Abstract
The vacuole in the yeast Saccharomyces cerevisiae plays a number of essential roles, and to provide some of these required functions the vacuole harbors at least seven distinct proteases. These proteases exhibit a range of activities and different classifications, and they follow unique paths to arrive at their ultimate, common destination in the cell. This review will first summarize the major functions of the yeast vacuole and delineate how proteins are targeted to this organelle. We will then describe the specific trafficking itineraries and activities of the characterized vacuolar proteases, and outline select features of a new member of this protease ensemble. Finally, we will entertain the question of why so many proteases evolved and reside in the vacuole, and what future research challenges exist in the field.
Keywords: protease, S. cerevisiae, hydrolysis, autophagy, endocytosis, metalloprotease, Vps10, CPY, secretory pathway, Pff1
Structure, Function, and Protein Trafficking Itineraries Associated with the Vacuole in the Yeast Saccharomyces cerevisiae
The vacuole in most yeasts can occupy ~20% of cell volume. It is functionally similar to the plant vacuole and the lysosome in higher eukaryotes, with an important role in the degradation of macromolecules, nutrient storage, protein homeostasis, and detoxification.1,2 This organelle was first described in the 1930s from light microscopy images of plant cells showing large “empty” (vacuus, in Latin) structures within the cytosol.3 The vacuole in yeast is host to numerous proteases, lipases, nucleases, and transporters,1,4,5 and is a dynamic organelle that undergoes fusion and fission events in response to the cell cycle and environmental cues. Logarithmically growing cells have a multi-lobed vacuole, while in stationary phase cells, or cells starved of a carbon source, these lobes fuse to produce a single spherical vacuolar structure.6 The vacuole is also a slightly acidic environment (pH ~6.2) compared with the cytosol (pH ~7.2) in logarithmically growing yeast.7,8 In the remainder of this review, we will refer to “yeast” to indicate the organism Saccharomyces cerevisiae, unless otherwise indicated.
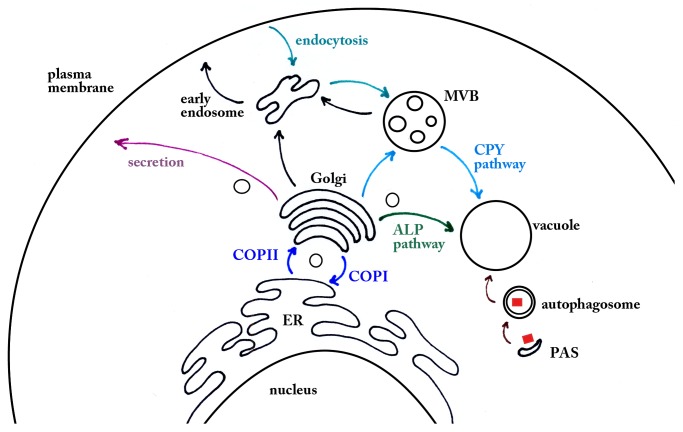
A host of proteins define the function of this organelle. Approximately 200 of the 6,000 yeast genes are annotated as encoding proteins having vacuolar localization, and 27% of those are described as having transporter function.2 The remaining proteins possess functions associated with macromolecular hydrolysis, membrane fusion, protein sorting and targeting to the vacuole, and acidification.9,10 Most of these proteins are targeted to the vacuole via the secretory pathway, and by definition the proteins are first translocated into the endoplasmic reticulum (ER). COPII vesicles then ferry the vacuole-targeted proteins to the Golgi apparatus. In the Golgi, the carbohydrate residues of secretory proteins are modified and extended.11,12 It is at this point that cargo proteins reach a crossroad in the secretory pathway; they can be retained in the Golgi, targeted to the plasma membrane, or trafficked to the vacuole. Cargo traffics from the Golgi to the cell surface either directly or indirectly via an endosomal intermediate,13 and for the latter trafficking pathway the adaptor protein complex 1 (AP-1) plays an important role.14 Similarly, Golgi-to-vacuolar trafficking can be direct, requiring adaptor protein complex 3 (AP-3)15,16 or indirect through a multivesicular body (MVB) intermediate.17-19 These two trafficking intervals, referred to as the alkaline phosphatase (ALP) and carboxypeptidase Y (CPY) pathways, respectively, are named for model cargos used in their characterization. Genetic screens that monitor CPY maturation and trafficking have been instrumental in identifying proteins required for vacuolar protein sorting and maintenance of vacuolar morphology (i.e., the VPS genes).20-23
In the initial stages of the CPY pathway, cargo proteins such as carboxypeptidase S (CPS), Kex2, and the CPY-Vps10 complex exit the Golgi in vesicles associated with clathrin and the adaptor proteins Gga1 and Gga2.24-27 Efficient cargo sorting into the MVB and vacuolar lumen is partially dependent on the phosphatidylinositol 3,5-bisphosphate composition of endosomal and vacuolar membranes.28,29 In addition, for cargos such as CPS, MVB sorting is controlled by protein ubiquitination.30-33 Specifically, the ubiquitin ligase Rsp5 appends K63-linked polyubiquitin chains on protein cargo, which promotes MVB trafficking.30,31,33,34 The endosomal sorting complex required for transport 0 (ESCRT 0) recognizes ubiquitinated cargo, and together with ESCRTs 1–3, drives cargo incorporation into and formation of intralumenal vesicles.29,35 To deliver intralumenal vesicles, the MVB docks and fuses with the vacuole, an event that requires the homotypic fusion and vacuole protein sorting (HOPS) complex, Rab family GTPase, Ypt7, and SNARE proteins.36-38 The HOPS complex is also needed for vesicles bearing AP-3 to dock and fuse to the vacuole in the ALP pathway, which does not require clathrin and bypasses the MVB completely.39 These vacuolar trafficking pathways are summarized in Figure 1.
Figure 1. Protein trafficking and vacuolar sorting pathways. Cellular compartments are labeled in black and secretory pathways discussed in this review are highlighted and color-coded. Of particular note are the vacuolar sorting pathways: the CPY pathway, shown in light blue; the ALP pathway, shown in green; and the Cvt pathway, shown in brown (see text for details). This image was adapted from Bowers et al. 2005.17
The Yeast Vacuole as a Site of Protein Degradation
The vacuole is host to numerous exopeptidases and endopeptidases that contribute to what is often considered the vacuole’s principle function, protein degradation.40 The vacuole maintains protein homeostasis under physiological conditions by degrading senescent, superfluous, and damaged proteins and organelles. Vacuole function is also vital under conditions of nutrient stress, when cell growth and proliferation are downregulated and proteins must be broken down so that their constituent amino acids can be recycled.41 Impressively, degradation of up to ~85% of the cell’s intracellular protein content has been observed during nutrient starvation.42 Nutrient stress is encountered when yeast reach stationary phase, or when they undergo sporulation. Some of the cues known to trigger nutrient stress include carbon and nitrogen starvation, and to a lesser extent when there is a lack of essential amino acids, nucleotides, and sulfates.43,44 This next section summarizes the various degradation functions attributed to the vacuole, and the distinct routes that deliver proteins to this organelle for degradation.
Autophagy
Autophagy, also known as macroautophagy, is a process whereby bulk cytoplasmic material and organelles are isolated in a double-membrane vesicle known as an autophagosome.45 The autophagosome fuses with the vacuole, where its contents are degraded and the constituent components are recycled. This mechanism is conserved throughout eukaryotes, and is used to maintain cellular homeostasis under physiological conditions by eliminating long-lived proteins and damaged organelles. Autophagy can also be induced under conditions of nutrient stress and during specific stages in the cell cycle, such as in stationary phase or during sporulation.46,47 Autophagy is mediated by over 30 autophagy-related proteins, known as the Atg proteins.48 The Atg proteins act during a series of sequential steps: induction of autophagy, cargo recognition, vesicle nucleation, expansion and completion of the autophagosome, Atg protein recycling, fusion of the autophagosome with the vacuole, vesicle breakdown, and macromolecular recycling.
