 9 |
| D133Hα-D134H | 3.3 | 5.8 | 2.2 | 3.2 | 4.0 | 3.9 |
| Q54Hα-H | 4.8 | 15 | 3.5 | 2.7 | 7 | 3 |
| D6Hα-Hβ1 | 8 | 14 | 10 | |||
| D6Hα-Hβ2 | 8 | 14 | 12 | |||
| W109/110/113Hα-Hβ (ω1/ω2:4.57/3.01ppm) | 7 | 6 | 7 | |||
| W109/110/113Hα-Hβ (ω1/ω2:4.57/2.87ppm) | 7 | 5 | 7 | |||
| N106Hα-Hβ1 | 7 | 14 | 13 | |||
| N106Hα-Hβ2 | 6 | 10 | 15 | |||
It is apparent from the buildup curves in Figure 5 that the side chains of a residue, M96 and its conformer (M96Hα-Hβ2 and M96*Hα-Hβ), belonging to the linker region are associated with unaveraged dipolar couplings, which could be because of their interaction with DPC molecules as discussed earlier. A stronger dipolar coupling and/or higher buildup rate is also observed for K39 (K39Hε-Hδ and K39Hε-Hβ) that is located in the helical region of cytb5 and, as a consequence, it may be rigid due to H-bonding interactions and therefore could avoid the averaging of the dipolar couplings associated with this residue. Additionally, it can undergo restricted motion due to electrostatic interactions. Also, the side chains of residues such as R52, E53 and D71 (R52Hδ−Q54Hγ2, E53Hα-Hγ2 and D71Hα-Hβ2) in a alpha-helix are shown to have weak dipolar interactions possibly due to weak electrostatic interactions. Interestingly, a stronger dipolar coupling is observed for the side chain of a C-terminal residue (D134) in the presence of micelles. This could be due to its interaction with micelles that results in a hindered motion of its side chain.
RFDR reveals the membrane interaction of residues in the transmembrane region of cytb5
The alpha-helical structure of the transmembrane region of cytb5 incorporated in magnetically-aligned DMPC/DHPC bicelles and its orientation in the membrane were previously determined using a 2D HIMSELF (Heteronuclear Isotropic Mixing leading to Spin Exchange via the Local Field) or HERSELF (HEteronuclear Rotating frame Spin Exchange via the Local Field)51 static solid-state NMR experiment.33,52 In our earlier report on the micelle-associated structure of rabbit cytb5 (PDB # 2M33), the tryptophan residues (W109/110/113) were predicted to be at the edge of transmembrane domain based on the observed broad indole side chain NH resonances along with additional peaks due to chemical exchange in the 1H-15N TROSY-HSQC (Transverse Relaxation-Optimized SpectroscopY - Heteronuclear Single Quantum Coherence) spectrum.30,40 However, no cross-peaks were observed for these residues in 2D 1H-1H NOESY and 3D HSQC-NOESY spectra recorded at 900 MHz. Analogous to this, we did not observe these resonances in the 2D NOESY spectrum obtained under MAS at 600 MHz (Figure 4). In contrast, some well-resolved resonances from the side chains of the transmembrane residues (Figure 6) were observed in the RFDR experiment. Resonance assignment based on the isotropic chemical shifts of AMX spin system indicated that these cross-peaks correspond to the side chains of D104, S105, N106, S107, S108, W109, W110, N112 and W113 located at the end of the linker region and/or the beginning of the transmembrane region of cytb5. These residues may interact with DPC molecules that could motionally-restrict the side chains to result in the observed dipolar couplings. Furthermore, this study also confirms our earlier prediction that tryptophan rings (W109/110/113) are in fact at the edge of the transmembrane domain of cytb5 in DPC micelles. This finding is cross validated further by comparing with the RFDR measurement on cytb5 in the absence of DPC micelles (Figure 6). Since the presence of DPC micelles restricts the motion of the transmembrane region, it should be highly mobile in the absence of the DPC molecules as indicated by the almost negligible dipolar recoupling observed for most of the resonances (Figure 6). Hence, in the present study, the RFDR experiment provides a direct evidence for the interaction of transmembrane residues of cytb5 with DPC micelles. It can be seen from the buildup curves (Figure 6B) that the observed dipolar couplings for these residues are weaker in comparison to that of the Hα and side chain regions. The dynamic parameters for these residues are extracted from the theoretical simulation and are given in Table 1. It is also interesting to note that the RFDR cross-peaks from the side chains of N-terminal (D6) and C-terminal (D134) residues appear both in the presence and absence of DPC micelles; however, their intensities are weaker in the absence of DPC micelles. This observation clearly suggests that the C-terminal residue is comparatively more rigid in the presence of micelles as discussed above. Similarly, the isotropic motion of the side chain of D6 is restricted probably due to electrostatic interactions with nearby charged residues as pointed out earlier.
Figure 6.
