Abstract
Wild barley Hordeum spontaneum (L.) shows a wide geographic distribution and ecological diversity. A key question concerns the spatial scale at which genetic differentiation occurs and to what extent it is driven by natural selection. The Levant region exhibits a strong ecological gradient along the North–South axis, with numerous small canyons in an East–West direction and with small-scale environmental gradients on the opposing North- and South-facing slopes. We sequenced 34 short genomic regions in 54 accessions of wild barley collected throughout Israel and from the opposing slopes of two canyons. The nucleotide diversity of the total sample is 0.0042, which is about two-thirds of a sample from the whole species range (0.0060). Thirty accessions collected at ‘Evolution Canyon' (EC) at Nahal Oren, close to Haifa, have a nucleotide diversity of 0.0036, and therefore harbor a large proportion of the genetic diversity. There is a high level of genetic clustering throughout Israel and within EC, which roughly differentiates the slopes. Accessions from the hot and dry South-facing slope have significantly reduced genetic diversity and are genetically more distinct from accessions from the North-facing slope, which are more similar to accessions from other regions in Northern Israel. Statistical population models indicate that wild barley within the EC consist of three separate genetic clusters with substantial gene flow. The data indicate a high level of population structure at large and small geographic scales that shows isolation-by-distance, and is also consistent with ongoing natural selection contributing to genetic differentiation at a small geographic scale.
Keywords: wild barley, DNA sequence variation, population structure, Evolution Canyon
Introduction
The sessile nature of plants implies a strong selective pressure to adapt to local environmental conditions. Plant species with a large geographic distribution range over a diversity of environments are particularly suited for studying local adaptation. Wild barley, Hordeum spontaneum (L.), the ancestor of cultivated barley, occurs naturally from North Africa through the Fertile Crescent to the Himalayan region (Harlan and Zohary, 1966). The Fertile Crescent is the center of its distribution and most likely the region where cultivated barley H. vulgare was domesticated (Badr et al., 2000). Wild barley occupies a variety of local habitats that differ by soil type, temperature, precipitation and altitude. This ecological diversity is reflected in the genetic population structure in the natural species range (Morrell et al., 2003; Morrell and Clegg, 2007). On a smaller geographic scale, both phenotypic and genetic variation of wild barley are highly variable along an environmental gradient in Israel, which comprises a transition from cool and moist Mediterranean climate in the North to the arid desert climate in the Negev (Nevo et al., 1979; Volis et al., 2002a,2002c; Vanhala et al., 2004). Common garden experiments suggested local adaptation to different levels of rainfall that affected traits like flowering time, seed size, seed germination and others (Volis et al., 2002b; Hubner et al., 2012). The genetic population structure overlaps with the major ecogeographic regions of the Levant area and may result from adaptation to different climatic regions (Hubner et al., 2009, 2012, 2013).
In addition to large-scale ecological gradients, there are also local sites with very strong environmental differences. Throughout the Levant, valleys with an East–West orientation are found in which the North-facing slope is characterized by a cooler and moist climate, whereas the South-facing slope shows xeric conditions, higher temperatures and a much stronger exposure to solar irradiation (Pavlicek et al., 2003; Nevo, 2009). The ‘Evolution Canyon' (EC) in Nahal Oren, close to Haifa, harbors numerous plant, animal and microbial species, which exhibit significant phenotypic and genetic differentiation between the two slopes, and suggests a strong and constant differential selection pressure to abiotic stress within a very small geographical distance (Nevo, 2009). Wild barley is among the plant species with strong interslope genetic variation and diversity levels.
Although genetic and phenotypic differentiation between contrasting environments may reflect local adaptation, it is also influenced by neutral processes such as isolation-by-distance (IBD), physical barriers that suppress gene flow or extinction–recolonization dynamics typical for metapopulations (Hanski and Gaggioti, 2004). The latter process is expected to occur frequently in predominately self-fertilizing plant species, such as wild barley, because a single seed is sufficient to establish a local population. Given that many plant species evolved mechanisms to promote seed dispersal by wind or animals, the extinction–recolonization dynamics may be high within short geographic distances and lead to a breakdown of population structure (Pannell and Charlesworth, 1999). For this reason, studies of plant genetic differentiation need to account for micro- and macroscale effects as well as model the demographic history of a species to allow tests of local adaptation. The sequencing of multiple short genomic fragments is a powerful approach to infer the demographic history of a species, to estimate population parameters and to identify genes that were targets of selection or linked with selective sweeps (Nordborg et al., 2005; Schmid et al., 2005).
The aim of this study was to investigate the population structure of wild barley along the North–South gradient in Israel on two different geographical scales by the analysis of DNA sequence polymorphism at multiple loci. We investigated whether similar patterns of geographic population structure can be observed with DNA sequence data as with microsatellites or other marker types (Nevo et al., 1979,2005; Owuor et al., 1997; Kalendar et al., 2000; Hubner et al., 2012). Furthermore, we tested whether DNA sequencing of short genomic fragments confirms the high level of population differentiation of wild barley within the Nahal Oren Canyon that was observed in previous studies and inferred whether it is consistent with neutral demographic processes or local adaptation to the different slopes of the ‘EC'. We sequenced short genomic regions that were derived from expressed sequence tags (EST) of cultivated barley in a collection of wild barley from the ‘EC' at Nahal Oren and a North–South gradient across Israel. We found a high level of nucleotide variation and strong population structure in wild barley at both spatial levels, which result from the joint effects of demographic history, IBD and, possibly, local adaptation.
Materials and methods
Plant materials
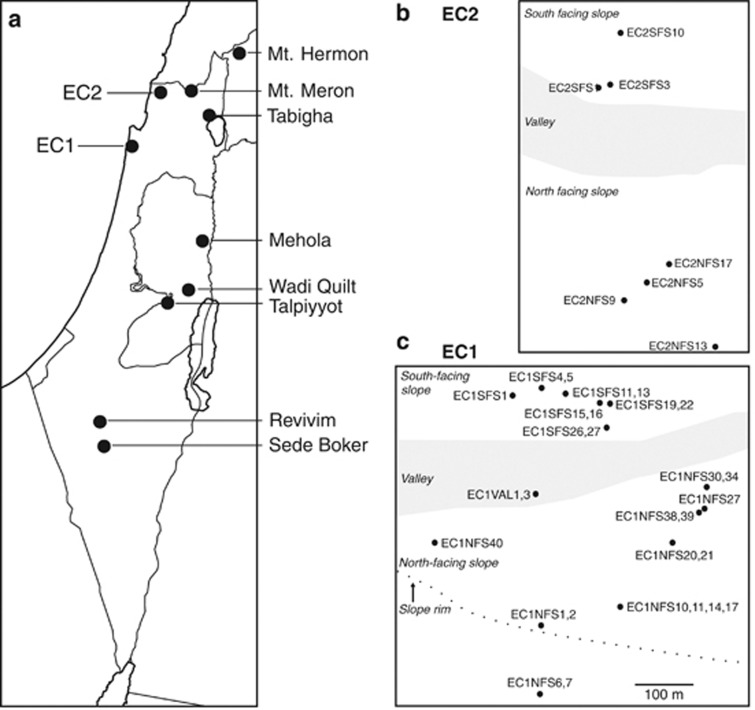
Natural accessions of wild barley were obtained from two sources. Thirty accessions from the ‘EC', Nahal Oren (EC1), and seven accessions from the ‘EC', Keziv Galilee (EC2), were collected by KJS in May 2005. All of these accessions were georeferenced with their Global Positioning System data. An additional set of 17 accessions was collected in eight regions by Nevo et al. (1979), which is stored in the gene bank of the Institute of Evolution in Haifa. The sampling locations are shown in Figure 1 and accessions are listed in Supplementary Table S1.