A notable type of selective autophagy is known as the cytoplasm-to-vacuole targeting (Cvt) pathway. In contrast to macroautophagy, the Cvt pathway occurs during nutrient-rich conditions and is a constitutive process.49 The Cvt pathway controls the selective transport of homooligomeric vacuolar proteins. Only three substrates in yeast are known to be targeted to the vacuole by this pathway: aminopeptidase 1 (Ape1), α-mannosidase 1 (Ams1), and aminopeptidase 4 (Ape4). Ape1 and Ams1 are resident vacuolar proteins, whereas Ape4 is cytosolic under physiological conditions, but targeted to the vacuole in a Cvt-dependent manner during nutrient starvation conditions.50 These substrates are synthesized in the cytoplasm, where they homooligomerize and bind to Atg19. Atg19 acts as a receptor for Cvt substrates.50-53 The autophagosomes associated with the Cvt pathway are smaller, and take approximately 10 times longer to form than those associated with autophagy.52,54
The Cvt pathway may be an important alternative trafficking mechanism to target vacuolar resident proteins that are otherwise damaging to secretory pathway residents. Alternatively, the Cvt pathway may facilitate transport of stable oligomeric proteins that are too large to traverse the vacuolar membrane via transporters. Finally, the Cvt pathway may facilitate vacuolar localization of membrane-associated proteins, such as Ams1, which lack a signal sequence to direct their translocation into the secretory pathway (Fig. 1).53
Endocytosis
Protein composition at the cell surface is carefully regulated. Proteins that become damaged or that may be detrimental are removed by endocytosis and often traffic to the vacuole, where they are degraded. Plasma membrane-to-vacuole trafficking was first observed in yeast by monitoring lucifer yellow, a fluorescent dye that initially localizes to the cell surface but gradually accumulates in the vacuole after endocytosis.55 Subsequent genetic screens were instrumental in identifying components of the endocytic machinery,56-58 and more recently, cutting edge microscopy has generated a detailed temporal and spatial map of clathrin-mediated endocytosis (CME).59-62 CME is by far the best characterized endocytic pathway, requiring the orchestration of > 50 proteins in yeast. The details of CME are the focus of many excellent recent reviews,63-66 but in brief this process is characterized by: 1) arrival of early endocytic and coat proteins, including Ede1, Syp1 and clathrin; 2) cargo recruitment, and the arrival of actin nucleation promoting factors, such as Pan1 and Las17, which is the WASP homolog; 3) regulated branched actin assembly by Arp2/3 and association of additional actin nucleation regulators, including Myo3 and Myo5; 4) membrane invagination, which is driven by actin polymerization and allows for recruitment of the amphiphysins Rvs161 and Rvs167 that aid in constricting the neck of the tubule; and 5) vesicle scission, which may involve the dynamin related protein Vps1, vesicle uncoating, and actin disassembly.59-61 It is important to note that the organization and timing of endocytic events described in yeast is now thought to be largely conserved in mammalian cells.67,68 In addition to the well-characterized CME pathway, a clathrin-independent endoctyic pathway (CIE), analogous to the RhoA CIE pathway in mammals, was also recently identified in yeast.69
Endocytosis and vacuolar sorting of cell surface proteins requires cargo ubiquitination. Pioneering studies of the yeast G-protein coupled receptors and nutrient permeases showed that ligand- or nutrient-induced ubiquitination precedes protein endocytosis.70-72 The ubiquitin ligase Rsp5 is required for ubiquitination,72-75 but most membrane cargos do not bind the ligase directly. Instead, adaptor proteins, such as the α-arrestins, recruit Rsp5 to specific membrane cargo, often in a signal-induced fashion and stimulate endocytosis.76-80 Ubiquitinated cargo proteins, once removed from the cell surface, are sorted to endosomes and enter the MVB in an ESCRT-dependent manner.29,35,81 Thus, there is convergence of the endocytic route to the vacuole with the Golgi-to-vacuolar sorting pathway, described above. Many studies have examined protein stability in cells harboring deletions of the Pep4 protease, which prevents maturation of the vacuolar proteases (see below).82,83 These efforts have helped determine the vacuolar trafficking itineraries for specific cargo.71,72,84,85
The Vacuolar Proteases
The yeast vacuole contains seven characterized vacuolar proteases, one of which is a transmembrane protease. Among all of these, three are metalloproteases, three are serine proteases, and one is an aspartyl protease. The vacuolar proteases include three aminopeptidases, two carboxypeptidases, and two endopeptidases. This section describes the known characteristics and function of these proteases, which are summarized in Table 1. Quite recently, we reported on an eighth vacuolar protease, which is a transmembrane metalloprotease and is the product of the YBR074w gene (Pff1) (also see below).86 However, its protease activity has yet to be demonstrated.
Table 1.Saccharomyces cerevisiae vacuolar proteases. Only those vacuolar resident proteases are shown whose activities and functions have been established experimentally (see text for details).
| Protease | eGene | Activity | Proteolytically activated by | Trafficking pathway | Function | Known P1 site amino acids |
|---|---|---|---|---|---|---|
| Proteinase A | PEP4 | aspartyl endoprotease | Pep4 and Prb1 | secretory | Initiator of protease activation cascade; protein degredation | Phe, Leu, Tyr, Trp, Thr, Asn, Gln, Glu, Lys, Ala, Ile |
| Proteinase B | PRB1 | serine endoprotease | Pep4 and Prb1 | secretory | protease activation; protease degredation | Leu, Arg, Phe, Tyr, Gln, Lys |
| Carboxypeptidase Y | PRC1 | serine carboxypeptidase | Pep4 and Prb1 | CPY pathway | peptide degredation | Ala, Gly, Val, Leu, Ile, Met, Phe |
| Carboxypeptidase S | CPS1 | Zinc metalloprotease | Prb1 | CPY pathway | peptide degredation | Gly, Leu |
| Aminopeptidase I | APE1 | Zinc metalloprotease | Prb1 | Cvt pathway | glutathione degredation | Leu, Cys/Gly |
| Aminopeptidase Y | APE3 | metalloprotease | Prb1 | secretory | unknown | Pro, Ala, Leu, Met, Phe, Tyr, Ser, Lys, Arg |
| Dipeptidylaminopeptidase B | DAP2 | serine dipeptidase | none | CPY pathway | unknown | Xaa-Ala, Xaa-Pro |
The MEROPS peptidase database (http://merops.sanger.ac.uk) classifies known proteases into evolutionarily related groups, which will be referenced in the following discussion. A protease clan refers to proteases derived from a single common ancestor, and clans are subdivided into families. A protease family refers to a sub-group of proteases that share sequence similarity, either throughout the entire protein sequence or only within the catalytic domain.87 Protease families are assigned a letter referring to the catalytic mechanism of the protease: M for metalloprotease, S for serine protease, and A for aspartyl protease. The catalytic designation is followed by a numerical family identifier.88 Another important aspect of nomenclature refers to the site of cleavage of the substrate. The amino acid on the N-terminal side of the hydrolyzed peptide bond is referred to as P1, whereas the amino acid on the C-terminal side is referred to as P1’. Similarly, the site on the proteolytic enzyme known to bind to the P1 residue is referred to as S1, and the site recognizing the P1’ residue is referred to as S1’.
In brief, serine proteases comprise a large family whose enzymatic activity is mediated by a characteristic catalytic triad consisting of Asp, His, and Ser. The members, substrates and catalytic mechanism of the serine type proteases is reviewed in Hedstrom et al.89 In contrast, metalloproteases depend on metal ions, especially Zn2+, for their catalytic function.90 The aspartyl proteases are characterized instead by two catalytic Asp residues mediating an acid-base hydrolysis reaction.91 The structure and function of aspartyl proteases was reviewed by Dunn.92
It is important to highlight the fact that early studies of vacuolar proteases led to seminal discoveries in cell biology. First, many vacuolar proteases, including CPY, CPS, and Ape1, are translated as precursors that are proteolytically cleaved to become mature, active proteases in the vacuole. In addition to proteolysis, CPY and CPS are glycosylated in the secretory pathway, altering their molecular weight. Thus, progressive protease maturation is a marker that can pinpoint protease location during vacuolar transit.40,82 (The reader is referred to figure 2 in ref. 40 for a cartoon outlining the maturation events associated with the vacuolar proteases.)