(A) Superimposed 2D 1H-1H RFDR spectra of cytb5 in the presence (black) and absence (blue) of DPC micelles recorded at a mixing time of 50 ms. (B) Simulated buildup curves for the RFDR cross-peak intensities measured from the side chain resonances of residues in the transmembrane region of cytb5 in the presence of DPC micelles. All other experimental details are as given in Figure 5 caption.
To better understand the sources of unaveraged 1H-1H dipolar couplings present in the micelle-associated cytb5, it is important to discuss the interactions between the protein and DPC micelles used in this study. It is known that there are approximately 50 DPC molecules per micelle (molecular weight ~19.5 kDa),53 which gives a total molecular weight of ~36.2 kDa for the micelle-associated cytb5 system. On the basis of molecular size, the micelle-associated protein sample that does not form any aggregates should be suitable for structural studies using solution NMR spectroscopy. However, depending on the folding of the protein and its interaction with micelles, the difference in the time scale of motion for different domains of the protein could result in a difference in their transverse relaxation rates. This is indeed the case as demonstrated by our previous study on a well-folded micelle-associated cytb5.30 As an example, the transmembrane domain of micelle-associated cytb5 undergoes slow motion (ms or slower) in comparison to the soluble domain that undergoes fast motion (µs or faster). While the high-resolution structure of the soluble domain of cytb5 incorporated in DPC micelles was solved by traditional solution NMR experiments, the residues in the transmembrane domain of the protein were not observable due to a significant line broadening caused by their restricted motion in the hydrophobic region of micelles.30 On the other hand, as demonstrated in this study, MAS overcomes the line broadening effects and thus enables the observation of resonances from the transmembrane residues. While the motionally-unaveraged 1H-1H dipolar couplings, that are not completely suppressed due to the restricted motion of cytb5 in DPC micelles under static conditions, are suppressed by MAS, and are recoupled by the 2D RFDR experiment as shown in this study. It is worth mentioning here that that 1H-1H RFDR cross-peaks for some of the residues were not detected from the transmembrane region of micelle-associated cytb5, which could be due to the stronger 1H-1H dipolar couplings. RFDR experiments performed at shorter mixing times and/or higher spinning speeds might be suitable to address this issue.
