Abstract
Cancer cell motility and invasiveness are fundamental characteristics of the malignant phenotype and are regulated through diverse signaling networks involving kinases and transcription factors. This study establishes an estrogen receptor (ERα)/MAPK (ERK5)/cofilin (CFL1) network that specifies the degree of breast cancer cell aggressiveness through coupling of actin reorganization and hormone receptor-mediated transcription. Using dominant negative and constitutively active forms, as well as small molecule inhibitors of ERK5 and MEK5, it was revealed that hormone activation of estrogen receptor-α determined the subcellular localization of ERK5, which functions as a co-regulator of ERα-dependent gene transcription. Notably, ERK5 acted in concert with the actin remodeling protein, CFL1, and upon hormone exposure both localized to active nuclear transcriptional hubs as verified by immunofluorescence and proximity ligation assays. Both ERK5 and CFL1 facilitated PAF1 recruitment to the RNA Pol II complex and both were required for regulation of gene transcription. By contrast, in cells lacking ERα, ERK5 and CFL1 localized to cytoplasmic membrane regions of high actin remodeling, promoting cell motility and invasion, thereby revealing a mechanism likely contributing to the generally poorer prognosis of ERα-negative breast cancer patients. Thus, this study uncovers the dynamic interplay of nuclear receptor-mediated transcription and actin reorganization in phenotypes of breast cancer aggressiveness.
Implications
Identification of the ER/ERK5/CFL1 axis suggests new prognostic biomarkers and novel therapeutic avenues to moderate cancer aggressiveness.
Keywords: estrogen receptor, protein kinase ERK5, cofilin, breast cancer, cell motility and invasion, gene transcription
INTRODUCTION
Elucidation of the factors and networks that regulate cancer cell motility and invasiveness is fundamental to understanding the malignant phenotype and may also highlight biomarkers of disease and reveal opportunities for the development of novel targeted therapies to moderate cancer aggressiveness. The nuclear hormone receptor, estrogen receptor alpha (ERα), present in two-thirds of human breast cancers, is a master regulator of the phenotypic properties of these cancers. It is considered the single most crucial predictor of breast cancer prognosis and is targeted by endocrine therapies, which are generally well tolerated and avoid the morbidity associated with radiation and chemotherapy (1). Molecular subtyping of breast cancers has revealed that ERα-containing tumors are generally less aggressive and that patients with ER-positive cancers have a better prognosis and overall survival. Although many studies have documented that ER directly regulates over 3% of protein-encoding genes and indirectly regulates many more (2–6), the mechanisms by which this hormone-regulated transcription factor controls cell phenotype and reduces cell invasiveness remain unclear. To address this, we have examined the involvement of protein kinases in modulating ER activity.
The importance of kinases in cancer biology is well known, as increased kinase activity through phosphorylation, mutation or elevated expression is often observed in tumors and is associated with a less good clinical outcome for breast cancer patients (7–11). However, the cellular processes underlying the interplay between ERα and protein kinase pathways and the manner by which ERα maintains and controls these pathways and their phenotypic outcomes are poorly understood. Our previous genome-wide analyses revealed the importance of the MAPK signaling pathway and the involvement and direct binding of ERK2 along with ERα at enhancers of many estrogen-regulated genes that control cell proliferation (12).
In the current studies, we have identified the protein kinase ERK5 and Cofilin (CFL1), an actin-severing protein required for actin cytoskeleton reorganization (13), as two interacting factors that are moved into the nucleus and recruited to the transcription start site (TSS) of estrogen-stimulated genes upon hormone treatment of ERα-containing breast cancer cells. Notably, in breast cancer cells that lack ERα, ERK5 and CFL1 remain outside the nucleus and increase cell motility and invasiveness. Thus, by eliciting nuclear localization of ERK5 and CFL1, thereby diminishing their co-localization to regions of high actin remodeling, ERα is playing a critical role in maintaining the lower metastatic activity characteristic of many ERα-positive breast cancers. These novel findings reveal a transcription factor-mediated regulatory mechanism that modulates cancer cell aggressiveness, through relocalization of two key factors, ERK5 and CFL1, highlighting the crucial cross-talk between ERα-driven nuclear events and the actin cytoskeleton and suggesting alternative opportunities for targeted therapies.
MATERIAL AND METHODS
Cell Culture, siRNA, Adenovirus, and Ligand Treatments
MCF-7, BT474, T47D, MDA-MB-453, MDA-MB-468, and SKBR3 cells were obtained from and grown as recommended by American Type Culture Collection. For experiments with E2 treatment, cells were maintained in the corresponding phenol red-free medium for at least 5 days prior to use. Cell lines were authenticated by checking ERα expression by q-PCR and western blot analysis, checking responsiveness to E2 in ERα (+) cell lines and monitoring morphology of the cells (compared to the images reported by ATCC). Recombinant adenoviruses were obtained from Applied Biological Materials and used to generate cells expressing dominant negative ERK5 (ERK5-DN) or dominant negative MEK5 (MEK5-DN). ERK5 knockdown utilized the smart pool of 4 siRNAs from Dharmacon and were performed as described (12) with 20 nM siCtrl or siERK5 for 72 h, and resulted in knockdown of ERK5 mRNA and protein by greater than 80%.
Western Blotting, ChIP and ChIP-Re-ChIP Assays
Western blot analysis used specific antibodies for ERα (HC-20, Santa Cruz); ERK2 (D-2, Santa Cruz); pERK5 (Upstate, 07-507); and total ERK5 (Bethyl, A302-655A) and was performed as described (14).
ChIP was carried out as described (15, 16). Antibodies used were for pERK5 (Upstate, 07-507, sc-135760), phospho-PXSP motif (2325, Cell Signaling), CFL1 (D3F9) rabbit (5175S, Cell Signaling), pSer5 RNA Pol II (Santa Cruz, sc-47701), PAF1 (sc-74795), SPT5 (sc-28678), Cyclin T1 (sc-8127) and WDR61 (sc-100897). ChIP DNA was isolated using QIAGEN PCR purification kit and used for ChIP-seq analysis and quantitative real-time PCR. ChIP-reChIP experiments were done following the same ChIP protocol. After the first pull down, immunoprecipitated material was recovered with 10mM dithiothreitol in IP buffer at 37°C for 30 min, diluted, and submitted to a second round of immunoprecipitation. Quantitative real-time PCR (qPCR) was used to calculate recruitment to the regions studied, as described before (16).
ChIP-seq Analysis and Clustering
The ChIP DNA was prepared into libraries according to Illumina Solexa ChIP-seq sample processing methods, and single read sequencing was performed using the Illumina HiSeq 2000 (15). Sequences generated were mapped uniquely onto the human genome (hg18) by Bowtie2 (17). MACS (Model-based Analysis of ChIP-Seq) algorithm (18) was used to identify enriched peak regions (default settings) with a p-value cutoff of 6.0e-7 and FDR of 0.01.
The seqMINER density array method with a 500-bp window in both directions was used for the generation of clusters, i.e., groups of loci having similar compositional features (15, 19). BED files for each cluster were used for further analysis with Galaxy Cistrome (20) integrative analysis tools (Venn diagram, conservation, CEAS).
GeneChip mRNA Transcriptional Profiling Microarrays
Total RNA was used to generate cRNA, which was labeled with biotin as recommended by Affymetrix. The biotin-labeled cRNA was hybridized to Affymetrix U133 plus 2.0 GeneChips and analyzed using Affymetrix processing software. CEL files were processed using GeneSpring GX 11.0 software (Agilent) to obtain fold-change and p-value with Benjamini and Hochberg multiple test correction for each gene for each treatment relative to the vehicle control (15). We considered genes with fold-change > 1.8 and p-value < 0.05 as statistically significant, differentially expressed. Data is available from GEO with the accession number GSE53394.
Motif and GO Category Analysis
Overrepresented GO biological processes were determined by the web-based DAVID Bioinformatics Resources database (18), GeneSpring and web-based GREAT software (21). Motifs enrichment analysis was done using Seqpos (21).
Immunofluorescence Microscopy, Proximity Ligation Assay and Data Analysis
Cells treated with Vehicle (0.1% EtOH) or 10 nM E2 were washed in PBS, fixed on glass coverslips and then incubated with antibodies against ERα (F10, Santa Cruz), CFL1 (AbCam, ab54532), pSer5 RNA Pol II (Santa Cruz,47701), or PAF1 (sc-74795). Next day, cells were incubated with Goat anti mouse Alexa 568 or goat anti rabbit Alexa 488 secondary antibodies. Slides were mounted and stained using Prolong Gold antifade with 4,6-diamidino-2-phenylindole (DAPI, Molecular Probes) to identify the nuclei.