Perhaps the best-characterized example of a protease that follows these events is CPY. The CPY signal sequence targets the nascent protein to the ER and is then removed. ER lumenal CPY has a propeptide, which blocks protease activity and aids in CPY folding and glycosylation to generate p1-CPY.93,94 Further glycosylation in the Golgi forms the higher molecular weight p2-CPY species. Finally, in the vacuole Pep4 cleaves the CPY propeptide to produce the lower molecular weight, mature CPY enzyme.95 Defects in CPY processing were instrumental in identifying proteins involved in protein quality control, glycosylation, and trafficking pathways between the ER and vacuole. When Golgi-to-vacuolar trafficking is impaired, CPY is secreted into the periplasmic space, and several genetic screens cleverly exploited this feature to identify proteins required for this trafficking interval.20,22,23,96-98 For example, the CPY sorting receptor, Vps10, which is required for Golgi-to-MVB trafficking, was first identified in one such screen, and additional sorting receptors have since been characterized.99,100
Unlike CPY, which requires the Vps10 receptor for Golgi-to-MVB trafficking, carboxypeptidase S (CPS) is produced as a precursor (pCPS) that is a type-II membrane protein.40,101 Once pCPS reaches the vacuole Pep4 or Prb1 cleave the transmembrane anchor to generate mature CPS (mCPS), which is a soluble peptidase in the vacuolar lumen.101 Therefore, CPS maturation can also be tracked by monitoring changes in the protein’s molecular weight.101,102 Studies using CPS demonstrated that Rsp5-mediated ubiquitination of cargos and recognition of the ubiquitinated species by the ESCRTs drives sorting to the vacuolar lumen.29,31,102,103 Efficient ubiquitination and segregation of CPS to the vacuolar lumen also requires adaptor proteins Tul1 and Bsd2, each of which contain L/PPxY motifs that recruit the Rsp5 ubiquitin ligase to CPS.104,105 This paradigm of ubiquitin ligase recruitment to substrates in the secretory pathway by adaptors is broadly conserved.34
In stark contrast to CPY and CPS, aminopeptidase 1 (referred to as Ape1 or API) does not enter the secretory pathway and lacks a signal sequence motif.106 While CPY and CPS transit to the vacuole in ~6 min, Ape1 requires over 40 min to mature.106,107 As noted above, Ape1 uses the Cvt pathway.52,106,108 The Cvt pathway requires similar machinery to the autophagy pathway and represents a de novo mechanism for vesicle formation in the cytoplasm.109,110
Soluble vacuolar proteases
Proteinase A (PrA), encoded by the PEP4 gene, is a monomeric 42 kDa aspartyl endoprotease of the A1 family of proteases. PrA is targeted to the vacuole via the secretory pathway and is a key enzyme in the vacuolar protease activation cascade.1,40,111 Many vacuolar hydrolases, including PrA, are initially produced as inactive precursor forms, known as zymogens.112 Zymogens are not activated until they have been delivered to the vacuole, where an inhibitory propeptide is removed by proteolysis. This allows hydrolase activity to be sequestered within the vacuole, thereby protecting residents of the secretory pathway from proteolytic damage. The vacuolar proteases, proteinase B (PrB), carboxypeptidase Y (CPY), and aminopeptidase I (API), which are discussed below, each depend on PrA for proteolytic activation. Therefore, PrA mutant strains are deficient in the activity of these proteases as well.40
PrA is initially synthesized as an inactive precursor referred to as preproPrA. PreproPrA is 405 amino acids in length and has a molecular mass of 52 kDa. PreproPrA has a hydrophobic 22 amino acid signal sequence that directs its translocation into the ER, where it is modified by N-linked glycans at Asn67 and Asn266.113,114 The signal peptide is cleaved by the signal peptidase to produce proPrA, which is then transported to the Golgi. The carbohydrate residues of proPrA are extended by mannosylation in the Golgi. ProPrA is then recognized by the vacuolar protein sorting receptor, Vps10, which targets it to the vacuole.115 Either within the transport vesicle or the vacuole, proPrA undergoes autocatalytic activation, whereby a 54 amino acid propeptide is removed to produce active PrA.116 Active PrA was shown in early work to preferentially cleave substrates between two hydrophobic amino acids, with Phe, Leu, and Glu at the P1 site and Phe, Ile, Leu, and Ala at the P1’ site.117,118 PrA activity extending to polar and charged residues has also been observed, suggesting that PrA may be a more general protease.119
Proteinase B (PrB) is a serine endoprotease of the S8 family and is encoded by PRB1. The zymogen preproPrB is initially synthesized as a 76 kDa polypeptide that is translocated into the ER where its 20 amino acid signal peptide is removed and the resulting polypeptide is modified by a single N-linked glycan.120 PreproPrB undergoes autocatalytic cleavage in the ER, removing an N-terminal 260 amino acid propeptide.121 To facilitate the proper folding of proPrB, the ER-resident protein, Pbn1, acts as a chaperone during this process,122,123 and proPrB is further modified by O-linked glycans in the Golgi.120 The N-terminal propeptide is thought to remain non-covalently associated with proPrB, partially inhibiting its enzymatic activity until proPrB is targeted to the vacuole. Here, the N-terminal propeptide is degraded by PrA, which activates the enzyme.123 In the vacuole a C-terminal propeptide (~30 amino acids) of proPrB is also cleaved by PrA to produce a 37 kDa PrB species. The final cleavage event is mediated autocatalytically by PrB, removing a C-terminal peptide modified by a single N-linked glycan, and yielding the mature 31 kDa PrB.120 Intriguingly, while disrupting PrA activity leads to accumulation of proPrB and the N-terminal propeptide, proPrB retains residual catalytic activity.40
Information on the substrate specificity of PrB comes from studies of PrB isolated from both S. cerevisiae and C. albicans. Both artificial peptides and non-yeast proteins, such as insulin, were used to test PrB substrate specificity in these studies.124,125 Based on these studies, PrB has broad substrate specificity that depends on amino acid context. The protease cleaves substrates at Arg, Leu, Tyr, Phe, Lys, or Asp in the P1 site, and Phe, Tyr, Val, Gly, Leu, Glu, or His in the P1’ site.124,125
Carboxypeptidase Y (CPY) is a serine carboxypeptidase of the S10 family, and is encoded by PRC1 in yeast. As outlined above, CPY is initially synthesized as a ~60 kDa precursor that is 532 amino acids in length. The protein is translocated into the ER where a 20 amino acid signal peptide is removed by signal peptidase and four N-linked glycan residues are appended to produce a 67 kDa form called p1-CPY.126 CPY protein folding in the ER involves the formation of five disulfide bonds and is facilitated by a 91 amino acid propeptide, which acts as an intramolecular chaperone.127 The p1-CPY protein is delivered to the Golgi where glycan residues are extended to produce the 69 kDa species, referred to as p2-CPY. In the Golgi, p2-CPY is recognized by Vps10 for delivery to the MVB via a Gln-Arg-Pro-Leu recognition sequence in the propeptide region.40,99,126 In the vacuole, the propeptide of CPY is removed by the sequential action of PrA and PrB to produce active CPY.1,126
CPY contains a catalytic triad characteristic of serine proteases, which is comprised of Ser146, His397, and Asp338.126 The enzyme is active at low pH and high salt concentrations, which are characteristic of the vacuolar environment. Substrates of CPY are recognized via their C-terminal carboxyl group, which associates with CPY by hydrogen bonding near the S1’ binding pocket. The S1’ subsite in the substrate binding pocket is relatively large and can recognize both hydrophobic and hydrophilic residues. However, the hydrophobic S1 subsite exhibits greater specificity toward hydrophobic amino acids by virtue of being lined with bulky Tyr residues and having a Leu at the bottom of the binding pocket.126
Carboxypeptidase S (CPS) is a zinc-dependent metallo-carboxypeptidase of the M20 family, and is encoded by CPS1. As introduced above, CPS is synthesized as a ~64 kDa precursor containing a membrane sequence spanning amino acids 20 through 40 that is inserted into the ER membrane such that CPS is oriented with its C-terminus facing the lumen.128 The CPS membrane-bound precursor is glycosylated and transits through the Golgi before being targeted to the vacuole via the CPY pathway.33 Once in the vacuole, CPS is processed by PrB and is released into the vacuolar lumen. Both 74 kDa and 77 kDa mature forms of CPS are observed, representing CPS modified by 2 or 3 N-linked glycans, respectively. It is interesting to note that the membrane-bound form of CPS also exhibits proteolytic activity.101
CPS has partially overlapping substrate specificity with CPY, contributing ~60% of the enzymatic activity required to hydrolyze the synthetic dipeptide Cbz-Gly-Leu, where Cbz is the amino protecting group benzyloxycarbonyl.129 In a prc1Δ strain, CPS is required for growth on media when Cbz-Gly-Leu is the sole source of nitrogen and Leu.128 CPS has been found to play a role in sporulation efficiency. Specifically, disrupting PrB activity produces a partial defect in sporulation, but when PrB activity is disrupted together with CPY and CPS activity yeast are unable to sporulate.129
