Abstract
Mitochondrial diseases are heterogeneous, multi-systemic disorders for which mechanistic understanding is limited. To investigate common downstream effects of primary respiratory chain dysfunction on global gene expression and pathway regulation, we reanalyzed transcriptome datasets from all publicly available studies of respiratory dysfunction resulting from genetic disorders, acute pathophysiologic processes, or environmental toxins. A general overview is provided of the bioinformatic processing of transcriptome data to uncover biological insights into in vivo and in vitro adaptations to mitochondrial dysfunction, with specific examples discussed from a variety of independent cell, animal, and human tissue studies. To facilitate future community efforts to cohesively mine these data, all reanalyzed transcriptome datasets were deposited into a publicly-accessible central web archive. Our own integrated meta-analysis of these data identified several commonly dysregulated genes across diverse mitochondrial disease etiologies, models, and tissue types. Overall, transcriptome analyses provide a useful means to survey cellular adaptation to mitochondrial diseases.
Keywords: Oxidative phosphorylation (OXPHOS), Gene expression, Microarray, RNAseq
1. Introduction
The mitochondrial respiratory chain (RC) functions at the core of oxidative metabolism to convert reducing equivalents generated from cellular nutrients into chemical energy in the form of adenosine triphosphate (ATP). In the process, the RC plays an essential role in establishing the reduced and oxidized (redox) balance of nicotinamide adenine dinucleotide (NAD+), which regulates hundreds of cellular reactions and serves as an acetylation precursor. The RC also generates reactive oxygen species (ROS), whose balance can variably serve as secondary messengers to influence cell signaling or generate damage to cellular DNA, lipids, and proteins. RC function further lies at the hub of the intermediary metabolism network, cellular calcium regulation, and apoptosis. Given the wide-reaching implications of these core cellular functions, it is not unexpected that RC dysfunction will produce a host of changes across diverse aspects of cellular physiologic and metabolic processes. The challenge lies in accurately and concisely characterizing the specific nature or degree of such effects in any particular disease state, tissue, or cellular condition.
A ‘transcriptome’ refers to all RNA classes (mRNA, miRNA, etc.), forms (unspliced, degraded, etc.), and locations (nucleus, mitochondria). Transcriptome profiling has emerged as a potent means by which to investigate the downstream, or “retrograde”, effects of primary RC dysfunction, as can variably result from a genetic etiology, acute pathophysiologic process, or environmental toxin. Widespread transcriptome analysis became possible with the advent of microarray technologies. While microarray analyses can utilize different methodologies and platforms to measure RNA abundance, they generally provide consistent results that define major differential expression (DE) between any two conditions at the levels of genes and pathways (Zhang et al., 2010). Recent progress in massively parallel sequencing technologies has now made RNAseq an even more comprehensive and sensitive tool than microarrays to simultaneously interrogate more sophisticated aspects of the transcriptome, such as alternative splice forms, allele-specific expression, and a broader dynamic range to accurately delineate low-level and high-level gene expression.
To investigate common downstream effects of primary RC dysfunction on global gene expression and pathway regulation, we reanalyzed transcriptome datasets from all publicly available studies of respiratory dysfunction resulting from genetic disorders, acute pathophysiologic processes, or environmental toxins. We accessed the data from Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/), which is the major public repository for transcriptome data. However, the reusability of GEO data has generally been impeded by inconsistent data processing and gene annotation between independent studies. To address this problem, we downloaded and subjected to a common processing and annotation protocol data from more than 30 GEO datasets that were previously generated from a wide variety of primary RC disease cell, tissue, and animal models. Each individual data set was reanalyzed to calculate gene-level DE by comparing samples that were divided into two groups, as based on their having either normal or impaired RC function. Since no results had previously been reported at the integrated metabolic pathway-level DE for many of the datasets, we also applied gene set enrichment analysis (GSEA) to investigate the overall DE of canonical metabolic pathways in each dataset (Subramanian et al., 2005). All data from these reanalyses were deposited at a public web archive (http://goo.gl/nOGWC2), along with a complete description of the detailed data processing and analysis pipeline used. A brief overview of the major data processing and analysis steps used for transcriptome analysis is graphically depicted in Figure 1.
