Abstract
Background
Several bacterial plant pathogens colonize their hosts through the secretion of effector proteins by a Type III protein secretion system (T3SS). The role of T3SS in bacterial pathogenesis is well established but whether this system is involved in multicellular processes, such as bacterial biofilm formation has not been elucidated. Here, the phytopathogen Xanthomonas citri subsp. citri (X. citri) was used as a model to gain further insights about the role of the T3SS in biofilm formation.
Results
The capacity of biofilm formation of different X. citri T3SS mutants was compared to the wild type strain and it was observed that this secretion system was necessary for this process. Moreover, the T3SS mutants adhered proficiently to leaf surfaces but were impaired in leaf-associated growth. A proteomic study of biofilm cells showed that the lack of the T3SS causes changes in the expression of proteins involved in metabolic processes, energy generation, exopolysaccharide (EPS) production and bacterial motility as well as outer membrane proteins. Furthermore, EPS production and bacterial motility were also altered in the T3SS mutants.
Conclusions
Our results indicate a novel role for T3SS in X. citri in the modulation of biofilm formation. Since this process increases X. citri virulence, this study reveals new functions of T3SS in pathogenesis.
Keywords: Xanthomonas citri subsp. citri, Biofilm, T3SS, Proteomics
Background
The bacterial genus Xanthomonas comprises a number of Gram-negative plant pathogenic bacteria that cause a variety of severe plant diseases [1]. Xanthomonas citri subsp. citri, the phytopathogen causing citrus canker, invades host plant tissues entering through stomata or wounds and then colonizes the apoplast of fruit, foliage and young stems, causing raised corky lesions and finally breaking the epidermis tissue due to cell hyperplasia, thus allowing bacterial dispersal to other plants [2].
Persistent and severe disease can lead to defoliation, dieback and fruit drop, reducing yields and causing serious economic losses [3]. To date, no commercial citrus cultivars are resistant to citrus canker and current control methods are insufficient to manage the disease [3]. Thus, there is a need to study the infection process in order to enable the development of new tools for disease control. Furthermore, the study of X. citri-citrus interactions has been used as a model to provide new advances in the understanding of plant-pathogen interactions [1].
The Type III protein secretion system (T3SS) is conserved in many Gram-negative plant and animal pathogenic bacteria [4]. The T3SS is subdivided into (i) the non-flagellar T3SS (T3aS) involved in the assembly of the injectisome or hypersensitive response and pathogenicity (Hrp) pilus, and (ii) the flagellar T3SS (T3bS), responsible for assembly of the flagellum [5]. The T3SS spans both bacterial membranes and is associated with an extracellular filamentous appendage, termed ‘needle’ in animal pathogens and ‘Hrp pilus’ in plant pathogens, which is predicted to function as a protein transport channel to the host-pathogen interface [4]. Translocation of effector proteins across the host membrane requires the presence of the T3SS translocon, a predicted protein channel that consists of bacterial Type III-secreted proteins [6].
A number of surface appendages, such as conjugative pili, flagella, curli, and adhesins have been shown to play a role in biofilm formation [7,8]. The role of T3SS as an effector protein delivery machine is well established, however, whether this secretion system participates in multicellular processes such as biofilm formation remains unanswered. Several studies concluded that T3SS is only necessary for pathogenicity and that expression of this secretion system is repressed in biofilm-growing bacteria. For example, Pseudomonas aeruginosa PA14 sadRS mutant strains that cannot form biofilms have enhanced expression of T3SS genes, while a P. aeruginosa PA14 T3SS mutant exhibits enhanced biofilm formation compared to wild type strain [9]. Furthermore, in Yersinia pseudotuberculosis, it has been shown that the T3SS needle blocks biofilm formation in the model host Caenorhabditis elegans[10]. In contrast, other studies highlighted the role of T3SS in bacterial biofilm formation. Microarray experiments performed in P. aeruginosa cystic fibrosis epidemic strain AES-2 showed expression of T3SS encoding genes up-regulated in biofilms as compared to planktonic bacteria [11]. In the plant pathogen Erwinia chrysanthemi, it has been shown that the T3SS pilus is involved in the aggregative multicellular behavior that leads to pellicle formation [12]. The enterohemorrhagic Escherichia coli O157 has a well-defined T3SS, termed E. coli Type III secretion system 1 (ETT1), which is involved in attachment and effacement and is critical for virulence. This strain also has a gene cluster potentially encoding an additional T3SS (ETT2) [13]. Studies on an ETT2 deletion mutant strain showed that although ETT2 is not responsible for protein secretion, it is involved in biofilm formation and hence in virulence [13]. Recently, it has been shown that the Salmonella enterica serovar Typhimurium T3SS secretion system SPI-1 is involved in the formation of an adherent biofilm and cell clumps in the culture media [14]. Taken together, the evidence suggests that T3SS may play a role in bacterial biofilm formation.
In X. citri, biofilm formation is required for optimal virulence as revealed by several reports with different bacterial mutants. For instance, X. citri mutants that are unable to biosynthesize molecules needed for biofilm formation such as exopolysaccharide (EPS), an adhesin protein and the lipopolysaccharide show a reduced virulence [15-17]. Consistent with this, X. citri infection is reduced by foliar application of compounds that are able to inhibit X. citri biofilm formation [18]. The role of X. citri T3SS in pathogenicity is well known since T3SS mutants are unable to grow in host plants indicating that X. citri T3SS is responsible for the secretion of effector proteins [19]. Taking into account that biofilm formation is a requirement for X. citri to achieve full virulence, we have characterized the ability of a T3SS mutant to form biofilms and by performing a proteomic analysis we have identified differentially expressed proteins with a view to obtain a greater understanding of this process.
Results
The T3SS contributes to X. citri in vitro biofilm formation
In order to study the role of the T3SS in X. citri biofilm formation, a X. citri T3SS mutant in the hrpB operon termed hrpB− mutant [19] was characterized in their ability to form a biofilm compared to the wild type strain. The hrpB− mutant was previously obtained by single crossover plasmid integration in the region that comprises the 3′ end of hrpB5 and the 5′ region of the ATPase hrcN[19] (Additional file 1: Figure S1A). Here, in order to complement this strain, hrpB5 and hrcN were cloned in frame in the expression plasmid pBBR1MCS-5 under the control of the lacZ promoter [20]. However, the resulting strain did not restore biofilm formation or pathogenicity (data not shown) suggesting that downstream genes of the hrpB operon, hrpB7 and hrcT, may be also affected in the hrpB− mutant due to polarity effects (Additional file 1: Figure S1A). Therefore, the entire region containing hrpB5, hrcN, hrpB7 and hrcT was cloned in the pBBR1MCS-5 vector (Additional file 1: Figure S1A) and the resulting strain (hrpB−c) was tested for its ability to trigger HR in non-host plants and disease in citrus leaves (Additional file 1: Figure S1B and S1C). As expected, the HR response in non-host plants was similar for the hrpB−c strain and X. citri (Additional file 1: Figure S1B). In host tissue infections, the hrpB−c strain did cause lesions, though it was less virulent than X. citri, showing a reduction in water soaking and in canker lesion formation (Additional file 1: Figure S1C). A partial complementation was also observed by RT-qPCR assays of CsLOB1. This gene encodes a protein that is a member of the Lateral Organ Boundaries (LOB) gene family of transcription factors whose expression is induced by the X. citri TAL effector protein PthA4 [21,22]. As expected, in leaves infected with X. citri, an induction of CsLOB1 was observed, the hrpB− mutant did not induce the expression of this gene suggesting that this mutant is not secreting PthA4 and the hrpB−c strain induced CsLOB1 expression albeit at lower levels than X. citri probably due a lower amount of PthA4 secreted by this strain (Additional file 1: Figure S1D). Given of the possibility that bacteria may be loosing the plasmids during the host plant assays, bacteria were extracted from plant tissues and quantified at different times using appropriate antibiotics and no loss of plasmid was observed even 30 days after infiltrations (data not shown). Therefore, this partial complementation may be due to the fact that these genes are expressed under the lacZ promoter and that expression levels are likely to be somewhat different from those of the endogenous genes. This proposition is supported by recent work that shows that lac promoter-driven expression of hrpB1 only partially complemented the hrpB1 mutant phenotype in susceptible plants, while complete complementation was observed for HR in pathogen resistant plants [23]. For the biofilms assays, first the strains were cultured statically in 24-well PVC plates in XVM2. After seven days of growth, X. citri and hrpB−c strain were able to form mature biofilms with a conformation similar of that previously observed for X. citri strain [16], while the hrpB− mutant showed impaired biofilm formation (Figure 1A). Next, the strains were grown statically in borosilicate glass tubes in XVM2 medium for seven days. Staining of bacterial cells with the specific crystal violet (CV) stain showed that under these conditions X. citri and hrpB−c strain can form biofilms of cells that adhere to the glass surface forming a thick ring at the air–liquid surface–interface of the culture medium, whereas the hrpB− mutant was altered in its ability to form such a structure. Instead, the hrpB− mutant formed only a narrow ring of cells (Figure 1B). CV staining of X. citri and hrpB−c strains was over nine times greater than that of the hrpB− mutant (p < 0.05) (Figure 1C), thereby confirming a reduction in the capacity of biofilm formation for the mutant. Since the hrpB− mutant is a polar mutant, in order to discern whether the hrpB5-hrcT genes or the ‘Hrp pilus’ are involved in the process of biofilm formation, the hrpD− and hrpF− mutants previously obtained were analyzed [19] (Additional file 1: Figure S1A). These two mutants, like the hrpB− mutant, were impaired in biofilms formation (Figure 1A, 1B and 1C). All strains showed similar growth rates in XVM2 medium under agitation, with a generation time of 200 min, indicating that mutations of hrp genes do not impair growth of the hrp mutants in vitro (data not shown). Further, differences in statically growing cells were analyzed by confocal laser scanning microscopy using X. citri and hrpB− strains transformed with a pBBR1MCS-5 vector that carries a copy of the gfp gene (pBBR1MCS-5EGFP). The analysis showed that X. citri formed large clusters of aggregated cells that were not observed in the hrpB− mutant (Figure 2). Moreover, serial images taken at 0.5 μm-distance (vertical z-stack) covering the entire well length revealed that X. citri formed thick bacterial biofilms of about 250 μm deep, while the hrpB− mutant formed narrower unstructured biofilms of 50 μm in length (Figure 2).