The proximity ligation assay (PLA) was performed using the Duolink In Situ kit (Olink Bioscience) according to the manufacturer’s instructions. Briefly, cells were prepared as for to immunofluorescence analysis. Overnight incubation with primary antibodies was followed by hybridizaton with two PLA probes at 37°C for 1 h, and then by ligation for 15 min and amplification for 90 min at 37°C. A cover slip was mounted on each slide and image acquisition and analysis conducted. Samples were imaged using a 63×/1.4 Oil DIC M27 objective in a Zeiss LSM 700 or 710 laser scanning confocal microscope. Images were obtained in a sequential manner using a 488 Ar (10 mW) laser line for ERK5, pERK5 or PLA signal (500–550 nm emission) and 555 nm (10 mW) laser line for phalloidin, SC-35 or ERα (600–650 nm emission). The individual channels were obtained using a sequential scanning mode to prevent bleed-through of the excitation signal. Laser power, gain and offset were kept constant across the samples and scanned in a high resolution format of 512×512 or 1024×1024 pixels with 2/4 frames averaging. Further quantification of the images used Fiji software (http://fiji.sc/wiki/index.php/Fiji) (22). Briefly, images were converted to 8 bit for segmentation for each channel, and images were background subtracted using a rolling-ball method, with a pixel size of 100 and segmented using the DAPI channel.
Cell Proliferation, Soft Agar Colony Formation, Migration, and Invasion Assays
For proliferation assays, cells were seeded at 1000 cells/well in 96-well plates. On the second day, the cells were treated with Veh or 10 nM E2 (Day 0) in the presence or absence of 1 uM XMD8-92 or 10 uM BIX-02189 and again at Day 2. Proliferation was assessed using WST-1 reagent (Roche) as described (23). Invasion assays used BDBioCoat Matrigel invasion chambers (BD Biosciences) with 10% fetal bovine serum as chemoattractant in the lower chamber as described (23). Motility and soft agar colony formation assays were performed as described (7, 23).
Tumor Data Sets and Data Analysis
log2 median-centered intensity expression values for signature genes were obtained from Oncomine database. Hierarchical clustering of data was performed and displayed using Cluster3.0 and Java Tree View software for analysis and visualization. Patients were stratified according to average expression value of the genes in the signature and the top 30% and bottom 30% were used for computation of Kaplan-Meier curves by the Cox-Mantel log-rank test and Gehan-Breslow-Wilcoxon Test in Graphpad Prism.
RESULTS
ERK5 is a novel kinase recruited to the transcription start site (TSS) of 17β-estradiol (E2) regulated genes
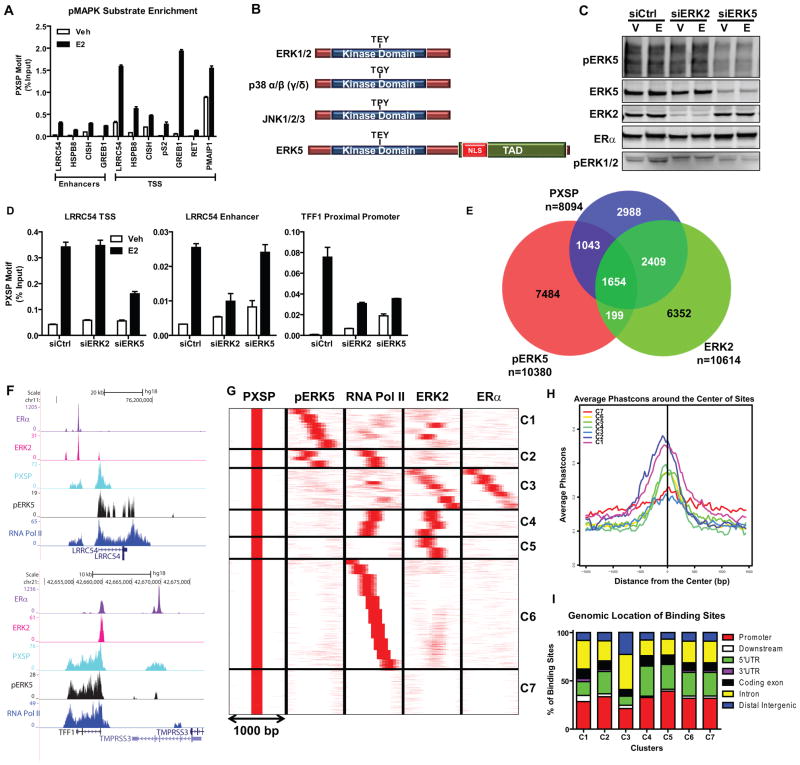
To characterize MAPK signaling networks underlying ERα-mediated transcriptional regulation, we monitored MAPK activity on chromatin through ChIP assays by using an antibody that recognizes phosphorylated MAPK targets at the PXSP consensus motif (pMAPK substrates). Upon E2 treatment, pMAPK substrates were more highly enriched at the TSS of E2 target genes compared to the intragenic enhancers (Fig. 1a). Since we previously showed that ERK2 is responsible for activity at enhancers but not at the TSS (12), the data in Fig. 1A suggests that a different MAPK is responsible for phosphorylating targets at the TSS of E2-regulated genes.
Figure 1. ERK5 is a novel kinase recruited to the transcription start site (TSS) of hormone-activated ERα-regulated genes.
(a) ChIP-qPCR of pMAPK substrates recruitment to regulatory regions of various E2-regulated genes in MCF-7 cells. Cells were treated with vehicle or 10 nM E2 for 45 min. After cross-linking, pMAPK substrates/DNA complexes were immunoprecipitated. Immunoprecipitated DNA levels were measured by qPCR and % input calculated. Results (mean ± SD) are from three independent experiments.
(b) Domains of typical MAPK family members. NLS, nuclear localization sequence. TAD, transcription activation domain.
(c) Validation of selective ERK2 or ERK5 knockdowns. Cells were transfected with siCtrl, siERK2, or siERK5 for 72h. Protein levels were verified by Western blotting.
(d) pMAPK substrates recruitment and impact of knockdown of ERK2 or pERK5 on recruitment to gene regulatory regions. Chromatin was prepared from cells transfected with siCtrl, siERK2 or siERK5 for 72h and then treated with Veh or E2 for 45 min. DNA/Protein complexes were immunoprecipitated and levels were measured by qPCR and % input calculated. Results (mean ± SD) are from two independent experiments.
(e) Venn diagram depicting overlapping pMAPK substrates, ERK2 and pERK5 cistromes after MCF-7 treatment with E2.
(f) Track view of ERα, ERK2, pMAPK substrates, pERK5 and RNA Pol II ChIP-seq density profiles after E2 treatment shown using the UCSC genome browser centered on the LRRC54 and TFF1 genes. Cells were treated with E2 for 45 min and protein/DNA complexes were immunoprecipitated and ChIP-sequencing performed. Sequences were mapped uniquely onto the human genome (hg18). MACS algorithm was used to identify enriched peak regions (default settings) with p-value cutoff of 6.0e-7 and FDR of 0.01.
(g) Cluster analysis of ChIP-Seq data for pMAPK substrates (PXSP), pERK5, RNA Pol II, ERK2 and ERα after E2 treatment using SeqMINER software in 1000 bp window; pMAPK substrates cistrome was used as the reference.
(h) Conservancy of binding sites from each cluster identified in Fig. 1g. Conservancy plots were generated with CEAS software using a 3000 bp window.
(i) Location of binding sites in each cluster (C1–C7) relative to annotated genes, determined using CEAS software.
ERK1/2, together with p38, JNKs and ERK5 are members of the family of typical MAPKs. These kinases share high sequence homology at the N-terminal region (24, 25). Strikingly, however, ERK5 harbors a unique C-terminal transactivation domain and nuclear localization sequence (NLS) (26) (Fig. 1b). Therefore, we tested whether ERK5 was the kinase responsible for E2-mediated protein phosphorylation at the TSS of ER target genes by knocking down either ERK5 or ERK2 in MCF-7 cells. We did not observe any compensation between these two kinases after knockdown (Fig. 1c). ChIP assay demonstrated that, whereas ERK2 knockdown abrogated the increase in pMAPK substrates at the enhancer regions, ERK5 knockdown reduced them at the TSS of the LRRC54 gene (Fig. 1d, 1f upper panel). TFF1, which has a proximal promoter ERα binding site, showed decreased pMAPK substrate recruitment after knockdown of both kinases, consistent with the recruitment of ERα and RNA Pol II to this site (Fig. 1d, 1f lower panel).