Figure 1.
Geographic location of sampling sites of 54 accessions from 10 locations. (a) Distribution of the sampling sites in Israel. The 24 accessions from the eight locations listed on the right side are from Nevo et al. (1979). (b) Sampling sites in EC2 at Lower Nahal Keziv (Western Upper Galilee) and (c) in EC1 at Lower Nahal Oren (Mt. Carmel). The shaded areas indicate the valley bottom separating the NFS from the SFS.
Selection of genes for sequencing
Protein-coding genes were selected from cultivated barley ESTs as sequencing targets. To avoid the inclusion of duplicated genes, single-copy ESTs were identified and used for primer design. All available barley EST sequences in HarvEST Version 1.44 (Close et al., 2004) and 41 923 unigenes that were assembled from ESTs under low-stringency conditions were exported into a FASTA-formatted file. These unigenes were compared against each other using the BLAST program, and only those unigenes with an e-value >10−5 to the second best hit were retained because they likely represent single-copy genes or highly diverged paralogous genes in the barley genome. From this subset, 100 unigenes were selected and submitted to primer design with the Primer3 program (Rozen and Skaletsky, 2000). As the exon–intron structure of the selected genes was unknown, pairs of primers were designed to amplify a protein-coding region and were then tested with a subset of the genomic DNAs. A total of 34 primer pairs that resulted in a single band and high-quality sequences were used for the sequencing study. The list of selected primer pairs and corresponding genes are available as Supplementary Table S2.
Sequencing and sequence analysis
Plants were grown from seeds under well-watered conditions in the greenhouse. DNA was extracted from young leaves using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany). Gene fragments were amplified from 50 ng of the DNA with an initial denaturation time of 10 min, and 35 cycles of 1 min at 95 °C, 1 min at 55–60 °C (depending on the annealing temperature of individual primer pairs) and for 2 min at 72 °C. Polymerase chain reaction fragments were purified with a QIAquick PCR Purification Kit (Qiagen) and Sanger-sequenced on an ABI3730xl capillary sequencer (Life Technologies, Carlsbad, CA, USA). The sequencing data were quality trimmed and assembled using the PHRED/PHRAP/CONSED package (Ewing et al., 1998; Gordon et al., 1998,2001). The resulting contigs of each fragment were aligned with CLUSTALW (Thompson et al., 1994). Each polymorphism was visually inspected using the original chromatograms with the CONSED sequence editor. Except for the visual inspection, the whole process was conducted automatically using a pipeline of Python scripts and a PostgreSQL database developed in previous sequencing projects (Schmid et al., 2005). To annotate the 34 amplified gene fragments, we conducted homology searches against barley ‘high-confidence' (HC) genes CDS and Morex whole-genome sequence (Mayer et al., 2012).
Analysis of sequence diversity
We calculated standard population genetic statistics such as the number of haplotypes, the number of segregating single-nucleotide polymorphisms (SNPs), gene diversity (Nei, 1987), nucleotide variation and Tajima's D (Tajima, 1989) of each gene fragment with DNASP v.5.10.01 (Librado and Rozas, 2009). Critical values of Tajima's D and the minimum number of recombination events (Rm) (Hudson and Kaplan, 1985) were tested with coalescent simulations of a standard neutral model using estimated values of θ and ρ as simulation parameters. The minimum number of recombination events (Rh or Rs) was analyzed with RecMin (Myers and Griffiths, 2003).
Inference of population structure
Population structure was inferred with STRUCTURE v.2.3.3 (Pritchard et al., 2000; Falush et al., 2003) by encoding haplotypes as alleles. Clusters were inferred under an admixture model (without prior information on the location) assuming correlated allele frequencies. Different numbers of clusters, K, were analyzed with 10 runs each, consisting of 200 000 burn-in cycles and 500 000 sampling replications. To infer the most likely number of clusters, we used the posterior probability or maximum-likelihood estimate, (Pr(X|K)), and the clusteredness index, which quantifies the extent to which an individual belongs to a single cluster rather than to multiple clusters (Rosenberg et al., 2005). We also used the INSTRUCT program, which simultaneously estimates the inbreeding coefficient and population structure of self-fertilizing species (Gao et al., 2007). However, no significant differences to STRUCTURE were found, and, for this reason, the INSTRUCT results are not reported. Population structure was also investigated with a discriminant analysis of principal components (DAPCs) (Jombart et al., 2010) using the adegenet and ade4 R packages. The same haplotype data sets were used for the STRUCTURE and DAPC analyses. The DAPC method clusters genetic distances from a principal components analysis with a K-means algorithm and infers the optimal number of clusters with the Bayesian information criterion.
Neighbor-joining trees were constructed with MEGA v.5 (Tamura et al., 2011) using the average pairwise FST distance of all gene fragments. Neighbor-joining trees were mapped to sampling sites with GenGIS v.1.08 (Parks et al., 2009). We analyzed the hierarchical population structure with an analysis of molecular variation (AMOVA) (Excoffier et al., 1992) and the F-statistics (Weir and Cockerham, 1984) as implemented in ARLEQUIN v.3.5.1.2 (Excoffier and Lischer, 2010). FST values were calculated for each individual gene based on haplotype counts. Hudson's Snn statistic was calculated (Hudson, 2000), but results were similar to FST values (Supplementary Table S5), and only FST values were used in further analyses. In the AMOVA analysis, total genetic variation among groups, within groups and within populations was assessed. Different hypotheses of genetic structure, panmixia and variable number (2–5) of subpopulations were statistically tested by 20 000 permutations of haplotypes between populations under the null hypothesis of no genetic population structure for each gene fragment. The pairwise distance matrixes used for AMOVA and F-statistics analyses were calculated with the best-fit nucleotide substitution model, as determined by jModeltest (Posada, 2008).
To investigate IBD effects on population genetic structure, a Mantel test (Mantel, 1967) was carried out with the ade4 R package based on 10 000 random permutations of average genetic and geographical distance matrices. The genetic distance between wild barley accessions was calculated with the dnadist program from the PHYLIP package v.3.69 (Felsenstein, 2005). The pairwise genetic distance between accessions at each gene fragment was first calculated using the best-fit model for nucleotide substitution as indicated above. The average pairwise genetic distance between accessions was then calculated from the pairwise genetic distances of all gene fragments. The geographical distance matrix between pairs of locations was calculated using latitude and longitude data of each accession.