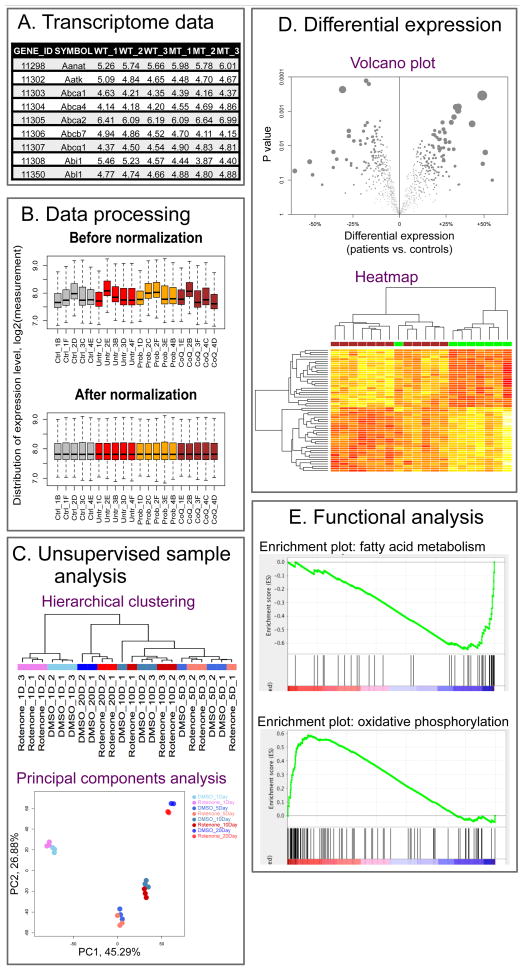
Figure 1. Graphic synopsis of the common analytic process applied for transcriptome dataset analysis.
(A) Transcriptome Data. Transcriptome datasets are usually represented as a data matrix. Each row or column corresponds to a single gene or sample, respectively. (B) Data Processing. Raw data should be properly processed before data analysis. Normalization is a crucial step to correct for inter-sample differences in data distribution that could result from systematic biases, such as total RNA amount and microarray hybridization efficiency. (C) Unsupervised Sample Analysis. Samples are grouped according to their transcriptome data characteristics instead of known sample features, such as disease type or mutation. Hierarchical clustering evaluates the similarity of samples based on their position in the clustering tree, where outliers tend to form their own sub-trees. Principal component analysis (PCA) is most useful for identifying known and unknown factors affecting the transcriptome, as shown in this example where the duration of chemical perturbation is the dominant factor. (D) Differential Expression. Tests of differential gene expression are most commonly performed between two sample groups. Group differences of all genes can be summarized in a Volcano plot to show both the magnitude (x-axis) and statistical significance (y-axis) of the difference. In contrast, a heatmap is often used to visualize selected genes (y-axis) that have the most significant difference between samples (x-axis). (E) Functional Analysis. Functional analyses, such as we performed using GSEA, explore the overall change of genes within the same gene set, such as a KEGG-defined pathway or Gene Ontology (GO) term. In this example, genes of the fatty acid metabolism pathway were generally downregulated while those in the OXPHOS pathway were upregulated. Each vertical black bar represents a single gene in the given gene set. The location of the bar along the x-axis corresponds to its relative position in all genes after they were ranked according to their DE between two sample groups (as indicated by the red-to-blue color bar at the bottom). The green line is the running enrichment score, whose skewness determines the significance of enrichment of the DE genes within a given pathway.