We then compared the ChIP-seq cistromes for ERK5, ERK2 and pMAPK substrates after E2 treatment of cells to obtain a genome-wide characterization of the ERα/MAPK network (Fig. 1e). Nearly 30% of pMAPK substrate binding sites overlapped with pERK5 binding sites. Notably, less than 20% of pERK5 sites overlapped with ERK2 binding sites in the presence of E2 (Fig. 1e).
To obtain a better picture of the kinase landscape in the cells, we utilized a clustering approach using seqMINER (19), which identifies those binding sites that share a similar pattern of factor localization within a 1000-bp window (15). Comparison of pMAPK substrate, pERK5, RNA Pol II, ERK2, and ERα cistromes (Fig. 1g) revealed seven distinct clusters (C1–C7) showing different extents of conservation (Fig. 1h) and different genomic localization (Fig. 1i). For example, cluster 2 (C2) is characterized by increased pMAPK substrate, ERK5 and RNA Pol II recruitment, while lacking ERα and ERK2 binding; as expected, this cluster contained binding sites associated with the TSS of E2-regulated genes. Moreover, C2 had the highest conservancy score among the seven clusters.
As shown in Supplementary Fig. 1, we validated hormone-dependent recruitment of pERK5 to these binding sites by ChIP assays. Cluster C3, which exclusively contained ERα and ERK2 binding sites, mapped to enhancer regions of the E2-regulated genes and had the lowest conservation across vertebrates. Clusters C4–C7 showed distinct patterns of factor binding. Thus, our clustering analysis revealed different modes of modulation of E2-mediated transcriptional events by ERK5 at the TSS and by ERK2 and ERα at enhancers.
ERK5 binds to the TSS of genes required for E2-mediated cell proliferation
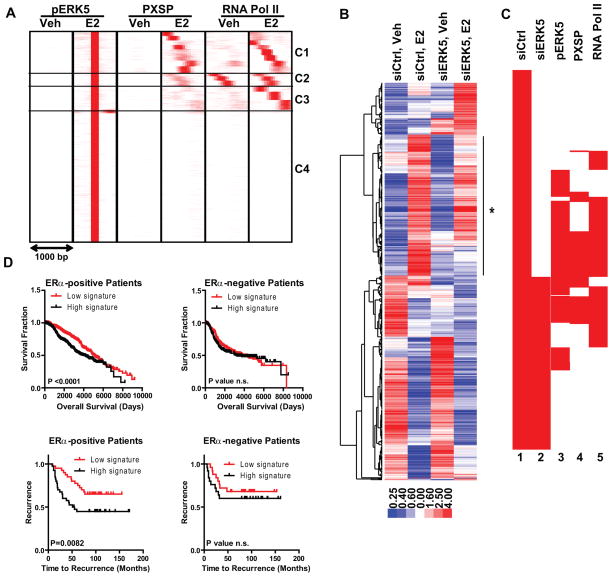
To further elucidate ERK5 association with E2-mediated gene regulation, we compared the cistromes for pERK5, pMAPK substrates and RNA Pol II in the presence and absence of E2, using the E2-induced pERK5 cistrome as the reference (Fig. 2a). This analysis revealed that there was minimal recruitment of pERK5, pMAPK substrates and RNA Pol II before addition of hormone, suggesting that pERK5 marked E2-induced transcriptional events. Only at cluster C2 did we observe constitutive presence of RNA Pol II, possibly marking paused genes.
Figure 2. ERK5 binds to and regulates genes required for E2-mediated cell proliferation and predicts clinical outcome in patients with ERα-positive breast cancers.
(a) Cluster analysis of ChIP-Seq data for pERK5, pMAPK substrates, and RNA Pol II after Veh (0.1% EtOH) or 10 nM E2 treatment of MCF-7 cells using SeqMINER software in 1000 bp window and pERK5-E2 cistrome as the reference.
(b) Cluster diagram of genes impacted by ERK5 knockdown. MCF-7 cells were treated with siCtrl or siERK5 for 72h prior to treatment with vehicle or E2 for 24h. Affymetrix gene expression microarrays were analyzed using LIMMA and Tightcluster software. The cluster map is visualized using Treeview Java. Expression level is indicated below. Line with star at right indicates gene cluster associated with reduced E2 stimulation after ERK5 knockdown.
(c) Clustering of chromatin coordinates of the TSS region of E2-regulated genes in siCtrl or siERK5 cells (lanes 1,2) and comparison of chromatin binding sites for pMAPK substrates, pERK5, and RNA Pol II (lanes 3,4,5). Density arrays were generated using Enrichment Based Method option of SeqMINER software. Data were clustered using Cluster3 software and visualized using Java Treeview software.
(d) Survival and time to recurrence analysis using our ERK5 modulated gene list and patient gene expression and survival/recurrence data from (upper panels) Curtis (50) and Wang (51) datasets (lower panels) obtained from the Oncomine database. Data is shown for patients with ERα-positive (left) and ERα-negative (right) breast cancers.
To understand the functional outcomes of ERK5 recruitment and its effect on E2-mediated cell processes, we performed gene expression microarray analysis in MCF-7 cells after ERK5 or control siRNA transfection. Hierarchical clustering of modulated transcripts revealed that E2 regulation of many genes was lost when ERK5 level was reduced (Fig. 2b). 296 genes that were stimulated with E2 treatment in siCtrl cells were no longer modulated when ERK5 was reduced, and by Gene Ontology analysis these genes were associated with cell cycle control (Supplementary Table 1 and Supplementary Fig. 2a, left). Interestingly, 277 new genes became regulated only in siERK5 cells, suggesting a repressive role for ERK5 in modulating transcription of these genes, which were associated with immune function and apoptosis (Supplementary Table 1). E2-mediated repression of 397 genes was lost with ERK5 knockdown, whereas 151 genes were now further repressed by E2 when ERK5 was knocked down (Supplementary Fig. 2a, right). We also generated scatter plots which showed that the direction of regulation by E2 did not change with depletion of ERK5, meaning that if a gene is regulated in both cell backgrounds, it will always change in the same direction (Supplementary Fig. 2b). Overall, this analysis revealed an activator role for ERK5 in control of genes regulating cell proliferation and a repressor role in processes associated with immune function and apoptosis.
To better correlate ERK5 binding with E2-mediated gene regulation, we generated BED files for the TSS of E2-regulated genes in siCtrl or siERK5 cells and monitored pERK5, pMAPK substrates and RNA Pol II recruitment to these sites (Fig. 2c). Notably, ERK5 knockdown blocked regulation of 54% of E2-stimulated genes that had pERK5 binding at their TSS, emphasizing that recruitment and activity of ERK5 are good indicators of E2 target gene regulation.
Examination of ERK5-regulated genes and their association with breast cancer patient clinical outcome
To delineate the translational importance of ERK5 modulated genes, we monitored levels of these genes in large scale breast tumor datasets. E2-upregulated genes that harbored pERK5 binding sites and that were blocked by ERK5 knockdown were associated with cell cycle progression, specifically with the S-phase of the cell cycle (Supplementary Table 1). Overexpression of these genes predicted a very poor outcome in 29 breast cancer datasets obtained from Oncomine. When we compared survival curves of breast cancer patients stratified according to ERα status and expression of all the genes from our list (Supplementary Table 2), we observed that low expression of this gene signature predicted better outcome only in ERα-positive patients, consistent with the identification of these genes as being E2-modulated in MCF-7 cells. Analysis of expression of our ERK5 signature genes in two such large primary breast cancer datasets is shown in Fig. 2d. These findings emphasize the importance of ERK5 in E2-mediated regulation of genes in ERα-positive breast cancers and mark ERK5 transcriptional targets as a predictor of outcome in patients with ERα-positive breast cancers.