Analysis of gene flow
Population models with gene flow between the slopes of EC1 were investigated with MIGRATE v.3.2.7 (Beerli 2006; Beerli and Palczewski 2010), which is based on a full maximum-likelihood coalescent analysis, to estimate the population mutation parameter θ=4Neμ, where Ne is the effective population size and μ is the mutation rate per site, and the mutation-scaled migration rate, M=m/μ, where m is the migration rate per generation. We evaluated three different models: Model 1 assumes a single panmictic population at EC1 with parameter θ; Model 2 consists of two populations corresponding to North-facing slope (NFS) and South-facing slope (SFS) with parameters θ1 and θ2 and an exchange of migrants with rates with mutation scaled migration rate, M1→2 from NFS to SFS and M2→1 from SFS to NFS per generation; and Model 3 assumes three populations with parameters θ1, θ2 and θ3 that freely exchange migrants with rate M (i.e., in total six mutation scaled migration rates, M1→2, M1→3, M2→1, M2→3, M3→1 and M3→2 among three populations) per generation. Accessions were grouped into one (all EC1), two (NFS and SFS) and three (based on STRUCTURE inference with K=3) populations, and parameters were estimated by Bayesian inference. We used the observed value of θ=0.004 and FST estimates as a starting point for θ and M. The mutation rates were estimated separately for each gene fragment from the data. The Bayesian analysis assumed a uniform prior distribution for θ with a minimum of 0.0 and a maximum of 0.1, and for the migration rate M with a minimum of 0.0 to a maximum of 20 000. Each MCMC search was based on a single long chain that was replicated three times. A total of 10 000 samples were recorded every 200 steps, and from the total of six million sampled trees, 10% (600 000) were discarded per replicate as burn-in for each gene. To select the most likely model, the natural log Bayes factor (Kass and Raftery, 1995; Beerli and Palczewski, 2010) based on the thermodynamic integration estimator was used.
Model analysis with approximate Bayesian computation
Coalescent simulations were performed with the ABCtoolbox package (Wegmann et al., 2010) based on three population clusters in EC1 that were inferred with STRUCTURE and Migrate-n. We compared three different models that differed in migration rates between populations but were identical for the remaining parameters. Model 1 assumed no migration among populations, Model 2 allowed migration among populations at a migration rate m sampled from a uniform distribution of 0 and 1 and Model 3 allowed migration between populations at rate m sampled from a uniform distribution of 0 and 0.05. We assumed that the three populations split 100 generations ago. The samples were drawn from sample sizes of 10 each for all three populations, and 34 genes with 427 bp were drawn from each sampled population. The parameters were estimated using simulations sampled from a log-uniform distribution of the current population size (N_NOW) and an ancestral population size (N_ANCESTRAL) ranging from 3 to 4, mutation as a uniform distribution between 1 × 10−9 and 1 × 10−5 and θ=2Nμ, where N is the effective population size (N_NOW) and μ the mutation parameter. For all three models, we simulated 50 000 data sets using the standard sampling approach implemented in the ABCsampler program. The postsampling regression adjustment method of Leuenberger and Wegmann (2010) in the ABCestimator program was used to obtain data for the posterior probability estimation by retaining the 5000 simulations with the smallest Euclidean distance to the observed data. The posterior distributions of the parameters were estimated by applying the locally weighted multivariate regression method of Beaumont et al. (2002) implemented in the abcEst program (Excoffier et al., 2005). Three summary statistics were used: average number of pairwise difference (π; Nei 1987), number of segregating sites (S) and Tajima's D (Tajima, 1989). Model selection was based on the Bayes factor, which was calculated as the ratio of the marginal density of the models.
Results
Selection of sequenced regions
Of 100 primer pairs tested, 34 were retained for the analysis of all individuals (Supplementary Table S2). BLAST searches revealed that 25 (73.5%) fragments were homologous to 27 barley ‘HC' genes and 32 (94.1%) fragments were similar to the Morex whole-genome sequences (Supplementary Table S3). Almost all fragments with hits against barley ‘HC' genes and Morex whole-genome sequence were single-copy genes—only two fragments against each of these databases with more than one best hit. On the basis of best BLAST hits against barley HC genes and Morex whole-genome sequence, 28 (82.4%) gene fragments were mapped onto barley chromosomes.
Patterns of nucleotide variation
The 34 gene fragments were sequenced in 54 wild barley accessions from Israel. After quality control, on average, sequences from 46 accessions were included in alignments of gene fragments (Table 1). The minimum number of sequences per fragment was 25 and the maximum was 54. There was no correlation between the number of sequences per candidate gene and nucleotide diversity, θ (P=0.07) or Tajima's D (P=0.93), indicating that missing sequences did not significantly affect further analyses. The 34 genes cover a region of 14 530 bp, and the whole data set consists of 670 391 bp. The shortest and longest fragments have a length of 101 bp and 791 bp, respectively, with an average length of 427 bp. On average, six haplotypes per gene fragment were observed. A total of 255 SNPs (polymorphic sites) with an average of eight SNPs per gene fragment were detected. All SNPs were biallelic, and hence the infinite sites model was used in the further analyses. Among coding SNPs, 64 were synonymous and 75 non-synonymous polymorphisms. The average estimate of nucleotide diversity per base pair, π, was 0.0042 (Table 1 and Figure 2a), and 0.0094 for synonymous and 0.0029 for non-synomyous SNPs, respectively. The average Tajima's D was −0.248, with a range from −2.017 to 3.284. The latter gene does not show a significant hit to annotated barley genes, but is homologous to the TRIUR3_14272 gene from Triticum urartu (BLAST e-value 3e−69), which encodes a heparanase-like protein 2. We also tested with coalescent simulations whether observed Tajima's D and haplotype diversity were consistent with a standard neutral model with no recombination and constant size. With respect to Tajima's D, 12 gene fragments (35%) showed a significant deviation from neutrality (Supplementary Table S4). Using different methods, recombination events were detected only at six gene fragments, with a maximum of four recombination events at UG23391. For this reason, recombination variation among genes and samples was not considered further in this study.
Table 1. Summary of π, θW and Tajima's D values for different groups of accessions.
| Group of accessions | Mean number of accessions per locus | π/bp ( × 10−3) | θW/bp ( × 10−3) | Tajima's D | ||
|---|---|---|---|---|---|---|
| |
|
Mean |
95% CI |
Mean |
95% CI |
Mean (per locus) |
| Total set | 46 | 4.17 | 2.54–5.82 | 4.06 | 2.88–5.23 | −0.263 |
| Non-EC1 | 21 | 3.74 | 2.18–5.23 | 3.88 | 2.64–5.12 | −0.277 |
| EC1 | 26 | 3.64 | 2.15–5.13 | 3.08 | 1.97–4.16 | 0.309 |
| EC1NFS | 15 | 3.88 | 2.14–5.62 | 3.31 | 1.96–4.66 | 0.596 |
| EC1SFS | 9 | 2.16 | 1.07–3.24 | 2.28 | 1.17–3.39 | −0.212 |
Abbreviations: CI, confidence interval; EC1, Evolution Canyon 1; EC1NFS, EC1 North-facing slope; EC1SFS, EC1 South-facing slope; non-EC1, all accessions except fromEC1.
Mean values and 95% CI are calculated from all 34 gene fragments.
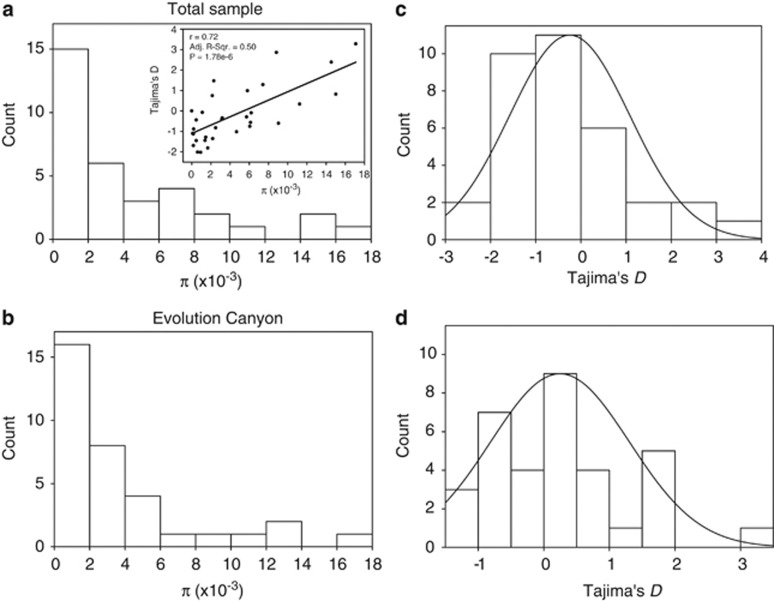
Figure 2.
