Abstract
The Biomarkers of Exposure to ARsenic (BEAR) pregnancy cohort in Gómez Palacio, Mexico was recently established to better understand the impacts of prenatal exposure to inorganic arsenic (iAs). In the present study, we examined a subset (n=40) of newborn cord blood samples for microRNA (miRNA) expression changes associated with in utero arsenic exposure. Levels of iAs in maternal drinking water (DW-iAs) and maternal urine were assessed. Levels of DW-iAs ranged from below detectable values to 236 μg/L (mean=51.7 μg/L). Total arsenic in maternal urine (U-tAs) was defined as the sum of iAs and its monomethylated and dimethylated metabolites (MMAs and DMAs, respectively) and ranged from 6.2 to 319.7 μg/L (mean=64.5 μg/L). Genome-wide miRNA expression analysis of cord blood revealed 12 miRNAs with increasing expression associated with U-tAs. Transcriptional targets of the miRNAs were computationally predicted and subsequently assessed using transcriptional profiling. Pathway analysis demonstrated that the U-tAs-associated miRNAs are involved in signaling pathways related to known health outcomes of iAs exposure including cancer and diabetes mellitus. Immune response-related mRNAs were also identified with decreased expression levels associated with U-tAs, and predicted to be mediated in part by the arsenic-responsive miRNAs. Results of this study highlight miRNAs as novel responders to prenatal arsenic exposure that may contribute to associated immune response perturbations.
Keywords: arsenic, prenatal, epigenetics, microRNAs, gene expression
INTRODUCTION
More than 100 million people around the globe are currently exposed to elevated levels of arsenic that are clearly linked to disease [Uddin and Huda, 2011]. Levels of arsenic above the World Health Organization's (WHO) recommended limit of 10 μg/L [WHO, 2006] have been detected in drinking water sources in several areas throughout the world, including but not limited to Bangladesh, India, the United States, Vietnam, and Mexico [ATSDR, 2007]. Inorganic arsenic is a known carcinogen with target sites including the liver, lung, prostate, skin, and urinary bladder [NTP, 2011]. Exposure to arsenic has also been associated with a variety of other non-cancer health effects, including adverse effects on memory and intellectual function, heart disease, liver hypertrophy, diabetes, and respiratory system disease [Kapaj et al., 2006].
Prenatal and early-life exposure to inorganic arsenic represents a global health issue. Inorganic arsenic is known to induce toxic effects on the developing fetus where even modest levels (<100 μg/L in urine) have been associated with decreased birth weight, decreased head and chest circumferences [Rahman et al., 2009], and increased risk of infection in infants [Rahman et al., 2011]. In addition to the immediate health effects of exposure, early-life exposure to arsenic is also associated with increased risk for disease later in life including both cancer and non-cancer endpoints [Yuan et al., 2010; Dauphiné et al., 2011; Smith et al., 2012; Naujokas et al., 2013]. Because of the serious health impacts resulting from inorganic arsenic exposure during critical times of development, elucidating the biological mechanisms that underlie these effects is of utmost importance.
Prenatal exposure to arsenic has been related to alterations in gene expression profiles in both rodents [Liu et al., 2004; Liu et al., 2006] and humans [Fry et al., 2007]. For instance, mice exposed transplacentally to inorganic arsenic display altered expression levels of oncogenes, tumor suppressor genes, and stress-related genes in the liver [Liu et al., 2004; Liu et al., 2006]. In humans, gene expression changes associated with inflammatory signaling pathways are altered in cord blood samples from newborns prenatally exposed to varying levels of arsenic in Thailand [Fry et al., 2007]. It is hypothesized that various epigenetic mechanisms may regulate transcriptional changes resulting from prenatal arsenic exposure.
MicroRNAs (miRNAs) represent part of the epigenome that play critical roles in regulating gene expression. These small RNA molecules are ~22 nucleotides in length and are partially complementary (usually to the 3’-untranslated region (3’-UTR)) to one or more target mRNAs [Friedman et al., 2009]. By base pairing to target mRNAs, miRNAs can cause mRNA degradation and/or translational repression [Friedman et al., 2009]. In humans, miRNAs are estimated to regulate between 30 and 60% of all protein-coding genes [Lewis et al., 2005; Friedman et al., 2009] and are involved in the regulation of virtually every cellular process including apoptosis, proliferation, and cellular differentiation [Calin and Croce, 2006]. Altered miRNA expression profiles have been associated with various diseases, including infectious diseases [Sonkoly et al., 2008] and cancers [Calin and Croce, 2006]. Circulating levels of placental-derived miRNAs in maternal serum have even been implicated as biomarkers for fetal congenital heart defects [Yu et al., 2011].
While altered miRNA expression has been thoroughly evaluated in the context of several diseases, the relationship between in utero exposure to arsenic and miRNA expression profiles is unstudied [Bailey and Fry, 2012]. In utero exposure to other toxicants and miRNA expression disruption has been studied to a limited extent. For example, prenatal exposure to alcohol is associated with altered miRNA signaling linked to teratogenesis in the fetal mouse brain [Wang et al., 2009]. The present study is the first to evaluate the potential impact of prenatal exposure to arsenic on miRNA expression profiles in the cord blood of newborns.
We assessed the impact of prenatal exposure to arsenic on genome-wide miRNA expression profiles and their association with mRNA levels in the Biomarkers of Exposure to ARsenic (BEAR) prospective pregnancy cohort. This cohort includes residents from Gómez Palacio, located in the state of Durango in the Lagunera region of Northern Mexico. More than 450,000 people are exposed to levels of inorganic arsenic in drinking water that exceed 50 μg/L in Mexico [Bundschuh et al., 2012]. Adverse health effects associated with inorganic arsenic exposure have been identified in Lagunera residents, including skin lesions [Valenzuela et al., 2009] and diabetes mellitus [Del Razo et al., 2011]. Here, we show that prenatal arsenic exposure is associated with altered miRNA expression levels in newborn cord blood. These miRNAs were analyzed in the context of mRNA transcriptional profiles and were found to be associated with innate and adaptive immune response signaling pathways in the newborn.
MATERIALS AND METHODS
Study subjects and subcohort selection
This study was approved by the Institutional Review Boards at the University of North Carolina at Chapel Hill (#10-1583) and the Universidad Juárez del Estado de Durango. A total of 200 pregnant women residing in Gómez Palacio, State of Durango, Mexico, were recruited at the General Hospital of Gómez Palacio to participate in the BEAR prospective pregnancy cohort. Requirements for participation in the study included a one year minimum residence in the Gómez Palacio region, which included urban locations of Gómez Palacio and Tlahualilo and their surrounding rural locations. Participants were also confirmed as having a singleton, intrauterine pregnancy without pregnancy complications such as eclampsia or preeclamsia. Study participants were also required to have good overall health status (e.g. no signs of chronic or acute disease). Each participant gave written, informed consent to participate and agreed to provide urine samples, drinking water samples, and to donate umbilical cord blood at delivery. Participants completed detailed questionnaires in order to obtain information on time at residence, socioeconomic factors, including age and education, and other co-exposure factors, including alcohol consumption and smoking status. Questionnaires also gathered information on potential sources of arsenic exposure, including sources of water used for drinking and cooking. Because analyses were carried out after delivery, women could not be informed of their arsenic exposure during pregnancy; however, the women were informed within three months of delivery.
The present study focuses on miRNA expression profiles and utilizes 40 samples obtained from mother-newborn pairs selected from the larger cohort (n=200). The subcohort was selected to include subjects exposed to varying levels of arsenic as determined by both total arsenic in maternal urine (U-tAs) and inorganic arsenic in drinking water (DW-iAs), prioritizing samples representing both lower levels and higher levels of exposure. Of the 40 subjects in the subcohort, samples from 38 subjects were also used to examine transcriptional profiles associated with arsenic exposure.
Determination of DW-iAs and U-tAs
Within four weeks of newborn delivery, water samples were collected from the participants’ stated main source of drinking water by a social worker or member of the research team. Water samples were collected from bottled water or municipally-supplied tap water collected from the subject's kitchen. The concentrations of DW-iAs were measured at the Faculty of Medicine, Universidad Juárez del Estado de Durango, Gómez Palacio, Durango, Mexico using hydride generation-atomic absorption spectrometry (HG-AAS) system as described previously [Le and Ma, 1998; Devesa et al., 2004]. The limit of detection (LOD) for DW-iAs was 0.456 μg/L.