ERK5 signaling regulates hormone-dependent tumorigenicity of ERα-positive breast cancer cells
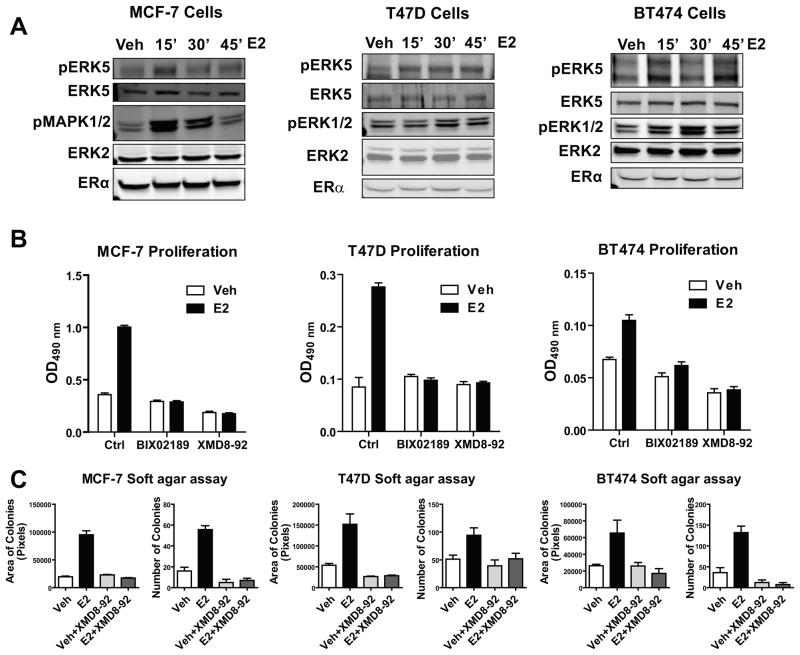
Since we observed association between ERK5 expression and E2-regulated genes linked to cell cycle progression, we examined the impact of ERK5 on E2-stimulated cell proliferation in several ERα-positive breast cancer models. ERK5 was rapidly and transiently activated upon addition of E2 in all 3 cell lines: MCF-7, T47D and BT474 (Fig. 3a). Notably, the ERK5 small molecule inhibitor (XMD8-92) (27) and the upstream kinase MEK5 inhibitor (BIX-02189) completely blocked E2-mediated cell proliferation without affecting ERα protein levels (Fig. 3b and Supplementary Figure 3). Moreover, in anchorage-independent growth assays, ERK5 inhibitor was very efficient in blocking E2-stimulated colony formation (Fig. 3c), suggesting a pro-proliferative role for ERK5 in ERα-containing breast cancer cells.
Figure 3. ERK5 modulated proliferation and tumorigenicity of ERα-positive breast cancer cell lines.
(a) Time course of ERK5 activation by E2. MCF-7, T47D and BT474 cells were treated with 10 nM E2 for the indicated times. Protein was harvested and subjected to SDS-PAGE analysis. pERK5, ERK5, pMAPK 1/2, ERK2, and ERα antibodies were used for Western blot analysis.
(b) Impact of ERK5 or MEK5 inhibitors on E2-stimulated cell proliferation. Cells were treated with 0.1% ethanol vehicle (open bars) or 10 nM E2 (black bars) in the presence or absence of 10 μM BIX02189 (MEK5 inhibitor) or 1 μM XMD8-92 (ERK5 inhibitor). Cell numbers were examined using the MTS assay at day 4. Values are the mean ± SD from at least 2 independent experiments.
(c) Anchorage-independent growth of ERα-positive breast cancer cells in the presence or absence of ERK5 inhibitor. Cells were plated at 10000 cells/well in 6 well plates in 0.4% softagar. The following day (day 0) cells were treated with vehicle or 10 nM E2 in the presence or absence of 1 μM XMD8-92. Treatments were repeated after 7 days. At 14 days, area of colonies and number of colonies were calculated using Fiji software. Values are mean ± SD from 2 independent experiments with 3 replicates.
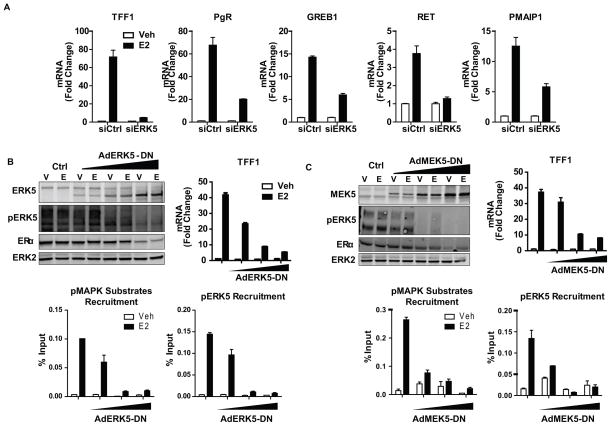
ERK5 kinase activity and activation by upstream MEK5 are required for the recruitment of ERK5 to chromatin and enrichment of pMAPK substrates at the TSS of hormone-regulated genes
To dissect the mechanism of ERK5 action, we knocked down ERK5 in MCF-7 cells and monitored the effect of this perturbation on the E2-stimulation of genes having ERK5 binding sites. ERK5 knockdown either completely abrogated or dampened E2 gene stimulation (Fig. 4a). To test the requirement of kinase activity of ERK5, or ERK5 activation by upstream MEK5 for kinase recruitment and gene activation, we infected MCF-7 cells with adenovirus containing a dominant negative ERK5 (ERK5-DN) (Fig. 4b) or a dominant negative MEK5 (MEK5-DN) (Fig. 4c) construct. With overexpression of either of these dominant negative kinases, we observed a dose-dependent decrease in gene stimulation, enrichment of pMAPK substrates and pERK5 recruitment, implying that activation of ERK5 by MEK5 as well as ERK5 kinase activity are both required for chromatin-related functions of ERK5. Interestingly, overexpression of a constitutively active MEK5 was not sufficient to drive ERK5 to the TSS of E2 regulated genes or increase mRNA output of these genes, suggesting that ERα activation is required for ERK5 activity at the chromatin (Supplementary Figure 4).
Figure 4. Activation of ERK5 by MEK5 and kinase activity of ERK5 are required for estrogen stimulation of gene expression.
(a) Impact of ERK5 knockdown on E2-stimulated gene regulation. MCF-7 cells were transfected with 20 nM siGENOME reagent for Ctrl or ERK5 for 72h and then treated with vehicle or 10 nM E2 for 24h. Total RNA was isolated, reverse transcribed and expression of several E2-stimulated genes was examined using qPCR. Values are mean ± SD from 3 independent experiments.
(b, c) Impact of dominant negative ERK5 (panel b) or dominant negative MEK5 (panel c) on gene stimulation, chromatin occupancy of pMAPK substrates and pERK5 recruitment to E2-stimulated genes. MCF-7 cells were infected with AdCMV (Ctrl) or increasing amounts of AdERK5-DN or AdMEK5-DN for 24h and then treated with vehicle or 10 nM E2 for 45 min for ChIP and western blot or 24h for gene expression analysis.
ERK5 localizes to transcription hubs in the nucleus upon cell treatment with E2
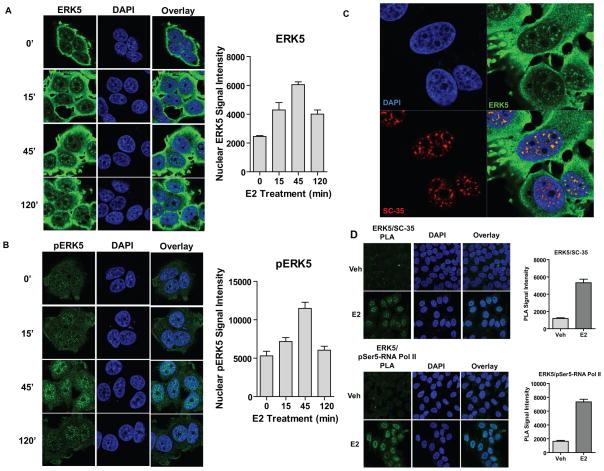
To further characterize the crosstalk between ERα and ERK5, we monitored by confocal microscopy the intracellular localization of total ERK5 and phosphorylated ERK5 in several ERα-positive breast cancer cell lines. In the absence of hormone treatment, ERK5 was localized throughout the cell, particularly outside the nucleus, but some nuclear-localized ERK5 appeared in a speckled pattern reminiscent of splicing factors and transcription hubs. This speckled pattern in the nucleus increased with time of E2 treatment, reaching maximum at 45 minutes (Fig. 5a). The same pattern was observed for pERK5, and it showed more clearly the E2-induced nuclear localization also peaking at 45 minutes (Fig. 5b). Since we observed a speckled nuclear pattern for both ERK5 and pERK5, we performed co-immunolabeling with antibodies for ERK5 and SC-35, a factor known to be part of splicing speckles and transcription hubs (28). Indeed, we observed that these two proteins colocalized in the nucleus after hormone treatment (Fig. 5c). In addition, we utilized a proximity ligation assay (PLA) (29) to assess interaction of ERK5 with several markers of transcriptional hubs. As seen in Fig. 5d, we detected an increase in PLA signal upon E2 treatment for interaction of ERK5 with SC-35, consonant with the colocalization findings, and also for ERK5 and pSer5-RNA Pol II, which is consistent with our ChIP-Seq data that showed overlap between RNA Pol II and ERK5 at the TSS of E2-regulated genes.
Figure 5. ERK5 is recruited to transcription factories upon cell treatment with E2.