Figure 1.
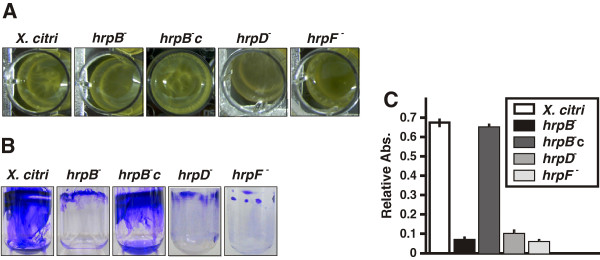
Biofilm assays for X. citri, the hrp mutants and the hrpB−c strain. Representative photographs of biofilm formation assays for X. citri, hrp mutants and hrpB−c strains grown statically in 24-well PVC plates (A) or in borosilicate glass tubes (B) for seven days in XVM2 medium. (C) Quantification of biofilm formation by CV stain measured spectrophotometrically (Abs. at 600 nm). Relative Abs. indicates: CV Abs. 600 nm/Planktonic cells Abs. 600 nm. Values represent the mean from seven tubes for each strain. Error bars indicate the standard deviation.
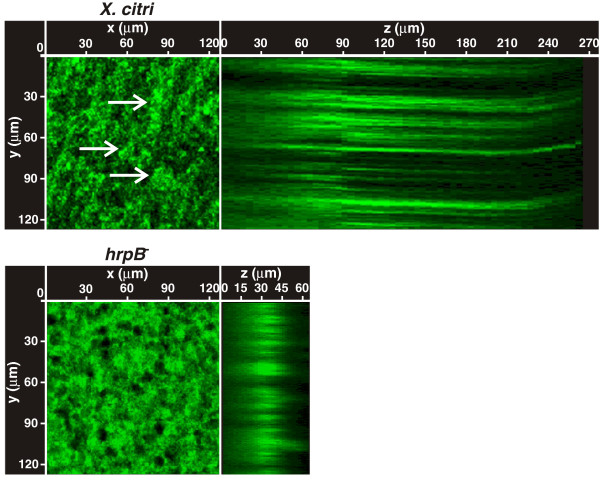
Figure 2.
Confocal laser scanning microscopy analysis of X. citri and hrpB− strains grown statically. GFP-expressing X. citri and hrpB− strains cultured statically in vitro were analyzed by confocal laser scanning microscopy, serial images were taken at 0.5 μm distances (vertical z-stack). Z represents the ZX axis projected images. At the XY images, white arrows point to X. citri clusters of aggregated cells.
The T3SS is not required for attachment to host tissue but is necessary for X. citri biofilm formation on the leaf surface
The role of X. citri T3SS in bacterial adherence, like attachment to plant tissue, was evaluated by quantitative measurement of CV staining of adhered cells to leaf tissues. X. citri wild type and all the hrp mutants adhered proficiently and to the same extent to leaf tissues (p < 0.05) (Figure 3A), indicating that T3SS is not involved in leaf surface attachment. In order to analyze biofilm growth of GFP-expressing X. citri and hrpB− strains on host leaf surfaces, bacterial drops were spread over the abaxial surface of citrus leaves and growth was examined confocal laser scanning microscopy. Under these conditions, X. citri cells grew and formed biofilm structures over the entire area of the drops on the leaf surface, with a higher density of cells accumulated at the border forming a circle (Figure 3B). The hrpB− mutant growth was limited compared to X. citri, forming only small cell cumuli at the center and a narrower border circle. Further examination of the 0.5 μm stacks at the circle borders showed that X. citri formed a thicker bacterial biofilm of about 20 μm, while the hrpB− mutant formed a narrower border of about 7.5 μm. These results indicate that the absence of the T3SS negatively affects biofilm formation.
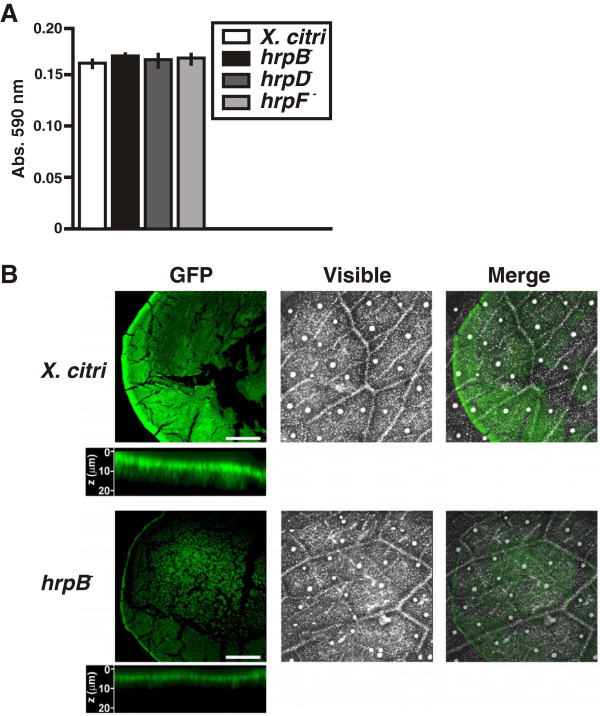
Figure 3.
Adherence of the hrp mutants to citrus leaf tissues and confocal laser scanning microscopy analysis on citrus leaves of X. citri and hrpB− strains. (A) Quantitative measurement of the CV retained by X. citri and hrp mutant strains adhered to abaxial leaf surfaces. Values represent the means of 20 quantified stained drops for each strain. Error bars indicate standard deviations. (B) Representative photographs of confocal laser scanning microscopy analysis of GFP-expressing X. citri and hrpB− cells grown on leaf surfaces. Below each of the fluorescent photographs of both strains, the ZX axis projected images accumulated over serial imaging taken at 0.5 μm distances (z-stack) are shown. Scale bars: 0.5 mm.
T3SS is required for X. citri leaf-associated survival
The expression profiles of genes involved in T3SS formation such as hrpG and hrpX, encoding for the two regulators of the hrp cluster [24], and hrpE, the major structural component of the ‘Hrp pilus’ [25] were evaluated in X. citri cells recovered from leaf surfaces at different times by RT-qPCR assays. A significant induction of the expression of these genes (p < 0.05) was detected after two days post-spraying of the bacteria on leaf surfaces (Figure 4A). Next, populations of the different strains were quantified at different times post-spraying on citrus leaf surfaces. One week after initial inoculation, the population size of X. citri decreased by almost one order of magnitude. Under these conditions, X. citri cannot enter through the tissue and replicate due to the thickness of the citrus leaf cuticle [16]. As a consequence, bacterial cell numbers remained relatively steady throughout the subsequent three weeks of growth. The population size of X. citri was nearly one order of magnitude higher at every time point analyzed (p < 0.05) as compared to the hrp mutants (Figure 4B). The population of the hrpB−c did not achieve X. citri levels, but was ever higher than that of the hrp mutants (Figure 4B). These results support the idea that the T3SS is necessary for X. citri colonization of the phyllosphere, which may be due, at least partly, to the role of T3SS in X. citri biofilm formation.
Figure 4.
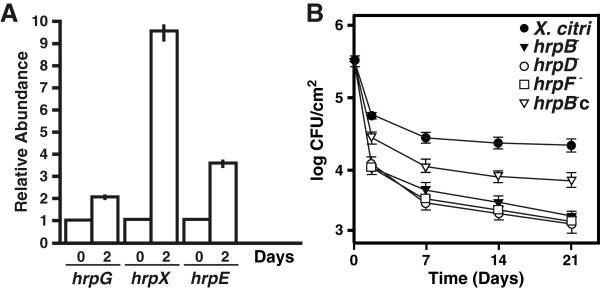
Analysis of T3SS gene expression in leaf-associated grown X. citri and survival of X. citri, hrp mutants and hrpB−c cells associated to leaves. (A) RT-qPCR to determine hrpG, hrpX and hrpE expression levels in X. citri grown associated to leaves. Bars indicate the expression levels of the T3SS genes at two days of leaf-associated growth relative to time 0. Values are the means of four biological replicates with three technical replicates each. (B)X. citri, hrp mutants and hrpB−c strains leaf-associated survival on citrus leaves. Values represent an average of four leaves assayed for each strain. Error bars indicate the standard deviation.