Maternal spot urine samples were collected at the time of delivery, immediately placed in a cryovial, and stored in liquid nitrogen. Samples were shipped at -80°C to the University of North Carolina at Chapel Hill (Chapel Hill, NC) for analysis. The specific gravity (SG) of each urine sample was measured using a handheld refractometer (Reichert TX 400 #13740000; Reichert Inc., Depew, NY). The major arsenical species, specifically iAs and its monomethylated and dimethylated metabolites (MMAs and DMAs), were measured using HG-AAS with cryotrapping [Devesa et al., 2004; Hernandez-Zavala et al., 2009]. The LOD for urinary iAs, MMAs, and DMAs were 0.2 ng/ml, 0.1 ng/ml, and 0.1 ng/ml, respectively. U-tAs was defined as the SG-adjusted sum of iAs, MMAs (trivalent + pentavalent monomethylated arsenicals), and DMAs (trivalent + pentavalent dimethylated arsenicals). Each urine sample was adjusted using the following formula: (mean measured SG-1)/(individual measured SG-1) [Nermell et al., 2008], where the overall mean SG was 1.014 g/ml of the larger cohort. Levels of arsenic in drinking water and urine that were below the LOD were converted to values according to the formula: LOD/(√2) [Del Razo et al., 2011]. The relationship between DW-tAs and U-tAs was determined using the statistical package SAS 9.3 (SAS Institute Inc., Cary, NC).
Cord blood collection and RNA extraction
Cord blood samples were collected from the newborns immediately following delivery. To preserve RNA integrity, blood samples were collected using PreAnalytix PaxGene RNA tubes and extracted using the PAXgene RNA Kit, per standard protocol (Qiagen, Valencia, CA). RNA was quantified with a Nanodrop 1000 spectrophotometer (Thermo Scientific, Waltham, MA) and the integrity verified with a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Isolated RNA used for microarray analysis were amplified and labeled using the NuGEN Ovation Pico WTA System V2 and Encore Biotin Module, respectively (NuGEN, San Carlos, CA).
Assessment of genome-wide miRNA and mRNA expression profiles
To assess whether prenatal exposure to arsenic modifies the expression levels of miRNAs in newborn cord blood, RNA isolated from 40 cord blood samples were labeled and hybridized to the Agilent Human miRNA Microarray, based off miRBase v16.0. Microarray results were extracted using Agilent Feature Extraction software. Data were analyzed for quality assessment and quality control, where no chip/batch/positional effects were present. The signal intensities of each miRNA represented on the array were averaged across probesets and data processed using quantile normalization.
To assess the influence of prenatal arsenic exposure on mRNA expression levels, cord blood RNA samples were assessed using microarray analysis. RNA samples were labeled and hybridized to the Affymetrix GeneChip® Human Gene 2.0 ST Array. Of the 40 cord blood samples that were assessed at the miRNA level, 38 were also assessed at the gene expression level. To analyze the mRNA microarray results, data were first normalized by robust multi-chip average [Irizarry et al., 2003].
The resulting miRNA and mRNA expression data were analyzed in similar manner. Specifically, the association between U-tAs levels and miRNA or mRNA expression levels was evaluated using a regression model, where U-tAs (log-transformed) was the independent variable and cord blood miRNA and mRNA expression levels were the dependent variables. The regression model included covariates that are plausibly related to cord blood miRNA or mRNA expression levels, specifically: mother's age (continuous variable), mother's smoking status (categorical variable), newborn sex (categorical variable), and a ratio of newborn weight / gestational age (continuous variable). Differential expression was defined as a significant association between miRNA or mRNA expression levels and maternal U-tAs, where two requirements were set: (i) miRNA probes were above background signals, and (ii) p-value < 0.01 (Analysis of covariance (ANCOVA) model). For quality filtering, microarray probes with signal intensities less than the median signal across five or more samples were removed. This resulted in the reduction from 1347 to 922 miRNAs and from 53,617 to 31,491 probes for the mRNA analysis. Fold changes were calculated using the following metric: (average miRNA expression levels of the highest exposed quartile (n=10)) / (average miRNA expression levels of the lowest exposed quartile (n=10)). The miRNA microarray analysis required an additional stringency requirement where fold change in expression was ≥ 1.5 or ≤ -1.5. Microarray analyses were calculated using Partek® Genomics Suite™ software (St. Louis, MO). A q-value estimate was calculated and is reported. All miRNAs and mRNAs that passed the statistical filters above were identified as significantly associated with U-tAs.
In order to confirm our statistical assessment of microarray data, an additional permutation-based analysis was performed. Normalized miRNA and mRNA expression estimates were modeled using the same variables as used in the previous statistical assessment: U-tAs (log-transformed), mother's age, mother's smoking status, newborn sex, and a ratio of newborn weight/gestational age, which were used as additive terms in a linear model. A reduced model excluding the U-tAs variable was also fit to each expression estimate. The difference in deviance statistics was used to estimate the effect of U-tAs by improvement in fit of the full model relative to the reduced model. Sample labels were permuted one hundred times, and the same statistic was estimated for each feature during permutation following the scheme outlined in Tusher et al. [Tusher et al., 2001]. The resulting multivariable deviation (mvaD) statistical values represent the difference in deviance between the full linear model, with all covariates, and the reduced linear model, where the U-tAs variable is removed. Statistical results for this test are reported with 1/mvaD values, defined as the multivariable deviation score. Decreasing multivariable deviation scores indicate increasing variation in expression explained by the U-tAs variable, independent of all covariates. Microarray data have been submitted to National Center for Biotechnology Information (NCBI) Gene Expression Omnibus repository [Edgar et al., 2002] and are available under accession numbers GSE48353 and GSE48354 (series GSE48355) (www.ncbi.nlm.nih.gov/geo).
Comparison of U-tAs-associated miRNAs/genes to immune cell-specific miRNAs/genes
It is established that environmental exposures can cause shifts in immune cell populations [Jadhav et al., 2007]. In order to identify whether the U-tAs-associated changes in expression levels of miRNAs and genes simply represent changes in immune cell populations, the U-tAs-associated miRNAs and genes were compared against previously published lists of immune cell subset-specific miRNAs and genes. For the miRNA assessment, the U-tAs-associated miRNAs identified in the present study were compared to a list of 18 miRNAs that are specifically expressed in blood cell subsets (e.g. neutrophils, eosinophils, T-cells, monocytes, and dendritic cells) [Allantaz et al., 2012]. For the gene expression assessment, the U-tAs-associated genes identified in the present study were compared to a list of 1135 genes previously identified as specifically expressed in blood cell subsets (e.g. B-cells, T-cells, CD8+ T-cells, granulocytes, and lymphocytes) [Palmer et al., 2006].
Predicting transcriptional targets of U-tAs-associated miRNAs
Transcriptional targets of the miRNAs associated with U-tAs were first predicted in silico. The Ingenuity Knowledge Database (Ingenuity Systems®, Redwood City, CA) was queried for experimentally validated miRNA-mRNA interactions, as well as computationally predicted interactions based on TargetScan algorithms. Over 600,000 predicted miRNA-mRNA interactions from TargetScan were queried, where interactions were based on algorithms that identify potential matches between 3’-untranslated mRNA regions and miRNA seed sequences [Whitehead, 2012]. The resulting interactions were filtered for high predicted confidence, defined as those with TargetScan total context plus scores < - 0.4. The total context plus score controls for factors influencing miRNA targeting including miRNA binding site type and location, local adenine and uracil content, supplementary pairing, target site abundance, and seed-pairing stability [Garcia et al., 2011]. Many experimentally validated miRNA-mRNA interactions were also included in the in silico prediction. TarBase5.0 was included in the queried database and contains a manually curated collection of more than 1300 experimentally supported miRNA targets [Papadopoulos et al., 2009]. In addition, miRecords was included in the predicted analysis, which is a database comprising over 2600 experimentally validated miRNA-mRNA interactions [Xiao et al., 2009]. Transcriptional targets of U-tAs-associated miRNAs were subsequently filtered to include those that were identified as U-tAs-associated and those with an inverse relationship in expression. To further establish the relationship between U-tAs-associated miRNAs and their predicted mRNA targets, correlation analyses were performed. The z-score normalized expression levels of the U-tAs-associated miRNAs were correlated to the z-score normalized expression levels of the U-tAs-associated target mRNAs using the Spearman Rank Correlation test (Spotfire® TIBCO Software).