Aminopeptidase I (Ape1) is a zinc-dependent metallo-aminopeptidase of the M18 family, encoded by the APE1 gene in yeast. Ape1 is synthesized as 61 kDa precursor known as preApe1, which contains a 45 amino acid N-terminal helix-loop-helix domain that is required for vacuolar localization. PreApe1 homooligomerizes in the cytoplasm, forming a dodecamer with a molecular mass of 372 kDa that then utilizes the Cvt pathway.130,131 The preApe1 complex is recognized by its receptor, Atg19, which interacts with Atg11 to tether preApe1 complex to the site of autophagosome formation, known as the Phagophore Assembly Site (PAS).132 Next, the preApe1 complex is encapsulated into the Cvt vesicle that fuses with the vacuole. Exposure to the acidic pH of the vacuolar lumen results in disassembly of the preApe1 complex to preApe1 dodecamers.131 Finally, PrB cleaves the N-terminal propeptide in preApe1 to produce the active 50 kDa enzyme.132,133
ApeI is also known as Lap4 because it was initially characterized as a Leu aminopeptidase in a screen for yeast mutants defective for the ability to hydrolyze the synthetic substrate Leu β-naphthylamide.108 Ape1 was also shown to mediate resistance to Cd2+, which is otherwise toxic and induces oxidative stress.134 Yeast sequester Cd2+ in the vacuole as a glutathione (GSH) S-conjugate. The GSH is recycled from the vacuole by the action of gamma-glutamyltranspeptidase (Ecm38), which hydrolyzes Glu, leaving a Cys-Gly dipeptide, which is thought to be further degraded by Ape1.134 Thus, Ape1 is required for GSH homeostasis, and defects in this process lead to Cd2+ sensitivity. However, more recent work has identified an alternative GSH degradation pathway involving the cytosolic dipeptidase Dug1, which is a member of the M20 family of metalloproteases. This study demonstrated that Ape1 was not required for growth of a met15Δ strain on media in which GSH was the only source of sulfur.135
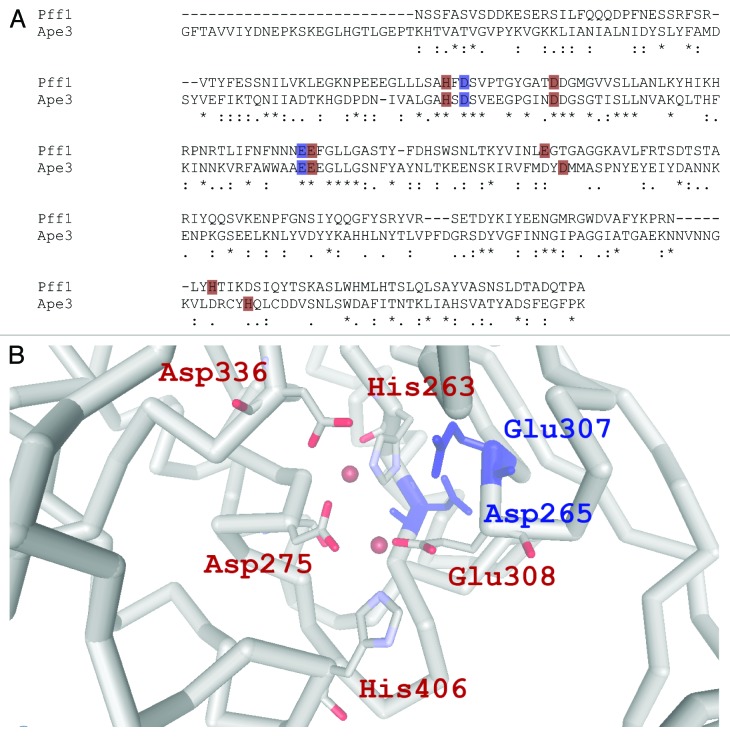
Aminopeptidase Y (Ape3), encoded by the gene APE3, is a metallo-aminopeptidase of the same M28 family of proteases as the recently discovered, putative vacuolar protease, Ybr074w (see below). Like YBR074w, the APE3 locus is found on chromosome II. Global pairwise sequence alignment of Ape3 and Ybr074w, shown in Figure 2, reveals 5.7% amino acid sequence identity, while comparison of the M28 protease domains indicates ~22% sequence identity including conservation of catalytic residues.136 Ape3 is synthesized as a 60 kDa precursor bearing a 21 amino acid signal sequence directing its translocation into the ER.137 The signal sequence is cleaved in the ER, and the Ape3 precursor is targeted to the Golgi, where—as evident with several of the other enzymes discussed above—it is recognized by Vps10 for vacuole targeting.115 Also similar to some of the other proteases, a 35 amino acid N-terminal propeptide is cleaved by PrB in the vacuole, producing mature Ape3.138 The mature Ape3 protein exists in both a 70 kDa and 75 kDa form that differ in the extent of glycan modification, as Ape3 has eight acceptor sites for N-linked glycans but only 5–7 sites are used.138
Figure 2. (A) Sequence alignment of Ape3 (MEROPS accession: MER001288) and Pff1 (MEROPS accession: MER001911), two vacuole-resident yeast metalloproteases. A global sequence alignment was performed using ClustalW default parameters.142 Only the M28 domains of Ape3 and Ybr074 are shown according to the MEROPS defined protease domain boundaries. (B) Active site of the M28 metalloprotease, AM-1 Aminopeptidase, from Aneurinibacillus sp. strain AM-1 (MEROPS accession: MER100667; protease domain: Gly173-Arg436). Of the M28 metalloproteases whose crystal structures have been solved, the protease domain amino acid sequence of AM-1 Aminopeptidase is most similar to that of the yeast Pff1 (MEROPS accession: MER001911; protease domain: Asn96-Ala341). The MEROPS defined protease domain boundaries of AM-1 and Pff1 show 19.2% identity when compared by global sequence alignment using ClustalW default parameters.142 Catalytic residues are labeled in blue and metal coordinating residues are labeled in red in both this panel and in part (A). Zinc ions are depicted as red spheres. This image was captured from PDB ID: 2EK9 using 3D Molecule Viewer of Vector NTI Advance, Version 11.5.2 (Life Technologies).
The enzymatic activity of Ape3 was characterized by Yasuhara et al. using synthetic peptides with a C-terminal 4-methylcoumaryl-7-amide (MCA) protecting group.138 Ape3 exhibits a preference for cleaving the basic residue Lys. However, it also cleaves N-terminal Pro, Ala, Leu, Met, Ser, Phe, and Tyr.138 Interestingly, the hydrolysis of amino acid-MCAs and dipeptides was enhanced in the presence of Co2+, while that of dipeptidyl-MCAs and larger unmodified peptides was inhibited. Although M28 family metalloproteases commonly bind two catalytic zinc ions, the authors suggested that zinc is inhibitory to Ape3 proteolytic activity.138
Membrane-bound vacuolar proteases
Dipeptidylaminopeptidase B (Dap2) is serine protease in the S9 family. Dap2 has a hydrophobic transmembrane segment positioned 30 amino acids from the N-terminus, which serves both as an ER-targeting sequence and membrane anchor. Dap2 is a type II membrane protein, having a cytosolic N-terminus and a prominent lumenal C-terminus. The protein is initially synthesized as a 93 kDa protein, 818 amino acids in length, and is modified in the ER with between 5 and 8 N-linked glycans, which undergo minimal extension in the Golgi. This results in a final ~120 kDa product. Unlike other vacuolar proteases, Dap2 is not proteolytically processed in the vacuole.40,139 Although the substrate specificity of Dap2 is unknown, a Golgi-resident homolog, dipeptidylaminopeptidase A (Ste13) processes repeating X-Ala dipeptides from the yeast α factor mating pheromone.140 Disrupting Ste13 function prevents maturation of α factor and results in sterile MATα cells. This defect in mating was partially restored when Dap2 was overexpressed ~10-fold in a MATα ste13 mutant strain.140
Finally, we recently identified Protease in FXNA-related Family 1 (Pff1) as a vacuolar membrane protein that is predicted to possess nine transmembrane segments.86 Pff1 harbors a conserved M28 protease domain (Fig. 2) that faces the vacuolar lumen, as determined based on the accessibility of this domain when exogenously added protease was added to a crude vesicle preparation. The vacuolar residence of Pff1 was evidenced by the analysis of several different tagged forms of the protein,86 and because the native protein was identified when highly enriched vacuolar membranes were subjected to mass spectrometry analysis.10 Although the specific function and substrate-specificity of Pff1 has yet to be elucidated, a whole genomic analysis of proteins that respond to the absence of the PFF1 (YBR074w) gene product is consistent with the protein playing a role in vacuolar function. Interestingly, a putative mammalian homolog of Pff1 exists.141 However, the mammalian protein was instead found to reside in the ER and appears to be involved in ovarian development. In the future, it will be exciting to determine whether the mammalian enzyme acts on select substrates that traffic through the secretory pathway, and whether Pff1 in yeast exhibits substrate specificity in the vacuole.
Conclusions
While the substrate specificities of many of the vacuolar proteases have been defined using synthetic substrates, significantly less information is available on the action of these proteases on endogenous, bona fide substrates. In principle, substrates might be identified through yeast genetic screens using mutant forms of the corresponding protease. But, because the absence of a protease is often masked by the activity of a functionally redundant enzyme, many of the proteases described in this review were instead obtained by analyzing activities for defined, synthetic substrates, or simply by virtue of sequence gazing. One view is that vacuolar proteases are indiscriminant enzymes because their major requirement in this organelle is to simply process full-length proteins to short peptides and amino acids, which can serve as precursors for ongoing protein synthesis and/or for metabolic, energy-generating processes. If this is the case, one might posit that the proteases evolved to ensure the efficient degradation of any protein substrate that enters the vacuole. Nevertheless, it remains possible that select proteases do exhibit substrate specificity. If so, do these proteases respond to certain physiological conditions requiring vacuole-dependent degradation or processing of distinct substrates? Are there functional characteristics that differentiate soluble vacuolar proteases from membrane-bound vacuolar proteases? Perhaps membrane-bound proteases associate more readily with membrane-bound substrates, but even this question has not been addressed. Given the growing appreciation that proteases can dictate so many critical cellular “decisions,” and combined with the development of genomic and proteomic tools that permit a better understanding of cellular protein networks, it is likely that much remains to be learned about the myriad proteases that provide house-keeping functions in the vacuole.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by development funds from the Department of Cell Biology (University of Pittsburgh) and grant DA014204–11 (awarded to A. Sorkin, University of Pittsburgh) from the National Institutes of Health for A.F.O, and by grant GM75061 and DK79307 (The Pittsburgh Center for Kidney Research) from the National Institutes of Health to J.L.B.