Proteomic analysis of statically cultured X. citri and hrpB− strains
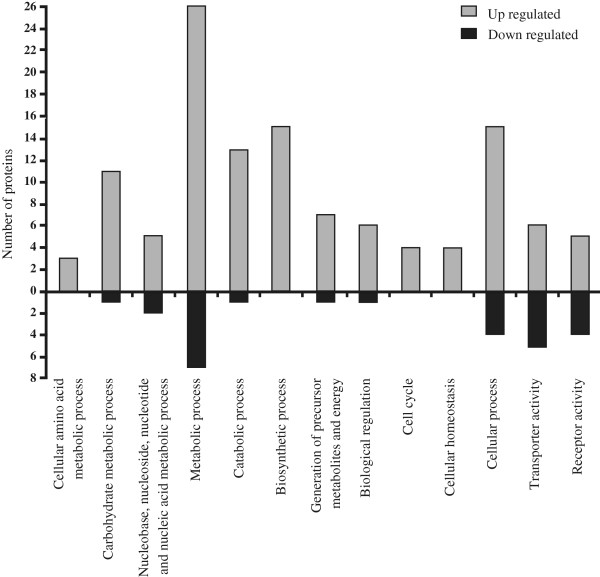
In order to gain new insights about the role of T3SS in biofilm formation, a proteomic analysis was performed to identify differentially expressed proteins between X. citri and the hrpB− mutant grown statically. A total of 49 differentially expressed protein spots were detected of which 32 were up- and 17 down-regulated in the hrpB- mutant in comparison to X. citri (Table 1). Identified proteins were used to determine enriched GO categories in biological processes and molecular function. The main enriched categories for the up- and down-regulated proteins with an average fold change of minimum ± 1.5 and p value < 0.05 in the hrpB− mutant relative to X. citri were represented graphically (Figure 5). The categories that showed a major enrichment in the up-regulated proteins in the hrpB- mutant include ‘metabolic process’, ‘catabolic process’, ‘biosynthetic process’ and ‘generation of precursor metabolites and energy’. Moreover, ‘cell cycle’, ‘cellular homeostasis’ and ‘cellular process’ were categories enriched in up-regulated proteins in this mutant. Most of the identified proteins in the categories of ‘transporter activity’ or ‘receptor activity’ belong to different classes of outer membrane proteins (OMPs) such as: FadL (XAC0019), that allows the passage of fatty acids [26], OmpW (XAC3664), involved in the transport of small hydrophilic molecules across the bacterial outer membrane [27] and RpfN (XAC2504), which was reported to play a role in carbohydrate transport [28]. In these categories also several TonB-dependent transporters (TBDTs), which are outer membrane transporters involved in the active uptake and/or in signal transduction [29], as well as two Oar (OmpA-related) proteins were detected as differentially expressed between the two strains.
Table 1.
Differentially expressed protein spots between X. citri and hrpB − strains statically cultured in XVM2 with a change abundance of minimum 1.5 fold and p value of < 0.05 (ANOVA)
| X. citri gene no. | Protein name | MOWSE score | Accession no. | Predicted MW/p I | Observed MW/p I | Peptide match/coverage | Fold change compared to WT |
|---|---|---|---|---|---|---|---|
| 01 Metabolism | |||||||
| 01.01 Amino acid metabolism | |||||||
| XAC0125 |
Aspartate/tyrosine/aromatic aminotransferase |
350 |
Q8PR41_XANAC |
43.3/5.72 |
49.0/4.8 |
19/38% |
1.9 |
| XAC4034 |
Shikimate 5-dehydrogenase |
297 |
AROE_XANAC |
29.9/4.93 |
30.0/5.9 |
19/17% |
2.4 |
| XAC2717 |
Tryptophan synthase subunit b |
31 |
TRPB_XANAC |
43.3/5.88 |
53.0/4.6 |
2/4% |
7.5 |
| XAC3709 |
Tryptophan repressor binding protein |
48 |
Q8PGA8_XANAC |
20.0/6.40 |
10.0/4.4 |
3/17% |
−1.6 |
| 01.02 Nitrogen, sulfur and selenium metabolism | |||||||
| XAC0554 |
NAD(PH) nitroreductase |
208 |
Y554_XANAC |
21.0/5.83 |
18.0/4.7 |
14/38% |
4.6 |
| 01.03 Nucleotide/nucleoside/nucleobase metabolism | |||||||
| XAC1716 |
CTP-synthase |
125 |
PYRG_XANAC |
61.7/5.91 |
67.0/4.5 |
14/21% |
3.5 |
| 01.05 C-compounds and carbohydrate metabolism | |||||||
| XAC2077 |
Succinate dehydrogenase flavoprotein subunit |
192 |
Q8PKT5_XANAC |
65.8/5.89 |
66.0/4.6 |
20/25% |
2.2 |
| XAC1006 |
Malate dehydrogenase |
1054 |
MDH_XANAC |
34.9/5.37 |
45.0/5.4 |
55/50% |
−1.8 |
| XAC3579 |
Phosphohexose mutases (XanA) |
98 |
Q8PGN7_XANAC |
49.1/5.29 |
54.0/5.6 |
7/10% |
1.7 |
| XAC3585 |
DTP-glucose 4,6-dehydratase |
235 |
Q8PGN1_XANAC |
38.6/5.86 |
48.0/4.7 |
12/17% |
2.1 |
| XAC0612 |
Cellulase |
245 |
Q8PPS3_XANAC |
51.6/5.76 |
57.0/4.9 |
23/32% |
2.6 |
| XAC3225 |
Transglycosylase |
178 |
Q8PHM6_XANAC |
46.2/5.89 |
53.0/4.8 |
14/22% |
−1.6 |
| 01.06 Lipid, fatty acid and isoprenoid metabolism | |||||||
| XAC3300 |
Putative esterase precursor (EstA) |
96 |
Q8PHF7_XANAC |
35.9/6.03 |
62.0/6.2 |
3/4% |
−3.1 |
| XAC1484 |
Short chain dehydrogenase precursor |
104 |
Q8PME5_XANAC |
26.0/5.97 |
30.0/4.4 |
5/9% |
2.2 |
| 01.06.02 Membrane lipid metabolism | |||||||
| XAC0019 |
Outer membrane protein (FadL) |
167 |
Q8PRE4_XANAC |
47.3/5.18 |
46.0/6.1 |
8/10% |
−10.0 |
| XAC0019 |
Outer membrane protein (FadL) |
79 |
Q8PRE4_XANAC |
47.3/5.18 |
35.0/6.0 |
7/13% |
−6.2 |
| 01.20 Secondary metabolism | |||||||
| XAC4109 |
Coproporphyrinogen III oxidase |
46 |
HEM6_XANAC |
34.6/5.81 |
37.0/4.9 |
8/19% |
1.5 |
| 02 Energy | |||||||
| 02.01 Glycolysis and gluconeogenesis | |||||||
| XAC1719 |
Enolase |
90 |
ENO_XANAC |
46.0/4.93 |
55.0/5.9 |
7/13% |
1.7 |
| XAC3352 |
Glyceraldehyde-3-phosphate dehydrogenase |
267 |
Q8PHA7_XANAC |
36.2/6.03 |
46.0/4.4 |
24/28% |
2.6 |
| XAC2292 |
UTP-glucose-1-phosphate uridylyltransferase (GalU) |
92 |
Q8PK83_XANAC |
32.3/5.45 |
38.0/5.3 |
13/30% |
4.2 |
| 02.07 Pentose phosphate pathway | |||||||
| XAC3372 |
Transketolase 1 |
85 |
Q8PH87_XANAC |
72.7/5.64 |
69.0/4.9 |
5/7% |
5.0 |
| 02.11 Electron transport and membrane-associated energy conservation | |||||||
| XAC3587 |
Electron transfer flavoprotein a subunit |
50 |
Q8PGM9_XANAC |
31.8/4.90 |
34.0/5.5 |
6/14% |
2.3 |
| 10 Cell cycle and DNA processing | |||||||
| 10.03 Cell cycle |
|
|
|||||
| XAC1224 |
Cell division topological specificity factor (MinE) |
33 |
MINE_XANAC |
9.6/5.37 |
12.0/4.9 |
1/14% |
2.7 |
| 10.03.03 Cytokinesis/septum formation and hydrolysis | |||||||
| XAC1225 |
Septum site-determining protein (MinD) |
143 |
Q8PN48_XANAC |
28.9/5.32 |
34.0/5.6 |
19/26% |
2.3 |
| 11 Transcription | |||||||
| XAC0996 |
DNA-directed RNA polymerase subunit a |
104 |
RPOA_XANAC |
36.3/5.58 |
33.0/5.0 |
5/7% |
−4.3 |
| XAC0966 |
DNA-directed RNA polymerase subunit b |
150 |
RPOC_XANAC |
155.7/7.82 |
35.0/4.6 |
16/8% |
−3.3 |
| 14 Protein fate (folding, modification and destination) | |||||||
| 14.01 Protein folding and stabilization | |||||||
| XAC0542 |
60 kDa chaperonin (GroEL) |
199 |
CH60_XANAC |
57.1/5.05 |
41.0/5.5 |
15/27% |
−11.2 |
| 16 Protein with binding function or cofactor requirement | |||||||
| XAC2726 |
Adenine-specific methylase (Dam methylase) |
84 |
Q8PJ19_XANAC |
33.9/4.70 |
41.0/6.2 |
10/12% |
−1.9 |
| XAC1362 |
GTN reductase |
44 |
Q8PMR4_XANAC |
39.4/5.37 |
50.0/5.3 |
7/10% |
2.3 |
| XAC3664 |