Nucleotide variation in wild barley collection across Israel and in EC1. (a and b) Distribution of nucleotide diversity, π and Tajima's D in the total sample (n=54). (c and d) Distributions for the sample in EC1 (n=30). The inset in (a) shows the correlation between nucleotide diversity and Tajima's D in the total sample.
Nucleotide variation in the Nahal Oren Canyon (EC1)
Accessions from EC1 were analyzed together (n=30) and separately for the NFS(n=17) and SFS (n=11) (Figure 2b and Supplementary Figure S1). Using a Wilcoxon's signed-rank test, we compared the pairwise mean difference between the EC1 and non-EC1 samples across all genes. We found significant differences for θW (P=0.013), number of haplotypes (P=0.02) and number of SNPs (P=0.02), but not for π (P=0.34), haplotype diversity (P=0.46) and Tajima's D (P=0.1). Similarly, we compared the NFS and SFS accessions for different population parameters, and hence significant interslope differences were observed for almost all parameters such as number of haplotypes (P<0.001), haplotype diversity (P<0.001), π (P=0.01), SNPs (P=0.006) and Tajima's D (P=0.01), but not for θw (P=0.26).
Analysis of geographic population structure
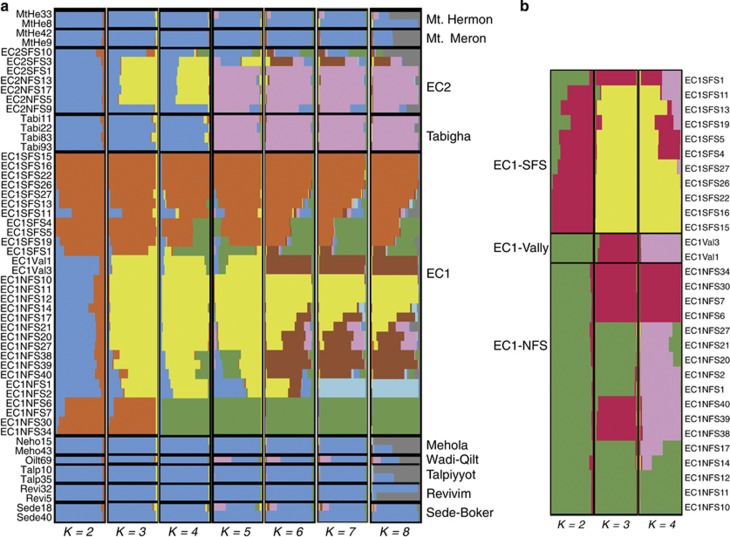
We first inferred genetic population structure with STRUCTURE. The complete set of 54 accessions clustered into eight populations based on maximum-likelihood values (K=8, ln L=−1232.52), and into three populations with the clusteredness index (0.856). Figure 3 shows the graphical display of inferred clusters for K=2–8. The key results of this analysis were: (1) a clustering of Northern and Southern accessions, (2) a grouping of EC1NFS accessions with other Northern accessions and (3) the genetic difference of EC1SFS from other accessions. If EC1 accessions were clustered separately, they grouped into seven (ln L=−41.73) or three groups (clusteredness index=0.94). At K=3, the NFS accessions clustered into two groups, whereas almost all SFS accessions clustered as a single group. Furthermore, the two valley accessions and a single accession from SFS (EC1SFS1) clustered with the NFS accessions from the bottom and top sites of the slope (Figure 3b). In general, the EC1SFS accessions appeared highly distinct from the NFS and valley accessions (Figure 4). A DAPCs clustered the 54 accessions into four groups (Supplementary Figure S2). As with STRUCTURE, the SFS accessions from EC1 were genetically most distant from the other clusters.
Figure 3.
Inference of population structure in wild barley from Israel with STRUCTURE. (a) Population structure in the complete sample of 54 accessions for K=2–8. The left side of the panel shows the names of individual accessions, and the right side the geographic origin. (b) Structure of 30 accessions from EC1 for K=2–4. In both figures, the sampling locations are ordered along the North–South orientation. A full color version of this figure is available at the Heredity journal online.
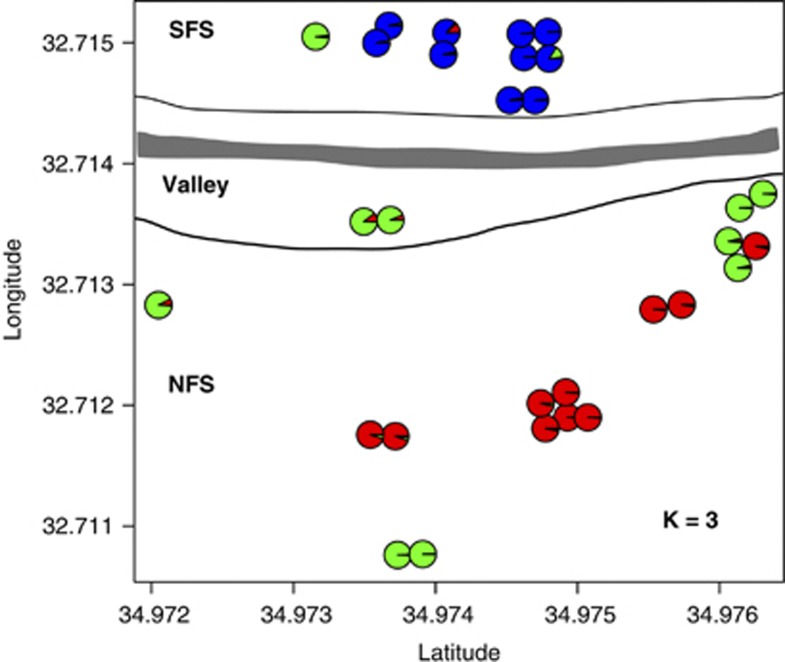
Figure 4.
Spatial distribution of accessions in EC1 based on their Global Positioning System (GPS) coordinates and the STRUCTURE analysis with K=3. The pie diagram indicates the relative proportion to which each of the three accessions was assigned proportionally to one of the three clusters. The line of dashes outlines the area of the valley bottom, and the gray line shows the road crossing through EC1. A full color version of this figure is available at the Heredity journal online.
Genetic and geographic differentiation
To characterize the extent and geographic pattern of genetic differentiation, we estimated pairwise FST values of a priori groups representing the sampling locations from haplotype data. Average pairwise FST values between collection sites based on the average of 34 gene fragments genes indicated significant genetic differentiation (Supplementary Table S5). The highest and lowest levels of genetic differentiation were observed between EC1SFS and Mehola (FST=0.3648) and Talpiyyot and Wadi–Quilt (FST=0.0385), respectively. In general, accessions from EC1 (particularly from SFS), Mt Meron and Tabigha regions showed the highest differentiation to other sites. Within EC1, there was a moderate differentiation between SFS and NFS (FST=0.118, 95% confidence interval (CI)=0.063–0.173). The low genetic differentiation was also confirmed by a hierarchical AMOVA in which intraslope genetic variation (63.08%) was about fourfold higher than interslope genetic variation (16.34% Supplementary Table S6). At similar partitioning of variation within and between sites was observed at all sampling locations throughout Israel, where genetic variation within and between sampling locations was 68.05% and 25.94%, respectively.