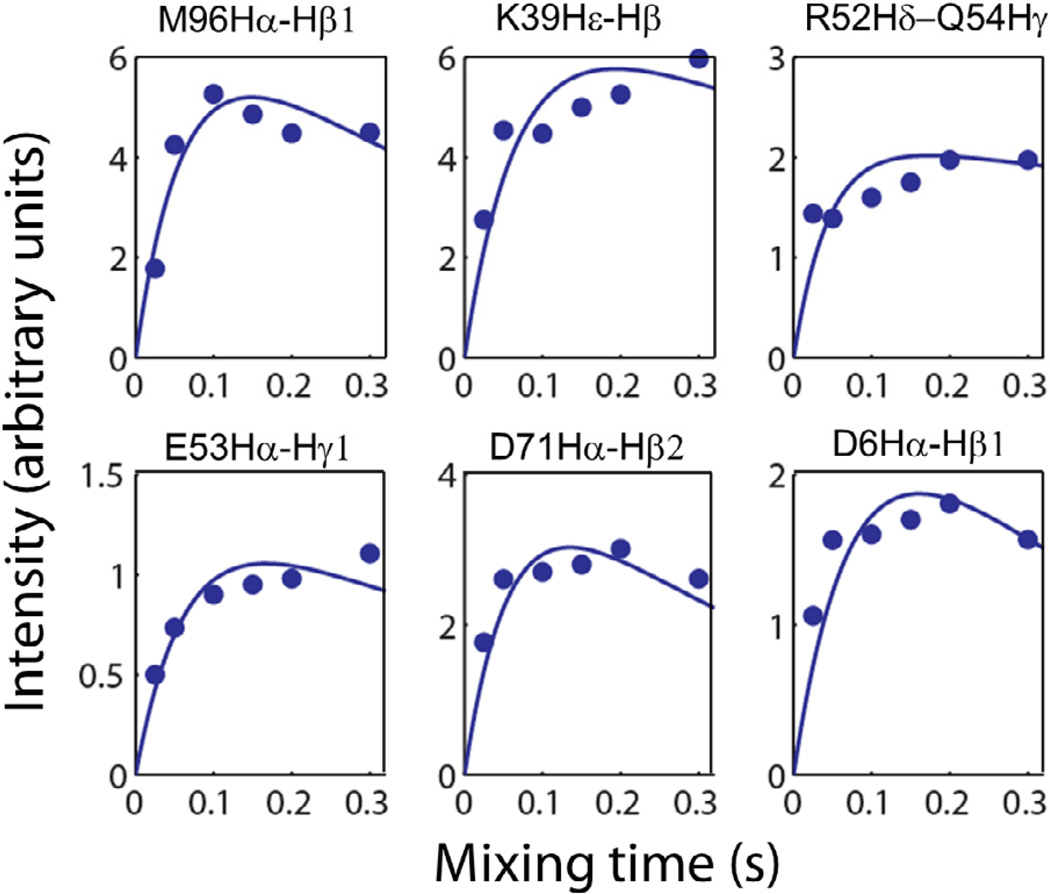
To further confirm the above observations, we measured the buildup rates for the RFDR cross-peaks detected in the absence of DPC micelles (Figure 7); superimposed 2D RFDR spectra displaying alpha-side-chain 1H-1H correlations obtained at different mixing times are shown in Figure S2 of the Supporting Information. It can be seen from Figure 7 that the maximum intensity for these cross-peaks could only be obtained at longer mixing times resulting in slower buildup rates in comparison to that observed from the micelle-associated cytb5 (refer to the relaxation parameters in Table S1 of the Supporting Information). This observation suggests that cytb5 in the absence of DPC micelles is more flexible than in association with DPC micelles. Importantly, this observation (in Figure 7) is also similar to what is observed from NOESY buildup curves for micelle-associated cytb5 wherein the 1H-1H cross-peaks arise only due to incoherent NOE; which is in contrast to the observations in RFDR where the magnetization exchange occurs via both incoherent NOE effect and coherent (recoupled) 1H-1H dipolar interactions. In other words, these results suggest that the 1H-1H dipolar couplings are suppressed by the isotropic tumbling of cytb5 in the absence of DPC micelles and therefore the magnetization exchange is mostly due to incoherent NOE effect. Notably, this also accounts for the reason why a recoupling RFDR sequence should work on micelle-associated molecules.
Figure 7.
The simulated buildup curves for the RFDR 1H-1H cross-peak intensities observed in Hα and side chain regions for selected residues of cytb5 in the absence of DPC micelles.
Dipolar recoupling for aromatic protons
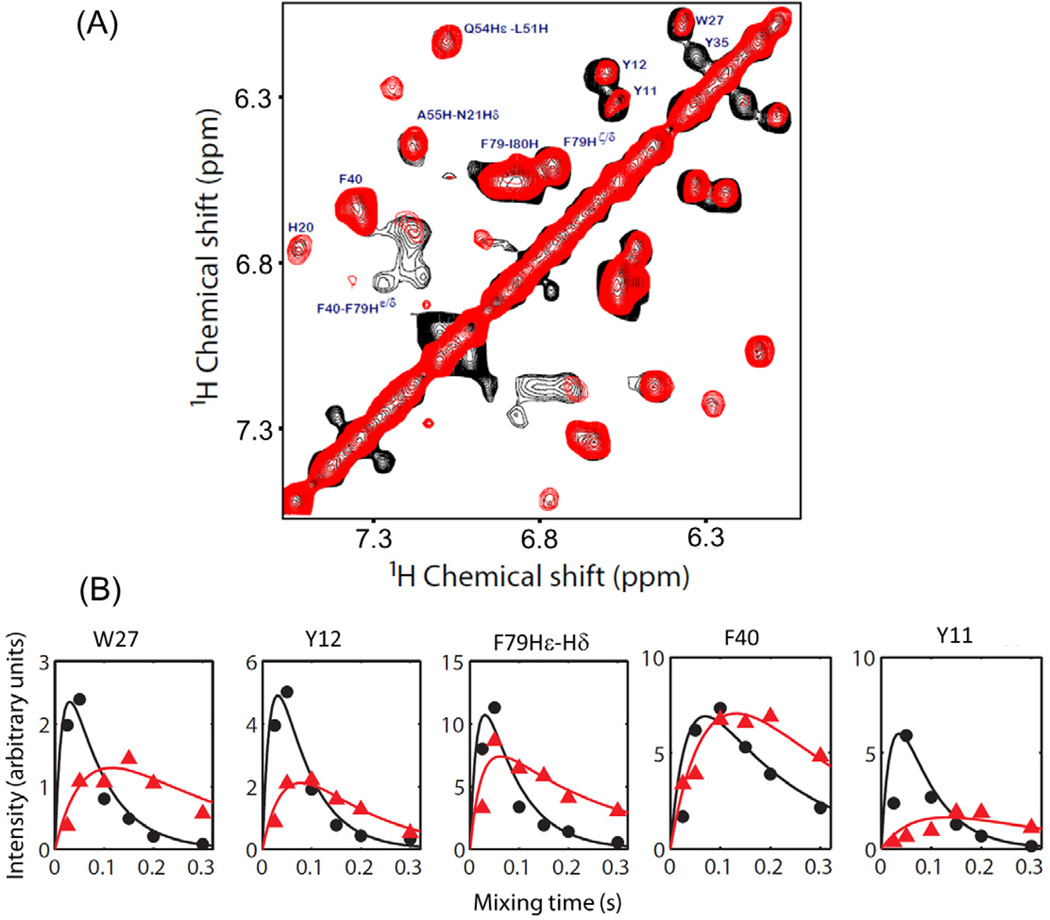
Since an aromatic side chain could undergo a restricted motion depending on its location in the protein sequence and secondary structure, we investigated the ability of RFDR experiments to recouple dipolar couplings associated with aromatic protons. Superimposed 2D 1H-1H RFDR and NOESY spectra for aromatic side chain resonances are shown in Figure 8. Results from Figure 8 indicate that the residues containing aromatic groups such as Y11, Y12, W27, Y35 and F79 exhibit higher cross-peak intensity in the RFDR spectrum than that in the NOESY spectrum. In contrast, residue F40 seems to have negligible intensity difference from the two experiments.