OmpW family outer membrane protein precursor |
226 |
Q8PN48_XANAC |
23.8/4.97 |
28.0/6.2 |
12/13% |
2.3 |
| 30 Cellular communication/Signal transduction mechanism | |||||||
| XAC0291 |
Oar protein ( TonB-dependent transporter) |
50 |
Q8PQN2_XANAC |
107.9/5.29 |
108.0/5.7 |
2/1% |
4.3 |
| XAC2672 |
Oar protein ( TonB-dependent transporter) |
280 |
Q8PJ70_XANAC |
117.4/5.10 |
90.0/5.9 |
19/18% |
2.4 |
| XAC4273 |
TonB-dependent transporter |
100 |
Q8PJL0_XANAC |
109.2/5.14 |
90.0/5.6 |
3/3% |
2.8 |
| XAC1143 |
TonB-dependent transporter |
576 |
Q8PND0_XANAC |
87.7/5.21 |
70.0/6.1 |
30/33% |
1.7 |
| XAC3050 |
TonB-dependent transporter |
596 |
Q8PI48_XANAC |
105.8/4.76 |
64.0/6.2 |
30/16% |
−3.0 |
| XAC3444 |
TonB-dependent transporter |
1280 |
Q8PH16_XANAC |
103.2/4.79 |
90.0/6.3 |
84/37% |
3.9 |
| XAC3168* |
TonB-dependent transporter |
98 |
Q8PHT1_XANAC |
87.3/5.20 |
59.0/6.0 |
3/3% |
−3.1 |
| XAC3166* |
TonB-dependent transporter |
410 |
Q8PHT3_XANAC |
84.5/4.95 |
69.0/6.1 |
22/18% |
−2.9 |
| XAC3489 |
TonB-dependent transporter |
685 |
Q8PGX3_XANAC |
88.9/4.93 |
69.0/5.9 |
40/24% |
−1.7 |
| XAC1413 |
Outer membrane protein assembly factor BamA |
135 |
Q8PML3_XANAC |
87.6/5.53 |
88.0/5.4 |
13/15% |
2.8 |
| 32 Cell rescue, defense and virulence | |||||||
| XAC2504* |
Regulator of pathogenicity factors (RpfN) |
271 |
Q8PJM6_XANAC |
41.3/5.98 |
49.0/4.4 |
21/16% |
−4.8 |
| XAC0907 |
Alkyl hydroperoxide reductase subunit C |
240 |
O06464_XANAC |
20.6/6.15 |
20.0/4.2 |
28/61% |
1.3 |
| 32.07 Cellular detoxification | |||||||
| XAC1474 |
Glutathione transferase |
39 |
Q8PMF5_XANAC |
23.9/6.06 |
22.0/4.7 |
4/8% |
1.7 |
| 34 Interaction with the environment |
|
|
|
|
|
|
|
| 34.01 Homeostasis |
|
|
|
|
|
|
|
| XAC1149 |
Bacterioferritin |
100 |
Q8PNC4_XANAC |
21.2/4.71 |
20.0/6.3 |
6/20% |
2.1 |
| XAC0493 |
Bacterioferritin |
152 |
Q8PQ38_XANAC |
18.3/4.80 |
12.0/6.5 |
19/43% |
2.5 |
| XAC1533 |
Dihydrolipoamide dehydrogenase |
336 |
Q8PM99_XANAC |
50.5/5.80 |
59.0/4.6 |
34/47% |
4.0 |
| 42 Biogenesis of cellular components |
|
|
|
|
|
|
|
| XAC1230 |
Putative membrane protein |
71 |
Q8PN43_XANAC |
43.1/6.88 |
24.0/4.4 |
4/11% |
−3.5 |
| 99 Unclassified proteins |
|
|
|
|
|
|
|
| XAC1262 |
Protein of unknown function (Aminopeptidase) |
121 |
Q8PN12_XANAC |
63.4/5.85 |
68.0/4.6 |
13/15% |
5.3 |
| XAC1344 | Protein of unknown function (CcmA) | 67 | Q8PMT2_XANAC | 18.7/5.45 | 23.0/5.7 | 4/18% | −1.7 |
*Protein spots 240 and 398 were previously named “ferric enterobactin receptor” are now classified as TonB-dependent transporter, while protein spot 31 previously identified as “carbohydrate selective porin” and is now classified as Regulator of pathogenicity factors.
Figure 5.
Gene ontology (GO) terms enriched in differentially expressed proteins between X. citri and hrpB− static cells. Proteins up-regulated and down-regulated in the hrpB− mutant relative to X. citri in the main enriched categories are shown. The GO enrichment analysis was performed using Blast2GO.
The lack a T3SS enhances X. citri EPS production and decreases bacterial motility
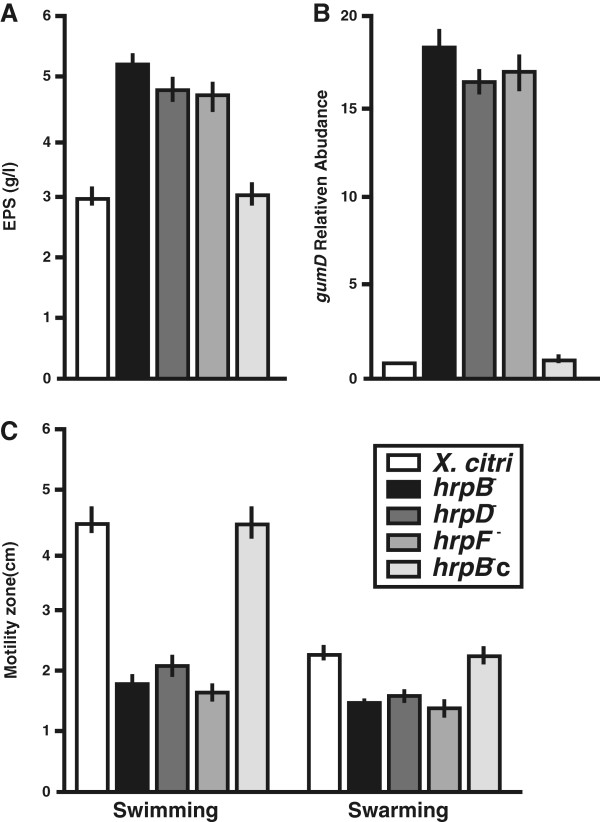
The proteomic assay detected an over-expression of the enzymes XanA and GalU in the hrpB− mutant compared to X. citri (Table 1). The XanA enzyme encodes a bifunctional phosphoglucomutase/phosphomannomutase involved in the synthesis of both glucose 1-phosphate and mannose 1-phosphate. Glucose 1-phosphate is then converted to UDP-glucose by GalU and mannose 1-phosphate to GDP-mannose by mannose 1-phosphate guanylyltransferase. These nucleotide sugars are directly implicated in EPS synthesis [30,31]. Production of EPS was measured in X. citri, the hrp mutants and the hrpB−c strains and results showed that EPS production in these mutants was over 1.7 times that in X. citri and hrpB−c strain (p < 0.05) (Figure 6A). Additionally, the expression of gumD, a gene encoding a protein of the EPS biosynthetic pathway, was analyzed by RT-qPCR in all the strains. The results showed that the transcript levels of gumD were over 17 times higher in hrp mutant strains as compared to X. citri and the hrpB−c strain (p < 0.05) (Figure 6B). Moreover, the proteomic analysis also showed a down-regulation of the outer membrane protein XAC0019 in the hrpB− mutant (Table 1) and recently, it has been shown that this protein is necessary for X. citri swimming [32]. Furthermore, CcmA that is required for bacterial motility [33,34] was also down-regulated in the hrpB− mutant (Table 1). Therefore, bacterial motility was assayed for the hrp mutants and results showed that X. citri and the hrpB−c strain moved about 2.5 and 1.25 further in swimming and swarming plates respectively, than the hrp mutants (p < 0.05) (Figure 6C) (Additional file 2: Figure S2).
Figure 6.
EPS production and bacterial motility assays in X. citri, the hrp mutants and the hrpB−c strains. (A) Quantification of EPS present in the supernatant fraction of cultures of the different strains. Quadruplicate measurements were made for each strain and an average of all measurements was obtained. Error bars indicate standard deviations. (B) RT-qPCR assay to determine gumD expression of the different stains relative to X. citri. Values are the means of four biological replicates with three technical replicates each. (C) Quantification of bacterial swimming and swarming motility. Results are the average of the motility zones of 16 Petri dishes per strain. Error bars indicate the standard deviation.
Discussion
The role of T3SS in bacterial pathogenesis as a machine involved in effector protein delivery is well established, however, little is known about other functions in bacterial behavior that this system may have. Given that biofilm formation is required for X. citri to achieve full virulence, we used X. citri as a model to gain further insights into the functional role of T3SS in biofilm formation. By comparing the capacity of biofilm formation of three T3SS mutants and X. citri and also performing a proteomic assay with the hrpB− mutant, which revealed differentially expressed proteins between both strains, we demonstrated that T3SS is involved in biofilm formation in X. citri.