A neighbor-joining tree based on a matrix of pairwise FST values partially overlapped with the population cluster inferred by STRUCTURE (Supplementary Figure S3). Accessions were clustered according to their geographical origin except for the two populations from the North, Mt Hermon and Mt Meron, which were grouped with accessions from the South and EC1, respectively. Mantel tests suggested that the geographic distribution of genetic variation was also influenced from IBD effects at local and regional scales, because a weak, but significant, correlation (r=0.12, P<0.0001) was observed among all 54 wild barley accessions across Israel and the accessions from EC1 (r=0.24, P<0.0001; Supplementary Figure S4).
Gene flow in EC1
To infer the extent of gene flow between slopes in EC1, we compared three population models. A model selection test suggested that Model 3 with three populations (inferred by STRUCTURE based on the clusteredness statistic) and free bidirectional migration was most strongly supported by the data (Table 2). The parameter estimates for Model 3 revealed similar values of average (per gene) mutation-scaled population size parameter θM estimates for all three populations. Likewise, the average number of immigrants per generation per locus, 4Nm, were all highly similar to each other and do not differ significantly (Table 3). To further differentiate among possible migration rates between the three subpopulations, we compared three models that allowed no, limited (m=0.05) and free migration (m=1) by coalescent simulations and approximate Bayesian computation (ABC). The model with limited migration was strongly supported by the data as the Bayes factor was > 100 (Table 4 and Supplementary Table S7). Prior and posterior probability distributions of parameters estimated in the ABC analysis are shown in Supplementary Figure S5.
Table 2. Likelihood-based comparison of three different demographic models for Evolution Canyon 1.
| Model | 4Nm (95% CI) | Log marginal likelihood | LBF | Model probability | Model rank | |||
|---|---|---|---|---|---|---|---|---|
| 1 | — | −21 719.76 | −64.28 | 1.2 × 10−28 | 3 | |||
| 2 | NFS | SFS | ||||||
| NFS | 26 (20–32) | −21 665.20 | −9.72 | 6.0 × 10−5 | 2 | |||
| SFS | 32 (19–46) | |||||||
| 3 | Pop. 1 | Pop. 2 | Pop. 3 | 0 | 0 | 1 | ||
| Pop. 1 | 17 (12–23) | 17 (13–21) | ||||||
| Pop. 2 | 19 (14–25) | 18 (14–22) | −21 655.48 | |||||
| Pop. 3 | 20 (14–25) | 18 (13–23) | ||||||
Abbreviations: LBF, log Bayes factors; NFS, North-facing slope; SFS, South-facing slope.
The estimation of migration rates (4Nm=M × θM of the recipient population) is based on 34 gene fragments. The estimation was conducted with Migrate-n and is based on Bayesian inference and model comparison using LBF, marginal log-likelihood differences, as approximated by thermodynamic integration. The model with the highest marginal log-likelihood (Model 3) was used as a reference model to calculate LBF and model probability. Model 1 assumes a single panmictic population at EC1 and Model 2 separate populations on the NFS and SFS. Model 3 assumes three models in which accessions are grouped according to the STRUCTURE analysis with K=3. Free migration among populations was assumed for Models 2 and 3. Parameters in all three models were estimated using a uniform prior θ of 0–0.1 and M (mutation scaled migration rate) of 0–30 000.
Table 3. Summary of observed and estimated nucleotide variation at EC1.
| Model | Population |
Observed data ( × 10−3) |
ML estimate ( × 10−3) |
ABC estimatea
( × 10−3) |
|---|---|---|---|---|
| θW/bp (95% CI) | θM/bp (95% CI) | πABC/bp (95% CI) | ||
| 1 | EC1 | 3.08 (2.01–4.15) | 13.18 (8.15–18.21) | |
| 2 | NFS | 3.31 (2.01–4.61) | 2.65 (1.60–3.71) | |
| SFS | 2.28 (1.21–3.35) | 2.22 (1.72–2.72) | ||
| 3 | Pop. 1 | 2.03 (1.02–3.04) | 1.63 (1.05–2.21) | 2.1 (0–4.3) |
| Pop. 2 | 2.84 (1.38–4.30) | 1.45 (0.09–1.97) | 2.5 (0.8–3.3) | |
| Pop. 3 | 1.54 (0.58–2.49) | 1.39 (1.07–1.70) | 1.8 (0–1.4) |
Abbreviations: ABC, approximate Bayesian computation; CI, confidence interval; EC1, Evolution Canyon 1; ML, maximum-likelihood; NFS, North-facing slope; SFS, South-facing slope.
Mean values from 34 gene fragments for three different population models based on ML or ABC estimation.
On the basis of the coalescent simulation with the best Model 3.
Table 4. Parameter estimates of three models of population structure and migration in EC1.
| Parameter | Population | Obs. data (95% CI) | Sim. Model 1 (95% CI) | Sim. Model 2 (95% CI) | Sim. Model 3 (95% CI) |
|---|---|---|---|---|---|
| π/bp ( × 10−3) | Pop. 1 | 2.3 (1.1–3.6) | 1.9 (0.8–3.6) | 2.2 (0.8–3.6) | 2.1 (0–4.3) |
| Pop. 2 | 3.3 (1.6–5) | 2.5 (1.1–4.3) | 2.4 (1.1–3.9) | 2.5 (0.8–3.3) | |
| Pop. 3 | 1.4 (0.6–2.3) | 1.1 (0–2.9) | 2.1 (0.8–3.4) | 1.8 (0–1.4) | |
| S | Pop. 1 | 2.5 (1–3.9) | 2.5 (1–4) | 2.6 (1–4) | 2.4 (0–5) |
| Pop. 2 | 2.8 (1.4–4.1) | 2.4 (1–4) | 2.6 (1–4) | 2.7 (1–5) | |
| Pop. 3 | 1.6 (0.6–2.5) | 1.2 (0–3) | 2.6 (1–4) | 2.2 (0–5) | |
| Tajima's D | Pop. 1 | 0.19 (−0.01–0.44) | 0.53 (−0.69–1.46) | 0.20 (−0.69–1.30) | 0.18 (−0.69–1.03) |
| Pop. 2 | 0.37 (0.08–0.65) | 0.69 (−0.51–1.74) | 0.34 (−0.51–1.34) | 0.36 (−0.51–1.30) | |
| Pop. 3 | −0.08 (−0.38–0.23) | 0.24 (1.11–1.34) | −0.00 (−0.76–0.83) | −0.02 (−1.04–0.83) | |
| Marginal density | 1.69e−21 | 8.99e−39 | 2.51e−13 | ||
| Bayes factor | M12>100 | M21<1 | M31>100 | ||
| M13<1 | M31<1 | M32>100 |
Abbreviations: π, average nucleotide diversity; ABC, approximate Bayesian computation; CI, confidence interval; EC1, Evolution Canyon 1; Obs., observed; S, average number of segregating sites; sim., simulated.