Network, pathway, and functional enrichment analysis
Network analysis was performed to understand the systems-level response to prenatal arsenic exposure, and to uncover which responses are possibly mediated via epigenetic (e.g. miRNA) regulation. For this analysis, arsenic-associated miRNAs and mRNAs were overlaid onto a global molecular interaction network. Here, networks were algorithmically constructed based on connectivity, as enabled through Ingenuity Pathway Analysis (Ingenuity Systems®). Canonical pathways and gene sets corresponding to biological functions / disease signatures within the constructed networks were then identified using the right-tailed Fisher's Exact test, as performed previously [Fry et al., 2007; Rager et al., 2013]. Over-represented pathways and gene sets were defined as those that contain more targets than expected by chance.
Confirming miRNA and mRNA microarray results using RT-PCR
To confirm the miRNA and mRNA microarray results, U-tAs associated changes in expression were validated using real-time reverse transcriptase polymerase chain reaction (RT-PCR). Specifically, cord blood samples from subjects representing the highest (n=5) and lowest (n=5) levels of U-tAs were selected for RT-PCR. Samples were plated in technical duplicate. For miRNA validation, TaqMan® MicroRNA Primer Assays (ID 4427975) for hsa-miR-107 (No. 000443), hsa-miR-26b (No. 000407), and the U6 housekeeping miRNA (No. 001973) were used with the TaqMan® Small RNA Assays PCR kit (Applied Biosystems, Carlsbad, CA). The MyCyler Thermal Cycler (Bio-Rad, Hercules, CA) was used for the reverse transcription step, and the Stratagene Mx3005P QPCR System (Agilent Technologies) was used for the real-time amplification step. The resulting RT-PCR cycle times were normalized against the U6 housekeeping miRNA. For the mRNA validation, QuantiTect Primer Assays were used with QuantiTect SYBR® Green PCR kits (Qiagen) and the Stratagene Mx3005P QPCR System (Agilent Technologies). Specifically, dynamin 2 (DNM2) (Cat. No. QT00037072), mitogen-activated protein kinase kinase kinase 3 (MAP3K3) (Cat. No. QT00007665), and phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange 1 (PREX1) (Cat. No. QT00027188) were evaluated for changes in mRNA expression. Resulting RT-PCR cycle times were normalized against the β-actin housekeeping gene. For both miRNA and miRNA analyses, fold changes in expression were calculated using ΔΔ cycle time values [Livak and Schmittgen, 2001]. Statistical significance was calculated using ANOVA (high vs. low U-tAs) (Partek®). The Spearman Rank Correlation test was used to correlate RT-PCR results with the microarray signal intensities.
RESULTS
Characteristics of the BEAR cohort and subcohort
The BEAR pregnancy cohort consists of 200 women and their newborns recruited in Gómez Palacio, Mexico. In the present study we focus on miRNA expression analysis coupled with transcriptional profiling utilizing 40 newborn cord blood samples obtained from the larger BEAR cohort. The samples used in the present analysis were selected to include subjects exposed to varying levels of arsenic as determined by both DW-iAs and U-tAs. The levels of all measured arsenicals (iAs, MMAs, and DMAs) in U-tAs showed variation, where 20 cord blood samples represented maternal U-tAs below 25 μg/L, and 20 were above 25 μg/L (total range=6.2 to 319.7 μg/L). The mean concentration of U-tAs was 64.5 μg/L (median=25.2 μg/L) and none of the U-tAs levels were below detection. The DW-iAs levels ranged between <LOD (0.456 μg/L) and 236 μg/L (mean=51.7 μg/L, median=17.1 μg/L) (Table I). Of the drinking water samples collected from the subcohort, approximately half (n=21, 52.5%) had DW-iAs levels that exceeded the WHO standard (10 μg/L), ranging between 16.4 and 236 μg/L (Table I). Within the subcohort, 11 of the 40 DW-iAs samples had levels < LOD, where the U-tAs levels for these 11 subjects ranged between 7.3 and 16.2 μg/L. Although these samples were collected postpartum, it has been demonstrated that arsenic levels in drinking water show little temporal variability [Slotnick et al., 2006]. Furthermore, this population is stable with the average time living in current residence of 17 years (range=1 to 39 years). Based on these two factors, the DW-iAs levels likely reflect prepartum arsenic exposure through drinking water.
The levels of DW-iAs and U-tAs were significantly correlated in both the larger BEAR cohort (r=0.51, p<0.001) and the subcohort (r=0.87, p<0.001). More detailed demographic characteristics of the women and their newborns are provided in Supporting Information, Table SI.
Prenatal exposure to arsenic is associated with miRNA expression changes
In order to determine whether prenatal exposure to arsenic is associated with the altered expression levels of miRNAs within human cord blood, RNAs from cord blood samples were assessed using the Agilent Human miRNA Microarray, developed using miRBase v16.0. This array measures the expression levels of over 1300 human miRNAs. Microarray analysis revealed that maternal U-tAs is associated with the differential expression of 12 miRNAs in newborn cord blood (see Supporting Information, Table SII). All 12 miRNAs, namely let-7a, miR-107, miR-126, miR-16, miR-17*, miR-195, miR-20a, miR-20b, miR-26b, miR-454, miR-96, and miR-98, showed increased expression levels associated with U-tAs. Supporting the regression model, an additional permutation-based analysis showed that all of the 12 U-tAs-associated miRNAs had a multivariable deviation score < 0.0001 (see Supporting Information, Table SII). The 12 miRNAs were selected based on their linear regression p-values and permutation-based scores and were not filtered using q-values. To control for possible white blood cell shifts, these miRNAs were compared to those known to be immune cell subset-specific [Allantaz et al., 2012]. None of the 12 miRNAs identified here belong to the cell shift-associated miRNAs.
Disease-related signaling is associated with miRNAs
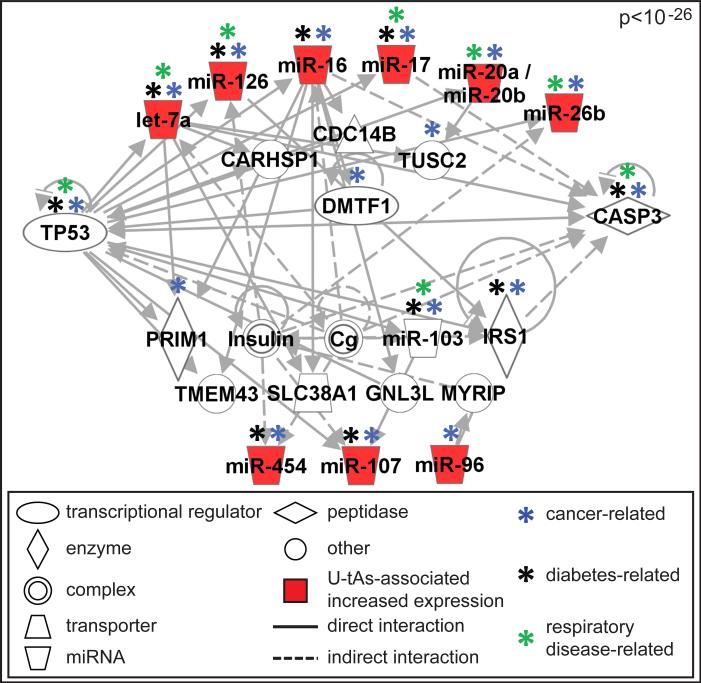
Network analysis of the 12 U-tAs-associated miRNAs generated one significant network (p<10-26) containing known interactions between miRNAs, proteins, and other molecules (Fig. 1). This signaling network was enriched for several biological functions and disease signatures. The most significantly associated functions/diseases were cancer (p=7.1x10-13), reproductive system disease (p=3.3x10-12), connective tissue disorders (p=6.1x10-10), inflammatory disease (p=6.1x10-10), inflammatory response (p=6.1x10-10), organismal injury and abnormalities (p=6.1x10-10), respiratory disease (p=6.1x10-10), gastrointestinal disease (p=4.7x10-9), hepatic system disease (p=4.7x10-9), endocrine system disorders (1.6x10-7), and metabolic disease (e.g. diabetes mellitus) (p=3.0x10-7). Of note, the network containing U-tAs-associated miRNAs includes 16 cancer-associated molecules, 8 respiratory-disease associated molecules and 10 diabetes mellitus-associated molecules, three diseases with known association with iAs exposure (Fig. 1).