Figure 8.
(A) Superimposed 2D 1H-1H RFDR (black) and NOESY (red) spectra of cytb5 incorporated in DPC micelles recorded at a mixing time of 50 ms and 200 ms, respectively, showing the chemical shift correlation of aromatic protons. (B) Simulated buildup curves and experimentally measured cross-peaks intensities for aromatic side chains obtained from RFDR (black) and NOESY (red) experiments. All other experimental details are as given in Figure 5 caption.
In addition, observed dipolar couplings are weaker for most of the aromatic protons in comparison to Hα and side chain regions (Figure 8B). It can be seen from the buildup rates that aromatic protons of F40 are associated with a smaller dipolar coupling in comparison to Y11, Y12, W27 and F79 residues. On the basis of our recently reported backbone amide-15N chemical shift anisotropy (CSA) measurements, residue F40 is associated with a smaller CSA span than Y11, Y12, Y35, W27 and F79.39 In other words, F40 was found to be more flexible than Y11, Y12, Y35, W27 and F79. This finding corroborates well with the present result wherein 1H-1H dipolar coupling for aromatic protons of F40 is averaged out due to flexibility while other residues exhibit unaveraged dipolar couplings. The buildup curve for aromatic protons of Y35 is not shown in Figure 8B as its RFDR cross peak intensity reached the maximum at a mixing time of 50 ms and decayed entirely with a further increase in the mixing time due to fast relaxation. Superimposed 2D 1H-1H RFDR and NOESY spectra showing aromatic 1H-1H correlations obtained using different mixing times (25–300 ms) are shown in Figure S3 of the supporting information.
Dipolar couplings associated with amide-protons cannot be recoupled
An overlay of 2D amide 1H-1H RFDR and NOESY spectra for cytb5 incorporated in DPC micelles is shown in Figure 9. It is interesting that the 2D NOESY spectrum reveals more cross-peaks with higher intensities in comparison to the RFDR spectrum.
Figure 9.
(A) 2D 1H-1H RFDR (black) and NOESY (red) spectra for the amide-side chain proton region of cytb5 incorporated in DPC micelles recorded using a mixing time of 50 ms (RFDR) and 200 ms (NOESY) under 2.7 kHz MAS. Spinning sidebands are denoted by SSB. (B) Simulation of the variation of cross-peak intensities measured for amide-protons resonances from the RFDR (black) and NOESY (red) spectra. All other details are as given in Figure 5 caption.
This observation is attributed to a constant exchange of amide protons with water that average out coherent dipolar interactions associated with amide protons. But, if this is the only explanation for this observation, then RFDR intensities should be very similar to that of NOESY as the magnetization transfer related to amide protons in the RFDR experiment is only due to incoherent cross-relaxation effect. As can be seen from Figure 9, this is obviously not the case.
The amide-proton cross-peak intensities measured from the RFDR spectra are, in fact, lower than that obtained from the NOESY spectra. It is probably due to the loss of amide-proton magnetization to surrounding spins via solvent exchange. This finding is not surprising since the major contribution to the RFDR cross-peak intensity comes from the spin diffusion driven process originating from the coherent dipolar interaction among protons. Therefore, in cases where solvent exchange of protons is highly plausible, 2D NOESY measurement could be a better option than RFDR to obtain 1H-1H correlations; however, there could be few exceptions to this. For example, the RFDR intensity for a helical residue, Q54, is slightly higher than the NOESY intensity despite that its amide proton is most likely to be exchanging with water (Figure 9B); but the overall intensity is considerably lower than that observed for protons in the side chain region.
Conclusion
There is considerable interest in the development of proton-based solid-state NMR experiments to study the structure and dynamics of membrane-bound proteins. Though solution NMR experiments are routinely used to solve high-resolution structures of relatively small-size membrane proteins embedded in isotropic phases like detergent micelles or isotropic bicelles, the use of MAS experiments on these samples to recouple motionally-unaveraged 1H-1H dipolar couplings could provide additional valuable constraints (long-range distance) to improve the accuracy of structures solved by NMR spectroscopy. In addition, such residual dipolar couplings measured by recoupling MAS solid-state NMR techniques could be used to characterize the slow motions and the aggregation process that may not be easily determined from solution NMR experiments. To demonstrate the feasibility of recoupling dipolar couplings and using proton-based solid-state MAS experiments, we successfully carried out 2D 1H-1H RFDR and NOESY experiments under MAS on an unlabeled cytb5 incorporated in perdeuterated DPC micelles.