Footnotes
Previously published online: www.landesbioscience.com/journals/cellularlogistics/article/28023
References
- 1.Broach JR, Pringle JR, Jones EW. The Molecular and cellular biology of the yeast Saccharomyces. Cold Spring Harbor, N.Y.: Cole Spring Harbor Laboratory Press, 1991. [Google Scholar]
- 2.Li SC, Kane PM. The yeast lysosome-like vacuole: endpoint and crossroads. Biochim Biophys Acta. 2009;1793:650–63. doi: 10.1016/j.bbamcr.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marty F. Plant vacuoles. Plant Cell. 1999;11:587–600. doi: 10.1105/tpc.11.4.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Veses V, Richards A, Gow NA. Vacuoles and fungal biology. Curr Opin Microbiol. 2008;11:503–10. doi: 10.1016/j.mib.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 5.Klionsky DJ, Herman PK, Emr SD. The fungal vacuole: composition, function, and biogenesis. Microbiol Rev. 1990;54:266–92. doi: 10.1128/mr.54.3.266-292.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wiemken A, Matile P, Moor H. Vacuolar dynamics in synchronously budding yeast. Arch Mikrobiol. 1970;70:89–103. doi: 10.1007/BF00412200. [DOI] [PubMed] [Google Scholar]
- 7.Brett CL, Kallay L, Hua Z, Green R, Chyou A, Zhang Y, Graham TR, Donowitz M, Rao R. Genome-wide analysis reveals the vacuolar pH-stat of Saccharomyces cerevisiae. PLoS One. 2011;6:e17619. doi: 10.1371/journal.pone.0017619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Preston RA, Murphy RF, Jones EW. Assay of vacuolar pH in yeast and identification of acidification-defective mutants. Proc Natl Acad Sci U S A. 1989;86:7027–31. doi: 10.1073/pnas.86.18.7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sarry JE, Chen S, Collum RP, Liang S, Peng M, Lang A, Naumann B, Dzierszinski F, Yuan CX, Hippler M, et al. Analysis of the vacuolar luminal proteome of Saccharomyces cerevisiae. FEBS J. 2007;274:4287–305. doi: 10.1111/j.1742-4658.2007.05959.x. [DOI] [PubMed] [Google Scholar]
- 10.Wiederhold E, Gandhi T, Permentier HP, Breitling R, Poolman B, Slotboom DJ. The yeast vacuolar membrane proteome. Mol Cell Proteomics. 2009;8:380–92. doi: 10.1074/mcp.M800372-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Yan A, Lennarz WJ. Unraveling the mechanism of protein N-glycosylation. J Biol Chem. 2005;280:3121–4. doi: 10.1074/jbc.R400036200. [DOI] [PubMed] [Google Scholar]
- 12.Goto M. Protein O-glycosylation in fungi: diverse structures and multiple functions. Biosci Biotechnol Biochem. 2007;71:1415–27. doi: 10.1271/bbb.70080. [DOI] [PubMed] [Google Scholar]
- 13.Harsay E, Schekman R. A subset of yeast vacuolar protein sorting mutants is blocked in one branch of the exocytic pathway. J Cell Biol. 2002;156:271–85. doi: 10.1083/jcb.200109077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valdivia RH, Baggott D, Chuang JS, Schekman RW. The yeast clathrin adaptor protein complex 1 is required for the efficient retention of a subset of late Golgi membrane proteins. Dev Cell. 2002;2:283–94. doi: 10.1016/S1534-5807(02)00127-2. [DOI] [PubMed] [Google Scholar]
- 15.Cowles CR, Odorizzi G, Payne GS, Emr SD. The AP-3 adaptor complex is essential for cargo-selective transport to the yeast vacuole. Cell. 1997;91:109–18. doi: 10.1016/S0092-8674(01)80013-1. [DOI] [PubMed] [Google Scholar]
- 16.Stepp JD, Huang K, Lemmon SK. The yeast adaptor protein complex, AP-3, is essential for the efficient delivery of alkaline phosphatase by the alternate pathway to the vacuole. J Cell Biol. 1997;139:1761–74. doi: 10.1083/jcb.139.7.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowers K, Stevens TH. Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 2005;1744:438–54. doi: 10.1016/j.bbamcr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 18.Vida TA, Graham TR, Emr SD. In vitro reconstitution of intercompartmental protein transport to the yeast vacuole. J Cell Biol. 1990;111:2871–84. doi: 10.1083/jcb.111.6.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vida TA, Huyer G, Emr SD. Yeast vacuolar proenzymes are sorted in the late Golgi complex and transported to the vacuole via a prevacuolar endosome-like compartment. J Cell Biol. 1993;121:1245–56. doi: 10.1083/jcb.121.6.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bankaitis VA, Johnson LM, Emr SD. Isolation of yeast mutants defective in protein targeting to the vacuole. Proc Natl Acad Sci U S A. 1986;83:9075–9. doi: 10.1073/pnas.83.23.9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banta LM, Robinson JS, Klionsky DJ, Emr SD. Organelle assembly in yeast: characterization of yeast mutants defective in vacuolar biogenesis and protein sorting. J Cell Biol. 1988;107:1369–83. doi: 10.1083/jcb.107.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raymond CK, Howald-Stevenson I, Vater CA, Stevens TH. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol Biol Cell. 1992;3:1389–402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rothman JH, Stevens TH. Protein sorting in yeast: mutants defective in vacuole biogenesis mislocalize vacuolar proteins into the late secretory pathway. Cell. 1986;47:1041–51. doi: 10.1016/0092-8674(86)90819-6. [DOI] [PubMed] [Google Scholar]
- 24.Abazeed ME, Fuller RS. Yeast Golgi-localized, gamma-Ear-containing, ADP-ribosylation factor-binding proteins are but adaptor protein-1 is not required for cell-free transport of membrane proteins from the trans-Golgi network to the prevacuolar compartment. Mol Biol Cell. 2008;19:4826–36. doi: 10.1091/mbc.E07-05-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonifacino JS. The GGA proteins: adaptors on the move. Nat Rev Mol Cell Biol. 2004;5:23–32. doi: 10.1038/nrm1279. [DOI] [PubMed] [Google Scholar]
- 26.Costaguta G, Duncan MC, Fernández GE, Huang GH, Payne GS. Distinct roles for TGN/endosome epsin-like adaptors Ent3p and Ent5p. Mol Biol Cell. 2006;17:3907–20. doi: 10.1091/mbc.E06-05-0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dell’Angelica EC, Puertollano R, Mullins C, Aguilar RC, Vargas JD, Hartnell LM, Bonifacino JS. GGAs: a family of ADP ribosylation factor-binding proteins related to adaptors and associated with the Golgi complex. J Cell Biol. 2000;149:81–94. doi: 10.1083/jcb.149.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gary JD, Wurmser AE, Bonangelino CJ, Weisman LS, Emr SD. Fab1p is essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J Cell Biol. 1998;143:65–79. doi: 10.1083/jcb.143.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Odorizzi G, Babst M, Emr SD. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 1998;95:847–58. doi: 10.1016/S0092-8674(00)81707-9. [DOI] [PubMed] [Google Scholar]
- 30.Helliwell SB, Losko S, Kaiser CA. Components of a ubiquitin ligase complex specify polyubiquitination and intracellular trafficking of the general amino acid permease. J Cell Biol. 2001;153:649–62. doi: 10.1083/jcb.153.4.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reggiori F, Pelham HR. Sorting of proteins into multivesicular bodies: ubiquitin-dependent and -independent targeting. EMBO J. 2001;20:5176–86. doi: 10.1093/emboj/20.18.5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beck T, Schmidt A, Hall MN. Starvation induces vacuolar targeting and degradation of the tryptophan permease in yeast. J Cell Biol. 1999;146:1227–38. doi: 10.1083/jcb.146.6.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lauwers E, Jacob C, André B. K63-linked ubiquitin chains as a specific signal for protein sorting into the multivesicular body pathway. J Cell Biol. 2009;185:493–502. doi: 10.1083/jcb.200810114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lauwers E, Erpapazoglou Z, Haguenauer-Tsapis R, André B. The ubiquitin code of yeast permease trafficking. Trends Cell Biol. 2010;20:196–204. doi: 10.1016/j.tcb.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 35.Babst M. A protein’s final ESCRT. Traffic. 2005;6:2–9. doi: 10.1111/j.1600-0854.2004.00246.x. [DOI] [PubMed] [Google Scholar]