2. Transcriptome profiling of cellular effects in genetic and pharmacologic-based cellular and animal models of primary mitochondrial RC function
Retrograde effects of RC dysfunction have been evaluated by transcriptome profiling in a wide range of experimental animal and cellular models that were variably generated by genetic means (eg, retroviral gene transfection or RNAi interference) or chemical exposure to dysregulate the expression of individual mitochondria-localized proteins and enable identification of the downstream responses to primary RC dysfunction. However, caution must be used when interpreting transcriptome data to assess potential functional consequences of any primary metabolic deficiency. In particular, while identifying significant DE of genes or pathways is suggestive that a cellular response has been invoked, it does not clarify whether that response achieved a particular functional effect. For example, although many RC disease models produce secondary upregulation of genes directly involved in oxidative phosphorylation (OXPHOS), this transcriptional response may not necessarily be fully effective in restoring electron flux, the mitochondrial membrane potential, and ATP generation. Exploring the specific components of a differentially regulated pathway that drive a particular result is also important, as some pre-defined ‘pathways’ may lump together both anabolic and catabolic aspects of metabolism and potentially mask DE of that pathway toward one key direction. Thus, transcriptome profiling can implicate specific biological pathways that are dysregulated by RC disease, but alone is insufficient to determine whether a specific transcriptional response is physiologically adaptive or maladaptive.
In vivo effects of RC dysfunction has been studied by transcriptome profiling in several animal models. Models have variably used either genetic mutations in specific RC complex protein subunits or chemical compounds that directly inhibit specific RC complexes, such as rotenone (complex I (CI) inhibitor) or oligomycin (complex V (CV) inhibitor). The dosage and duration of a chemical exposure can be precisely manipulated to gain insight into RC inhibition duration, degree, and cell-type specific effects. For example, adult C. elegans worms exposed to rotenone for a period of 1 to 20 days showed a transcriptome response that was reported to be most evident after a 1 day exposure and then diminish with prolonged exposure (Schmeisser, 2013). Our reanalysis of that dataset confirmed that transcriptome changes following different rotenone treatment durations were poorly correlated (r = 0.03): the mTOR signaling pathway was significantly downregulated after 1 day of rotenone exposure but then gradually normalized with prolonged exposure; the proteasome pathway was downregulated until 10 days and then showed significant upregulation by day 20 of rotenone exposure; and ribosomal proteins stayed upregulated while WNT pathway expression stayed downregulated regardless of rotenone exposure duration. Interestingly, exposing C. elegans to rotenone for 20 days resulted in similar transcriptome changes as were seen in the C. elegans gas-1(fc21) strain that harbors a genetic missense mutation in a nuclear-encoded CI protein subunit (Falk et al., 2008), where both models showed significant upregulation of nutrient metabolism pathways. These findings exemplify the types of insights that may be gained into in vivo adaptation by studying the effects in model animals of pharmacologic RC inhibitors with varying concentration and time-course experiments.
Chemical inhibitors are also useful to characterize the in vitro, cell type-specific transcriptome response to primary RC dysfunction. For example, exposure of human neuroblastoma cells to two rotenone concentrations (5 nMol and 50 nMol) for 1 or 4 weeks (Cabeza-Arvelaiz and Schiestl, 2012) revealed that while the higher concentration induced a globally greater degree of DE, transcriptome responses at both concentrations were positively correlated (r = 0.70). Comparison of the different durations of rotenone treatment, however, revealed a globally reversed pattern of DE at 4 weeks compared to 1 week, particularly at the higher rotenone concentration (r = −0.60). Furthermore, unlike the attenuation in transcriptome changes that was seen in rotenone-treated adult C. elegans worms, cultured neuroblastoma cells responded to longer rotenone exposure with a greater degree of DE. Rotenone effects have also been studied in mouse embryonic fibroblasts (MEFs) derived from genetic models of mitochondrial disease, such as the HtrA2 knock-out (KO) mice (Moisoi et al., 2009). Although HtrA2 KO cells exhibited a stronger response to rotenone exposure than did normal MEFs, their global pattern of DE was positively correlated (r = 0.60). For example, the proteasome pathway was among the most significantly downregulated KEGG pathways by rotenone treatment in both HtrA2 KO and control MEF cells. These results are suggestive that the specific response to rotenone-induced CI inhibition depends on cell type, underlying genetic background, duration of RC inhibition, as well as in vivo versus in vitro status that may be influenced by a variety of factors including differences in oxygen tension, nutrient availability, and tissue-specific energy demand.