Fig 1. Molecular network associated with U-tAs-associated miRNAs.
The network displays known interactions between miRNAs with U-tAs-associated increased expression (red molecules) and molecules related to the miRNAs (white molecules). Signaling molecules related to cancer, diabetes, and respiratory disease are also noted.
mRNA expression profiles are associated with U-tAs
In order to assess the influence of prenatal arsenic exposure on mRNA signaling in cord blood, a transcriptomics-based analysis was carried out using Affymetrix Human Gene 2.0 ST arrays. This array measures the relative expression levels of over 25,000 genes across the genome. A total of 334 transcripts (represented by 537 probe sets) were identified as differentially expressed and associated with U-tAs (see Supporting Information, Table SIII), as determined by linear regression p-values. Of these U-tAs-associated transcripts, 110 displayed increased expression levels and 224 displayed decreased expression levels. Supporting the regression model, an additional permutation-based analysis showed that the majority of the 334 U-tAs-associated mRNAs (n=294, 88%) had a multivariable deviation score < 0.05 (see Supporting Information, Table SIII).
Of the 334 U-tAs-associated mRNAs, only seven were in common with known immune cell-specific genes (n=1135) [Palmer et al., 2006]. Namely, the seven genes were Fc fragment of IgG, receptor, transporter, alpha (FCGRT), metastasis associated lung adenocarcinoma transcript 1 (MALAT1), pleckstrin homology domain containing, family G (with RhoGef domain) member 3 (PLEKHG3), paxillin (PXN), RAB43, member RAS oncogene family (RAB43), RER1 retention in endoplasmic reticulum 1 homolog (S. cerevisiae) (RER1), and transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) (TAP1). The minimal overlap between the U-tAs-associated mRNAs and known immune cell-specific suggests that the disrupted expression profiles are not the result of changes in immune cell populations.
Comparison to previous prenatal arsenic transcriptomic study
As prenatal arsenic exposure has been shown to alter mRNA signaling previously [Fry et al., 2007], we set out to compare the gene expression patterns across cohorts. When U-tAs association in the present study is considered at p<0.05, the number of overlapping genes between the current study and Fry et al. (2007) is 28 (see Supporting Information, Table IV). Eleven of these 28 genes display the same directionality in expression level in arsenic exposed vs. unexposed subjects across cohorts. Among these 28 genes is dual specificity phosphatase 1 (DUSP1), identified previously as a potential gene biomarker of prenatal arsenic exposure [Fry et al., 2007].
Some U-tAs-associated mRNAs are likely regulated by miRNAs
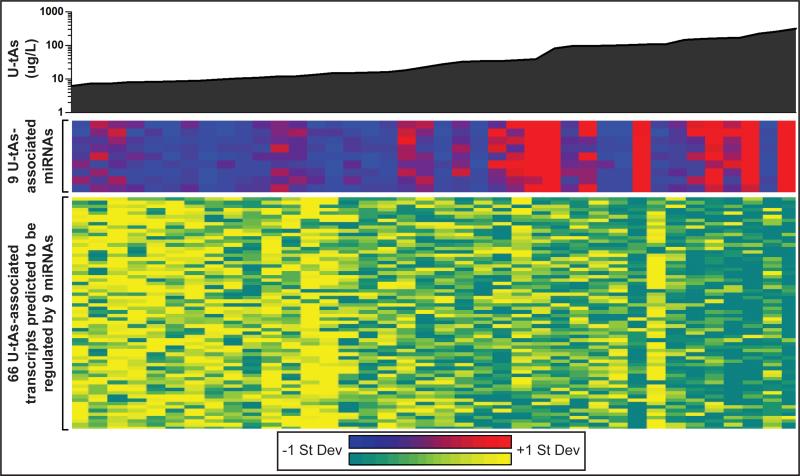
In the present study we aimed to gain insight into the potential impact miRNAs may have on gene expression in human cord blood from newborns with varying levels of prenatal iAs exposure. Thus, interactions between U-tAs-associated miRNAs and differentially expressed mRNAs were predicted in silico. The analysis was carried out using a database comprising previous experimental findings as well as computational predictions largely based on sequence matches between 3’-untranslated mRNA regions and miRNA seed sequences. These predictions revealed that of the 334 U-tAs-associated mRNAs, 66 (20%) are likely regulated by 9 U-tAs-associated miRNAs (Fig. 2, see Supporting Information, Table SV).
Fig 2. Prenatal arsenic exposure (U-tAs) is associated with the differential expression of miRNAs and mRNAs.
Heat maps display the relative expression levels of 9 U-tAs-associated miRNAs (middle) predicted to regulate 66 U-tAs-associated mRNAs (bottom) in newborn cord blood. Expression levels are z-score normalized across rows. For miRNAs, red shading indicates relatively higher expression levels associated with U-tAs, and blue shading indicates relatively lower expression levels. For mRNAs, yellow shading indicates relatively higher expression levels associated with U-tAs, and turquoise indicates relatively lower expression levels.
Immune response signaling is associated with the transcriptional responses
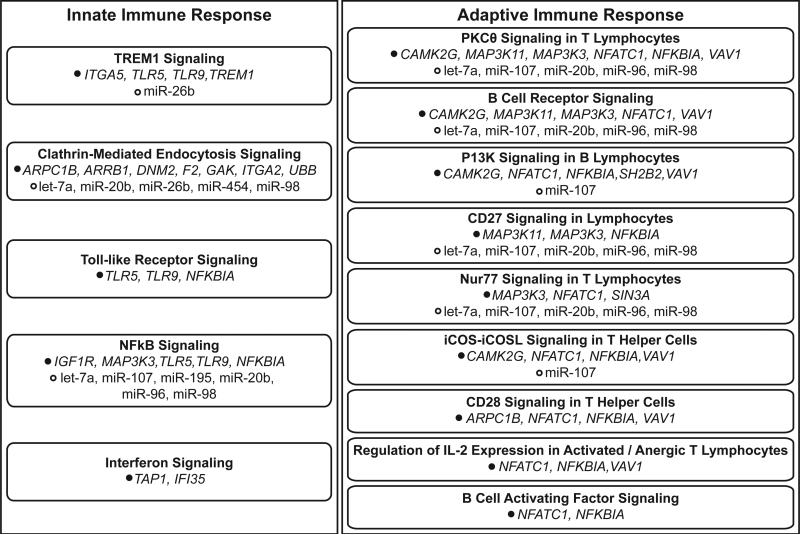
In order to evaluate the potential effects of in utero arsenic exposure at the systems level, enriched canonical signaling pathways were evaluated for the 334 U-tAs-associated mRNAs. A total of 42 canonical pathways were significantly (p<0.10) over-represented amongst the networks constructed using the U-tAs-associated transcripts. Of these 42 canonical pathways, 14 are directly involved in innate or adaptive immune response signaling (Fig. 3). Pathways involved in innate immune response signaling include TREM1 signaling (p=0.007), clathrin-mediated endocytosis signaling (p=0.010), toll-like receptor signaling (p=0.035), nuclear factor kappa B (NFκB) signaling (p=0.062), and interferon signaling (p=0.069). These pathways were composed of genes with decreased expression associated with maternal U-tAs, including DNM2, IFI35 (interferon-induced protein 35), ITGA5 (integrin, alpha 5 (fibronectin receptor, alpha polypeptide), NFκBIA, TLR5 (toll-like receptor 5), and TLR9 (toll-like receptor 9). Pathways involved in adaptive immune response signaling include protein kinase-c-theta (PKCθ) signaling in T lymphocytes and B cell receptor signaling (p=0.004), among others. These immune response signaling pathways included proteins encoded by MAP3K3 and NFκBIA, among others. Nine of these immune response signaling pathways include proteins encoded by U-tAs-associated mRNAs that are predicted to be regulated by U-tAs-associated miRNAs (Fig. 3).
Fig 3. Summary of innate and adaptive immune response pathways associated with transcriptional responses to prenatal arsenic exposure.