To date the involvement of X. citri T3SS in bacterial attachment and survival on leaf tissue has not been studied. In this work we observed that the adherence of different T3SS mutants to host cell tissue was not altered. Studies in several pathogenic bacteria, such as Salmonella typhimurium[35], E. coli[36,37] and the plant pathogen P. syringae[38] revealed that mutants unable to produce T3SS appendages become affected in their interactions with host cells. However, in the phytopathogen Ralstonia solanacearum, it has been shown that the lack of a T3SS pilus does not affect attachment to plant cells [39], and this is consistent with our observation that adherence of X. citri to the host tissue was not affected by the absence of a functional T3SS. In addition, we determined that T3SS is required for X. citri survival on citrus leaves and that T3SS genes are expressed while bacteria reside on the plant surface. Expression of T3SS genes on the leaf surface was also detected in Xanthomonas euvesicatoria cells suggesting a role for T3SS in epiphytic survival of the bacteria [40]. In a recent report, it was revealed that the survival of Pseudomonas syringae T3SS-deficient strains on leaf surfaces is reduced, supporting a role of T3SS and effector proteins in the promotion of epiphytic bacterial survival [41]. Our results suggest that T3SS plays a role in X. citri leaf-associated survival on the leaf surface by enabling biofilm formation.
The proteomic study revealed differentially expressed proteins between X. citri and the hrpB− mutant strain and GO analysis detected enrichment of up-regulated proteins in different metabolic processes and generation of energy in the hrpB− mutant. Similarly, in a previous proteomic study, these categories were also enriched with up-regulated proteins in X. citri planktonic cells compared to biofilm, suggesting a slower metabolism and reduction in aerobic respiration in the X. citri biofilm [42]. Therefore, the higher expression of proteins involved in these processes in the hrpB− mutant compared to X. citri may be caused by the lack of biofilm formation of the mutant.
It is remarkable that among the differentially expressed proteins between the mutant and the wild type strain, some have been previously characterized as involved in biofilm formation in X. citri or in other pathogenic bacteria. Such is the case of DNA-directed RNA polymerase subunit β [32], tryptophan synthase [43], GroEL [44,45], FadL [32,42,46] and several TBDTs [42,47]. Interestingly, high intracellular L-tryptophan concentration prevents biofilm formation and triggers degradation of mature biofilm in E. coli[43]. The proteomic assay showed that tryptophan synthase (XAC2717) was up-regulated, while the tryptophan repressor binding protein (XAC3709) was down-regulated in hrpB− strain suggesting a link also between tryptophan metabolism and biofilm formation in X. citri. Another example is the outer membrane protein XAC0019 that displays high homology to the fatty acid transport porin FadL. Pseudomonas fluorescens mutants in the fadL gene showed defects in their ability to develop a biofilm on a abiotic surfaces leading to the suggestion that long chain fatty acids bind to FadL thereby altering surface hydrophobicity, and adhesion characteristics [46]. Consistent with this, a recent work showed that a X. citri mutant in XAC0019 displays reduced capacity to form a biofilm [32] and its expression is increased during X. citri biofilm formation [42]. In the present study, XAC0019 protein was down-regulated in the hrpB− mutant impaired in biofilm formation, reinforcing the role of this protein in this process.
Enzymes involved in EPS production XanA and GalU, [30,31] were up-regulated in the hrpB− mutant. Consistently, all the hrp mutant analyzed in this work produced larger amounts of EPS in comparison with X. citri and also had higher expression levels of gumD. Recent reports have shown that X. citri galU mutant strain is not pathogenic and also loses its capacity to form a biofilm due to a reduction in EPS production [30,32], and that a X. citri xanA mutant has an altered capacity for biofilm formation [47]. Although, the hrp mutants are impaired in biofilm formation, these mutants produce more EPS than X. citri. This interesting result open new hypotheses about the link between T3SS and EPS production, thus further studies are needed to unravel this issue. In other pathogens, such as P. aeruginosa, T3SS gene expression is coordinated with many other cellular activities including motility, mucoidy, polysaccharide production, and also biofilm formation [48].
Bacterial motility was impaired in the hrp mutants and consistently, proteins known as involved in these processes such as the outer membrane protein XAC0019 [32] and the bactofilin CcmA [33,34] were down-regulated in the hrpB− mutant. Besides, swarming motility was less affected than swimming in the hrp mutants compared with X. citri. This may be due to the fact that in X. citri swarming motility depends on flagella and also on the amount of EPS secreted [16], and since these mutants over-produced EPS swarming was less affected than swimming.
This work demonstrated that in X. citri T3SS is involved in multicellular processes such as motility and biofilm formation. Furthermore, our results suggest that T3SS may also have an important role in modulating adaptive changes in the cell, and this is supported by the altered protein expression when this secretion system is not present. It was previously shown that an E. coli O157 strain mutant in the additional T3SS named ETT2 is impaired in biofilm formation [13]. It was also suggested that deletion of ETT2 might cause structural alterations of the membrane modifying bacterial surface properties, thus affecting bacteria-bacteria interactions or the interaction with host cells [13]. Further, it was proposed that these structural alterations could trigger a signal that activates differential gene expression and/or protein secretion [13]. In line with this, we propose that in X. citri the ‘Hrp pilus’ structure per se, or its interaction with a solid surface, stabilizes the outer membrane structure, hence the lack of T3SS may trigger membrane remodeling itself. These membrane modifications in turn may change the pattern of protein expression, leading to the impairment of cellular processes directly related to bacterial virulence including biofilm formation. Another possibility is that the ‘Hrp pilus’ may function like an attachment device or flagellum. Future studies are likely to add further insights into the exact role and modes of operation of X. citri ‘Hrp pilus’ in biofilm formation and motility.
Conclusions
This work demonstrates that the presence of T3SS in X. citri, besides its participation in the secretion of effector proteins is also required for biofilm formation, motility and survival on leaf tissue revealing novel functions for this secretion system in X. citri. In biofilm formation, T3SS may have an important role in modulating adaptive changes that lead to this process. Some of these changes are revealed by variations in proteins involved in metabolic processes, energy generation, EPS production and bacterial motility as well as in outer membrane proteins between the wild type strain and the T3SS mutant. In summary, the present study reveals novel contributions of this protein secretion system to bacterial virulence.
Methods
Bacterial strains, culture conditions and media
X. citri strain Xac99-1330 was isolated from C. sinensis and kindly provided by Blanca I. Canteros (INTA Bella Vista, Argentina). The hrpB− mutant was constructed in previous work [19]. Here, hrpB−c complemented strain was constructed by cloning the region from hrpB5 to hrcT in the replicative plasmid pBBR1MCS-5 [20] under the control of the lacZ promoter. This region was amplified from X. citri genomic DNA with the oligonucleotides: HrpB5F-Hind (5′ ATAGAAGCTTCATGCGTCTCTGGTTGAGGTC 3′) and HrcTR-Bam (5′ ATCAGGATCCTCAGTGCGACGCGGCTCTCT 3′) and cloned into pBBR1MCS-5 previously digested with the restriction enzymes HindIII and BamHI. The resulting construction was electroporated into the hrpB− strain and the complemented mutant strain was selected by for gentamicin antibiotic resistance. For confocal laser scanning microscopy analyses, a GFP-expressing hrpB− strain was obtained. To this end, the coding sequence for EGFP from the broad-host-range vector pBBR1MCS-2EGFP [16] was digested with BamHI and XbaI and ligated in frame with the LacZ-α-peptide of the pBBR1MCS-5 vector [20] previously digested with the same enzymes, rendering the plasmid pBBR1MCS-5EGFP. E. coli S17-1 cells transformed with this plasmid were conjugated with the hrpB− strain and the cells carrying the plasmid pBBR1MCS-5EGFP were selected for Gm resistance. All strains were grown at 28°C in Silva Buddenhagen (SB) medium [16] or in XVM2 medium [49]. Antibiotics were used at the following final concentrations: 25 μg/ml ampicillin (Amp), 5 μg/ml gentamicin (Gm) and 40 μg/ml kanamycin (Km).
Plant material
Orange (Citrus sinensis cv. Valencia) was used as the host plant for X. citri. All plants were grown in a growth chamber with incandescent light at 28°C with a photoperiod of 16 h.
Biofilm assays
For biofilms development, bacteria were grown in SB with shaking until exponential growth phase and then diluted 1:10 in fresh XVM2 medium containing appropriate antibiotics. A 2 ml aliquot of diluted bacterial suspension was placed in borosilicate glass tubes or in 24-well PVC plates and incubated statically for seven days at 28°C. The quantification of biofilm formation by CV staining was carried out as previously described [50]. Briefly, the culture medium was decanted and the absorbance of planktonic cells was measured at 600 nm using a UV-visible spectrophotometer (Synergy 2 Reader, BioTek). After washing the tubes three times with distilled water (dH2O) during 10 min with gentle agitation, the remaining attached cells were incubated for 10 min at 60°C and stained with 0.1% (w/v) CV for 30 min at room temperature. Excess CV stain was removed by washing under running tap water. The CV stain was solubilized by the addition of 1.5 ml ethanol:acetone (80:20, v/v) to each tube and quantified by measuring the absorbance at 600 nm. The relative absorbance (Relative abs.) was calculated as: CV Abs. 600 nm/Planktonic cells Abs. 600 nm. Values represent the mean from seven tubes for each strain, data were statistically analyzed using one-way analysis of variance (ANOVA) (p < 0.05).