The variations of experimentally measured cross-peak intensities with the mixing time compare very well with our theoretical simulations and enabled the extraction of NMR parameters to understand the dynamics of micelle-associated cytb5. The longitudinal relaxation time (T1*) for the RFDR cross-peaks are found to be much shorter than that for the NOESY cross-peaks, thus accounting for an accelerated magnetization transfer via the recoupled 1H-1H dipolar couplings. Our results demonstrate that the residual 1H-1H dipolar couplings associated with Hα and side chain protons could be easily recoupled by RFDR and their measurement could be valuable in structural studies of micelle-associated proteins. Results from the RFDR measurement provides a direct evidence for the interaction between the side chains of the transmembrane residues with DPC detergent molecules, which is observed through the appearance of additional resonances in the RFDR spectra that is otherwise not possible due to their fast spin-spin relaxation. Our results also show that the coherent dipolar interactions associated with amide-protons are suppressed by the fast exchange between amide and water protons. While the results presented here can provide qualitative information about the structural constraints in the form of 1H-1H dipolar couplings or distances, further studies are needed to obtain quantitative estimation of the constraints. One of the major challenges to overcome is to accurately model the multiple 1H spin network to extract structural constraints. Although our measurements were on a micelle-associated cytb5 from a 600 MHz spectrometer under slow MAS, we strongly believe that multidimensional MAS solid-state NMR experiments utilizing RFDR performed at higher magnetic fields and higher spinning speeds can be of greater applicability over commonly employed methods for 3D structure determination of unlabeled membrane proteins and amyloid fibrils.
Supplementary Material
Highlights.
2D 1H/1H RFDR MAS spectra of an unlabeled cytochrome-b5 are presented.
Dipolar couplings are recoupled by RFDR-MAS for aromatic and side chain protons.
Residues interacting with membrane are revealed.
Dipolar couplings with amide-protons are suppressed due to exchange with water.
T1 values of 1H resonances observed in RFDR are shorter than that in NOESY.
Acknowledgment
This research was supported by funds from NIH (GM084018 and GM095640 to A.R.). We would like to thank Rui Huang and Joshua Dameron for a valuable discussion.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supporting Information Available
Figures for 2D 1H-1H RFDR and NOESY pulse sequences, superimposed 2D 1H-1H RFDR spectra recorded at different mixing times for cytb5 in the presence and absence of DPC micelles as well as superimposed 2D 1H-1H NOESY and RFDR spectra recorded at different mixing times for cytb5 incorporated in DPC micelles. Table for relaxation parameters obtained from the simulation of experimentally measured cross-peak intensities of cytb5 in the absence of DPC micelles.
References
- 1.Shahid SA, Bardiaux B, Franks WT, Krabben L, Habeck M, van Rossum BJ, Linke D. Membrane-Protein Structure Determination by Solid-State NMR Spectroscopy of Microcrystals. Nat. methods. 2012;9:1212–1217. doi: 10.1038/nmeth.2248. [DOI] [PubMed] [Google Scholar]
- 2.Laage S, Marchetti A, Sein J, Pierattelli R, Sass HJ, Grzesiek S, Lesage A, Pintacuda G, Emsley L. Band-Selective 1H-13C Cross-Polarization in Fast Magic Angle Spinning Solid-State NMR Spectroscopy. J. Am. Chem. Soc. 2008;130:17216–17217. doi: 10.1021/ja805926d. [DOI] [PubMed] [Google Scholar]
- 3.Wang S, Munro RA, Shi L, Kawamura I, Okitsu T, Wada A, Kim SY, Jung KH, Brown LS, Ladizhansky V. Solid-State Nmr Spectroscopy Structure Determination of a Lipid-Embedded Heptahelical Membrane Protein. Nat. methods. 2013;10:1007–1012. doi: 10.1038/nmeth.2635. [DOI] [PubMed] [Google Scholar]