(a, b) Subcellular localization of total ERK5 (panel a) or pERK5 (panel b) after E2 treatment. MCF-7 cells were treated with 10 nM E2 for indicated times (0–120 min) and immunofluorescence microscopy was performed with an antibody specific to ERK5 or pERK5. Nuclei were stained with DAPI. Right panel: quantification of nuclear ERK5 or pERK5 per cell (3 different fields were scored in each experiment n= at least 50 cells). Values are mean ± SD from 2 independent experiments.
(c) Colocalization of ERK5 and SC-35 at transcription factories. MCF-7 cells treated with 10 nM E2 for 45 min and immunofluorescence microscopy was performed with antibodies specific to ERK5 and SC-35. Nuclei were stained with DAPI. Values are mean ± SD from 2 independent experiments
(d) Interaction between ERK5 and SC-35 and between ERK5 and pSer5 RNA Pol II. Cells treated with E2 for 45 min and PLA was performed with specific antibodies. Nuclei were stained with DAPI. Right panels: quantification of nuclear ERK5/SC-35 or ERK5/RNA Pol II complex formation per cell (an average of 30 cells were scored) by PLA. Values are mean ± SD from 3 independent experiments.
Nuclear localization of ERK5 is dependent on ERα, and CFL1 links the actin cytoskeleton to ERK5 and ER signaling in the nucleus
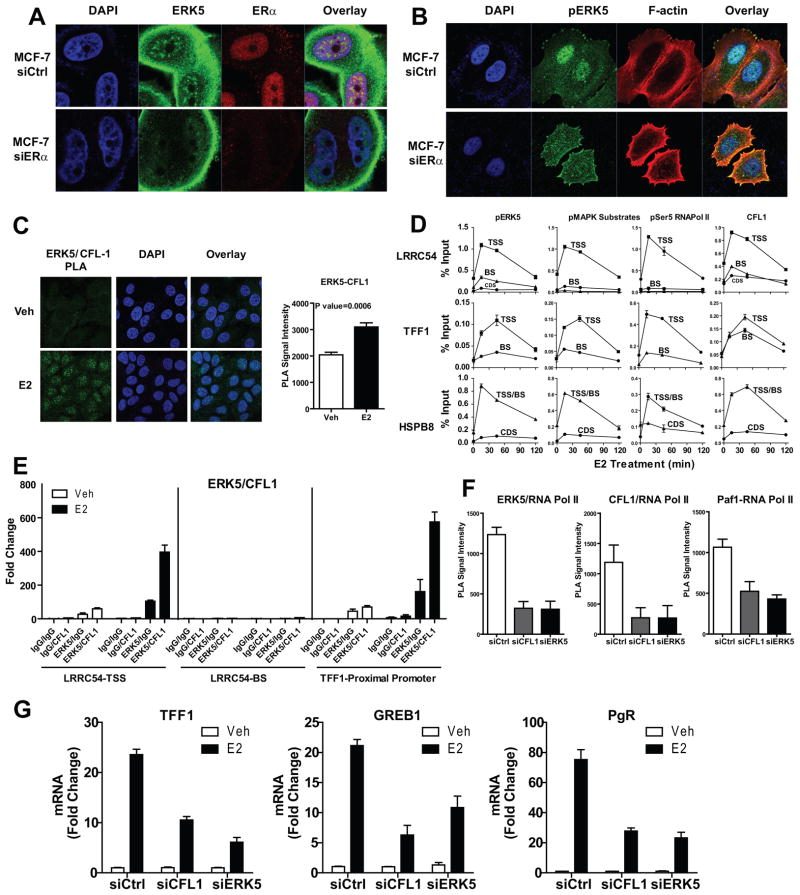
To test whether the presence of ERK5 in the nucleus is dependent upon ERα, we knocked down ERα and found that this abrogated localization of pERK5 in the nucleus, supporting the idea that ERα is the main determinant of ERK5 nuclear localization in ERα-positive breast cancer cells (Fig. 6a, 6b, Supplementary Fig. 5). Of note, knockdown of ERα increased localization of pERK5 to actin bundles near the leading edge of MCF-7 cells, to the cell-cell interface in BT474 and to stress fibers in T47D cells, which are all actin-rich structures. These findings highlighted an interesting link between ERα, pERK5 and the actin cytoskeleton, and prompted us to investigate a possible role for actin reorganization modulator proteins in the ERα/ERK5 axis. We focused on CFL1, a member of the Cofilin/ADF family of actin-severing proteins that is critical for formation of cell protrusions that adhere to extracellular matrix (30, 31), define the direction of motility (31–33) and initiate cell crawling leading to cell motility and increased invasiveness (34). We first monitored the intracellular localization of CFL1. PLA demonstrated an E2-dependent interaction of CFL1 and ERK5 (Fig. 6c). Immunofluorescence microscopy after treatment with the antiestrogen fulvestrant or ERK5 inhibitor XMD 8–92 showed nuclear localization of CFL1 to be ERα and ERK5 activity-dependent in ERα-positive cancer cells (Supplementary Fig. 6).
Figure 6. ERK5 nuclear localization is dependent on ERα, and CFL1 links the actin cytoskeleton to ERK5 and ERα signaling in the nucleus.
(a) Loss of ERK5 localization to the nucleus in the absence of ERα. MCF-7 cells were transfected with 20 nM siCtrl or siERα for 72 h and then treated with 10 nM E2 for 45 min. Immunofluorescence microscopy was performed with antibodies specific to ERK5 and ERα. Nuclei were stained with DAPI.
(b) Loss of pERK5 localization to the nucleus in the absence of ERα. Cells treated as in panel a, and immunofluorescence microscopy performed with antibody specific to pERK5. F-actin was stained with Alexa568-phalloidin and nuclei with DAPI.
(c) Interaction of ERK5 with CFL1 in the presence of E2. Cells treated with E2 for 45 min and PLA was performed with antibodies specific to ERK5 and CFL1. Nuclei were stained with DAPI. Right panel: quantification of nuclear ERK5/CFL1 complex formation per cell (an average of 40 cells were scored) by PLA. Mean ± SD from 3 independent experiments.
(d) ChIP assay monitoring time course of recruitment of pERK5, pMAPK substrates, pSer5 RNA Pol II, and CFL1 to regulatory regions of E2-stimulated genes. Cells were treated with 10 nM E2, chromatin was cross-linked and precipitated DNA subjected to qPCR analysis. Values are mean ± SD from 3 independent experiments. TSS, transcription start site; BS, binding sites; CDS, coding sequence.
(e) ERK5 and CFL1 are recruited together to the TSS but not distal enhancer regions of E2 regulated genes. MCF-7 cells were treated with 10 nM E2 for 45 min, ERK5/DNA complexes were immunoprecipitated using specific ERK5 antibody or mouse IgG as a negative control. The complexes were eluted and subjected to a second immunoprecipitation with either CFL1 or rabbit IgG antibody. Immunoprecipitated DNA levels were measured by qPCR, and fold change over IgG/IgG pull-down was calculated.
(f) E2-mediated ERK5- RNA Pol II, CFL1-RNA Pol II, and PAF1-RNA Pol II interactions. MCF-7 cells transfected with 20 nM siCtrl, siERK5 or siCFL1 for 72h were treated with E2 for 45 min and PLA performed with specific antibodies. Quantification of nuclear ERK5/RNA Pol II or CFL1/RNA Pol II or PAF1/RNA Pol II complex formation per cell (an average of 30 cells were scored) by PLA used Fiji software. Mean ± SD from 3 independent experiments.
(g) Impact of knockdown of CFL1 or ERK5 on E2-stimulated genes. Cells transfected with 20 nM siGENOME reagent for Ctrl, CFL1 or ERK5 for 72h and then treated with vehicle or E2 for 24h. Total RNA was isolated, reverse transcribed and expression of several E2-stimulated genes examined by qPCR. Mean ± SD from 2 independent experiments.
Very interestingly, ChIP assays demonstrated that active ERK5, active RNA Pol II and CFL1 were all recruited to the TSS of E2 target genes with the same kinetics, and that this followed the pattern of enrichment of pMAPK substrates (Fig. 6d). ChIP-re-ChIP analysis showed that ERK5 and CFL1 are in the same complex that is recruited to the TSS but not to distal enhancers of ERα regulated genes (Fig. 6e). PLA demonstrated that E2-mediated RNA Pol II interaction with ERK5, CFL1, or the transcription elongation factor PAF1 was dependent upon the presence of both CFL1 and ERK5 (Fig. 6f). Moreover, knockdown of CFL1 mimicked ERK5 knockdown in dampening E2 stimulation of target genes (Fig. 6g). To examine the function of ERK5 in E2-ERα-mediated transcriptional effects, we overexpressed ERK5-DN and compared recruitment of several factors involved in transcription (Supplementary Fig. 7). ChIP assays demonstrated that ERK5-DN overexpression caused a loss of E2-stimulated recruitment of Polymerase Associated Factor-1 (PAF1) and an increase in chromatin recruitment of SPT5, a repressor of transcription elongation, whereas recruitments of RNA Pol II, WDR61, another component of the PAF1 complex, and the kinase Cyclin T1 were not affected. Thus, our findings uncover a novel mechanism for ERK5 and CFL1 function at chromatin, affecting transcription through recruitment of PAF1 and shedding of SPT5.