3. Central signaling mediators regulate physiologic effects of mitochondrial RC dysfunction
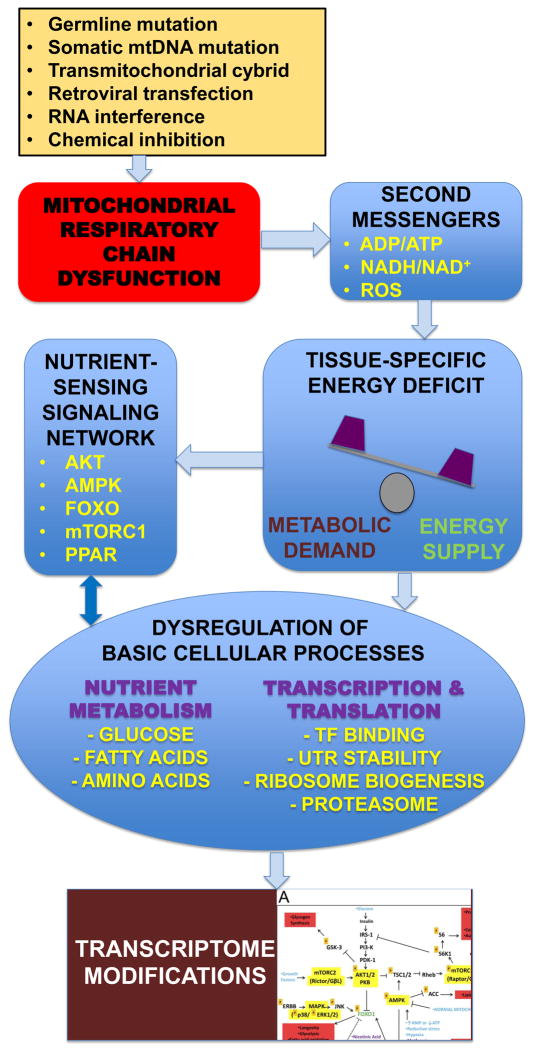
While transcriptome response to RC dysfunction clearly is influenced by a range of variables, the identification of concordant gene changes and recurrent pathway-level DE results across independent studies and models are suggestive that a “central hub” exists that converges upstream signals from RC dysfunction to modulate downstream cellular responses. The integrated nutrient-sensing signaling network (NSSN) centered on the AKT/mTORC pathways appears to be one such central mediator of cellular response to RC dysfunction (Zhang et al., 2013). Major regulatory NSSN nodes include AMPK (low energy sensor), mTORC1 (cell growth regulator by balancing cytosolic protein synthesis and autophagy), SREBP (lipid homeostasis), FOXO1 (glucose homeostasis), and PPAR family transcription factors (lipid metabolism), as well as YY1/PGC1α (mitochondrial ribosome biogenesis) and HIF1α (hypoxia response) transcription factors. The involvement of the integrated NSSN in regulating cellular response to RC dysfunction was first revealed by our recent study of skeletal muscle and fibroblasts from a heterogeneous group of human subjects with documented RC enzyme deficiency and/or known pathogenic mutations (Zhang, Tsukikawa, 2013). The most significantly upregulated gene in muscle from the cohort of RC disease patients relative to controls was RHEB, a direct mediator of mTORC1 activity, with significant upregulation of the ribosome biogenesis pathway. Interestingly, ribosomal protein genes showed significant differential expression depending on whether they encoded cytosolic ribosomal proteins (downregulated in RC disease muscle) or mitochondrial ribosomal proteins (upregulated in RC disease muscle). In addition, target genes of FOXO1 and PPAR family transcription factors were upregulated and downregulated, respectively, in RC disease muscle. Validation of these findings by additional studies of gene expression and phosphoprotein activity in human cells treated with direct RC chemical inhibitors similarly implicated this network of signaling mediators in coordinating the integrated cellular response to primary RC dysfunction. Remarkably, this integrated network of signaling pathways showed significant DE in the reversed direction (r = −0.49) in fibroblast cell lines (Zhang, Tsukikawa, 2013). Such dependence of transcriptome response on tissue type and/or nutrient supply highlights the importance of characterizing changes in the integrated NSSN to understand, and potentially modulate, the wide-spread physiologic alterations that occur in RC disease (Figure 2).