- 4.Tang M, Comellas G, Rienstra CM. Advanced Solid-State NMR Approaches for Structure Determination of Membrane Proteins and Amyloid Fibrils. Acc. Chem. Res. 2013;46:2080–2088. doi: 10.1021/ar4000168. [DOI] [PubMed] [Google Scholar]
- 5.Linser R, Dasari M, Hiller M, Higman V, Fink U, Lopez del Amo JM, Markovic S, Handel L, Kessler B, Schmieder P, Oesterhelt D, Oschkinat H, Reif B. Proton-Detected Solid-State NMR Spectroscopy of Fibrillar and Membrane Proteins. Angew. Chem. 2011;50:4508–4512. doi: 10.1002/anie.201008244. [DOI] [PubMed] [Google Scholar]
- 6.Nielsen NC, Malmendal A, Vosegaard T. Techniques and Applications of NMR to Membrane Proteins (Review) Mol. Membr. Biol. 2004;21:129–141. doi: 10.1080/09687680410001693679. [DOI] [PubMed] [Google Scholar]
- 7.McDermott A. Structure and Dynamics of Membrane Proteins by Magic Angle Spinning Solid-State NMR. Ann. Rev. Biophys. 2009;38:385–403. doi: 10.1146/annurev.biophys.050708.133719. [DOI] [PubMed] [Google Scholar]
- 8.Tycko R. NMR at Low and Ultralow Temperatures. Acc. Chem. Res. 2013;46:1923–1932. doi: 10.1021/ar300358z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Paepe G. Dipolar Recoupling in Magic Angle Spinning Solid-State Nuclear Magnetic Resonance. Ann. Rev. Phys. Chem. Vol 63. 2012;63:661–684. doi: 10.1146/annurev-physchem-032511-143726. [DOI] [PubMed] [Google Scholar]
- 10.Huang KY, Siemer AB, McDermott AE. Homonuclear Mixing Sequences for Perdeuterated Proteins. J. Magn. Reson. 2011;208:122–127. doi: 10.1016/j.jmr.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou DH, Nieuwkoop AJ, Berthold DA, Comellas G, Sperling LJ, Tang M, Shah GJ, Brea EJ, Lemkau LR, Rienstra CM. Solid-State NMR Analysis of Membrane Proteins and Protein Aggregates by Proton Detected Spectroscopy. J. Biomol. NMR. 2012;54:291–305. doi: 10.1007/s10858-012-9672-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou DH, Shah G, Cormos M, Mullen C, Sandoz D, Rienstra CM. Proton-Detected Solid-State NMR Spectroscopy of Fully Protonated Proteins at 40 Khz Magic-Angle Spinning. J. Am. Chem. Soc. 2007;129:11791–11801. doi: 10.1021/ja073462m. [DOI] [PubMed] [Google Scholar]
- 13.Huber M, Bockmann A, Hiller S, Meier BH. 4D Solid-State NMR for Protein Structure Determination. Phys. Chem. Chem. Phys. 2012;14:5239–5246. doi: 10.1039/c2cp23872a. [DOI] [PubMed] [Google Scholar]
- 14.Huber M, Hiller S, Schanda P, Ernst M, Bockmann A, Verel R, Meier BH. A Proton-Detected 4D Solid-State NMR Experiment for Protein Structure Determination. ChemPhysChem. 2011;12:915–918. doi: 10.1002/cphc.201100062. [DOI] [PubMed] [Google Scholar]
- 15.Linser R, Bardiaux B, Higman V, Fink U, Reif B. Structure Calculation from Unambiguous Long-Range Amide and Methyl 1H-1H Distance Restraints for a Microcrystalline Protein with Mas Solid-State NMR Spectroscopy. J. Am. Chem. Soc. 2011;133:5905–5912. doi: 10.1021/ja110222h. [DOI] [PubMed] [Google Scholar]
- 16.Nishiyama Y, Malon M, Gan Z, Endo Y, Nemoto T. Proton-Nitrogen-14 Overtone Two-Dimensional Correlation NMR Spectroscopy of Solid-Sample at Very Fast Magic Angle Sample Spinning. J. Magn. Reson. 2013;230:160–164. doi: 10.1016/j.jmr.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 17.Ward ME, Shi L, Lake E, Krishnamurthy S, Hutchins H, Brown LS, Ladizhansky V. Proton-Detected Solid-State Nmr Reveals Intramembrane Polar Networks in a Seven-Helical Transmembrane Protein Proteorhodopsin. J. Am. Chem. Soc. 2011;133:17434–17443. doi: 10.1021/ja207137h. [DOI] [PubMed] [Google Scholar]
- 18.Morris VK, Linser R, Wilde KL, Duff AP, Sunde M, Kwan AH. Solid-State NMR Spectroscopy of Functional Amyloid from a Fungal Hydrophobin: A Well-Ordered Beta-Sheet Core Amidst Structural Heterogeneity. Angew. Chem. 2012;51:12621–12625. doi: 10.1002/anie.201205625. [DOI] [PubMed] [Google Scholar]
- 19.Asami S, Rakwalska-Bange M, Carlomagno T, Reif B. Protein-RNA Interfaces Probed by 1H-Detected MAS Solid-State NMR Spectroscopy. Angew. Chem. 2013;52:2345–2349. doi: 10.1002/anie.201208024. [DOI] [PubMed] [Google Scholar]