- 36.Balderhaar HJ, Ungermann C. CORVET and HOPS tethering complexes - coordinators of endosome and lysosome fusion. J Cell Sci. 2013;126:1307–16. doi: 10.1242/jcs.107805. [DOI] [PubMed] [Google Scholar]
- 37.Bröcker C, Kuhlee A, Gatsogiannis C, Balderhaar HJ, Hönscher C, Engelbrecht-Vandré S, Ungermann C, Raunser S. Molecular architecture of the multisubunit homotypic fusion and vacuole protein sorting (HOPS) tethering complex. Proc Natl Acad Sci U S A. 2012;109:1991–6. doi: 10.1073/pnas.1117797109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lachmann J, Ungermann C, Engelbrecht-Vandré S. Rab GTPases and tethering in the yeast endocytic pathway. Small GTPases. 2011;2:182–6. doi: 10.4161/sgtp.2.3.16701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Angers CG, Merz AJ. HOPS interacts with Apl5 at the vacuole membrane and is required for consumption of AP-3 transport vesicles. Mol Biol Cell. 2009;20:4563–74. doi: 10.1091/mbc.E09-04-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Den Hazel HB, Kielland-Brandt MC, Winther JR. Review: biosynthesis and function of yeast vacuolar proteases. Yeast. 1996;12:1–16. doi: 10.1002/(SICI)1097-0061(199601)12:1<1::AID-YEA902>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 41.Thumm M. Structure and function of the yeast vacuole and its role in autophagy. Microsc Res Tech. 2000;51:563–72. doi: 10.1002/1097-0029(20001215)51:6<563::AID-JEMT6>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 42.Teichert U, Mechler B, Müller H, Wolf DH. Lysosomal (vacuolar) proteinases of yeast are essential catalysts for protein degradation, differentiation, and cell survival. J Biol Chem. 1989;264:16037–45. [PubMed] [Google Scholar]
- 43.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–73. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 44.Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992;119:301–11. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mari M, Tooze SA, Reggiori F. The puzzling origin of the autophagosomal membrane. F1000 Biol Rep. 2011;3:25. doi: 10.3410/B3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reggiori F, Klionsky DJ. Autophagy in the eukaryotic cell. Eukaryot Cell. 2002;1:11–21. doi: 10.1128/EC.01.1.11-21.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hopper AK, Magee PT, Welch SK, Friedman M, Hall BD. Macromolecule synthesis and breakdown in relation to sporulation and meiosis in yeast. J Bacteriol. 1974;119:619–28. doi: 10.1128/jb.119.2.619-628.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–9. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 49.Khalfan WA, Klionsky DJ. Molecular machinery required for autophagy and the cytoplasm to vacuole targeting (Cvt) pathway in S. cerevisiae. Curr Opin Cell Biol. 2002;14:468–75. doi: 10.1016/S0955-0674(02)00343-5. [DOI] [PubMed] [Google Scholar]
- 50.Yuga M, Gomi K, Klionsky DJ, Shintani T. Aspartyl aminopeptidase is imported from the cytoplasm to the vacuole by selective autophagy in Saccharomyces cerevisiae. J Biol Chem. 2011;286:13704–13. doi: 10.1074/jbc.M110.173906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scott SV, Baba M, Ohsumi Y, Klionsky DJ. Aminopeptidase I is targeted to the vacuole by a nonclassical vesicular mechanism. J Cell Biol. 1997;138:37–44. doi: 10.1083/jcb.138.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baba M, Osumi M, Scott SV, Klionsky DJ, Ohsumi Y. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J Cell Biol. 1997;139:1687–95. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hutchins MU, Klionsky DJ. Vacuolar localization of oligomeric alpha-mannosidase requires the cytoplasm to vacuole targeting and autophagy pathway components in Saccharomyces cerevisiae. J Biol Chem. 2001;276:20491–8. doi: 10.1074/jbc.M101150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kraft C, Reggiori F, Peter M. Selective types of autophagy in yeast. Biochim Biophys Acta. 2009;1793:1404–12. doi: 10.1016/j.bbamcr.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 55.Riezman H. Endocytosis in yeast: several of the yeast secretory mutants are defective in endocytosis. Cell. 1985;40:1001–9. doi: 10.1016/0092-8674(85)90360-5. [DOI] [PubMed] [Google Scholar]
- 56.Munn AL, Stevenson BJ, Geli MI, Riezman H. end5, end6, and end7: mutations that cause actin delocalization and block the internalization step of endocytosis in Saccharomyces cerevisiae. Mol Biol Cell. 1995;6:1721–42. doi: 10.1091/mbc.6.12.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wendland B, McCaffery JM, Xiao Q, Emr SD. A novel fluorescence-activated cell sorter-based screen for yeast endocytosis mutants identifies a yeast homologue of mammalian eps15. J Cell Biol. 1996;135:1485–500. doi: 10.1083/jcb.135.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Raths S, Rohrer J, Crausaz F, Riezman H. end3 and end4: two mutants defective in receptor-mediated and fluid-phase endocytosis in Saccharomyces cerevisiae. J Cell Biol. 1993;120:55–65. doi: 10.1083/jcb.120.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaksonen M, Toret CP, Drubin DG. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell. 2005;123:305–20. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 60.Kukulski W, Schorb M, Kaksonen M, Briggs JA. Plasma membrane reshaping during endocytosis is revealed by time-resolved electron tomography. Cell. 2012;150:508–20. doi: 10.1016/j.cell.2012.05.046. [DOI] [PubMed] [Google Scholar]
- 61.Newpher TM, Smith RP, Lemmon V, Lemmon SK. In vivo dynamics of clathrin and its adaptor-dependent recruitment to the actin-based endocytic machinery in yeast. Dev Cell. 2005;9:87–98. doi: 10.1016/j.devcel.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 62.Idrissi FZ, Grötsch H, Fernández-Golbano IM, Presciatto-Baschong C, Riezman H, Geli MI. Distinct acto/myosin-I structures associate with endocytic profiles at the plasma membrane. J Cell Biol. 2008;180:1219–32. doi: 10.1083/jcb.200708060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boettner DR, Chi RJ, Lemmon SK. Lessons from yeast for clathrin-mediated endocytosis. Nat Cell Biol. 2012;14:2–10. doi: 10.1038/ncb2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Godlee C, Kaksonen M. Review series: From uncertain beginnings: initiation mechanisms of clathrin-mediated endocytosis. J Cell Biol. 2013;203:717–25. doi: 10.1083/jcb.201307100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kaksonen M, Toret CP, Drubin DG. Harnessing actin dynamics for clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2006;7:404–14. doi: 10.1038/nrm1940. [DOI] [PubMed] [Google Scholar]
- 66.Weinberg J, Drubin DG. Clathrin-mediated endocytosis in budding yeast. Trends Cell Biol. 2012;22:1–13. doi: 10.1016/j.tcb.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Doyon JB, Zeitler B, Cheng J, Cheng AT, Cherone JM, Santiago Y, Lee AH, Vo TD, Doyon Y, Miller JC, et al. Rapid and efficient clathrin-mediated endocytosis revealed in genome-edited mammalian cells. Nat Cell Biol. 2011;13:331–7. doi: 10.1038/ncb2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Taylor MJ, Perrais D, Merrifield CJ. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 2011;9:e1000604. doi: 10.1371/journal.pbio.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Prosser DC, Drivas TG, Maldonado-Báez L, Wendland B. Existence of a novel clathrin-independent endocytic pathway in yeast that depends on Rho1 and formin. J Cell Biol. 2011;195:657–71. doi: 10.1083/jcb.201104045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hicke L, Riezman H. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell. 1996;84:277–87. doi: 10.1016/S0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- 71.Galan JM, Moreau V, Andre B, Volland C, Haguenauer-Tsapis R. Ubiquitination mediated by the Npi1p/Rsp5p ubiquitin-protein ligase is required for endocytosis of the yeast uracil permease. J Biol Chem. 1996;271:10946–52. doi: 10.1074/jbc.271.18.10946. [DOI] [PubMed] [Google Scholar]