Figure 2. Schematic overview of how central signaling mediators regulate physiologic cellular effects of primary mitochondrial RC dysfunction.
Primary RC dysfunction can result from genetic mutation and/or environmental induction. Impaired mitochondrial RC function causes energy imbalance and triggers a cellular response that is both tissue-specific and nutrient-dependent. Basic cellular processes including nutrient metabolism, transcription, and protein translation are significantly disrupted, likely through modulation of the integrated NSSN. The overall cellular response to mitochondrial RC dysfunction can be partially characterized by transcriptome profiling.
RC dysfunction also impacts pathways regulating basic cellular processes, such as those involved in regulating the life cycle of RNA and protein. Upregulation of almost all processes related to this umbrella categorization, such as the spliceosome, aminoacyl-tRNA synthesis, and protein export, in skeletal muscle from the cohort of patients with human RC disease suggests that an accelerated pace of RNA and protein metabolism occurs in RC dysfunction. It is unclear to what extent NSSN dysregulation accounts for these changes in basic RNA and protein metabolism, although the involvement of mTORC1 is supported by downregulation of these same pathways in MEF cells exposed to the chemical rapamycin that inhibits mTORC1 (Cunningham et al., 2007). In addition, RNAseq and advanced microarray platforms now enable examination of RNA processing and decay dynamics. For example, data generated from exon-level microarray analysis of RC disease patient muscle revealed that the 5′-untranslated regions (UTRs) of transcripts are generally shortened to accelerate protein translation, whereas the 3′-UTRs that include AU-rich elements are more slowly degraded to stabilize mRNAs (Zhang, Tsukikawa, 2013). More widespread use of increasingly sophisticated massively parallel sequencing methodologies to profile transcriptome and chromatin modifications will enable further clarification of the specific nature of global gene dysregulation that occurs in primary RC dysfunction.
4. Human mitochondrial disease tissue transcriptome investigations and meta-analysis
Identifiying the transcriptome response to mitochondrial dysfunction in human mitochondrial RC disease samples is complicated by the plethora of disease etiologies and potential experimental confounders. For meaningful transcriptome interpretation in human samples, it is critical to carefully select RC disease subjects and well-matched controls, preferably in large cohorts. While we recently showed a genetically heterogeneous group of primary RC disease subjects share a common “transcriptome signature” at the pathway level, gene-level DE showed great inter-subject variability for many individual genes (Zhang, Tsukikawa, 2013). Both common and subtype-specific DE was similarly observed in another study that compared skeletal muscle specimens from controls to those from three RC disease subgroups (mitochondrial DNA (mtDNA) deletion, m.3243A>G mtDNA mutation without clinical symptoms, and m.3243A>G mtDNA mutation in individuals clinically manifesting the mitochondrial encephalomyopathy and lactic acid syndrome, (MELAS)) (Crimi et al., 2005). Upon reanalysis of these muscle transcriptome data by GSEA, the aminoacyl-tRNA biosynthesis and ribosome biogenesis pathways were found to be commonly upregulated across all three of these RC disease subgroups and in our heterogeneous RC disease cohort (Zhang, Tsukikawa, 2013).