Estimates were obtained by ABC. Models differed only by the migration rate among populations. Accessions were assigned to one of the three populations with STRUCTURE. Bayes factors were calculated as the marginal density of one model divided by the other model. S and Tajima's D are calculated as averages per locus.
Discussion
The key results of our sequence survey are the presence of a substantial population structure in the whole sampling area as well as a significant level of genetic differentiation between the two slopes in the EC at Nahal Oren. Overall, our results are consistent with earlier studies that used different samples and genetic markers, but there are also substantial differences as discussed below.
High level of genetic variation in wild barley across Israel
The average total nucleotide diversity, π, was 0.0042 and is about two-third of the variation in 18 genes sequenced in a wild barley collection from the Western part of the Fertile Crescent (π=0.0060; Morrell and Clegg, 2007). Hence, a substantial proportion of the total genetic variation of wild barley is present in a small geographic region relative to the total distribution range. As expected, levels of variation at individual loci were highly variable, and in total were not consistent with a standard neutral model of sequence variation as indicated, for example, by an average negative Tajima's D value. Both the level of DNA sequence variation and the frequency distribution of polymorphisms as measured with Tajima's D are similar to other self-fertilizing wild plants such as Arabidopsis thaliana (Schmid et al., 2005). Divergent haplotypes at highly polymorphic loci may result from unrecognized gene duplication polymorphisms, which we tried to exclude by selecting putative single copy genes. Alternatively, both theoretical (Nordborg, 2000) and empirical studies showed that the existence of divergent haplotypes is not uncommon in predominately self-fertilizing plant species (Cork and Purugganan, 2005; Nordborg et al., 2005; Schmid et al., 2005). In addition, highly polymorphic genes may represent local adaptation along environmental gradients or the presence of balancing selection. Although neutrality tests such as Tajima's D suggested that some loci evolved under natural selection, the short regions sequenced and the effects of demographic history on neutrality tests call for a cautious interpretation of such tests. Given the low level of linkage disequilibrium in wild barley (Morrell et al., 2005) as well as the small number of loci sampled from a large genome, it seems unlikely that the genomic fragments comprise targets of recent strong selection.
Geographical population structure of wild barley across Israel
We inferred the population structure in wild barley across Israel using haplotype data extracted from all 34 gene fragments. The methods used suggested different numbers of clusters (K=3 or K=8 with STRUCTURE and K=4 with DAPC), similar to previous analyses using other marker types (Hubner et al., 2009, 2012). The present sample is unbalanced because more than half of the accessions originated from EC1, and both clustering methods separate primarily the accessions from the different slopes within EC1 for lower values of K, which demonstrates the high diversity present in EC1 accessions. In contrast, EC2 accessions did not cluster by slope, but were grouped with the Tabigha population in all three clustering approaches (STRUCTURE-, DAPC- and Fst-based neighbor-joining tree). Geographical proximity, similar ecogeographic factors and, possibly, similar patterns of natural selection may explain this relationship. Also, EC1 accessions grouped together with the other accessions from North Israel, whereas the accessions from the sampling sites in Central or Southern Israel were clustered separately. This grouping reflects the ecogeographic differences between the mesic North and xeric South and corresponds with the clustering into Mediterranean (Northern), coastal and desert wild barley (Hubner et al., 2009, 2012). As an exception, the Mt. Hermon accessions from Northern Israel are grouped together with the Southern accessions. A coclustering of Northern and Southern populations was also observed by Hubner et al. (2012) and is supported by the Mantel test, which shows a decreasing genetic distance for large geographical distances. These populations may reflect gene flow through seed dispersal by humans or animals, or a different distribution of genotypes in historical times.
Despite a population structure that is consistent with the ecogeographic structure, there is a high level of shared genetic variation between regions. The hierarchical AMOVA indicates a threefold greater variation within sampling sites than between sampling sites (Supplementary Table S6). Hence, variation within wild barley populations at a given geographical region or microsite significantly contribute to the overall genomic variation in wild barley across Israel, which is indicative of a high proportion of shared ancestral polymorphisms or recent gene flow. Similar results were previously reported for different collections of wild barley from Israel based on AFLP, simple sequence repeat (SSR) and SNP markers (Hubner et al., 2012). The latter study proposed that gene flow explains the high proportion of intrapopulation variation.
Genetic variation of wild barley in EC1
In the ‘EC' at Nahal Oren (EC1), the average nucleotide variation, π, was 87% of the whole sample and was 1.8-fold higher among NFS than SFS accessions. The three clusters obtained with STRUCTURE differ in their level of genetic diversity, and the NFS accessions are about two times more diverse than the SFS accessions. Most previous studies of genetic diversity of wild barley in EC1 found a higher level of genetic diversity at SFS than NFS based on allozymes, DNA sequences of single genes (Yang et al., 2009) and copy number of BARE-1 retrotransposons (Kalendar et al., 2000). In contrast, REMAP (Retrotransposons-Microsatellite Amplified Polymorphism) analyses revealed more genotypes in the NFS than the SFS (Kalendar et al., 2000). Likewise, SSRs revealed a higher genetic diversity on the NFS than the SFS (Nevo et al., 2005). The low SSR diversity of SFS accessions was explained with extreme microclimatic conditions such as high solar radiation, temperature and drought conditions on the SFS that select against high SSR diversity. Such a selectionist view appears too simplistic as the diversity of neutral markers such as SSRs is mainly influenced by genetic drift, whose extent depends on demographic parameters and the mating system. The analysis of demographic models in this study suggests that DNA sequence variation in wild barley is strongly influenced by neutral demographic effects. Furthermore, the reduction of SSR diversity by direct selection of alleles or by indirect selection via hitchhiking appears unlikely given the low level of genome-wide LD in wild barley (Morrell et al., 2005).
The difference of our data to most previous studies likely results from the marker type and the sampling scheme. Earlier studies of EC1 were based on collections from seven fixed stations along a linear transect across the canyon (three on each side of the slopes and one at valley). In contrast, we randomly sampled from different sites across the canyon that also included the seven fixed stations to obtain a more comprehensive sample of genetic variation within slopes. In addition, a large number of short genomic regions analyzed provide evidence for heterogeneous patterns of genomic diversity among loci. For example, SFS accessions showed higher values for π than NFS at six polymorphic loci, although the average value of diversity was lower for SFS than NFS accessions. Hence, an inference based on one or several candidate genes may be misleading owing to the random nature of the evolutionary process.
Genetic differentiation among EC1 accessions
Accessions mainly clustered by the two slopes, which are the key physiographic feature of the canyon. Most NFS accessions are genetically similar to other accessions from North Israel, whereas most SFS accessions are genetically distinct from the other accessions of this study. Wild barley is a highly self-fertilizing (>98%) species (Brown et al., 1978; Abdel-Ghani et al., 2004), in which gene flow by pollen and/or seed dispersal is potentially limited (Volis et al., 2010). A higher level of genetic variation between rather than within local populations is expected. The strongest differentiation in EC was found with three populations (K=3) and the spatial distribution of these clusters is largely consistent with the slopes in EC1, suggesting that the orientation of the Canyon and the presence of a road separating the slopes have the strongest effect on the geographic distribution of genotypes either by inferring with neutral processes such as migration or by differential natural selection of genotypes on both slopes. Alternatively, the observed structure may result from IBD. IBD within short distances may reflect a high degree of kinship of neighboring stands or an unequal distribution of sampling sites (sampling bias). IBD confounds the inference of population structure (Meirmans, 2012), but two observations suggest that the observed structure in EC1 is not an artifact of IBD. First, IBD is present within a short distance (up to 100 m), but not at larger distances within EC1 (Supplementary Figure S4), because some accessions collected at the rim of NFS with a South-Eastern exposure and edaphic conditions similar to SFS are also genetically similar to SFS accessions (Figure 4). Second, SFS accessions are genetically distinct compared with NFS accessions, because NFS accessions show moderate (FST=0.078–0.0188) and SFS accessions strong (FST=0.181–0.365) differentiation to other regions sampled. Despite differences in marker type and sampling strategy, our results are in agreement with most previous studies: see the review by Nevo (2006, 2009).