Confocal analysis of biofilm architecture
In vitro biofilm of the GFP-expressing hrpB− mutant and X. citri previously constructed [16] grown in 24-well PVC plates in XVM2 medium were analyzed after seven days by confocal laser scanning microscopy (Nikon Eclipse TE-2000-E2). For biofilms assays on leaf surfaces, overnight cultures of both GFP-expressing strains grown in XVM2 medium were centrifuged, washed and resuspended in phosphate buffer (pH 7.0) to the same OD600 and 20 μl of each bacterial suspension were applied on abaxial leaf surfaces. These biofilms were also analyzed after seven days by confocal laser scanning microscopy (Nikon Eclipse TE-2000-E2).
Adhesion assays
The adhesion capacity to leaf surfaces was measured as described previously [16]. Overnight cultures of the different strains in XVM2 medium were centrifuged to recover cell pellets, washed and resuspended in phosphate buffer (pH 7.0) to the same optic density measured at 600 nm (OD600). Then, 20 μl of each bacteria suspension were place on abaxial leaf surfaces and incubated for 6 h at 28°C in a humidified chamber. After washing the non-adhered cells, bacteria were stained with CV, the CV stain was extracted from the bacterial drops with 95% (v/v) ethanol by pipetting up and down with a 20 μl micropipette. Quantification of the extracted CV stain was carried out by measuring the absorbance at 590 nm as described above. For each strain 20 stained drops were quantified and data were statistically analyzed using one-way ANOVA (p < 0.05).
Quantification of leaf-associated survival
Leaf-associated fitness was evaluated as previously described [51]. Briefly, overnight cultures in SB medium were centrifuged to recover bacteria cell pellets, washed and resuspended in 10 mM phosphate buffer (pH 7.0) at a concentration of 109 CFU/ml. These bacterial suspensions were sprayed onto leaves until each leaf surfaces were uniformly covered. Old citrus leaves were used since the greater thickness of the cuticles of these leaves naturally render the leaves resistant to bacterial entry (unpublished results). Four different leaves were inoculated with each strain, leaves were photographed and the surfaces were quantified using the software Image-Pro (Media Cybernetics). Leaves were collected on different days post-inoculation and transferred to borosilicate glass flasks containing 10 mM potassium phosphate buffer (pH 7.0). Flasks were submerged in a sonicator (Branson model #5510) for 10 min. Subsequently, each flask was vortexed for 5 sec, bacteria were recovered by centrifugation and serial dilutions were plated on SB plates containing Ap to count bacterial colonies. Results were expressed in CFU/cm2 of inoculated leaves. Values represent an average of four leaves assayed for each strain, the data were statistically analyzed using one-way ANOVA (p < 0.05).
RNA preparation and RT-qPCR
Total RNA from bacterial cultures grown at the indicated conditions and from bacteria recovered from leaves at the indicated times were isolated using TRIzol® reagent (Invitrogen), according to the manufacturer’s instructions. The RT-qPCRs were performed as previously described [52] with the specific oligonucleotides detailed in Additional file 3: Table S1. As a reference gene, a fragment of 16S rRNA (XAC3896) was amplified using the same RT-qPCR conditions. To control that no bacterial DNA contamination was present in the samples, the same PCR reactions were carried out without retrotranscription and non amplification was observed. To ascertain the absence of plant RNA in bacterial samples controls with plant actin primers were carried out (data not shown). Values were normalized by the internal reference (Ctr) according to the equation ΔCt = Ct – Ctr, and quantified as 2–ΔCt. A second normalization using a control (time = 0 days) (Ctc), ΔΔCt = Ct – Ctc, producing a relative quantification: 2–ΔΔCt[53]. Values are the means of four biological replicates with three technical replicates each. Results were analyzed by Student t-test (p < 0.05) and one-way ANOVA (p < 0.05).
Protein extraction and resolubilization for the proteomic analysis
Biofilms of statically grown bacterial cultures were obtained as previously described [42]. After seven days of static growth, the XVM2 medium was carefully removed and biofilms were collected by pipetting, transferred to a new tube and pelleted by centrifugation prior to protein extraction. Biofilm proteins were extracted and resuspended in urea buffer (9 M urea, 2 M thiourea and 4% (w/v) 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS)) with vigorous vortexing at room temperature. Concentration of total protein extracts was estimated using a modified Bradford assay [54] and using bovine serum albumin as standard. Protein extracts were prepared from three biological replicates for each strain.
Proteomic analyses
Total proteins from biofilm cells were extracted and labeled using the fluorescent cyanine three-dye strategy (CyDyes; GE Healthcare), as described in [42]. X. citri and hrpB− protein samples were labeled with Cy3 and Cy5, respectively, according to manufacturer’s instructions. Protein extractions were performed from three independent biological samples, and two technical replicate gels for each experiment were run. Protein separation, quantification by two-dimensional-difference in-gel electrophoresis (2D-DIGE), comparative analysis and protein identification were also carried out as previously described [42]. Normalized expression profile data were used to statistically assess changes in protein spot expression. Differentially expressed protein spots between the two groups were calculated using the Student t-test with a critical p-value ≤ 0.05 and the permutation-based method to avoid biased results that may arise within replicate gels if spot quantities are not normally distributed. The adjusted Bonferroni correction was applied for false discovery rate (FDR) to control the proportion of false positives in the result set. Principal component analysis was performed to determine samples and spots that contributed most to the variance and their relatedness. Protein spots with a minimum of 1.5 fold change and p values < 0.05 only were considered as significantly differentially expressed between the two strains.
Quantification of EPS production
Quantification of EPS production was performed as previously described [55]. Briefly, bacterial strains were cultured to the stationary growth phase in 50 ml of SB liquid medium supplemented with 1% (w/v) glucose in 250 ml flasks, using an orbital rotating shaker at 200 rpm at 28°C. Cells were removed by centrifugation at 2,500 × g for 30 min at room temperature, and the supernatant fluids were separately supplemented with KCl at 1% (w/v) and 2 volumes of 96% (v/v) ethanol and then incubated for 30 min at -20°C to promote EPS precipitation. Precipitated crude EPS were collected, dried and weighed. Results were expressed in grams per culture liter. Quadruplicate measurements were made for each strain and an average of all measurements was obtained, data were statistically analyzed using one-way ANOVA (p < 0.05).
Swimming and swarming assays
Swimming and swarming motility were measured as previously described [16]. The SB plates fortified with 0.3% (w/v) or 0.7% (w/v) agar respectively were centrally inoculated with 5 μl of 1 × 107 CFU/ml cultures in exponential growth phase. Inoculated Petri dishes were then incubated in a humidity chamber for two days at 28°C and the motility zones were measured. Results are the average of the motility zones of sixteen Petri dishes per strain. Data was statistically analyzed using one-way ANOVA (p < 0.05).
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JO and NG conceived the project and designed the experiments. TZ, LT, CM, GGS, CGG, FAF and NG designed and performed the experiments. All authors contributed to the analysis and interpretation of the data and LT, CM, CG, JO and NG wrote the manuscript. All authors read and approved the manuscript.
Supplementary Material
Characterization of the hrpB− complemented strain on HR and pathogenicity. (A) Schematic organization of the hrp cluster of X. citri that was constructed based on the X. citri subsp. citri strain 306 genome sequence [1]. Boxes correspond to ORFs, arrows indicate orientation of the hrp operons. The hrp, hpa and hrc genes are indicated. Dotted boxes indicated the genomic regions replaced by mutagenesis. Bellow of the scheme, the black box represents the genomic fragment cloned in pBBR1MCS-5 to complement the hrpB− mutant strain. (B) Bacterial suspensions of X. citri, the hrpB− mutant and the hrpB−c strains were inoculated at 108 CFU/ml into the intercellular spaces of fully expanded tomato, cotton and pepper leaves. A representative photograph of a leaf is shown after 1 day of inoculation. (C) As in B, bacterial suspensions at 107 CFU/ml were inoculated into the intercellular spaces of fully expanded citrus leaves. A representative photograph of a leaf is shown after 8 days of inoculation. (D) RT-qPCR to determine CsLOB1 expression levels in leaves after 48 hours of infection with X. citri, the hrpB− mutant and hrpB−c strain. Bars indicate the expression levels relative to buffer infiltrations. Values are the means of four biological replicates with three technical replicates each.
Swimming and swarming assays. Representative photographs of Petri dishes with X. citri, the hrp mutants and the hrpB−c strain after 2 days of inoculation. Scale bars: 10 mm.
Oligonucleotides used in RT-qPCR assays.
Contributor Information
Tamara Zimaro, Email: zimaro@ibr-conicet.gov.ar.
Ludivine Thomas, Email: ludivine.thomas@kaust.edu.sa.
Claudius Marondedze, Email: claudius.marondedze@kaust.edu.sa.
Germán G Sgro, Email: sgro@ibr-conicet.gov.ar.