To evaluate for a consistent gene-level response to primary RC disease, we performed a meta-analysis comprised of 8 RC disease transcriptome datasets, 12 RC disease patient-control pairwise comparisons, and 99 RC disease subjects of diverse disease subtypes. The 2 genes yielding the most significant meta-analysis p values were MTX2 (p=1.8E-9), a membrane protein involved in mitochondrial protein import, and NGLY1 (p = 8.2E-9), a deglycosylase enzyme involved in the degradation of misfolded proteins. Other top-ranked genes include MTX3 (#3), LAMTOR5 (#10, MAPK/mTOR activator), MFN1 (#22, mitofusin), XPOT (#35, tRNA exportin) and several aminoacyl-tRNA synthetases. A complete list of datasets studied and meta-analysis results is available at the online data archive (http://goo.gl/nOGWC2). The general categories of datasets studied and key points related to the diverse models and samples from which these datasets were derived is provided in the Supplemental File. Overall, our preliminary meta-analysis demonstrates that statistical power can be achieved by analysis across datasets significantly beyond that achievable by the analysis of individual data sets.
5. Future outlook for applying transcriptome profiling in finer scale RC disease models, as biomarkers, and in therapeutic monitoring of human mitochondrial disease
Future transcriptome profiing in RC disease models and human subjects will increasingly utilize RNAseq as massively parallel sequencing technology advances and costs continue to fall. RNAseq permits finer resolution that possible by microarray analysis (Zhang, Gasser, 2010) into cell-type specific variation that may be relevant to better understanding effect of RC disease, such as in single cell or muscle fiber analyses. Cellular adaptation to different degrees of mitochondrial inhibition is also possible to interrogate by studying varying degrees of pharmacologic RC inhibition or varying heteroplasmy load in transmitochondrial cybrid cell lines that share a common nuclear background and only differ in their heteroplasmy levels of a specific mtDNA mutation (Chae et al., 2013, Trounce and Wallace, 1996), although it is possible that deviant energy metabolism in the host cancer cell line that is used to generate cybrids might influence the relative transcriptome findings in that particular cell line compared to changes in other cell lines. Comparison of the cellular transcriptome response to chemical RC inhibitors relative to genetic mutations in mitochondrial structural, enzymatic, or regulatory proteins will also be possible at different developmental stages, as through analyses in human embryonic stem cells created by transferring a somatic cell nucleus into an oocyte (Tachibana et al., 2013). Substantial insight into tissue-specific responses will also likely be gained through transcriptome comparison in mitochondrial disease patient-derived nuclear or mtDNA gene mutation cell lines that have been converted to an induced pluripotent stem cell (iPSC) line and terminally differentiated into different cell lineages (such as neuron or myocyte) (Grskovic et al., 2011, Skowron et al., 2014). Recent studies suggest it may also be possible to study the transcriptome response in patient blood cell derived terminal cell lineages directly converted by acid treatment without first developing an iPSC line (Obokata et al., 2014). Further, RNAseq will enable exploration of the role of additional RNA species, such as microRNAs, in causing or modifying cellular response to primary RC disease. It is also likely that integration of transcriptome data with investigations at the increasingly approachable levels of epigenomics, proteomics, and post-translational modifications will provide unique insight into the different levels and efficacy of cellular dysregulation that occurs in RC disease.
Transcriptome profiling will also be increasingly useful both to develop expression-level RC disease biomarkers and to monitor effects of RC disease therapies by simultaneously interrogating diverse aspects of cellular physiology and metabolism in a quantitative fashion in both mammalian models and in human patients. For example, prophylactic probucol (but not coenzyme Q10) therapy prevented a focal-segmental glomerulosclerosis like renal disease in pdss2 mutant coenzyme Q deficient mice by mechanisms that transcriptome profiling revealed to involve reversal of dysregulation in diverse nutrient metabolism pathways and PPAR targets (Falk et al., 2011, Zhang, Gasser, 2010). Similarly, coenzyme Q10 supplementation was shown to partially normalize the transcriptome of CoQ10–deficient human patients (Fernandez-Ayala et al., 2013). Thus, it is likely that transcriptome profiling in blood or cell lines obtained from a given individual and treated in vitro with specific therapeutic agent(s) will permit development of personalized therapeutic regimens to direct their future medical management. Finally, transcriptome profiling of blood or tissue samples from patients over time or in clinical treatment trials can potentially be used to quantify an individual patient’s mitochondrial disease progression and/or provide an objective indicator by which to monitor therapeutic effect.