Gene flow within EC1
Gene flow counteracts the formation of population structure over short geographic distances, such as in EC1. Rare outcrossing events through pollen dispersal and seed dispersal by humans, birds or other animals within and between slopes in EC1 appear very likely. Even over large geographical distances throughout Israel, high levels of gene flow (up to Nem=8.47) were documented in wild barley (Hubner et al., 2012). The gene flow models we used to estimate interslope and/or interpopulation immigrants by Migrate-n (NemNFS→SFS=26 and NemSFS→NFS=32) and ABC therefore agree with this finding and suggest the presence of gene flow among EC1 accessions.
In summary, we observed a strong genetic differentiation of wild barley over two geographic scales in Israel. A larger and more comprehensive sample of accessions from EC (EC1) confirmed the presence of strong genetic differentiation at this site. Although the observed patterns are similar to previous studies, and consistent with local adaptation, the analysis of neutral demographic models suggested a strong demographic contribution to the observed structure, which does not support a simple selectionist explanation, particularly in EC1. Differences from previous studies indicate an effect of marker types, collections and temporal changes in the population (Nevo et al., 2012). Future studies of wild barley adaptation should be carried out with additional samples around microsites such as the EC and be combined them with large systematic collections like the Barley1K (Hubner et al., 2009) to put the local patterns of genetic variation into a broader geographic context.
Data archiving
Sequence data were submitted to NCBI GenBank under accession numbers KC471630–KC473154.
Acknowledgments
We are grateful to Claudia Wenderoth and Domenica Schnabelrauch for the technical help, and to Andreas Weber for the greenhouse plant cultivation. This work was funded by the Minerva Foundation short-term fellowship and a FORMAS Grant to KJS. EN thanks the Ancell Teicher Research Foundation of Genetics and Molecular Evolution. A Beiles, T Günther and S Hübner provided comments on the manuscript. The constructive comments of two reviewers are gratefully acknowledged.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Abdel-Ghani AH, Parzies HK, Omary A, Geiger HH. Estimating the outcrossing rate of barley landraces and wild barley populations collected from ecologically different regions of Jordan. Theor Appl Genet. 2004;109:588–595. doi: 10.1007/s00122-004-1657-1. [DOI] [PubMed] [Google Scholar]
- Badr A, Muller K, Schafer-Pregl R, El Rabey H, Effgen S, Ibrahim HH, et al. On the origin and domestication history of barley (Hordeum vulgare) Mol Biol Evol. 2000;17:499–510. doi: 10.1093/oxfordjournals.molbev.a026330. [DOI] [PubMed] [Google Scholar]
- Beaumont M, Zhang W, Balding D. Approximate Bayesian computation in population genetics. Genetics. 2002;162:2025–2035. doi: 10.1093/genetics/162.4.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli P. Comparison of Bayesian and maximum-likelihood inference of population genetic parameters. Bioinformatics. 2006;22:341–345. doi: 10.1093/bioinformatics/bti803. [DOI] [PubMed] [Google Scholar]
- Beerli P, Palczewski M. Unified framework to evaluate panmixia and migration Direction among multiple sampling locations. Genetics. 2010;185:313–U463. doi: 10.1534/genetics.109.112532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AHD, Zohary D, Nevo E. Outcrossing rates and heterozygosity in natural populations of Hordeum spontaneum (Koch) in Israel. Heredity. 1978;41:49–62. [Google Scholar]
- Close TJ, Wanamaker SI, Caldo RA, Turner SM, Ashlock DA, Dickerson JA, et al. A new resource for cereal genomics: 22K barley GeneChip comes of age. Plant Physiol. 2004;134:960–968. doi: 10.1104/pp.103.034462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cork JM, Purugganan MD. High-diversity genes in the Arabidopsis genome. Genetics. 2005;170:1897–1911. doi: 10.1534/genetics.104.036152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl M, Green P. Basecalling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Estoup A, Cornuet JM. Bayesian analysis of an admixture model with mutations and arbitrarily linked markers. Genetics. 2005;169:1727–1738. doi: 10.1534/genetics.104.036236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Res. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes—Application to human mitochondrial–DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) v3.6. Distributed by the Author. Department of Genome Sciences, University of Washington: Seattle, WA, USA; 2005. [Google Scholar]
- Gao H, Williamson S, Bustamante CD. A Markov chain Monte Carlo approach for joint inference of population structure and inbreeding rates from multilocus genotype data. Genetics. 2007;176:1635–1651. doi: 10.1534/genetics.107.072371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Gordon D, Desmarais C, Green P. Automated finishing with Autofinish. Genome Res. 2001;11:614–625. doi: 10.1101/gr.171401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanski I, Gaggioti O.2004Metapopulation biology: past, present, and future Ecology, Genetics, and Evolution of MetapopulationsIn: Hanski I, Gaggiotti O (eds) Elsevier Acadamic Press:New York, NY, USA [Google Scholar]
- Harlan JR, Zohary D. Distribution of wild wheats and barley. Science. 1966;153:1074–1080. doi: 10.1126/science.153.3740.1074. [DOI] [PubMed] [Google Scholar]
- Hubner S, Bdolach E, Ein-Gedy S, Schmid KJ, Korol A, Fridman E. Phenotypic landscapes: phenological patterns in wild and cultivated barley. J Evol Biol. 2013;26:163–174. doi: 10.1111/jeb.12043. [DOI] [PubMed] [Google Scholar]
- Hubner S, Gunther T, Flavell A, Fridman E, Graner A, Korol A, et al. Islands and streams: clusters and gene flow in wild barley populations from the Levant. Mol Ecol. 2012;21:1115–1129. doi: 10.1111/j.1365-294X.2011.05434.x. [DOI] [PubMed] [Google Scholar]
- Hubner S, Hoffken M, Oren E, Haseneyer G, Stein N, Graner A, et al. Strong correlation of wild barley (Hordeum spontaneum) population structure with temperature and precipitation variation. Mol Ecol. 2009;18:1523–1536. doi: 10.1111/j.1365-294X.2009.04106.x. [DOI] [PubMed] [Google Scholar]
- Hudson RR. A new statistic for detecting genetic differentiation. Genetics. 2000;155:2011–2014. doi: 10.1093/genetics/155.4.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Kaplan NL. Statistical properties of the number of recombination events in the history of a sample of DNA-sequences. Genetics. 1985;111:147–164. doi: 10.1093/genetics/111.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T, Devillard S, Balloux F. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 2010;11:94. doi: 10.1186/1471-2156-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalendar R, Tanskanen J, Immonen S, Nevo E, Schulman AH. Genome evolution of wild barley (Hordeum spontaneum) by BARE-1 retrotransposon dynamics in response to sharp microclimatic divergence. Proc Natl Acad Sci USA. 2000;97:6603–6607. doi: 10.1073/pnas.110587497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass RE, Raftery AE. Bayes factors. J Am Stat Assoc. 1995;90:773–795. [Google Scholar]