U-tAs-associated mRNAs are listed according to pathway involvement, alongside miRNAs that are predicted to regulate the mRNAs.
miRNA expression correlates with mRNA expression levels
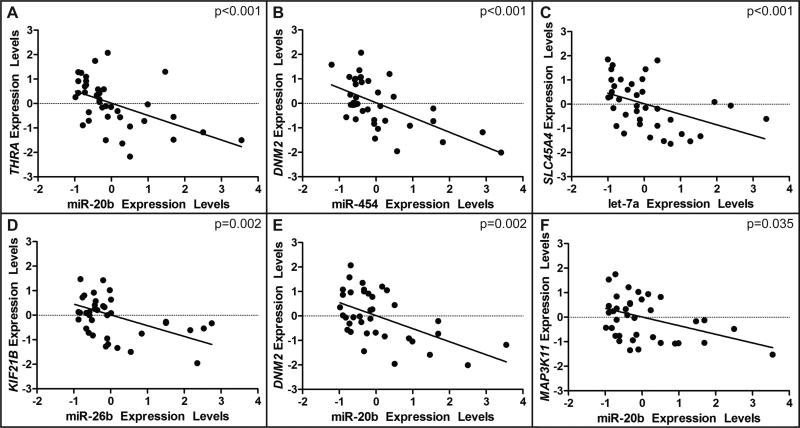
The expression levels of 9 U-tAs-associated miRNAs computationally predicted to regulate U-tAs-associated mRNAs were correlated to the expression levels of their predicted mRNA targets. The predicted interactions regulated by the 9 U-tAs-associated miRNAs were those detailed previously in the results section. Out of the 108 total predicted miRNA-mRNA expression pairings, 36 of the pairings (33%) had correlations where p<0.05 (Fig. 4, see Supporting Information, Table SV). MiRNAs can influence protein expression via multiple manners and are not limited to mRNA degradation. These correlation results suggest that miRNAs may be regulating mRNA expression levels through mechanisms other than mRNA degradation, and warrant the investigation of the limitations of interaction databases currently available. The significantly correlated miRNA-mRNA pairings include mRNAs involved in innate and adaptive immune response signaling, including DNM2 and MAP3K11 (Fig. 4).
Fig. 4. The expression levels of miRNAs are correlated with target mRNAs.
The four most significantly correlated miRNA-mRNA expression pairings are plotted for the subcohort (A-D), as well as some example miRNA-mRNA expression pairings for those related to immune cell signaling (E,F). Expression levels (10-ΔΔCt for RT-PCR and absolute intensity for microarray) are z-score normalized.
RT-PCR results
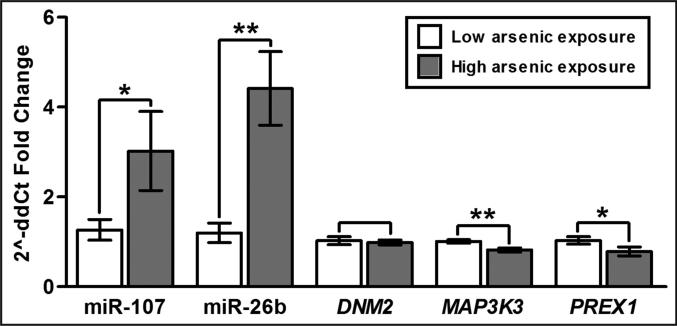
To confirm the microarray results, arsenic-associated changes in miRNA and mRNA expression were tested using RT-PCR with samples from a subset of the mother-newborn pairs. Specifically, cord blood samples from the five mothers with the highest levels of U-tAs were used for RT-PCR alongside samples from the five mothers with the lowest levels of U-tAs. A total of 10 subjects were used for RT-PCR due to sample availability. To confirm the miRNA microarray results, two miRNAs, miR-107 and miR-26b, were selected for RT-PCR analysis, as these miRNAs were involved in the disease-associated signaling network (Fig. 1) and likely play an important role in multiple immune response-related pathways (Fig. 3). The increased expression of miR-107 was verified through RT-PCR where p=0.07 (high U-tAs vs. low U-tAs) (Fig. 5). The increased expression of miR-26b was also verified through RT-PCR where p=0.001 (Fig. 5). There was a positive correlation between RT-PCR and microarray results for miR-107 (p=0.15, r=0.49) and miR-26b (p=0.02, r=0.72).
Fig 5. RT-PCR validation of the miRNAs and mRNAs.
(*) indicates p<0.10, and (**) indicates p<0.01 when comparing low versus high arsenic-exposed samples. Error bars represent standard error mean.
To validate the mRNA microarray results, the expression levels of DNM2 and MAP3K3, selected for their role in innate/adaptive immune signaling (Fig. 3), and PREX1 were measured. Supporting the microarray findings, an arsenic-associated decrease in expression was apparent with MAP3K3 (p=0.005) and PREX1 (p=0.078) through RT-PCR analysis (Fig. 5). DNM2 expression was found to be decreased in the high arsenic exposure samples using RT-PCR, but this reduction was not statistically significant (p=0.717) (Fig. 5). This difference in significance between microarray and RT-PCR results for DNM2 may have resulted from the smaller number of subjects used in the RT-PCR analysis (n=10) versus microarray analysis (n=40). There was a positive correlation between RT-PCR and microarray results for DNM2 (p=0.19, r=0.46), MAP3K3 (p=0.03, r=0.69), and PREX1 (p=0.02, r=0.73).
DISCUSSION
Inorganic arsenic is currently poisoning the drinking water of individuals around the globe including pregnant women and infants. Early-life exposure to arsenic is of particular concern as it is associated with increased risk for both cancer and non-cancer endpoints later in life [Yuan et al., 2010; Dauphiné et al., 2011; Smith et al., 2012; Naujokas et al., 2013]. In spite of this considerable impact on human health, the biological mechanisms that underlie these latent health effects are largely unknown. Prenatal arsenic exposure in humans is associated with altered gene expression profiles [Fry et al., 2007] as well as shifts in cytokine levels in cord blood [Ahmed et al., 2011]. These changes are likely regulated by various cellular mechanisms that impact transcription, including changes in the epigenome. In support of this, prenatal arsenic exposure has been associated with epigenetic changes such as 5-methyl cytosine DNA methylation [Pilsner et al., 2012; Koestler et al., 2013]. Here we present evidence that prenatal arsenic exposure impacts miRNA expression profiles, another key component of the epigenetic machinery [Bailey and Fry, 2012].
We employed an integrative approach to highlight the relationships between prenatal arsenic exposure, epigenetic events, and genomic signaling. Human cord blood samples were collected through the auspices of the BEAR pregnancy cohort in Gómez Palacio, Mexico. The cord blood samples were collected from newborns exposed in utero to varying levels of arsenic, as measured using DW-iAs and maternal U-tAs, where tAs was defined as the sum of iAs and its monomethylated and dimethylated metabolites (MMAs and DMAs). The hypothesis to be tested was that prenatal arsenic exposure impacts the expression profiles of an interrelated set of miRNAs and mRNAs, and that these likely influence pathway signaling related to arsenic-induced disease.
The DW-iAs in our study ranged from <LOD (0.456 μg/L) to 236.0 μg/L, with a mean of 51.7 μg/L (median=17.1 μg/L). Within the subcohort, approximately half (n=21) of the drinking water samples exceeded the WHO recommended limit of 10 μg/L [WHO, 2006]. Levels of DW-iAs from these samples that exceeded the WHO recommended limit ranged between 16.4 and 236.0 μg/L. Thus, while our study, along with many previous studies of arsenic exposure, demonstrates dangerously high levels of arsenic in the drinking water of subpopulations outside of the United States [ATSDR, 2007], this research also has significant implications for human health within the United States. Specifically, some of the women in the BEAR cohort had drinking water with iAs levels comparable to measures within the United States. For instance, drinking water arsenic levels in New Hampshire have been found to be 5.2 μg/L (median=3.7 μg/L), but levels ranged up to 67.5 μg/L [Farzan et al., 2013]. In Idaho, the mean iAs concentration in drinking water in 59 wells was measured as 32.6 μg/L (median=14.6 μg/L), and the iAs level in most of the wells (85%) exceeded the WHO recommendation of 10 μg/L [Hagan, 2004]. In North Carolina, of 63,000 wells tested for arsenic, 7712 (12%) had detectable levels ranging from 1 to 806 μg/L and 1436 (2.3%) had arsenic levels that exceeded the WHO standard [Sanders et al., 2012].