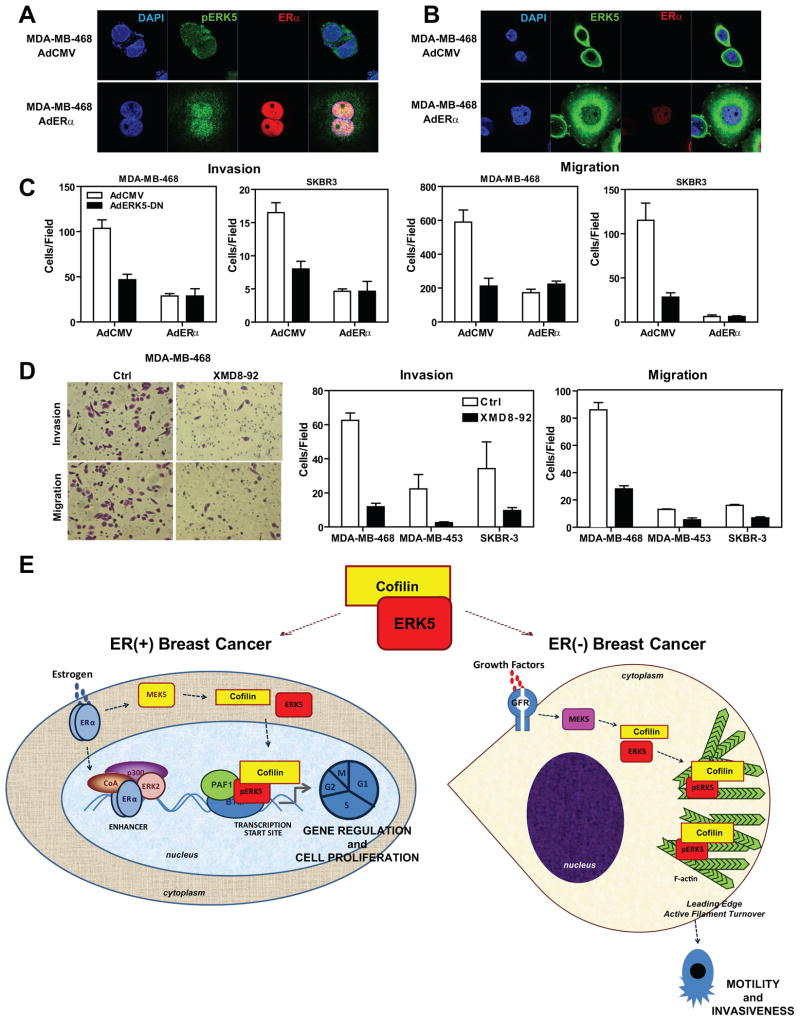
Expression of ERα in ER-negative cell lines induces nuclear localization of pERK5 and, coupled with ERK5 inhibition, abrogates cell migration and invasion
Since we observed an ERα-dependent switch of ERK5 intracellular localization in ERα-positive cells, we then performed the reverse experiments in ERα-negative cell lines. We introduced ERα by adenoviral infection into three ERα-negative breast cancer cell lines, MDA-MB-468, MDA-MB-453 and SKBR3. Introduction of ERα restored the nuclear localization of active ERK5 and localization of ERK5 to transcription factories in all three cell lines, while decreasing localization of ERK5 with F-actin in the cytoplasm (Figures 7a and 7b, Supplementary Figs.8 and 9). Moreover we observed increased cell spreading, a cytoskeletal feature suggesting that ERα causes major rearrangements in the actin cytoskeleton. In monitoring the impact of ERα introduction and ERK5 inhibition on cell motility and invasiveness of these cells, we observed that ERα or ERK5-DN greatly impaired the migration and invasiveness of these breast cancer cells (Fig. 7c). Similarly, use of the ERK5 inhibitor XMD8-92 also blocked migration and invasion of all three cell lines (Fig. 7d).
Figure 7. Expression of ERα in ERα-negative cell lines induces nuclear localization of pERK5, and ERK5 inhibition in these cells abrogates cell migration and invasion.
(a) Localization of pERK5 before and after ERα introduction into ERα-negative breast cancer cells. MDA-MB-468 cells were infected with AdCMV or AdERα for 96 h and immunofluorescence microscopy was performed with antibody specific to pERK5 and ERα. Nuclei were stained with DAPI.
(b) Localization of total ERK5 before and after ERα introduction to ERα-negative cells. MDA-MB-468 cells were treated as described in panel a, and immunofluorescence microscopy was performed with antibody specific to ERK5 and ERα. Nuclei were stained with DAPI.
(c) Impact of ERα introduction or ERK5 inhibition on invasion and migration of ERα-negative cells. MDA-MB-468 and SKBR3 cells were infected with AdCMV or AdERα for 72h and then reinfected with AdCMV or AdERK5-DN for 24h. Cells were then seeded on the upper chamber of transwell system for invasion assays (left two panels) or migration assays were performed (right two panels). Values are mean ± SD from 2 independent experiments.
(d) Impact of small molecule inhibitor of ERK5 (1 μM XMD8-92) on invasion and migration of ERα-negative cells. MDA-MB-468, MDA-MB-453 and SKBR3 cells were seeded on the upper chamber of transwell system for invasion assays (left two panels) or migration assays were performed (right two panels). Values are mean ± SD from 2 independent experiments.
(e) Model depicting modulation of the phenotypic properties of ERα(+) and ERα(−) breast cancer cells by ERK5 and CFL1. The two cell contexts are illustrated, ERα(+) on the left and ERα(−) on the right. In ERα(+) cells, treatment with hormone elicits recruitment of pERK5, CFL1, and PAF1 to the TSS of E2-regulated genes and regulation of gene expression, especially of genes that control cell proliferation. In ERα(−) cells, ERK5 and CFL1 stay in the cytoplasm and localize to sites of active actin turnover, thus contributing to motility and invasiveness of these ERα-negative cells.
DISCUSSION
In this work, we have uncovered a previously unknown relationship between a nuclear hormone receptor (ERα) and the MAPK family member, ERK5, which, working along with cofilin, an actin remodeling protein, control cell migration and invasiveness. Intriguingly, our studies in cells containing or lacking ERα, revealed that hormone activation of ERα determines the intracellular localization of the kinase and cofilin, directing their binding at the transcription start site of estrogen-regulated genes to control gene expression and removing them from cytoplasmic regions of high actin remodeling, thereby reducing cancer cell motility and invasiveness. This dynamic interplay between nuclear receptor-mediated transcription and actin reorganization links these two key cell regulatory processes to phenotypes of cancer indolence or aggressiveness, and may underlie the generally lower metastatic potential and better prognosis characteristic of many ERα-containing breast cancers.
These findings are schematized in the model in Fig. 7e. In ERα-containing breast cancer cells, ERα is directly involved in modulating ERK5 and CFL1 intracellular localization, directing them for control of transcription of target genes together with basal transcription machinery and excluding them from their cytoplasmic pro-metastatic actions (Fig. 7e, left). This nuclear function of ERK5 and CFL1 thereby redirected and reduced their localization and activity at the actin cytoskeleton, decreasing the invasive potential of ERα-positive cells. In ERα-negative cells, which lack this regulatory mechanism, ERK5 and CFL1 are localized mainly to the actin cytoskeleton where they modulate actin turnover and increase cell invasiveness (Fig. 7e, right). Notably, we were able to reverse this phenotype by either re-expressing ERα, or by using ERK5 inhibitors, which could provide a new and potentially valuable therapeutic approach for reducing the aggressive and metastatic activity that is characteristic of many ERα-negative tumors. In this regard, it is of interest that inhibition of ERK5 has been reported to sensitize breast cancer cells to the action of therapeutic agents, such as anti-HER2 (35), suggesting that ERK5 represents a target of interest that might improve the efficacy of currently used cancer treatment agents. In addition, our findings suggest that monitoring the intracellular localization of ERK5 should prove to be a useful biomarker for disease prognosis and for predicting responsiveness to therapies.