- 72.Springael JY, André B. Nitrogen-regulated ubiquitination of the Gap1 permease of Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:1253–63. doi: 10.1091/mbc.9.6.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dunn R, Hicke L. Multiple roles for Rsp5p-dependent ubiquitination at the internalization step of endocytosis. J Biol Chem. 2001;276:25974–81. doi: 10.1074/jbc.M104113200. [DOI] [PubMed] [Google Scholar]
- 74.Dunn R, Hicke L. Domains of the Rsp5 ubiquitin-protein ligase required for receptor-mediated and fluid-phase endocytosis. Mol Biol Cell. 2001;12:421–35. doi: 10.1091/mbc.12.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hein C, Springael JY, Volland C, Haguenauer-Tsapis R, André B. NPl1, an essential yeast gene involved in induced degradation of Gap1 and Fur4 permeases, encodes the Rsp5 ubiquitin-protein ligase. Mol Microbiol. 1995;18:77–87. doi: 10.1111/j.1365-2958.1995.mmi_18010077.x. [DOI] [PubMed] [Google Scholar]
- 76.Lin CH, MacGurn JA, Chu T, Stefan CJ, Emr SD. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell. 2008;135:714–25. doi: 10.1016/j.cell.2008.09.025. [DOI] [PubMed] [Google Scholar]
- 77.Nikko E, Sullivan JA, Pelham HR. Arrestin-like proteins mediate ubiquitination and endocytosis of the yeast metal transporter Smf1. EMBO Rep. 2008;9:1216–21. doi: 10.1038/embor.2008.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Becuwe M, Vieira N, Lara D, Gomes-Rezende J, Soares-Cunha C, Casal M, Haguenauer-Tsapis R, Vincent O, Paiva S, Léon S. A molecular switch on an arrestin-like protein relays glucose signaling to transporter endocytosis. J Cell Biol. 2012;196:247–59. doi: 10.1083/jcb.201109113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Merhi A, André B. Internal amino acids promote Gap1 permease ubiquitylation via TORC1/Npr1/14-3-3-dependent control of the Bul arrestin-like adaptors. Mol Cell Biol. 2012;32:4510–22. doi: 10.1128/MCB.00463-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.O’Donnell AF, Huang L, Thorner J, Cyert MS. A calcineurin-dependent switch controls the trafficking function of α-arrestin Aly1/Art6. J Biol Chem. 2013;288:24063–80. doi: 10.1074/jbc.M113.478511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nikko E, Marini AM, André B. Permease recycling and ubiquitination status reveal a particular role for Bro1 in the multivesicular body pathway. J Biol Chem. 2003;278:50732–43. doi: 10.1074/jbc.M306953200. [DOI] [PubMed] [Google Scholar]
- 82.Jones EW. Three proteolytic systems in the yeast saccharomyces cerevisiae. J Biol Chem. 1991;266:7963–6. [PubMed] [Google Scholar]
- 83.Ammerer G, Hunter CP, Rothman JH, Saari GC, Valls LA, Stevens TH. PEP4 gene of Saccharomyces cerevisiae encodes proteinase A, a vacuolar enzyme required for processing of vacuolar precursors. Mol Cell Biol. 1986;6:2490–9. doi: 10.1128/mcb.6.7.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Berkower C, Loayza D, Michaelis S. Metabolic instability and constitutive endocytosis of STE6, the a-factor transporter of Saccharomyces cerevisiae. Mol Biol Cell. 1994;5:1185–98. doi: 10.1091/mbc.5.11.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schandel KA, Jenness DD. Direct evidence for ligand-induced internalization of the yeast alpha-factor pheromone receptor. Mol Cell Biol. 1994;14:7245–55. doi: 10.1128/mcb.14.11.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hecht KA, Wytiaz VA, Ast T, Schuldiner M, Brodsky JL. Characterization of an M28 metalloprotease family member residing in the yeast vacuole. FEMS Yeast Res. 2013;13:471–84. doi: 10.1111/1567-1364.12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rawlings ND, Barrett AJ. Evolutionary families of peptidases. Biochem J. 1993;290:205–18. doi: 10.1042/bj2900205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rawlings ND, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2012;40:D343–50. doi: 10.1093/nar/gkr987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hedstrom L. Serine protease mechanism and specificity. Chem Rev. 2002;102:4501–24. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- 90.Rawlings ND, Barrett AJ. Evolutionary families of metallopeptidases. Methods Enzymol. 1995;248:183–228. doi: 10.1016/0076-6879(95)48015-3. [DOI] [PubMed] [Google Scholar]
- 91.Tang J, Wong RN. Evolution in the structure and function of aspartic proteases. J Cell Biochem. 1987;33:53–63. doi: 10.1002/jcb.240330106. [DOI] [PubMed] [Google Scholar]
- 92.Dunn BM. Structure and mechanism of the pepsin-like family of aspartic peptidases. Chem Rev. 2002;102:4431–58. doi: 10.1021/cr010167q. [DOI] [PubMed] [Google Scholar]
- 93.Winther JR, Sørensen P. Propeptide of carboxypeptidase Y provides a chaperone-like function as well as inhibition of the enzymatic activity. Proc Natl Acad Sci U S A. 1991;88:9330–4. doi: 10.1073/pnas.88.20.9330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stevens T, Esmon B, Schekman R. Early stages in the yeast secretory pathway are required for transport of carboxypeptidase Y to the vacuole. Cell. 1982;30:439–48. doi: 10.1016/0092-8674(82)90241-0. [DOI] [PubMed] [Google Scholar]
- 95.Zubenko GS, Park FJ, Jones EW. Mutations in PEP4 locus of Saccharomyces cerevisiae block final step in maturation of two vacuolar hydrolases. Proc Natl Acad Sci U S A. 1983;80:510–4. doi: 10.1073/pnas.80.2.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bonangelino CJ, Chavez EM, Bonifacino JS. Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:2486–501. doi: 10.1091/mbc.02-01-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8:4936–48. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rothman JH, Howald I, Stevens TH. Characterization of genes required for protein sorting and vacuolar function in the yeast Saccharomyces cerevisiae. EMBO J. 1989;8:2057–65. doi: 10.1002/j.1460-2075.1989.tb03614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Marcusson EG, Horazdovsky BF, Cereghino JL, Gharakhanian E, Emr SD. The sorting receptor for yeast vacuolar carboxypeptidase Y is encoded by the VPS10 gene. Cell. 1994;77:579–86. doi: 10.1016/0092-8674(94)90219-4. [DOI] [PubMed] [Google Scholar]
- 100.Whyte JR, Munro S. A yeast homolog of the mammalian mannose 6-phosphate receptors contributes to the sorting of vacuolar hydrolases. Curr Biol. 2001;11:1074–8. doi: 10.1016/S0960-9822(01)00273-1. [DOI] [PubMed] [Google Scholar]
- 101.Spormann DO, Heim J, Wolf DH. Biogenesis of the yeast vacuole (lysosome). The precursor forms of the soluble hydrolase carboxypeptidase yscS are associated with the vacuolar membrane. J Biol Chem. 1992;267:8021–9. [PubMed] [Google Scholar]
- 102.Katzmann DJ, Babst M, Emr SD. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106:145–55. doi: 10.1016/S0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- 103.Katzmann DJ, Sarkar S, Chu T, Audhya A, Emr SD. Multivesicular body sorting: ubiquitin ligase Rsp5 is required for the modification and sorting of carboxypeptidase S. Mol Biol Cell. 2004;15:468–80. doi: 10.1091/mbc.E03-07-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hettema EH, Valdez-Taubas J, Pelham HR. Bsd2 binds the ubiquitin ligase Rsp5 and mediates the ubiquitination of transmembrane proteins. EMBO J. 2004;23:1279–88. doi: 10.1038/sj.emboj.7600137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Reggiori F, Pelham HR. A transmembrane ubiquitin ligase required to sort membrane proteins into multivesicular bodies. Nat Cell Biol. 2002;4:117–23. doi: 10.1038/ncb743. [DOI] [PubMed] [Google Scholar]
- 106.Klionsky DJ, Cueva R, Yaver DS. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J Cell Biol. 1992;119:287–99. doi: 10.1083/jcb.119.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hasilik A, Tanner W. Biosynthesis of the vacuolar yeast glycoprotein carboxypeptidase Y. Conversion of precursor into the enzyme. Eur J Biochem. 1978;85:599–608. doi: 10.1111/j.1432-1033.1978.tb12275.x. [DOI] [PubMed] [Google Scholar]
- 108.Trumbly RJ, Bradley G. Isolation and characterization of aminopeptidase mutants of Saccharomyces cerevisiae. J Bacteriol. 1983;156:36–48. doi: 10.1128/jb.156.1.36-48.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Harding TM, Hefner-Gravink A, Thumm M, Klionsky DJ. Genetic and phenotypic overlap between autophagy and the cytoplasm to vacuole protein targeting pathway. J Biol Chem. 1996;271:17621–4. doi: 10.1074/jbc.271.30.17621. [DOI] [PubMed] [Google Scholar]