- 20.Knight MJ, Webber AL, Pell AJ, Guerry P, Barbet-Massin E, Bertini I, Felli IC, Gonnelli L, Pierattelli R, Emsley L, Lesage A, Herrmann T, Pintacuda G. Fast Resonance Assignment and Fold Determination of Human Superoxide Dismutase by High-Resolution Proton-Detected Solid-State MAS NMR Spectroscopy. Angew. Chem. 2011;50:11697–11701. doi: 10.1002/anie.201106340. [DOI] [PubMed] [Google Scholar]
- 21.Knight MJ, Felli IC, Pierattelli R, Bertini I, Emsley L, Herrmann T, Pintacuda G. Rapid Measurement of Pseudocontact Shifts in Metalloproteins by Proton-Detected Solid-State NMR Spectroscopy. J. Am. Chem. Soc. 2012;134:14730–14733. doi: 10.1021/ja306813j. [DOI] [PubMed] [Google Scholar]
- 22.Asami S, Reif B. Proton-Detected Solid-State NMR Spectroscopy at Aliphatic Sites: Application to Crystalline Systems. Acc. Chem. Res. 2013;46:2089–2097. doi: 10.1021/ar400063y. [DOI] [PubMed] [Google Scholar]
- 23.Durr UH, Soong R, Ramamoorthy A. When Detergent Meets Bilayer: Birth and Coming of Age of Lipid Bicelles. Prog. Nucl. Magn. Reson. Spectrosc. 2013;69:1–22. doi: 10.1016/j.pnmrs.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bennett AE, Ok JH, Griffin RG, Vega S. Chemical-Shift Correlation Spectroscopy in Rotating Solids - Radio Frequency-Driven Dipolar Recoupling and Longitudinal Exchange. J. Chem. Phys. 1992;96:8624–8627. [Google Scholar]
- 25.Bennett AE, Rienstra CM, Griffiths JM, Zhen WG, Lansbury PT, Griffin RG. Homonuclear Radio Frequency-Driven Recoupling in Rotating Solids. J. Chem. Phys. 1998;108:9463–9479. [Google Scholar]
- 26.Kumar A, Wagner G, Ernst RR, Wuthrich K. Buildup Rates of the Nuclear Overhauser Effect Measured by Two-Dimensional Proton Magnetic-Resonance Spectroscopy - Implications for Studies of Protein Conformation. J. Am. Chem. Soc. 1981;103:3654–3658. [Google Scholar]
- 27.Raya J, Bianco A, Furrer J, Briand JP, Piotto M, Elbayed K. Proton Dipolar Recoupling in Resin-Bound Peptides under High-Resolution Magic Angle Spinning. J. Mag. Reson. 2002;157:43–51. doi: 10.1006/jmre.2002.2573. [DOI] [PubMed] [Google Scholar]
- 28.Aucoin D, Camenares D, Zhao X, Jung J, Sato T, Smith SO. High-Resolution 1H MAS RFDR NMR of Biological Membranes. J. Mag. Reson. 2009;197:77–86. doi: 10.1016/j.jmr.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramamoorthy A, Xu J. 2D 1H/1H RFDR and NOESY NMR Experiments on a Membrane-Bound Antimicrobial Peptide under Magic Angle Spinning. J. Phys. Chem. B. 2013;117:6693–6700. doi: 10.1021/jp4034003. [DOI] [PubMed] [Google Scholar]
- 30.Ahuja S, Jahr N, Im SC, Vivekanandan S, Popovych N, Le Clair SV, Huang R, Soong R, Xu J, Yamamoto K, Nanga RP, Bridges A, Waskell L, Ramamoorthy A. A Model of the Membrane-Bound Cytochrome b5-Cytochrome P450 Complex from NMR and Mutagenesis Data. J. Bio. Chem. 2013;288:22080–22095. doi: 10.1074/jbc.M112.448225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Durr UH, Waskell L, Ramamoorthy A. The Cytochromes P450 and b5 and Their Reductases--Promising Targets for Structural Studies by Advanced Solid-State NMR Spectroscopy. Biochim. Biophys. Acta. 2007;1768:3235–3259. doi: 10.1016/j.bbamem.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 32.Xu JD, Soong R, Im SC, Waskell L, Ramamoorthy A. Inept-Based Separated-Local-Field NMR Spectroscopy: A Unique Approach to Elucidate Side-Chain Dynamics of Membrane-Associated Proteins. J. Am. Chem. Soc. 2010;132:9944–9947. doi: 10.1021/ja103983f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamamoto K, Gildenberg M, Ahuja S, Im SC, Pearcy P, Waskell L, Ramamoorthy A. Probing the Transmembrane Structure and Topology of Microsomal Cytochrome-P450 by Solid-State NMR on Temperature-Resistant Bicelles. Sci. Reports. 