There is no standard for what constitutes safe urinary arsenic levels during pregnancy, and other studies of pregnancy cohorts have reported extremely variable levels of maternal urinary arsenicals. Here we report a mean maternal U-tAs level, measured at time of delivery, of 64.5 μg/L (median=25.2 μg/L, range=6.2-319.7 μg/L). Comparably, in a New Hampshire cohort, mean maternal U-tAs at 24-28 weeks gestation was 6.0 ± 7.5 μg/L (median=3.7 μg/L) and a reported maximum was 58.3 μg/L [Farzan et al., 2013], considerably lower than the current study's maximum U-tAs of ~320 μg/L. In Bangladesh, a mean U-tAs at 30 weeks gestation of 166 ± 196 μg/L (median=80 μg/L, range=2-1440 μg/L) was reported [Rahman et al., 2011]. These findings are likely related to levels of DW-iAs that women are consuming during pregnancy and supported by the positive relationship between DW-iAs and U-tAs in the BEAR cohort.
We find that prenatal arsenic exposure is associated with the altered expression levels of miRNAs and mRNAs in newborn cord blood. Statistical assessment included both a regression model as well as a permutation-based analysis. Data were not filtered using a stringent q-value, an issue that is not uncommon in human subject-based assessment using genome-wide testing. Rather, p-values were used for prioritized analysis as in [Rastogi et al., 2013]. In addition, gene-specific analyses were used to ensure the finding that prenatal arsenic exposure modifies miRNA and mRNA expression profiles in newborn cord blood.
Maternal U-tAs levels were associated with the increased expression of 12 miRNAs, many of which have known roles in cancer and inflammatory response. While miRNAs have not been previously assessed in the cord blood of arsenic-exposed populations, 6 of the 12 U-tAs-associated miRNAs had altered expression in human umbilical vein endothelial cells exposed to inorganic arsenic (20 μM NaAsO2) in vitro [Li et al., 2012]. In a separate study, 6 of the 12 U-tAs-associated miRNAs had altered expression in acute promyelocytic leukemia NB4 cells exposed to 2 μM arsenic trioxide [Ghaffari et al., 2012]. Comparing results across the current study and the two aforementioned studies identifies three common miRNAs: let-7a, miR-16, and miR-20b. All three of these miRNAs have known association to carcinogenesis [Lui et al., 2007; Cascio et al., 2010]. Furthermore, the altered expression of miR-107 and miR-126 has been associated with diabetes mellitus [Guay et al., 2011], a known iAs-associated disease [Del Razo et al., 2011]. The perturbations in key miRNAs may, therefore, represent plausible biological mechanisms underlying arsenic-induced disease and warrant further investigation.
In addition to miRNAs, a set of 334 genes with differential expression associated with maternal U-tAs was identified. These genes were compared to a genomic signature from a study of prenatal arsenic exposure in the Ron Pibul district of Thailand [Fry et al., 2007]. While both cohorts have varying levels of arsenic exposure, in general, levels of DW-iAs in the Ron Pibul region are reported to be higher than those in the current study [Mandal and Suzuki, 2002]. Comparing the genomic datasets, there was overlap at the gene level including DUSP1, one of the proposed biomarkers of prenatal arsenic exposure [Fry et al., 2007]. DUSP1 is primarily involved in the regulation of both broad and local inflammatory response, influencing MAPK (mitogen-activated protein kinase) and JNK (c-Jun N-terminal kinase) signaling [Lang et al., 2006; Liu et al., 2007]. There were also overlaps at the level of biological pathways and functions with both studies demonstrating the enrichment of genes involved in cytokine activity, immune response, inflammatory response, and stress, including NFκB and Interleukin players. Thus at the pathway level there are commonalities demonstrating dysregulation of key signaling pathways in newborns exposed to prenatal arsenic exposure.
We present novel evidence for the dysregulation of genes that play a role in inflammation signaling and in both the innate and adaptive immune system. Genes related to the innate immune system showed decreased expression levels associated with maternal U-tAs in a dose-response manner. These include genes involved in TREM1 signaling (e.g. ITGA5), endocytosis signaling (e.g. DNM2), toll-like receptor signaling (e.g. TLR5, TLR9), NFκB signaling (e.g. NFκBIA), and interferon signaling (e.g. IFI35). Genes related to the adaptive immune systems were also found to have decreased expression levels associated with U-tAs. These include genes involved in PKCθ signaling in T lymphocytes (e.g. MAP3K3, NFκBIA) and B cell receptor signaling (e.g. MAP3K3). Our data support prior studies demonstrating arsenic-responsive inflammatory signaling in the mouse model [Kozul et al., 2009] as well as in vitro [Yager et al., 2013]. Inflammation during the prenatal period has been associated with later life health effects such as lung development and altered risks of lung disease [Kramer et al., 2009]. While these changes were measured in cord blood, the altered inflammation-associated changes are likely similar in the placenta, as previously shown [Ahmed et al., 2011]. Therefore, the changes observed in cord blood likely serve as a proxy for similar biological perturbations occurring in the placenta that could mediate arsenic-induced birth outcomes.
The finding that prenatal arsenic is associated with altered expression of genes that play a role in innate and adaptive immunity is particularly interesting as such exposure has been correlated with specific health outcomes indicative of a repressed immune system. These adverse immune-related outcomes include reduced thymic size and function in the newborn [Raqib et al., 2009; Ahmed et al., 2012], decreased levels of trophic factors (namely lactotransferrin and IL-7) [Raqib et al., 2009], reduced placental T cell count and impaired production of naïve T cells in the fetal thymus [Ahmed et al., 2011; Ahmed et al., 2012], elevated placental inflammatory cytokines [Ahmed et al., 2011], and increased morbidity from infectious diseases during infancy [Raqib et al., 2009; Rahman et al., 2011; Farzan et al., 2013]. Specifically, infants exposed to elevated inorganic arsenic levels in utero had an increased risk of developing lower respiratory tract infections [Rahman et al., 2011]. Similarly, there was a positive correlation between acute respiratory infections in male infants and maternal urinary inorganic arsenic during gestation in rural Bangladesh [Raqib et al., 2009]. Most recently, the New Hampshire Birth Cohort Study highlighted that prenatal inorganic arsenic exposure was correlated with an increased risk of an infant developing an infection [Farzan et al., 2013].
The results from the present study suggest that miRNAs play a role in mediating, at least in part, the immune-associated genomic response in cord blood. Some of the miRNAs that are predicted to regulate the U-tAs-associated mRNA transcripts are established as mediators within the immune system. For instance, miR-20a has been shown to inhibit T-cell activation genes involved in monocyte-macrophage differentiation [Cox et al., 2010]. MiRNAs are known regulators of the immune system [Baltimore et al., 2008] and have important functions in physiological responses to environmental exposures [Bailey and Fry, 2013]. Future studies will be needed to confirm these findings and would benefit from increased sample size, separate geographic locations of validation cohorts, and other biological indicators of effect. Specifically it will be important to identify whether the arsenic-associated miRNA changes directly cause mRNA transcriptional changes, and whether these results in changes in protein functionality. The relationship between miRNA expression profiles in cord blood and iAs metabolites should be established. Finally, the relationships between arsenic-induced genomic and epigenomic changes should ultimately be analyzed as they pertain to health outcomes in newborns.
In conclusion, these results highlight a novel mechanism by which prenatal arsenic exposure impacts cell signaling in newborns. We demonstrate that prenatal arsenic exposure is associated with altered expression of miRNAs that likely regulate genomic signaling in newborn cord blood. This signaling was found to be related to innate and adaptive immune response. These findings contribute to a growing body of literature aimed at elucidating the molecular basis of arsenic-associated disease and reveal potential targets for therapeutic intervention.
ACKNOWLEDGEMENTS
This research was supported by the National Institutes of Health, specifically from the National Institute of Environmental Health Sciences (ES007018, ES019315, ES010126).
Footnotes
STATEMENT OF AUTHOR CONTRIBUTIONS
RCF designed the study and applied for Research Ethics Board approval. GGV and MRA recruited the patients and collected the data with epidemiological oversight from AFO. MS, JC, and CD assessed iAs and its metabolites in urine samples. LS oversaw technical protocols related to RNA sample handling. JER analyzed the data and prepared draft figures and tables. JSP performed statistical analyses. JER, RCF, KAB, and SKM prepared the manuscript draft with important intellectual input from LS, JEL, ZD, AFO, MRA, MS, and GGV. All authors approved the final manuscript. RCF had complete access to the study data.