- 110.Scott SV, Hefner-Gravink A, Morano KA, Noda T, Ohsumi Y, Klionsky DJ. Cytoplasm-to-vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole. Proc Natl Acad Sci U S A. 1996;93:12304–8. doi: 10.1073/pnas.93.22.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Westphal V, Marcusson EG, Winther JR, Emr SD, van den Hazel HB. Multiple pathways for vacuolar sorting of yeast proteinase A. J Biol Chem. 1996;271:11865–70. doi: 10.1074/jbc.271.20.11865. [DOI] [PubMed] [Google Scholar]
- 112.van den Hazel HB, Kielland-Brandt MC, Winther JR. Autoactivation of proteinase A initiates activation of yeast vacuolar zymogens. Eur J Biochem. 1992;207:277–83. doi: 10.1111/j.1432-1033.1992.tb17048.x. [DOI] [PubMed] [Google Scholar]
- 113.Mechler B, Müller M, Müller H, Meussdoerffer F, Wolf DH. In vivo biosynthesis of the vacuolar proteinases A and B in the yeast Saccharomyces cerevisiae. J Biol Chem. 1982;257:11203–6. [PubMed] [Google Scholar]
- 114.Parr CL, Keates RA, Bryksa BC, Ogawa M, Yada RY. The structure and function of Saccharomyces cerevisiae proteinase A. Yeast. 2007;24:467–80. doi: 10.1002/yea.1485. [DOI] [PubMed] [Google Scholar]
- 115.Jørgensen MU, Emr SD, Winther JR. Ligand recognition and domain structure of Vps10p, a vacuolar protein sorting receptor in Saccharomyces cerevisiae. Eur J Biochem. 1999;260:461–9. doi: 10.1046/j.1432-1327.1999.00176.x. [DOI] [PubMed] [Google Scholar]
- 116.Rupp S, Hirsch HH, Wolf DH. Biogenesis of the yeast vacuole (lysosome). Active site mutation in the vacuolar aspartate proteinase yscA blocks maturation of vacuolar proteinases. FEBS Lett. 1991;293:62–6. doi: 10.1016/0014-5793(91)81153-Y. [DOI] [PubMed] [Google Scholar]
- 117.Kondo H, Shibano Y, Amachi T, Cronin N, Oda K, Dunn BM. Substrate specificities and kinetic properties of proteinase A from the yeast Saccharomyces cerevisiae and the development of a novel substrate. J Biochem. 1998;124:141–7. doi: 10.1093/oxfordjournals.jbchem.a022072. [DOI] [PubMed] [Google Scholar]
- 118.Dreyer T. Substrate specificity of proteinase yscA from saccharomyces cerevisiae. Carlsberg Res Commun. 1989;54:85–97. doi: 10.1007/BF02908301. [DOI] [PubMed] [Google Scholar]
- 119.Barrett AJ, Rawlings ND, Woessner JF. Handbook of proteolytic enzymes. Amsterdam; Boston, MA: Elsevier Academic Press, 2004. [Google Scholar]
- 120.Moehle CM, Dixon CK, Jones EW. Processing pathway for protease B of Saccharomyces cerevisiae. J Cell Biol. 1989;108:309–25. doi: 10.1083/jcb.108.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nebes VL, Jones EW. Activation of the proteinase B precursor of the yeast Saccharomyces cerevisiae by autocatalysis and by an internal sequence. J Biol Chem. 1991;266:22851–7. [PubMed] [Google Scholar]
- 122.Subramanian S, Woolford CA, Drill E, Lu M, Jones EW. Pbn1p: an essential endoplasmic reticulum membrane protein required for protein processing in the endoplasmic reticulum of budding yeast. Proc Natl Acad Sci U S A. 2006;103:939–44. doi: 10.1073/pnas.0505570103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Naik RR, Jones EW. The PBN1 gene of Saccharomyces cerevisiae: an essential gene that is required for the post-translational processing of the protease B precursor. Genetics. 1998;149:1277–92. doi: 10.1093/genetics/149.3.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Farley PC, Shepherd MG, Sullivan PA. The purification and properties of yeast proteinase B from Candida albicans. Biochem J. 1986;236:177–84. doi: 10.1042/bj2360177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kominami E, Hoffschulte H, Leuschel L, Maier K, Holzer H. The substrate specificity of proteinase B from baker’s yeast. Biochim Biophys Acta. 1981;661:136–41. doi: 10.1016/0005-2744(81)90092-9. [DOI] [PubMed] [Google Scholar]
- 126.Jung G, Ueno H, Hayashi R. Carboxypeptidase Y: structural basis for protein sorting and catalytic triad. J Biochem. 1999;126:1–6. doi: 10.1093/oxfordjournals.jbchem.a022408. [DOI] [PubMed] [Google Scholar]
- 127.Tang B, Nirasawa S, Kitaoka M, Hayashi K. The role of the N-terminal propeptide of the pro-aminopeptidase processing protease: refolding, processing, and enzyme inhibition. Biochem Biophys Res Commun. 2002;296:78–84. doi: 10.1016/S0006-291X(02)00838-0. [DOI] [PubMed] [Google Scholar]
- 128.Spormann DO, Heim J, Wolf DH. Carboxypeptidase yscS: gene structure and function of the vacuolar enzyme. Eur J Biochem. 1991;197:399–405. doi: 10.1111/j.1432-1033.1991.tb15924.x. [DOI] [PubMed] [Google Scholar]
- 129.Wolf DH, Ehmann C. Carboxypeptidase S- and carboxypeptidase Y-deficient mutants of Saccharomyces cerevisiae. J Bacteriol. 1981;147:418–26. doi: 10.1128/jb.147.2.418-426.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kim J, Scott SV, Oda MN, Klionsky DJ. Transport of a large oligomeric protein by the cytoplasm to vacuole protein targeting pathway. J Cell Biol. 1997;137:609–18. doi: 10.1083/jcb.137.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Adachi W, Suzuki NN, Fujioka Y, Suzuki K, Ohsumi Y, Inagaki F. Crystallization of Saccharomyces cerevisiae aminopeptidase 1, the major cargo protein of the Cvt pathway. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007;63:200–3. doi: 10.1107/S1744309107005441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Morales Quinones M, Winston JT, Stromhaug PE. Propeptide of aminopeptidase 1 protein mediates aggregation and vesicle formation in cytoplasm-to-vacuole targeting pathway. J Biol Chem. 2012;287:10121–33. doi: 10.1074/jbc.M111.311696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Schu P. Aminopeptidase I enzymatic activity. Methods Enzymol. 2008;451:67–78. doi: 10.1016/S0076-6879(08)03206-0. [DOI] [PubMed] [Google Scholar]
- 134.Adamis PD, Mannarino SC, Riger CJ, Duarte G, Cruz A, Pereira MD, Eleutherio EC. Lap4, a vacuolar aminopeptidase I, is involved in cadmium-glutathione metabolism. Biometals. 2009;22:243–9. doi: 10.1007/s10534-008-9160-9. [DOI] [PubMed] [Google Scholar]
- 135.Ganguli D, Kumar C, Bachhawat AK. The alternative pathway of glutathione degradation is mediated by a novel protein complex involving three new genes in Saccharomyces cerevisiae. Genetics. 2007;175:1137–51. doi: 10.1534/genetics.106.066944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Myers EW, Miller W. Optimal alignments in linear space. Comput Appl Biosci. 1988;4:11–7. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- 137.Nishizawa M, Yasuhara T, Nakai T, Fujiki Y, Ohashi A. Molecular cloning of the aminopeptidase Y gene of Saccharomyces cerevisiae. Sequence analysis and gene disruption of a new aminopeptidase. J Biol Chem. 1994;269:13651–5. [PubMed] [Google Scholar]
- 138.Yasuhara T, Nakai T, Ohashi A. Aminopeptidase Y, a new aminopeptidase from Saccharomyces cerevisiae. Purification, properties, localization, and processing by protease B. J Biol Chem. 1994;269:13644–50. [PubMed] [Google Scholar]
- 139.Roberts CJ, Pohlig G, Rothman JH, Stevens TH. Structure, biosynthesis, and localization of dipeptidyl aminopeptidase B, an integral membrane glycoprotein of the yeast vacuole. J Cell Biol. 1989;108:1363–73. doi: 10.1083/jcb.108.4.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Julius D, Blair L, Brake A, Sprague G, Thorner J. Yeast alpha factor is processed from a larger precursor polypeptide: the essential role of a membrane-bound dipeptidyl aminopeptidase. Cell. 1983;32:839–52. doi: 10.1016/0092-8674(83)90070-3. [DOI] [PubMed] [Google Scholar]
- 141.Garcia-Rudaz C, Luna F, Tapia V, Kerr B, Colgin L, Galimi F, Dissen GA, Rawlings ND, Ojeda SR. Fxna, a novel gene differentially expressed in the rat ovary at the time of folliculogenesis, is required for normal ovarian histogenesis. Development. 2007;134:945–57. doi: 10.1242/dev.02795. [DOI] [PubMed] [Google Scholar]
- 142.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]