- Leuenberger C, Wegmann D. Bayesian computation and model selection without likelihoods. Genetics. 2010;184:243–252. doi: 10.1534/genetics.109.109058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- Mayer KF, Waugh R, Brown JW, Schulman A, Langridge P, Platzer M, International Barley Genome Sequencing C et al. A physical, genetic and functional sequence assembly of the barley genome. Nature. 2012;491:711–716. doi: 10.1038/nature11543. [DOI] [PubMed] [Google Scholar]
- Meirmans PG. The trouble with isolation by distance. Mol Ecol. 2012;21:2839–2846. doi: 10.1111/j.1365-294X.2012.05578.x. [DOI] [PubMed] [Google Scholar]
- Morrell PL, Clegg MT. Genetic evidence for a second domestication of barley (Hordeum vulgare) east of the Fertile Crescent. Proc Natl Acad Sci USA. 2007;104:3289–3294. doi: 10.1073/pnas.0611377104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrell PL, Lundy KE, Clegg MT. Distinct geographic patterns of genetic diversity are maintained in wild barley (Hordeum vulgare ssp. spontaneum) despite migration. Proc Natl Acad Sci USA. 2003;100:10812–10817. doi: 10.1073/pnas.1633708100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrell PL, Toleno DM, Lundy KE, Clegg MT. Low levels of linkage disequilibrium in wild barley (Hordeum vulgare ssp. spontaneum) despite high rates of self-fertilization. Proc Natl Acad Sci USA. 2005;102:2442–2447. doi: 10.1073/pnas.0409804102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers SR, Griffiths RC. Bounds on the minimum number of recombination events in a sample history. Genetics. 2003;163:375–394. doi: 10.1093/genetics/163.1.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. Columbia University Press: New York, NY, USA; 1987. [Google Scholar]
- Nevo E. ‘Evolution canyon': a microcosm of life's evolution focusing on adaptation and speciation. Israel J Ecol Evol. 2006;52:485–506. [Google Scholar]
- Nevo E. Evolution in action across life at ‘Evolution Canyons', Israel. Trends Evol Biol. 2009;1:e3. [Google Scholar]
- Nevo E, Beharav A, Meyer RC, Hackett CA, Forster BP, Russell JR, et al. Genomic microsatellite adaptive divergence of wild barley by microclimatic stress in 'Evolution Canyon', Israel. Biol J Linnean Soc. 2005;84:205–224. [Google Scholar]
- Nevo E, Fu YB, Pavlicek T, Khalifa S, Tavasi M, Beiles A. Evolution of wild cereals during 28 years of global warming in Israel. Proc Natl Acad Sci USA. 2012;109:3412–3415. doi: 10.1073/pnas.1121411109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevo E, Zohary D, Brown A, Haber M. Genetic diversity and environmental associations of wild barley, Hordeum spontaneum in Israel. Evolution. 1979;33:815–833. doi: 10.1111/j.1558-5646.1979.tb04737.x. [DOI] [PubMed] [Google Scholar]
- Nordborg M. Linkage disequilibrium, gene trees and selfing: an ancestral recombination graph with partial self-fertilization. Genetics. 2000;154:923–929. doi: 10.1093/genetics/154.2.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M, Hu TT, Ishino Y, Jhaveri J, Toomajian C, Zheng H, et al. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 2005;3:e196. doi: 10.1371/journal.pbio.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owuor ED, Fahima T, Beiles A, Korol A, Nevo E. Population genetic response to microsite ecological stress in wild barley, Hordeum spontaneum. Mol Ecol. 1997;6:1177–1187. [Google Scholar]
- Pannell JR, Charlesworth B. Neutral genetic diversity in a metapopulation with recurrent local extinction and recolonization. Evolution. 1999;53:664–676. doi: 10.1111/j.1558-5646.1999.tb05362.x. [DOI] [PubMed] [Google Scholar]
- Parks DH, Porter M, Churcher S, Wang SW, Blouin C, Whalley J, et al. GenGIS: a geospatial information system for genomic data. Genome Res. 2009;19:1896–1904. doi: 10.1101/gr.095612.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlicek T, Shron D, Kravchenko V, Saaroni H, Nevoa E. Microclimatic interslope differences underlying biodiversity contrasts in ‘Evolution Canyon', Mt. Carmel, Israel. Israel J Earth Sci. 2003;52:1–9. [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA, Mahajan S, Ramachandran S, Zhao CF, Pritchard JK, Feldman MW. Clines, clusters, and the effect of study design on the inference of human population structure. PLoS Genet. 2005;1:660–671. doi: 10.1371/journal.pgen.0010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ.2000Primer3 on the WWW for general users and for biologist programmers Bioinformatics Methods and Protocols: Methods in Molecular BiologyIn: Misener SKaS (ed), Humana Press: Totowa, NJ, USA; 365–386. [DOI] [PubMed] [Google Scholar]
- Schmid KJ, Ramos-Onsins S, Ringys-Beckstein H, Weisshaar B, Mitchell-Olds T. A multilocus sequence survey in Arabidopsis thaliana reveals a genome-wide departure from a neutral model of DNA sequence polymorphism. Genetics. 2005;169:1601–1615. doi: 10.1534/genetics.104.033795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Petersen D, Petersen N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. Clustal-W—improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhala TK, van Rijn CPE, Buntjer J, Stam P, Nevo E, Poorter H, et al. Environmental, phenotypic and genetic variation of wild barley (Hordeum spontaneum) from Israel. Euphytica. 2004;137:297–309. [Google Scholar]
- Volis S, Mendlinger S, Turuspekov Y, Esnazarov U. Phenotypic and allozyme variation in Mediterranean and desert populations of wild barley, Hordeum spontaneum Koch. Evolution. 2002;56:1403–1415. doi: 10.1111/j.0014-3820.2002.tb01453.x. [DOI] [PubMed] [Google Scholar]
- Volis S, Mendlinger S, Ward D. Adaptive traits of wild barley plants of Mediterranean and desert origin. Oecologia. 2002;133:131–138. doi: 10.1007/s00442-002-0999-0. [DOI] [PubMed] [Google Scholar]
- Volis S, Mendlinger S, Ward D. Differentiation in populations of Hordeum spontaneum Koch along a gradient of environmental productivity and predictability: plasticity in response to water and nutrient stress. Biol J Linn Soc. 2002;75:301–312. [Google Scholar]
- Volis S, Zaretsky M, Shulgina I. Fine-scale spatial genetic structure in a predominantly selfing plant: role of seed and pollen dispersal. Heredity. 2010;105:384–393. doi: 10.1038/hdy.2009.168. [DOI] [PubMed] [Google Scholar]
- Wegmann D, Leuenberger C, Neuenschwander S, Excoffier L. ABCtoolbox: a versatile toolkit for approximate Bayesian computations. BMC Bioinform. 2010;11:116. doi: 10.1186/1471-2105-11-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Yang Z, Zhang T, Bolshoy A, Beharav A, Nevo E. Adaptive microclimatic structural and expressional dehydrin 1 evolution in wild barley, Hordeum spontaneum, at ‘Evolution Canyon', Mount Carmel, Israel. Mol Ecol. 2009;18:2063–2075. doi: 10.1111/j.1365-294X.2009.04140.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.