ERK5, a member of the MAPK family, shows highest sequence homology to ERK1 and ERK2, yet it contains an extra C-terminal transactivation domain and a NLS, which renders this kinase a very attractive candidate for regulating both nuclear and cytoplasmic events (36). ERK5 is present in most human breast tumors and is overexpressed in 20% of them (35). We observed that high expression of a gene signature rich in genes regulated by ERα and ERK5 and possessing binding sites for ERK5 was strongly correlated with a shorter time to recurrence and with reduced survival in patients with ERα-positive breast cancers, consistent with clinical findings that overexpression of ERK5 is strongly associated with decreased disease-free survival (35).
Our observations present new insights into chromatin-tethered MAPKs (24) and, for the first time, document a role for ERK5 directly on chromatin, actively modulating hormone-regulated gene expression programs. In addition, we identified ERα as the key factor in regulating the dynamic nuclear versus cytoplasmic localization of this kinase and in linking CFL1, an actin reorganizing protein, directly to ERK5 by showing that CFL1 was also recruited to the TSS of E2-regulated genes and that its nuclear localization was abrogated after treatment with ERK5 or ERα inhibitors.
As an actin-severing protein and regulator of actin dynamics, CFL1 is essential for the regulation of actin polymerization and depolymerization during cell migration and metastasis of tumor cells (37). Its expression is associated with the formation, stabilization, and maturation of invadopodia (31). Loss of the tumor suppressor PTEN, a frequent alteration in breast cancer, results in alterations in activation of CFL1, impacting the cell cytoskeletal phenotype (38). Hence, it is not surprising that there is accumulating evidence that CFL1 is a predictor of prognosis in several cancer types (39–42) and is a driver and biomarker of breast cancer metastasis (32, 33, 35). Our findings identified CFL1 as a requirement for ERK5 nuclear localization and for ERK5 and PAF1 interaction with RNA Pol II, and also documented CFL1 to be a crucial regulator of E2-mediated gene stimulation.
The paradigm we have observed for hormonal regulation by ERα, ERK5 and CFL1, and involvement of the actin cytoskeleton, may be utilized more generally in signaling induced by extracellular factors such as serum and DNA damage in a variety of cell types to alter the subcellular localization and functional activities of actin-regulated transcription factors. These include serum response factor (SRF) and its coregulators myocardin-related transcription factors (MRTFs and MAL), and the junction-mediating and regulatory protein JMY, a transcriptional coactivator of p53 (43–46). Our study is the first to reveal the dynamic interplay between hormone receptor-mediated transcription and the actin cell skeleton to regulate cancer phenotypes. Our findings also uncover the pivotal roles of the protein kinase ERK5 and cofilin in estrogen-induced receptor regulation, contributing to the increasing evidence for involvement of actin-dependent mechanisms, and MAPKs, in enhancing coordinated transcription of target genes and signaling by nuclear receptors and their coregulators (12, 47–49).
In summary, our study identifies an ERα-ERK5-CFL1 network that modulates the degree of breast cancer cell aggressiveness through coupling of actin reorganization and nuclear hormone receptor-mediated transcription. Our findings highlight possible new biomarkers for disease state and for predicting therapy outcomes, and they suggest that targeting ERK5 in combination with endocrine therapies in ERα-positive breast cancers might provide more effective treatment. In particular, our work emphasizes the promise of combined re-expression of ERα along with ERK5 inhibition in ERα-negative tumors, thereby decreasing their invasive potential and hopefully improving patient clinical outcome. This novel paradigm for regulation of cell behavior might apply more broadly to other nuclear receptors and transcription factors controlling cancer cell aggressiveness, and could form a new conceptual basis for future research.
Supplementary Material
Acknowledgments
We thank Alvaro Hernandez, Mark Band, and the staff of the Roy J. Carver and W.M. Keck Biotechnology Centers at the University of Illinois at Urbana-Champaign for their assistance.
Grant Support
This research was supported by grants from The Breast Cancer Research Foundation (BSK) and by NIH grants T32 ES007326 (ZME) and P50AT006268 (BSK) from the National Center for Complementary and Alternative Medicines (NCCAM), the Office of Dietary Supplements (ODS) and the National Cancer Institute (NCI).
Footnotes
Conflict of Interest Statement: The authors declare that they have no conflict of interest.
Accession Numbers and Data Availability
Gene expression data are available from the GEO Database under accession number GSE53394. ChIP-Seq data for ERK5, PXSP motif-containing proteins, and RNA Pol II are given as BED files in Table S3.
Supplementary Information accompanies the paper.
Disclosure of Potential Conflicts of Interest
The authors declare that they have no conflict of interest.
Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NCCAM, ODS, NCI, or the National Institutes of Health.
Authors’ Contributions
Conception and design: Z. Madak-Erdogan, B. Katzenellenbogen
Development of methodology: Z. Madak-Erdogan, R. Ventrella, L. Petry, B. Katzenellenbogen
Acquisition of Data: Z. Madak-Erdogan, R. Ventrella, L. Petry
Analysis and interpretation of data: (e.g., statistical analysis, biostatistics, computational analysis): Z. Madak-Erdogan, R. Ventrella, L. Petry, B. Katzenellenbogen
Writing, review, and/or revision of the manuscript: Z. Madak-Erdogan, R. Ventrella, L. Petry, B. Katzenellenbogen
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): Z. Madak-Erdogan, B. Katzenellenbogen
Study supervision: Z. Madak-Erdogan, B. Katzenellenbogen
References
- 1.Katzenellenbogen BS, Frasor J. Therapeutic targeting in the estrogen receptor hormonal pathway. Semin Oncol. 2004;31:28–38. doi: 10.1053/j.seminoncol.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 2.Frasor J, Danes JM, Komm B, Chang KC, Lyttle CR, Katzenellenbogen BS. Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology. 2003;144:4562–74. doi: 10.1210/en.2003-0567. [DOI] [PubMed] [Google Scholar]
- 3.Frasor J, Stossi F, Danes JM, Komm B, Lyttle CR, Katzenellenbogen BS. Selective estrogen receptor modulators: discrimination of agonistic versus antagonistic activities by gene expression profiling in breast cancer cells. Cancer Res. 2004;64:1522–33. doi: 10.1158/0008-5472.can-03-3326. [DOI] [PubMed] [Google Scholar]
- 4.Hah N, Danko CG, Core L, Waterfall JJ, Siepel A, Lis JT, et al. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145:622–34. doi: 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481:389–93. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soulez M, Parker MG. Identification of novel oestrogen receptor target genes in human ZR75-1 breast cancer cells by expression profiling. J Mol Endocrinol. 2001;27:259–74. doi: 10.1677/jme.0.0270259. [DOI] [PubMed] [Google Scholar]
- 7.Bergamaschi A, Christensen BL, Katzenellenbogen BS. Reversal of endocrine resistance in breast cancer: interrelationships among 14-3-3zeta, FOXM1, and a gene signature associated with mitosis. Breast Cancer Res. 2011;13:R70. doi: 10.1186/bcr2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Creighton CJ, Hilger AM, Murthy S, Rae JM, Chinnaiyan AM, El-Ashry D. Activation of mitogen-activated protein kinase in estrogen receptor alpha-positive breast cancer cells in vitro induces an in vivo molecular phenotype of estrogen receptor alpha-negative human breast tumors. Cancer Res. 2006;66:3903–11. doi: 10.1158/0008-5472.CAN-05-4363. [DOI] [PubMed] [Google Scholar]
- 9.Osborne CK, Schiff R. Mechanisms of endocrine resistance in breast cancer. Annu Rev Med. 2011;62:233–47. doi: 10.1146/annurev-med-070909-182917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schiff R, Massarweh S, Shou J, Osborne CK. Breast cancer endocrine resistance: how growth factor signaling and estrogen receptor coregulators modulate response. Clin Cancer Res. 2003;9:447S–54S. [PubMed] [Google Scholar]
- 11.Shou J, Massarweh S, Osborne CK, Wakeling AE, Ali S, Weiss H, et al. Mechanisms of tamoxifen resistance: increased estrogen receptor-HER2/neu cross-talk in ER/HER2-positive breast cancer. J Natl Cancer Inst. 2004;96:926–35. doi: 10.1093/jnci/djh166. [DOI] [PubMed] [Google Scholar]