Cecilia G Garofalo, Email: garofalo@ibr-conicet.gov.ar.
Florencia A Ficarra, Email: ficarra@ibr-conicet.gov.ar.
Chris Gehring, Email: christoph.gehring@kaust.edu.sa.
Jorgelina Ottado, Email: ottado@ibr-conicet.gov.ar.
Natalia Gottig, Email: gottig@ibr-conicet.gov.ar.
Acknowledgements
We thank Rodrigo Vena for assistance with the confocal microscopy facility, Microquin for the culture media, Catalina Anderson (INTA Concordia, Argentina), Gastón Alanis and Rubén Díaz Vélez (Proyecto El Alambrado) for the citrus plants, Sebastián Graziati and Diego Aguirre for plant technical assistance and the Proteomics laboratory from the Biosciences core laboratory, King Abdullah University of Science and Technology, for providing the facility and equipment for gel electrophoresis and mass spectrometry analyses.
This work was supported by grants from the Argentine Federal Government: ANPCyT (PICT2010-1507 to NG and PICT2010-0300 to JO) and CONICET (PIP2010-2012 to JO and NG), the Fundación Josefina Prats to CGG and FAF. JO and NG are staff members and TZ, GGS, CGG and FAF are fellows of the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET, Argentina).
References
- da Silva AC, Ferro JA, Reinach FC, Farah CS, Furlan LR, Quaggio RB, Monteiro-Vitorello CB, van Sluys MA, Almeida NF, Alves LM, DoAmaral AM, Bertolini MC, Camargo LE, Camarotte G, Cannavan F, Cardozo J, Chambergo F, Ciapina LP, Cicarelli RM, Coutinho LL, Cursino-Santos JR, El-Dorry H, Faria JB, Ferreira AJ, Ferreira RC, Ferro MI, Formighieri EF, Franco MC, Greggio CC, Gruber A. et al. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature. 2002;417(6887):459–463. doi: 10.1038/417459a. [DOI] [PubMed] [Google Scholar]
- Brunings AM, Gabriel DW. Xanthomonas citri: breaking the surface. Mol Plant Pathol. 2003;4(3):141–157. doi: 10.1046/j.1364-3703.2003.00163.x. [DOI] [PubMed] [Google Scholar]
- Graham JH, Gottwald TR, Cubero J, Achor DS. Xanthomonas axonopodis pv. citri: factors affecting successful eradication of citrus canker. Mol Plant Pathol. 2004;5(1):1–15. doi: 10.1046/j.1364-3703.2004.00197.x. [DOI] [PubMed] [Google Scholar]
- He SY, Nomura K, Whittam TS. Type III protein secretion mechanism in mammalian and plant pathogens. Biochim Biophys Acta. 2004;1694(1–3):181–206. doi: 10.1016/j.bbamcr.2004.03.011. [DOI] [PubMed] [Google Scholar]
- Desvaux M, Hebraud M, Henderson IR, Pallen MJ. Type III secretion: what’s in a name? Trends Microbiol. 2006;14(4):157–160. doi: 10.1016/j.tim.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Buttner D. Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant- and animal-pathogenic bacteria. Microbiol Mol Biol Rev. 2012;76(2):262–310. doi: 10.1128/MMBR.05017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey ME, O’Toole GA. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64(4):847–867. doi: 10.1128/MMBR.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagnot C, Zorgani MA, Astruc T, Desvaux M. Proteinaceous determinants of surface colonization in bacteria: bacterial adhesion and biofilm formation from a protein secretion perspective. Front Microbiol. 2013. In press. doi:10.3389/fmicb.2013.00303. [DOI] [PMC free article] [PubMed]
- Kuchma SL, Connolly JP, O’Toole GA. A three-component regulatory system regulates biofilm maturation and type III secretion in Pseudomonas aeruginosa. J Bacteriol. 2005;187(4):1441–1454. doi: 10.1128/JB.187.4.1441-1454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson S, Goldstone RJ, Joshua GW, Chang CY, Patrick HL, Camara M, Wren BW, Williams P. Biofilm development on Caenorhabditis elegans by Yersinia is facilitated by quorum sensing-dependent repression of type III secretion. PLoS Pathog. 2011;7(1):e1001250. doi: 10.1371/journal.ppat.1001250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manos J, Arthur J, Rose B, Bell S, Tingpej P, Hu H, Webb J, Kjelleberg S, Gorrell MD, Bye P, Harbour C. Gene expression characteristics of a cystic fibrosis epidemic strain of Pseudomonas aeruginosa during biofilm and planktonic growth. FEMS Microbiol Lett. 2009;292(1):107–114. doi: 10.1111/j.1574-6968.2008.01472.x. [DOI] [PubMed] [Google Scholar]
- Yap MN, Yang CH, Barak JD, Jahn CE, Charkowski AO. The Erwinia chrysanthemi type III secretion system is required for multicellular behavior. J Bacteriol. 2005;187(2):639–648. doi: 10.1128/JB.187.2.639-648.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideses D, Gophna U, Paitan Y, Chaudhuri RR, Pallen MJ, Ron EZ. A degenerate type III secretion system from septicemic Escherichia coli contributes to pathogenesis. J Bacteriol. 2005;187(23):8164–8171. doi: 10.1128/JB.187.23.8164-8171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings ME, Quick LN, Ubol N, Shrom S, Dollahon N, Wilson JW. Characterization of Salmonella type III secretion hyper-activity which results in biofilm-like cell aggregation. PLoS One. 2012;7(3):e33080. doi: 10.1371/journal.pone.0033080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigano LA, Siciliano F, Enrique R, Sendin L, Filippone P, Torres PS, Questa J, Dow JM, Castagnaro AP, Vojnov AA, Marano MR. Biofilm formation, epiphytic fitness, and canker development in Xanthomonas axonopodis pv. citri. Mol Plant Microbe Interact. 2007;20(10):1222–1230. doi: 10.1094/MPMI-20-10-1222. [DOI] [PubMed] [Google Scholar]
- Gottig N, Garavaglia BS, Garofalo CG, Orellano EG, Ottado J. A filamentous hemagglutinin-like protein of Xanthomonas axonopodis pv. citri, the phytopathogen responsible for citrus canker, is involved in bacterial virulence. PLoS One. 2009;4(2):e4358. doi: 10.1371/journal.pone.0004358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang N. The wxacO gene of Xanthomonas citri ssp. citri encodes a protein with a role in lipopolysaccharide biosynthesis, biofilm formation, stress tolerance and virulence. Mol Plant Pathol. 2011;12(4):381–396. doi: 10.1111/j.1364-3703.2010.00681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang N. Foliar application of biofilm formation inhibiting compounds enhances control of citrus canker caused by Xanthomonas citri subsp. citri. Phytopathology. 2014;104(2):134–142. doi: 10.1094/PHYTO-04-13-0100-R. [DOI] [PubMed] [Google Scholar]
- Dunger G, Arabolaza AL, Gottig N, Orellano EG, Ottado J. Participation of Xanthomonas axonopodis pv. citri hrp cluster in citrus canker and non-host plant responses. Plant Pathol. 2005;54(6):781–788. doi: 10.1111/j.1365-3059.2005.01284.x. [DOI] [Google Scholar]
- Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM 2nd, Peterson KM. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166(1):175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- Pereira AL, Carazzolle MF, Abe VY, de Oliveira ML, Domingues MN, Silva JC, Cernadas RA, Benedetti CE. Identification of putative TAL effector targets of the citrus canker pathogens shows functional convergence underlying disease development and defense response. BMC Genomics. 2014;15(1):157. doi: 10.1186/1471-2164-15-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Zhang J, Jia H, Sosso D, Li T, Frommer WB, Yang B, White FF, Wang N, Jones JB. Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proc Natl Acad Sci U S A. 2014;111(4):E521–E529. doi: 10.1073/pnas.1313271111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausner J, Hartmann N, Lorenz C, Buttner D. The periplasmic HrpB1 protein from Xanthomonas binds to peptidoglycan and to components of the type III secretion system. Appl Environ Microbiol. 2013;79(20):6312–6324. doi: 10.1128/AEM.01226-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengelnik K, van den Ackerveken G, Bonas U. HrpG, a key hrp regulatory protein of Xanthomonas campestris pv. vesicatoria is homologous to two-component response regulators. Mol Plant Microbe Interact. 1996;9(8):704–712. doi: 10.1094/MPMI-9-0704. [DOI] [PubMed] [Google Scholar]
- Weber E, Ojanen-Reuhs T, Huguet E, Hause G, Romantschuk M, Korhonen TK, Bonas U, Koebnik R. The type III-dependent Hrp pilus is required for productive interaction of Xanthomonas campestris pv. vesicatoria with pepper host plants. J Bacteriol. 2005;187(7):2458–2468. doi: 10.1128/JB.187.7.2458-2468.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black PN. Primary sequence of the Escherichia coli fadL gene encoding an outer membrane protein required for long-chain fatty acid transport. J Bacteriol. 1991;173(2):435–442. doi: 10.1128/jb.173.2.435-442.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H, Patel DR, Tamm LK, van den Berg B. The outer membrane protein OmpW forms an eight-stranded beta-barrel with a hydrophobic channel. J Biol Chem. 2006;281(11):7568–7577. doi: 10.1074/jbc.M512365200. [DOI] [PubMed] [Google Scholar]