Supplementary Material
Acknowledgments
Funding: This work was funded in part by the National Institutes of Health (R03-DK082446, R01- HD065858-01A1, and P30-HD026979).
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
The content of the article has not been influenced by the sponsors.
The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Cabeza-Arvelaiz Y, Schiestl RH. Transcriptome analysis of a rotenone model of parkinsonism reveals complex I-tied and -untied toxicity mechanisms common to neurodegenerative diseases. PloS one. 2012;7:e44700. doi: 10.1371/journal.pone.0044700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo S, Jain M, Xie X, Sheth SA, Chang B, Goldberger OA, et al. Systematic identification of human mitochondrial disease genes through integrative genomics. Nature genetics. 2006;38:576–82. doi: 10.1038/ng1776. [DOI] [PubMed] [Google Scholar]
- Chae S, Ahn BY, Byun K, Cho YM, Yu MH, Lee B, et al. A systems approach for decoding mitochondrial retrograde signaling pathways. Science signaling. 2013;6:rs4. doi: 10.1126/scisignal.2003266. [DOI] [PubMed] [Google Scholar]
- Cizkova A, Stranecky V, Mayr JA, Tesarova M, Havlickova V, Paul J, et al. TMEM70 mutations cause isolated ATP synthase deficiency and neonatal mitochondrial encephalocardiomyopathy. Nature genetics. 2008;40:1288–90. doi: 10.1038/ng.246. [DOI] [PubMed] [Google Scholar]
- Crimi M, Bordoni A, Menozzi G, Riva L, Fortunato F, Galbiati S, et al. Skeletal muscle gene expression profiling in mitochondrial disorders. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2005;19:866–8. doi: 10.1096/fj.04-3045fje. [DOI] [PubMed] [Google Scholar]
- Cui L, Jeong H, Borovecki F, Parkhurst CN, Tanese N, Krainc D. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450:736–40. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- Falk MJ, Polyak E, Zhang Z, Peng M, King R, Maltzman JS, et al. Probucol ameliorates renal and metabolic sequelae of primary CoQ deficiency in Pdss2 mutant mice. EMBO molecular medicine. 2011;3:410–27. doi: 10.1002/emmm.201100149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk MJ, Zhang Z, Rosenjack JR, Nissim I, Daikhin E, Nissim I, et al. Metabolic pathway profiling of mitochondrial respiratory chain mutants in C. elegans. Molecular genetics and metabolism. 2008;93:388–97. doi: 10.1016/j.ymgme.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Ayala DJ, Guerra I, Jimenez-Gancedo S, Cascajo MV, Gavilan A, Dimauro S, et al. Survival transcriptome in the coenzyme Q10 deficiency syndrome is acquired by epigenetic modifications: a modelling study for human coenzyme Q10 deficiencies. BMJ open. 2013:3. doi: 10.1136/bmjopen-2012-002524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley LW, Lee J, Souza A, Desquiret-Dumas V, Bullock K, Rowe GC, et al. Skeletal muscle transcriptional coactivator PGC-1alpha mediates mitochondrial, but not metabolic, changes during calorie restriction. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:2931–6. doi: 10.1073/pnas.1115813109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freije WA, Mandal S, Banerjee U. Expression profiling of attenuated mitochondrial function identifies retrograde signals in Drosophila. G3. 2012;2:843–51. doi: 10.1534/g3.112.002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grskovic M, Javaherian A, Strulovici B, Daley GQ. Induced pluripotent stem cells--opportunities for disease modelling and drug discovery. Nature reviews Drug discovery. 2011;10:915–29. doi: 10.1038/nrd3577. [DOI] [PubMed] [Google Scholar]