- 12.Madak-Erdogan Z, Lupien M, Stossi F, Brown M, Katzenellenbogen BS. Genomic collaboration of estrogen receptor alpha and extracellular signal-regulated kinase 2 in regulating gene and proliferation programs. Mol Cell Biol. 2011;31:226–36. doi: 10.1128/MCB.00821-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bravo-Cordero JJ, Magalhaes MA, Eddy RJ, Hodgson L, Condeelis J. Functions of cofilin in cell locomotion and invasion. Nat Rev Mol Cell Biol. 2013;14:405–15. doi: 10.1038/nrm3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stossi F, Madak-Erdogan Z, Katzenellenbogen BS. Macrophage-elicited loss of estrogen receptor-alpha in breast cancer cells via involvement of MAPK and c-Jun at the ESR1 genomic locus. Oncogene. 2012;31:1825–34. doi: 10.1038/onc.2011.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Madak-Erdogan Z, Charn TH, Jiang Y, Liu ET, Katzenellenbogen JA, Katzenellenbogen BS. Integrative genomics of gene and metabolic regulation by estrogen receptors alpha and beta, and their coregulators. Mol Syst Biol. 2013;9:676. doi: 10.1038/msb.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Madak-Erdogan Z, Katzenellenbogen BS. Aryl hydrocarbon receptor modulation of estrogen receptor alpha-mediated gene regulation by a multimeric chromatin complex involving the two receptors and the coregulator RIP140. Toxicol Sci. 2012;125:401–11. doi: 10.1093/toxsci/kfr300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model–based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye T, Krebs AR, Choukrallah MA, Keime C, Plewniak F, Davidson I, et al. seqMINER: an integrated ChIP-seq data interpretation platform. Nucleic Acids Res. 2011;39:e35. doi: 10.1093/nar/gkq1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu T, Ortiz JA, Taing L, Meyer CA, Lee B, Zhang Y, et al. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol. 2011;12:R83. doi: 10.1186/gb-2011-12-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim K, Madak-Erdogan Z, Ventrella R, Katzenellenbogen BS. A MicroRNA196a2* and TP63 Circuit Regulated by Estrogen Receptor-alpha and ERK2 that Controls Breast Cancer Proliferation and Invasiveness Properties. Horm Cancer. 2013;4:78–91. doi: 10.1007/s12672-012-0129-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klein AM, Zaganjor E, Cobb MH. Chromatin-tethered MAPKs. Curr Opin Cell Biol. 2013 doi: 10.1016/j.ceb.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turjanski AG, Vaque JP, Gutkind JS. MAP kinases and the control of nuclear events. Oncogene. 2007;26:3240–53. doi: 10.1038/sj.onc.1210415. [DOI] [PubMed] [Google Scholar]
- 26.Barsyte-Lovejoy D, Galanis A, Clancy A, Sharrocks AD. ERK5 is targeted to myocyte enhancer factor 2A (MEF2A) through a MAPK docking motif. Biochem J. 2004;381:693–9. doi: 10.1042/BJ20031940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Q, Deng X, Lu B, Cameron M, Fearns C, Patricelli MP, et al. Pharmacological inhibition of BMK1 suppresses tumor growth through promyelocytic leukemia protein. Cancer Cell. 2010;18:258–67. doi: 10.1016/j.ccr.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu Y, Plutz M, Belmont AS. Hsp70 gene association with nuclear speckles is Hsp70 promoter specific. J Cell Biol. 2010;191:711–9. doi: 10.1083/jcb.201004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 30.Chan AY, Bailly M, Zebda N, Segall JE, Condeelis JS. Role of cofilin in epidermal growth factor-stimulated actin polymerization and lamellipod protrusion. J Cell Biol. 2000;148:531–42. doi: 10.1083/jcb.148.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamaguchi H, Lorenz M, Kempiak S, Sarmiento C, Coniglio S, Symons M, et al. Molecular mechanisms of invadopodium formation: the role of the N-WASP-Arp2/3 complex pathway and cofilin. J Cell Biol. 2005;168:441–52. doi: 10.1083/jcb.200407076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghosh M, Song X, Mouneimne G, Sidani M, Lawrence DS, Condeelis JS. Cofilin promotes actin polymerization and defines the direction of cell motility. Science. 2004;304:743–6. doi: 10.1126/science.1094561. [DOI] [PubMed] [Google Scholar]
- 33.Meira M, Masson R, Stagljar I, Lienhard S, Maurer F, Boulay A, et al. Memo is a cofilin-interacting protein that influences PLCgamma1 and cofilin activities, and is essential for maintaining directionality during ErbB2-induced tumor-cell migration. J Cell Sci. 2009;122:787–97. doi: 10.1242/jcs.032094. [DOI] [PubMed] [Google Scholar]
- 34.Wang W, Eddy R, Condeelis J. The cofilin pathway in breast cancer invasion and metastasis. Nat Rev Cancer. 2007;7:429–40. doi: 10.1038/nrc2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Montero JC, Ocana A, Abad M, Ortiz-Ruiz MJ, Pandiella A, Esparis-Ogando A. Expression of Erk5 in early stage breast cancer and association with disease free survival identifies this kinase as a potential therapeutic target. PLoS One. 2009;4:e5565. doi: 10.1371/journal.pone.0005565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim KY, Levin DE. Mpk1 MAPK association with the Paf1 complex blocks Sen1-mediated premature transcription termination. Cell. 2011;144:745–56. doi: 10.1016/j.cell.2011.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang W, Mouneimne G, Sidani M, Wyckoff J, Chen X, Makris A, et al. The activity status of cofilin is directly related to invasion, intravasation, and metastasis of mammary tumors. J Cell Biol. 2006;173:395–404. doi: 10.1083/jcb.200510115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vitolo MI, Boggs AE, Whipple RA, Yoon JR, Thompson K, Matrone MA, et al. Loss of PTEN induces microtentacles through PI3K-independent activation of cofilin. Oncogene. 2013;32:2200–10. doi: 10.1038/onc.2012.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muller CB, de Barros RL, Castro MA, Lopes FM, Meurer RT, Roehe A, et al. Validation of cofilin-1 as a biomarker in non-small cell lung cancer: application of quantitative method in a retrospective cohort. J Cancer Res Clin Oncol. 2011;137:1309–16. doi: 10.1007/s00432-011-1001-5. [DOI] [PubMed] [Google Scholar]
- 40.Pastor MD, Nogal A, Molina-Pinelo S, Melendez R, Salinas A, Gonzalez De la Pena M, et al. Identification of proteomic signatures associated with lung cancer and COPD. J Proteomics. 2013;89:227–37. doi: 10.1016/j.jprot.2013.04.037. [DOI] [PubMed] [Google Scholar]
- 41.Peng XC, Gong FM, Zhao YW, Zhou LX, Xie YW, Liao HL, et al. Comparative proteomic approach identifies PKM2 and cofilin-1 as potential diagnostic, prognostic and therapeutic targets for pulmonary adenocarcinoma. PLoS One. 2011;6:e27309. doi: 10.1371/journal.pone.0027309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang ZL, Miao X, Xiong L, Zou Q, Yuan Y, Li J, et al. CFL1 and Arp3 are biomarkers for metastasis and poor prognosis of squamous cell/adenosquamous carcinomas and adenocarcinomas of gallbladder. Cancer Invest. 2013;31:132–9. doi: 10.3109/07357907.2012.756113. [DOI] [PubMed] [Google Scholar]
- 43.Medjkane S, Perez-Sanchez C, Gaggioli C, Sahai E, Treisman R. Myocardin-related transcription factors and SRF are required for cytoskeletal dynamics and experimental metastasis. Nat Cell Biol. 2009;11:257–68. doi: 10.1038/ncb1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miralles F, Posern G, Zaromytidou AI, Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–42. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- 45.Vartiainen MK, Guettler S, Larijani B, Treisman R. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science. 2007;316:1749–52. doi: 10.1126/science.1141084. [DOI] [PubMed] [Google Scholar]
- 46.Zuchero JB, Belin B, Mullins RD. Actin binding to WH2 domains regulates nuclear import of the multifunctional actin regulator JMY. Mol Biol Cell. 2012;23:853–63. doi: 10.1091/mbc.E11-12-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu Q, Kwon YS, Nunez E, Cardamone MD, Hutt KR, Ohgi KA, et al. Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proc Natl Acad Sci U S A. 2008;105:19199–204. doi: 10.1073/pnas.0810634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang W, Ghisletti S, Saijo K, Gandhi M, Aouadi M, Tesz GJ, et al. Coronin 2A mediates actin-dependent de-repression of inflammatory response genes. Nature. 2011;470:414–8. doi: 10.1038/nature09703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Madak-Erdogan Z, Kieser KJ, Kim SH, Komm B, Katzenellenbogen JA, Katzenellenbogen BS. Nuclear and extranuclear pathway inputs in the regulation of global gene expression by estrogen receptors. Mol Endocrinol. 2008;22:2116–27. doi: 10.1210/me.2008-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–52. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–9. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.