- Chan YY, Chua KL. The Burkholderia pseudomallei BpeAB-OprB efflux pump: expression and impact on quorum sensing and virulence. J Bacteriol. 2005;187(14):4707–4719. doi: 10.1128/JB.187.14.4707-4719.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postle K. TonB system, in vivo assays and characterization. Methods Enzymol. 2007;422:245–269. doi: 10.1016/S0076-6879(06)22012-3. [DOI] [PubMed] [Google Scholar]
- Guo Y, Sagaram US, Kim JS, Wang N. Requirement of the galU gene for polysaccharide production by and pathogenicity and growth in planta of Xanthomonas citri subsp. citri. Appl Environ Microbiol. 2010;76(7):2234–2242. doi: 10.1128/AEM.02897-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koplin R, Arnold W, Hotte B, Simon R, Wang G, Puhler A. Genetics of xanthan production in Xanthomonas campestris: the xanA and xanB genes are involved in UDP-glucose and GDP-mannose biosynthesis. J Bacteriol. 1992;174(1):191–199. doi: 10.1128/jb.174.1.191-199.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamud F, Homem RA, Conforte VP, Yaryura PM, Castagnaro AP, Marano MR, Morais do Amaral A, Vojnov AA. Identification and characterization of biofilm formation-defective mutants of Xanthomonas citri subsp. citri. Microbiology. 2013;159(9):1911–1919. doi: 10.1099/mic.0.064709-0. [DOI] [PubMed] [Google Scholar]
- Hay NA, Tipper DJ, Gygi D, Hughes C. A novel membrane protein influencing cell shape and multicellular swarming of Proteus mirabilis. J Bacteriol. 1999;181(7):2008–2016. doi: 10.1128/jb.181.7.2008-2016.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopala SV, Titz B, Goll J, Parrish JR, Wohlbold K, McKevitt MT, Palzkill T, Mori H, Finley RL Jr, Uetz P. The protein network of bacterial motility. Mol Syst Biol. 2007;3:128. doi: 10.1038/msb4100166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginocchio CC, Olmsted SB, Wells CL, Galan JE. Contact with epithelial cells induces the formation of surface appendages on Salmonella typhimurium. Cell. 1994;76(4):717–724. doi: 10.1016/0092-8674(94)90510-X. [DOI] [PubMed] [Google Scholar]
- Ebel F, Podzadel T, Rohde M, Kresse AU, Kramer S, Deibel C, Guzman CA, Chakraborty T. Initial binding of Shiga toxin-producing Escherichia coli to host cells and subsequent induction of actin rearrangements depend on filamentous EspA-containing surface appendages. Mol Microbiol. 1998;30(1):147–161. doi: 10.1046/j.1365-2958.1998.01046.x. [DOI] [PubMed] [Google Scholar]
- Knutton S, Rosenshine I, Pallen MJ, Nisan I, Neves BC, Bain C, Wolff C, Dougan G, Frankel G. A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 1998;17(8):2166–2176. doi: 10.1093/emboj/17.8.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roine E, Wei W, Yuan J, Nurmiaho-Lassila EL, Kalkkinen N, Romantschuk M, He SY. Hrp pilus: an hrp-dependent bacterial surface appendage produced by Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci U S A. 1997;94(7):3459–3464. doi: 10.1073/pnas.94.7.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gijsegem F, Vasse J, Camus JC, Marenda M, Boucher C. Ralstonia solanacearum produces hrp-dependent pili that are required for PopA secretion but not for attachment of bacteria to plant cells. Mol Microbiol. 2000;36(2):249–260. doi: 10.1046/j.1365-2958.2000.01851.x. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Callaway EM, Jones JB, Wilson M. Visualisation of hrp gene expression in Xanthomonas euvesicatoria in the tomato phyllosphere. Eur J Plant Pathol. 2009;124:379–390. doi: 10.1007/s10658-008-9423-x. [DOI] [Google Scholar]
- Lee J, Teitzel GM, Munkvold K, del Pozo O, Martin GB, Michelmore RW, Greenberg JT. Type III secretion and effectors shape the survival and growth pattern of Pseudomonas syringae on leaf surfaces. Plant Physiol. 2012;158(4):1803–1818. doi: 10.1104/pp.111.190686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimaro T, Thomas L, Marondedze C, Garavaglia BS, Gehring C, Ottado J, Gottig N. Insights into Xanthomonas axonopodis pv. citri biofilm through proteomics. BMC Microbiol. 2013;13:186. doi: 10.1186/1471-2180-13-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazaki J, Furukawa S, Ogihara H, Morinaga Y. L-Tryptophan prevents Escherichia coli biofilm formation and triggers biofilm degradation. Biochem Biophys Res Commun. 2012;419(4):715–718. doi: 10.1016/j.bbrc.2012.02.085. [DOI] [PubMed] [Google Scholar]
- Lemos JA, Luzardo Y, Burne RA. Physiologic effects of forced down-regulation of dnaK and groEL expression in Streptococcus mutans. J Bacteriol. 2007;189(5):1582–1588. doi: 10.1128/JB.01655-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka T, Furukawa T, Matsumoto-Mashimo C, Yamane K, Sugimori C, Nambu T, Mori N, Nishikawa H, Walker CB, Leung KP, Fukushima H. Gene expression profile and pathogenicity of biofilm-forming Prevotella intermedia strain 17. BMC Microbiol. 2009;9:11. doi: 10.1186/1471-2180-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lima Pimenta A, di Martino P, le Bouder E, Hulen C, Blight MA. In vitro identification of two adherence factors required for in vivo virulence of Pseudomonas fluorescens. Microbes Infect. 2003;5(13):1177–1187. doi: 10.1016/j.micinf.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Li J, Wang N. Genome-wide mutagenesis of Xanthomonas axonopodis pv. citri reveals novel genetic determinants and regulation mechanisms of biofilm formation. PLoS One. 2011;6(7):e21804. doi: 10.1371/journal.pone.0021804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz MR, King JM, Yahr TL. Intrinsic and extrinsic regulation of type III secretion gene expression in Pseudomonas Aeruginosa. Front Microbiol. 2011;2:89. doi: 10.3389/fmicb.2011.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengelnik K, Marie C, Russel M, Bonas U. Expression and localization of HrpA1, a protein of Xanthomonas campestris pv. vesicatoria essential for pathogenicity and induction ofthe hypersensitive reaction. J Bacteriol. 1996;178(4):1061–1069. doi: 10.1128/jb.178.4.1061-1069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signaling pathways: a genetic analysis. Mol Microbiol. 1998;28(3):449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- Dunger G, Relling VM, Tondo ML, Barreras M, Ielpi L, Orellano EG, Ottado J. Xanthan is not essential for pathogenicity in citrus canker but contributes to Xanthomonas epiphytic survival. Arch Microbiol. 2007;188(2):127–135. doi: 10.1007/s00203-007-0227-8. [DOI] [PubMed] [Google Scholar]
- Sgro GG, Ficarra FA, Dunger G, Scarpeci TE, Valle EM, Cortadi A, Orellano EG, Gottig N, Ottado J. Contribution of a harpin protein from Xanthomonas axonopodis pv. citri to pathogen virulence. Mol Plant Pathol. 2012;13(9):1047–1059. doi: 10.1111/j.1364-3703.2012.00814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Ramagli LS. Quantifying protein in 2-D PAGE solubilization buffers. Methods Mol Biol. 1999;112:99–103. doi: 10.1385/1-59259-584-7:99. [DOI] [PubMed] [Google Scholar]
- Becker A, Katzen F, Puhler A, Ielpi L. Xanthan gum biosynthesis and application: a biochemical/genetic perspective. Appl Microbiol Biotechnol. 1998;50(2):145–152. doi: 10.1007/s002530051269. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characterization of the hrpB− complemented strain on HR and pathogenicity. (A) Schematic organization of the hrp cluster of X. citri that was constructed based on the X. citri subsp. citri strain 306 genome sequence [1]. Boxes correspond to ORFs, arrows indicate orientation of the hrp operons. The hrp, hpa and hrc genes are indicated. Dotted boxes indicated the genomic regions replaced by mutagenesis. Bellow of the scheme, the black box represents the genomic fragment cloned in pBBR1MCS-5 to complement the hrpB− mutant strain. (B) Bacterial suspensions of X. citri, the hrpB− mutant and the hrpB−c strains were inoculated at 108 CFU/ml into the intercellular spaces of fully expanded tomato, cotton and pepper leaves. A representative photograph of a leaf is shown after 1 day of inoculation. (C) As in B, bacterial suspensions at 107 CFU/ml were inoculated into the intercellular spaces of fully expanded citrus leaves. A representative photograph of a leaf is shown after 8 days of inoculation. (D) RT-qPCR to determine CsLOB1 expression levels in leaves after 48 hours of infection with X. citri, the hrpB− mutant and hrpB−c strain. Bars indicate the expression levels relative to buffer infiltrations. Values are the means of four biological replicates with three technical replicates each.
Swimming and swarming assays. Representative photographs of Petri dishes with X. citri, the hrp mutants and the hrpB−c strain after 2 days of inoculation. Scale bars: 10 mm.
Oligonucleotides used in RT-qPCR assays.