2013;3:2556. doi: 10.1038/srep02556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soong R, Smith PES, Xu JD, Yamamoto K, Im SC, Waskell L, Ramamoorthy A. Proton-Evolved Local-Field Solid-State NMR Studies of Cytochrome b5 Embedded in Bicelles, Revealing Both Structural and Dynamical Information. J. Am. Chem. Soc. 2010;132:5779–5788. doi: 10.1021/ja910807e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu J, Durr UH, Im SC, Gan Z, Waskell L, Ramamoorthy A. Bicelle-Enabled Structural Studies on a Membrane-Associated Cytochrome b5 by Solid-State MAS NMR Spectroscopy. Angew. Chem. 2008;47:7864–7867. doi: 10.1002/anie.200801338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamamoto K, Durr UH, Xu J, Im SC, Waskell L, Ramamoorthy A. Dynamic Interaction between Membrane-Bound Full-Length Cytochrome P450 and Cytochrome b5 Observed by Solid-State Nmr Spectroscopy. Sci. Reports. 2013;3:2538. doi: 10.1038/srep02538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pandey MK, Ramamoorthy A. Quantum Chemical Calculations of Amide-15N Chemical Shift Anisotropy Tensors for a Membrane-Bound Cytochrome-b5. J. Phys. Chem. B. 2013;117:859–867. doi: 10.1021/jp311116p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pandey MK, Vivekanandan S, Ahuja S, Huang R, Im SC, Waskell L, Ramamoorthy A. Cytochrome-P450-Cytochrome-b5 Interaction in a Membrane Environment Changes 15N Chemical Shift Anisotropy Tensors. J. Phys. Chem. B. 2013;117:13851–13860. doi: 10.1021/jp4086206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pandey MK, Vivekanandan S, Ahuja S, Pichumani K, Im SC, Waskell L, Ramamoorthy A. Determination of 15n Chemical Shift Anisotropy from a Membrane- Bound Protein by NMR Spectroscopy. J. Phys. Chem. B. 2012;116:7181–7189. doi: 10.1021/jp3049229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vivekanandan S, Ahuja S, Im SC, Waskell L, Ramamoorthy A. H, C and N Resonance Assignments for the Full-Length Mammalian Cytochrome b5 in a Membrane Environment. Biomolecular NMR assignments. 2013 doi: 10.1007/s12104-013-9528-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Braunschweiler L, Ernst RR. Coherence Transfer by Isotropic Mixing - Application to Proton Correlation Spectroscopy. J. Magn. Reson. 1983;53:521–528. [Google Scholar]
- 42.Rucker SP, Shaka AJ. Broad-Band Homonuclear Cross Polarization in 2D NMR Using DIPSI-2. Mol. Phys. 1989;68:509–517. [Google Scholar]
- 43.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRpipe: A Multidimensional Spectral Processing System Based on Unix Pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 44.Goddard TD, Kneller DG. Sparky 3. San Francisco: University of California; [Google Scholar]
- 45.Ishii Y. C-13-C-13 Dipolar Recoupling under Very Fast Magic Angle Spinning in Solid-State Nuclear Magnetic Resonance: Applications to Distance Measurements, Spectral Assignments, and High-Throughput Secondary-Structure Determination. J. Chem. Phys. 2001;114:8473–8483. [Google Scholar]
- 46.Mcconnell HM. Reaction Rates by Nuclear Magnetic Resonance. J. Chem. Phys. 1958;28:430–431. [Google Scholar]
- 47.Bloom M, Reeves LW, Wells EJ. Spin Echoes and Chemical Exchange. J. Chem. Phys. 1965;42 1615-&. [Google Scholar]
- 48.Bain AD. Chemical Exchange in NMR. Prog. Nucl. Magn. Reson. Spectrosc. 2003;43:63–103. [Google Scholar]
- 49.Taylor DM, Ramamoorthy A. Coherence Transfer through Homonuclear Dipolar Coupling in an Unoriented Two Spin-1/2 Solid-State System. J. Mol. Struct. 2002;602:115–124. [Google Scholar]
- 50.Taylor DM, Ramamoorthy A. Analysis of Dipolar-Coupling-Mediated Coherence Transfer in a Homonuclear Two Spin-1/2 Solid-State System. J. Magn. Reson. 1999;141:18–28. doi: 10.1006/jmre.1999.1893. [DOI] [PubMed] [Google Scholar]
- 51.Dvinskikh SV, Yamamoto K, Ramamoorthy A. Heteronuclear Isotropic Mixing Separated Local Field NMR Spectroscopy. J. Chem. Phys. 2006;125:34507. doi: 10.1063/1.2212939. [DOI] [PubMed] [Google Scholar]
- 52.Durr UH, Yamamoto K, Im SC, Waskell L, Ramamoorthy A. Solid-State NMR Reveals Structural and Dynamical Properties of a Membrane-Anchored Electron-Carrier Protein, Cytochrome b5. J. Am. Chem. Soc. 2007;129:6670–6671. doi: 10.1021/ja069028m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kallick DA, Tessmer MR, Watts CR, Li CY. The Use of Dodecylphosphocholine Micelles in Solution NMR. J. Mag. Reson. Ser. B. 1995;109:60–65. doi: 10.1006/jmrb.1995.1146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.