REFERENCES
- Ahmed S, Ahsan KB, Kippler M, Mily A, Wagatsuma Y, Hoque AM, Ngom PT, El Arifeen S, Raqib R, Vahter M. In utero arsenic exposure is associated with impaired thymic function in newborns possibly via oxidative stress and apoptosis. Toxicol Sci. 2012;129(2):305–314. doi: 10.1093/toxsci/kfs202. [DOI] [PubMed] [Google Scholar]
- Athmed S, Mahabbat-e Khoda S, Rekha RS, Gardner RM, Ameer SS, Moore S, Ekström EC, Vahter M, Raqib R. Arsenic-associated oxidative stress, inflammation, and immune disruption in human placenta and cord blood. Environ Health Perspect. 2011;119(2):258–264. doi: 10.1289/ehp.1002086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allantaz F, Cheng DT, Bergauer T, Ravindran P, Rossier MF, Ebeling M, Badi L, Reis B, Bitter H, D'Asaro M, Chiappe A, Sridhar S, Pacheco GD, Burczynski ME, Hochstrasser D, Vonderscher J, Matthes T. Expression profiling of human immune cell subsets identifies miRNA-mRNA regulatory relationships correlated with cell type specific expression. PLoS One. 2012;7(1):e29979. doi: 10.1371/journal.pone.0029979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ATSDR Agency for Toxic Substances and Disease Registry. Toxicological Profile for Arsenic. 2007:7440–382. i–500. CAS#. [PubMed] [Google Scholar]
- Bailey KA, Fry RC. Arsenic-induced changes to the epigenome. In: Sahu SC, editor. Toxicology and Epigenetics. Wiley; West Sussex, United Kingdom: 2012. pp. 149–190. [Google Scholar]
- Bailey KA, Fry RC. Environmental Toxicants and Perturbation of miRNA Signaling. In: Sahu SC, editor. microRNAs in Toxicology and Medicine. John Wiley & Sons, Ltd.; 2013. doi: 10.1002/9781118695999.ch2. [Google Scholar]
- Baltimore D, Boldin MP, O'Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol. 2008;9(8):839–845. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- Bundschuh J, Litter MI, Parvez F, Román-Ross G, Nicolli HB, Jean JS, Liu CW, López D, Armienta MA, Guilherme LR, Cuevas AG, Cornejo L, Cumbal L, Toujaguez R. One century of arsenic exposure in Latin America: a review of history and occurrence from 14 countries. Sci Total Environ. 2012;429:2–35. doi: 10.1016/j.scitotenv.2011.06.024. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Cascio S, D'Andrea A, Ferla R, Surmacz E, Gulotta E, Amodeo V, Bazan V, Gebbia N, Russo A. miR-20b modulates VEGF expression by targeting HIF-1 alpha and STAT3 in MCF-7 breast cancer cells. J Cell Physiol. 2010;224(1):242–249. doi: 10.1002/jcp.22126. [DOI] [PubMed] [Google Scholar]
- Cox MB, Cairns MJ, Gandhi KS, Carroll AP, Moscovis S, Stewart GJ, Broadley S, Scott RJ, Booth DR, Lechner-Scott J. ANZgene Multiple Sclerosis Genetics Consortium. MicroRNAs miR-17 and miR-20a inhibit T cell activation genes and are under-expressed in MS whole blood. PLoS One. 2010;5(8):e12132. doi: 10.1371/journal.pone.0012132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauphiné DC, Ferreccio C, Guntur S, Yuan Y, Hammond SK, Balmes J, Smith AH, Steinmaus C. Lung function in adults following in utero and childhood exposure to arsenic in drinking water: preliminary findings. Int Arch Occup Environ Health. 2011;84(6):591–600. doi: 10.1007/s00420-010-0591-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Razo LM, García-Vargas GG, Valenzuela OL, Castellanos EH, Sánchez-Peña LC, Currier JM, Drobná Z, Loomis D, Stýblo M. Exposure to arsenic in drinking water is associated with increased prevalence of diabetes: a cross-sectional study in the Zimapán and Lagunera regions in Mexico. Environ Health. 2011;10:73. doi: 10.1186/1476-069X-10-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devesa V, Del Razo LM, Adair B, Drobná Z, Waters SB, Hughes MF, Stýblo M, Thomas DJ. Comprehensive analysis of arsenic metabolites by pH-specific hydride generation atomic absorption spectrometry. Journal of Analytical Atomic Spectrometry. 2004;19(11):1460–1467. [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzan SF, Korrick S, Li Z, Enelow R, Gandolfi AJ, Madan J, Nadeau K, Karagas MR. In utero arsenic exposure and infant infection in a United States cohort: A prospective study. Environ Res. 2013;126:24–30. doi: 10.1016/j.envres.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry RC, Navasumrit P, Valiathan C, Svensson JP, Hogan BJ, Luo M, Bhattacharya S, Kandjanapa K, Soontararuks S, Nookabkaew S, Mahidol C, Ruchirawat M, Samson LD. Activation of inflammation/NF-kappaB signaling in infants born to arsenic-exposed mothers. PLoS Genet. 2007;3(11):e207. doi: 10.1371/journal.pgen.0030207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia DM, Baek D, Shin C, Bell GW, Grimson A, Bartel DP. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol. 2011;18(10):1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaffari SH, Bashash D, Dizaji MZ, Ghavamzadeh A, Alimoghaddam K. Alteration in miRNA gene expression pattern in acute promyelocytic leukemia cell induced by arsenic trioxide: a possible mechanism to explain arsenic multi-target action. Tumour Biol. 2012;33(1):157–172. doi: 10.1007/s13277-011-0259-1. [DOI] [PubMed] [Google Scholar]
- Guay C, Roggli E, Nesca V, Jacovetti C, Regazzi R. Diabetes mellitus, a microRNA-related disease? Transl Res. 2011;157(4):253–264. doi: 10.1016/j.trsl.2011.01.009. [DOI] [PubMed] [Google Scholar]
- Hagan EF. Ground Water Quality Technical Brief Statewide Ambient Ground Water Quality Monitoring Program Arsenic Speciation Results (2002 & 2003) Idaho Department of Water Resources; 2004. [Google Scholar]
- Hernandez-Zavala A, Drobna Z, Styblo M, Thomas DJ. Analysis of arsenical metabolites in biological samples. Curr Protoc Toxicol. 2009;42(4.33):1–4. doi: 10.1002/0471140856.tx0433s42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31(4):e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadhav SH, Sarkar SN, Ram GC, Tripathi HC. Immunosuppressive effect of subchronic exposure to a mixture of eight heavy metals, found as groundwater contaminants in different areas of India, through drinking water in male rats. Arch Environ Contam Toxicol. 2007;53(3):450–458. doi: 10.1007/s00244-006-0177-1. [DOI] [PubMed] [Google Scholar]
- Kapaj S, Peterson H, Liber K, Bhattacharya P. Human health effects from chronic arsenic poisoning--a review. J Environ Sci Health A Tox Hazard Subst Environ Eng. 2006;41(10):2399–2428. doi: 10.1080/10934520600873571. [DOI] [PubMed] [Google Scholar]
- Koestler DC, Avissar-Whiting M, Houseman EA, Karagas MR, Marsit CJ. Differential DNA Methylation in Umbilical Cord Blood of Infants Exposed to Low Levels of Arsenic in Utero. Environ Health Perspect. 2013;121(8):971–977. doi: 10.1289/ehp.1205925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozul CD, Hampton TH, Davey JC, Gosse JA, Nomikos AP, Eisenhauer PL, Weiss DJ, Thorpe JE, Ihnat MA, Hamilton JW. Chronic exposure to arsenic in the drinking water alters the expression of immune response genes in mouse lung. Environ Health Perspect. 2009;117(7):1108–1115. doi: 10.1289/ehp.0800199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer BW, Kallapur S, Newnham J, Jobe AH. Prenatal inflammation and lung development. Semin Fetal Neonatal Med. 2009;14(1):2–7. doi: 10.1016/j.siny.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang R, Hammer M, Mages J. DUSP meet immunology: dual specificity MAPK phosphatases in control of the inflammatory response. J Immunol. 2006;177(11):7497–7504. doi: 10.4049/jimmunol.177.11.7497. [DOI] [PubMed] [Google Scholar]