- Mende S, Royer L, Herr A, Schmiedel J, Deschauer M, Klopstock T, et al. Whole blood genome-wide expression profiling and network analysis suggest MELAS master regulators. Neurological research. 2011;33:638–55. doi: 10.1179/1743132810Y.0000000016. [DOI] [PubMed] [Google Scholar]
- Moisoi N, Klupsch K, Fedele V, East P, Sharma S, Renton A, et al. Mitochondrial dysfunction triggered by loss of HtrA2 results in the activation of a brain-specific transcriptional stress response. Cell death and differentiation. 2009;16:449–64. doi: 10.1038/cdd.2008.166. [DOI] [PubMed] [Google Scholar]
- Mormeneo E, Jimenez-Mallebrera C, Palomer X, De Nigris V, Vazquez-Carrera M, Orozco A, et al. PGC-1alpha induces mitochondrial and myokine transcriptional programs and lipid droplet and glycogen accumulation in cultured human skeletal muscle cells. PloS one. 2012;7:e29985. doi: 10.1371/journal.pone.0029985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obokata H, Wakayama T, Sasai Y, Kojima K, Vacanti MP, Niwa H, et al. Stimulus-triggered fate conversion of somatic cells into pluripotency. Nature. 2014;505:641–7. doi: 10.1038/nature12968. [DOI] [PubMed] [Google Scholar]
- Peng M, Falk MJ, Haase VH, King R, Polyak E, Selak M, et al. Primary coenzyme Q deficiency in Pdss2 mutant mice causes isolated renal disease. PLoS genetics. 2008;4:e1000061. doi: 10.1371/journal.pgen.1000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeisser S, Priebe S, Groth M, Monajembashi S, Hemmerich P, Guthke R, Platzer M, Ristow M. Neuronal ROS signaling rather than AMPK/sirtuin-mediated energy sensing links dietary restriction to lifespan extension. Molecular Metabolism. 2013;2:92–102. doi: 10.1016/j.molmet.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowron K, Tomsia M, Czekaj P. An experimental approach to the generation of human embryonic stem cells equivalents. Molecular biotechnology. 2014;56:12–37. doi: 10.1007/s12033-013-9702-4. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Amato P, Sparman M, Gutierrez NM, Tippner-Hedges R, Ma H, et al. Human embryonic stem cells derived by somatic cell nuclear transfer. Cell. 2013;153:1228–38. doi: 10.1016/j.cell.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trounce I, Wallace DC. Production of transmitochondrial mouse cell lines by cybrid rescue of rhodamine-6G pre-treated L-cells. Somatic cell and molecular genetics. 1996;22:81–5. doi: 10.1007/BF02374379. [DOI] [PubMed] [Google Scholar]
- Vianna CR, Huntgeburth M, Coppari R, Choi CS, Lin J, Krauss S, et al. Hypomorphic mutation of PGC-1beta causes mitochondrial dysfunction and liver insulin resistance. Cell metabolism. 2006;4:453–64. doi: 10.1016/j.cmet.2006.11.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voets AM, Huigsloot M, Lindsey PJ, Leenders AM, Koopman WJ, Willems PH, et al. Transcriptional changes in OXPHOS complex I deficiency are related to anti-oxidant pathways and could explain the disturbed calcium homeostasis. Biochimica et biophysica acta. 2012;1822:1161–8. doi: 10.1016/j.bbadis.2011.10.009. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Gasser DL, Rappaport EF, Falk MJ. Cross-platform expression microarray performance in a mouse model of mitochondrial disease therapy. Molecular genetics and metabolism. 2010;99:309–18. doi: 10.1016/j.ymgme.2009.10.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Tsukikawa M, Peng M, Polyak E, Nakamaru-Ogiso E, Ostrovsky J, et al. Primary respiratory chain disease causes tissue-specific dysregulation of the global transcriptome and nutrient-sensing signaling network. PloS one. 2013;8:e69282. doi: 10.1371/journal.pone.0069282. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.