- Le XC, Ma M. Short-column liquid chromatography with hydride generation atomic fluorescence detection for the speciation of arsenic. Anal Chem. 1998;70(9):1926–1933. doi: 10.1021/ac971247q. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li X, Shi Y, Wei Y, Ma X, Li Y, Li R. Altered expression profiles of microRNAs upon arsenic exposure of human umbilical vein endothelial cells. Environ Toxicol Pharmacol. 2012;34(2):381–387. doi: 10.1016/j.etap.2012.05.003. [DOI] [PubMed] [Google Scholar]
- Liu J, Xie Y, Ducharme DM, Shen J, Diwan BA, Merrick BA, Grissom SF, Tucker CJ, Paules RS, Tennant R, Waalkes MP. Global gene expression associated with hepatocarcinogenesis in adult male mice induced by in utero arsenic exposure. Environ Health Perspect. 2006;114(3):404–411. doi: 10.1289/ehp.8534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Xie Y, Ward JM, Diwan BA, Waalkes MP. Toxicogenomic analysis of aberrant gene expression in liver tumors and nontumorous livers of adult mice exposed in utero to inorganic arsenic. Toxicol Sci. 2004;77(2):249–257. doi: 10.1093/toxsci/kfh055. [DOI] [PubMed] [Google Scholar]
- Liu Y, Shepherd EG, Nelin LD. MAPK phosphatases--regulating the immune response. Nat Rev Immunol. 2007;7(3):202–212. doi: 10.1038/nri2035. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lui WO, Pourmand N, Patterson BK, Fire A. Patterns of known and novel small RNAs in human cervical cancer. Cancer Res. 2007;67(13):6031–6043. doi: 10.1158/0008-5472.CAN-06-0561. [DOI] [PubMed] [Google Scholar]
- Mandal BK, Suzuki KT. Arsenic round the world: a review. Talanta. 2002;58(1):201–235. [PubMed] [Google Scholar]
- Naujokas MF, Anderson B, Ahsan H, Aposhian HV, Graziano JH, Thompson C, Suk WA. The broad scope of health effects from chronic arsenic exposure: update on a worldwide public health problem. Environ Health Perspect. 2013;121(3):295–302. doi: 10.1289/ehp.1205875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nermell B, Lindberg AL, Rahman M, Berglund M, Persson LA, El Arifeen S, Vahter M. Urinary arsenic concentration adjustment factors and malnutrition. Environ Res. 2008;106(2):212–218. doi: 10.1016/j.envres.2007.08.005. [DOI] [PubMed] [Google Scholar]
- NTP National Toxicology Project 12th Report on Carcinogens. Rep Carcinog. 2011;12:iii–499. [Google Scholar]
- Palmer C, Diehn M, Alizadeh AA, Brown PO. Cell-type specific gene expression profiles of leukocytes in human peripheral blood. BMC Genomics. 2006;7:115. doi: 10.1186/1471-2164-7-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos GL, Reczko M, Simossis VA, Sethupathy P, Hatzigeorgiou AG. The database of experimentally supported targets: a functional update of TarBase. Nucleic Acids Res. 2009;37:D155–158. doi: 10.1093/nar/gkn809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilsner JR, Hall MN, Liu X, Ilievski V, Slavkovich V, Levy D, Factor-Litvak P, Yunus M, Rahman M, Graziano JH, Gamble MV. Influence of prenatal arsenic exposure and newborn sex on global methylation of cord blood DNA. PLoS One. 2012;7(5):e37147. doi: 10.1371/journal.pone.0037147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rager JE, Moeller BC, Doyle-Eisele M, Kracko D, Swenberg JA, Fry RC. Formaldehyde and epigenetic alterations: microRNA changes in the nasal epithelium of nonhuman primates. Environ Health Perspect. 2013;121(3):339–344. doi: 10.1289/ehp.1205582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A, Vahter M, Ekström EC, Persson LÅ. Arsenic exposure in pregnancy increases the risk of lower respiratory tract infection and diarrhea during infancy in Bangladesh. Environ Health Perspect. 2011;119(5):719–724. doi: 10.1289/ehp.1002265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A, Vahter M, Smith AH, Nermell B, Yunus M, El Arifeen S, Persson LA, Ekström EC. Arsenic exposure during pregnancy and size at birth: a prospective cohort study in Bangladesh. Am J Epidemiol. 2009;169(3):304–312. doi: 10.1093/aje/kwn332. [DOI] [PubMed] [Google Scholar]
- Raqib R, Ahmed S, Sultana R, Wagatsuma Y, Mondal D, Hoque AM, Nermell B, Yunus M, Roy S, Persson LA, Arifeen SE, Moore S, Vahter M. Effects of in utero arsenic exposure on child immunity and morbidity in rural Bangladesh. Toxicol Lett. 2009;185(3):197–202. doi: 10.1016/j.toxlet.2009.01.001. [DOI] [PubMed] [Google Scholar]
- Rastogi D, Suzuki M, Greally JM. Differential epigenome-wide DNA methylation patterns in childhood obesity-associated asthma. Sci Rep. 2013;3:2164. doi: 10.1038/srep02164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders AP, Messier KP, Shehee M, Rudo K, Serre ML, Fry RC. Arsenic in North Carolina: public health implications. Environ Int. 2012;38(1):10–16. doi: 10.1016/j.envint.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotnick MJ, Meliker JR, Nriagu JO. Effects of time and point-of-use devices on arsenic levels in Southeastern Michigan drinking water, USA. Sci Total Environ. 2006;369(1-3):42–50. doi: 10.1016/j.scitotenv.2006.04.021. [DOI] [PubMed] [Google Scholar]
- Smith AH, Marshall G, Liaw J, Yuan Y, Ferreccio C, Steinmaus C. Mortality in young adults following in utero and childhood exposure to arsenic in drinking water. Environ Health Perspect. 2012;120(11):1527–1531. doi: 10.1289/ehp.1104867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonkoly E, Ståhle M, Pivarcsi A. MicroRNAs and immunity: novel players in the regulation of normal immune function and inflammation. Semin Cancer Biol. 2008;18(2):131–140. doi: 10.1016/j.semcancer.2008.01.005. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin R, Huda NH. Arsenic poisoning in bangladesh. Oman Med J. 2011;26(3):207. doi: 10.5001/omj.2011.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela OL, Drobná Z, Hernández-Castellanos E, Sánchez-Peña LC, García-Vargas GG, Borja-Aburto VH, Stýblo M, Del Razo LM. Association of AS3MT polymorphisms and the risk of premalignant arsenic skin lesions. Toxicol Appl Pharmacol. 2009;239(2):200–207. doi: 10.1016/j.taap.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LL, Zhang Z, Li Q, Yang R, Pei X, Xu Y, Wang J, Zhou SF, Li Y. Ethanol exposure induces differential microRNA and target gene expression and teratogenic effects which can be suppressed by folic acid supplementation. Hum Reprod. 2009;24(3):562–579. doi: 10.1093/humrep/den439. [DOI] [PubMed] [Google Scholar]
- Whitehead . TargetScanHuman: Prediction of microRNA targets, release 6.2. Whitehead Institute for Biomedical Research; Mar 5, 2012. 2013. http://www.targetscan.org/ [Google Scholar]
- WHO . First addendum to 3rd addition. Vol. 1. World Health Organization; WHO Press; Geneva, Switzerland: 2006. Guidelines for drinking water quality. [Google Scholar]
- Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37:D105–110. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yager JW, Gentry PR, Thomas RS, Pluta L, Efremenko A, Black M, Arnold LL, McKim JM, Wilga P, Gill G, Choe KY, Clewell HJ. Evaluation of gene expression changes in human primary uroepithelial cells following 24-hr exposures to inorganic arsenic and its methylated metabolites. Environ Mol Mutagen. 2013;54(2):82–98. doi: 10.1002/em.21749. [DOI] [PubMed] [Google Scholar]
- Yu Z, Han S, Hu P, Zhu C, Wang X, Qian L, Guo X. Potential role of maternal serum microRNAs as a biomarker for fetal congenital heart defects. Med Hypotheses. 2011;76(3):424–426. doi: 10.1016/j.mehy.2010.11.010. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Marshall G, Ferreccio C, Steinmaus C, Liaw J, Bates M, Smith AH. Kidney cancer mortality: fifty-year latency patterns related to arsenic exposure. Epidemiology. 2010;21(1):103–108. doi: 10.1097/EDE.0b013e3181c21e46. [DOI] [PubMed] [Google Scholar]